Abstract
Protein nanocages have emerged as an important engineering platform for biotechnological and biomedical applications. Among naturally occurring protein cages, encapsulin nanocompartments have recently gained prominence due to their favorable physico-chemical properties, ease of shell modification, and highly efficient and selective intrinsic protein packaging capabilities. Here, we expand encapsulin function by designing and characterizing encapsulins for concurrent RNA and protein encapsulation in vivo. Our strategy is based on modifying encapsulin shells with nucleic acid-binding peptides without disrupting the native protein packaging mechanism. We show that our engineered encapsulins reliably self-assemble in vivo, are capable of efficient size-selective in vivo RNA packaging, can simultaneously load multiple functional RNAs, and can be used for concurrent in vivo packaging of RNA and protein. Our engineered encapsulation platform has potential for codelivery of therapeutic RNAs and proteins to elicit synergistic effects and as a modular tool for other biotechnological applications.
Keywords: encapsulin, protein cage, nanocompartment, RNA, drug delivery
Graphical Abstract
INTRODUCTION
Over the past decade, protein nanocages have gained much attention for various biotechnological and biomedical applications due to their unique and desirable properties.1-3 Their biological origin makes them inherently biocompatible and allows facile genetic functionalization, while their defined shell-like structure enables the creation of multifunctional and atomically defined nano-devices by modifying both their inner and outer surfaces. Furthermore, established recombinant protein production strategies make protein-based nanostructures simple to produce, purify, and scale.
Based on these favorable properties, significant effort has been dedicated toward engineering protein nanocages like bacterial microcompartments (BMCs),4,5 lumazine synthase,6,7 ferritin,8,9 virus-like particles (VLPs),10,11 and computationally designed protein shells.12 For example, BMCs have been engineered as catalytic nanoreactors13-15 and molecular scaffolds,16 while lumazine synthase has been utilized for molecular display,17-19 RNA packaging and delivery,20,21 and enzyme encapsulation.22,23 Ferritins and VLPs have long been used for in vitro bionanotechnology, biomaterials research, and therapeutics delivery.8-11 More recently, designed protein assemblies have similarly been explored for related applications.20,24-26
Among naturally occurring protein nanocages, encapsulins have emerged as an alternative and attractive engineering platform for applications in medicine, catalysis, and nano-technology.27-33 Encapsulins are self-assembling icosahedral protein compartments composed of a single type of shell protomer possessing the HK97 phage-like fold.34-36 They can assemble into T1 (60 subunits, ca. 24 nm),36-39 T3 (180 subunits, ca. 32 nm),40,41 and T4 (240 subunits, ca. 42 nm)42 shells and are widely distributed throughout both the bacterial and archaeal domains.43,44 Encapsulins have been proposed to play diverse roles in cellular metabolism including iron storage,42 redox stress resistance,45 and sulfur metabolism.38 Their key feature is the ability to selectively encapsulate dedicated cargo proteins in vivo.36 All native cargo proteins contain N- or C-terminal domains38 or targeting peptides (TPs) necessary for efficient cargo loading during shell self-assembly.46,47 This feature—a dedicated and modular protein loading mechanism—has been widely utilized to package non-native cargo proteins into the encapsulin shell via simple genetic fusion of TPs to proteins of interest.48-50
Engineered encapsulins have shown potential as nanoreactors, 50,51 drug delivery systems,52 imaging agents,53-55 and immunotherapies.30,56,57 Straightforward genetic and chemical shell modification allows small-molecule conjugation,33 peptide loop insertion,58 pore modification,50,59,60 and fusion of protein domains to the N- and C-terminus of the encapsulin protomer.50,52,56,61 Recently, engineered encapsulins capable of triggered reversible disassembly, enabling in vitro cargo loading and stimulus-responsive cargo release, have also been reported.62
A topic of particular current interest is the selective packaging and delivery of nucleic acids inside protein-based cages.1,63 Engineering such systems has shed light on the evolution and function of viruses and allowed the creation of nonviral systems mimicking select virus characteristics.21,64-66 The ability to encapsulate nucleic acids in vivo may provide novel approaches for RNA regulation and cytosolic sampling21 with broad implications for RNA biology.67-70 RNA- and DNA-based therapeutics have tremendous clinical potential.71,72 However, their broad application has been hampered by poor pharmacokinetic properties,73 difficulty in overcoming cell membranes,74 susceptibility to nucleases, inherent immunogenicity, and rapid clearance from the body.75,76 These challenges could be overcome by engineering efficient nucleic acid delivery systems, with many different approaches and materials already having been employed toward achieving this goal.73,77-79 Due to their desirable properties and engineerability, protein cages, in general, and encapsulins, in particular, represent a promising alternative strategy for nucleic acid packaging and delivery. In addition, expanding encapsulin function toward nucleic acid encapsulation would allow for the concurrent sequestration and colocalization of proteins and nucleic acids. This may enable the future codelivery of two types of functional macromolecules acting in either an orthogonal or a synergistic manner.
Here, we engineer and characterize encapsulins as novel nano-encapsulation platforms for simultaneous RNA and protein packaging in vivo, laying the foundation for their future use as targeted codelivery systems.
RESULTS AND DISCUSSION
Design and Initial Characterization of an Encapsulin-Based In Vivo RNA Encapsulation System.
We set out to design robust and modular encapsulin-based nanocages for the in vivo sequestration of RNA, without disturbing their native protein loading capabilities. Furthermore, the three naturally occurring assembly states of encapsulins—T1, T3, and T4—were exploited to design a range of RNA packaging nanocages with different dimensions. This strategy allows the exploration of the influence of luminal volume and charge on in vivo RNA and protein loading. In particular, the three established encapsulin systems from Thermotoga maritima (TmT1), Myxococcus xanthus (MxT3), and Quasibacillus thermotolerans (QtT4) were chosen as engineering scaffolds (Figure 1A). Although the Mx encapsulin primarily forms T3 shells, a recent study showed that in the absence of native protein cargo, about 36% of shells assemble into small T1-sized encapsulins with a diameter of 18 nm (MxT1).40 This feature was used to additionally explore the RNA packaging capacity of MxT1.
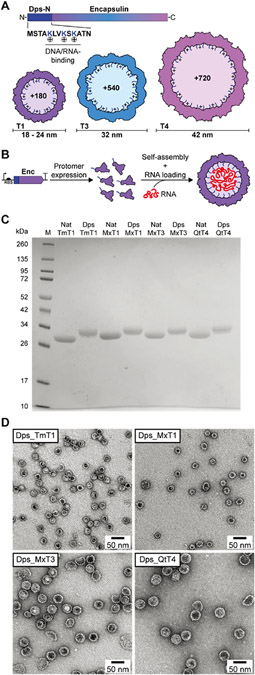
Figure 1.
Design and initial characterization of the encapsulin-based in vivo RNA encapsulation system. (A) Schematic of the engineered encapsulin protomer sequence with the N-terminally fused nucleic acid-binding peptide Dps-N. The three different sizes (T1, T3, and T4) of engineered encapsulins (Dps_Encs) created in this study are shown, highlighting the additional luminal positive charges introduced by Dps-N. T: triangulation number (T-number). (B) Schematic outlining the principle of in vivo nucleic acid encapsulation by Dps_Encs. Enc: encapsulin. (C) SDS-PAGE analysis and comparison of purified native (Nat) and engineered (Dps) encapsulins used in this study. Encapsulins are labeled by their organism of origin and T-number. Tm: Thermotoga maritima, Mx: Myxococcus xanthus, Qt: Quasibacillus thermotolerans. (D) Negative-stain TEM micrographs of all four purified Dps_Encs used in this study.
To imbue encapsulins with the ability to bind and encapsulate nucleic acid, we genetically fused the Escherichia coli Dps-N peptide (MSTAKLVKSKATN)—originating from the DNA-binding protein from starved cells (Dps)80—to the N-terminus of the encapsulin protomer via a flexible six-residue linker (GGSGGS), yielding our Dps_Enc fusion constructs. Dps-N consists of the 13 N-terminal residues of Dps and includes three positively charged lysines (Figure 1A). It is able to bind to both DNA and RNA, likely via the electrostatic interaction of the positively charged lysine residues with the negatively charged DNA/RNA phosphate backbone. Dps-N was specifically chosen for our fusion constructs due to its broad specificity and prior successful use as a nucleic acid-binding peptide.81 In assembled Tm, Mx, and Qt encapsulins, the N-termini of all protomers are pointed toward the shell interior. Therefore, in engineered Dps_Encs, three additional positive charges per protomer will be introduced to the encapsulin lumen, resulting in overall charge increases of + 180 (T1), +540 (T3), and +720 (T4) for our fusion constructs (Figure 1A). This increased positive charge of the shell interior will drive the encapsulation of RNA during shell self-assembly (Figure 1B). We envisioned that Dps_Encs would allow in vivo packaging of native or overexpressed RNAs while minimizing concurrent DNA packaging. This is due to the relatively small size of encapsulins and the fact that generally, no DNA molecules small enough to be encapsulated inside encapsulin shells are present inside cells. Furthermore, the broad specificity of Dps-N could in the future also be used for the flexible in vitro loading of variable nucleic acids, both RNA and DNA.
Dps_Encs and unmodified native Tm, Mx, and Qt controls (Nat_Encs) were produced in E. coli and purified through a combination of polyethylene glycol (PEG) precipitation, ion exchange chromatography (IEC), and size-exclusion chromatography (SEC). SDS-PAGE analysis of purified Dps_Encs and Nat_Encs was used to confirm sample homogeneity (Figure 1C). Further analyses using negative-stain transmission electron microscopy (TEM), dynamic light scattering (DLS), and analytical SEC indicated that all Dps_Encs formed stable shells with similar size and appearance compared to the corresponding Nat_Encs (Figures 1D and S1). These results confirm the feasibility of our novel Dps-N fusion designs and highlight the ease of luminal encapsulin shell modification without disturbing shell self-assembly.
In Vivo Nucleic Acid Encapsulation and Resistance toward Nuclease Digestion.
After confirming the proper assembly of all Dps_Enc designs, we next focused on their nucleic acid encapsulation capacity and ability to protect encapsulated nucleic acid from nuclease digestion (Figure 2A). Native agarose gel electrophoresis with both protein and nucleic acid staining showed that all purified Dps_Encs contained significantly more nucleic acid than the respective Nat_Enc controls (Figure 2B, middle). To exclude nonspecific nucleic acid binding to the outside of encapsulin shells, Benzonase treatments and IEC were carried out during all purifications. These results confirm that Dps-N fusion does indeed confer nucleic acid encapsulation capacity to all of our engineered encapsulin shells, further supported by direct A260/A280 measurements of purified samples, and increased A260/A280 signal observed during SEC (Figure S1).
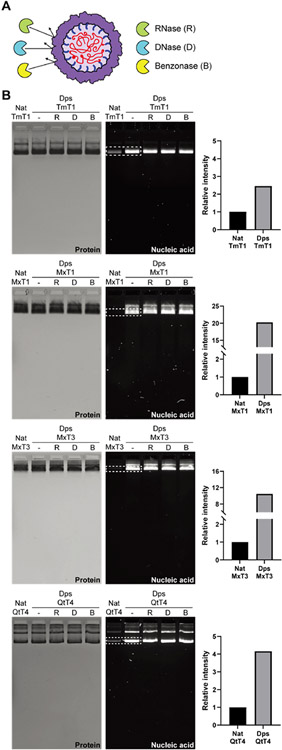
Figure 2.
In vivo nucleic acid encapsulation inside Dps_Encs and resistance toward nuclease digestion. (A) Schematic of Dps_Encs protecting in vivo encapsulated nucleic acids from nucleases. (B) Native agarose gel electrophoresis of Nat_Encs and Dps_Encs before and after nuclease treatment. Gels were stained with Coomassie blue to visualize protein (left) or GelRed to visualize nucleic acids (middle). On the right, the relative intensity of nucleic acid bands (dotted box) normalized by protein amount is shown, highlighting the encapsulation capacity difference between Nat_Encs and Dps_Encs.
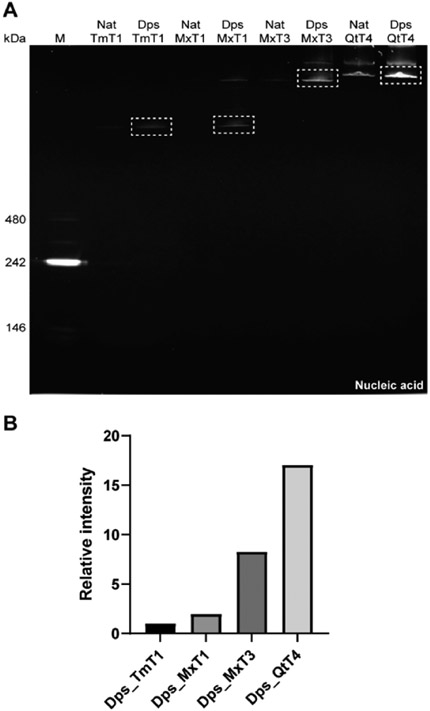
Dps_MxT1 and Dps_MxT3 showed the highest relative nucleic acid packaging capacity with 20- and 11-fold increases in signal when compared to their native forms (Figure 2B, right). Dps_TmT1 and Dps_QtT4 yielded more moderate signal increases of 2.4- and 4-fold, respectively. These discrepancies in increased nucleic acid loading capacity over native encapsulins are partially caused by the larger background signals observed for Nat_TmT1 and Nat_QtT4. For both Nat_Encs and Dps_Encs, several minor higher molecular weight bands could be observed. We confirmed via tryptic digest and mass spectrometry that all observed bands represent the respective Nat_Encs or Dps_Encs (Figure S2). Higher molecular weight bands are likely a result of partial aggregation of encapsulin shells during gel electrophoresis.
Next, purified Dps_Encs were treated with DNase, RNase, or Benzonase. No reduction in the intensity of nucleic acid bands was observed for any of our fusion constructs, confirming that the encapsulin shell can effectively protect encapsulated nucleic acid from nuclease digestion (Figure 2B, middle). The protective role of encapsulin shells is likely due to the physical sequestration of nucleic acid inside a protein barrier. Encapsulin shells possess small pores at the 5-, 3-, and 2-fold symmetry axes with diameters ranging from 2 to 7 Å,40,42,59 which is too small to allow nuclease access to the shell interior. Furthermore, encapsulin shells are generally very stable and once formed can only be disassembled under harsh nonphysiological conditions, thus making them excellent containers for protecting labile nucleic acids.
Analysis of Encapsulated Nucleic Acid Content and Size-Selective RNA Packaging.
To identify the type and size distribution of encapsulated nucleic acid, we first extracted the total nucleic acid contents from purified Dps_Encs and subjected them to differential nuclease treatment using DNase, RNase, or Benzonase (Figure 3).
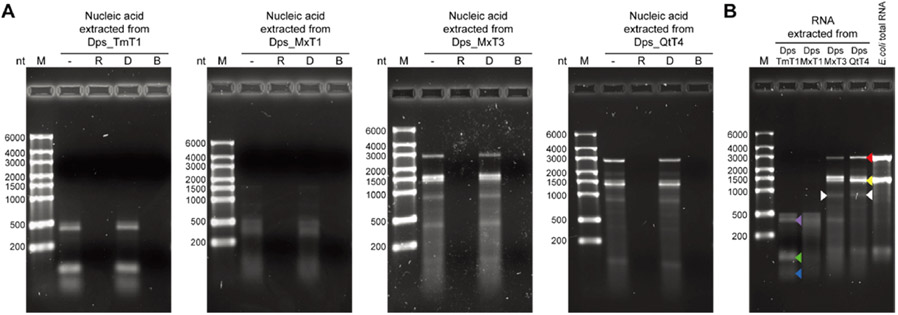
Figure 3.
Analysis of encapsulated nucleic acid content and size-selective RNA packaging of Dps_Encs. (A) Native agarose gel electrophoresis of nucleic acid extracted from purified Dps_Encs treated with either RNase (R), DNase (D), or Benzonase (B). Only nucleic acid treated with DNase retained its integrity, whereas RNase and Benzonase treatment led to complete digestion. (B) Native agarose gel electrophoresis of RNA extracted from Dps_Encs along with E.coli total RNA showing the size selectivity of differently sized Dps_Encs. A differential band for Dps_MxT3 and Dps_QtT4—absent in E. coli total RNA or T1 Dps_Encs—is highlighted by white arrows and putatively represents the respective Dps_Enc mRNA. Colored arrows indicate the following: 23S rRNA (red), 16S rRNA (yellow), tmRNA (purple), 5S rRNA (green), and tRNA (blue).
Exposure to DNase had no effect on any of the extracted nucleic acid samples, while RNase and Benzonase treatment resulted in complete digestion (Figure 3A). This clearly indicates that nucleic acid encapsulated in Dps_Encs is exclusively RNA, thus confirming our initial design for in vivo RNA packaging based on the fact that generally, only DNA molecules too large to be encapsulated inside encapsulin shells exist in cells, specifically chromosomes and plasmids.
We further compared the size distribution of RNA extracted from purified Dps_Encs. RNA encapsulation capacity was found to be size-selective and proportional to shell size, with smaller encapsulins showing a lower upper size limit for RNA compared to larger shells (Figure 3B). Specifically, Dps_TmT1 and Dps_MxT1 exhibited a maximum size of encapsulated RNA of ~500 nt, whereas the larger Dps_MxT3 and Dps_QtT4 shells were able to encapsulate RNA of up to ~3000 nt in length. Comparison of extracted RNA with E. coli total RNA indicated that for Dps_TmT1 and Dps_MxT1, tRNAs (blue arrow), 5S rRNA (green arrow), and tmRNA (purple arrow) likely made up a substantial part of sequestered RNA, while for Dps_MxT3 and Dps_QtT4, 16S (yellow arrow) and 23S (red arrow) rRNA were found to be the main RNA species (Figure 3B). This result is in accordance with rRNA and tRNA generally representing the majority of available RNA inside cells. Because the molecular size of RNA depends on its ability to form secondary structures, with many functional RNAs even able to adopt stable and compact 3D folds, RNAs larger than the observed size limits could potentially be encapsulated in Dps_Encs as well. Furthermore, differential bands of ~1000 nt in length, absent in E.coli total RNA and T1 Dps_Encs, were observed for Dps_MxT3 and Dps_QtT4 (Figure 3B, white arrows). Given that the mRNA size of Dps_MxT3 and Dps_QtT4 transcripts is ~1000 nt and that they would have been overexpressed, these differential bands likely represent Dps_MxT3 and Dps_QtT4 mRNA. Overall, these results indicate that Dps_Encs are able to encapsulate RNA in a size-selective manner within a relevant size range for future applications including the delivery of siRNAs which often are between 20 and 25 nt in length. We also observed that RNA abundance plays an important role in determining encapsulation efficiency.
Analysis of RNA Packaging Capacity.
To determine the relative RNA packaging capacities of our Dps_Encs per encapsulin shell, we performed native polyacrylamide gel electrophoresis (PAGE), loading the same normalized amount of encapsulin shells per lane for all Dps_Encs (Figure 4A). We found that RNA packaging capacity per shell increases with shell diameter (Figure 4B). Larger Dps_Encs have larger volumes for RNA packaging and contain more Dps-N-fused protomers, which result in an increased number of positive luminal charges. This indicates that RNA encapsulation capacity for Dps_Encs correlates with the overall number of charges and the available shell volume, rather than approximate luminal charge density which would be maximal for Dps_MxT1 (Figure S3).
Figure 4.
RNA packaging capacity analysis of differently sized Nat_Encs and Dps_Encs. (A) Native PAGE gel analysis of Nat_Encs and Dps_Encs stained with GelRed to visualize RNA where equivalent amounts of Dps_Encs are loaded (normalized to the number of Dps_Enc shells per lane) across all lanes for comparative analysis. (B) Relative intensity of RNA bands (dotted boxes in A) showing that per Dps_Enc shell, RNA packaging capacity increases with the shell size of Dps_Encs.
Simultaneous In Vivo Packaging of Two Functional RNAs.
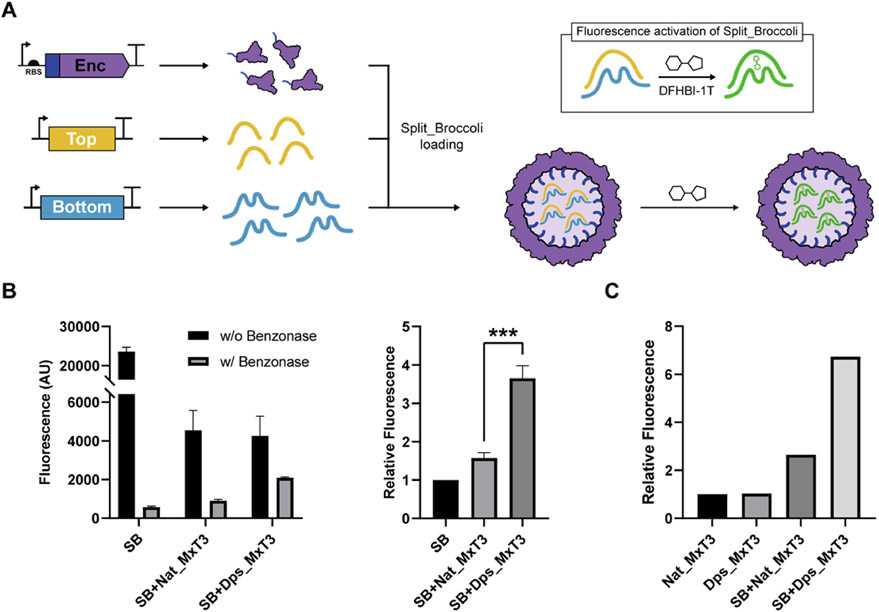
To expand the utility of Dps_Encs, we sought to test if multiple nonendogenous functional RNAs could be copackaged at the same time and be protected from nucleases. We utilized the split fluorogenic aptamer Split_Broccoli (SB)82 and coexpressed its two RNAs—Top (97 nt) and Bottom (153 nt)—together with Dps_MxT3 (Figures 5A and S4). Dps_MxT3 was used due to its overall favorable performance, combining a high upper size limit for RNA, low background, and high loading capacity. In initial experiments, Benzonase was added to cleared cell lysates—from cells expressing SB alone, SB + Nat_MxT3, or SB + Dps_MxT3—to remove free SB, highlight the protective role of encapsulin shells, and allow the detection of encapsulated SB via the addition of the small-molecule SB binding partner DFHBI-1T, yielding a fluorescence readout (Figure 5B). The highest SB fluorescence signal was observed for SB + Dps_MxT3 (Figure 5B), indicating that Dps_MxT3 successfully packaged both SB RNAs, protected them from nuclease digestion, and allowed access of the small-molecule DFHBI-1T to the shell interior. To confirm these experiments,Nat_MxT3 and Dps_MxT3 were purified alone or from cells coexpressing SB, followed by incubation with DFHBI-1T. Again, substantially higher SB fluorescence signal was observed for Dps_MxT3, confirming our initial results (Figure 5C).
Figure 5.
Simultaneous in vivo packaging of two functional Split_Brocolli (SB) RNAs using Dps_MxT3. (A) Schematic outlining the expression strategy and fluorescence activation of the SB aptamer (box). After in vivo packaging of both SB RNAs, SB fluorescence can be activated by the addition of the small fluorogenic molecule DFHBI-1T. SB is composed of the Top (yellow) and Bottom (blue) RNAs. (B) Fluorescence measurements (left) of DFHBI-1T-supplemented cell lysates—from cells expressing SB, SB + Nat_MxT3, or SB + Dps_MxT3—with or without prior Benzonase treatment. Relative fluorescence (right) of Benzonase-treated cell lysates normalized by the fluorescence of Benzonase-treated SB cell lysate. Data are shown as mean values, while error bars represent standard deviations from three independent experiments. (***P = 0.0005, two-sided unpaired t-test.) (C) Relative fluorescence of purified and DFHBI-1T-supplemented Nat_MxT3 and Dps_MxT3 that were expressed in cells with or without concurrent SB expression. Relative fluorescence was normalized based on the fluorescence of the Nat_MxT3 control.
Concurrent RNA and Protein Packaging In Vivo.
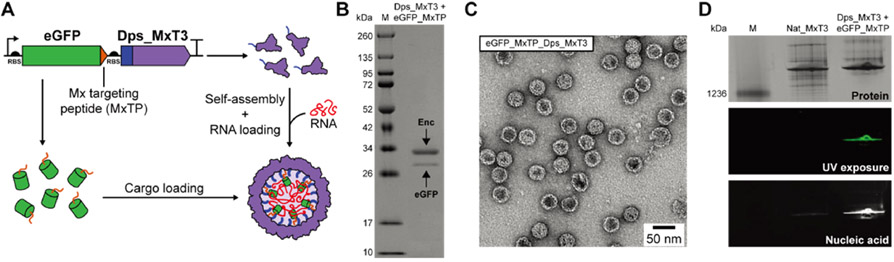
To design a system for the simultaneous in vivo packaging of both RNA and protein, we sought to combine the newly engineered ability of our Dps_Encs to encapsulate RNA with encapsulins’ native capacity for specific protein encapsulation. As a proof of concept, the Mx targeting peptide (MxTP, PEKRLTVGSLRR) with a flexible six-residue linker (GGSGGS) was genetically fused to the C-terminus of eGFP and cloned immediately upstream of the Dps_MxT3 gene for coexpression (Figure 6A). SDS-PAGE analysis of purified Dps_MxT3 confirmed the successful in vivo loading and copurification of MxTP-tagged eGFP (Figure 6B). Protein cargo loading for Dps_MxT3 was found to be comparable with Nat_MxT3 (Figure S5). Negative-stain TEM analysis further confirmed that eGFP-loaded Dps_MxT3 particles still formed homogeneous shells, very similar in size and appearance to Nat_MxT3 (Figures 6C and S1). To test the concurrent loading of both eGFP and RNA, native PAGE analysis was performed on purified eGFP-loaded Dps_MxT3 shells. Compared to Nat_MxT3, eGFP-loaded Dps_MxT3 exhibited substantially higher RNA signal intensity and eGFP fluorescence of the high-molecular-weight encapsulin band. Coelution of RNA and eGFP signals confirms successful copackaging of RNA and a specific heterologously expressed protein in vivo. Also, as expected, the encapsulation of eGFP diminished the amount of encapsulated RNA as shown by comparison of the A260/A280 ratios of Dps_MxT3 and eGFP_MxTP_Dps_MxT3 (Figure S1). Overall, this novel ability could be useful for future biomedical delivery applications where therapeutic effects may be potentiated by the synergistic action of codelivered functional RNAs and proteins.
Figure 6.
Concurrent in vivo RNA and protein packaging using Dps_MxT3. (A) Schematic of Dps_MxT3 concurrently packaging RNA and eGFP. RNA is packaged through interaction with the fused Dps-N peptide, while eGFP is loaded through specific targeting peptide (MxTP) interaction. (B) SDS-PAGE analysis of purified eGFP- and RNA-loaded Dps_MxT3. (C) Negative-stain TEM micrograph of purified eGFP- and RNA-loaded Dps_MxT3. (D) Native PAGE analysis showing Nat_MxT3 and eGFP- and RNA-loaded Dps_MxT3 via Coomassie blue staining (top), UV exposure for eGFP fluorescence analysis (middle), and GelRed staining for RNA detection (bottom).
CONCLUSIONS
Protein nanocage engineering has the potential to significantly contribute innovative solutions to challenging problems across various fields, including catalysis, nanotechnology, and medicine. In particular, innovative nanocage designs can yield high-performing tools for biomedical delivery applications. Delivery modalities based on protein cages have so far focused on safely and efficiently transporting protein- or RNA-based therapeutics into target cells.20,68,83,84 However, the possibility of combining two types of specific therapeutic macromolecules—protein and RNA—in a single nanocage design has barely been explored.85 One advantage of codelivering, in principle, any combination of therapeutic RNAs and proteins of interest to cells includes the potential for targeting multiple intracellular target classes, for example, mRNA and protein, at the same time. For example, combining siRNA and antibodies against the same target in a single nanocage-based delivery vehicle may lead to improved suppression due to dual action at both the mRNA and protein level. The release of cargo from protein nanocages can be promoted through inducing disassembly of the nanocage. After disassembly, especially for RNA cargo, high cytosolic concentrations of RNA—particularly tRNA—will compete for Dps-N binding, thus liberating the delivered RNA cargo.20 Furthermore, it has been proposed that low pH in the late endosome can also promote the release of encapsulated nucleic acids.85 A second advantage of copackaging RNA and protein in a single nanocage is that it leads to ensured codelivery to each target cell, whereas concurrent administration of the same therapeutics via separate delivery methods has the potential to lead to heterogeneous populations of singly and doubly targeted cells.86
To address the challenge of copackaging multiple types of functional macromolecules in a single nanocage design, we have developed encapsulin-based protein cages, called Dps_Encs, capable of concurrently packaging functional RNAs and specific proteins of interest in vivo. Importantly, all Dps_Encs efficiently protected encapsulated RNA from nuclease digest, which is one of the main challenges of RNA-based therapeutics (Figure 3).87 Four different sizes of Dps_Encs were created, namely, MxT1 (18 nm, luminal volume: ~905 nm3), TmT1 (24 nm, ~3054 nm3), MxT3 (32 nm, ~9203 nm3), and QtT4 (42 nm, ~24,429 nm3). The available luminal volume spans more than an order of magnitude and correlates well with the observed RNA loading capacity per shell (Figure 4). We further showed that Dps_MxT3 is capable of colocalizing and protecting two functional RNAs, the split aptamer Split_Broccoli, and importantly, that the SB binding partner DFHBI-1T can access the shell interior, likely via the 5-, 3-, or 2-fold pores natively present in MxT3 (Figure 5). Finally, we showed that Dps_MxT3 retained the ability to specifically sequester a TP-tagged coexpressed cargo protein while simultaneously packaging RNA (Figure 6).
While most protein cage systems to date rely on in vitro packaging of cargo,88-91 requiring separate purification as well as disassembly and reassembly steps, our encapsulin-based Dps_Encs can copackage RNA and specific proteins in vivo in a single step. This in situ assembly of functional nanocages simplifies purification and avoids nonphysiological in vitro conditions, often necessary for disassembly and cargo loading of other protein nanocages.88-91 One challenge of in vivo cargo loading is the potential copackaging of unwanted molecules, including endogenous RNA and protein. However, the intrinsic specificity of encapsulins for packaging coexpressed TP-tagged proteins has been extensively used to assemble highly homogeneous cargo-loaded cages with minimal nonspecific loading.46-50 After purification, in vivo eGFP-loaded Dps_MxT3 showed minimal background of non-TP-tagged proteins (Figure 6B). In future encapsulin-based nanocage designs, Dps-N could easily be replaced with RNA-binding peptides or domains that bind RNA in a sequence-specific manner.92-94 Functional RNAs could then be tagged with this packaging RNA sequence likely resulting in sequence-selective in vivo RNA loading. Another potential advantage of nanocages that can be loaded in vivo is their potential use in living therapeutics.95 In living therapeutics, engineered bacteria are used as a drug delivery modality to reach a target site of interest. Once at the target, bioactive molecules can be continuously produced locally by the bacteria, leading to increased therapeutic effects with minimal systemic side effects.96,97 Thus, nanocage systems that do not require in vitro assembly could be locally assembled in vivo and released.96,98 Finally, Dps_Encs could themselves be imbued with cell targeting capabilities through the genetic fusion of cell penetrating peptides or similar targeting systems to the encapsulin C-terminus exposed on the shell exterior.33,52
In sum, the Dps_Encs developed in this study lay the foundation for using encapsulins for the codelivery of therapeutic RNA and protein with the ultimate goal of eliciting homogeneous synergistic effects at a single cell level. This work further highlights the versatility of encapsulins as modular and robust tools with broad potential applicability for different biomedical and biotechnological applications.
EXPERIMENTAL SECTION
Molecular Biology and Cloning.
All constructs used in this study, except Dps-N-fused encapsulins (Dps_Encs), were ordered from Integrated DNA Technologies (IDT) as E. coli codon-optimized gBlocks (Table S1). Genes for Dps_Encs were obtained through overhang PCR using the native encapsulin genes as templates, adding Dps-N in the process (Table S2). All genes except Split_Broccoli (SB) were cloned into the pETDuet-1 vector, while SB was inserted into pCDFDuet-1 using Gibson Assembly. Top and Bottom of SB separated with a 270 bp spacer were inserted into a single pCDFDuet-1 vector (Table S2). E. coli BL21 (DE3) cells were transformed with the assembled plasmids via electroporation and were confirmed through Sanger sequencing (Eurofins Scientific).
Protein Expression and Purification.
All expression experiments were carried out using lysogeny broth (LB) medium supplemented with the appropriate selection marker [100 mg/mL ampicillin (pETDuet-1), 50 mg/mL spectinomycin (pCDFDuet-1), or both]. 500 mL of fresh LB medium was inoculated 1:100 using a 5 mL overnight culture, grown at 37 °C to OD600 of 0.4–0.5, and then induced with 0.1 mM IPTG. After induction, cultures were grown at 30 °C overnight for ca. 18 h and harvested via centrifugation (8000 g, 10 min, 4 °C). The resulting cell pellets were frozen and stored at −20 °C until further use.
Frozen cell pellets were resuspended in 5 mL/g (wet cell mass) of Tris buffer (20 mM Tris, 150 mM NaCl, pH 7.5). Lysis components [lysozyme (0.5 mg/mL), Benzonase nuclease (25 units/mL), MgCl2 (1.5 mM), and SIGMAFAST EDTA-free protease inhibitor cocktail (one tablet per 100 mL)] were added, and cells were incubated on ice for 15 min. Samples were then sonicated at 55% amplitude and a pulse time of 10 s on and 20 s off for 5 min total (Model 120 Sonic Dismembrator, Fisher Scientific). After sonication, samples were clarified by centrifugation (10,000 g, 15 min, 4 °C). To the supernatant, NaCl and PEG-8000 were added to a final concentration of 0.5 M and 10%, respectively, and incubated on ice for 40 min, followed by centrifugation (8000 g, 10 min, 4 °C). The supernatant was removed, and the pellet was resuspended in 3 mL of Tris buffer (pH 7.5) and filtered using a 0.2 μm syringe filter.
The filtered sample was subjected to SEC using a Sephacryl S-500 16/60 column and Tris buffer (pH 7.5) at a flow rate of 1 mL/min. Fractions were evaluated using SDS-PAGE and encapsulin-containing fractions were combined, concentrated, and dialyzed using Amicon filter units (100 kDa MWCO) and Tris buffer without NaCl (20 mM Tris, pH 7.5). The low salt sample was then loaded on a HiPrep DEAE FF 16/10 Ion Exchange column at a flow rate of 3 mL/min to remove nucleic acid contamination. Encapsulin-containing fractions were concentrated, centrifuged (10,000 g, 10 min, 4 °C), and then subjected to SEC using a Superose 6 10/300 GL column and Tris buffer (pH 7.5) at a flow rate of 0.5 mL/min. Purified proteins were stored in Tris buffer (pH 7.5) at 4 °C until further use.
Transmission Electron Microscopy.
Encapsulin samples for negative-stain TEM were diluted to 0.15 mg/mL in Tris buffer (pH 7.5). Gold grids (200-mesh coated with a Formvar–carbon film, EMS) were made hydrophilic by glow discharge at 5 mA for 60 s (easiGlow, PELCO). 4 μL of sample was added to the grid and incubated for 1 min, wicked with filter paper, and washed with 0.75% uranyl formate before staining with 0.75% uranyl formate for 1 min. Stain was removed using filter paper, and the grid was dried for at least 20 min before imaging. TEM micrographs were captured using a Morgagni transmission electron microscope at 100 keV at the University of Michigan Life Sciences Institute.
DLS Analysis.
All sizing and polydispersity measurements were carried out on an Uncle instrument (Unchained Labs) at 15 °C in triplicate. All encapsulin samples were adjusted to 0.5 mg/mL of monomer using Tris buffer (pH 7.5), centrifuged (10,000 g, 10 min, 4 °C), and then immediately analyzed via DLS.
RNA Extraction.
RNA was extracted from purified Dps_Enc samples via phenol–chloroform extraction, followed by ethanol precipitation. Phenol:chloroform:isoamyl alcohol (25:24:1, pH 8) was used for phenol–chloroform extraction, and after ethanol precipitation, the desalted nucleic acid extracts were dissolved in TEN buffer (Tris 10 mM, EDTA 1 mM, pH 8) and stored at −80 °C. E. coli total RNA was purchased from ThermoFisher Scientific (AM7940). Quantification of RNA was carried out using a Nanodrop Spectrophotometer from ThermoFisher Scientific, Inc. (USA).
Nuclease Challenge of Extracted RNA and RNA-Loaded Dps_Encs.
DNase (ThermoFisher Scientific, EN0521), RNase (ThermoFisher Scientific, EN0531), and Benzonase (Sigma Aldrich, E8263) were used for nuclease challenge experiments of extracted RNA and RNA-loaded Dps_Encs. For all nuclease incubation experiments, 1 μL (1.5 μL) of DNase, RNase, and Benzonase was added to 9 μL (13.5 μL) of extracted RNA (RNA-loaded Dps_Encs) samples (final concentration: 10, 5, and 25 U/mL, respectively), followed by 30 min incubation at 37 °C.
Native Gel Electrophoresis.
Native Agarose Gel Electrophoresis.
3% native agarose gels were used to determine the nucleic acid encapsulation capacity of Dps_Encs and to demonstrate the nuclease resistance of Dps_Encs shell. 1× TAE buffer was used to make agarose gels. The amount of Dps_Enc loaded per lane was adjusted for each Dps_Enc encapsulin so as to easily visualize nucleic acid signal after GelRed staining, while corresponding Nat_Encs were loaded at equal amounts for direct comparison. Per lane, 15 μL of sample was loaded with an additional 2 μL of 70% (v/v) aqueous glycerol. Gel electrophoresis was carried out using 1× TAE buffer at a constant voltage of 90 V for 35 min. Gels were first stained with GelRed to visualize nucleic acids and then stained with Coomassie blue to visualize proteins. Nucleic acid encapsulation capacity of Nat_Encs and Dps_Encs was compared via gel densitometry, first, normalizing the intensity of nucleic acid bands by their corresponding protein band (N/P). Then, N/P values of Dps_Encs were normalized by N/P values of the corresponding Nat_Encs for comparison. Band intensities of nucleic acid and protein were measured using Fiji/ImageJ v2.1.0/1.53c.
2% native agarose gels were used for nucleic acid extracted from purified Dps_Encs. The extracted nucleic acid was incubated with nucleases and loaded on the gels along with undigested nucleic acid for comparison. Per lane, 10 μL of sample was loaded with an additional 10 μL of 2× RNA loading buffer. Gel electrophoresis was carried out in 1× TAE buffer at a constant voltage of 125 V for 25–30 min. The gel was stained with GelRed to visualize nucleic acids.
Native Polyacrylamide Gel Electrophoresis.
All native polyacrylamide gel electrophoresis analyses were conducted in an Invitrogen XCell SureLock using NativePAGE 3 to 12% bistris mini protein gels with 1× NativePAGE Anode Buffer and 1× NativePAGE Cathode Buffer. 850 fmol of encapsulin shells was loaded per lane to maintain equivalent amounts of shells across all lanes for comparative analysis. The number of shells was calculated as follows: # of shells = [protein concentration (mg/mL)]/[protomer Mw (g/mol) × # of protomer per shell]. Protein concentration was measured by A280 using Nanodrop, and absorption coefficient was calculated for each Dps_Enc based on the protomer sequence. Native PAGE gels were run at a constant voltage of 150 V for 1 h, followed by an additional 1 h run at 250 V at 4 °C. Gels were then stained, first with GelRed for nucleic acid visualization and then with Coomassie blue for protein detection. For eGFP_MxTP_Dps_MxT3, the gel was first exposed to UV light for eGFP visualization before staining with GelRed and Coomassie blue.
To quantify and compare the amount of RNA loaded in each Dps_Enc encapsulin, gel densitometry of GelRed-stained gels was carried out using Fiji/ImageJ v2.1.0/1.53c. Pixel intensities of bands were background subtracted, yielding final overall intensities per band for comparisons.
Split_Broccoli (SB) Fluorescence Experiments.
50 mL of fresh LB medium containing appropriate antibiotic(s) was inoculated using an overnight 1 mL culture of either SB-, SB + Nat_MxT3−, or SB + Dps_MxT3-expressing cells. Cultures were grown at 37 °C to an OD600 of 0.4–0.5, then induced with 0.2 mM IPTG, and further grown for 5 h at 30 °C. As a control, 50 mL of E.coli BL21 (DE3) without transformed plasmids was similarly grown in LB at 30 °C for 5 h. Harvested cells were resuspended in 5 mL of Tris buffer (pH 7.5) and sonicated at 55% amplitude and a pulse time of 10 s on and 20 s off for 3 min 30 s total. Lysates were clarified by centrifugation (10,000 g, 15 min, 4 °C), and supernatants were filtered using 0.2 μm syringe filters. For each sample, two 100 μL aliquots were prepared. To one of the two aliquots of each sample, 14.7 μL of DFHBI-1T (final concentration: 1 mM) was added and incubated at 37 °C for 40 min, followed by fluorescence measurements using a Synergy H1 plate reader configured with filter sets for green fluorescence (λex = 472 nm, λem = 507 nm). For the other aliquot, 1 μL of MgCl2 (final concentration: 1.5 mM) and 1 μL of Benzonase (250 units) were added and incubated overnight at room temperature. The following day, 15 μL of DFHBI-1T (final concentration: 1 mM) was added and incubated at 37 °C for 40 min, followed by fluorescence analysis. For fluorescence measurements, 25 μL of each sample was loaded per well in triplicate into a black-flat bottom 384-well plate. Background fluorescence from the control E.coli BL21(DE3) sample without plasmid was subtracted from all samples, yielding final fluorescence intensities.
SB fluorescence was also measured using purified Nat_MxT3, Dps_MxT3, SB + Nat_MxT3, and SB + Dps_MxT3 samples. To 75 μL of each sample containing 5 pmol of capsid, 2 μL of DFHBI-1T (final concentration: 200 μM) was added and incubated at 37 °C for 40 min, followed by fluorescence analysis as described above. As background, 75 μL of Tris buffer (pH 7.5) was used and subtracted from the fluorescence signal of each sample.
Protein Identification.
In-gel digestion with trypsin was performed using a robot (ProGest, DigiLab) with the following protocol: (a) washed with 25 mM ammonium bicarbonate, followed by acetonitrile, (b) reduced with 10 mM dithiothreitol at 60 °C, (c) alkylated with 50 mM iodoacetamide at RT, (d) digested with sequencing grade trypsin (Promega) at 37 °C for 4 h, and (e) quenched with formic acid, and the supernatant was analyzed directly without further processing.
Half of each digested sample was analyzed by nano LC–MS/MS with a Waters M-Class HPLC system interfaced to a ThermoFisher Fusion Lumos mass spectrometer. Peptides were loaded on a trapping column and eluted over a 75 μm analytical column at 350 nL/min; both columns were packed with Luna C18 resin (Phenomenex). The mass spectrometer was operated in a data-dependent mode, with the Orbitrap operating at 60,000 FWHM and 15,000 FWHM for MS and MS/MS, respectively. The instrument was run with a 3 s cycle for MS and MS/MS.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge funding from the NIH (R35GM133325) (T.W.G.) and a Korean Government Scholarship (S.K.). We thank Dr. Henriette Remmer from the U-M Proteomics and Peptide Synthesis Core for project consultation and analysis of proteomics data. The proteomics experiments were conducted at MS Bioworks LLC.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.2c00391.
TEM images, DLS data, SEC results of Nat_Encs and Dps_Encs and absolute amount of extracted RNA from Dps_Encs; mass spectrometry results of minor higher molecular weight bands from Native PAGE analysis for Nat_Encs and Dps_Encs; change in luminal charge and approximate luminal surface charge density upon Dps-N fusion in engineered Dps_Encs; SDS-PAGE analysis comparing the expression level of Nat_MxT3 and Dps_MxT3 in the context of SB coexpression; protein cargo loading comparison of Dps_MxT3 and Nat_MxT3; protein sequences of all proteins used in this study; and DNA sequences of primers and the Split_Broccoli aptamer used in this study (PDF)
The authors declare no competing financial interest.
Contributor Information
Seokmu Kwon, Department of Chemical Engineering, University of Michigan, Ann Arbor, Michigan 48109, United States.
Tobias W. Giessen, Department of Biological Chemistry and Department of Biomedical Engineering, University of Michigan Medical School, Ann Arbor, Michigan 48109, United States
REFERENCES
- (1).Edwardson TGW; Levasseur MD; Tetter S; Steinauer A; Hori M; Hilvert D Protein Cages: From Fundamentals to Advanced Applications. Chem. Rev 2022, 122, 9145–9197. [DOI] [PubMed] [Google Scholar]
- (2).Giessen TW; Silver PA Encapsulation as a Strategy for the Design of Biological Compartmentalization. J. Mol. Biol 2016, 428, 916–927. [DOI] [PubMed] [Google Scholar]
- (3).Bhaskar S; Lim S Engineering protein nanocages as carriers for biomedical applications. NPG Asia Mater. 2017, 9, No. e371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Planamente S; Frank S Bio-engineering of bacterial micro-compartments: a mini review. Biochem. Soc. Trans 2019, 47, 765–777. [DOI] [PubMed] [Google Scholar]
- (5).Plegaria JS; Kerfeld CA Engineering nanoreactors using bacterial microcompartment architectures. Curr. Opin. Biotechnol 2018, 51, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Azuma Y; Edwardson TGW; Hilvert D Tailoring lumazine synthase assemblies for bionanotechnology. Chem. Soc. Rev 2018, 47, 3543–3557. [DOI] [PubMed] [Google Scholar]
- (7).Wei Y; Kumar P; Wahome N; Mantis NJ; Middaugh CR Biomedical Applications of Lumazine Synthase. J. Pharm. Sci 2018, 107, 2283–2296. [DOI] [PubMed] [Google Scholar]
- (8).Jutz G; van Rijn P; Santos Miranda B; Böker A Ferritin: a versatile building block for bionanotechnology. Chem. Rev 2015, 115, 1653–1701. [DOI] [PubMed] [Google Scholar]
- (9).Song N; Zhang J; Zhai J; Hong J; Yuan C; Liang M Ferritin: A Multifunctional Nanoplatform for Biological Detection, Imaging Diagnosis, and Drug Delivery. Acc. Chem. Res 2021, 54, 3313–3325. [DOI] [PubMed] [Google Scholar]
- (10).Hill BD; Zak A; Khera E; Wen F Engineering Virus-like Particles for Antigen and Drug Delivery. Curr. Protein Pept. Sci 2017, 19, 112–127. [DOI] [PubMed] [Google Scholar]
- (11).Nooraei S; Bahrulolum H; Hoseini ZS; Katalani C; Hajizade A; Easton AJ; Ahmadian G Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnology 2021, 19, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Stupka I; Heddle JG Artificial protein cages – inspiration, construction, and observation. Curr. Opin. Struct. Biol 2020, 64, 66–73. [DOI] [PubMed] [Google Scholar]
- (13).Lawrence AD; Frank S; Newnham S; Lee MJ; Brown IR; Xue WF; Rowe ML; Mulvihill DP; Prentice MB; Howard MJ; Warren MJ Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth. Biol 2014, 3, 454–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Liang M; Frank S; Lünsdorf H; Warren MJ; Prentice MB Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E. coli. Biotechnol. J 2017, 12, No. 1600415. [DOI] [PubMed] [Google Scholar]
- (15).Kirst H; Ferlez BH; Lindner SN; Cotton CAR; Bar-Even A; Kerfeld CA Toward a glycyl radical enzyme containing synthetic bacterial microcompartment to produce pyruvate from formate and acetate. Proc. Natl. Acad. Sci. U. S. A 2022, 119, No. e2116871119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhang G; Johnston T; Quin MB; Schmidt-Dannert C Developing a Protein Scaffolding System for Rapid Enzyme Immobilization and Optimization of Enzyme Functions for Biocatalysis. ACS Synth. Biol 2019, 8, 1867–1876. [DOI] [PubMed] [Google Scholar]
- (17).Wei Y; Wahome N; VanSlyke G; Whitaker N; Kumar P; Barta ML; Picking WL; Volkin DB; Mantis NJ; Middaugh CR Evaluation of lumazine synthase from Bacillus anthracis as a presentation platform for polyvalent antigen display. Protein Sci. 2017, 26, 2059–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Komatsu Y; Terasaka N; Sakai K; Mihara E; Wakabayashi R; Matsumoto K; Hilvert D; Takagi J; Suga H De novo peptide grafting to a self-assembling nanocapsule yields a hepatocyte growth factor receptor agonist. iScience 2021, 24, No. 103302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Levasseur MD; Mantri S; Hayashi T; Reichenbach M; Hehn S; Waeckerle-Men Y; Johansen P; Hilvert D Cell-Specific Delivery Using an Engineered Protein Nanocage. ACS Chem. Biol 2021, 16, 838–843. [DOI] [PubMed] [Google Scholar]
- (20).Edwardson TGW; Mori T; Hilvert D Rational Engineering of a Designed Protein Cage for siRNA Delivery. J. Am. Chem. Soc 2018, 140, 10439–10442. [DOI] [PubMed] [Google Scholar]
- (21).Azuma Y; Edwardson TGW; Terasaka N; Hilvert D Modular Protein Cages for Size-Selective RNA Packaging in Vivo. J. Am. Chem. Soc 2018, 140, 566–569. [DOI] [PubMed] [Google Scholar]
- (22).Azuma Y; Hilvert D Enzyme Encapsulation in an Engineered Lumazine Synthase Protein Cage. Methods Mol. Biol 2018, 1798, 39–55. [DOI] [PubMed] [Google Scholar]
- (23).Azuma Y; Bader DLV; Hilvert D Substrate Sorting by a Supercharged Nanoreactor. J. Am. Chem. Soc 2018, 140, 860–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Edwardson TGW; Levasseur MD; Hilvert D The OP Protein Cage: A Versatile Molecular Delivery Platform. Chimia (Aarau) 2021, 75, 323–328. [DOI] [PubMed] [Google Scholar]
- (25).Cannon KA; Park RU; Boyken SE; Nattermann U; Yi S; Baker D; King NP; Yeates TO Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering. Protein Sci. 2020, 29, 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Harcus TE; Gluckman M; Pontzer H; Raichlen DA; Marlowe FW; Siegfried WR; Macdonald IAW; Call J; Fischer J; Stryjewski KF; Quader S; Sorenson MD; Boogert N; Davies N; Flower T; Jamie G; Magrath R; Rendall D; Ruxton G; Sorensen M; Wood B; David C Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 2016, 353, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Giessen TW Encapsulins: Microbial nanocompartments with applications in biomedicine, nanobiotechnology and materials science. Curr. Opin. Chem. Biol 2016, 34, 1–10. [DOI] [PubMed] [Google Scholar]
- (28).Giessen TW; Silver PA Engineering carbon fixation with artificial protein organelles. Curr. Opin. Biotechnol 2017, 46, 42–50. [DOI] [PubMed] [Google Scholar]
- (29).Groaz A; Moghimianavval H; Tavella F; Giessen TW; Vecchiarelli AG; Yang Q; Liu AP Engineering spatiotemporal organization and dynamics in synthetic cells. WIREs: Nanomed. Nanobiotechnol 2021, 13, No. e1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lagoutte P; Mignon C; Stadthagen G; Potisopon S; Donnat S; Mast J; Lugari A; Werle B Simultaneous surface display and cargo loading of encapsulin nanocompartments and their use for rational vaccine design. Vaccine 2018, 36, 3622–3628. [DOI] [PubMed] [Google Scholar]
- (31).Giessen TW; Silver PA Converting a Natural Protein Compartment into a Nanofactory for the Size-Constrained Synthesis of Antimicrobial Silver Nanoparticles. ACS Synth. Biol 2016, 5, 1497–1504. [DOI] [PubMed] [Google Scholar]
- (32).Lee TH; Carpenter TS; D’haeseleer P; Savage DF; Yung MC Encapsulin carrier proteins for enhanced expression of antimicrobial peptides. Biotechnol. Bioeng 2020, 117, 603–613. [DOI] [PubMed] [Google Scholar]
- (33).Moon H; Lee J; Min J; Kang S Developing genetically engineered encapsulin protein cage nanoparticles as a targeted delivery nanoplatform. Biomacromolecules 2014, 15, 3794–3801. [DOI] [PubMed] [Google Scholar]
- (34).Giessen TW Encapsulins. Annu. Rev. Biochem 2022, 91, 353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Andreas MP; Giessen TW Large-scale computational discovery and analysis of virus-derived microbial nanocompartments. Nat. Commun 2021, 12, 4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sutter M; Boehringer D; Gutmann S; Günther S; Prangishvili D; Loessner MJ; Stetter KO; Weber-Ban E; Ban N Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol 2008, 15, 939–947. [DOI] [PubMed] [Google Scholar]
- (37).Ross J; McIver Z; Lambert T; Piergentili C; Bird JE; Gallagher KJ; Cruickshank FL; James P; Zarazúa-Arvizu E; Horsfall LE; Waldron KJ; Wilson MD; Logan Mackay C; Baslé A; Clarke DJ; Marles-Wright J Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment. Sci. Adv 2022, 8, No. eabj4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Nichols RJ; Lafrance B; Phillips NR; Radford DR; Oltrogge LM; Valentin-Alvarado LE; Bischoff AJ; Nogales E; Savage DF Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism. Elife 2021, 10, No. e59288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Lončar N; Rozeboom HJ; Franken LE; Stuart MCA; Fraaije MW Structure of a robust bacterial protein cage and its application as a versatile biocatalytic platform through enzyme encapsulation. Biochem. Biophys. Res. Commun 2020, 529, 548–553. [DOI] [PubMed] [Google Scholar]
- (40).Eren E; Wang B; Winkler DC; Watts NR; Steven AC; Wingfield PT Structural characterization of the Myxococcus xanthus encapsulin and ferritin-like cargo system gives insight into its iron storage mechanism. Structure 2022, 30, 551–563.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Akita F; Chong KT; Tanaka H; Yamashita E; Miyazaki N; Nakaishi Y; Suzuki M; Namba K; Ono Y; Tsukihara T; Nakagawa A The Crystal Structure of a Virus-like Particle from the Hyperthermophilic Archaeon Pyrococcus furiosus Provides Insight into the Evolution of Viruses. J. Mol. Biol 2007, 368, 1469–1483. [DOI] [PubMed] [Google Scholar]
- (42).Giessen TW; Orlando BJ; Verdegaal AA; Chambers MG; Gardener J; Bell DC; Birrane G; Liao M; Silver PA Large protein organelles form a new iron sequestration system with high storage capacity. Elife 2019, 8, No. e46070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Giessen TW; Silver PA Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat. Microbiol 2017, 2, 17029. [DOI] [PubMed] [Google Scholar]
- (44).Tracey JC; Coronado M; Giessen TW; Lau MCY; Silver PA; Ward BB The Discovery of Twenty-Eight New Encapsulin Sequences, Including Three in Anammox Bacteria. Sci. Rep 2019, 9, 20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Lien KA; Nichols RJ; Cassidy-Amstutz C; Dinshaw K; Knight M; Singh R; Eltis LD; Savage DF; Stanley SA A nanocompartment system contributes to defense against oxidative stress in Mycobacterium tuberculosis. Elife 2021, 10, No. e74358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Altenburg WJ; Rollins N; Silver PA; Giessen TW Exploring targeting peptide-shell interactions in encapsulin nanocompartments. Sci. Rep 2021, 11, 4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Cassidy-Amstutz C; Oltrogge L; Going CC; Lee A; Teng P; Quintanilla D; East-Seletsky A; Williams ER; Savage DF Identification of a Minimal Peptide Tag for in Vivo and in Vitro Loading of Encapsulin. Biochemistry 2016, 55, 3461–3468. [DOI] [PubMed] [Google Scholar]
- (48).Lau YH; Giessen TW; Altenburg WJ; Silver PA Prokaryotic nanocompartments form synthetic organelles in a eukaryote. Nat. Commun 2018, 9, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Sigmund F; Massner C; Erdmann P; Stelzl A; Rolbieski H; Desai M; Bricault S; Wörner TP; Snijder J; Geerlof A; Fuchs H; de Angelis MH; Heck AJR; Jasanoff A; Ntziachristos V; Plitzko J; Westmeyer GG Bacterial encapsulins as orthogonal compartments for mammalian cell engineering. Nat. Commun 2018, 9, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Jenkins MC; Lutz S Encapsulin Nanocontainers as Versatile Scaffolds for the Development of Artificial Metabolons. ACS Synth. Biol 2021, 10, 857–869. [DOI] [PubMed] [Google Scholar]
- (51).Lohner P; Zmyslia M; Thurn J; Pape JK; Gerasimaitė R; Keller-Findeisen J; Groeer S; Deuringer B; Süss R; Walther A; Hell SW; Lukinavičius G; Hugel T; Jessen-Trefzer C Inside a Shell—Organometallic Catalysis Inside Encapsulin Nanoreactors. Angew. Chem., Int. Ed 2021, 60, 23835–23841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Van de Steen A; Khalife R; Colant N; Mustafa Khan H; Deveikis M; Charalambous S; Robinson CM; Dabas R; Serna SE; Catana DA; Pildish K; Kalinovskiy V; Gustafsson K; Frank S Bioengineering bacterial encapsulin nanocompartments as targeted drug delivery system. Synth. Syst. Biotechnol 2021, 6, 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Gabashvili AN; Vodopyanov SS; Chmelyuk NS; Sarkisova VA; Fedotov KA; Efremova MV; Abakumov MA Encapsulin based self-assembling iron-containing protein nanoparticles for stem cells mri visualization. Int. J. Mol. Sci 2021, 22, 12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Efremova MV; Bodea SV; Sigmund F; Semkina A; Westmeyer GG; Abakumov MA Genetically encoded self-assembling iron oxide nanoparticles as a possible platform for cancer-cell tracking. Pharmaceutics 2021, 13, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Zhang Y; Wang X; Chu C; Zhou Z; Chen B; Pang X; Lin G; Lin H; Guo Y; Ren E; Lv P; Shi Y; Zheng Q; Yan X; Chen X; Liu G Genetically engineered magnetic nanocages for cancer magneto-catalytic theranostics. Nat. Commun 2020, 11, 5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Kanekiyo M; Bu W; Joyce MG; Meng G; Whittle JRR; Baxa U; Yamamoto T; Narpala S; Todd JP; Rao SS; McDermott AB; Koup RA; Rossmann MG; Mascola JR; Graham BS; Cohen JI; Nabel GJ Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 2015, 162, 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Choi B; Moon H; Hong SJ; Shin C; Do Y; Ryu S; Kang S Effective Delivery of Antigen-Encapsulin Nanoparticle Fusions to Dendritic Cells Leads to Antigen-Specific Cytotoxic T Cell Activation and Tumor Rejection. ACS Nano 2016, 10, 7339–7350. [DOI] [PubMed] [Google Scholar]
- (58).Michel-Souzy S; Hamelmann NM; Zarzuela-Pura S; Paulusse JMJ; Cornelissen JJLM Introduction of Surface Loops as a Tool for Encapsulin Functionalization. Biomacromolecules 2021, 22, 5234–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Adamson LSR; Tasneem N; Andreas MP; Close W; Jenner EN; Szyszka TN; Young R; Cheah LC; Norman A; MacDermott-Opeskin HI; O’Mara ML; Sainsbury F; Giessen TW; Lau YH Pore structure controls stability and molecular flux in engineered protein cages. Sci. Adv 2022, 8, No. eabl7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Williams EM; Jung SM; Coffman JL; Lutz S Pore Engineering for Enhanced Mass Transport in Encapsulin Nanocompartments. ACS Synth. Biol 2018, 7, 2514–2517. [DOI] [PubMed] [Google Scholar]
- (61).Choi H; Eom S; Kim HU; Bae Y; Jung HS; Kang S Load and Display: Engineering Encapsulin as a Modular Nanoplatform for Protein-Cargo Encapsulation and Protein-Ligand Decoration Using Split Intein and SpyTag/SpyCatcher. Biomacromolecules 2021, 22, 3028–3039. [DOI] [PubMed] [Google Scholar]
- (62).Jones JA; Cristie-David AS; Andreas MP; Giessen TW Triggered Reversible Disassembly of an Engineered Protein Nanocage. Angew. Chem., Int. Ed 2021, 60, 25034–25041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Comas-Garcia M; Colunga-Saucedo M; Rosales-Mendoza S; Comas-Garcia M The Role of Virus-Like Particles in Medical Biotechnology. Mol. Pharmaceutics 2020, 17, 4407–4420. [DOI] [PubMed] [Google Scholar]
- (64).Terasaka N; Azuma Y; Hilvert D Laboratory evolution of virus-like nucleocapsids from nonviral protein cages. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 5432–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Edwardson TGW; Hilvert D Virus-Inspired Function in Engineered Protein Cages. J. Am. Chem. Soc 2019, 141, 9432–9443. [DOI] [PubMed] [Google Scholar]
- (66).Tetter S; Terasaka N; Steinauer A; Bingham RJ; Clark S; Scott AJP; Patel N; Leibundgut M; Wroblewski E; Ban N; Stockley PG; Twarock R; Hilvert D Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein. Science 2021, 372, 1220–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Fang PY; Ramos LMG; Holguin SY; Hsiao C; Bowman JC; Yang HW; Williams LD Functional RNAs: Combined assembly and packaging in VLPs. Nucleic Acids Res. 2017, 45, 3519–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Choi KM; Kim K; Kwon IC; Kim IS; Ahn HJ Systemic delivery of siRNA by chimeric capsid protein: Tumor targeting and RNAi activity in vivo. Mol. Pharmaceutics 2013, 10, 18–25. [DOI] [PubMed] [Google Scholar]
- (69).Qazi S; Miettinen HM; Wilkinson RA; McCoy K; Douglas T; Wiedenheft B Programmed Self-Assembly of an Active P22-Cas9 Nanocarrier System. Mol. Pharmaceutics 2016, 13, 1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Ponchon L; Catala M; Seijo B; el Khouri M; Dardel F; Nonin-Lecomte S; Tisné C Co-expression of RNA-protein complexes in Escherichia coli and applications to RNA biology. Nucleic Acids Res. 2013, 41, No. e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Edvard Smith CI; Zain R Therapeutic oligonucleotides: State of the art. Annu. Rev. Pharmacol. Toxicol 2019, 59, 605–630. [DOI] [PubMed] [Google Scholar]
- (72).Lundin KE; Gissberg O; Smith CIE Oligonucleotide Therapies: The Past and the Present. Hum. Gene Ther 2015, 26, 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Whitehead KA; Langer R; Anderson DG Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov 2009, 8, 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Dowdy SF Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol 2017, 35, 222–229. [DOI] [PubMed] [Google Scholar]
- (75).Gantier MP; Williams BRG The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 2007, 18, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Iversen F; Yang C; Dagnæs-Hansen F; Schaffert DH; Kjems J; Gao S Optimized siRNA-PEG conjugates for extended blood circulation and reduced urine excretion in mice. Theranostics 2013, 3, 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Seeman NC DNA Nanotechnology at 40. Nano Lett. 2020, 20, 1477–1478. [DOI] [PubMed] [Google Scholar]
- (78).Lehto T; Ezzat K; Wood MJA; el Andaloussi S Peptides for nucleic acid delivery. Adv. Drug Deliv. Rev 2016, 106, 172–182. [DOI] [PubMed] [Google Scholar]
- (79).Ding Y; Jiang Z; Saha K; Kim CS; Kim ST; Landis RF; Rotello VM Gold nanoparticles for nucleic acid delivery. Mol. Ther 2014, 22, 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Calhoun LN; Kwon YM Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: A review. J. Appl. Microbiol 2011, 110, 375–386. [DOI] [PubMed] [Google Scholar]
- (81).Park C; Jin Y; Kim YJ; Jeong H; Seong BL RNA-binding as chaperones of DNA binding proteins from starved cells. Biochem. Biophys. Res. Commun 2020, 524, 484–489. [DOI] [PubMed] [Google Scholar]
- (82).Alam KK; Tawiah KD; Lichte MF; Porciani D; Burke DH A Fluorescent Split Aptamer for Visualizing RNA-RNA Assembly in Vivo. ACS Synth. Biol 2017, 6, 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Wang Y; Uchida M; Waghwani HK; Douglas T Synthetic Virus-like Particles for Glutathione Biosynthesis. ACS Synth. Biol 2020, 9, 3298–3310. [DOI] [PubMed] [Google Scholar]
- (84).Kaczmarczyk SJ; Sitaraman K; Young HA; Hughes SH; Chatterjee DK Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. U. S. A 2011, 108, 16998–17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Banskota S; Raguram A; Suh S; Du SW; Davis JR; Choi EH; Wang X; Nielsen SC; Newby GA; Randolph PB; Osborn MJ; Musunuru K; Palczewski K; Liu DR Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 2022, 185, 250–265.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Glassman PM; Muzykantov VR Pharmacokinetic and pharmacodynamic properties of drug delivery systems. J. Pharmacol. Exp. Ther 2019, 370, 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Kaczmarek JC; Kowalski PS; Anderson DG Advances in the delivery of RNA therapeutics: From concept to clinical reality. Genome Med. 2017, 9, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Herbert FC; Brohlin OR; Galbraith T; Benjamin C; Reyes CA; Luzuriaga MA; Shahrivarkevishahi A; Gassensmith JJ; Gassensmith JJ Supramolecular Encapsulation of Small-Ultrared Fluorescent Proteins in Virus-Like Nanoparticles for Noninvasive in Vivo Imaging Agents. Bioconjugate Chem. 2020, 31, 1529–1536. [DOI] [PubMed] [Google Scholar]
- (89).Tagit O; de Ruiter MV; Brasch M; Ma Y; Cornelissen JJLM Quantum dot encapsulation in virus-like particles with tuneable structural properties and low toxicity. RSC Adv. 2017, 7, 38110–38118. [Google Scholar]
- (90).Zhang J; Cheng D; He J; Hong J; Yuan C; Liang M Cargo loading within ferritin nanocages in preparation for tumor-targeted delivery. Nat. Protoc 2021, 16, 4878–4896. [DOI] [PubMed] [Google Scholar]
- (91).Stupka I; Azuma Y; Biela AP; Imamura M; Scheuring S; Pyza E; Woźnicka O; Maskell DP; Heddle JG Chemically induced protein cage assembly with programmable opening and cargo release. Sci. Adv 2022, 8, No. eabj9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Austin RJ; Xia T; Ren J; Takahashi TT; Roberts RW Designed arginine-rich RNA-binding peptides with picomolar affinity. J. Am. Chem. Soc 2002, 124, 10966–10967. [DOI] [PubMed] [Google Scholar]
- (93).Hyun S; Lee KH; Han A; Yu J An RNA aptamer that selectively recognizes symmetric dimethylation of arginine 8 in the histone H3N-terminal peptide. Nucleic Acid Ther. 2011, 21, 157–163. [DOI] [PubMed] [Google Scholar]
- (94).Puglisi JD; Chen L; Blanchard S; Frankel AD Solution structure of a bovine immunodeficiency virus Tat-TAR peptide-RNA complex. Science 1995, 270, 1200–1203. [DOI] [PubMed] [Google Scholar]
- (95).Cubillos-Ruiz A; Guo T; Sokolovska A; Miller PF; Collins JJ; Lu TK; Lora JM Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov 2021, 20, 941–960. [DOI] [PubMed] [Google Scholar]
- (96).Gurbatri CR; Lia I; Vincent R; Coker C; Castro S; Treuting PM; Hinchliffe TE; Arpaia N; Danino T Engineered probiotics for local tumor delivery of checkpoint blockade nano-bodies. Sci. Transl. Med 2020, 12, No. eaax0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Leventhal DS; Sokolovska A; Li N; Plescia C; Kolodziej SA; Gallant CW; Christmas R; Gao J; James MJ; Abinfuentes A; Momin M; Bergeron C; Fisher A; Miller PF; West KA; Lora JM Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun 2020, 11, 2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Bar-Zion A; Nourmahnad A; Mittelstein DR; Shivaei S; Yoo S; Buss MT; Hurt RC; Malounda D; Abedi MH; Lee-Gosselin A; Swift MB; Maresca D; Shapiro MG Acoustically triggered mechanotherapy using genetically encoded gas vesicles. Nat. Nanotechnol 2021, 16, 1403–1412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.