Abstract
Keratinophilic fungi are mostly soil-inhabiting organisms with occasional infections in humans and animals. Even though most dermatophytes are host-adapted, cross-species infections are common by zoophilic and geophilic dermatophytes. N. nana is considered an etiological agent of ringworm in pigs but has also been isolated from other animals, including humans. However, it also possesses many characteristics of geophilic dermatophytes including the ability to grow in soil. N. nana produces characteristic pear-shaped macroconidia and usually exhibits an ectothrix pattern of hair infection. It has been isolated from dermatitis lesions as well as from soil. N. nana infections in pigs are not of much concern as far as economy or health is concerned. But it has been associated with onychomycosis and gonathritis in humans, which are significant in human medicine. The shift in the predominance of dermatophytes in humans and the ability to evolve into a potential tinea pathogen necessitates more understanding of the physiology and genetics of N. nana. In this review, we have attempted a detailed analysis of the studies about N. nana, emphasizing growth and cultural characters, physiology, isolation, infection in humans and animals, molecular characterization and antifungal susceptibility.
Keywords: Microsporum nanum, Nannizzia nana, Dermatophytes, Cross-species infection, Ringworm, Skin disease
Introduction
Dermatophytes are molds that are keratinophilic and keratinolytic in nature and are associated with tinea or ringworm in humans and animals, respectively. Dermatophytes are thought to be evolved into facultative or obligatory parasites from soil-inhabiting keratinophiles [1]. Earlier, dermatophytes were classified into three genera: Microsporum, Trichophyton, and Epidermophyton. Recently, based on a multilocus phylogenetic analysis dermatophytes are divided into seven genera: Trichophyton, Epidermophyton, Nannizzia, Paraphyton, Lophophyton, Microsporum, and Arthroderma [2]. They are also classified based on their ecological preferences: zoophiles, anthropophiles, and geophiles. Zoophilic dermatophytes like M. canis, T. verrucosum mainly affect animals, and anthropophilic dermatophytes like T. rubrum, T. tonsurans have an affinity toward humans. Geophilic dermatophytes such as N. gypsea can grow and multiply in the soil without any host preference but occasionally produces diseases in both humans and animals. The anthropophiles and zoophiles are considered to be evolved from soil-dwelling geophiles [3]. Dermatophytes are present throughout the globe, including regions with harsh climatic conditions [4], and have been isolated from animals [5, 6], humans [7], and soil [8]. The presence of dermatophytes in the soil can be interpreted in two ways: soil-inhabiting geophilic dermatophytes with the ability to propagate in the soil [9] or those shed from the host animal and remaining dormant in the soil within the tissue debris for a particular period [10]. In both ways, soil can act as a source of infection to the host with spores of actively multiplying geophilic dermatophytes and dormant spores of zoophilic/anthropophilic dermatophytes [11, 12].Certain species of dermatophytes can infect humans through contact with animals and soil [13]. N. nana is one such dermatophyte that can infect humans from soil and their preferred host, pigs [14–16]. Even though the occurrence and severity of ringworm infection due to N. nana is not a matter of serious concern in the current scenario, it has the potential to evolve into a predominant pathogen due to its geophilic and zoophilic nature [17, 18].
The present review aimed to analyze the studies about N. nana, emphasizing growth and cultural characters, physiology, isolation, infection in humans and animals, molecular characterization, and antifungal susceptibility.
Nannizzia nana–geophile or zoophile?
Microsporum nanum was first isolated from an inflammatory lesion of the scalp of an 8-year-old boy. The isolate was initially identified as a dwarf form of Microsporum gypseum and named M. gypseum var nanum [19]. Later, a second isolate showing similar properties to the first one was isolated and thus, a separate species from M. gypseum named M. nanum was accepted [20]. A recent phylogenetic study has reclassified M. nanum into Nannizzia nana [2]. Earlier, the genus Nannizzia was introduced to include only the teleomorphs or sexual states of dermatophytes. But based on molecular studies, it has now considered as a separate clade and the genus Nannizzia includes several geophilic and zoophilic species, namely N. gypea, N. fulva, N. aenygmaticum, N. corniculata, N. duboisii, N. incurvata, N. persicolor, and N. praecox [21]. In contrast, the genus Microsporum, in which N. nana was earlier grouped, includes three species namely M. canis, M. audouinii, and M. ferrugineum [2].
The ecological preference of N. nana is still debated and has been classified both as a geophile and pig-adapted zoophile in various literatures [17, 18]. Even though it is known that certain dermatophytes have developed an affinity towards a particular host, the mechanism associated with host adaptability is not clearly understood. However, the species of dermatophyte involved, the host, and the environment may play a role in the process of developing affinity or adaptability. Dermatophytes were initially soil saprophytes, and during the evolutionary course, some species started utilizing keratin from the living host. Thus, species such as N. nana and Trichophyton quinckeanum started colonizing and utilizing the keratin substrate of pigs and mice, respectively living close to the soil [18]. Otčenašek and Dvořak, 1975 [17] categorized N. nana as a ‘species strongly specialized (susiophilic) paratrophic (pathogenic)’ dermatophyte. However, N. nana can also survive in the soil as a saprophyte, not as a contaminant. Macroconidia of N. nana have been isolated from the supernatant of soil collected from pig lots [22] which indicate that N. nana can multiply and sporulate in the soil as a saprophyte and thus can be considered a geophilic dermatophyte that has a specific affinity for swine and swine habitats [22]. In addition, the abundance of macroconidia production decreases in the order of geophilic, zoophilic, and anthropophilic dermatophytes [18]. N. gypsea produces abundant ellipsoidal macroconidia with rounded ends [23]. N. nana produces a large number of pear-shaped macroconidia in cultures, a characteristic feature of geophilic dermatophyte.
Geophilic dermatophytes can utilize the keratinous substrates present in the soil and propagate through various modes of reproduction. Zoophilic dermatophytes exhibit poor saprophytic growth and remain in the soil in a dormant state on the contaminated hair or scabs from the host. Whole genome sequence analysis has revealed that geophilic dermatophytes have their genome enriched with genes of virulence and metabolism, many of which have been lost in the zoophilic and anthropophilic dermatophytes during the course of adaptation to their respective hosts [24]. Since N. nana can survive both in soil and host body, the genome repertoire will be between a zoophile and geophile to maintain the genes required for survival in both soil and host body. Even though most dermatophytes exhibits variations in their ecological preferences, most of them secrete similar types of enzymes; however, the amount of enzyme production varies with each species [25]. The enzyme production can vary with the ambient temperature also. The maximum keratinase production in geophiles occurs at around 30 ºC whereas zoophiles and anthropophiles exhibit maximum keratinase production at a temperature range of 35–40 ºC, which indicates the host adaptation [26]. So, it can be assumed that N. nana, which can survive in both soil and pigs, may have different enzyme production profiles while propagating as a saprophyte and as a parasite.
The development of affinity towards a particular host keratin was considered a major reason for the host adaptability of dermatophytes [27]. Apart from the enzymes required for keratinolysis, dermatophytes also secrete a variety of other proteinases. The host skin and superficial structures will contain proteins other than keratin, which the dermatophyte will also degrade. Geophilic dermatophytes such as N. gypsea produce more enzyme elastase than zoophiles and anthropophiles, which will degrade the elastin present in human and animal skin [28]. This high elastase production can be attributed to the severe inflammation associated with geophilic dermatophytes. It should also be noted that N. nana produces severe inflammatory reaction in humans [14] while producing mild inflammation in pigs indicating its geophilic nature and adaptation to pigs, respectively. Other than the enzyme production profile, the attachment of dermatophyte propagule to host tissue also plays a very important role in colonizing the host tissue. Dermatophytes may vary in the number and types of adhesion proteins present, which may also influence host adaptability [24]. Thus, in the course of evolution, dermatophytes developed an affinity towards particular host tissue determined by the keratin content and associated protein components and the binding ability of the dermtaophyte propagule. The soil inhabitants may also have played some role in the evolution of the dermatophytes from geophilic to parasitic life [29, 30]. Soil-dwelling keratinophilic fungi like Chrysosporium keratinophilum produces anti-dermatophytic substances and might have influenced the evolution of dermatophytes into host-adapted zoophiles and anthropophiles [30]. In view of these facts, it can be assumed that N. nana has developed an affinity towards pig tissue while retaining the genes required for survival in the soil as a saprophyte. Since N. nana exhibits the properties of both geophiles and zoophiles it can be considered a link between these two classes of dermatophytes.
Isolation of N. nana from animals, humans, and soil
N. nana is considered to prefer pigs as their major host and infection in pigs has been reported in Europe [31], North America [32], and New Zealand [33]. It has also been attributed to the outbreaks of ringworm in Iberian pigs in Spain [34]. Apart from pigs, N. nana has been isolated from healthy and diseased domestic animals across the globe (Table 1). The isolation of N. nana from healthy animals indicates their ability to survive in a dormant form in the host body or the animal might have acquired it as a contaminant from the soil. Chances of acquiring N. nana infection are higher in animals having close contact with soil, especially pigs inhabiting regions. This again underlines the dual nature of N. nana to propagate in the soil while maintaining a special affinity towards pig. Since N. nana can grow as a saprophyte, it can survive for an extended period once introduced into the soil. In general, the prevalence of N. nana infection is low in animals compared to other zoophiles and geophiles. Even in pigs the rate of isolation of N. nana is low. This reduced isolation rate of N. nana from pigs may be due to the less evident clinical signs that may go unnoticed. Hence, N. nana can be considered a low virulent pathogen in pigs.
Table 1.
Reports of Nannizzia nana isolated from animals
| Country | Healthy animals or animals with skin lesions | No. of animals positive for N. nana | Reference |
|---|---|---|---|
| New Zealand | Skin lesions | 4 pigs | [5] |
| Jordan | Healthy animals | 1 cow, 2 rabbits and 1 cat | [108] |
| Jordan | Healthy animals | 4 sheeps | [109] |
| Nigeria | Skin lesions | 7 pigs | [110] |
| Western Turkey | Skin lesions |
2 dogs 6 cats |
[111] |
| Eastern Turkey | Healthy animals | 3 van cats | [112] |
| Egypt | Skin lesions | 4 camels | [113] |
Ringworm in humans due to N. nana has been reported in various countries like Brazil [35–38], Canada [39], Africa [40], South Australia [41], India [42, 43], Romania [44], and Victoria [16]. Pig rearing has been considered the major infection source in humans [45]. Close contact with the animals will help transmit N. nana from symptomatic and asymptomatic pigs. At the same time, cases of N. nana infection have been reported in humans with no contact with pigs, indicating the soil propagating ability of N. nana [15]. However, ringworm infection in humans from pigs or soil is rare compared to other dermatophyte infections. A survey for the prevalence of dermatophytoses in humans for a period of 3 years (1979–1981) in the USA could isolate N. nana only three times during the study period [46]. Similarly, the survey conducted in the USA from 1982 to 1984 and 1993 to 1995 in human patients isolated zero and one isolate of N. nana during the study period, respectively [47, 48]. A low prevalence of N. nana infection in humans has also been reported from other parts of the world. A study on the prevalence of dermatophytoses in humans over 9 months conducted in Sana’a, Yemen Republic, identified 157 out of 1100 patients exhibiting clinical signs of tinea corporis, tinea capitis, tinea cruris, tinea unguium, and tinea pedis as positive for dermatophytes [49]. However, only two patients suffering from tinea corporis yielded N. nana on culturing. In a 2-year study conducted in Southern Taiwan among 375 patients with onychomycosis, the etiological agents of onychomycosis were identified as dermatophytes (227), Candida species (1180), and non-dermatophytic molds (30) [50]. Two of the onychomycosis cases were found to be caused by N. nana. A retrospective study identifying the etiology of onychomycosis in humans detected 1 out of 4220 cases of onychomycosis due to N. nana [51]. Even though these two studies reported very low prevalence of N. nana in humans; it also indicates the ability of N. nana to affect nails. The low isolation rate of N. nana from humans cannot be attributed to a single reason. Misdiagnosis of N. nana infection can be a prime reason as the etiological agent is not well studied and characterized [15]. Restricted rearing of pigs and lack of exposure to pig habitat or soil due to better hygienic practices in modern society can also be a reason. At the same time, there are reports of transmission of N. nana infection to humans from cows indicating the ability of N. nana as a potential pathogen to humans with multiple sources of infection [52].
N. nana has been isolated from the soil of different countries across the globe. But their prevalence as soil saprophytes is low as compared to other geophilic dermatophytes (Table 2). It has been isolated from various geographical regions such as glacier banks, valleys, and mountains stipulating its ability to survive in soil of different compositions and temperatures [4, 53]. In many studies, the presence of pigs has been attributed to the isolation of N. nana from the soil [53]. Limited information is available on the nutrient requirement and conditions required for the growth and development of N. nana in the soil. The presence of organic matter is inevitable for survival in the soil as N. nana has not been isolated from sandy soil and hot arid regions [54, 55]. Soil pH is major factor, which influences the growth of keratinophilic fungi, including dermatophytes. N. nana has been isolated from soil with a slightly alkaline pH (7.02–9.06) but not from acidic soil [10]. But N. gypsea has been isolated from soils with a pH range of 5.77–8.31, specifying its ability to survive in a wider range of pH unlike N. nana. The moisture content of the soil is another critical factor for the growth of geophilic dermatophytes. N. gypsea could grow in less than 5% moisture content, and soil with moisture content greater than 20% will not favor its growth [56]. Since N. nana can survive in the soil, and share characters of geophiles, it can be attributed that the moisture requirement will be more or less similar to N. gypsea. In general, dermatophytes capable of propagating in the soil are osmotolerant, have urease activity, are independent of the exogenous sources of vitamins and amino acids, and constitutively secrete protease enzymes [57]. Since N. nana survives and multiplies in soil, it might be harboring these intrinsic properties of a geophile, still extrinsic factors such as presence of pigs, pH, organic content, and moisture of the soil along with climatic conditions also significantly influence the survival of N. nana in the soil.
Table 2.
Reports of Nannizzia nana isolated from soil
Growth and physiology
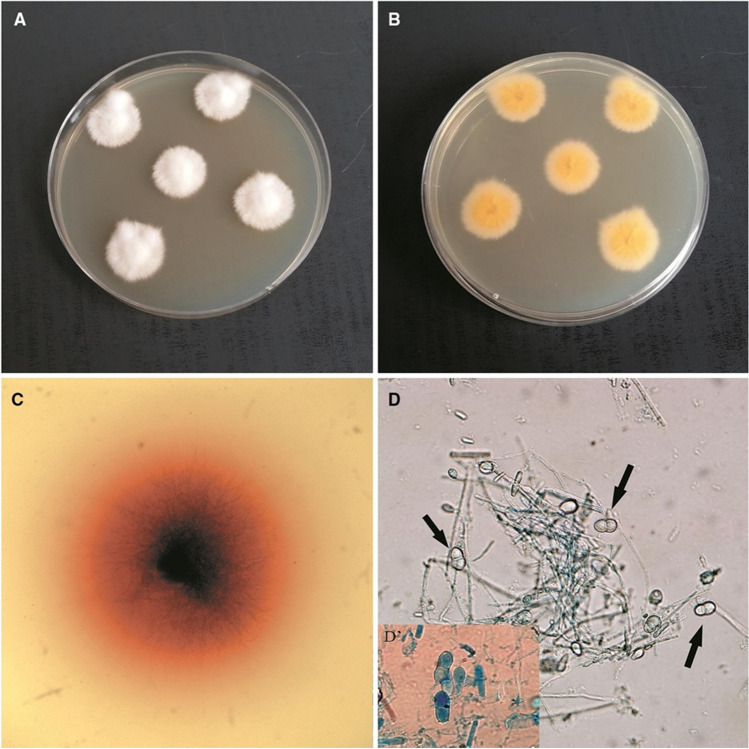
N. nana can be isolated from infected hair/ skin scrapings using Sabouraud’s Dextrose Agar supplemented with cycloheximide and chloramphenicol. Penicillin and Streptomycin have also been used to prevent the growth of bacterial contaminants [19]. The growth usually appears around the 4th day of incubation [19]. The colonies will be initially whitish cottony with an orangish reverse side but later, around the 11th day, become granular and buff-colored with a brownish-red reverse side (Fig. 1A–C). Macroconidia are abundantly produced, but few microconidia are observed. Microscopically, the macroconidia (13–29 × 8–13 µm) are usually clavate to obovoid shaped with one or two septa and are thin verrucose walled with projections and truncate base [21] (Fig. 1D, D’, Fig. 2). Many macroconidia may be borne on a single stalk. Spherical, elliptical and aseptate macroconidia may also be produced. Microconidia (3–7 × 2–3 µm) usually will be ovoidal or clavate in shape. The macroscopic and microscopic characteristics of the colonies do not alter with the agar used for isolation [19]. Racquet hyphae, pectinate hyphae, and intercalary chlamydospores can also be observed microscopically. N. nana can also be characterized by various laboratory and biochemical tests such as growth in dermatophyte test medium (DTM), ability to grow in the presence of cycloheximide and 5% NaCl, urease production, lipase activity in tween 80 opacity test medium, milk hydrolysis in bromocresol purple-milk solids-glucose agar (BCP-MS-G agar), beta-hemolysis in blood agar and ability to grow at 37 ºC [21]. N. nana isolates produce reddish discoloration in DTM and can grow in presence of cycloheximide and 5% NaCl. Most of the isolates produce urease enzyme and exhibit lipase activity. They give variable results in milk hydrolysis and produce beta-haemolysis after 2 weeks of incubation. Most isolates can grow at 37 ºC, depicting its thermo tolerance [15]. Thus, the colony morphology, abundance in macroconidia production, and the enzyme activity of N. nana classify it more towards the geophilic group. But, the ability to grow at 37 ºC indicates their thermotolerance and infectivity towards mammalian skin as certain geophiles such as Arthroderma uncinatum grow well at 25 ºC but are unable to grow at 34 ºC or above [24].
Fig. 1.
Micro- and macroscopic morphology of Nannizzia nana. (A) Obverse of N. nana culture. (B) Reverse of N. nana culture. (C) Reddish discoloration on Dermatophyte Test medium due to N. nana growth. (D) Micro-morphology of N. nana, arrows indicate characteristic macroconidia (× 400) (D’) Macroconidia stained with lactophenol blue (× 1000).
Reproduced from Gnat et al. 2020 [15] under a Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/)
Fig. 2.
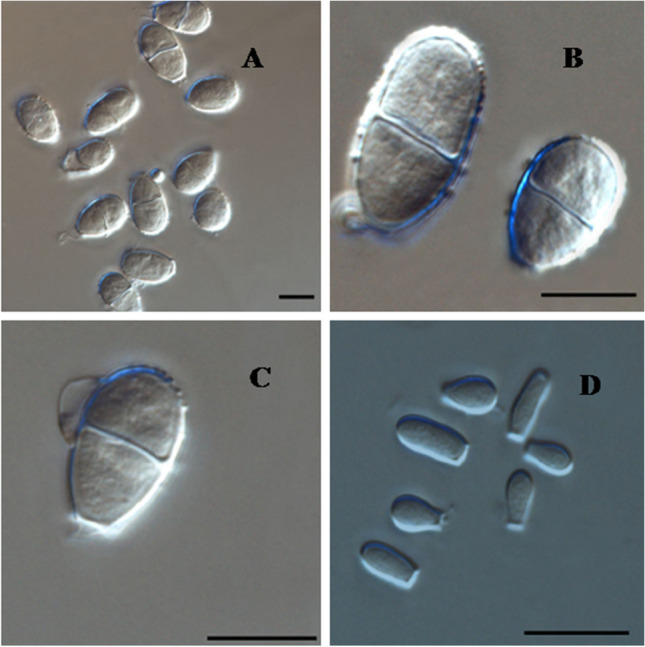
Conidia produced by N. nana. (A–C) Macroconidia. (B) Microconidia. Scale bars = 10 μm.
Reproduced from Dukik et al. 2020 [21] under a Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0)
The preferred substrate for the growth of dermatophytes is the keratinous structures present in the host body or the soil. Most zoophiles and anthropophiles degrade keratin slower than geophilic dermatophytes [58], indicating the ability of the geophiles to assimilate nutrients efficiently and grow faster. Keratin is a fibrous protein present in mammals, birds, and reptiles [59]. There are two types of keratin-alpha keratin (which contain alpha helix) and beta keratin (which contains beta pleated sheets). Most of the keratinous structures are a mixture of alpha and beta keratin with varying proportions. Keratin also contains disulfide bridges providing rigid structure. There are variations in the amino acid composition of keratin in hair, epidermis, and epithelium [60]. Also, the degradability of the host keratin depends on the cystine content, which determines the hardness of the tissue. N. nana produces abundant growth in hair and fair growth in the toenails [19]. Even though both hair and toenails are made up of hard keratin, the difference in the affinity of N. nana towards both may be due to the presence of proteins other than keratin [61]. N. nana also exhibits differential preference toward nutrients as it can utilize aspartic acid, arginine, citrulline, alanine, ornithine, histidine, and proline but cannot utilize phenylalanine, cysteine, hydroxyproline, and methionine [62]. It can utilize a limited number of carbohydrates as a carbon source and will not grow in starch-containing media [62].Thus N. nana exhibits selectivity in its nutrition. However, it can utilize keratin from different hosts apart from pig keratin. It was found that N. nana grew equally on human, horse, and pig hair [22]. Also, N. nana can utilize keratin from chicken feathers [63]. So, it can be considered that N. nana retains the ability to utilize keratin from different hosts like geophiles, but has a slighter affinity toward pig tissue. As all recognized dermatophytes produce keratinases, the preference towards pigs may be due to the presence or absence of other proteins and the physiological conditions prevailing in the pig skin surface. This has to be validated by expression studies of proteases and analyzing the enzyme production profile both in the soil and host tissue.
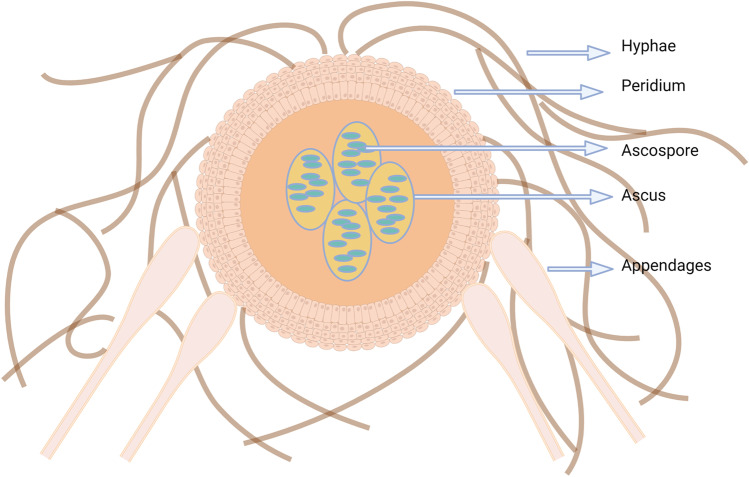
Most recognized dermatophytes exist in two sexual states: anamorphic (asexual or imperfect) and teleomorphic (sexual or perfect). Adaptation to the host has a critical impact on the sexual dimorphism of dermatophytes. Most anthropophilic dermatophytes reproduce asexually as they now possess one mating type due to the selection pressure during the adaptation process [64]. Even though zoophiles have both mating types; particular mating type predominance indicates the gradual shift to asexual reproduction [64]. But geophiles possess both mating types in equal proportion and reproduce both by sexual and asexual methods [65]. Dawson and Gentles, 1962 [66] studied sexual reproduction in N. nana and proposed Nannizia obtusa as the perfect stage. They produced cleistothecia (Fig. 3) by crossing a Cuban and Korean isolate of N. nana and identified it as heterothallic. Fewer cleistothecia are produced and require a longer incubation period and lower temperature. The sexual stages are characterized by echinulate peridial cells, special appendages at terminal peridial cells, absence of disarticulation of peridial cells at maturity, dichotomous and verticillate peridial hyphae, and obtuse angle between the branches and main hyphae. Asci are sub-globose with eight-spored, and ascospores are lenticular in shape [66]. The sex-determining region in the genome of dermatophytes is the mating type locus (MAT). There are two idiomorphs of MAT, namely MAT1-1 and MAT1-2 [67]. But the idiomorphs in N. nana have not been determined. The type and frequency of the idiomorphs among the N. nana population have to be studied to understand whether the distribution pattern is similar to that of geophiles or imbalanced, as in zoophiles.
Fig. 3.
Schematic representation of cleistothecia. Image created using Biorender.com
Clinical signs and lesions
The inflammatory response associated with dermatophyte infection is mainly towards the proteases secreted by these fungi. However, a cutaneous immune response may also develop toward the cell wall components. Interestingly, the dermatophytes exhibit variations in the composition of their cell wall. The cell wall polysaccharide composition of geophilic dermatophytes was studied by Guarro et al. 1993 [68]. Glucomannan and glucan-chitin complex was identified as the major polysaccharide fraction of the cell wall of geophilic dermatophytes. N. nana exhibits a slight deviation in the cell wall composition compared to geophiles. Xylose was absent in the water-soluble fraction of the cell wall in N. nana, when compared to N. gypsea. Also, the galactose to glucose ratio in the water-soluble fraction was high in N. nana whereas the ratio was low in N. gypsea. These variations in the cell wall composition may reflect in the inflammatory response towards N. nana and N. gypsea in the host.
The pattern of hair infection varies with the species of dermatophyte involved. N. nana usually causes ectothrix infection of hair, but favic and endothrix type of hair infection has also been reported [19, 69]. Electron microscopic examination of hairs infected with N. nana has revealed an ectothrix type of infection [70], which is the common mode of hair infection by N. nana. It gives variable results in Woods lamp analysis but majority being Woods lamb negative [19, 22, 71]. This may be due to the strain variations in the ability to metabolize tryptophan or false positive results. N. nana can produce wedge-shaped penetrations in human hair [45], signifying its ability to produce perforating organs, a characteristic feature of geophiles. But in an experimental infection on human models, the hairs were enmeshed with hyphae without any penetration [20]. These variations in the infection pattern of hair and tryptophan metabolism indicate heterogeneity among the population of N. nana.
The clinical course and lesions produced by N. nana depend on the host affected. In pigs, N. nana produces lesions mainly in the shoulder, flank, udder, neck, and behind the ears. Brownish crust formation without any signs of alopecia and pruritis is seen primarily in pigs [22, 72]. Lesions originate as small dark circles with crusts and progress in a circular fashion. Later, these lesions will coalesce and cover larger body regions [34]. Physiological status, sex, and age may influence the outcome of N. nana infection in swine [34]. Exudates formation is not common in N. nana infection but has been reported in pigs, suggesting the difference in the virulence pattern of N. nana strains [73]. In sows, ringworm infection due to N. nana can be complicated by acanthosis [74]. Unlike N. nana infection, other dermatophytes can evoke more severe inflammatory responses in pigs [75].The pigs infected with M. canis exhibit alopecia and pruritis [76]. Roughening of the body surface can be seen in Trichophyton mentagrophytes infection in pigs [77]. The difference in clinical signs and inflammatory response produced in pigs by N. nana and other dermatophytes emphasizes the adaptation of N. nana towards swine. All known dermatophytes produce keratinases [78]. Even though the amino acid sequence of the proteases is conserved among the dermatophytes, they secrete variable amounts of the same enzyme [25].The process of keratinolysis starts with sulfitolysis where the disulfide bonds are destroyed [79]. This is followed by keratin proteolysis carried out by the sequential action of endopeptidases such as fungalysin and subtilisins and exoproteases such as aminopeptidases and carboxypeptidases [79]. Apart from the enzymes associated with keratinolyis, dermatophytes may vary in the secretion of other proteases such as elastase, which determines the inflammatory nature of dermatophytosis. It has been identified that dermatophytes produce less proteases in the host to which they are adapted but produce a large amount of enzymes in other host species [80]. The protein secretion profile will depend on the habitat of the dermatophyte, and it, in turn determines the level of inflammatory reaction in the host [81]. Thus, N. nana qualitatively and quantitatively produces enzymes in pigs in such a way to avoid severe inflammatory reactions and thereby survive for a longer period.
N. nana can also infect hosts other than pigs but vary in the inflammatory response. In an experimental infection study, the human model exhibited erythematous pustular lesions, whereas in kittens and guinea pigs, scaly and crusty lesions were noticed [19]. Variation was also observed in the duration of regression of the lesions. The lesion started regressing after 5 weeks in humans, whereas the lesion lasted for only 2 weeks in kittens and guinea pigs. Histopathological study of N. nana infected guinea pig skin revealed hyperkeratosis of the epidermis, spongiosis, and excoriations with normal hair follicles, dermis, and subcutaneous tissue. Thus, even though N. nana can infect heterologous hosts more intense inflammation may be produced in species distantly related to pigs. But it was also found that N. nana was unable to infect monkeys [19], indicating the requirement of specific physiological conditions or protein adhesins to establish the infection in the host body. In humans, anthropophilic dermatophytes produce low-intensity inflammation, zoophiles produce moderate inflammation and geophiles produce high-intensity inflammation [82]. N. nana produces more intense skin infections in humans than in pigs. The common lesions in humans include erythema, pruritis, scale formation, and lustreless and brittle hair [19, 71]. But it has also been associated with more severe conditions such as mycoses of the submandibular region and associated exudative gonarthrtis in humans [52]. Pustules and hyperkeratotic crust were observed in the chin area, along with inflammation of submandibular lymphatic glands. The association of N. nana with these clinical signs was confirmed by isolating the organism from the clinical samples. N. nana can also be associated with tinea corporis [7] and inflammatory tinea facei and tinea cruris [14]. Severe itching with a burning sensation has been reported in humans [15]. Even though nail infections are not common in N. nana it has been associated with total dystrophic onychomycosis and recurrent onychomycosis [15, 51]. Onychomycosis due to N. nana may be due to simple invasion by mechanical action [83] or the ability of certain strains to degrade nail tissue. Thus, the clinical spectrum of N. nana infection in humans is wider than that in pigs and has the potential to emerge as a major cause of tinea in the due course of time.
Molecular characterization
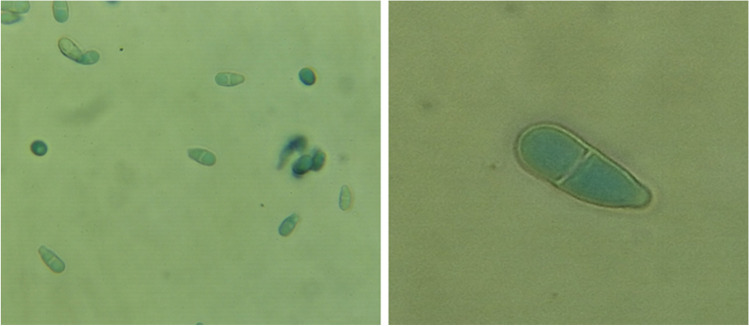
Most dermatophytes can be identified by their morphological and microscopic characteristics. But, the emergence of genotypes and strains associated with a particular geographical region and antifungal resistance has necessitated the molecular characterization of dermatophytes [84]. For example, N. nana can be distinguished easily by the microscopic examination and identification of the characteristic pear-shaped macroconidia. But, certain non-dermatophyte fungi, such as Trichothecium roseum, also have morphologically similar macroconidia, often resulting in misidentification [85] (Fig. 4). Also, heterogeneity has been detected among the strains of N. nana [15], which further emphasizes the need for molecular characterization of N. nana.
Fig. 4.
Conidia of Trichothecium roseum stained with lactophenol cotton blue
The polymerase chain reaction (PCR) is a highly sensitive molecular technique that can detect the presence of microbial genome in clinical samples even at very low concentrations. PCR has been standardized to detect the presence of dermatophytes from clinical samples such as nail and skin scrapings [86]. Compared to culture, the high sensitivity of PCR has made it highly useful in the fast and accurate diagnosis of dermatophytosis in conjunction with clinical signs [86]. PCR-based identification of N. nana is not commonly used, but it has been included as a study organism in assessing various types of PCR [87, 88] and PCR-based techniques [89, 90]. Restriction enzyme digestion of amplified products of ITS region and topoisomerase II gene produced N. nana specific band patterns [89, 91, 92]. Arbitrarily primed PCR (AP-PCR) or random amplified polymorphic DNA (RAPD) PCR which can be used for species and strain identification has also been found useful to distinguish N. nana from other common dermatophytes [90, 93]. Recently, PCR has been employed to detect the strain variations among N. nana. Melting profile PCR (MP-PCR) of 3 N. nana isolates detected two different electrophoretic profiles [15]. One isolate was obtained from a man (lesions were in the neck) and two from women (lesions were in the feet and nails, respectively). Interestingly, the isolate from the women shared a similar electrophoretic profile. This study indicates the presence of genetic diversity among N. nana isolates and the need to develop a molecular assay to identify those variants that may be associated with particular traits, host preference, virulence, or geographical distribution.
MALDI-TOF mass spectrometry is an advanced molecular technique that helps to identify organisms or biomolecules based on the ions produced from the analytes. A small amount of fungal colony is needed for MALDI-TOF MS identification. Fungal identification can be made in 3 to 6 days, including the period required for the isolate to produce sufficient growth in the culture plate for protein extraction [94]. MALDI-TOF has been optimized for identifying dermatophytes by many researchers [95, 96]. However, limited studies have been done on the MALDI-TOF identification of N. nana. In one study, MALDI-TOF was able to correctly identify two clinical isolates of N. nana up to the genus and species level [97]. One of the major disadvantages of MALDI-TOF-based identification is the pre-requirement of a sufficient database [98]. Also, the mass spectra produced by the dermatophyte may show variations in individual peaks and peak intensity depending on the media in which they have grown [94, 99]. The requirement of costly sophisticated equipment and skilled personnel also limits the use of MALDI-TOF.
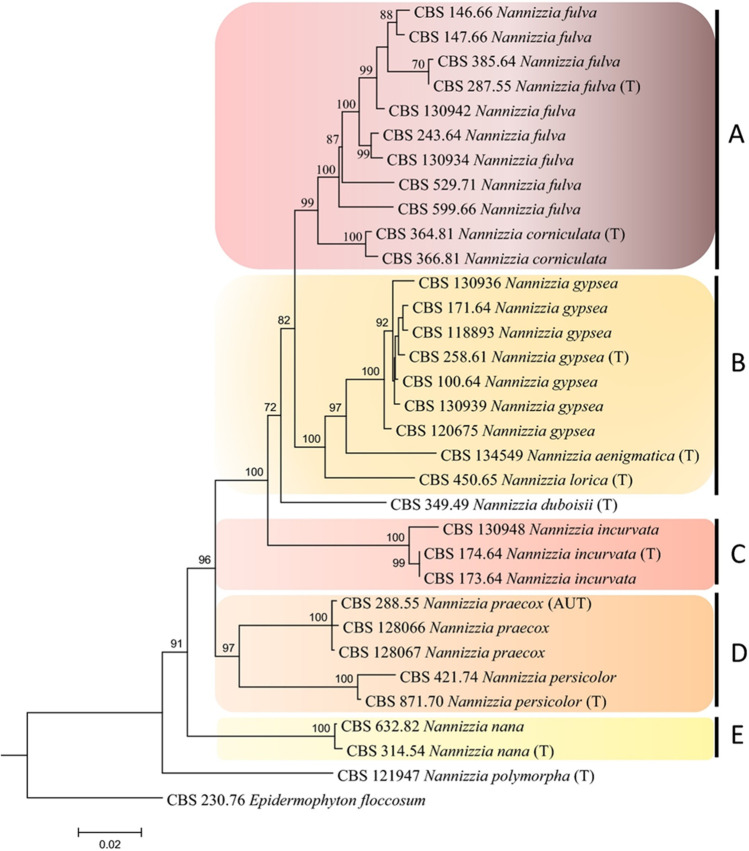
Some of the closely related dermatophyte species have been grouped into species complexes like N. gypsea complex (N. gypsea, N. fulva, and N. incurvata) [115], and T. mentagrophytes complex (T. mentagrophytes, T. interdigitale, T. erinacei, T quinckeanum, and T. benhamiae) [100]. The members within the complexes are difficult to be distinguished by morphological or conventional laboratory tests. Sequencing genomic regions and subsequent phylogentic studies will help us differentiate the species and strains. For example, members of T. mentagrophytes complex are difficult to differentiate morphologically but can be identified based on the sequence analysis of genomic regions such as ITS [100]. Earlier, N. nana was grouped in the N. gypsea cluster based on the ITS region analysis [101]. But when additional genomic regions were exploited, N. nana was found to be genetically distinct. A phylogenetic tree based on TEF-1α and BT2 sequences positioned N. nana as a separate branch owing to interspecies diversity [102, 103]. Recently, a phylogenetic study based on the ITS, LSU, beta-tubulin, translation elongation factor, and 60S ribosomal protein L10 has identified five clades among Nannizzia species, with N. nana forming a separate clade with a sole member [21] (Fig. 5). Interestingly, N. nana took an ancestral position to all species of Nannizzia studied. This indicates N. nana is harboring the ancestral characteristics of dermatophytes even after developing host affinity towards pigs. Whole genome sequence analysis of N. nana isolates will give a clear picture of the genome repertoire. Also, identifying beta-tubulin and TEF genotypes indicates intra-species variation among N. nana, which warrants detailed genomic studies.
Fig. 5.
Maximum likelihood concatenated tree of Nannizzia species based on ITS LSU, TUB2, TEF3, and RP 60S L1. AUT: authentic strain. T: type strains. A to E: different clades within genus Nannizzia.
Reproduced from Dukik et al. 2020 [21] under the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/)
Antifungal susceptibility
Dermatophyte infections are characterized by their chronic nature and the requirement for prolonged antifungal therapy. They exhibit species variations in antifungal susceptibility, so susceptibility testing must be done before initiating anti-dermatophytic treatment. A limited spectrum of antifungal drugs is available for the treatment of dermatophytosis. The most commonly used drugs are azoles such as ketoconzaole, voriconazole, and itraconazole, allylamines such as terbinafine, echinocandins such as caspofungin and micafungin, polyene drugs such as amphotericin B, pyridinone derivatives such as ciclopirox and non-polyene drugs such as griseofulvin. These antifungal drugs vary in their mechanism of action and target enzyme/ cellular structure affected [104]. Azole drugs and allylamine interrupt the ergosterol synthesis by inhibiting the 14α lanosterol demethylase and squalene epoxidase, respectively. Polyenes will increase cellular permeability by binding to ergosterol whereas griseofulvin will disrupt microtubule polymerization and depolymerization. Echinocandins inhibit the β-(1, 3)-D-glucan biosynthesis and ciclopirox acts by irreversibly binding to the cellular structures. Most of the antifungals are found to be effective against N. nana [105, 106]. In many studies, the minimum inhibitory concentration (MIC) of the antifungal drugs has been determined for N. nana, and the values were in the normal range of susceptibility. Single isolate of N. nana was included in a study and the MIC for voriconazole and fluconazole was found to be 0.125 µg/ml and 4 µg/ml, respectively [105]. In another study, fluconazole exhibited higher MIC against different strains of dermatophytes but was highly effective against N. nana [106]. The MICs of the antifungal drugs for N. nana were voriconazole < 0.125–0.25 µg/ml, itraconazole 0.03 µg/ml, terbinafine 0.125–001 µg/ml, fluconazole 4 µg/ml, ketoconazole 0.5–0.1 µg/ml, and griseofulvin 4–8 µg/ml. Perera et al. 2001[107] studied the antifungal susceptibility of 19 species of dermatophyte to voriconazole, itraconazole, terbinafine, fluconazole, ketoconazole, and griseofulvin [107]. Two strains of N. nana were included into the study. The geometric mean MIC for N. nana were, voriconazole 0.18 µg/ml, itraconazole 0.03 µg/ml, terbinafine 0.03 µg/ml, fluconazole 4 µg/ml, ketoconazole 0.71 µg/ml, and griseofulvin 5.66 µg/ml. All these studies indicate the susceptibility of N. nana to the most common available antifungal drugs. This may be due to the low frequency of occurrence and limited exposure of N. nana isolates to these drugs. Clinically, N. nana infection has been treated with a high success rate. Enilconazole solution (0.2%) has been used to treat N. nana-infected lactating sows and disinfection of surroundings [34].Total dystrophic onychomycosis due to N. nana has been treated successfully with itraconazole [51]. Three cases of N. nana infection were treated successfully with oral terbinafine and topical ketoconazole [15]. Even though the medication was the same, the duration was different in each case. Thus, the success of therapy may depend on other factors such as the patient’s age and the region of infection.
Conclusion
Increased frequency of atypical tinea is being reported in humans. Zoophilic dermatophytes like M. canis and geophilic dermatophytes like N. gypsea are among those agents associated with atypical manifestations. The exact role of animals in transmitting dermatophytes to humans is still debated. N. nana having the characteristics of both geophiles and zoophiles with a special affinity towards swines can thus be considered a potential agent for human dermatophytosis. Limited studies on this pathogen are a setback in understanding its exact physiology and virulence. Whole genome sequencing of a few isolates will enable us to decipher its genomic repertoire and help us understand the genetic difference from other zoophiles, geophiles, and anthropophiles through comparative genomics technologies. Such studies will indisputably give hints on the host adaptation mechanism of dermatophytes also.
Acknowledgements
We thank Indian Council of Medical Research for granting a project on dermatophytes titled 'Unraveling the Pheno-Genotyping Linking and Distrubution Dynamics of Trichophyton mentagrophytes among Human and Animals'. Figure 3 is created using Biorender.com.
Authors’ contributions
SSN and A both conceptualized the idea and wrote the review article along with the valuable inputs from all other authors. All authors read, revised, and approved the final manuscript.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Data availability
Not applicable.
Code availability
Not applicable.
Declarations
Ethics approval and consent to participate
The authors confirm that no ethical approval was required.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sonu S. Nair, Email: sonunair001@gmail.com
Abhishek, Email: abhivbm@gmail.com.
References
- 1.Novak EK, Galgoczy J. Notes on dermatophytes of soil origin. Mycopathologia et mycologiaapplicata. 1966;28(4):289–296. doi: 10.1007/BF02145099. [DOI] [Google Scholar]
- 2.de Hoog GS, Dukik K, Monod M, Packeu A, Stubbe D, Hendrickx M, Kupsch C, Stielow JB, Freeke J, Göker M, Rezaei-Matehkolaei A. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia. 2017;182(1–2):5–31. doi: 10.1007/s11046-016-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gräser Y, El Fari M, Vilgalys R, Kuijpers AFA, De Hoog GS, Presber W, Tietz HJ (1999) Phylogeny and taxonomy of the family Arthrodermataceae(dermatophytes) using sequence analysis of the ribosomal ITS region. Med Mycol 37(2):105–114 [PubMed]
- 4.Deshmukh SK. Incidence of dermatophytes and other keratinophilic fungi in the glacier bank soils of the Kashmir valley India. Mycologist. 2002;16(4):165–167. doi: 10.1017/S0269915X0200407X. [DOI] [Google Scholar]
- 5.Carman MG, Rush-Munro FM, Carter ME. Dermatophytes isolated from domestic and feral animals. N Z Vet J. 1979;27(7):136–144. doi: 10.1080/00480169.1979.34628. [DOI] [PubMed] [Google Scholar]
- 6.Bontems O, Fratti M, Salamin K, Guenova E, Monod M. Epidemiology of Dermatophytoses in Switzerland According to a Survey of Dermatophytes Isolated in Lausanne between 2001 and 2018. Journal of Fungi. 2020;6(2):95. doi: 10.3390/jof6020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yerga AL, Rodríguez-Nevado I, Recio JF, Álvarez AJC, Durán DDAF, Eseverri EG (2007) Infeccióncutánea de Microsporum nanum. Semergen: revistaespañola de medicina de familia (3):159–160
- 8.Jain M, Shukla PK, Srivastava OP. Keratinophilic fungi and dermatophytes in Lucknow soils with their global distribution:*KeratinophilePilze und DermatophytenimErdboden von Lucknow und ihrweltweitesVorkommen. Mycoses. 1985;28(3):148–153. doi: 10.1111/j.1439-0507.1985.tb02108.x. [DOI] [PubMed] [Google Scholar]
- 9.Chmel L, Buchvald J (1970) Ecology and transmission of Microsporum gypseum from soil to man. Sabouraudia 8(2):149–156 [PubMed]
- 10.Pontes ZBVDS, Oliveira ACD, Guerra FQS, Pontes LRDA, Santos JPD. Distribution of dermatophytes from soils of urban and rural areas of cities of Paraiba State, Brazil. Rev Inst Med Trop Sao Paulo. 2013;55(6):377–383. doi: 10.1590/S0036-46652013000600002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain N, Sharma M. Distribution of dermatophytes and other related fungi in Jaipur city, with particular reference to soil pH. Mycoses. 2011;54(1):52–58. doi: 10.1111/j.1439-0507.2009.01751.x. [DOI] [PubMed] [Google Scholar]
- 12.Oyeka CA, Okoli I. Isolation of dermatophytes and non-dermatophytic fungi from soil in Nigeria. Mycoses. 2003;46(8):318–320. doi: 10.1046/j.1439-0507.2003.00899.x. [DOI] [PubMed] [Google Scholar]
- 13.Romano C, Massai L, Gallo A, Fimiani M (2009) Microsporum gypseum infection in the Siena area in 2005–2006. Mycoses 52(1):67–71 [DOI] [PubMed]
- 14.Roller JA, Westblom TU (1986) Microsporum nanuminfection in hog farmers. J Am Acad Dermatol 15(5):935–939 [DOI] [PubMed]
- 15.Gnat S, Łagowski D, Nowakiewicz A, Dyląg M (2020) Unusual dermatomycoses caused by Nannizzia nana: the geophilic origin of human infections. Infection 48(3):429–434. 10.1007/s15010-020-01416-5 [DOI] [PMC free article] [PubMed]
- 16.Kelly R, Searls S (1977) Microsporum nanum infection in Victoria. Australas J Dermatol 18(3):137–138 [DOI] [PubMed]
- 17.Otčenašek M, Dvořak J. Ecological classification of dermatophytes. Mycoses. 1975;18(10):425–434. doi: 10.1111/j.1439-0507.1975.tb03521.x. [DOI] [PubMed] [Google Scholar]
- 18.Aly R. Ecology and epidemiology of dermatophyte infections. J Am Acad Dermatol. 1994;31(3):S21–S25. doi: 10.1016/S0190-9622(08)81262-5. [DOI] [PubMed] [Google Scholar]
- 19.Fuentes CA, Aboulafia R, Vidal RJ (1954) A dwarf form of Microsporum gypseum. J Investig Dermatol 23(1):51–61 [DOI] [PubMed]
- 20.Fuentes CA (1956) A new species of Microsporum. Mycologia 48(4)
- 21.Dukik K, de Hoog GS, Stielow JB, Freeke J, van den Ende BG, Vicente VA, Menken SBJ, Ahmed SA (2020) Molecular and phenotypic characterization of Nannizzia (Arthrodermataceae). Mycopathologia 185(1):9–35. 10.1007/s11046-019-00336-9 [DOI] [PubMed]
- 22.Ajello L, Varsavsky E, Ginther OJ, Bubash G (1964) The natural history of Microsporum nanum. Mycologia 56(6):873–884
- 23.Souza BDS, Sartori DS, Andrade CD, Weisheimer E, Kiszewski AE (2016) Dermatophytosis caused by Microsporum gypseum in infants: report of four cases and review of the literature. An Bras Dermatol 91:823–825 [DOI] [PMC free article] [PubMed]
- 24.Zheng H, Blechert O, Mei H, Ge L, Liu J, Tao Y, Li D, de Hoog GS, Liu W (2020) Assembly and analysis of the whole genome of Arthroderma uncinatum strain T10, compared with Microsporum canis and Trichophyton rubrum. Mycoses 63(7):683–693 [DOI] [PubMed]
- 25.Giddey K, Favre B, Quadroni M, Monod M. Closely related dermatophyte species produce different patterns of secreted proteins. FEMS Microbiol Lett. 2007;267(1):95–101. doi: 10.1111/j.1574-6968.2006.00541.x. [DOI] [PubMed] [Google Scholar]
- 26.Sharma A, Chandra S, Sharma M. Difference in keratinase activity of dermatophytes at different environmental conditions is an attribute of adaptation to parasitism. Mycoses. 2012;55(5):410–415. doi: 10.1111/j.1439-0507.2011.02133.x. [DOI] [PubMed] [Google Scholar]
- 27.Nardoni S, Rocchigiani G, Papini RA, Veneziano V, Brajon G, Martini M, Salari F, Mancianti F (2016) Dermatophytosis in donkeys (Equus asinus) due to Microsporum racemosum, an unusual geophilic agent. Medical mycology case reports 12:8–10 [DOI] [PMC free article] [PubMed]
- 28.Sharifzadeh A, Shokri H, Khosravi AR. In vitro evaluation of antifungal susceptibility and keratinase, elastase, lipase and DN ase activities of different dermatophyte species isolated from clinical specimens in Iran. Mycoses. 2016;59(11):710–719. doi: 10.1111/myc.12521. [DOI] [PubMed] [Google Scholar]
- 29.Shankar SG, Ranganathan S, Ranjith MS, Vijayalakshmi GS. Did earthworms contribute to the parasitic evolution of dermatophytes? Mycoses. 2002;45(9–10):399–401. doi: 10.1046/j.1439-0507.2002.00772.x. [DOI] [PubMed] [Google Scholar]
- 30.Gokulshankar S, Ranjithsingh AJA, Ranjith MS, Ranganathan S, Palaniappan R (2005) Role of Chrysosporium keratinophillum in the parasitic evolution of dermatophytes. Mycoses 48(6):442–446 [DOI] [PubMed]
- 31.Morganti L, Bianchedi M, Ajello L, Padhye A (1976) First European report of swine infection by Microsporum nanum. Mycopathologia 59(3):179–182 [DOI] [PubMed]
- 32.Bubash GR, Ginther OJ, Ajello L. Microsporumnanum: First recorded isolation from animals in the United States. Science. 1964;143(3604):366–367. doi: 10.1126/science.143.3604.366. [DOI] [PubMed] [Google Scholar]
- 33.Smith JMB and Steffert IJ (1966) Microsporum nanum in New Zealand pigs. New Zealand veterinary journal 14(7) [DOI] [PubMed]
- 34.García-Sánchez A, Bazán J, de Mendoza JH, Martínez R, Sánchez S, de Mendoza MH. Outbreak of ringworm in a traditional Iberian pig farm in Spain. Mycoses. 2011;54(2):179–181. doi: 10.1111/j.1439-0507.2009.01776.x. [DOI] [PubMed] [Google Scholar]
- 35.Londero AT, Benevenga JP (1972) Human infection by Microsporum nanum in Brazil. Rev Inst Med Trop Sao Paulo 14(6):388–391 [PubMed]
- 36.De Camargo RM, Silvares MR, Carvalho CR, Dillon NL, Marques SA (1992) Microsporum nanum A fourth report of human infection in Brazil. Revista do Instituto de Medicina Tropical de Sao Paulo 34(6):581 [PubMed]
- 37.Camargo RMP, Silva NAS, Marques SA, Stolf HO, Dillon NL (1984) Report of the second human infection case by Microsporum nanum, in Brazil. Rev Inst Med Trop Sao Paulo 26(3):165–169 [DOI] [PubMed]
- 38.Camargo RM, Silvares MRC, Carvalho CR, Dillon NL, Marques SA (1992) Report of the fourth human infection case by Microsporum nanum in Brazil. Rev Inst Med Trop Sao Paulo 34(6):581–585 [PubMed]
- 39.Carmichael JW, Reid JF (1962) Microsporum nanum infection in Alberta. Mycopathologia et mycologiaapplicata 17(4):325–326
- 40.Ponnighaus JM, Warndorff D, Port G (1995) Microsporum nanum-a report from Malawi (Africa) Microsporum nanum-einBerichtaus Malawi (Afrika). Mycoses 38(3–4):149–150 [DOI] [PubMed]
- 41.TurnerKAMINSK TWGW (1976) Microsporum nanum infection in South Australia. Med J Aust 1(20):743–744 [PubMed]
- 42.Garg AK, Mulay DN (1972) Isolation of Microsporum nanum from man in India. Hindustan Antibiot Bull 14(3):137–139 [PubMed]
- 43.Ranganathan S, Menon T, Balajee SAM (1997) Isolation of Microsporum nanum from a patient with tinea corporis in Madras India. Mycoses 40(5–6):229–230 [DOI] [PubMed]
- 44.Alteraş I (1970) First case of tinea infection by Microsporum nanum in Romania. Mycoses 13(9):447–450 [DOI] [PubMed]
- 45.O’Keeffe MF (1973) A report of three human infections due to Microsporum nanum. Australas J Dermatol 14(2):73–74 [DOI] [PubMed]
- 46.Sinski JT, Flouras K. A survey of dermatophytes isolated from human patients in the United States from 1979 to 1981 with chronological listings of worldwide incidence of five dermatophytes often isolated in the United States. Mycopathologia. 1984;85(1–2):97–120. doi: 10.1007/BF00436709. [DOI] [PubMed] [Google Scholar]
- 47.Sinski JT, Kelley LM. A survey of dermatophytes isolated from human patients in the United States from 1982 to 1984. Mycopathologia. 1987;98(1):35–40. doi: 10.1007/BF00431015. [DOI] [PubMed] [Google Scholar]
- 48.Weitzman I, Chin NX, Kunjukunju N, Della-Latta P. A survey of dermatophytes isolated from human patients in the United States from 1993 to 1995. J Am Acad Dermatol. 1998;39(2):255–261. doi: 10.1016/S0190-9622(98)70085-4. [DOI] [PubMed] [Google Scholar]
- 49.Mahmoud AL. A study of dermatophytoses in Sana'a Yemen Republic. Mycoses. 2002;45(3–4):105–108. doi: 10.1046/j.1439-0507.2002.00729.x. [DOI] [PubMed] [Google Scholar]
- 50.Chi CC, Wang SH, Chou MC. The causative pathogens of onychomycosis in southern Taiwan. Mycoses. 2005;48(6):413–420. doi: 10.1111/j.1439-0507.2005.01152.x. [DOI] [PubMed] [Google Scholar]
- 51.Martínez E, Ameen M, Tejada D, Arenas R (2014) Microsporum spp onychomycosis disease presentation, risk factors and treatment responses in an urban population. Brazilian J Infectious Diseas 18(2):181–186 [DOI] [PMC free article] [PubMed]
- 52.Ratka P (1985) Microsporidgonitis caused by Microsporum nanum. Mycopathologia 92(1):45–47 [DOI] [PubMed]
- 53.Shaqra QA, Al-Jamaien H, Al Zoubi M. Isolation of soil dermatophytes from three distinct geographic locations in Jordan. Fungal Ecology. 2012;5(2):274–276. doi: 10.1016/j.funeco.2011.07.003. [DOI] [Google Scholar]
- 54.Anane S. Epidemiological investigation of keratinophilic fungi from soils of Djerba (Tunisia) J de mycologiemédicale. 2012;22(3):225–229. doi: 10.1016/j.mycmed.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 55.Deshmukh SK, Mandeel QA, Verekar SA. Keratinophilic fungi from selected soils of Bahrain. Mycopathologia. 2008;165(3):143–147. doi: 10.1007/s11046-007-9067-y. [DOI] [PubMed] [Google Scholar]
- 56.Ranganathan S, Balajee SAM (2000) Microsporum gypseum complex in Madras India. Mycoses 43(5):177–180 [DOI] [PubMed]
- 57.Summerbell RC. Form and function in the evolution of dermatophytes. Bilbao, Spain: Biology of dermatophytes and other keratinophilic fungi. RevistaIberoamericana de Micologia; 2000. pp. 30–43. [Google Scholar]
- 58.Kunert J. Physiology of keratinophilic fungi. RevistaIberoamericana de Micologia. 2000;1:77–85. [Google Scholar]
- 59.Meyers MA, Chen PY, Lin AYM, Seki Y. Biological materials: structure and mechanical properties. Prog Mater Sci. 2008;53(1):1–206. doi: 10.1016/j.pmatsci.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 60.Strnad P, Usachov V, Debes C, Gräter F, Parry DA, Omary MB. Unique amino acid signatures that are evolutionarily conserved distinguish simple-type, epidermal and hair keratins. J Cell Sci. 2011;124(24):4221–4232. doi: 10.1242/jcs.089516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rice RH. Proteomic analysis of hair shaft and nail plate. J Cosmet Sci. 2011;62(2):229. [PMC free article] [PubMed] [Google Scholar]
- 62.Koehne GW (1962) Nutrition of three species of Microsporum. Mycopathologia et mycologiaapplicata 18(3):199–206 [DOI] [PubMed]
- 63.Wawrzkiewicz K, Wolski T, Łobarzewski J. Screening the keratinolytic activity of dermatophytes in vitro. Mycopathologia. 1991;114(1):1–8. doi: 10.1007/BF00436684. [DOI] [PubMed] [Google Scholar]
- 64.Kosanke S, Hamann L, Kupsch C, Garcia SM, Chopra A, Gräser Y. Unequal distribution of the mating type (MAT) locus idiomorphs in dermatophyte species. Fungal Genet Biol. 2018;118:45–53. doi: 10.1016/j.fgb.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 65.Metin B, Heitman J. She loves me, she loves me not: on the dualistic asexual/sexual nature of dermatophyte fungi. Mycopathologia. 2020;185(1):87–101. doi: 10.1007/s11046-019-00390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dawson CO, Gentles JC (1962) The perfect states of Keratinomyces ajelloi van-Breuseghem, Trichophyton terrestre Durie & Frey and Microsporum nanum Fuentes. Sabouraudia 1(1):49–57 [PubMed]
- 67.Metin B, Heitman J. Sexual reproduction in dermatophytes. Mycopathologia. 2017;182(1–2):45–55. doi: 10.1007/s11046-016-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guarro J, Cano J, Leal JA, Gómez-Miranda B, Bernabé M. Composition of the cell wall polysaccharides in some geophilic dermatophytes. Mycopathologia. 1993;122(2):69–77. doi: 10.1007/BF01103602. [DOI] [Google Scholar]
- 69.Brocksins JM (1961) Microsporum nanum: A Cause of Tinea Capitis: A Case Report. Arch Dermatol 84(3):504–505
- 70.Wenyan Z, Mingyu X, Li H, Shaoxi W, Qing Z, Bian Z (1987) Electron microscopic observation on infected hairs of kerion caused by Microsporum nanum. Int J Dermatol 26(10):641–644 [DOI] [PubMed]
- 71.Mullins JF, Willis CJ, Bergeron JR, Johnson DA, Stone OJ (1966) Microsporum nanum: a review of the literature and a report of two cases. Arch Dermatol 94(3):300–303 [DOI] [PubMed]
- 72.Long JR, Brandenburg AC, Oliver PG (1972) Case report Microsporum nanum: a cause of porcine ringworm in Ontario. Canadian Veterinary J 13(7):164 [PMC free article] [PubMed]
- 73.Dodd DC, Newlin RW, Niksch GR (1965) Infection of Swine with Microsporum nanum. J Am Vet Med Assoc 146(5):486–489 [PubMed]
- 74.Carter GR, Glenn MW. Ringworm with complimenting acanthpsis in Swine. J Am Vet Med Assoc. 1966;149(1):42–45. [PubMed] [Google Scholar]
- 75.Chermette R, Ferreiro L, Guillot J. Dermatophytoses in animals. Mycopathologia. 2008;166(5–6):385–405. doi: 10.1007/s11046-008-9102-7. [DOI] [PubMed] [Google Scholar]
- 76.Cabo JG, Asensio MB, Rodriguez FG, Lázaro JA (1995) An outbreak of dermatophytosis in pigs caused by Microsporum canis. Mycopathologia 129(2):79–80 [DOI] [PubMed]
- 77.Cabo JG, Cequiel ML, Aisa CS, Arribas MV (1988) Dermatophytosis of pigs by Trichophyton mentagrophytes. Mycopathologia 101(3):161–164 [DOI] [PubMed]
- 78.Rippon JW (1988) Medical mycology. The pathogenic fungi and the pathogenic actinomycetes (No. Ed. 3). WB Saunders company
- 79.Mercer DK, Stewart CS. Keratin hydrolysis by dermatophytes. Medical mycology. 2019;57(1):13–22. doi: 10.1093/mmy/myx160. [DOI] [PubMed] [Google Scholar]
- 80.Rippon JW (1983) Host specificity in dermatophytoses. In Proceedings of the 8th Congress of the International Society for Human and Animal Mycology, 1983. Massey University Press. India
- 81.Monod M. Secreted proteases from dermatophytes. Mycopathologia. 2008;166(5):285–294. doi: 10.1007/s11046-008-9105-4. [DOI] [PubMed] [Google Scholar]
- 82.Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2008;51:2–15. doi: 10.1111/j.1439-0507.2008.01606.x. [DOI] [PubMed] [Google Scholar]
- 83.Raubitschek F, Maoz R. Invasion of nails in vitro by certain dermatophytes. J Invest Dermatol. 1957;28:261–267. doi: 10.1038/jid.1957.30. [DOI] [PubMed] [Google Scholar]
- 84.Kano R, Kimura U, Kakurai M, Hiruma J, Kamata H, Suga Y et al (2020) Trichophyton indotineae sp. nov: a new highly terbinafine-resistant anthropophilic dermatophyte species. Mycopathologia 185(6):947–58 [DOI] [PubMed]
- 85.Yin H, Zhou JB, Chen YL, Ren L, Qin N, Xing YL, Zhao XJ (2022) Morphology, phylogeny, and pathogenicity of Trichothecium, Alternaria, and Fusarium species associated with panicle rot on Chenopodium quinoa in Shanxi Province China. Plant Pathology 71(2):344–360
- 86.Uchida T, Makimura K, Ishihara K, Goto H, Tajiri Y, Okuma M, Fujisaki R, Uchida K, Abe S, Iijima M. Comparative study of direct polymerase chain reaction, microscopic examination and culture-based morphological methods for detection and identification of dermatophytes in nail and skin samples. J Dermatol. 2009;36(4):202–208. doi: 10.1111/j.1346-8138.2009.00624.x. [DOI] [PubMed] [Google Scholar]
- 87.Okeke CN, Tsuboi R, Kawai M, Hiruma M, Ogawa H. Isolation of an intron-containing partial sequence of the gene encoding dermatophyte actin (ACT) and detection of a fragment of the transcript by reverse transcription-nested PCR as a means of assessing the viability of dermatophytes in skin scales. J Clin Microbiol. 2001;39(1):101–106. doi: 10.1128/JCM.39.1.101-106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pounder JI, Williams S, Hansen D, Healy M, Reece K, Woods GL. Repetitive-sequence-PCR-based DNA fingerprinting using the Diversilab system for identification of commonly encountered dermatophytes. J Clin Microbiol. 2005;43(5):2141–2147. doi: 10.1128/JCM.43.5.2141-2147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanbe T, Suzuki Y, Kamiya A, Mochizuki T, Kawasaki M, Fujihiro M, Kikuchi A (2003) Species-identification of dermatophytes Trichophyton, Microsporum and Epidermophyton by PCR and PCR-RFLP targeting of the DNA topoisomerase II genes. J Dermatol Sci 33(1):41–54 [DOI] [PubMed]
- 90.Liu D, Coloe S, Baird R, Pedersen J (1997) Rapid differentiation of Microsporumdermatophytes based on arbitrarily primed PCR amplification. Opportunistic pathogens 9(1):3–6
- 91.Leon-Mateos A, Paredes-Suárez C, Pereiro M Jr, Toribio J (2006) Study of the ITS region in an atypical isolate and comparison with six species of Microsporum. Mycoses 49(6):452–456 [DOI] [PubMed]
- 92.Rezaei-Matehkolaei A, Makimura K, Shidfar MR, Zaini F, Eshraghian MR, Jalalizand N, Nouripour-Sisakht S, Hosseinpour L, Mirhendi H. Use of single-enzyme PCR-restriction digestion barcode targeting the internal transcribed spacers (ITS rDNA) to identify dermatophyte species. Iran J Public Health. 2012;41(3):82. [PMC free article] [PubMed] [Google Scholar]
- 93.Liu D, Coloe S, Baird R, Pedersen J. Molecular determination of dermatophyte fungi using the arbitrarily primed polymerase chain reaction. Br J Dermatol. 1997;137(3):351–355. doi: 10.1046/j.1365-2133.1997.18481941.x. [DOI] [PubMed] [Google Scholar]
- 94.L’Ollivier C, Cassagne C, Normand AC, Bouchara JP, Contet-Audonneau N, Hendrickx M, Fourquet P, Coulibaly O, Piarroux R, Ranque S. A MALDI-TOF MS procedure for clinical dermatophyte species identification in the routine laboratory. Med Mycol. 2013;51(7):713–720. doi: 10.3109/13693786.2013.781691. [DOI] [PubMed] [Google Scholar]
- 95.Nenoff P, Erhard M, Simon JC, Muylowa GK, Herrmann J, Rataj W, Gräser Y. MALDI-TOF mass spectrometry–a rapid method for the identification of dermatophyte species. Med Mycol. 2013;51(1):17–24. doi: 10.3109/13693786.2012.685186. [DOI] [PubMed] [Google Scholar]
- 96.Theel ES, Hall L, Mandrekar J, Wengenack NL. Dermatophyte identification using matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol. 2011;49(12):4067–4071. doi: 10.1128/JCM.01280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Panda A, Ghosh AK, Mirdha BR, Xess I, Paul S, Samantaray JC, Srinivasan A, Khalil S, Rastogi N, Dabas Y. MALDI-TOF mass spectrometry for rapid identification of clinical fungal isolates based on ribosomal protein biomarkers. J Microbiol Methods. 2015;109:93–105. doi: 10.1016/j.mimet.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 98.Rychert J (2019) Benefits and limitations of MALDI-TOF mass spectrometry for the identification of microorganisms. J Infectiol Epidemiol 2(4)
- 99.Erhard M, Hipler UC, Burmester A, Brakhage AA, Wöstemeyer J. Identification of dermatophyte species causing onychomycosis and tinea pedis by MALDI-TOF mass spectrometry. Exp Dermatol. 2008;17(4):356–361. doi: 10.1111/j.1600-0625.2007.00649.x. [DOI] [PubMed] [Google Scholar]
- 100.Frías-De-León MG, Martínez-Herrera E, Atoche-Diéguez CE, González-Cespón JL, Uribe B, Arenas R, Rodríguez-Cerdeira C (2020) Molecular identification of isolates of the Trichophyton mentagrophytes complex. Int J Med Sci 17(1):45 [DOI] [PMC free article] [PubMed]
- 101.Makimura K, Tamura Y, Murakami A, Kano R, Nakamura Y, Hasegawa A, Uchida K, Yamaguchi H (2001) Cluster analysis of human and animal pathogenic Microsporum species and their teleomorphic states, Arthroderma species, based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1. Microbiol Immunol 45(3):209–216 [DOI] [PubMed]
- 102.Rezaei-Matehkolaei A, Mirhendi H, Makimura K, de Hoog GS, Satoh K, Najafzadeh MJ, Shidfar MR. Nucleotide sequence analysis of beta tubulin gene in a wide range of dermatophytes. Med Mycol. 2014;52(7):674–688. doi: 10.1093/mmy/myu033. [DOI] [PubMed] [Google Scholar]
- 103.Mirhendi H, Makimura K, de Hoog GS, Rezaei-Matehkolaei A, Najafzadeh MJ, Umeda Y, Ahmadi B. Translation elongation factor 1-α gene as a potential taxonomic and identification marker in dermatophytes. Med Mycol. 2015;53(3):215–224. doi: 10.1093/mmy/myu088. [DOI] [PubMed] [Google Scholar]
- 104.Houšť J, Spížek J, Havlíček V. Antifungal drugs. Metabolites. 2020;10(3):106. doi: 10.3390/metabo10030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carrillo-Muñoz AJ, Giusiano G, Guarro J, Quindós G, Guardia C, Del Valle O, Rodríguez V, Estivill D, Cárdenes CD (2007) In vitro activity of voriconazole against dermatophytes, Scopulariopsis brevicaulis and other opportunistic fungi as agents of onychomycosis. Int J Antimicrob Agents 30(2):157–161 [DOI] [PubMed]
- 106.Fernández-Torres B, Carrillo AJ, Martın E, Del Palacio A, Moore MK, Valverde A, Serrano M, Guarro J. In vitro activities of 10 antifungal drugs against 508 dermatophyte strains. Antimicrob Agents Chemother. 2001;45(9):2524–2528. doi: 10.1128/AAC.45.9.2524-2528.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perea S, Fothergill AW, Sutton DA, Rinaldi MG. Comparison of in vitro activities of voriconazole and five established antifungal agents against different species of dermatophytes using a broth macrodilution method. J Clin Microbiol. 2001;39(1):385–388. doi: 10.1128/JCM.39.1.385-388.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ali-Shtayeh MS, Arda HM, Hassouna M, Shaheen SF. Keratinophilic fungi on the hair of cows, donkeys, rabbits, cats, and dogs from the West Bank of Jordan. Mycopathologia. 1988;104(2):109–121. doi: 10.1007/BF00436936. [DOI] [PubMed] [Google Scholar]
- 109.Ali-Shtayeh MS, Arda HM, Hassouna M, Shaheen SF. Keratinophilic fungi on sheep hairs from the West Bank of Jordan. Mycopathologia. 1989;106(2):95–101. doi: 10.1007/BF00437087. [DOI] [PubMed] [Google Scholar]
- 110.Nweze EI. Dermatophytoses in domesticated animals. Rev Inst Med Trop Sao Paulo. 2011;53(2):94–99. doi: 10.1590/S0036-46652011000200007. [DOI] [PubMed] [Google Scholar]
- 111.Seker E, Dogan N. Isolation of dermatophytes from dogs and cats with suspected dermatophytosis in Western Turkey. Prev Vet Med. 2011;98(1):46–51. doi: 10.1016/j.prevetmed.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 112.Ilhan Z, Karaca M, Ekin IH, Solmaz H, Akkan HA, Tutuncu M. Detection of seasonal asymptomatic dermatophytes in Van cats. Brazilian J Microbiol. 2016;47(1):225–230. doi: 10.1016/j.bjm.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mahmoud ALE. Dermatophytes and other associated fungi isolated from ringworm lesions of camels. Folia Microbiol. 1993;38(6):505–508. doi: 10.1007/BF02814404. [DOI] [PubMed] [Google Scholar]
- 114.Ramesh VM, Hilda A. Incidence of keratinophilic fungi in the soil of primary schools and public parks of Madras city India. Mycopathologia. 1998;143(3):139. doi: 10.1023/A:1006945012620. [DOI] [PubMed] [Google Scholar]
- 115.Dolenc-Voljč M, Gasparič J. Human infections with Microsporum gypseum complex (Nannizzia gypsea) in Slovenia. Mycopathologia. 2017;182(11):1069–1075. doi: 10.1007/s11046-017-0194-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.