Abstract
The meat and egg of goose is one of the main components of human food supply. The improvement of goose egg production is particularly important for the increasing human population. However, limited information is available about the effective molecular markers and mechanisms of egg production in goose. In this study, we jointly utilized the data of genome resequencing in different egg production Sichuan white goose and transcriptome at different follicle development stages to identified the molecular markers and mechanisms of egg production. The coefficient of variation of individual egg production in Sichuan white goose population is 0.42 to 0.49. Fifty individuals with the highest (laying 365 days egg number, LEN365 = 79–145) and 50 individuals with the lowest (LEN365 = 8–48) egg production were divided into high and low egg production groups. Based on whole-genome sequencing data of the selected samples, 36 SNPs (annotation novel.12.470, CELF2, ATP1A1, KCNJ6, RAB4A, UST, REV3L, DHX15, CAVN2, SLC5A9, Cldn5, MRPS23, and Tspan2) associated with the LEN365 were identified, involving multiple pathways such as metabolism and endocrinology. Notably, 5 SNPs located in the exon9 of ATP1A1 were identified by GWAS analysis. The association analysis with LEN365 showed the phenotypic variance explained of this haplotype consisting of 5 SNPs is 20.51%. Through transcriptome data analysis, we found the expression of ATP1A1 in the granular layers was increased in the stage of small yellow follicle to large yellow follicle (LYF) and LYF to F5, while decreased in F2 to F1. For the first time, we report the haplotype region formed by 5 SNPS on exon9 of ATP1A1 is associated with egg production in goose and involved in follicle selection and maturation processes.
Key words: goose, egg production, GWAS, RNA-seq
INTRODUCTION
The global demand for geese is growing rapidly as the World Health Organization (WHO) listed them as the best meat when presenting a new global health strategy at its 113th session. However, compared with poultry such as chicken and duck, the low egg production of goose (20–60 eggs / year) greatly restricts the development of the industry. Egg production in geese is mainly determined by the development (Hu et al., 2020; Yuan et al., 2021) and selection (Li et al., 2019, 2020) of follicle, and these 2 biological processes are co-regulated by endocrine (Du et al., 2020), nutritional (Pan et al., 2014; Wei et al., 2019), environmental (Manser, 1996; Vandana et al., 2020), disease (Johnson and Giles, 2013), and genetic factors (Dana et al., 2011). The mechanisms of these regulatory factors are different and interact with each other. Due to the diverse influencing factors and low heritability (Emamgholi Begli et al., 2021), the genetic progress of egg production in goose obtained by the traditional phenotypic selection method is slow.
Rapid progress has been made in identifying loci controlling egg production in poultry and applying them to marker-assisted selection with the development of genome sequencing technology. More than 600 QTL loci related to egg production in chicken have been identified (Hu et al., 2019), and the functions of some major genes have been gradually resolved (Rubin et al., 2010). A study in the F2 of mallard and Pekin ducks have report the candidate gene of reproduction (Liu et al., 2021). In recent years, the single cage breeding technology of goose has gradually matured, and found that the individual egg production of geese varied greatly through individual egg production record. Then, some SNPs associated with the egg production of Sichuan white goose (Gao, et al., 2021) and Lion head goose (Zhao et al., 2019) has been identified. However, the 2 studies identified different markers, indicating that different populations may have different molecular markers related to goose egg production.
On the other hand, the researchers found some candidate genes were related to goose egg production by used transcriptome sequencing technology to compare the tissue at different laying levels (Wu, et al., 2020; Ouyang et al., 2022), physiological stages (Ouyang et al., 2020; Qin et al., 2021), and follicles development stages. However, whether these candidate genes have reliable molecular markers is unknown. In other words, there is still a lack of candidate molecular markers and pathways for identified goose egg production by combined analysis of multiple genomics. Therefore, we jointly GWAS data of different egg production Sichuan white goose and RNA-seq data of different follicle development stages to identified the molecular markers and mechanisms of egg production in this study.
MATERIALS AND METHODS
Ethics Approval and Consent to Participate
All geese were obtained from the Waterfowl Breeding Experimental Farm of Sichuan Agricultural University. All experimental procedures that involved in animal manipulation were approved by the Institutional Animal Care and Use Committee (IACUC) of Sichuan Agricultural University (Chengdu Campus, Sichuan, China), approval number: 20160067.
Sample Collection and DNA Extraction
A total of 200 female Sichuan white geese hatched in the same batch were reared in single cages after the end of the brood period. All geese were raised under same environment (single cage and light duration is 16 h one day) and fed the same diet (free feeding) with free access to water. The laying performance of 200 geese was recorded daily until laying 365 d. The egg number of 365-day-old was record as EN365, the egg number of laying 365 d was record as LEN365, and the egg production frequency was record as EF365 and LEF365. Blood samples were collected from the wings of all geese using vacuum tubes containing ethylenediaminetetraacetic acid. These blood samples were used for DNA extraction (50 individuals with the highest and lowest LEN365 values). Genomic DNA was extracted from the blood tissues of Sichuan white geese with dneasy blood and tissue Kit (Qiagen, Valencia, CA). The quality and quality of DNA were evaluated by Agilent 2200 tapestation (Agilent Technologies, Palo Alto, CA), qubit 2.0 fluorometer (Thermo Fisher Scientific, Wilmington, DE) and agarose gel electrophoresis.
Whole-Genome Resequencing and Variation Calling
We use the genobaits DNA SEQ library prep kit to construct a resequencing Library of qualified DNA (50 individuals with the highest and lowest LEN365 values, respectively), and then use qubit2.0 for preliminary quantification. We use qPCR to accurately quantify the effective concentration of the library to ensure the quality of the library. After the library passed the quality inspection, it was sequenced using BGI MGI-2000/MGI-T7 sequencing platform, and the sequencing mode was PE150. The original sequencing data for this study can be found in the Sequence Read Ar-chive (https://www.ncbi.nlm.nih.gov/sra) at NCBI with the BioProject ID: PRJNA883750. Then, Fastqc software was used to detect the original data. According to the quality of the original data, we used the NGS QC toolkit (Patel and Jain, 2012) to quality control the original sequencing data and remove the residual primer and linker sequences. At the same time, the bases with a sequencing read length of less than 70% and a sequencing quality of more than 20 were also removed, and Fastqc software was used to test the data after quality control. Subsequently, the data after quality control were aligned to the chromosomes level Sichuan white goose we assembled using the 'bwa-k40-m-r' parameter of BWA-MEM software (Li, 2013) (data not released). Samtools software was used (Li et al., 2009) to convert the mapping results into BAM format and filter unmapped and non-uniquely mapped reads. The variation information was obtained according to the best practice workflow recommended by GATK (v4.1.7.0) software (McKenna et al., 2010). In brief, SNPs calling was performed for each sample using the HaplotypeCaller module and GVCF model using GATK software, and the combined GVCF file was used to carry out a joint genotyping step for the comprehensive variation among all samples. The “SelectVariants” of GATK software was used to selected SNPs and Indels. SNPs with missing rate > 0.1, sequencing depth < 4, minor allele frequency (MAF) < 0.05 or GQ < 5 were also excluded, and the remaining were used for subsequent analysis.
Population Structure Analysis
In this study, Plink software (Purcell et al., 2007) was used to calculate the eigenvalues and eigenvector values of each principal component. Then, the ggplot2 package of R was used to plot PCA using PC1, PC2, and PC3 values. In addition, Admixture software (V1.3) (Alexander et al., 2009) was used to infer the population structure, and the number of clusters (K value) of the sample was assumed to be 1 to 15, and then clustering was performed. The optimal number of clusters is determined according to the cross-validation error rate, and the K value with the smallest cross-validation error rate corresponds to the optimal number of clusters. At the same time, GCTA software (V1.92.4) (Yang et al., 2011) was used for kinship analysis to obtain the kinship matrix between pairwise samples, and R was used to draw the heat map.
Genome-Wide Association Analysis
GWAS was performed using Genome-wide Efficient Mixed-Model Association (GEMMA) software (Zhou and Stephens, 2012). The population structure corresponding to no covariate and Admixture optimal K value and the relationship matrix were analyzed as covariates. The qqman software package in R software was used to visualize the Manhattan and Q-Q maps of GWAS results. The optimal analysis model was selected according to the Q-Q plot. P<0.05/N+1 (9.35E-09) and 10E-05 was set as the thresholds of significant and potential correlation with traits, respectively, and variant information was annotated into gene names. LDblockShow software (Dong et al., 2021) was used to analysis the of candidate gene. The structural diagram of genes was visualized by IBS software (Liu et al., 2015). The IGV software was used to visualize the genotype of ATP1A1. Functional analysis was used KOBAS 3.0 online (http://kobas.cbi.pku.edu.cn/kobas3/?t=1) (Xie et al., 2011).
Bioinformatics Analysis of RNA-seq Data
The RNA-seq data was download from previous study in NCBI PRJNA506334 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA506334) and PRJNA552525 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA552525). The clean reads were obtained after the filtration of low-quality reads using standard quality control by FastaQC software. Clean reads were mapped to the Anser cygnoides domestication reference genome (data being published) using the HISAT2 (version 2.2.1) software (Kim et al., 2015). The output SAM (sequencing alignment/mapping) file was converted to a BAM (binary alignment/mapping) file and sorted using SAMtools (version 1.10) (Li et al., 2009). Subsequently, the expression of each transcript was calculated by featureCounts (version 1.6.0) (Liao et al., 2014) and the readcounts of ATP1A1 was extracted.
RESULTS
Analysis of Egg Production Data of Sichuan White Geese
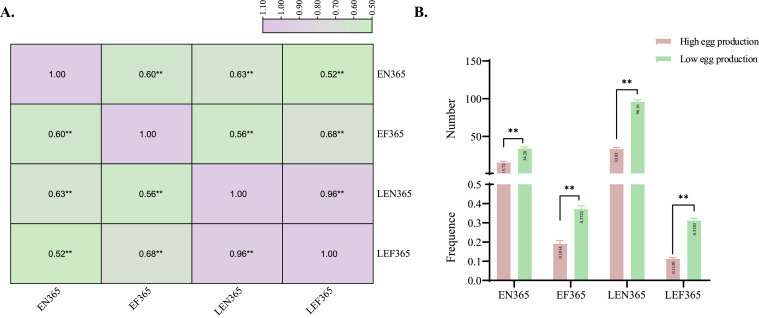
By the end of data collection in this experiment, 195 geese were survived, of which 188 had begun to lay eggs. The average EN365 is 25.69, the average EF365 is 0.29, and the coefficient variation (CV) is 0.49 and 0.45, respectively. And, the average LEN365 was 64.46, the average LEF365 was 0.21, and the CV was 0.42 and 0.43, respectively. As shown in Figure 1A, the results of correlation analysis showed that the four traits were all positively correlated. The LEN365 larger than 79 was divided into high egg production group (LEN365 = 79–145, N = 50), and lower than 48 was divided into low egg production group (LEN365 = 8–48, N = 50). As shown in Figure 1B, there was a significant difference between the 2 groups in EN365, EF365, LEN365, and LEF365.
Figure 1.
Correlation analysis (A) and comparison between high and low egg production groups (B) of four egg production traits in Sichuan white goose.
Population Structure Analysis and the Selection of Model
The basic information of genome re-sequence data was shown in Supplementary Table 1. A total of 5,347,972 SNPs were obtained after filtering. PCA analysis was conducted to obtain the variance interpretation rate of each PC and the score matrix of samples in each PC. The results of PC1, PC2, and PC3 of all individuals are shown in Figure 2A. Population structure is a common issue in GWAS analysis, which refers to the large difference of genetic background among different populations. In studies with large samples, the population structure will cause false positive and false negative results. In this study, Admixture software was used to simulate the stratification of the population when K = 1–15. As shown in Figure 2B, there was a minimum cross-validation error rate when K = 1 (0.66292), indicating that the population had no obvious stratification. The genetic relationship is defined as the relative value of the genetic similarity between 2 specific materials and that between any materials. In GWAS, in order to avoid false positives caused by possible small families, the relationship matrix is often added to the GWAS model as a random effect covariate matrix (K matrix). Based on the SNP markers obtained after screening, we obtained the affinity matrix between 2 samples. The heat map drawn with the affinity matrix is shown in Figure 2C. In order to confirm the best model of GWAS in this study, we used GEMMA software to calculate the common models of GWAS. In this study, K = 1 is the optimal K value, so only Q-Q charts without covariates and kinship matrix are shown as covariates. As shown in Figure 2D, the model without covariates has low significance in the lower left corner of the scatter plot, and the observed P values of these sites should be consistent with the expected values. However, most points of the model with covariates in the relationship matrix are below the diagonal, indicating that the observed P values of most sites are less than the expected values, which may be caused by the over correction of P values due to the unreasonable model (Figure 2E). Therefore, the best model of this study is the logistic regression model without covariates.
Figure 2.
Population structure analysis and the GWAS model comparison. (A) Principal component analysis of population genome-wide SNP. (B) Line chart of cross validation error. (C) Kinship heat map of individuals in population. (D) Q-Q plot of GWAS analysis using model without covariates. (E) Q-Q plot of GWAS analysis using model with kinship matrix as covariate.
Identify SNPs Related to Egg Production of Sichuan White Geese by GWAS
As shown in Figure 3A, using the optimal model, one SNP (Chr12: 19283739) significantly related to the LEN365 was identified and annotated to the downstream region of novel.12.470 gene. In addition, 36 SNPs potential associated with the LEN365 were identified and annotated with novel.12.470, CELF2, ATP1A1, KCNJ6, RAB4A, UST, REV3L, DHX15, CAVN2, SLC5A9, Cldn5, MRPS23, and Tspan2 (Table 1), involving multiple pathways such as metabolism and endocrinology (Figure 3B). Notably, 5 SNPs located in the exon9 of ATP1A1 were identified to be related to LEN365.
Figure 3.
The Manhattan plot of genome-wide association analysis by SNPs (A) and bubble plots of KEGG pathways enriched by the potential associated genes (B).
Table 1.
The annotation of potential SNPs related to LEN365.
| Chr | Position | P_wald | Alt | Ref | Annotation | Gene Id |
|---|---|---|---|---|---|---|
| 12 | 19283739 | 6.76516E-09 | G | A | Distal Intergenic | novel.12.470 |
| 12 | 19283743 | 1.38083E-08 | G | A | Distal Intergenic | novel.12.470 |
| 12 | 19282504 | 2.22321E-08 | T | C | Distal Intergenic | novel.12.470 |
| 12 | 19283709 | 2.44495E-08 | G | A | Distal Intergenic | novel.12.470 |
| 12 | 19282898 | 8.00825E-08 | T | A | Distal Intergenic | novel.12.470 |
| 12 | 19282903 | 8.00825E-08 | T | C | Distal Intergenic | novel.12.470 |
| 1 | 101632368 | 1.07272E-07 | A | G | Exon 9 | ATP1A1 |
| 12 | 19282307 | 2.1286E-07 | T | C | Distal Intergenic | novel.12.470 |
| 1 | 101632371 | 2.18728E-07 | C | T | Exon 9 | ATP1A1 |
| 1 | 101632386 | 2.24423E-07 | G | T | Exon 9 | ATP1A1 |
| 12 | 19283607 | 2.92242E-07 | C | T | Distal Intergenic | novel.12.470 |
| 1 | 101632377 | 6.6004E-07 | A | G | Exon 9 | ATP1A1 |
| 4 | 19170167 | 6.86078E-07 | G | A | Intron2 | DHX15 |
| 12 | 19283513 | 1.42251E-06 | T | C | Distal Intergenic | novel.12.470 |
| 3 | 52772215 | 2.18016E-06 | C | T | Intron1 | UST |
| 12 | 19282675 | 2.28861E-06 | C | T | Distal Intergenic | novel.12.470 |
| 7 | 4097228 | 3.00353E-06 | T | C | Intron7 | CAVN2 |
| 1 | 101641618 | 3.11451E-06 | A | G | Exon 18 | ATP1A1 |
| 12 | 19282615 | 3.62545E-06 | C | T | Distal Intergenic | novel.12.470 |
| 19 | 11490985 | 3.73786E-06 | A | C | Promoter (1-2kb) | MRPS23 |
| 26 | 392954 | 4.21723E-06 | A | G | Promoter (1-2kb) | Tspan2 |
| 3 | 72820629 | 4.22868E-06 | A | G | Intron1 | REV3L |
| 1 | 7710803 | 4.6221E-06 | G | C | Distal Intergenic | CELF2 |
| 12 | 19279685 | 5.14488E-06 | C | T | Intron2 | novel.12.470 |
| 8 | 24388477 | 5.5261E-06 | G | T | Distal Intergenic | SLC5A9 |
| 3 | 44398354 | 5.77172E-06 | A | G | Distal Intergenic | RAB4A |
| 12 | 19300377 | 6.16305E-06 | C | T | Distal Intergenic | novel.12.470 |
| 1 | 101632395 | 6.35856E-06 | T | C | Exon 9 | ATP1A1 |
| 3 | 44423996 | 7.56207E-06 | T | G | Distal Intergenic | RAB4A |
| 1 | 117591565 | 7.82765E-06 | C | T | Distal Intergenic | KCNJ6 |
| 12 | 19282117 | 8.54134E-06 | T | C | Distal Intergenic | novel.12.470 |
| 3 | 44376799 | 8.61107E-06 | A | G | Distal Intergenic | RAB4A |
| 12 | 19279630 | 8.97311E-06 | G | A | Intron2 | novel.12.470 |
| 3 | 44423548 | 9.086E-06 | C | T | Distal Intergenic | RAB4A |
| 15 | 7582115 | 9.63907E-06 | A | G | Distal Intergenic | Cldn5 |
| 12 | 19314129 | 9.84568E-06 | C | T | Distal Intergenic | novel.12.470 |
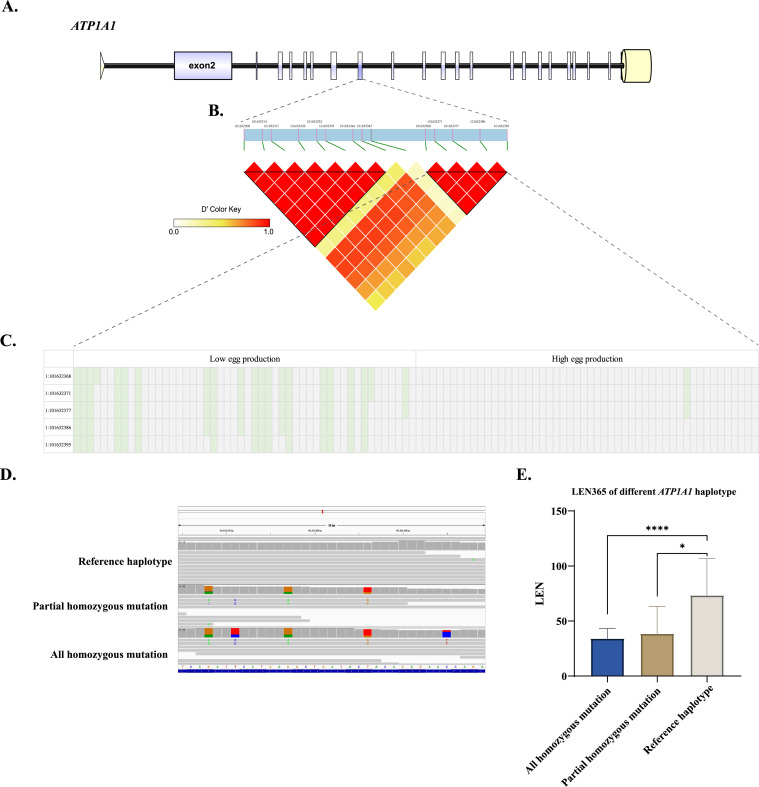
Five SNPs on Exon9 of ATP1A1 are Associated With Egg Production of Sichuan White Geese
ATP1A1 was highly conserved among different species, and the 2 mutations in the exon9 were nonsense mutations (Supplementary Figure 1). As shown in Supplementary Figure 2, all SNPs in ATP1A1 were analyzed for linkage disequilibrium (LD), and it was found the 5 SNPs in the exon9 of goose ATP1A1 were in one block (Figures 4A and 4B). As shown in Figure 4C, the haplotype showed the different distribution between high and low egg production goose. Then, we verified the accuracy of genome resequencing and supplemented the genotype data of another 100 individuals by PCR and sanger sequencing of the mutation region (Figure 4D). Furthermore, it was found that the LEN365 of the samples with reference haplotype is significantly higher than that of partial homozygous mutation haplotype, and is extremely significantly higher than that of all mutation haplotype (Figure 4E). Based on the regression equation, we found the phenotypic variance explained (PVE) of this haplotype is 20.51%.
Figure 4.
Verification, sequence, and functional analysis of 5 SNPs in the exon9 of goose ATP1A1. (A) The gene structural of goose ATP1A1. The modular with purple represents the exon, yellow represent the 5’ and 3’ sequence. (B) The LD analysis of ATP1A1 exon9. (C) Haplotype of 5 SNPs located in exon9 of ATP1A1 between high and low egg production goose. (D) IGV visualizes of different haplotype samples. (E) The comparison of LEN365 among different haplotype, *represents P < .05, ****represents P < .00001.
Transcriptome Analysis Revealed Changes in the Expression of ATP1A1 During Follicle Development
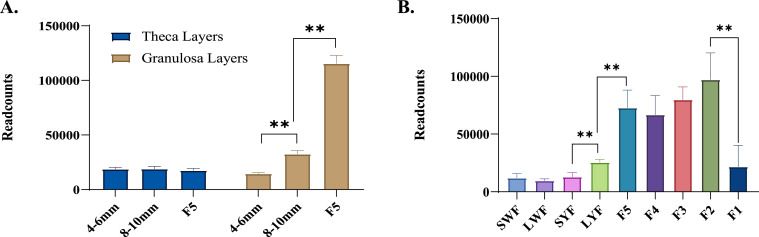
Through transcriptome analysis, we found that the expression of ATP1A1 in the theca layers of small yellow follicle (SYF), large yellow follicle (LYF) and F5 follicles did not change, but significantly increased in the granulosa layer during this process (Figure 5A). Meanwhile, the data from another project showed the same results among SYF, LYF, and F5 follicles granulosa. The expression of ATP1A1 in the follicle granulosa layer was not changed at the stage of small white follicle (SWF) to large white follicle (LWF), LWF to SYF, and F5 to F2. During follicle maturation stage (F2-F1), the expression of ATP1A1 decreased significantly.
Figure 5.
Expression trend and functional analysis of ATP1A1 in granulosa and theca layers of different follicle development stage in goose. (A) Expression trend of ATP1A1 in granulosa and theca layers of SYF, LYF, and F5 follicles in goose (Raw data from PRJNA506334). (B) Expression trend of ATP1A1 in granulosa layers of SWF, LWF, SYF, LYF, and F1-F5 follicles in goose (Raw data from PRJNA552525). ** represents |log2Foldchange|>1 and FDR < 0.05. The data was shown as Mean±SEM.
DISCUSSION
In the past few decades, the selection of goose egg production was based on the group due to the limitation of the feeding mode of geese. However, with the development of single cage technology, we have been able to achieve individual records of egg production in goose. The results of this study showed that the CV of EN365 and laying LEN365 were 0.49 and 0.42, respectively, indicating there were great differences among individuals within the breed. On the other hand, the correlation among EN365, LEN365, EF365, and LEF365 of Sichuan white geese is relatively high, and LEN365 with a longer statistical period may better reflect the real situation of individuals, while EN365 can be used for early selection.
In populations with simple genetic background and obvious segregation of traits, 36 SNPs were identified to be related with LEN365. The SNP on novel.12.470 has the highest correlation with LEN365 of Sichuan white goose. However, novel.12.470, as an un annotated gene, its role and the pathway need to be further explored. A SNP within the distal intergenic of CELF2 was significantly associated with LEN365. CELF2 can form complexes with different enzymes to play a role in APOB editing (Anant et al., 2001; Chen et al., 2007), and regulated by estradiol (Del Pino Sans et al., 2015). In addition, KCNJ6, a hormone related gene, was also identified to be related to LEN365. The mutation KCNJ6 can cause decrease of serum thyroid hormone level (Blum et al., 1999). KCNJ6 is also one of the specific subunits in GnRH neurons and a messenger regulating reproductive function (Constantin and Wray, 2018). UST (Cadwalader et al., 2012) and REV3L (Lange et al., 2016) play important roles in embryonic development. RAB4A has been reported to be related to the litter size of sheep (Esmaeili-Fard et al., 2021). RAB4A has been identified to be related to reproductive performance in geese and sheep, which may be due to its role in angiogenesis and proliferation related to follicular development (Fraser, 2006; Jopling et al., 2014).
It is worth noting that the 5 SNPS located in a linked block of the ATP1A1 exon9 were identified to be associated with goose egg production. As the encoding gene of Na+-K+-ATPase, ATP1A1 has a wide range of functions, as almost all cells depend on the proper functioning of the NA+-K+ pump to maintain ion, osmotic, and electrical homeostasis. Na+-K+-ATPase maintaining intracellular ion composition by transporting internal Na+ and external K+ across the cell membrane in opposite concentration gradients. The sequence mutation of ATP1A1 was found to be related to heat tolerance (Liu et al., 2011), Charcot Marie Tooth (Lassuthova et al., 2018), and other diseases. Our study firstly reported the mutations in the exon of ATP1A1 are related to the reproductive of goose. Histochemical localization of ATPase activity sites in the ovary has been studied in several mammalian species, such as guinea pigs, rabbits, and humans (Adams et al., 1966; Koudstaal and Jöbsis, 1974). Sangha et al. (1991) reported the histological changes of general ATPase activity during follicles formation, corpus luteum formation and regression in the rat ovary. They found that with the growth of follicles, the ATPase activity in oocytes and granulosa cells decreased, while moderate to strong ATPase activity was observed in corpus luteum. The activity of ATPase is the sum of the activities of Na+- K+-ATPase, Ca2+-ATPase, Mg2+-ATPase, and H+-ATPase. For the Na+-K+-ATPase, Ge and Spicer reported their immunoreactivity in stroma cells and membrane cells of rat ovary (Ge and Spicer, 1988). The expression of ATP1A1 in pig primordial follicles is weak, but the expression is significantly increased when the follicles at the pre-ovulation stage (Aljonaid et al., 2003). Meanwhile, the expression of ATP1A1 is also significantly increased during flatfish oocyte maturation (Tingaud-Sequeira et al., 2009). It is speculated that it is related to changes in permeability and metabolism during oocyte maturation. At the same time, ATP1A1 immunostaining increased in pig granulosa cells with the growth of follicles, which was consistent with our transcriptome results in goose granulosa cells. Mattioli et al. proved that the up regulation of ATP1A1 expression may be related to the depolarization of granulosa cells (Mattioli et al., 1990). This change in potential level may participate in the endocrine signal transduction mechanism, and hormone stimulation may make cells enter a new functional state. These studies provide auxiliary evidence for ATP1A1 to regulate the egg production of goose by participating in the development of follicles.
In conclusion, we have reported the CV of individual egg production in Sichuan white goose population is 0.42 to 0.49. 36 SNPs (annotation novel.12.470, CELF2, ATP1A1, KCNJ6, RAB4A, UST, REV3L, DHX15, CAVN2, SLC5A9, Cldn5, MRPS23, and Tspan2) associated with the egg production of Sichuan white geese were identified by the GWAS. The joint analysis of GWAS and transcriptome found that five SNPs in a linkage region of ATP1A1 exon9 may regulate the egg production of goose by participating in the follicle development process, and the PVE for the egg production of geese was 20.51%. These results play an important role in further improving the egg production of geese.
ACKNOWLEDGMENTS
This research was supported by China Agriculture Research System of MOF and MARA (CARS-42-4) and Key Technology Support Program of Sichuan Province (2021YFYZ0014) for the financial support
DISCLOSURES
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2023.102488.
Appendix. Supplementary materials
References
- Adams E.C., Hertig A.T., Foster S. Studies on guinea pig oocytes. II. Histochemical observations on some phosphatases and lipid in developing and in atretic oocytes and follicles. Am. J. Anat. 1966;119:303–339. doi: 10.1002/aja.1001190207. [DOI] [PubMed] [Google Scholar]
- Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljonaid A.A., Sato A., Asahara S., Maruo T. Abundant expression of sodium-potassium-activated adenosinetriphosphatase alpha 1 subunit in corpus luteum of porcine ovary. Endocrine. 2003;21:233–240. doi: 10.1385/ENDO:21:3:233. [DOI] [PubMed] [Google Scholar]
- Anant S., Henderson J.O., Mukhopadhyay D., Navaratnam N., Kennedy S., Min J., Davidson N.O. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J. Biol. Chem. 2001;276:47338–47351. doi: 10.1074/jbc.M104911200. [DOI] [PubMed] [Google Scholar]
- Blum M., Weickert C., Carrasco E. The weaver GIRK2 mutation leads to decreased levels of serum thyroid hormone: characterization of the effect on midbrain dopaminergic neuron survival. Exp. Neurol. 1999;160:413–424. doi: 10.1006/exnr.1999.7231. [DOI] [PubMed] [Google Scholar]
- Cadwalader E.L., Condic M.L., Yost H.J. 2-O-sulfotransferase regulates Wnt signaling, cell adhesion and cell cycle during zebrafish epiboly. Development. 2012;139:1296–1305. doi: 10.1242/dev.078238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Eggerman T.L., Patterson A.P. ApoB mRNA editing is mediated by a coordinated modulation of multiple apoB mRNA editing enzyme components. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G53–G65. doi: 10.1152/ajpgi.00118.2006. [DOI] [PubMed] [Google Scholar]
- Constantin S., Wray S. Nociceptin/Orphanin-FQ inhibits gonadotropin-releasing hormone neurons via G-Protein-gated inwardly rectifying potassium channels. eNeuro. 2018;5 doi: 10.1523/ENEURO.0161-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana N., Waaij E.H.V., Arendonk J.A.M.V. Genetic and phenotypic parameter estimates for body weights and egg production in Horro chicken of Ethiopia. Trop. Anim. Health Prod. 2011;43:21. doi: 10.1007/s11250-010-9649-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pino Sans J., Krishnan S., Aggison L.K., Adams H.L., Shrikant M.M., López-Giráldez F., Petersen S.L. Microarray analysis of neonatal rat anteroventral periventricular transcriptomes identifies the proapoptotic Cugbp2 gene as sex-specific and regulated by estradiol. Neuroscience. 2015;303:312–322. doi: 10.1016/j.neuroscience.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Dong S.S., He W.M., Ji J.J., Zhang C., Guo Y., Yang T.L. LDBlockShow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief Bioinform. 2021;22 doi: 10.1093/bib/bbaa227. [DOI] [PubMed] [Google Scholar]
- Du Y., Liu L., He Y., Dou T., Jia J., Ge C. Endocrine and genetic factors affecting egg laying performance in chickens: a review. Br. Poult. Sci. 2020;61:538–549. doi: 10.1080/00071668.2020.1758299. [DOI] [PubMed] [Google Scholar]
- Emamgholi Begli H., Schaeffer L.R., Abdalla E., Lozada-Soto E.A., Harlander-Matauschek A., Wood B.J., Baes C.F. Genetic analysis of egg production traits in turkeys (Meleagris gallopavo) using a single-step genomic random regression model. Genet. Sel. Evol. 2021;53:61. doi: 10.1186/s12711-021-00655-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili-Fard S.M., Gholizadeh M., Hafezian S.H., Abdollahi-Arpanahi R. Genome-wide association study and pathway analysis identify NTRK2 as a novel candidate gene for litter size in sheep. PLoS One. 2021;16 doi: 10.1371/journal.pone.0244408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H.M. Regulation of the ovarian follicular vasculature. Reprod. Biol. Endocrinol. 2006;4:18. doi: 10.1186/1477-7827-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Gao D., Zhao X., Xu S., Zhang K., Wu R., Yin C., Li J., Xie Y., Hu S., Wang Q. Genome-wide association study-based identification of SNPs and haplotypes associated with goose reproductive performance and egg quality. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.602583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z.H., Spicer S.S. Immunocytochemistry of ion transport mediators in the genital tract of female rodents. Biol. Reprod. 1988;38:439–452. doi: 10.1095/biolreprod38.2.439. [DOI] [PubMed] [Google Scholar]
- Hu S., Yang S., Lu Y., Deng Y., Li L., Zhu J., Zhang Y., Hu B., Hu J., Xia L., He H., Han C., Liu H., Kang B., Li L., Wang J. Dynamics of the transcriptome and accessible chromatin landscapes during early goose ovarian development. Front. Cell Dev. Biol. 2020;8:196. doi: 10.3389/fcell.2020.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.L., Park C.A., Reecy J.M. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic. Acids. Res. 2019;47:D701–D710. doi: 10.1093/nar/gky1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.A., Giles J.R. The hen as a model of ovarian cancer. Nat. Rev. Cancer. 2013;13:432–436. doi: 10.1038/nrc3535. [DOI] [PubMed] [Google Scholar]
- Jopling H.M., Odell A.F., Pellet-Many C., Latham A.M., Frankel P., Sivaprasadarao A., Walker J.H., Zachary I.C., Ponnambalam S. Endosome-to-plasma membrane recycling of VEGFR2 receptor tyrosine kinase regulates endothelial function and blood vessel formation. Cells. 2014;3:363–385. doi: 10.3390/cells3020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudstaal J., Jöbsis A.C. Enzyme histochemistry of the human ovary. Eur. J. Obstet. Gynecol. Reprod. Biol. 1974;4:S51–S57. [PubMed] [Google Scholar]
- Lange S.S., Tomida J., Boulware K.S., Bhetawal S., Wood R.D. The polymerase activity of mammalian DNA Pol ζ is specifically required for cell and embryonic viability. PLos Genet. 2016;12 doi: 10.1371/journal.pgen.1005759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassuthova P., Rebelo A.P., Ravenscroft G., Lamont P.J., Davis M.R., Manganelli F., Feely S.M., Bacon C., Brožková D., Haberlova J., Mazanec R., Tao F., Saghira C., Abreu L., Courel S., Powell E., Buglo E., Bis D.M., Baxter M.F., Ong R.W., Marns L., Lee Y.C., Bai Y., Isom D.G., Barro-Soria R., Chung K.W., Scherer S.S., Larsson H.P., Laing N.G., Choi B.O., Seeman P., Shy M.E., Santoro L., Zuchner S. Mutations in ATP1A1 cause dominant charcot-marie-tooth type 2. Am. J. Hum. Genet. 2018;102:505–514. doi: 10.1016/j.ajhg.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Chen R., Qin Q., Zhu H., Shi Z. Transcriptome analysis to unravel the gene expression profile of ovarian follicular development in Magang goose. J. Reprod. Dev. 2020;66:331–340. doi: 10.1262/jrd.2019-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 e-prints. [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Hu S., Wang Y., Deng Y., Yang S., Hu J., Li L., Wang J. mRNA and miRNA transcriptome profiling of granulosa and theca layers from geese ovarian follicles reveals the crucial pathways and interaction networks for regulation of follicle selection. Front. Genet. 2019;10:988. doi: 10.3389/fgene.2019.00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu H., Wang L., Guo Z., Xu Q., Fan W., Xu Y., Hu J., Zhang Y., Tang J., Xie M., Zhou Z., Hou S. Genome-wide association and selective sweep analyses reveal genetic loci for FCR of egg production traits in ducks. Genet. Sel. Evol. 2021;53:98. doi: 10.1186/s12711-021-00684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Xie Y., Ma J., Luo X., Nie P., Zuo Z., Lahrmann U., Zhao Q., Zheng Y., Zhao Y., Xue Y., Ren J. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 2015;31:3359–3361. doi: 10.1093/bioinformatics/btv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li D., Li H., Zhou X., Wang G. A novel SNP of the ATP1A1 gene is associated with heat tolerance traits in dairy cows. Mol. Biol. Rep. 2011;38:83–88. doi: 10.1007/s11033-010-0080-8. [DOI] [PubMed] [Google Scholar]
- Manser C.E. Effects of lighting on the welfare of domestic poultry: a review. Anim. Welfare (South Mimms, England) 1996;5:341–360. [Google Scholar]
- Mattioli M., Barboni B., Bacci M.L., Seren E. Maturation of pig oocytes: observations on membrane potential. Biol. Reprod. 1990;43:318–322. doi: 10.1095/biolreprod43.2.318. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Q., Hu S., Tang B., Hu B., Hu J., He H., Li L., Wang J. Comparative transcriptome analysis provides novel insights into the effect of lipid metabolism on laying of geese. Animals (Basel) 2022;12 doi: 10.3390/ani12141775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Q., Hu S., Wang G., Hu J., Zhang J., Li L., Hu B., He H., Liu H., Xia L. Comparative transcriptome analysis suggests key roles for 5-hydroxytryptamlne receptors in control of goose egg production. Genes. 2020;11:455. doi: 10.3390/genes11040455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.E., Liu Z.C., Chang C.J., Huang Y.F., Lai C.Y., Walzem R.L., Chen S.E. Feed restriction ameliorates metabolic dysregulation and improves reproductive performance of meat-type country chickens. Anim. Reprod. Sci. 2014;151:229–236. doi: 10.1016/j.anireprosci.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Patel R.K., Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Li X., Wang J., Sun G., Mu X., Ji R. Ovarian transcriptome profile from pre-laying period to broody period of Xupu goose. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C.J., Zody M.C., Eriksson J., Meadows J.R., Sherwood E., Webster M.T., Jiang L., Ingman M., Sharpe T., Ka S., Hallböök F., Besnier F., Carlborg O., Bed'hom B., Tixier-Boichard M., Jensen P., Siegel P., Lindblad-Toh K., Andersson L. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- Sangha G.K., Bilaspuri G.S., Guraya S.S. Histochemical changes in adenosine triphosphatase activity during folliculogenesis and corpus luteum formation and regression in the rat ovary. Eur. J. Morphol. 1991;29:285–290. [PubMed] [Google Scholar]
- Tingaud-Sequeira A., Chauvigné F., Lozano J., Agulleiro M.J., Asensio E., Cerdà J. New insights into molecular pathways associated with flatfish ovarian development and atresia revealed by transcriptional analysis. BMC Genomics. 2009;10:434. doi: 10.1186/1471-2164-10-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandana G.D., Sejian V., Lees A.M., Pragna P., Maloney S.K. Heat stress and poultry production: impact and amelioration. Int. J. Biometeorol. 2020;65:163–179. doi: 10.1007/s00484-020-02023-7. [DOI] [PubMed] [Google Scholar]
- Wei Z., Li P., Huang S., Lkhagvagarav P., Jia C. Identification of key genes and molecular mechanisms associated with low egg production of broiler breeder hens in ad libitum. BMC Genomics. 2019;20 doi: 10.1186/s12864-019-5801-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhao X., Chen L., Wang J., Duan Y., Li H., Lu L. Transcriptomic analyses of the hypothalamic-pituitary-gonadal axis identify candidate genes related to egg production in Xinjiang Yili Geese. Animals (Basel) 2020;10 doi: 10.3390/ani10010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C.Y., Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic. Acids. Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Hu S., Li L., Han C., Liu H., He H., Xia L., Hu J., Hu B., Ran M., Liu Y., Wang J. Lipidomics profiling of goose granulosa cell model of stearoyl-CoA desaturase function identifies a pattern of lipid droplets associated with follicle development. Cell Biosci. 2021;11:95. doi: 10.1186/s13578-021-00604-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Chen J., Zhang X., Xu Z., Lin Z., Li H., Lin W., Xie Q. Genome-wide association analysis reveals key genes responsible for egg production of lion head goose. Front. Genet. 2019;10:1391. doi: 10.3389/fgene.2019.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.