Abstract
Salmonellosis is a common foodborne zoonosis worldwide. The most common Salmonella serovar in humans is Salmonella enterica subsp. enterica serovar Enteritidis (50.3%) in the world. The main transmission route for S. Enteritidis is consumption of contaminated poultry products. Therefore, it is important to determine the diversity and spread of chicken-originated S. Enteritidis isolates in order to monitor and control salmonellosis. Pulsed-field gel electrophoresis (PFGE) and multiple locus variable number of tandem repeats analysis (MLVA) are frequently used for typing of S. Enteritidis isolates. This study aimed to determine the antimicrobial resistance (AMR) profiles and MLVA and PFGE genotypes of chicken-originated S. Enteritidis isolates. A total of 200 S. Enteritidis isolated from chicken broiler, layer, and breeder flocks from different locations in Turkey were investigated by Kirby–Bauer disk diffusion method, PFGE, and MLVA. The AMR test indicated that 57% of the S. Enteritidis isolates were susceptible to all antimicrobials, while 39% were resistant to at least one antimicrobial. The highest resistance (25%) was against ampicillin. Multi-drug resistance rate was low (21%) and mostly from broiler flocks (93%). All isolates were genotyped into 32 different PFGE genotypes (PT) and 34 different MLVA genotypes (MT). The dominant genotypes were PT6 (12.5%) and MT22 (50%). In specific sample groups, there was a correlation between genotypes, breeding type, geographic location, and isolation years of the isolates. There was no significant difference in the discrimination power of PFGE and MLVA. However, MLVA was more suitable for large sample groups and routine genotyping because it was easier, quicker, and less labor-intensive to use.
Keywords: AMR, Chicken, Genotype, MLVA, PFGE, Salmonella Enteritidis
Introduction
Salmonellosis is one of the leading foodborne infections worldwide [1]. The majority (99%) of human and animal salmonellosis infections are caused by S. enterica subsp. enterica. According to the EFSA 2019 zoonotic agents report, the most common Salmonella serovar in humans is Salmonella enterica subsp. enterica serovar Enteritidis (50.3%) [2]. Globally, S. Enteritidis is frequently reported as the most common cause of human salmonellosis [3]. The main transmission route for S. Enteritidis is consumption of contaminated eggs and poultry meat. Therefore, it is important to determine the phylogeny and epidemiology of chicken-originated S. Enteritidis isolates in order to understand the transmission routes and control salmonellosis [1, 4, 5].
Genotyping methods provide objective and reliable tools to determine the sources and transmission routes of S. Enteritidis outbreaks [6, 7]. The gold standard method, pulsed-field gel electrophoresis (PFGE), is frequently used for genotyping of S. Enteritidis [3]. However, PFGE is labor-intensive and time-consuming, so it is less suitable for genotyping large sample groups. In addition, it has limited discriminatory power for genotyping genetically homogeneous pathogens like S. Enteritidis because it is based on single endonucleotidase analysis. Discriminatory power also varies depending on the plasmids, transposons, and integrons in the bacterial DNA [3, 8]. Therefore, multiple locus variable number of tandem repeats analysis (MLVA) is usually recommended as an additional or an alternative genotyping method for determining the diversity of S. Enteritidis outbreaks. Some researchers have reported that MLVA provides stronger discrimination in genotyping of S. Enteritidis isolates that PFGE indicates to be highly clonal [6, 8]. Whole genome sequencing (WGS) is becoming a gold standard method for genotyping of Salmonella agents. In recent years, WGS has been widely used because of it provides rapid and effective discrimination between Salmonella isolates [7].
Antimicrobial resistance (AMR) is a concern for human and animal health globally. In recent years, the reported incidence and spread of antimicrobial resistant Salmonella have increased [9]. According to the World Health Organization’s (WHO) global report on surveillance of antimicrobial resistance, non-typhoidal multi-drug-resistant (MDR) Salmonella is considered a serious problem in many countries [10]. This microorganism which is transmitted to humans from contaminated food causes infections that are difficult to treat. Therefore, antimicrobial resistance monitoring of chicken-originated Salmonella is important for public and animal health [9, 10].
Accordingly, this study aimed to determine the AMR profiles and MLVA and PFGE genotypes of chicken-originated S. Enteritidis isolates. The epidemiological analysis was performed by evaluating the AMR, PFGE, and MLVA findings together with the breeding type, isolation year, and geographic location of the isolates. The study also aimed to evaluate the advantages and disadvantages of MLVA and PFGE for genotyping S. Enteritidis isolates.
Material and methods
Salmonella Enteritidis isolates and conventional serotyping
The S. Enteritidis isolates were derived from the culture collection at Ankara University, Faculty of Veterinary Medicine, Microbiology Department. A total of 200 isolates were used which were isolated from broiler, broiler breeder, layer, and layer breeder chicken flocks located in 16 different cities around Turkey, 2018–2020 (Table 1). S. Enteritidis ATCC13076 was used as the positive control strain. The isolates were stored in 20% glycerol at – 80 °C before being grown overnight at 37 °C in nutrient agar. All isolates were confirmed by conventional serotyping with commercial Salmonella polyvalent and monovalent O and H antisera (SSI, Denmark).
Table 1.
Distribution of S. Enteritidis isolates by breeding type and geographical location
| Region | City | Broiler | Broiler breeder | Layer | Layer breeder |
|---|---|---|---|---|---|
| Aegean | Afyon | 5 | |||
| Central Anatolia | Ankara | 1 | 7 | ||
| Aegean | Aydın | 1 | |||
| Marmara | Balıkesir | 53 | 1 | 1 | 1 |
| Marmara | Bilecik | 6 | |||
| Black Sea | Bolu | 12 | 1 | 1 | |
| Marmara | Bursa | 26 | 2 | ||
| Marmara | Çanakkale | 25 | 1 | ||
| Black Sea | Düzce | 2 | 1 | ||
| Aegean | İzmir | 18 | 1 | 1 | |
| Central Anatolia | Konya | 2 | |||
| Aegean | Kütahya | 2 | |||
| Aegean | Manisa | 11 | 1 | ||
| Marmara | Sakarya | 13 | 1 | ||
| Black Sea | Samsun | 2 | |||
| Aegean | Uşak | 1 | |||
| Total | 167 | 14 | 18 | 1 | |
Antimicrobial resistance test
The AMR test was performed using the Kirby–Bauer disk diffusion method on Mueller––Hinton agar [11] for the following 14 antimicrobials: ampicillin (AMP, 10 μg), gentamicin (CN, 10 μg), cefotaxime (CTX, 30 μg), cefoxitin (FOX, 30 μg), ceftazidime (CAZ, 30 μg), ceftriaxone (CRO, 30 μg), chloramphenicol (C, 30 μg), ciprofloxacin (CIP, 5 μg), meropenem (MEM, 10 μg), nalidixic acid (NA, 30 μg), sulfonamides (S, 300 μg), tetracycline (TE, 30 μg), trimethoprim (W, 5 μg), and trimethoprim-sulfamethoxazole (SXT, 25 μg) (Oxoid, UK). Escherichia coli ATCC25922 was used as the quality control strain. The isolates were classified as resistant, intermediate, or susceptible in accordance with Clinical and Laboratory Standard Institute (CLSI) guidelines [12]. MDR was defined as resistance to three or more drug classes [10].
Pulsed-field gel electrophoresis analysis
PFGE was performed according to the standard operating procedure for PulseNet PFGE of Salmonella Serotypes [13]. The restriction endonuclease XbaI (Thermo Scientific, USA) was used for the digestion of bacterial DNA, while separation was made using a contour-clamped homogeneous electric field (CHEF-DR III) apparatus (Bio-Rad, USA) in 1% agarose gel. Electrophoresis was performed as follows: pulse time 2.2–63.8 s, voltage 6 V, temperature 14 °C, and time 19 h. The PFGE patterns were clustered using GelCompar II software v.6.6 (Applied Maths, Belgium). Clustering was conducted using the unweighted pair group method with arithmetic averages (UPGMA) using the dice coefficient and a band position tolerance of 1.5%. PFGE genotypes (PT) were determined according to the Tenover criteria for bacterial strain typing [14].
Multiple locus variable number tandem repeat analysis
Bacterial DNA was extracted by boiling the bacterial suspensions at 100 °C for 10 min. MLVA was performed according to the standard operating procedure for PulseNet MLVA of S. Enteritidis [15]. Seven VNTR loci (SE1, SE2, SE3, SE5, SE6, SE8, and SE9) were amplified by multiplex PCR using specific fluorescent primers. The amplification was conducted by an initial denaturation at 95 °C for 5 min, 34 cycles of 94 °C for 20 s, 65 °C for 20 s, 72 °C for 20 s, and a final elongation at 72 °C for 5 min. The amplicons were analyzed by capillary electrophoresis using an ABI 3500 Genetic Analyzer (Applied Biosystems, USA). The allele types were assigned using GeneMapper software 5.0 (Applied Biosystems, USA). For each isolate, the MLVA genotypes (MT) were determined from the combination of VNTR loci alleles.
Results
Antimicrobial resistance test
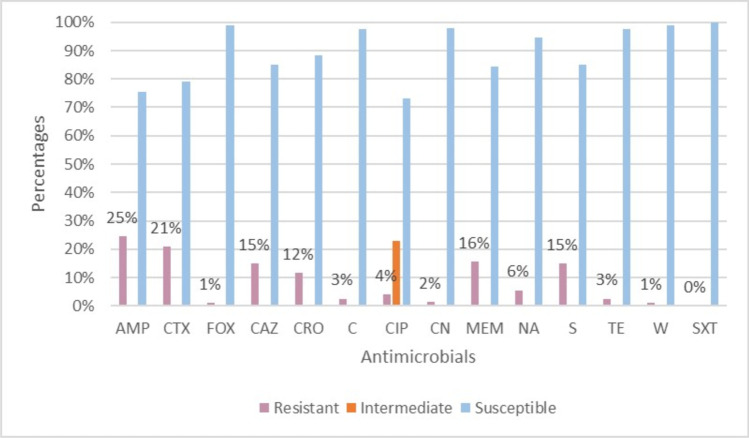
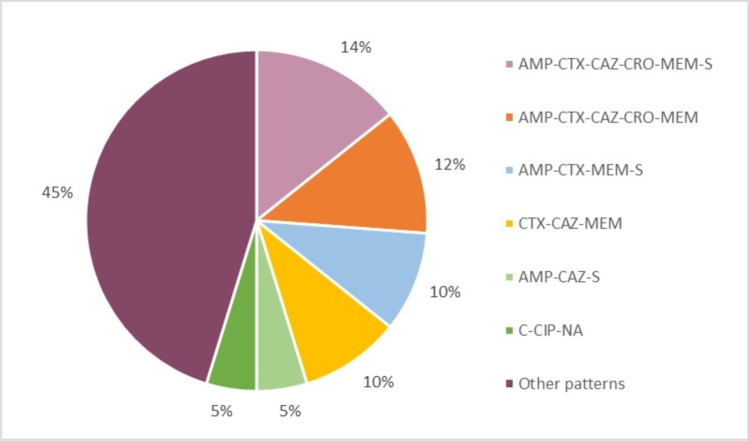
Among the S. Enteritidis isolates studied, 113 (57%, 113/200) were susceptible to all the tested antimicrobials, while 77 (39%, 77/200) were resistant to at least one antimicrobial. The highest resistance rate (25%, 49/200) was against AMP, while the highest intermediate resistance rate (23%, 46/200) was against CIP, and the highest susceptibility rate (100%, 200/200) was against SXT (Fig. 1). Forty two (21%, 42/200) of the S. Enteritidis isolates were MDR, of which most were of broiler origin (93%, 36/42). There were 25 different MDR patterns, of which the most frequent (14%, 6/42) was the AMP-CTX-CAZ-CRO-MEM-S hexa-resistant pattern, followed by AMP-CTX-CAZ-CRO-MEM (12%, 5/42), AMP-CTX-MEM-S (10%, 4/42), and CTX-CAZ-MEM (10%, 4/42) (Fig. 2). One isolate was resistant to nine antimicrobials, with an MDR pattern of AMP-CTX-CAZ-CRO-FOX-CIP-MEM-NA-S.
Fig. 1.
Percentage distribution of resistant, intermediate, and susceptible S. Enteritidis isolates. The percentages for resistant isolates are indicated by the numerical values
Fig. 2.
Percentage distribution of common MDR patterns in S. Enteritidis isolates
Pulsed-field gel electrophoresis analysis
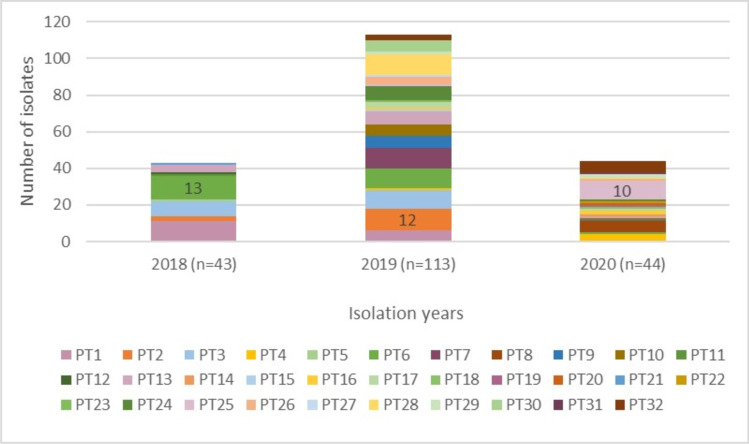
The S. Enteritidis isolates were grouped into XbaI-PFGE patterns with similarity indices ranging from 52.8 to 100% (Fig. 3). The isolates were divided into two major clusters, A and B. Cluster A was larger, with 57.5% (115/200) of the isolates. Analysis of the XbaI-PFGE patterns according to the Tenover criteria (1995) identified 32 different PFGE genotypes (PT1-PT32). The majority of genotypes were grouped in cluster B, with 20 different PTs (PT13-PT32). The isolates in cluster A showed less diversity in the number of PT (n = 12). The predominant PFGE genotype in all samples was PT6 (12.5%, 26/200). PT6 was also predominant in 2018 (n = 13), whereas PT2 was predominant in 2019 (n = 12) and PT25 in 2020 (n = 10) (Fig. 4).
Fig. 3.
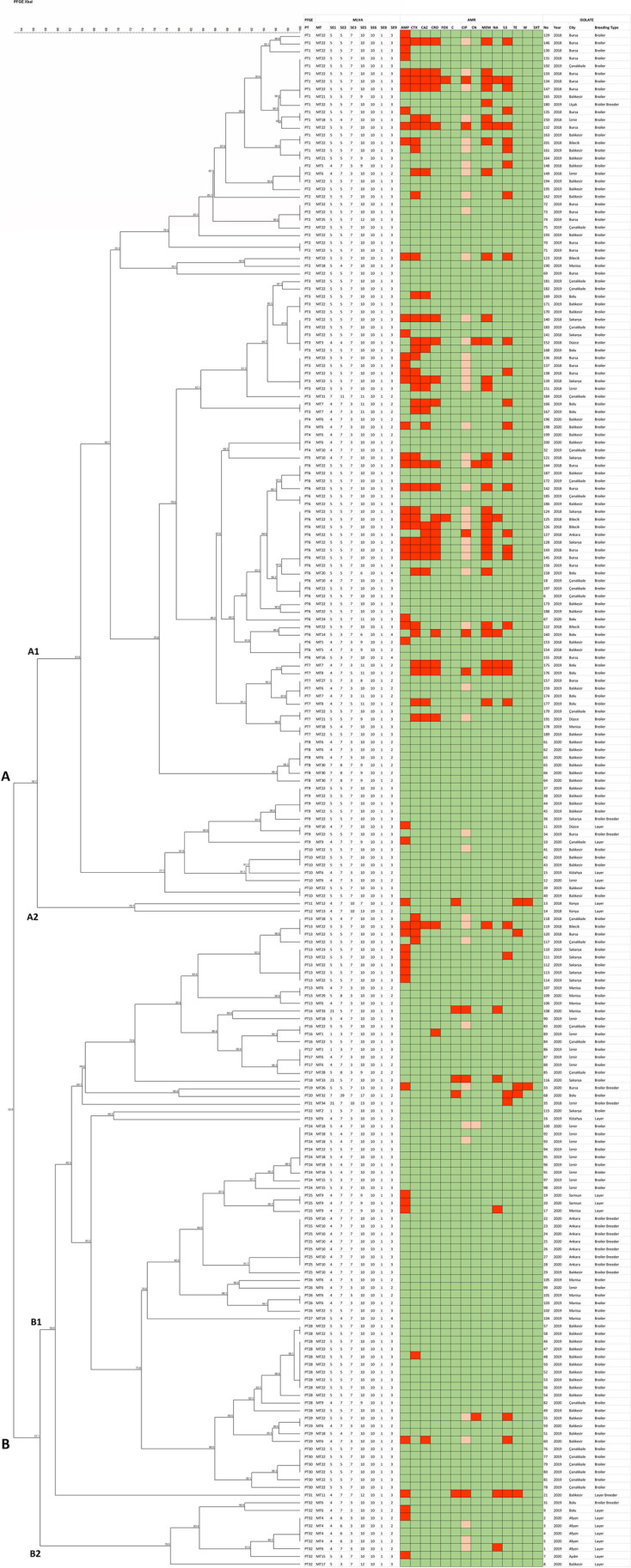
Dendrogram of AMR, PFGE, and MLVA results, and source details of S. Enteritidis strains. A red box represents high antimicrobial resistance; an orange box represents intermediate resistance; a green box represents susceptibility to antimicrobials
Fig. 4.
Distribution of S. Enteritidis PFGE genotypes by years
Multiple locus variable number tandem repeat analysis
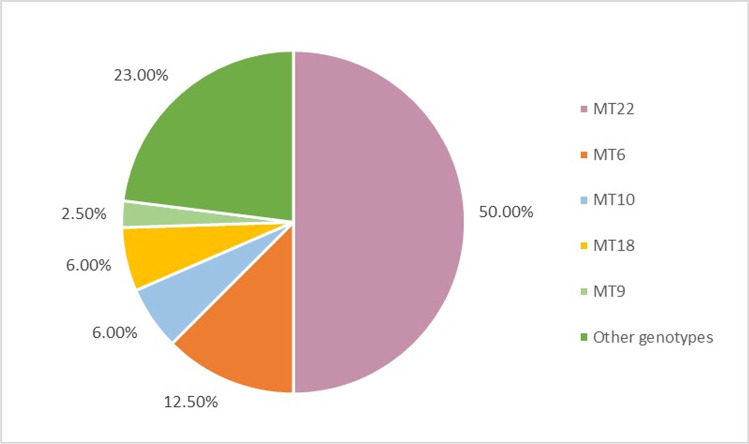
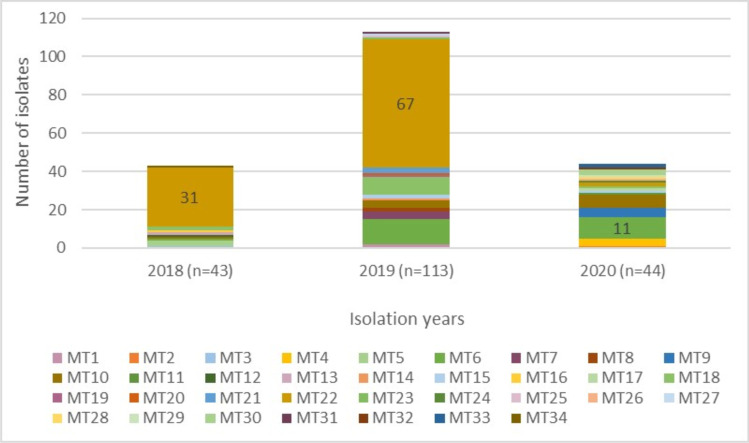
Thirty-four different MLVA genotypes (MT1-MT34) were detected (Fig. 3), of which 20 different MTs were represented by just one isolate. The predominant MT was MT22, with 50% (100/200) of all isolates, followed by MT6, with 12.5% (25/200) (Fig. 5). MT22 was the dominant MLVA genotype in 2018 and 2019 (n = 13 and n = 67, respectively), whereas it was MT6 in 2020 (n = 11) (Fig. 6). While the highest number of alleles was detected at SE5 locus, no diversity was observed at SE6 locus (Table 2).
Fig. 5.
Percentage distribution of common S. Enteritidis MLVA genotypes
Fig. 6.
Distribution of S. Enteritidis MLVA genotypes by years
Table 2.
Number of variants for seven VNTR loci in S. Enteritidis isolates
| VNTR locus | Repeat size (bp) | Number of variants |
|---|---|---|
| SE1 | 7 | 5 |
| SE2 | 7 | 8 |
| SE3 | 12 | 4 |
| SE5 | 6 | 9 |
| SE6 | 33 | 1 |
| SE8 | 86 | 2 |
| SE9 | 9 | 3 |
Discussion
S. Enteritidis is the most common Salmonella serovar causing foodborne outbreaks worldwide. The major source for S. Enteritidis infections is poultry products. The genotyping methods are frequently used to detect diversity and spread of S. Enteritidis isolates, such as PFGE and MLVA [3, 16]. In the present study, chicken-originated S. Enteritidis isolates were genotyped using MLVA and PFGE, while the AMR and MDR profiles were determined for all S. Enteritidis isolates. The AMR, MLVA, and PFGE results were compared in terms of the identified isolates’ breeding type, isolation year, and geographical location.
The spread of MDR Salmonella isolates causes failure in antimicrobial treatment of salmonellosis. MDR Salmonella is transmitted to humans via the food chain, especially from contaminated poultry products. Hence, the WHO includes Salmonella for monitoring on its priority pathogen list [16–18]. In this study, 57% of S. Enteritidis isolates were pan-susceptible to antimicrobials, while 77 isolates (39%) were resistant to at least one antimicrobial. This resistance rate (39%) was lower than those reported in studies from Thailand (86.8%), Iran (97.7%), and China (100%) [3, 16, 19].
The S. Enteritidis isolates in this study were most frequently resistant to AMP (25%). Previous studies in Turkey have reported varying rates. Kahraman et al. [20] reported 12.5% AMP resistance in Salmonella isolates, while the National Salmonella Control Program (NSCP) [21] found 12.3% average AMP resistance across all S. Enteritidis isolates, but 10% resistance in broiler isolates and 33.3% in layer isolates specifically. In the present study, AMP resistance rates in broiler and layer isolates were 23% and 50%, respectively. Thus, our findings are compatible with those from the NSCP in detecting higher AMP resistance in layer than broiler isolates. Considered together, our results indicate that both average AMP resistance and AMP resistance based on breeding types increased between 2018 and 2020. This may lead to failure in treatment of S. Enteritidis infections.
The present study also detected resistance to CTX (21%), CAZ (15%), and CRO (12%), whereas neither the NSCP [21] nor Guran et al. [22] reported resistance to cephalosporins (e.g., CTX, CAZ, and CRO) in chicken-originated Salmonella isolates in Turkey. However, our findings are compatible with Wei et al. [3], who reported “increased resistance to cephalosporins” in China. Therefore, monitoring and further studies focused on cephalosporin group antibiotics are needed.
The frequency of MDR S. Enteritidis detected in our study (21%) was lower than that in previous studies from South Korea (52.8%), Iran (68.2%), and China (81%) [3, 9, 16]. This may be because of the comprehensive NSCP strategies implemented to control S. Enteritidis in Turkey. In our study, AMP was the common antimicrobial in the four most frequent MDR patterns, which is compatible with Utrarachkij et al. [19].
PFGE is the current gold standard method for genotyping S. Enteritidis strains in outbreaks [3]. However, its ability to discriminate between genetically homogenous S. Enteritidis strains is limited [23]. Therefore, MLVA is commonly used as an additional or alternative genotyping method due to its greater discriminatory power for S. Enteritidis strains [8]. The strains in the present study fell into 32 different PT and 34 different MT groups, with no significant difference between the discrimination power of PFGE and MLVA. Mezal et al. [1] found 41 strains in the most common PFGE pattern by genotyping poultry and clinically originated S. Enteritidis strains with MLVA and PFGE, whereas they detected 14 different MLVA genotypes. They therefore concluded that PFGE has limited discriminatory power, so MLVA is more useful in genotyping of clinical and poultry-originated strains. Liu et al. [24] genotyped clinical-originated S. Enteritidis strains with MLVA and PFGE, and detected 29 different PFGE genotypes and 33 different MLVA genotypes, with no significant difference in the discrimination powers of MLVA and PFGE. However, their MLVA indicated that two different outbreaks had the same origin and were closely related epidemiologically.
Our results for AMR, PFGE, and MLVA were evaluated together using an XbaI-PFGE dendrogram. Firstly, clusters A and B were examined in terms of the strains’ breeding types. Broiler strains constituted 92% of the strains in cluster A, while cluster B included 37% of the broiler strains, 67% of the layer strains, and one laying breeder strain. Thus, the predominant cluster A had less diversity in terms of breeding types than cluster B.
Regarding the MDR S. Enteritidis strains, 26% were in the predominant PT genotype PT6, while 55% were in the predominant MT genotype MT22. This indicates that MDR was more common in the predominant genotypes. Both Fardsanei et al. [16] and Wei et al. [3] found no correlation between the S. Enteritidis strains’ AMR profiles and PFGE genotypes. In the present study, however, all the hexa-resistant strains with the dominant MDR profile AMP-CTX-CAZ-CRO-S-MEM-S were MT22. That is, there was a correlation between the predominant MLVA genotype and predominant MDR pattern. Regarding breeding type, the majority of MDR S. Enteritidis strains (93%) originated from broiler samples, indicating that AMR levels were quite high in S. Enteritidis strains of broiler chicken origin.
The genotyping results were also compared by geographic location, isolation year, and breeding type of the strains. Regarding all S. Enteritidis strains, there was no significant correlation between genotypes, AMR patterns, and geographic location, isolation year, and breeding type of strains. However, there were correlations for specific sample groups. All strains isolated from layer chickens located in Afyon were grouped in cluster B2. These strains were PT32 genotype isolated in the years of 2019 and 2020. Regarding the MLVA genotypes of these strains, the single 2019 strain was MT6, whereas the 2020 strains were MT4. MLVA was able to discriminate between the strains from the breeding type, geographical location, and PFGE genotype by isolation year. Moreover, the single repeat difference at the SE2 locus between the MT4 and MT6 genotypes showed that the strains were genetically related. Thus, MLVA was able to identify the genetic similarities of the strains while discriminating between them based on isolation year. The strains isolated from broiler breeders from Ankara were grouped in cluster B1. These strains were PT25 and MT10 genotypes, isolated in 2020. The analysis demonstrated that the strains with the same geographic location, isolation year, and breeding type also had the same genotype. One broiler strain from Ankara was grouped in cluster A1. This strain was PT6 and MT22. Finally, the PFGE and MLVA genotypes varied when the breeding type was changed. That is, PFGE and MLVA discriminated by breeding type for this sample group.
Overall, our findings indicated that MLVA was easier, quicker, and less labor-intensive to use than PFGE. PFGE was thought more difficult to implement as it involves many manual processes including the preparation of the chemicals. In addition, PFGE lacks a confirmation step, whereas MLVA results could be confirmed by electrophoresis before completing the analysis. Thus, MLVA is a suitable method for large sample groups. Regarding the VNTR loci, SE5 and SE2 were the most diverse in this study, which is compatible with Mezal et al. [1], Wei et al. [3], and Liu et al. [24]. Considering all studies, the most diverse locus was found between 2014 and 2020, which suggests that future MLVA genotyping studies can use the SE5 locus to differentiate closely related strains.
Conclusions
In this study, MLVA and PFGE were applied to genotype chicken-originated S. Enteritidis isolates. AMR profiles were investigated for all isolates. More than half (57%) of the S. Enteritidis isolates were pan-susceptible to antimicrobials. The most frequently detected resistance was against AMP (25%). Almost all MDR S. Enteritidis isolates (93%) came from broiler samples. The most frequent MDR pattern (14%) was the AMP-CTX-CAZ-CRO-MEM-S hexa-resistant pattern. All strains were genotyped into 32 different PFGE genotypes (PT) and 34 different MLVA genotypes (MT). No significant difference was observed in the discrimination power of the two methods. Among the chicken-originated S. Enteritidis strains circulating in our country, the dominant genotypes were PT6 and MT22. For specific sample groups, there was a correlation between the strains’ genotypes, breeding type, geographic location, and isolation year. Given that it is easy and quick to implement, and has high discriminatory power, MLVA was considered a more effective tool than PFGE for genotyping S. Enteritidis strains.
Acknowledgements
This study was summarized from the PhD dissertation of the first author.
Author contribution
SSI: Investigation, methodology, writing, and editing. HKM: Supervision and editing. All authors read and approved the final manuscript.
Funding
This study has been supported by Ankara University Scientific Research Projects Coordination Unit under grant number 21L0239004.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Responsible Editor: Mariza Landgraf
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mezal EH, Sabol A, Khan MA, Ali N, Stefanova R, Khan AA. Isolation and molecular characterization of Salmonella enterica serovar Enteritidis from poultry house and clinical samples during 2010. Food Microbiol. 2014;38:67–74. doi: 10.1016/j.fm.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 2.EFSA (European Food Safety Authority) The European Union One Health 2019 Zoonoses Report. EFSA J. 2021;19:6406. doi: 10.2903/j.efsa.2021.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei XY, You L, Wang D, Huang H, Li SJ, Wang DM. Antimicrobial resistance and molecular genotyping of Salmonella enterica serovar Enteritidis clinical isolates from Guizhou province of Southwestern China. PLoS ONE. 2019;14:1–14. doi: 10.1371/journal.pone.0221492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campioni F, Davis M, Medeiros MIC, Falcao JP, Shah DH. MLVA typing reveals higher genetic homogeneity among S. Enteritidis strains isolated from food, humans and chickens in Brazil in comparison to the North American strains. Int J Food Microbiol. 2013;162:174–181. doi: 10.1016/j.ijfoodmicro.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Hashemi A, Baghbani-Arani F, Ahmadiyan S, Ghavami-Nejad S. Multiple-locus variable-number tandem-repeat analysis in Salmonella isolates as an effective molecular subtyping method. Microb Pathog. 2017;113:11–16. doi: 10.1016/j.micpath.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Dewaele I, Rasschaert G, Bertrand S, Wildemauwe C, Wattiau P, Imberechts H, Herman L, Ducatelle R, De Reu K, Heyndrickx M. Molecular characterization of Salmonella Enteritidis: comparison of an optimized multi-locus variable-number of tandem repeat analysis (MLVA) and pulsed-field gel electrophoresis. Foodborne Pathog Dis. 2012;9:885–895. doi: 10.1089/fpd.2012.1199. [DOI] [PubMed] [Google Scholar]
- 7.Tang S, Orsi RH, Luo H, Ge C, Zhang G, Baker RC, Stevenson A, Wiedmann M. Assessment and comparison of molecular subtyping and characterization methods for Salmonella. Front Microbiol. 2019;10:1–17. doi: 10.3389/fmicb.2019.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrari RG, Panzenhagen PHN, Conte-Junior CA. Phenotypic and genotypic eligible methods for Salmonella typhimurium source tracking. Front Microbiol. 2017;8:1–15. doi: 10.3389/fmicb.2017.02587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang K, Wei B, Cha SY, Zhang JF, Park JY, Lee YJ, Jang HK, Kang M. The occurrence of antimicrobial-resistant Salmonella enterica in hatcheries and dissemination in an integrated broiler chicken operation in Korea. Animals. 2021;11:1–14. doi: 10.3390/ani11010154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez JE, Vinas MR, Herrera M, Moroni M, Gutkind GO, Mercado EC, Di Conza JA, Chacana PA. Molecular characterization and antimicrobial resistance profiles of Salmonella Heidelberg isolates from poultry. Zoonoses Public Health. 2021;68:309–315. doi: 10.1111/zph.12819. [DOI] [PubMed] [Google Scholar]
- 11.WHO (World Health Organization) (2018) Protocol for serotyping and antimicrobial susceptibility testing of Salmonella test strains.G00–06–001
- 12.CLSI (Clinical and Laboratory Standards Institute) (2018) Performance standards for antimicrobial susceptibility testing (M100). 28ed. Wayne, PA, USA
- 13.Centers for Disease Control and Prevention (2017) Standard operating procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf. Accessed 17 July 2021
- 14.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CLSI (Centers for Disease Control and Prevention) (2013) PulseNet standard operating procedure for PulseNet MLVA of Salmonella enterica serotype Enteritidis – Applied Biosystems Genetic Analyzer 3500 Platform. https://www.cdc.gov/pulsenet/pdf/st-abi-3500-508c.pdf. Accessed 17 Jul 2021
- 16.Fardsanei F, Soltan Dallal MM, Douraghi M, Memariani H, Bakhshi B, Zahraei Salehi T, Nikkhahi F. Antimicrobial resistance, virulence genes and genetic relatedness of Salmonella enterica serotype Enteritidis isolates recovered from human gastroenteritis in Tehran. Iran J Glob Antimicrob Resist. 2018;12:220–226. doi: 10.1016/j.jgar.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Di Marcantonio L, Janowicz A, Zilli K, Romantini R, Bilei S, Paganico D, Persiani T, Di Donato G, Di Giannatale E. Genomic comparison of Salmonella Enteritidis strains isolated from laying hens and humans in the Abruzzi region during 2018. Pathogens. 2020;9:1–9. doi: 10.3390/pathogens9050349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, Working WPPL. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 19.Utrarachkij F, Nakajima C, Siripanichgon K, Changkaew K, Thongpanich Y, Pornraungwong S, Suthienkul O, Suzuki Y. Genetic diversity and antimicrobial resistance pattern of Salmonella enterica serovar Enteritidis clinical isolates in Thailand. J Infect Chemother. 2016;22(4):209–215. doi: 10.1016/j.jiac.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Kahraman BB, Issa G, Kahraman T. Prevalence, antimicrobial resistance and molecular characterization of Salmonella spp. and Listeria monocytogenes isolated from chicken carcass. Kafkas Univ Vet Fak Derg. 2018;24(5):775–779. doi: 10.9775/kvfd.2018.19754. [DOI] [Google Scholar]
- 21.Republic of Turkey Ministry of Agriculture and Forestry (2018) National Salmonella Control Program (NSCP). 1–45
- 22.Guran HS, Ciftci R, Gursoy NC, Ozekinci T, Alali WQ. Prevalence of antibiotic-resistant Salmonella in retail organic chicken. Br Food J. 2020;122(4):1238–1251. doi: 10.1108/BFJ-10-2019-0790. [DOI] [Google Scholar]
- 23.Muvhali M, Smith AM, Rakgantso AM, Keddy KH. Investigation of Salmonella Enteritidis outbreaks in South Africa using multi-locus variable-number tandem-repeats analysis, 2013–2015. BMC Infect Dis. 2017;17:1–9. doi: 10.1186/s12879-017-2751-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Shi X, Li Y, Chen Q, Jiang M, Li W, Qiu Y, Lin Y, Jiang Y, Kan B, Sun Q, Hu Q. The evaluation and application of multilocus variable number tandem repeat analysis (MLVA) for the molecular epidemiological study of Salmonella enterica subsp. enterica serovar Enteritidis infection. Ann Clin Microbiol Antimicrob. 2016;15:1–9. doi: 10.1186/s12941-016-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]