Abstract
Single-cell technologies are currently widely applied to obtain a deeper understanding of the phenotype of single-cells in heterogenous mixtures. However, integrated multilayer approaches including simultaneous detection of mRNA, protein expression, and intracellular phospho-proteins are still challenging. Here, we combined an adapted method to in vitro–differentiate peripheral B-cells into antibody-secreting cells (ASCs) (i.e., plasmablasts and plasma cells) with integrated multi-omic single-cell sequencing technologies to detect and quantify immunoglobulin subclass-specific surface markers, transcriptional profiles, and signaling transduction pathway components. Using a common set of surface proteins, we integrated two multimodal datasets to combine mRNA, protein expression, and phospho-protein detection in one integrated dataset. Next, we tested whether ASCs that only seem to differ in its ability to secrete different IgM, IgA, or IgG antibodies exhibit other differences that characterize these different ASCs. Our approach detected differential expression of plasmablast and plasma cell markers, homing receptors, and TNF receptors. In addition, differential sensitivity was observed for the different cytokine stimulations that were applied during in vitro differentiation. For example, IgM ASCs were more sensitive to IL-15, while IgG ASC responded more to IL-6 and IFN addition. Furthermore, tonic BCR activity was detected in IgA and IgM ASCs, while IgG ASC exhibited active BCR-independent SYK activity and NF-κB and mTOR signaling. We confirmed these findings using flow cytometry and small molecules inhibitors, demonstrating the importance of SYK, NF-κB, and mTOR activity for plasmablast/plasma cell differentiation/survival and/or IgG secretion. Taken together, our integrated multi-omics approach allowed high-resolution phenotypic characterization of single cells in a heterogenous sample of in vitro–differentiated human ASCs. Our strategy is expected to further our understanding of human ASCs in healthy and diseased samples and provide a valuable tool to identify novel biomarkers and potential drug targets.
Keywords: single-cell phosphoproteomics, multimodal, transcriptomics, plasmacells, antibody-secreting cells
Abbreviations: ASCs, antibody-secreting cells; BCR, B-cell receptor; MOFA+, multi-omics factor analysis; PC, plasma cell; PB, plasmablast; IgM, immunoglobulin M; IgA, immunoglobulin A; IgG, immunoglobulin G; IgE, immunoglobulin E
Graphical Abstract
Highlights
-
•
In-vitro differentiation of human B-cells into antibody-secreting cells.
-
•
Simultaneous single-cell detection of (phospho)-proteins and mRNA through sequencing.
-
•
MOFA+ allows data integration and phenotyping of IgM, IgG, and IgA cells.
-
•
Ig-class–specific transcriptome and expression of homing protein markers.
-
•
Signaling activity differs between IgM, IgA, and IgG cells, including BCR and NF-κB.
In Brief
Multi-modal single-cell sequencing technology enabled quantifying the characteristics of human antibody-secreting cells (ASCs). Integrative analysis identified three classes of ASCs and their molecular features, including surface protein markers, phospho-proteins, and transcriptional profiles. Each Ig-class of IgM, IgA, and IgG shows a specific expression of homing receptors and protein markers. IgM and IgA ASCs have tonic BCR signaling, while IgG has more active signaling pathways, including SYK, mTOR, IL6, and NF-kB.
Upon activation, B-cells differentiate into plasmablasts (PBs) and long-lived memory antibody-secreting cells (ASCs) (1, 2). An extraordinary number of control mechanisms, at both the cellular and molecular levels, underlie the generation and maintenance of PBs and plasma cells (PCs) from their B-cell precursors (3). Aberrant PB and ASC activation/proliferation has been linked to several human diseases. In inflammatory and autoimmune diseases like systemic lupus erythematosus or IgA nephropathy, aberrant ASC activation is linked to disease etiology and manifestation (1). In cancer, multiple myeloma is a malignancy derived from a PC (4). A deeper insight into ASC phenotype can aid our understanding of such diseases and identify biomarkers of disease and potential novel drug targets.
Human PCs only represent ∼0.25% of total bone marrow cells and these cells home to different tissues in the human body, making extensive molecular analysis and functional studies challenging (5). Protocols for in vitro differentiation of human peripheral B-cells into ASCs (6, 7, 8) form the basis for phenotypic and functional studies (9, 10). Studies in mice have shown divergent gene expression profiles for the different Ig class PCs (11) and different expression profiles were observed comparing human class–switched germinal center B-cells from human tonsils as compared to nonclass–switched cells (12). However, bulk analysis obscures potential differences across ASCs secreting immunoglobulin M (IgM), A (IgA), E, (IgE), or G (IgG) antibodies. Single-cell technologies can characterize secreted antibody repertoire of individual cells (13, 14) and fully characterize the functional and transcriptional diversity of ASC (15).
Single-cell sequencing technologies have revolutionized our insight into the molecular phenotyping of many cell types of the human body. The combined quantification of mRNA and surface protein markers allows for additional (sub-)classification of cells (16, 17), while immune-detection by sequencing (18, 19) and droplet-based QuRIE-seq (20) allows quantification of intracellular phospho-proteins. These methods rely on the reversible fixation of cells and immunostaining with DNA-tagged antibodies, enabling RNA sequencing and quantification of (phospho-)proteins in a single assay. Unfortunately, our fixation-based methods did not result in high-quality mRNA libraries in primary immune-cells (20). To circumvent this technical challenge, we combined multimodal data from fixed and nonfixed cells. Using a common set of surface proteins, we were able to integrate mRNA and intracellular (phospho-) proteins detection. Applying this method to in vitro–differentiated human ASCs, we identified in this mixture of human differentiating B cells, PB and PCs distinctly different phenotypes of IgM-, IgA-, and IgG-expressing ASCs on both transcriptional and protein levels. More importantly, within these different Ig-secreting ASCs, we were able to unravel some of the active signal transduction and validated the importance of these signal transduction pathways using small molecule inhibitors.
Experimental Procedures
Antibody Labeling
Carrier-free antibodies (supplemental Tables S1 and S2) were conjugated to 5′-azide-oligos (supplemental Tables S3 and S4, Biolegio) as described in Supplementary text. In brief, dibenzocyclooctyne-S-S-N-hydroxysuccinimidyl-ester–functionalized antibodies were incubated with 3× molar access azide-oligo for 16 h at 4 °C. Unconjugated oligos were removed using 100K amicon centrifuge filters (Merck).
B-Cell Isolation and Culture
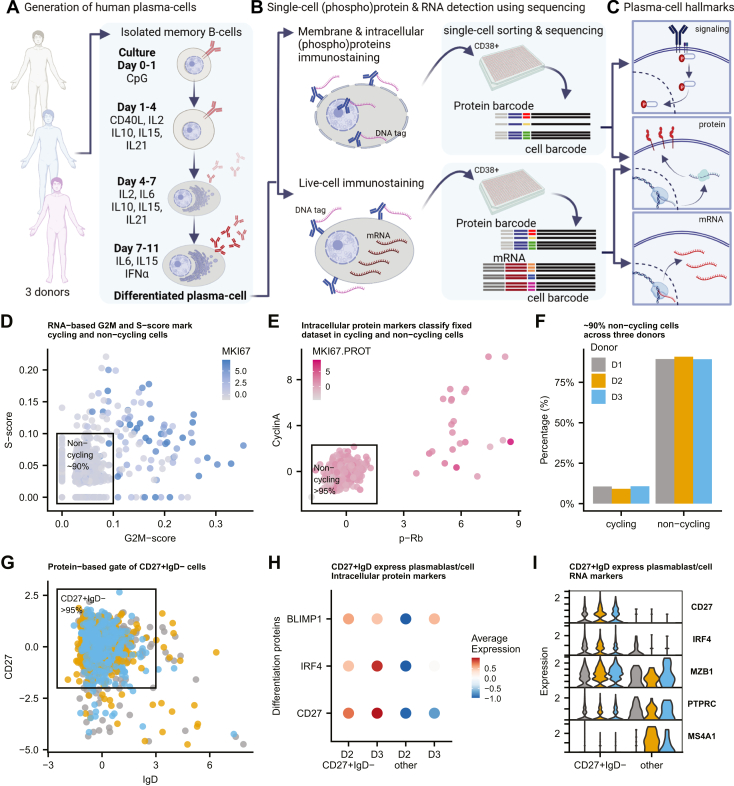
Total B-cells were isolated from Buffy coats that were obtained from healthy volunteers with written consent and ethics committee approval according to institutional guidelines (Sanquin). Subsequent differentiation cultures were performed in Iscove's Modified Dulbecco's Medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific), and 1% Penicillin-Streptomycin (Thermo Fisher Scientific). Purified B-cells were differentiated for 11 days as visualized in Figure 1A and described in detail in Supplementary text.
Fig. 1.
Experimental workflow for multi-modal single-cell analysis of in-vitro differentiated human antibody secreting cells.A, peripheral B-cells isolated from healthy human blood are cultured in the presence of indicated stimuli for 11 days. The protocol results in differentiated cells with ASC phenotype, capable of secreting antibodies. Created with BioRender.com. B, day 11 cells are divided into one sample for fixation, permeabilization (allowing intracellular immune-staining with DNA-tagged antibodies), and a second sample for live-cell immunostaining with DNA-tagged antibodies. CD38+ single-cells are sorted into 384-well plates for multi-modal library preparation. C, analyses of both samples with a common set of surface-proteins allows identification of molecular hallmarks: signaling (phospho-)proteins, protein levels, and gene expression. Images (A–C) were created with BioRender.com. D, RNA-based G2M-score and S-score based on predefined genesets is used for classification of each cells as noncycling or cycling. Color represents normalized RNA-expression of proliferation marker MKI67. E, classification of noncycling cells based on intracellular signal of p-Rb, Cyclin A, and Ki67. F, percentage classified cycling or noncycling cells per donor (N = 3). G, CD27+/IgD-gate to select for differentiated PCs. Color represent three donors (legend see panel C). H, average (scaled) expression of selected intracellular proteins. I, per-donor expression gene-expression of three differentiation markers (CD27, IRF4, and MZB1) and two genes downregulated during PC differentiation (Wilcox, p-val < 0.05) in gated versus other cells (panel D). Colors represent donors (legend in panel C). ASC, antibody-secreting cell; PC, plasma cell.
Antibody Staining and Cell Sorting
Day 11 cells were harvested and split in two tubes, one to be fixed and permeabilized and one for live cells analysis. Cells were immediately fixed by adding 2× concentrated fixative to the cell suspension 5 mM dithiobis (succinimidyl propionate) (Thermo Fisher Scientific) and 5 mM succinimidyl 3-(2-pyridyldithio)propionate (Thermo Fisher Scientific) in PBS (Thermo Fisher Scientific). Cells were incubated at RT for 45 min while gently shaking. Fixed cells were quenched and permeabilized with 100 mM Tris–HCl pH 7.5, 150 mM NaCl, and 0.1% Triton X100 (Thermo Fisher Scientific) for 10 min at RT. Cells were washed once and blocked for 45 min in 0.5× protein-free blocking buffer (Thermo Fisher Scientific) with 0.2 mg/ml dextran sulfate (Sigma Aldrich) and 0.5 U/ml RNasin plus (Promega) in PBS (Thermo Fisher Scientific). Cells were stained in blocking buffer containing DNA-tagged antibodies (supplemental Table S3) and CD38-PE-Cy7 (supplemental Table S5) for 1 h at RT. Following staining, cells were washed twice with the blocking buffer and resuspended in PBS containing 5 mg/ml bovine serum albumin (Thermo Fisher Scientific), 0.5 U/ml RNsing plus (Promega), and 0.1 μg/ml DAPI (Biolegend).
Nonfixed live cells were immediately washed twice in ice cold blocking buffer. Cells were stained in ice-cold blocking buffer containing DNA-tagged Abs (supplemental Table S4) and CD38-PE-Cy7 Ab (supplemental Table S5). Cells were incubated on ice in the dark for 20 min. Cells were washed three times in ice-cold blocking buffer and resuspended in blocking buffer containing 0.1 μg/ml DAPI (Biolegend).
Using the FACS Melody (BD Biosciences), cells were sorted single-cell in 384-wells PCR plates (Bio-Rad) containing 100 nl water containing 7.5 ng/μl unique Cellseq-2 primers (supplemental Table S6) and 5 μl mineral oil (Sigma Aldrich). Plates were stored at −80 °C until further use. Oligonucleotide sequences were adapted to allow sequencing of the transcripts/ARC sequences in read 1 and the cell barcode and UMI in read 2 (supplemental Table S4).
Library Preparation
Plates with sorted cells were thawed on ice. For fixed samples, cells were reverse-crosslinked, releasing RNA and antibody-barcodes as described by Gerlach et al. (21). Live cells were incubated for 5 min at 65 degrees. Reverse transcription was performed as described by Gerlach et al. (21). After pooling cells, the mRNA was separated from antibody barcodes using 0.6× AMPure XP magnetic beads (Beckman Coulter). Then, mRNA-sequencing libraries were generated as described by Gerlach et al. (21) using primers listed in supplemental Table S4. From the separated antibody barcodes sample, protein libraries were generated as described in detail in Supplementary text. mRNA and protein libraries were sequenced with NextSeq500.
Data Analysis
Bcl2fastq software (illumina) was used for data-demultiplexing. The CEL-seq2 pipeline (22) and CITE-seq-count (23) tool were used to create count tables of mRNA or protein libraries, respectively. Standard settings were used with some minor modifications. The CEL-seq2 pipeline was run using settings [min_bc_quality = 10, cut_length = 50]. Additionally, the read names for read 1 and read 2 were swapped to allow compatibility of our barcode design with the pipeline. The CITE-seq-count settings were kept at default with exception of [ max_error of Ab barcode = 1, umi_collpasing_dist = 1].
Extensive documentation of quality control, filtering (supplemental Fig. S3), and data-normalization and processing can be found at https://vanbuggenum.github.io/Multimodal-Plasmacell_manuscript/QC.html. In brief, cells were filtered based on either mRNA library quality (Livecell dataset: >300 genes, <5% mitochondrial counts per cell) or protein library (>1500 and <9000 protein counts, >40 different proteins per cell); Fixed dataset: >2500 and <20,000 protein counts, >65 proteins per cell. RNA counts were normalized using ‘single-cell transform’ (Seurat V4) (24). Then, normalized counts were scaled, while regressing out differences in % mitochondrial counts and total counts. Protein counts were normalized using the CLR method (24). Then, the normalized protein counts were scaled with regressing out variance in number of proteins detected and total counts.
Experimental Design and Statistical Rationale: Multi-Modal Analysis
To identify noncycling ASCs, first, a cell-cycle scoring for S-phase and G2M-phase was calculated using Ucell algorithm (25) using gene lists provided by the Seurat R-package (V4) (24). For the live dataset, a gate was determined based on these two scores. For the fixed dataset, p-Rb and Cyclin A were used to identify cycling cells instead of mRNA-based cell-cycle score. To determine differentiated phenotype, protein levels of CD27 and IgD were used for gating. These noncycling CD27+IgD-cells were kept for further analysis using multi-omics factor analysis (MOFA+) (26). In brief, cells were grouped per donor, and the model received four molecular modalities. Principal component analysis was performed using normalized and scaled counts of Ig proteins.
Compound Treatment of in-Vitro Differentiated ASC
Total B-cells were isolated and differentiated as described above. At Day 10, cells were retrieved and tubes of individual donors were pooled and reseeded to normalize any culture differences. Day 10 ASCs were treated with dimethyl sulfoxide (DMSO, Sigma-Aldrich), R406, idelalisib, IKK16, or rapamycin (Selleckchem) at 1 μM (0.01% DMSO) up to Day 12. On Day 12, cells were retrieved and resuspended. To assess the effect of compound treatment on cell health, viable absolute cell count was measured with 123count eBeads counting beads (Thermo Fisher Scientific) in combination with 7-AAD (Thermo Fisher Scientific) using the FACS Verse (BD Biosciences). Absolute counts were calculated according to manufacturer protocol: (Cell Count × eBead Volume)/(eBead Count × Cell Volume) × eBead concentration = Absolute Cell Count per ml.
On Day 10 (before treatment) and Day 12 (after treatment), 100,000 cells were retrieved for each condition and stained for CD19-PerCP/Cy5.5, IgD-BV510, CD27-FITC, CD38-PE-Cy7, CD138-APC, IgA-PE, and IgM-APC-Cy7 (supplemental Table S5). After staining, cells were resuspended in PBS containing 5 mg/ml bovine serum albumin (Thermo Fisher Scientific) and 0.1 μg/ml DAPI (Biolegend). To assess the effect of compound treatment on immunoglobulin-expressing subpopulations, frequency of IgA, IgM, and IgA/M double negative populations were determined on the total CD38+ population, PB population, and the PC population. Frequency was determined from the total viable population. Viable absolute cell count was calculated as follows: Absolute Cell Count per ml/100 × IgX population frequency = IgX cells per ml.
To assess the effect of compound treatment on immunoglobulin production, supernatant was retrieved on Day 10 (before treatment) and Day 12 (after treatment) for each condition. IgA, IgG, and IgM was detected in supernatant using Human IgA uncoated ELISA kit, Human IgG Total uncoated ELISA kit, and Human IgM Uncoated ELISA kit (Thermo Fisher Scientific) according to manufacturer’s protocol. Immunoglobulin concentration Day 10 was subtracted from immunoglobulin concentration on Day 12 to detect immunoglobulin production during compound treatment.
Results
Phenotypic MultiModal Single-Cell Analysis of Human CASCs
We adopted an in vitro differentiation protocol of human peripheral B-cells to generate human ASCs (6, 7, 8). Isolated B-cells from healthy donors were stimulated with CpG for 1 day. Then, in three phases, activated B-cells were differentiated into ASCs using a mixture of cytokines and CD40L (Fig. 1A). Characterization by flow cytometry showed that the procedure generated cells that displayed hallmarks of ASCs: IgD−CD27++ increased cell-size and gained expression of CD38 (27) (supplemental Fig. S1, A and B). CD38+ ASCs showed increased mRNA expression levels for CD38, BLIMP1, and XBP1 as compared to naïve B-cells, as well as downregulation of proliferation marker Ki-67 and naïve B-cell–specific transcription factor PAX-5 (supplemental Fig. S1C). Consistent with ASC differentiation, Ab secretion was detected for all three Ig-classes from day 7 onwards and was most strongly increased on day 11 (supplemental Fig. S1D).
ASCs were collected on Day 11 and further purified by CD38+ cell-sorting (supplemental Fig. S2). These CD38+ ASCs were used for high resolution characterization via a single-cell multimodal sequencing technology. To quantify signaling pathway activity (i.e., phosphorylation levels), surface markers, and mRNA expression on single-cell level, we developed a strategy based on CITE-seq (16), immuno-detection by sequencing (18, 19), RNA and immuno-detection (21), and droplet-based QuRIE-seq (20). To enable immunostaining, mRNA analysis, and intracellular immuno-detection on single-cell, we divided the ASCs from each donor into two portions before staining and sorting for multimodal single-cell analysis (Fig. 1B). One portion of ASCs was first stained for extracellular protein detection and immediately lysed and processed for single-cell library preparation, where using a CITE-seq–like sample preparation, extracellular protein and mRNA detection was done (referred to as “live” dataset). The second portion of ASCs was first fixed and permeabilized to allow intracellular immuno-detection by sequencing (referred to as “fixed” dataset). Importantly, both samples were costained with a common set of 17 surface antibodies including IgM, IgA, and IgG (supplemental Tables S1 and S2). We reasoned that such a common ‘bridge modality’ should allow the integration and matching of the two parallel datasets based on the expression of these surface proteins. Together, the workflow generates a dataset that includes information on ASC signaling pathway activity, protein levels, and mRNA expression at single-cell resolution (Fig. 1C).
Quality control and filtering of the sequencing data (including >300 genes per cell and <5% mitochondrial gene counts per cell, sample specific minimum and maximum UMI counts per cell for the protein libraries, see supplemental Fig. S3 for full details) resulted in a selection of a total of 1433 cells in the live-cell dataset and 1038 cells in the fixed-cell dataset from three individual donors. To confirm the terminal differentiation of the in-vitro–generated ASCs, we analyzed the cell-cycle state at both mRNA and protein-expression level. We categorized cells based on RNA-expression levels of cell-cycle genes, using the gene signature scoring method UCell (25) with G2M- and S-phase gene lists as an input. Based on the computed scores and expression of proliferation marker KI-67, >95% of cells were classified as noncycling (Fig. 1D), which was confirmed using intracellular protein expression of Cyclin A, p-Rb, and Ki-67 (Fig. 1E). All three donors showed comparable percentages of dividing versus nondividing cells (Fig. 1F and supplemental Fig. S4A).
ASCs, most specifically PBs and PCs, are characterized by IgD−CD27+CD38+ (27). Using the cell-sorted CD38+ cells, we compared IgD−CD27+ and IgD+CD27− cells (Fig. 1G and supplemental Fig. S4, B and C). CD27+IgD− cells showed differential protein levels of ASC transcription factors BLIMP1 and IRF4 (Fig. 1H), which was further confirmed with the observed increased expression (supplemental Fig. S4, D–F) of CD27, IRF4, and MZB1 genes (Fig. 1I) and downregulation of PTPRC (encoding CD45) and MS4A1 genes (Fig. 1I) as compared to CD27−IgD+ cells. Taken together, our analyses confirmed that the in vitro procedure generated differentiated ASCs. For subsequent analysis, only nonproliferating IgD−CD27+CD38+ cells were studied.
Single-Cell mRNA and Protein Analysis Reveals Ig-Class–Specific Phenotypes
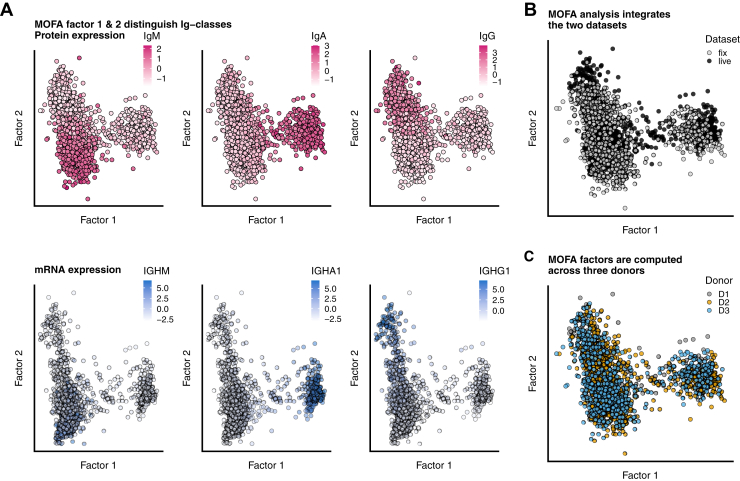
We analyzed the datasets using MOFA+ (26). This analysis identifies shared variance across multiple molecular modalities and allows integrated downstream analysis such as clustering. We supplied the model with four ‘modalities’: mRNA, fixed-protein, live-protein and common-protein, and grouped cells per donor to diminish donor-specific differences. (supplemental Fig. S5A). MOFA+ analysis computed nine factors, of which factor 1 and 2 explain the most variance within the common modality (supplemental Fig. S5B). IgM, IgG, and IgA proteins are the major contributing features to these factors, which suggests that factor 1 and 2 classify ASCs based on IgM, IgA, and IgG levels (Fig. 2A and supplemental Fig. S5, B and C). Protein expression of IgM, IgA, and IgG (Fig. 2A, upper panels in pink) correlated with the gene-expression levels of these Ig types (Fig. 2A, lower panels in blue). No variation of these factors was observed between the three individual donors or the live- and fixed-cell dataset (Fig. 2, B and C and supplemental Fig. S5C). Additionally, using shared-proteins as input, a UMAP representation of both the live and fixed cell dataset shows overlapping embedding of the two datasets (supplemental Fig. S5D). Based on the correlation with IgG, IgA, and IgM, factor 1 and 2 were used for k-means clustering, annotating individual cells as IgG, IgM, or IgA subclass (supplemental Fig. S5E). Using an independent analysis method (principal component analysis and clustering on Ig proteins), we confirmed the grouping of cells into Ig subclasses (supplemental Fig. S6). Interestingly, analyzing cells with all features excluding the Ig genes or proteins showed that even in that case, distinguishing characteristics are found between the ASC-secreting different Ig-classes (supplemental Fig. S7). Finally, we observe that all three donors displayed similar percentages of IgA, IgM, and IgG ASCs (supplemental Fig. S8).
Fig. 2.
Joined analysis of all modalities using Multi-Omics Factor Analysis reveals three major Ig-classes IgM, IgA, and IgG across datasets and donors.A, protein-levels (upper panels, pink color) and gene-expression (lower panels, blue color) of IgM, IgA, and IgG showing Factor 1 and Factor 2 split cells based on Ig-class. B, MOFA factors 1 and 2 split sample independent of dataset origin (fix or live cell sample). C, Factorplot colored by donor shows no donor-effects. MOFA, multi-omics factor analysis.
To dissect phenotypic differences between ASCs secreting IgM, IgG, and IgA, we identified proteins and genes with high loading values (weights) for factor 1 and 2, correlating with either IgA, IgG, or IgM (Fig. 3A and supplemental Fig. S9). First, we studied the abundance of ASC markers across the different Ig subclasses. All major population of human peripheral B cells can be identified using a small set of phenotypic markers: CD19, CD20, IgD, CD27, and CD38 (27). Using our analysis, we only study IgD-CD27+ cells that were purified by CD38+ cell-sorting on Day 11 after in vitro differentiation of peripheral B-cells. As shown in Figure 3B, differences exist between the expression levels of these phenotypic markers across different Ig subclasses. Where CD19 and CD20 are found expressed most highly on IgM ASCs, CD19 and CD20 show low signals on IgG and IgA ASCs. Studying the expression of CD138, as a marker of more mature PC, it is interesting to note that only IgG ASCs highly express CD138, suggesting that IgG ASCs have a more PC phenotype, where IgM and IgA ASCs have a more PB phenotype.
Fig. 3.
Exploration of protein and gene expression levels per Ig-classes IgM, IgA, and IgG highlights subclass-specific abundance of markers for B-cells, PC differentiation, and homing-receptors.A, scatterplot of loading values per protein modality. Highlighted with text are selected contributing B-cell markers, differentiation related proteins, and homing receptors. Gray arrows indicate direction of loadings contributing to the IgM, IgG, or IgA subclass. B, visualization of average expression of selected membrane proteins in IgM, IgG, and IgA cells. C, manually curated list of intracellular differentiation genes and transcription factor proteins. D, scaled protein levels of BCMA, BAFFR, and TACI. E, average expression of chemokine and adhesion-related surface proteins. For all panels, same legend applies presented as displayed in right lower corner. PC, plasma cell.
Also, when studying transcription and translation factors, IgG ASCs seem to reflect more strongly a PC phenotype with high levels of BLIMP1 on both mRNA (PRDM1) and protein level and IRF4 on mRNA (Fig. 3C). In accordance with literature, high levels of BLIMP1 coincide with low levels of PAX5 and Bcl6 protein. Interestingly, high levels of ell2 mRNA and c-myc and Tbet protein are also detected in the IgG ASCs as compared to IgA and IgM ASCs, where ell2 has been linked to IgG protein secretion in mice (28). In IgM ASCs, high levels of XBP1 protein are detected in the absence of PAX5 (Fig. 3C), where in IgA ASCs, high levels of PAX5 repress XBP1 protein levels. The low levels of BLIMP1 expression in both IgA and IgM ASCs and high BLC6 protein levels in IgM ASCs again suggest that IgM and IgA ASCs have a more PB phenotype. In IgA ASCs, also other PAX5-regulated genes such as CD79A, IRF8, and ID3 were highly expressed (Fig. 3C) and high expression was observed for RUNX2, a crucial factor in IgA differentiation of B-cells (29).
Different TNF-receptor family members have been implicated in B-cell and ASC differentiation. As shown in Figure 3D, highest expression of CD40 is found on IgM ASCs, consistent with its reported role in class-switching and survival (30). Expression of CD27 and its ligand CD70 appears to be mutually exclusive as described before, where high CD70 levels are observed on IgG ASCs as compared to IgM and IgA ASCs. Previously, CD70 signaling has been associated with specific downregulation of IgG, which is potentially explained by the differential expression of CD70 on IgG ASCs (31). Comparing expression of the BCMA, TACI, and BAFF-R indicates that BCMA is most highly expressed on IgA ASCs, while TACI is most highly expressed on IgG ASCs and BAFF-R is highest expressed on IgM ASCs (Fig. 3D). These observations are very well aligned with the observed defects in Ig production seen in TACI−/− KO mice (32) and low IgM and IgG levels in the BAFF-R−/− KO mice (33).
IgG PCs are thought to migrate to the bone marrow, while IgA PCs migrate to mucosal-associated lymphoid tissue (2). We next studied whether different Ig subclasses of ASCs are differently imprinted to mediate migration to these sites. Indeed, the bone marrow homing receptor CXCR4 was highly expressed on IgG ASCs of which the ligand, CXCL12 is expressed by bone marrow stromal cells (Fig. 3E) (34). Furthermore, CD44 and CD31 were highest expressed on IgG ASCs, which have been shown to be important for adhesion to the bone marrow stroma (Fig. 3E) (35). In contrast to IgG ASCs, IgA ASCs expressed integrins α4 (CD49d) and β1 and β7, which have been reported to direct IgA PCs to respiratory and intestine mucosal tissue, respectively (Fig. 3E) (36). IgM PCs can remain in the lymphoid organs or migrate to sites of infection or mucosal tissue, the latter supported by the moderate expression of integrins α4 and β7 (Fig. 3E) and CXCR5 (under direction of CD40) on IgM ASCs (37). Overall, our single-cell method revealed that ASCs expressing different isotype classes differentially express transcriptional factors, membrane proteins, and homing receptors.
Ig Subclasss Show Differential Activity of Cytokine Signaling Across Modalities
To establish the sensitivity for cytokine-induced signaling across IgM, IgA, and IgG ASCs, we analyzed receptor expression, intracellular phospho-proteins, and downstream gene expression of IL2/IL15, IL6, JAK/STAT, and IFNA pathways. As shown in Figure 4, IgM ASCs show elevated expression of IL-2RA, IL-2RB, IL-2RG mRNA, and high-affinity IL2 receptor (CD25) protein (Fig. 4, A and B) as well as moderate levels of pJAK1 and high levels of pSTAT5/6 (Fig. 4, C and D). Signal transduction is most likely driven by IL-15, as from Day 7, no IL-2 was added to the in vitro culture, and IL-15 is present between Day 7 to 11 (Fig. 4, A–D) (38). In contrast, IgG ASCs express high levels of IL-6R mRNA as well as the corresponding protein IL-6R (Fig. 4A). IL-6 signaling induces high levels of p-STAT3, which in literature is described as a key inducer of PRDM1 expression (39). In addition, high expression of IFNAR1 and IFNAR2 protein was detected in IgG ASCs (supplemental Fig. S4, A and B) and IFN-α signaling mediators were most abundantly detected in IgG ASCs with pJAK1, pSTAT1, and STAT1 gene expression (Fig. 4, C and D). Finally, IgA ASCs showed low JAK/STAT pathway activation and modest IL-6R and STAT3/6 expression, suggesting IgA PCs are less dependent or sensitive to these cytokines. Taken together, our analysis allowed us to observe two distinctly different cytokine-induced signaling profiles in PCs: IgM shows high IL-15 and STAT5/6 signaling, while IgG shows high IL-6/IFNR and STAT1/3 signaling (Fig. 4D).
Fig. 4.
Ig-class–specific cytokine, JAK-STAT, tonic B-cell receptor, and mTOR signaling activity.A and B, gene (dotplot) and protein expression (violin plots) of cytokine receptors. C, gene expression of STAT-signaling components. D, average phosphorylation levels of JAK-stat signaling components. E, UMAP representation of fixed dataset displaying annotated Ig-classes (left panel) and scaled phosphorylation levels of B-cell coreceptor CD79a (right panel). F, per subclass average signaling activity of selected B-cell receptor, mTOR-, and NFKb-related signaling components.
Tonic BCR Signaling and NF-κB/mTOR Show Different Activity in IgM, IgG, or IgA Cells
Differentiation of B cells in ASCs/PCs is accompanied by a switch from membrane to secreted Ig, thereby losing expression of the B-cell receptor (BCR) and its signal transduction. Recent studies in both mice and human PCs revealed that IgM and IgA PCs still express a BCR capable of signaling, suggesting these cells can still detect and respond to antigens (40, 41). To study whether evidence of active BCR signaling can be detected in the in vitro–differentiated human ASCs, we analyzed different elements of BCR pathway activation. Both IgA and IgM ASCs had high levels of phosphorylated CD79a, the immediate downstream signal transduction protein of the BCR (Fig. 4F). Also other downstream signal components of the BCR, pSRC, pBLNK, and pp38 were detected in IgM ASCs, while pSYK was detected in IgA ASCs (Fig. 4F). In contrast to IgM and IgA ASCs, and in accordance with recent reports, little or no activation of the BCR pathway was detected in IgG ASCs (Fig. 4F). In fact, we observed phosphorylation of Syk, mTOR, and observed NF-κB pathway activation as detected by pIKK and pP65, suggestive of an active BCR-independent signal transduction in IgG ASCs. Reversely, no or limited activation of NF-κB and/or mTOR was detected in IgA or IgM ASCs. Together, we find a unique phenotypic profile for each Ig-class across modalities that includes surface-protein expression, activity of distinct signalling pathways and mRNA expression (Fig. 5).
Fig. 5.
Highlights of human Ig-specific antibody secreting cell expression profiles and signaling pathway activity. Single-cell multi-omics analysis revealed three major Ig-classes among the in-vitro–differentiated human antibody-secreting cells (ASCs): IgM (left), IgG (middle), and IgA (right). Each Ig-class shows a unique combination of phenotypes across multiple molecular modalities: surface-protein expression of homing and cytokine receptors, signal pathway activity, and mRNA expression. Created with BioRender.com
To validate the findings from our integrated single-cell (phospho-)protein and RNA detection approach, we differentiated human ASCs following the same procedures and on Day 10, added different small molecule inhibitors. Flow cytometry was used as an independent technology to validate our proteomics findings. As described above, increasing numbers of PBs and PCs were detected from Day 10 onwards (Fig. 6A). When comparing PB and PC differentiation across the different Ig-secreting ASCs, again IgG ASCs seem to display a more PC phenotype where IgA and IgM ASCs show a more PB phenotype from Day 11 onwards (Fig. 6B). To understand the relevance of the proteomic observations made on Day 11, inhibitors targeting SYK (R406), PI3K (idelalisib), NF-κB (IKK16), and mTOR (rapamycin) activation were added on Day 10 and on Day 12, and PB, PC counts, and immunoglobulin production were determined for each of the Ig isotypes. The strong inhibition on downstream signal transduction in IgA, IgM, and IgG ASCs by R406, rapamycin, and IKK-16 was confirmed using phospho-flow cytometry (supplemental Fig. S10). As shown in Figure 6C, using DMSO-treated cells as a reference, PI3K inhibition using idelalisib similarly reduced PB and PC numbers across Ig-isotypes. Also looking at immunoglobulin production between Day 12 and 10 did not reveal strong effects of inhibition of PI3K (Fig. 6D). In contrast and fully in line with the obtained proteomics data, Syk inhibition using R406 showed a significant drop in IgA and IgG PBs and PCs, while not affecting IgM ASCs. Interestingly, SYK inhibition did not affect immunoglobulin secretion (Fig. 6D). Using our proteomics approach, mTOR and NF-κB signal transduction were found to be highest active in IgG ASCs. As shown in Figure 6C, NF-κB inhibition using IKK16 inhibited PB and PC numbers for IgG-secreting ASCs, while IKK16 inhibited IgA and IgM PB numbers, but not affecting IgM and enhancing IgA PCs numbers. Inhibition of NF-κB resulted in similar secretion of IgM, enhanced secretion of IgA, and reduced secretion of IgG (Fig. 6D). When studying the effect of mTOR inhibition by rapamycin, a strong reduction of PB and PCs were observed for all Ig-secreting ASCs. Interestingly, while strongest cell number reductions were observed for IgA and IgM ASCs than IgG-secreting ASCs, IgG secretion was most strongly inhibited than IgA and IgM.
Fig. 6.
Inhibition of plasmablast and plasma cell differentiation by SYK, PI3K, NF-κB, and mTOR inhibiton.A, peripheral B-cells isolated from healthy human blood were differentiated for 12 days. Using flow cytometry, fraction of plasmablast and plasma cells were determined on Day 0, 7, 10, 11, and 12 after differentiation. B, on Day 7, 10, 11, and 12, plasmablast and plasma cell fraction of IgM, IgA, and IgG ASCs was determined using flow cytometry. C, on Day 10 during ASC differentiation, inhibitors of SYK (R406), PI3K (idelalisib), NF-κB (IKK16), and mTOR (rapamycin) and the effect on plasmablast and plasma cell numbers was established using flow cytometry using addition of DMSO as a reference (indicated with dotted line). D, immunoglobulin production between Day 12 and 10 was detected using ELISA and presented is the subtraction of immunoglobulin detected on Day 12 versus 10. Production levels as detected for the control cultures (DMSO) are indicated by the dotted line. Statistical significance was determined relative to DMSO-treated cultures using Paired two tailed sample t test: ns = p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. ASC, antibody-secreting cell.
Discussion
In this study, we undertook integrated single-cell (phospho-)proteomics and RNA detection to phenotypically dissect human in vitro–differentiated PBs and PCs to understand differences that might be present between human IgA, IgM, and IgG ASCs. We employed an in vitro differentiation protocol to obtain large quantities of human ASCs from peripheral B-cells. Our multimodal single-cell analysis method detected the Ig subclass–specific expression of B cells phenotypic markers, transcription factors, TNF-receptor mediated survival receptors, and expression of niche-specific homing receptors in this bulk heterogenous sample of in vitro–differentiated noncycling IgD−CD27++CD38+ ASCs. More importantly, our method allowed simultaneous detection of active cytokine-induced, BCR and mTOR/AKT signal transduction that revealed that each Ig subclass has differential sensitivity towards these stimuli based on the differential expression of their receptors. Finally, we validated our proteomic findings using alternative methods and demonstrated the importance of the identified signal transduction pathways for differentiation, survival, and immunoglobulin secretion using small molecule inhibitors of SYK, PI3K, NF-κB, and mTOR.
To our knowledge, our study is the first to map, at the single-cell level, phenotypic markers and record active signal transduction in heterogeneous mixture of differentiating and antibody-secreting B cells. ASCs composed of PBs and PCs are crucially important to maintain circulating immunoglobulin levels and provide long-lasting immunity by their sometimes life-long survival in dedicated niches in the body. Aberrant PCs have been linked to human diseases like rheumatoid arthritis, systemic lupus erythematosu, IgA nephropathy, and multiple myeloma. A detailed molecular phenotyping and deep understanding of human ASC differentiation and PC survival is therefore important to understand the etiology of such diseases as well as to identify molecular targets for drug discovery.
Recently, studies using single-cell RNA-seq technologies have provided more understanding on the (heterogeneity in) phenotype of aberrant PCs (42, 43, 44, 45). However, these studies only provide insight in the mRNA expression of different cells, whereas in our study, we combine mRNA, extracellular, and intracellular (phospho-)protein information at the single-cell level. One of the current major challenges in the single-cell field is integrating datasets across samples and molecular modalities. Computational strategies encounter several considerations as how to define anchors, scalability, and handling missing data (46). Several of these challenges are being addressed by recently developed tools including MOFA+ (26), multiVI (47), COBOLT (48), StabMap (49), scMVP (50), and Bridge Integration (51). So-far, there was no illustration of integrating mRNA datasets with a comprehensive intracellular phospho-protein and transcription factor dataset using a common set of surface proteins. Here, we illustrate the successful use of the MOFA+ framework bridging single-cell mRNA quantification with detailed insight in signal pathway activity from the same cell (sub)type. In principle, as long as the phenotype (such as cell subtypes in this article) is being quantified in the shared molecular modality, any other associated phenotypes in separate datasets can be integrated and explored. Taken together, based on our analyses performed, integration of both datasets allowed us to gain detailed insight into human ASCs across three molecular modalities.
We detected, in a heterogenous mixture of differentiated ASCs, phenotypic markers that are differentially expressed across different Ig-secreting ASCs. These include PB and PC markers, TNF-receptors, and homing receptors. Our study uniquely allowed the simultaneous detection of active signal transduction at the moment of isolation of ASCs on Day 11 in the differentiation protocol. First, we detected active cytokine signaling that occurs as a result of the addition of the cytokines during the in vitro differentiation protocol. Here, we demonstrated that not all Ig-secreting ASCs are equally sensitive to the added cytokines as was evidenced by the differential sensitivity of IgM ASCs towards IL-15 (STAT5/6) versus IgG ASCs that demonstrated high sensitivity towards IL-6/IFN signaling. As demonstrated before, our method uses an unbiased and potentially unlimited set of antibodies for detection, allowing unbiased findings to occur. Here, we found that tonic BCR signaling seems to be active on Day 11 of in vitro differentiation in IgM and IgA ASCs, while in IgG ASCs, SYK, NF-κB, and mTOR signal transduction is active. Our findings were substantiated using an independent technology (flow cytometry) and studying the effect of small molecule inhibitors that target SYK, PI3K, NF-κB, and mTOR. In human and mice, IgM and IgA but not IgG ASCs have been shown to express a functional BCR that can activate intracellular signaling (40, 41), and tonic BCR signaling has been reported for pre- and naive B-cells as an important survival signal (52). The in vitro–differentiated IgM and IgA ASCs show all hallmarks of tonic BCR signaling, suggesting that these cells still rely on this pathway for survival. It has been proposed that CD45, a positive regulator of BCR signaling, triggers receptor tyrosine kinases to phosphorylate CD79a and CD79b, thereby initiating tonic signaling. This could explain the tonic signaling observed in this culture, as CD45 expression was higher in IgM and IgA cells than IgG cells (52). Also, the observed expression of BCL6 in IgM ASCs may positively affect tonic BCR signaling in B-cell lymphoma cells (53). In contrast, IgG ASCs displayed no p-CD79a or downstream signaling, but our methods did reveal strong phosphorylation of SYK, NF-κB, and mTOR pathway components. Apart from BCR-induced activation, SYK can be activated through different ITAM-containing receptors (54) and SYK may regulate activation of the mTOR pathway, supporting cell growth and proliferation (55, 56). Additionally, in IgG, we detected active IL-6 signaling. This signaling axis, in concert with the NF-κB and BLIMP1–XBP1 axis, has been described to induce mTOR activity and antibody production through regulating the unfolded protein response (20, 57). In fact, our study revealed that while all ASCs were strongly reduced in numbers by rapamycin treatment, IgG secretion by ASCs was most dramatically inhibited by rapamycin despite having the lowest effect on PB and PC numbers than IgA and IgM ASCs, confirming that mTOR activity is modulating immunoglobulin production and secretion. Taken together with the phenotypic markers (i.e., CD138, PAX5, ell2, homing-, and TNF receptors etc.) using our multimodal omics approach, we were able, in a single biological experiment, to reveal differential sensitivities to signal transduction in a heterogenous mixture of cells as evidenced by IgG ASCs defined by active IL-6, JAK-STAT, SYK, NF-κB versus IgA and IgM ASCs.
The rapidly expanding range of single-cell technologies provides quantitative information about different ‘layers’ in the cell but often not on multiple layers at the same time. We have demonstrated that integrating single-cell datasets on mRNA, surface markers, and intracellular (phospho-)protein via the quantification of a shared modality is a powerful strategy to characterize the transcriptional wiring and signaling pathway activity in in-vitro–differentiated primary cells. Our strategy yielded valuable insights into the molecular differences across modalities between human IgM, IgA, and IgG ASCs in heterogenous mixture of cells. We envision that an integrated single-cell multi-omic analysis pipeline is especially suitable for studying cells in the human peripheral blood mononuclear cells compartment, where the cell-type–specific response to a range of stimuli provides a deeper understanding of key processes in immunology or blood borne cancers and where detailed dissection of mode of action of pharmaceutical interventions will aid the development of novel therapeutic interventions.
Data Availability
Detailed code documentation of the analysis and generation of figures is available from https://vanbuggenum.github.io/Multimodal-Plasmacell_manuscript/. The fastq files and count are accessible through GEO (57) Series accession number GSE189953 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189953).
Supplemental data
This article contains supplemental data.
Conflict of interest
H. v. E. and A. v. E. are shareholders of Aduro Biotech (now Chinook Therapeutics). All other authors declare they have no competing interests.
Acknowledgments
We thank Klaas Mulder for providing 384-well plates with Celseq2 oligo’s, Marijke Baltissen for assistance with sequencing, and Dyah Karjosukarso for processing FASTQ files generating the count tables. We thank Nico Stam and Boris Rodenko for their contributions.
Funding and additional information
This study was supported (in part) by research funding from Aduro Biotech (E. v. B., W. J., P. V., M. J. M. H., A. v. E., and H. v. E.), Radboud University (L. J. A. W., W. T. S. H., and J. A. G. L. v. B.), Spinoza Grant (W. T. S. H.), VENI grant from the Dutch Research Council VI.Veni.202.228 (J. A. G. L. v. B.).
Author contributions
E. v. B., W. J., P. V., M. J. M. H., L. J. A. W., and J. A. G. L. v. B. investigation; E. v. B., W. J., P. V., and M. J. M. H. methodology; P. V., A. v. E., J. A. G. L. v. B., and H. v. E. conceptualization; J. A. G. L. v. B. formal analysis; P. V., W. T. S. H., and H. v. E. supervision; E. v. B., W. T. S. H., J. A. G. L. v. B., and H. v. E. writing–original draft.
Supplemental Data
References
- 1.Hiepe F., Radbruch A. Plasma cells as an innovative target in autoimmune disease with renal manifestations. Nat. Rev. Nephrol. 2016;12:232–240. doi: 10.1038/nrneph.2016.20. [DOI] [PubMed] [Google Scholar]
- 2.Tellier J., Nutt S.L. Plasma cells: the programming of an antibody-secreting machine. Eur. J. Immunol. 2019;49:30–37. doi: 10.1002/eji.201847517. [DOI] [PubMed] [Google Scholar]
- 3.Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 4.van de Donk N.W.C.J., Pawlyn C., Yong K.L. Multiple myeloma. Lancet. 2021;397:410–427. doi: 10.1016/S0140-6736(21)00135-5. [DOI] [PubMed] [Google Scholar]
- 5.Terstappen L.W.M.M., Johnsen S., Segers-Nolten I.M.J., Loken M.R. Identification and characterization of plasma cells in normal human bone marrow by high-resolution flow cytometry. Blood. 1990;76:1739–1747. [PubMed] [Google Scholar]
- 6.Jourdan M., De Boussac H., Viziteu E., Kassambara A., Moreaux J. In vitro differentiation model of human normal memory B cells to long-lived plasma cells. J. Vis. Exp. 2019 doi: 10.3791/58929. [DOI] [PubMed] [Google Scholar]
- 7.Jourdan M., Caraux A., De Vos J., Fiol G., Larroque M., Cognot C., et al. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood. 2009;114:5173–5181. doi: 10.1182/blood-2009-07-235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoua Maïga R., Bonnaure G., Tremblay Rochette J., Néron S. Human CD38hiCD138+ plasma cells can be generated in vitro from CD40-activated switched-memory B lymphocytes. J. Immunol. Res. 2014;2014:635108. doi: 10.1155/2014/635108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jourdan M., Cren M., Robert N., Bolloré K., Fest T., Duperray C., et al. IL-6 supports the generation of human long-lived plasma cells in combination with either APRIL or stromal cell-soluble factors. Leukemia. 2014;28:1647–1656. doi: 10.1038/leu.2014.61. [DOI] [PubMed] [Google Scholar]
- 10.Kassambara A., Herviou L., Ovejero S., Jourdan M., Thibaut C., Vikova V., et al. RNA-sequencing data-driven dissection of human plasma cell differentiation reveals new potential transcription regulators. Leukemia. 2021;35:1451–1462. doi: 10.1038/s41375-021-01234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price M.J., Hicks S.L., Bradley J.E., Randall T.D., Boss J.M., Scharer C.D. IgM, IgG, and IgA influenza-specific plasma cells express divergent transcriptomes. J. Immunol. 2019;203:2121–2129. doi: 10.4049/jimmunol.1900285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King H.W., Orban N., Riches J.C., Clear A.J., Warnes G., Teichmann S.A., et al. Single-cell analysis of human B cell maturation predicts how antibody class switching shapes selection dynamics. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abe6291. [DOI] [PubMed] [Google Scholar]
- 13.Eyer K., Doineau R.C.L., Castrillon C.E., Briseño-Roa L., Menrath V., Mottet G., et al. Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat. Biotechnol. 2017;35:977–982. doi: 10.1038/nbt.3964. [DOI] [PubMed] [Google Scholar]
- 14.Gérard A., Woolfe A., Mottet G., Reichen M., Castrillon C., Menrath V., et al. High-throughput single-cell activity-based screening and sequencing of antibodies using droplet microfluidics. Nat. Biotechnol. 2020;38:715–721. doi: 10.1038/s41587-020-0466-7. [DOI] [PubMed] [Google Scholar]
- 15.Broketa M., Bruhns P. Single-cell technologies for the study of antibody-secreting cells. Front. Immunol. 2022;12:821729. doi: 10.3389/fimmu.2021.821729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson V.M., Zhang K.X., Kumar N., Wong J., Li L., Wilson D.C., et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 18.van Buggenum J.A.G., Gerlach J.P., Tanis S.E.J., Hogeweg M., Jansen P.W.T.C., Middelwijk J., et al. Immuno-detection by sequencing enables large-scale high-dimensional phenotyping in cells. Nat. Commun. 2018;9:2384. doi: 10.1038/s41467-018-04761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Eijl R.A.P.M., van Buggenum J.A.G.L., Tanis S.E.J., Hendriks J., Mulder K.W. Single-cell ID-seq reveals dynamic BMP pathway activation upstream of the MAF/MAFB-program in epidermal differentiation. iScience. 2018;9:412–422. doi: 10.1016/j.isci.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivello F., van Buijtenen E., Matuła K., van Buggenum J.A.G.L., Vink P., van Eenennaam H., et al. Single-cell intracellular epitope and transcript detection reveals signal transduction dynamics. Cell Rep. Methods. 2021;1:100070. doi: 10.1016/j.crmeth.2021.100070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlach J.P., van Buggenum J.A.G., Tanis S.E.J., Hogeweg M., Heuts B.M.H., Muraro M.J., et al. Combined quantification of intracellular (phospho-)proteins and transcriptomics from fixed single cells. Sci. Rep. 2019;9:1469. doi: 10.1038/s41598-018-37977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimshony T., Senderovich N., Avital G., Klochendler A., de Leeuw Y., Anavy L., et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roelli P., Bbimber, Flynn B., Santiagorevale, Gui G. Hoohm/CITE-seq-Count: 1.4.2. Zenodo. 2019 doi: 10.5281/zenodo.2590196. [DOI] [Google Scholar]
- 24.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreatta M., Carmona S.J. UCell: robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 2021;19:3796–3798. doi: 10.1016/j.csbj.2021.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argelaguet R., Arnol D., Bredikhin D., Deloro Y., Velten B., Marioni J.C., et al. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. 2020;21:111. doi: 10.1186/s13059-020-02015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halliley J.L., Tipton C.M., Liesveld J., Rosenberg A.F., Darce J., Gregoretti I.V., et al. Long-lived plasma cells are contained within the CD19−CD38hiCD138+ subset in human bone marrow. Immunity. 2015;43:132–145. doi: 10.1016/j.immuni.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park K.S., Bayles I., Szlachta-McGinn A., Paul J., Boiko J., Santos P., et al. Transcription elongation factor ELL2 drives Ig secretory-specific mRNA production and the unfolded protein response. J. Immunol. 2014;193:4663–4674. doi: 10.4049/jimmunol.1401608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe K., Sugai M., Nambu Y., Osato M., Hayashi T., Kawaguchi M., et al. Requirement for Runx proteins in IgA class switching acting downstream of TGF-beta 1 and retinoic acid signaling. J. Immunol. 2010;184:2785–2792. doi: 10.4049/jimmunol.0901823. [DOI] [PubMed] [Google Scholar]
- 30.Kawabe T., Naka T., Yoshida K., Tanaka T., Fujiwara H., Suematsu S., et al. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 31.Arens R., Nolte M.A., Tesselaar K., Heemskerk B., Reedquist K.A., van Lier R.A.W., et al. Signaling through CD70 regulates B cell activation and IgG production. J. Immunol. 2004;173:3901–3908. doi: 10.4049/jimmunol.173.6.3901. [DOI] [PubMed] [Google Scholar]
- 32.Mantchev G.T., Cortesão C.S., Rebrovich M., Cascalho M., Bram R.J. TACI is required for efficient plasma cell differentiation in response to T-independent type 2 antigens. J. Immunol. 2007;179:2282–2288. doi: 10.4049/jimmunol.179.4.2282. [DOI] [PubMed] [Google Scholar]
- 33.Smulski C.R., Eibel H. BAFF and BAFF-receptor in B cell selection and survival. Front. Immunol. 2018;9:2285. doi: 10.3389/fimmu.2018.02285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radbruch A., Muehlinghaus G., Luger E.O., Inamine A., Smith K.G.C., Dörner T., et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat. Rev. Immunol. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 35.Cassese G., Arce S., Hauser A.E., Lehnert K., Moewes B., Mostarac M., et al. Plasma cell survival is mediated by synergistic effects of cytokines and adhesion-dependent signals. J. Immunol. 2003;171:1684–1690. doi: 10.4049/jimmunol.171.4.1684. [DOI] [PubMed] [Google Scholar]
- 36.Kunkel E.J., Butcher E.C. Plasma-cell homing. Nat. Rev. Immunol. 2003;3:822–829. doi: 10.1038/nri1203. [DOI] [PubMed] [Google Scholar]
- 37.Wei C., Chen Y., Xu L., Yu B., Lu D., Yu Y., et al. CD40 signaling promotes CXCR5 expression in B cells via noncanonical NF- κ B pathway activation. J. Immunol. Res. 2020;2020:1859260. doi: 10.1155/2020/1859260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ring A.M., Lin J.X., Feng D., Mitra S., Rickert M., Bowman G.R., et al. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat. Immunol. 2012;13:1187–1195. doi: 10.1038/ni.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diehl S.A., Schmidlin H., Nagasawa M., van Haren S.D., Kwakkenbos M.J., Yasuda E., et al. STAT3-mediated up-regulation of BLIMP1 is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J. Immunol. 2008;180:4805. doi: 10.4049/jimmunol.180.7.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto D., Montani E., Bolli M., Garavaglia G., Sallusto F., Lanzavecchia A., et al. A functional BCR in human IgA and IgM plasma cells. Blood. 2013;121:4110–4114. doi: 10.1182/blood-2012-09-459289. [DOI] [PubMed] [Google Scholar]
- 41.Blanc P., Moro-Sibilot L., Barthly L., Jagot F., This S., De Bernard S., et al. Mature IgM-expressing plasma cells sense antigen and develop competence for cytokine production upon antigenic challenge. Nat. Commun. 2016;7:13600. doi: 10.1038/ncomms13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gulla A., Anderson K.C. Multiple myeloma: the (r)evolution of current therapy and a glance into the future. Haematologica. 2020;105:2358–2367. doi: 10.3324/haematol.2020.247015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jang J.S., Li Y., Mitra A.K., Bi L., Abyzov A., van Wijnen A.J., et al. Molecular signatures of multiple myeloma progression through single cell RNA-Seq. Blood Cancer J. 2019;9:2. doi: 10.1038/s41408-018-0160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen Y.C., Zada M., Wang S.Y., Bornstein C., David E., Moshe A., et al. Identification of resistance pathways and therapeutic targets in relapsed multiple myeloma patients through single-cell sequencing. Nat. Med. 2021;27:491–503. doi: 10.1038/s41591-021-01232-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng H., Wang L., Li J., Luo S., Han Q., Su F., et al. Single-cell RNA-sequencing reveals distinct immune cell subsets and signaling pathways in IgA nephropathy. Cell Biosci. 2021;11:203. doi: 10.1186/s13578-021-00706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Argelaguet R., Cuomo A.S.E., Stegle O., Marioni J.C. Computational principles and challenges in single-cell data integration. Nat. Biotechnol. 2021;39:1202–1215. doi: 10.1038/s41587-021-00895-7. [DOI] [PubMed] [Google Scholar]
- 47.Ashuach T., Gabitto M.I., Jordan M.I., Yosef N. MultiVI: deep generative model for the integration of multi-modal data. bioRxiv. 2021 doi: 10.1101/2021.08.20.457057. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gong B., Zhou Y., Purdom E. Cobolt: integrative analysis of multimodal single-cell sequencing data. Genome Biol. 2021;22:351. doi: 10.1186/s13059-021-02556-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghazanfar S., Guibentif C., Marioni J.C. StabMap: mosaic single cell data integration using non-overlapping features. bioRxiv. 2022 doi: 10.1101/2022.02.24.481823. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li G., Fu S., Wang S., Zhu C., Duan B., Tang C., et al. A deep generative model for multi-view profiling of single-cell RNA-seq and ATAC-seq data. Genome Biol. 2022;23:20. doi: 10.1186/s13059-021-02595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao Y., Stuart T., Kowalski M., Choudhary S., Hoffman P., Hartman A., et al. Dictionary learning for integrative, multimodal, and scalable single-cell analysis. bioRxiv. 2022 doi: 10.1101/2022.02.24.481684. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monroe J.G. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat. Rev. Immunol. 2006;6:283–294. doi: 10.1038/nri1808. [DOI] [PubMed] [Google Scholar]
- 53.Juszczynski P., Chen L., O'Donnell E., Polo J.M., Ranuncolo S.M., Dalla-Favera R., et al. BCL6 modulates tonic BCR signaling in diffuse large B-cell lymphomas by repressing the SYK phosphatase, Ptprot. Blood. 2009;114:5315–5321. doi: 10.1182/blood-2009-02-204362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mócsai A., Ruland J., Tybulewicz V.L.J. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat. Rev. Immunol. 2010;106:387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fruchon S., Kheirallah S., Al Saati T., Ysebaert L., Laurent C., Leseux L., et al. Involvement of the Syk-mTOR pathway in follicular lymphoma cell invasion and angiogenesis. Leukemia. 2012;26:795–805. doi: 10.1038/leu.2011.248. [DOI] [PubMed] [Google Scholar]
- 56.Leseux L., Hamdi S.M., Al Saati T., Capilla F., Recher C., Laurent G., et al. Syk-dependent mTOR activation in follicular lymphoma cells. Blood. 2006;108:4156–4162. doi: 10.1182/blood-2006-05-026203. [DOI] [PubMed] [Google Scholar]
- 57.Benhamron S., Pattanayak S.P., Berger M., Tirosh B. mTOR activation promotes plasma cell differentiation and bypasses XBP-1 for immunoglobulin secretion. Mol. Cell. Biol. 2015;35:153–166. doi: 10.1128/MCB.01187-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed code documentation of the analysis and generation of figures is available from https://vanbuggenum.github.io/Multimodal-Plasmacell_manuscript/. The fastq files and count are accessible through GEO (57) Series accession number GSE189953 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189953).