Abstract
Particulate organic carbon settling through the marine water column is a key process that regulates the global climate by sequestering atmospheric carbon. The initial colonization of marine particles by heterotrophic bacteria represents the first step in recycling this carbon back to inorganic constituents—setting the magnitude of vertical carbon transport to the abyss. Here, we demonstrate experimentally using millifluidic devices that, although bacterial motility is essential for effective colonization of a particle leaking organic nutrients into the water column, chemotaxis specifically benefits at intermediate and higher settling velocities to navigate the particle boundary layer during the brief window of opportunity provided by a passing particle. We develop an individual-based model that simulates the encounter and attachment of bacterial cells with leaking marine particles to systematically evaluate the role of different parameters associated with bacterial run-and-tumble motility. We further use this model to explore the role of particle microstructure on the colonization efficiency of bacteria with different motility traits. We find that the porous microstructure facilitates additional colonization by chemotactic and motile bacteria, and fundamentally alters the way nonmotile cells interact with particles due to streamlines intersecting with the particle surface.
Keywords: Marine snow, chemotaxis, bacterial colonization, bacterial encounter, individual-based modeling
Significance Statement.
Bacteria in the ocean rely on ephemeral nutrient patches from sinking marine particles. Nevertheless, attaching to these structures is challenging as particle settling rates often exceed bacterial swimming velocities and the numerically dominant marine bacteria are nonmotile. Here, we quantify the importance of chemotaxis and motility for the efficient colonization of marine particles and find that although motility is the basic requirement for particle colonization, chemotaxis provides a clear advantage. We expand this analysis to consider highly heterogeneous particle structures and find a noticeable benefit for all motility traits by facilitating a direct encounter with the particle surface.
Introduction
Heterotrophic bacteria exert a fundamental role on ocean biogeochemistry by recycling dissolved and particulate organic matter back to inorganic constituents. The most abundant bacterial species in the marine environment are nonmotile (1) and rely on diffusive fluxes to consume the fairly homogeneous but low concentrations of dissolved and recalcitrant carbon in the bulk water (2). In contrast, particulate organic matter such as phytoplankton aggregates or fecal pellets (together termed “marine snow”) sink through the water column, resulting in a vertical flux of carbon from the surface mixed layer to the abyss. This process of sequestering atmospheric carbon through photosynthesis and subsequent transport to the ocean interior and sediments is termed the biological carbon pump, and is a key driver in regulating Earth's global carbon cycle (3). Compared to the surrounding bulk water, these particles can have orders of magnitude higher carbon and nutrient concentrations and act as microbial hotspots in an otherwise nutrient-poor marine environment (4). The intensified metabolic activity within sinking particles (4, 5) can drive rapid remineralization of the organic matter during settling and ultimately determine the efficiency of the organic carbon pump (6, 7). It has been shown that the particle-associated microbial community is distinct when compared to the surrounding, free-living community (8, 9) and is typically enriched in fast-growing copiotrophic bacteria (10–12) associated with motile and chemotactic (i.e., the ability to alter swimming patterns in response to chemical stimuli) behavior.
Marine particles exist across a spectrum of shapes and sizes (13). The fate of these particles is governed by their remineralization dynamics and settling velocity, which themselves are complex functions determined by their size, shape, and excess density (difference in average particle density compared to surrounding seawater) (14). Across the globe, particle size distributions are captured by a power law, suggesting that large particles (e.g., >3 mm) are rare whereas the numerically dominant particles are very small (<1 mm). As a result, some large particles (namely, fecal pellets) may settle at extremely fast rates of up to a few hundred meters per day (14). These fast-sinking particles play an important role in carbon sequestration, as their speed limits the available time for remineralization by heterotrophic bacteria. Nevertheless, the average particle in the ocean typically sinks very slowly (15), ranging between 1 and 10 m d−1. Yet, at the scale of a bacterial cell, particles settling at seemingly slow velocities may still outpace the swimming velocity of a bacterium. For instance, a modest settling velocity of 5 m d−1 translates to 58 µm s−1 and—especially when considering the stochastic random walk patterns frequently observed in flagellated bacterial species (16)—presents a scenario where many bacterial species cannot effectively chase fleeting particles. Therefore, colonization of marine particles must occur in a very short window of time during the bacteria-particle encounter when the particle passes by the bacterial cell.
The question of how bacterial cells colonize marine particles has been continually researched for more than two decades. In a seminal publication, Kiørboe and colleagues systematically observed how bacterial isolates colonize inert, spherical particles and found that flow significantly increases the colonization rate by motile cells (17). They furthermore unveiled that tumbling, a required trait of chemotactic bacteria, permits a more rapid colonization of synthetic particles enriched with nutrients compared to inert particles in still water. Using an agent-based model, Kiørboe and Jackson could show that the optimal parameters for particle colonization concerning tumbling frequency, swimming velocity, and chemotactic sensitivity are very close to the observed behavior of marine bacteria (18). However, revealing the mechanisms of how chemotaxis governs bacterial life in the ocean is challenging (19). The ability of bacterial cells to navigate toward nutrient-rich particle plumes has been demonstrated in laboratory microcosms using innovative microfluidic devices (20). Similar approaches have since been used to show that self-propulsion is required for bacterial cells to traverse the steep velocity gradients in close proximity of inert particle surfaces (21), or that their colonization efficiency depends on the physical shape of the cell (e.g., cocci, rods, and filaments) due to shear-driven reorientation (21, 22). Furthermore, the optimal foraging of bacteria on dynamically emerging synthetic nutrient patches in vitro (23) and the emergence of localized nutrient patches resulting from phytoplankton exudation, copepod death, and fecal pellets that induce a rapid chemotactic response have been studied in the absence of flow (24, 25). Finally, the prevalence of such chemotactic responses has been demonstrated in situ by deploying devices with microwells containing chemoattractant directly into marine environments and subsequently quantifying the number of cells relative to an inert chamber (26, 27). In summary, this extensive body of work has revealed that motility and chemotaxis toward naturally occurring chemical stimuli in the marine environment is prevalent and an important mechanism to traverse the velocity gradients in proximity of the particle surfaces or navigate towards the nutrient-rich particle plumes. However, the relative benefit of being chemotactic (in addition to motile) when colonizing particle surfaces in the presence of a continuous carbon source leaking from the particle under the influence of flow (i.e., of a sinking particle) has not been quantified experimentally.
In this study, we use a millifluidic experimental system and fluorescence microscopy (Fig. 1) to quantify how chemotaxis facilitates the colonization of synthetic marine particles that maintain a constant nutrient source (representative of leaky marine snow) during the brief window of time when the particle passes by the bacterial cell. By employing fluorescently tagged mutants that are either chemotactic, nonchemotactic but motile, or nonmotile, we can systematically distinguish the contribution of different motility traits to particle colonization across controlled flow conditions mimicking different settling velocities. We combine these experimental observations with computational predictions based on solving coupled Navier–Stokes and reaction-diffusion equations with bacterial cells represented as individual agents to explore how colonization efficiency varies with bacterial traits. We investigate how the microstructure of more realistic, porous marine snow (resulting from the aggregation of biogenic polymers and inorganic minerals or an organic particle being actively degraded) affects bacterial colonization, and furthermore, compare the porous particles with their rough-surface and smooth-surface counterparts.
Fig. 1.
Conceptual image of the experimental system and resulting nutrient plume. (a) The experimental system includes a millifluidic device (Polydimethylsiloxane—PDMS) on top of a glass microscope slide with a synthetic marine particle in the form of a 3 mm diameter agarose disk. Nutrients leak from the sterile reservoir (red) through the agarose disk into the flow chamber (dark blue) and mimics a constantly leaking particle. The flow chamber is sealed to the glass slide using two 3D-printed parts that are screwed together with the nutrient reservoir directly engineered inside the upper part. The synthetic marine particle that connects the two systems ensures a steady leak of nutrients from the agarose disk into the main flow chamber. (b) The total length of the flow chamber is 20 mm with a channel width of 5 mm. The diameter of the central expansion is 8 mm to accommodate the 3 mm agarose particle and keep the channel width constant at 5 mm across the whole channel length. The flow channel is 1 mm in depth with the agarose disk embedded and an additional 1 mm within the PDMS for structural stability (i.e., a total height of 2 mm). The highlighted area shows the approximate region visualized in panels c and d. (c) We imitate different settling velocities from 1 to 100 m d−1 using a syringe pump and visualize the resulting particle plume using fluorescein. (d) We use a digital twin approach (congruent geometry, flow, and nutrient conditions as in the experiments) to simulate the resulting flow pattern and particle plumes that are in good agreement with the experimental observations (panel c).
Results
Colonization of synthetic marine particles across motility traits
We use three genetically engineered strains of Pseudomonas aeruginosa PA14 in our experiments that constitutively express the same yellow fluorescent protein. A wild-type strain that is both motile and chemotactic (“chemotactic”), a chemotactic-deficient mutant P. aeruginosa PA14 cheZ::TnM that can nonetheless swim (“motile”), and a motility-deficient mutant P. aeruginosa PA14 ∆fliC (”nonmotile”). Motility and chemotaxis traits were confirmed by plate assay (Supplementary Fig. S1). By using mutants of the otherwise isogenic parent strain, we can isolate the influence of chemotaxis and/or motility on colonization and directly compare the fluorescence signals captured with the microscope (Fig. 2a and b). Due to the background fluorescence of the particle and the potential to bias the results through parameter choice, we refrained from converting the fluorescence signal to total cell number and only present the quantified fluorescent signal, which is directly proportional to cell abundance (Supplementary Fig. S2a). We observe and quantify the colonization of the particles by bacteria with different motility traits across three settling velocities that represent nearly suspended particles (1 m d−1), reasonable settling velocities for the majority of sinking particles (10 m d−1), and rapidly sinking, large particles that are numerically rare in the ocean but contribute overwhelmingly to the mass flux of the biological carbon pump (100 m d−1) (28). For all settling velocities, we observe a consistent pattern where the chemotactic strain shows the highest mean fluorescence intensity, followed by the motile strain, and lastly the nonmotile strain (Fig. 2b, statistical significance in Supplementary Table S1). In addition, the observed fluorescent intensity decreases for both the chemotactic and motile strains with increasing settling velocity (Fig. 2b). However, this decrease is less pronounced for the chemotactic strain compared to the motile strain (53, 80, and 138% higher fluorescent intensity than the motile strain for 1, 10, and 100 m d−1), suggesting that the chemotactic strain gains a competitive advantage over nonchemotactic motile cells at higher settling velocities due to the ability to rapidly respond during the ephemeral particle encounter. We further confirmed that the increase in colonization is indeed due to a chemotactic response by exchanging the chemoattractant with motility medium (i.e., lacking the chemoattractant), where we observed the same fluorescence for both the chemotactic and motile strains (Supplementary Fig. S2c). For the nonmotile strain, we observed no significant difference in fluorescent signal across settling velocities. However, since the fluorescent signal is higher when compared to a sterile particle (the dashed gray line in Fig. 2b), there is some colonization of the particles due to collision between the nonmotile cells and the particle surface.
Fig. 2.
Experimental colonization results and comparison to mathematical predictions. (a) We use the fluorescence intensity of the three mutant strains as a proxy for the number of cells that colonized the particle surface within a region of ±250 µm from the particle surface (delineated by the yellow dashed circles in the central image). These results show colonization for a simulated settling velocity of 10 m d−1 with a flow direction from left to right. (b) Quantification of the fluorescence signal as a proxy for the total number of colonized cells with pairwise comparisons between the strains for the same settling velocities and within the same strain across different settling velocities (* for P < 0.05, ** for P < 0.01). The dashed gray line indicates the mean fluorescent intensity of sterile particles. (c) Prediction of colonization using the numerical model, where the horizontal lines show the mean value across 10 replicates using the standard parameterization of bacterial motility and bars show the variation when strategically exploring the sensitivity across the parameter space in the chemotaxis and motility algorithm (Methods, Fig. 3).
Simulations show a consistent pattern of colonization efficiency when compared to the experimental results (Fig. 2c). Here and throughout the manuscript, the colonization efficiency is defined as the fraction of the inoculated cells that ultimately attached to the particle. The high number of simulated cells (average density of 25 cells µm−1 across the channel at the inoculation location) results in negligible variance among the 10 technical replicates. For this reason, we only visualized the mean value of the simulation replicates in Fig. 2c (horizontal bars) and superimposed the variation in colonization efficiency when exploring the parameter space of the motility and chemotaxis algorithms (further described below). All pairwise statistical tests between the different strains at the same settling velocity for the simulation replicates were significant with the exception between the chemotactic and motile at a settling velocity of 100 m d−1. Congruent to the experimental results, both the chemotactic and motile strains showed a decrease in colonization efficiency with increasing settling velocities (P < 0.01). The model predicts the highest benefit of chemotaxis at intermediate settling velocities with an increase in colonized cells of 31, 60, and 34% for 1 m d−1, 10 m d−1, and 100 m d−1, respectively.
Colonization efficiency is highest at intermediate settling velocities
We further explore the importance of the different motility and chemotaxis parameters for colonizing the particles (Fig. 3). For all combinations of swimming velocity and base run length, the diffusivity of the bacterial cells falls within the range of 0 (for nonmotile cells) to 1.3 × 10−8 m2 s−1 (fast cells with a long base run length). Diffusivities are calculated as 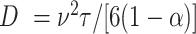 where
where 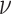 is the bacterial cell velocity [m s−1],
is the bacterial cell velocity [m s−1], 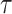 the base run length [s] and
the base run length [s] and  the cosine of the average angle between two runs (17). Overall, the simulated settling velocity and associated localized flow velocity governs the ability of bacteria to influence their trajectory and colonization capability (Fig. 3a). The intensity of the response increases with settling velocity as the greater flow around the particle warps the nutrient concentration field into steeper nutrient gradients. Particle velocity profoundly influences the importance of chemotactic sensitivity (Fig. 3b). Interestingly, for both the slow and rapid settling velocities (1 and 100 m d−1), the resulting colonization efficiency is primarily a function of the swimming velocity, while the chemotactic sensitivity only contributes marginally. In contrast, the chemotactic sensitivity governs the colonization efficiency at an intermediate settling efficiency, particularly for cells with higher swimming velocities. A similar pattern to the one described above emerges when comparing the interaction between the swimming velocity and the base run length of the chemotactic strain (Fig. 3c). These data suggest that the reduction in base run length is compensated by the reduction in tumbling probability via chemotaxis when colonizing the particles. This concept is further supported when comparing the colonization pattern of the chemotactic strain to that of the motile strain (Fig. 3d). In this case, a reduction in the base run length also results in a reduction of the colonization efficiency as cells fail to navigate the nutrient gradient in close proximity to the particle. In summary, the ability for bacterial cells to swim is fundamental to efficiently colonize particles at any settling velocity and chemotaxis plays an important role at intermediate settling velocities (10 m d−1) where the flow around the particle creates steep substrate gradients (in comparison to low settling velocities) but there remains sufficient time for the chemotactic behavior to navigate the particle boundary layer (in comparison to higher settling velocities where bacterial cells are rapidly swept around the particle).
the cosine of the average angle between two runs (17). Overall, the simulated settling velocity and associated localized flow velocity governs the ability of bacteria to influence their trajectory and colonization capability (Fig. 3a). The intensity of the response increases with settling velocity as the greater flow around the particle warps the nutrient concentration field into steeper nutrient gradients. Particle velocity profoundly influences the importance of chemotactic sensitivity (Fig. 3b). Interestingly, for both the slow and rapid settling velocities (1 and 100 m d−1), the resulting colonization efficiency is primarily a function of the swimming velocity, while the chemotactic sensitivity only contributes marginally. In contrast, the chemotactic sensitivity governs the colonization efficiency at an intermediate settling efficiency, particularly for cells with higher swimming velocities. A similar pattern to the one described above emerges when comparing the interaction between the swimming velocity and the base run length of the chemotactic strain (Fig. 3c). These data suggest that the reduction in base run length is compensated by the reduction in tumbling probability via chemotaxis when colonizing the particles. This concept is further supported when comparing the colonization pattern of the chemotactic strain to that of the motile strain (Fig. 3d). In this case, a reduction in the base run length also results in a reduction of the colonization efficiency as cells fail to navigate the nutrient gradient in close proximity to the particle. In summary, the ability for bacterial cells to swim is fundamental to efficiently colonize particles at any settling velocity and chemotaxis plays an important role at intermediate settling velocities (10 m d−1) where the flow around the particle creates steep substrate gradients (in comparison to low settling velocities) but there remains sufficient time for the chemotactic behavior to navigate the particle boundary layer (in comparison to higher settling velocities where bacterial cells are rapidly swept around the particle).
Fig. 3.
Sensitivity analysis of the motility and chemotaxis algorithms. (a) Simulated trajectory of individual bacterial cells at different settling velocities. Highlighted trajectories represent cells that colonized the particle in contrast to faded trajectories that are not successful. The ability of bacterial cells to influence their trajectory with self-propulsion becomes negligible at rapid particle settling velocities. (b) Colonization efficiency at different combinations of chemotactic sensitivity and cell swimming velocity for the chemotactic strain. (c) Colonization efficiency at different combinations of base run length and cell swimming velocity for the chemotactic strain. (d) Colonization efficiency at different combinations of base run length and cell swimming velocity for the motile strain. Dotted lines in panels b, c, and d represent the parameter values for P. aeruginosa in the experimental twin geometries and stochastic simulations. We used different scales (shown by the color bar) for the different settling velocities with markedly lower colonization at higher settling velocities for all parameter combinations.
Particle microstructure facilitates colonization by nonmotile cells.
In contrast to the simple disk-shape used in the experimental system, natural marine snow comes in myriad shapes and can be highly porous. To explore how these aspects change the colonization by bacteria with different motility traits, we extend our experimentally-validated modeling approach to simulate scenarios that are infeasible experimentally and more representative of heterogeneous marine particles and aggregates in the ocean. Geometries that support intra-particle advective fluxes may readily emerge through the process of aggregation of smaller particles due to differential settling velocities or following polymer lysis by actively degrading bacterial colonies that colonized the particle previously. We craft a cohort of 500 stochastically generated particle geometries ranging from small (100 µm diameter) to large (3 mm diameter) particles with target porosities between 0 (densely packed particles) and 0.7 (flocculent particles/aggregates). Each particle is composed of stochastically arranged subparticles that span between 10 and 100 µm diameter (that can overlap to form larger aggregates) and may permit interstitial advective flows (Figs. 4 and 5a). We used a larger domain for the porous particle simulations to ensure that there are no boundary effects on the flow around the particles. We validate that this assumption does not significantly change our results by comparing the large domain and experimental twin geometry for a 3 mm solid particle at the same simulated velocities used in the experimental results (Supplementary Fig. S3 and Table S2). We calculate the macroscopic settling velocity for each aggregate as a function of overall particle size and porosity (Methods). The distribution of Reynolds and Sherwood numbers, as well as visualizations of particles with equal size but dissimilar porosities are shown in Supplementary Fig. S4. By using an individual-based approach, we can relate the initial location and exact trajectory of each bacterium to the ability of the cell to find the particle. We observe no significant difference in model predictions that use the large domain compared to those that use an experimental twin geometry, with the exception of a lower colonization efficiency at slow settling velocities (1 m d−1). In this case, bacterial swimming velocity exceeds the flow velocity, and cells may actively swim away from the particle in the large domain. However, since predicted settling velocities for all porous particles (Eq. 2 in the methods section) exceed 20 m d−1, the difference in simulated domain geometry does not bias the colonization results.
Fig. 4.
Example stochastic particle geometries and concentration fields. The density and settling velocity of the stochastic particles are determined by the number of aggregated subparticles and overall particle size. These parameters further govern the resulting particle plume, which is visualized as the predicted concentration field around the particle. The localized flow field, nutrient concentration, and colonization by bacterial cells of the emphasized particle are highlighted in Fig. 5a.
Fig. 5.
Quantifying bacterial colonization of porous particles. (a) We use a cohort of 500 stochastically generated particles with varying sizes and porosities to simulate more realistic particle geometries. Porous particles have a similar macroscopic distribution of nutrient around them, but permit interstitial advective flows that change the localized flow field around the particles. The relationship between particle size, porosity, and settling velocity for the 500 stochastically generated particles is shown in Supplementary Fig. S5a. (b) Capture radius of the particles as a function of their settling velocity. The capture radius is the radial distance from the stagnation line that contains 95% of the cells that colonized the particle. Results agree well with the characteristic diffusion distance calculated as 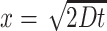 where t is the advective time from the inoculation point to the front of the particle and D is the diffusivity of motile or nonmotile cells. (c) Fraction of the inoculated cells that colonize the particle as a function of the particle porosity. (d) Increase in colonization by chemotactic cells compared to motile cells as a function of porosity and total flow through the particle.
where t is the advective time from the inoculation point to the front of the particle and D is the diffusivity of motile or nonmotile cells. (c) Fraction of the inoculated cells that colonize the particle as a function of the particle porosity. (d) Increase in colonization by chemotactic cells compared to motile cells as a function of porosity and total flow through the particle.
Interestingly, the capture radius of the stochastically generated particles for both the chemotactic and motile cells is a function of the particle settling velocity and is notably governed by bacterial random walk motility rather than chemotactic behavior as might be expected a priori (Fig. 5b). The capture radius is defined as the radial distance from the stagnation line (the streamline colliding with the stagnation point from which the other streamlines diverge) that contains 95% of the cells that colonized the particle. This is expected since there are no nutrient gradients upstream of the particle for the chemotactic cells to navigate, and further supported by the fact that we can predict the capture radius for both the chemotactic and motile cells via the characteristic diffusion distance for motile cells. For nonmotile cells, on the other hand, the observed capture radius is always larger than predicted by diffusive theory (equations shown in Fig. 5b), suggesting that the particle heterogeneity and interstitial advective fluxes may play a significant role for colonization. A higher particle porosity increases the colonization efficiency for all strains (Fig. 5c) and further benefits the chemotactic strain when compared to the motile strain (Fig. 5d). Higher porosity results in less flow being diverted around the particle and, consequentially, a diminished boundary layer and velocity gradient. The chemotactic cells benefit disproportionally from this phenomenon, as the lower flow velocity in close proximity of the particle permits ample time to chemotactically navigate the boundary layer. The relation between the colonization efficiency and additional particle properties is shown in Supplementary Fig. S5.
In addition, we can distinguish between the effects of particle porosity, roughness, and a smooth surface by creating two additional twin geometries for each of the 500 stochastic porous particles that use the same subparticle structure with a filled core (particle with a rough surface) or a filled core and smoothed surface (Supplementary Fig. S6). The chemotactic and motile strains generally show a 10-fold increase in colonization efficiency when colonizing a porous compared to a rough particle (Supplementary Fig. S6a). In comparison, the nonmotile strain shows a far wider fold increase distribution dependent on the specific particle heterogeneity. Although these results suggest that the nonmotile strains may benefit from particle porosity, the absolute number of nonmotile cells that colonize any particle is vanishingly small when compared to their chemotactic and motile counterparts.
Discussion
The benefit of chemotaxis and motility for bacterial colonization of marine snow
In this study, we visualized and quantified the colonization of marine particles by bacteria with different motility traits and highlighted the important influence of particle microstructures on the colonization efficiency. Our experimental system enabled the direct quantification of the colonization of a synthetic leaking particle by bacterial strains with different motility traits using fluorescence microscopy. A key improvement of this experimental system is the ability to mimic a leaking particle over a time period of multiple hours with controllable settling velocities. This system enables us to observe how chemotaxis and motility facilitate the colonization of marine particles under realistic flow and chemoattractant conditions, which was not possible with previous experimental approaches. Furthermore, by using mutants of P. aeruginosa (in contrast to different motile and nonmotile marine species) and tagging the strains with the same fluorescent protein, we can directly compare and isolate the influence of the different motility traits on the colonization efficiency across the settling velocities. Similar to previous observations for the colonization of curved surfaces by bacteria (21), our experiments and simulations confirm that motility is essential to colonize particles efficiently. Interestingly, we do not observe a more pronounced colonization leeward of the particle due to shear-induced reorientation as previously observed (21) and attribute this to the difference in particle size, imposed flow conditions and associated lower shear rates in our system. In addition to the fundamental requirement of being motile to colonize particles, we observe that chemotaxis benefits the bacterial cells when navigating the particle boundary layer, especially at intermediate and higher settling velocities where a rapid response is crucial (Fig. 2b and c). From a bacterial perspective, colonizing marine snow early on is especially important for fast-sinking particles where nutrients are abundant and support the rapid proliferation of surface-attached cells. However, early colonization does not guarantee success as they compete (or cooperate) with the already established community on the particle (29, 30) or struggle with changing environmental conditions that may result in a loss of diversity on the particles (31).
Choosing a single, tractable species for the experiments does not fully elucidate the breadth of diverse motility and chemotaxis mechanisms found in the oceans. Although P. aeruginosa is not a typical marine bacterium [albeit one found in the ocean (32–35)], it is a model organism for studying the chemotactic and motile behavior of singly flagellated cells (36) that are representative of the majority of the marine species (37). While P. aeruginosa [swimming velocity of 45 to 60 µm s−1 (21, 38)] is considered a rapid swimmer, marine bacteria can be similarly fast or even faster, frequently exceeding speeds over 100 µm s−1 and potentially reaching up to 445 µm s−1 when tracking nutrient patches (39). Indeed, bacterial swimming speed has previously been designated a key parameter for efficient chemotaxis, with an optimum as a function of the nutrient patch size (40). Bacterial motility in our simulations is modeled as a classic run-and-tumble algorithm with a uniform turn angle distribution to minimize bias to the computational results due to parameter choice. However, P. aeruginosa motility differs and follows a “run-reverse” or “run-reverse-pause” pattern with a different turn angle distribution (41). We strategically tested the influence of different turn angle distributions on the resulting colonization efficiency of the experimental twin geometry and found that the trend of reduced colonization efficiency as a function of the settling velocity and higher colonization efficiency by the chemotactic cells remains unchanged (Supplementary Fig. S7 and Table S3). To further explore the importance of different aspects in a bacterial run-and-tumble movement (such as the average run length, swimming velocity, or chemotactic sensitivity) to colonize the marine particles, we strategically explored the parameter space of the motility algorithm in the individual-based simulations. The bacterial swimming speed has the greatest influence on the colonization efficiency, in contrast to a change in the average run length that only marginally affects the colonization efficiency (Figs. 2c and 3). This should result in an advantage for cells that evolve faster swimming speeds when relying on particle-associated nutrients. For the parameter space exploration, we varied both the swimming velocity and chemotactic sensitivity over two orders of magnitude, whereas we only changed the average run length over one order of magnitude to remain within a physiologically meaningful realm (e.g., average run lengths of 100 s are unrealistic). Contrary to our initial expectation, varying the chemotactic sensitivity across two orders of magnitude had a lesser influence on the colonization efficiency when compared to the swimming speed (Fig. 2c). We attribute this to the fact that bacterial cells already need to be in close proximity of the particle to sense the chemoattractant, and the additional benefit of chemotaxis primarily comes into play when navigating the velocity gradient to the particle surface. Evidently, the chemotactic response of marine bacteria has to be very efficient in order to benefit from the brief time window where bacterial cells are within a nutrient gradient of a particle. Indeed, when compared to chemotaxis in E. coli (representing the most studied chemotactic system), marine bacteria show far more rapid chemotactic responses (20, 42) and other adaptions, such as a velocity-dependent motile behavior (42–44), that suggest an evolutionary adaptation to profit from these ephemeral opportunities.
Finally, even when getting to the surface of the particle, bacterial strains show different strategies for surface attachment beyond the dynamics explored here. For example, strains as closely related as P. aeruginosa PAO1 and PA14 show stark differences in their strategy to surface attachment and detachment (45). Even within the same bacterial population, the actual surface attachment strength cannot be deduced from population average observations as there is significant variability at the single cell level (46). These observations preclude directly translating our specific quantitative observations to the dynamics of particle attachment and detachment in the real ocean beyond the initial encounter. For this reason, we primarily focus on the initial encounter and attachment of the bacterial cells in the model (perfect sticking condition) and observe the bacterial cells after a fixed amount of liquid has flown past the particle in the experiments to assess the influence of the initial colonization of the particles rather than the subsequent dynamics that emerge due to differential attachment and detachment. As such, our results describe the maximum potential of colonization. Nevertheless, the initial encounter and attachment of bacterial cells to the particle is of fundamental importance since it is the first and necessary step of planktonic cells to colonize marine sinking particles, contribute to the remineralization of marine snow, and thus influence the biological carbon pump efficiency.
Particle heterogeneity and microstructure alters trait-specific interaction mechanisms
The geometry and structure of idealized disk-shaped synthetic particles are in stark contrast to natural particles, which can be highly heterogeneous at the microscale (13, 47, 48). In contrast to perfect spheres, the irregular shape creates steep velocity gradients and heterogeneous flow fields close to the particle surface (49), which affect the bacterial trajectory during colonization. In addition, the flocculent and porous appearance of natural marine particles (13) suggests the presence of interstitial advective flows that permeate the particles. However, direct measurement of particle permeation or diffusivity in natural marine snow is inherently difficult due to their fragile nature. Multiple studies suggest that mass transport in marine snow is solely dominated by diffusive fluxes with negligible convective interstitial flow. For instance, diffusivities measured in select diatom aggregates using a diffusivity microsensor did not suggest the presence of any substantial permeation (50). Furthermore, particle image velocimetry of flow around diatom aggregates created in roller tanks did not show any streamlines that penetrate the particles (49). In contrast, several studies promote the presence of interstitial flows in real marine snow or bacterial aggregates. For instance, a discrepancy between the predicted diffusive oxygen flux and experimentally observed oxygen consumption rates suggest the presence of advective oxygen flux into the particles and interstitial advective flow on the order of 40 µm s−1 (51). Similarly, bacterial aggregates in activated sludge, an analogous system to marine snow when comparing the fractal dimension (52, 53), show substantial interstitial flow despite being predominantly filled with bacterial exopolymers (54). Due to the lack of consensus on the presence of advective interstitial flow in marine particles, we study particles with a wide range of different porosities (Fig. 4), including their rough and smooth counterparts (Supplementary Fig. S6). Resolving the pore size distribution in marine particles is paramount since it fundamentally changes the mass transport within and around the particles. In addition, the permeation of marine snow profoundly changes the way these settling particles interact and scavenge smaller particles in their path. Scavenging efficiencies are 100 times higher for porous particles compared to their smooth counterparts, even when deflecting most of the flow around the particles (55). Interestingly, most of this scavenging is attributed to a thin boundary layer just interior to the particle surface. This is critical since the process of scavenging is therefore not dependent on total perfusion of the particle (55) and can still occur if the center of the particle restricts advective flows due to, for example, clogging of the interstitial pore space by transparent exopolymers (TEP) (56). These theoretical considerations are in good agreement with our experimental observations, where a higher particle porosity facilitates the colonization by all three strains (Fig. 5c). In summary, the permeability of marine snow (even if restricted to the surface) can play a principal role in shaping the colonization by chemotactic and motile cells (by reducing the total deflection of flow around the particles and reducing the velocity gradients), but importantly permits some colonization by nonmotile cells by allowing stream lines to intercept with the particle surface and thus passively increases the encounter between nonmotile cells and marine snow—a process that is nigh impossible in the absence of any porosity. To the best of our knowledge, microfluidic devices that trap porous marine snow but permit the addition of a constant chemoattractant source to systematically study their colonization by bacteria with different motility traits currently do not exist. Recent experimental observations using particle image velocimetry showed similar small-scale alterations of the flow field around particles of heterogeneous shapes when compared to spherical particles (49). Combined with our results, these observations suggest that even in the absence of any particle porosity that supports interstitial advective flows, the microscale heterogeneity of the particles can fundamentally alter the mechanisms of colonization for bacteria with different motility traits.
The ability of bacterial cells to exploit the nutrient-rich microenvironments presented by marine snow is a key process in global biogeochemistry. A recent study highlighted the importance of flow for the degradation dynamics of marine particulate organic matter (57). In this study, we look at the encounter and attachment of bacteria to marine particles (the sum of these processes describing successful colonization) that represent the first step of particle remineralization, and a mechanistic understanding of these processes is thus crucial to advance our predictive capabilities (58). Our study confirms that motility is the key trait that enables particle colonization, but further provides insights how chemotaxis facilitates particle colonization during the brief window of opportunity by enabling to rapidly navigate the particle boundary layer. We further highlight how the colonization by nonmotile cells is facilitated by the porous microstructure of marine snow and associated interstitial advective flows. Disentangling the interplay between the preferential colonization by motile cells guided by chemotaxis and the benefit of particle plumes for nonmotile oligotrophic bacteria in the planktonic community across the global oceans warrants continued study.
Methods
Bacterial strains and culture conditions
We performed the experiments with a motile and chemotactic wild-type P.aeruginosa PA14, a chemotaxis deficient mutant P. aeruginosa PA14 cheZ::TnM [deficient in dephosphorylation of the CheY protein, which transmits sensory signals to the flagella motors (59)], and a motility deficient mutant P. aeruginosa PA14 ∆fliC [deficient in production of flagellin, the primary component of the flagellum (60)]. Although we recognize that P.aeruginosa is not an archetypical marine species, it is found in many marine ecosystems (32–35) and represents a model organism for studying chemosensory responses and motile behavior of bacteria with a single, polar flagellum (36). We explore the relation between classical run-and-tumble motility and run-and-reverse or run-and-flick strategies that are prevailing in the marine environment (44) using the individual-based model. We confirmed the fluorescence and phenotype of the mutants using fluorescence microscopy and motility/chemotaxis assays (Supplementary Fig. S1). All strains were routinely kept in 25% glycerol at −80°C and grown in Lennox lysogeny broth (5 g L−1 NaCl, BD Life Sciences, further abbreviated LB) medium or on 1.5% agarose LB plates. All strains were tagged with the same yellow fluorescent protein (pUC18T-mini-Tn7T-Tp-eyfp, Addgene plasmid #65035) following the mini-Tn7 insertion protocol (61). After conjugation, 300 µg mL−1 trimethoprim (Trp) was used for routine growth and maintenance of all strains to ensure pure cultures.
Measurement of fluorescence and bacterial density in liquid culture
We used a Tecan Spark plate-reading fluorometer and spectrophotometer (Tecan Trading AG, Switzerland) to verify the monotonic and increasing relationship between the optical density of a liquid bacterial culture at 600 nm (further referred to as OD600) and their fluorescent intensity for all three strains (Supplementary Fig. S2a). We grew overnight cultures of the three strains in LB, after which we diluted each culture to an OD600 of 1. We then created a serial dilution (four technical replicates, five dilution steps) by diluting 20% of the culture in 80% fresh LB medium. We transferred 250 µL of the final diluted cultures into a 96-well plate and added two rows of controls containing sterile LB medium. We subsequently measured the absorbance at 600 nm and fluorescent intensity at 513 nm excitation and 530 nm emission wavelengths characteristic of eYFP for all wells.
Fabrication of millifluidic devices
We fabricated the millifluidic devices using Poly(dimethylsiloxane) (PDMS) and 3D printed molds. All 3D devices were printed using a Form3 stereolithography printer (Formlabs, Somerville, MA, USA) and clear resin (RS-F2-GPCL-04) at 25 µm resolution with Form Wash (20 min) and Form Cure (1 h at 60°C) for post processing. We exposed the mixed PDMS (Polydimethylsiloxane, DOW, CAS# 68083–19–2, 10:1 ratio of base to curing agent) to vacuum for 30 min at room temperature before pouring it into the 3D-printed molds. We subsequently exposed the molds a second time to vacuum for 30 min at room temperature before curing at 70°C for 24 h.
Colonization experiment using the leaky particle device
We grew the bacterial strains overnight in culture tubes containing 5 mL of LB medium (37°C, 220 rpm). We then diluted 200 µL of the overnight cultures with 20 mL fresh LB medium and regrew the cultures (37°C, 220 rpm) for 3 h into the exponential phase (typically reaching an OD600 between 0.5 and 1). We further diluted the cells to an OD600 of 0.1 and washed the cells twice by carefully centrifuging the cells at 1,000 × g for 10 min and subsequently resuspending the cells in a saline medium void of nutrients (20) (further called motility medium, containing 10 mM potassium phosphate monobasic (Sigma–Aldrich, CAS# 7778–77–0), 100 µM EDTA (Sigma–Aldrich, CAS# 6381–92–6), 10 mM NaCl (Sigma–Aldrich, CAS# 7647–14–5) and adjusted to pH 7.5 using 1 M NaOH) by gently shaking for 5 min on an orbital shaker at 100 rpm. We then added 1 mL of this culture to 10 mL of motility medium, resulting in ∼ 500,000 cells per mL, and transferred the final bacterial solution into a sterile syringe (Beckton–Dickinson, 10 mL slip tip). We verified that this procedure does not affect swimming capacity or shear off flagella.
We prepared synthetic marine particles using low-melting agarose as previously described by Smriga et al. 2021 (62). In short, we dissolved 15 mg of low melting agarose (Agarose low gelling temperature, Sigma–Aldrich, CAS# 39346–81–1) in 1 mL of motility medium that contained 10 µM each of L-serine and L-aspartate in a block heater at 80°C. After cooling for 3 min, we cast a 2 mm-thick slab of agarose using a 3 mL syringe (BD, 3 mL Luer lock) and a 22 G blunt needle (Industrial Dispensing Supplies) in between two microscope slides (VWR, Vistavision Microscope Slide, 75 x 50 x 1 mm) separated by 2 mm using additional glass slides. After solidifying at room temperature for 15 min, we punched the agarose discs from the agarose slab using 3 mm-diameter biopsy punches (Integra Miltex, Disposable Biopsy Punch, 3 mm), which we transferred into the experimental device previously filled with motility medium containing the two chemoattractant amino acids in the reservoirs (L-serine and L-aspartate). We verified that the bacterial cells are unable to grow in the motility medium when supplemented with the two amino acids (Supplementary Fig. S2b), confirming that the observed increase in fluorescent intensity is due to bacterial attachment alone and not growth during the experiment.
Fluorescent microscopy and image analysis
We used a Nikon Ti2-E inverted microscope equipped with an Andor Zyla 4.2 sCMOS camera and a SOLA Seii 365 Light Engine to visualize the colonized particles. The microscope was controlled using Nikon Elements (v.4). All images were captured in 16-bit with a 10x Plan Fluor DLL objective and an eYFP filter set for visualizing the bacterial cells (Chroma 49003, ex 500/20 nm, and em 535/30 nm). Each particle was visualized at 5 Z levels with 50 µm between the layers and the center located approximately at the middle of the channel (i.e., ∼500 µm from the bottom glass slide, which we located using the perfect focus system of the Nikon microscope). Each Z layer was captured as a 4 × 4 tile scan with a 20% overlap that was automatically stitched by the Nikon Elements software using the built-in blending algorithm. Individual images of the tile scans were captured at 50% illumination with a shutter speed of 800 ms for all particles and strains to ensure equal illumination of each particle and thus reliably quantify the colonized population. These settings resulted in an optimal dynamic range within the images for all strains and conditions tested and thus require the least amount of postprocessing.
All image analysis was performed in MATLAB R2021a (The Mathworks, Natick, MA, USA) using ND2Reader (63) to import the ND2 files produced by Nikon Elements. For each particle, we only considered an area ±250 µm around the particle edge for quantification. Fluorescence intensities in Fig. 2b are reported as mean pixel values across the whole area surrounding the particles without any further postprocessing to minimize the potential bias from image postprocessing.
Individual-based model of bacterial cells colonizing marine particles
We developed a mathematical framework to validate experimental results and gain further insights by combining computational fluid dynamics with reaction-diffusion modeling and an agent-based representation of motile bacterial cells. We used the FEATool Multiphysics software 1.15.2 (Precise Simulation Ltd.) in MATLAB to predict the flow and nutrient field around the marine particles and a custom algorithm to simulate nonmotile, motile, and chemotactic cells. Here, we first describe the individual-based representation of bacterial cells (which is identical for all geometries) and subsequently the specific geometries used for the different studies. The Navier–Stokes equations in combination with reaction-diffusion modeling were solved on a triangular mesh with a nonuniform resolution of 100 µm increasing to 10 µm at any boundary of the simulation for the experimental twin scenario, and 1 mm increasing to 10 µm for the large rectangular domain. Example geometries and grid meshes are shown in Supplementary Fig. S3. A list of model parameters and associated references for the standard conditions is shown in Supplementary Table S4.
Numerical simulation of bacterial motility
For all conditions and each replicate, we inoculate bacterial cells 6.5 mm upstream of the particle equator (i.e., 5 mm upstream of the leading edge of a 3 mm particle) uniformly random over a width of 4 mm. Each replicate simulates 100,000 cells of a single strain which results in an approximate inoculation density of 25 cells µm−1 across the channel. The model includes three modes of bacterial cell displacement: advective displacement by the fluid motion, active self-propulsion using a run-and-tumble mechanism, and random displacement due to Brownian motion. The displacement vector for each individual bacterial cell is determined by the local fluid flow field multiplied by the time step. All bacterial cells are additionally displaced by Brownian motion where the displacement vector is determined by the characteristic distance of diffusion for a particle of 1 µm diameter with the diffusion coefficient calculated using the Stokes–Einstein equation (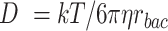 , with k the Boltzmann constant, T the temperature,
, with k the Boltzmann constant, T the temperature, 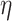 the dynamic viscosity and rbac the bacterial cell radius). At 20°C and a bacterial radius of 0.5 µm, each cell is displaced in a random direction by 92 nm in each numerical time step of 0.01 s. When simulating motile cells, bacterial cells additionally use self-propulsion at a constant velocity in a fixed direction. Bacterial cells choose a new direction at random with a probability of
the dynamic viscosity and rbac the bacterial cell radius). At 20°C and a bacterial radius of 0.5 µm, each cell is displaced in a random direction by 92 nm in each numerical time step of 0.01 s. When simulating motile cells, bacterial cells additionally use self-propulsion at a constant velocity in a fixed direction. Bacterial cells choose a new direction at random with a probability of 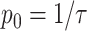 where p0 is the average tumbling probability per unit time [-] and
where p0 is the average tumbling probability per unit time [-] and 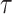 the average run length of the bacterial cells [s]. The total displacement of bacterial cells in one time step is then the sum of all displacement vectors. Chemotactic cells have the ability to reduce their tumbling frequency as a function of positive nutrient gradients (64), a feature that we capture in the model using the following Eq. 1:
the average run length of the bacterial cells [s]. The total displacement of bacterial cells in one time step is then the sum of all displacement vectors. Chemotactic cells have the ability to reduce their tumbling frequency as a function of positive nutrient gradients (64), a feature that we capture in the model using the following Eq. 1:
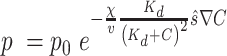 |
(1) |
Where p is the chemotactically adjusted tumbling probability [-], p0 the tumbling probability associated with the average run length 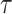 [-],
[-], 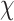 the chemotactic sensitivity [m2 s−1],
the chemotactic sensitivity [m2 s−1], 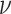 the bacterial swimming velocity [m s−1], Kd the receptor/ligand dissociation equilibrium constant [mol m−3], C the nutrient concentration [mol m−3],
the bacterial swimming velocity [m s−1], Kd the receptor/ligand dissociation equilibrium constant [mol m−3], C the nutrient concentration [mol m−3], 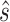 the normalized vector of swimming direction [m] and
the normalized vector of swimming direction [m] and 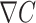 the nutrient gradient [mol m−4]. For positive nutrient gradients, Eq. 1 describes an exponential decay of the tumbling probability. When experiencing a negative nutrient gradient (i.e., the cell is swimming away from the particle or the plume), we set the tumbling probability to p0 (64, 65). The nutrient gradient is calculated based on the experienced change in nutrient concentrations over the past 0.3 s (66), where the current nutrient concentration is taken from the nutrient concentration at the bacterial location. If the cell tumbles, a new direction is chosen at random. Finally, if cells collide with the in silico marine particle, we assume a “perfect sticking” condition (i.e., if the bacterial trajectory intersects with the particle boundary within a time step, we assume an irreversible attachment). We selected a time step of 10 ms, which equates to a maximum advective travelling distance of 14 µm per time step at the highest settling velocity (which is below the smallest grid meshing and thus restricts numerical inaccuracies from the advection of cells). We set the total simulation time such that individual bacteria are advected a total of 60 mm even for the lowest flow of 1 m d−1 settling velocity.
the nutrient gradient [mol m−4]. For positive nutrient gradients, Eq. 1 describes an exponential decay of the tumbling probability. When experiencing a negative nutrient gradient (i.e., the cell is swimming away from the particle or the plume), we set the tumbling probability to p0 (64, 65). The nutrient gradient is calculated based on the experienced change in nutrient concentrations over the past 0.3 s (66), where the current nutrient concentration is taken from the nutrient concentration at the bacterial location. If the cell tumbles, a new direction is chosen at random. Finally, if cells collide with the in silico marine particle, we assume a “perfect sticking” condition (i.e., if the bacterial trajectory intersects with the particle boundary within a time step, we assume an irreversible attachment). We selected a time step of 10 ms, which equates to a maximum advective travelling distance of 14 µm per time step at the highest settling velocity (which is below the smallest grid meshing and thus restricts numerical inaccuracies from the advection of cells). We set the total simulation time such that individual bacteria are advected a total of 60 mm even for the lowest flow of 1 m d−1 settling velocity.
Experimental twin and no-boundary geometries
We mimic the experimental system using a digital twin approach. The domain has a total length and width of 20 mm and 5 mm, respectively, where a 3 mm particle is embedded in the center of an 8 mm spherical expansion in the center of the system to ensure the same channel width for the flow around the particle (to minimize flow acceleration and deceleration around the particle). Flow velocity is set as a parabolic profile at the inlet 10 mm upstream of the particle equator with a maximum flow velocity equal to the experimental condition (1, 10, and 100 m d−1). Due to the narrow channel (5 mm), bacterial cells are reflected by the boundary of the domain in case of collision whilst assuming a “perfect sticking” condition upon collision with the particle. We set a constant relative concentration boundary at the particle surface (C = 1) with a diffusion coefficient of 10−10 m2 s−1 that is representative of amino acids (67). We furthermore create a large rectangular domain (50 × 50 mm) with a 3 mm particle at the center to investigate the influence of the constrained geometry in the experimental twin condition on the local flow field around the particle, the associated nutrient plume, and colonization probability, and better link our experiments to the real ocean.
Porous particle geometry
Finally, we numerically simulate a cohort of 500 porous particles, including their rough and smooth alterations to strategically investigate the influence of porosity, roughness, and size on the colonization efficiency. Each particle is assigned a particle size at random between 100 µm and 3 mm and a target porosity between 0 and 0.7 (uniform distribution). We calculate a target number of subparticles that form the aggregate using an average subparticle diameter of 60 µm. We then create a stochastic geometry for each particle by choosing a location within the particle at random with a diameter between 20 and 100 µm for each of the subparticles. Sub-particles are allowed to overlap within the particles to generate a spectrum of shapes and sizes of subaggregates. The rough alteration of each particle includes a solid circular core with a diameter equal to the outermost subparticle center, and the smooth particle solely being this solid core (without any roughness). We calculate the resulting particle porosity using the particle surface area and total particle area. Each particle is located in a 5 by 5 cm domain where the settling velocity is calculated based on Eq. 2 (68):
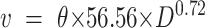 |
(2) |
With v the settling velocity [m d−1], 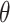 the particle porosity [-] and ESD the particle diameter [mm]. We set a constant relative nutrient concentration of 1 for each subparticle boundary.
the particle porosity [-] and ESD the particle diameter [mm]. We set a constant relative nutrient concentration of 1 for each subparticle boundary.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the reviewers and editors for their constructive comments improving the paper. We are also thankful to Dr. Bruce Heflinger for his generous support of this work.
Notes
Competing Interest: The authors declare no competing interests.
Contributor Information
Benedict Borer, Department of Earth, Atmospheric, and Planetary Sciences, Massachusetts Institute of Technology Cambridge, Cambridge, MA 02139, USA.
Irene H Zhang, Department of Earth, Atmospheric, and Planetary Sciences, Massachusetts Institute of Technology Cambridge, Cambridge, MA 02139, USA.
Amy E Baker, Department of Microbiology and Immunology, Geisel School of Medicine at Dartmouth, Hanover, NH 03755, USA.
George A O'Toole, Department of Microbiology and Immunology, Geisel School of Medicine at Dartmouth, Hanover, NH 03755, USA.
Andrew R Babbin, Department of Earth, Atmospheric, and Planetary Sciences, Massachusetts Institute of Technology Cambridge, Cambridge, MA 02139, USA.
Funding
This project was funded by a Swiss National Science Foundation Grant (P500PN_202842) awarded to BB, NIH Grant (R37_AI83256) awarded to GAO, and a Simons Foundation Grant (#622065) and NSF Grant (OCE-2142998) awarded to ARB.
Authors' Contributions
B.B. and A.R.B. developed the research question and designed the experiments. A.E.B., I.Z., and B.B. modified the bacterial strains. B.B. performed the experiments, developed the model, and analyzed the data. B.B., I.Z., G.A.O., and A.R.B. contributed to the writing of the paper, and all authors reviewed and approved the final version of the paper.
Preprints
This manuscript was posted as a preprint: https://doi.org/10.1101/2022.05.13.491378.
Data Availability
All modeling data are available on https://doi.org/10.5281/zenodo.7267650.
References
- 1. Giovannoni SJ, et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 309:1242–1245. [DOI] [PubMed] [Google Scholar]
- 2. Keil RG, Kirchman DL. 1999. Utilization of dissolved protein and amino acids in the northern Sargasso Sea. Aquat Microb Ecol. 18:293–300. [Google Scholar]
- 3. Hense I, Stemmler I, Sonntag S. 2017. Ideas and perspectives: climate-relevant marine biologically driven mechanisms in Earth system models. Biogeosciences. 14:403–413. [Google Scholar]
- 4. Kiørboe T. 2001. Formation and fate of marine snow: small-scale processes with large-scale implications. Sci Mar. 65:57–71. [Google Scholar]
- 5. Simon M, Grossart H-P, Schweitzer B, Ploug H. 2002. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol. 28:175–211. [Google Scholar]
- 6. Mari X, Passow U, Migon C, Burd AB, Legendre L. 2017. Transparent exopolymer particles: effects on carbon cycling in the ocean. Prog Oceanogr. 151:13–37. [Google Scholar]
- 7. Le Moigne FAC. 2019. Pathways of organic carbon downward transport by the oceanic biological carbon pump. Front Mar Sci. 6:634. [Google Scholar]
- 8. Rieck A, Herlemann DPR, Jürgens K, Grossart H-P. 2015. Particle-associated differ from free-living bacteria in surface waters of the Baltic Sea. Front Microbiol. 6:1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zäncker B, Engel A, Cunliffe M. 2019. Bacterial communities associated with individual transparent exopolymer particles (TEP). J Plankton Res. 41:561–565. [Google Scholar]
- 10. Polz MF, Hunt DE, Preheim SP, Weinreich DM. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos Trans R Soc Lond B Biol Sci. 361:2009–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lauro FM, et al. 2009. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci. 106: 15527–15533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yooseph S, et al. 2010. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 468:60–66. [DOI] [PubMed] [Google Scholar]
- 13. Flintrop CM, et al. 2018. Embedding and slicing of intact in situ collected marine snow. Limnol Oceanogr Methods. 16:339–355. [Google Scholar]
- 14. Laurenceau-Cornec EC, et al. 2020. New guidelines for the application of Stokes’ models to the sinking velocity of marine aggregates. Limnol Oceanogr. 65:1264–1285. [Google Scholar]
- 15. Bach LT, et al. 2012. An approach for particle sinking velocity measurements in the 3–400 μm size range and considerations on the effect of temperature on sinking rates. Mar Biol. 159:1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grognot M, Taute KM. 2021. More than propellers: how flagella shape bacterial motility behaviors. Curr Opin Microbiol. 61:73–81. [DOI] [PubMed] [Google Scholar]
- 17. Kiørboe T, Grossart H-P, Ploug H, Tang K. 2002. Mechanisms and rates of bacterial colonization of sinking aggregates. Appl Environ Microbiol. 68:3996–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kiørboe T, Jackson GA. 2001. Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol Oceanogr. 46:1309–1318. [Google Scholar]
- 19. Brumley DR, et al. 2020. Cutting through the noise: bacterial chemotaxis in marine microenvironments. Front Mar Sci. 7:527. [Google Scholar]
- 20. Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. 2008. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci. 105:4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Secchi E, et al. 2020. The effect of flow on swimming bacteria controls the initial colonization of curved surfaces. Nat Commun. 11:2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Słomka J, Alcolombri U, Secchi E, Stocker R, Fernandez VI. 2020. Encounter rates between bacteria and small sinking particles. New J Phys. 22:043016. [Google Scholar]
- 23. Yawata Y, Carrara F, Menolascina F, Stocker R. 2020. Constrained optimal foraging by marine bacterioplankton on particulate organic matter. Proc Natl Acad Sci. 117:25571–25579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blackburn N, Fenchel T, Mitchell J. 1998. Microscale nutrient patches in planktonic habitats shown by chemotactic bacteria. Science. 282:2254–2256. [DOI] [PubMed] [Google Scholar]
- 25. Smriga S, Fernandez VI, Mitchell JG, Stocker R. 2016. Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc Natl Acad Sci. 113:1576–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lambert BS, et al. 2017. A microfluidics-based in situ chemotaxis assay to study the behaviour of aquatic microbial communities. Nat Microbiol. 2: 1344–1349. [DOI] [PubMed] [Google Scholar]
- 27. Raina J-B, et al. 2022. Chemotaxis shapes the microscale organization of the ocean's microbiome. Nature. 605: 132–138. [DOI] [PubMed] [Google Scholar]
- 28. Guidi L, et al. 2008. Relationship between particle size distribution and flux in the mesopelagic zone. Deep Sea Res Part I. 55: 1364–1374. [Google Scholar]
- 29. Enke TN, et al. 2019. Modular assembly of polysaccharide-degrading marine microbial communities. Curr Biol.29: 1528–1535.e6. [DOI] [PubMed] [Google Scholar]
- 30. Ebrahimi A, Schwartzman J, Cordero OX. 2019. Cooperation and spatial self-organization determine rate and efficiency of particulate organic matter degradation in marine bacteria. Proc Nat Acad Sci USA. 116:23309–23316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baumas CMJ, et al. 2021. Mesopelagic microbial carbon production correlates with diversity across different marine particle fractions. ISME J. 15:1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khan NH, et al. 2007. Isolation of Pseudomonas aeruginosa from open ocean and comparison with freshwater, clinical, and animal isolates. Microb Ecol. 53:173–186. [DOI] [PubMed] [Google Scholar]
- 33. Khan NH, et al. 2008. Multilocus sequence typing and phylogenetic analyses of Pseudomonas aeruginosa isolates from the ocean. Appl Environ Microbiol.74:6194–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kimata N, Nishino T, Suzuki S, Kogure K. 2004. Pseudomonas aeruginosa isolated from marine environments in Tokyo Bay. Microb Ecol. 47:41–47. [DOI] [PubMed] [Google Scholar]
- 35. Jayakumar DA, Francis CA, Naqvi SWA, Ward BB. 2004. Diversity of nitrite reductase genes (nirS) in the denitrifying water column of the coastal Arabian Sea. Aquat Microb Ecol. 34:69–78. [Google Scholar]
- 36. Wadhwa N, Berg HC. 2022. Bacterial motility: machinery and mechanisms. Nat Rev Microbiol.20:161–173. [DOI] [PubMed] [Google Scholar]
- 37. Leifson E, Cosenza BJ, Murchelano R, Cleverdon RC. 1964. MOTILE MARINE BACTERIA I. J Bacteriol. 87:652–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Conrad JC, et al. 2011. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa.Biophys J. 100:1608–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barbara GM, Mitchell JG. 2003. Bacterial tracking of motile algae. FEMS Microbiol Ecol. 44:79–87. [DOI] [PubMed] [Google Scholar]
- 40. Blackburn N, Fenchel T. 1999. Influence of bacteria, diffusion and shear on micro-scale nutrient patches, and implications for bacterial chemotaxis. Mar Ecol Prog Ser. 189:1–7. [Google Scholar]
- 41. Tian M, Wu Z, Zhang R, Yuan J. 2022. A new mode of swimming in singly flagellated Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 119:e2120508119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xie L, Altindal T, Chattopadhyay S, Wu X-L. 2011. Bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc Natl Acad Sci. 108:2246–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Son K, Guasto JS, Stocker R. 2013. Bacteria can exploit a flagellar buckling instability to change direction. Nat Phys. 9:494–498. [Google Scholar]
- 44. Son K, Menolascina F, Stocker R. 2016. Speed-dependent chemotactic precision in marine bacteria. Proc Natl Acad Sci. 113:8624–8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee CK, et al. 2020. Social cooperativity of bacteria during reversible surface attachment in young biofilms: a quantitative comparison of Pseudomonas aeruginosa PA14 and PAO1. mBio. 11:e02644–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. El-Kirat-Chatel S, et al. 2017. Phenotypic heterogeneity in attachment of marine bacteria toward antifouling copolymers unraveled by AFM. Front Microbiol. 8:1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trudnowska E, et al. 2021. Marine snow morphology illuminates the evolution of phytoplankton blooms and determines their subsequent vertical export. Nat Commun. 12:2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Durkin CA, et al. 2022. Tracing the path of carbon export in the ocean though DNA sequencing of individual sinking particles. ISME J. 16:1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zetsche E-M, Larsson AI, Iversen MH, Ploug H. 2020. Flow and diffusion around and within diatom aggregates: effects of aggregate composition and shape. Limnol Oceanogr. 65:1818–1833. [Google Scholar]
- 50. Ploug H, Passow U. 2007. Direct measurement of diffusivity within diatom aggregates containing transparent exopolymer particles. Limnol Oceanogr. 52:1–6. [Google Scholar]
- 51. Ploug H, Hietanen S, Kuparinen J. 2002. Diffusion and advection within and around sinking, porous diatom aggregates. Limnol Oceanogr. 47:1129–1136. [Google Scholar]
- 52. Logan BE, Wilkinson DB. 1990. Fractal geometry of marine snow and other biological aggregates. Limnol Oceanogr. 35:130–136. [Google Scholar]
- 53. Kilps JR, Logan BE, Alldredge AL. 1994. Fractal dimensions of marine snow determined from image analysis of in situ photographs. Deep Sea Res Part I. 41:1159–1169. [Google Scholar]
- 54. Li XY, Yuan Y, Wang HW. 2003. Hydrodynamics of biological aggregates of different sludge ages: an insight into the mass transport mechanisms of bioaggregates. Environ Sci Technol. 37:292–299. [DOI] [PubMed] [Google Scholar]
- 55. Stolzenbach KD. 1993. Scavenging of small particles by fast-sinking porous aggregates. Deep Sea Res Part I. 40: 359–369. [Google Scholar]
- 56. Passow U. 2002. Transparent exopolymer particles (TEP) in aquatic environments. Prog Oceanogr. 55:287–333. [Google Scholar]
- 57. Alcolombri U, et al. 2021. Sinking enhances the degradation of organic particles by marine bacteria. Nat Geosci. 14:775–780. [Google Scholar]
- 58. Nguyen TTH, et al. 2022. Microbes contribute to setting the ocean carbon flux by altering the fate of sinking particulates. Nat Commun. 13:1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liberati NT, et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A. 103:2833–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lewis KA, et al. 2022. Nonmotile subpopulations of Pseudomonas aeruginosa repress flagellar motility in motile cells through a type IV Pilus- and Pel-Dependent mechanism.J Bacteriol. 204:e0052821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Choi K-H, Schweizer HP. 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa.Nat Protoc. 1:153–161. [DOI] [PubMed] [Google Scholar]
- 62. Smriga S, Ciccarese D, Babbin AR. 2021. Denitrifying bacteria respond to and shape microscale gradients within particulate matrices. Commun Biol. 4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zuo J. 2023. ND2 Reader. (https://github.com/JacobZuo/nd2reader), Github. Retrieved February 2, 2023. [Google Scholar]
- 64. Berg HC, Brown DA. 1972. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 239:500–504. [DOI] [PubMed] [Google Scholar]
- 65. RIVERO MA, Tranquillo RT, Buettner HM, LAUFFENBURGER DA. 1989. Transport models for chemotactic based on individual. Chem Eng Sci. 44:2881–2897. [Google Scholar]
- 66. Emonet T, Macal CM, North MJ, Wickersham CE, Cluzel P. 2005. AgentCell: a digital single-cell assay for bacterial chemotaxis. Bioinformatics.21:2714–2721. [DOI] [PubMed] [Google Scholar]
- 67. Ma Y, Zhu C, Ma P, Yu KT. 2005. Studies on the diffusion coefficients of amino acids in aqueous solutions. J Chem Eng Data. 50:1192–1196. [Google Scholar]
- 68. Iversen MH, Ploug H. 2010. Ballast minerals and the sinking carbon flux in the ocean: carbon-specific respiration rates and sinking velocity of marine snow aggregates. Biogeosciences. 7:2613–2624. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All modeling data are available on https://doi.org/10.5281/zenodo.7267650.







