Abstract
Animals display significant inter-species variation in the rate of embryonic development despite broad conservation of the overall sequence of developmental events. Differences in biochemical reaction speeds, including the rates of protein production and degradation, are thought to be responsible for species-specific rates of development [1–3]. However, the cause of differential biochemical reaction speeds between species remains unknown. Using pluripotent stem cells, we have established an in vitro system that recapitulates the two-fold difference in developmental rate between mouse and human embryos. This system provides a quantitative measure of developmental speed as revealed by the period of the segmentation clock, a molecular oscillator associated with the rhythmic production of vertebral precursors. Using this system, we showed that mass-specific metabolic rates scale with developmental rate and are therefore elevated in mouse cells compared to human cells. We further showed that reducing these metabolic rates by inhibiting the electron transport chain slowed down the segmentation clock by impairing the cellular NAD+/NADH redox balance and, further downstream, lowering the global rate of protein synthesis. Conversely, increasing the NAD+/NADH ratio in human cells by overexpression of the NADH oxidase LbNOX increased translation rate and accelerated the segmentation clock. These findings represent a starting point for the manipulation of developmental rate, with multiple translational applications including the acceleration of human PSCs differentiation for disease modeling and cell-based therapies.
The rate of embryonic development across animal taxa varies from species to species and is correlated with lifespan, body size, and other life history traits [4, 5]. In mammals, large-bodied species develop at slower rates and display increased lifespans (e.g. humans) compared to small-bodied animals (e.g. mice) [6]. Even though early mouse and human embryos undergo the same series of developmental steps and share a similar overall size, human embryos do so at rates 2-3 times slower [7]. The segmentation clock represents an ideal model to study developmental rate because its period is species-specific, temperature-sensitive, and it scales with the speed of embryonic development [1, 8, 9]. This clock consists of a molecular oscillator that operates in the precursors of the musculo-skeletal system in the presomitic mesoderm (PSM), where it controls somite formation periodicity [10].
In vitro model of developmental rate
We previously reported the establishment of pluripotent stem cell (PSC)-derived models of the mouse and human segmentation clocks [11]. We first developed an updated protocol to differentiate mouse and human PSCs towards PSM fate under identical conditions (Fig. 1a). Differentiation efficiency was remarkably high with 78.3 ± 1% of mouse and 96.5 ± 1.5% of human cells expressing the posterior PSM-specific marker MSGN1-Venus (Fig. 1b, Extended Data Fig. 1a). Mouse cells activated MSGN1-Venus with accelerated kinetics compared to human cells (1-2 days for mouse vs. 2-3 days for human) (Fig. 1b). The cell cycle duration was also shorter in mouse than in human PSM cells (13.9 ± 2 vs. 21.9 ± 3.6 hours) (Fig. 1c). Oscillations of the segmentation clock reporter HES7-Achilles were faster in mouse than in human PSM cells (2.6 ± 0.3 vs. 4.9 ± 0.3 hours) (Fig. 1d–e, Supplementary Videos 1–2) [11–14]. Importantly, the cell cycle time and segmentation clock period of PSC-derived mouse PSM cells did not significantly differ from primary PSM cells from E9.5 embryos (clock period: 2.5 ± 0.3 hours vs. 2.6 ± 0.1 hours, p=0.314; cell cycle: 13.9 ± 2 hours vs. 13.07 ± 1 hour, p=0.057) (Extended Data Fig. 1b–c) [15]. Together, these observations indicated an approximately two-fold difference in developmental rate between mouse and human PSM cells differentiating in vitro.
Figure 1.
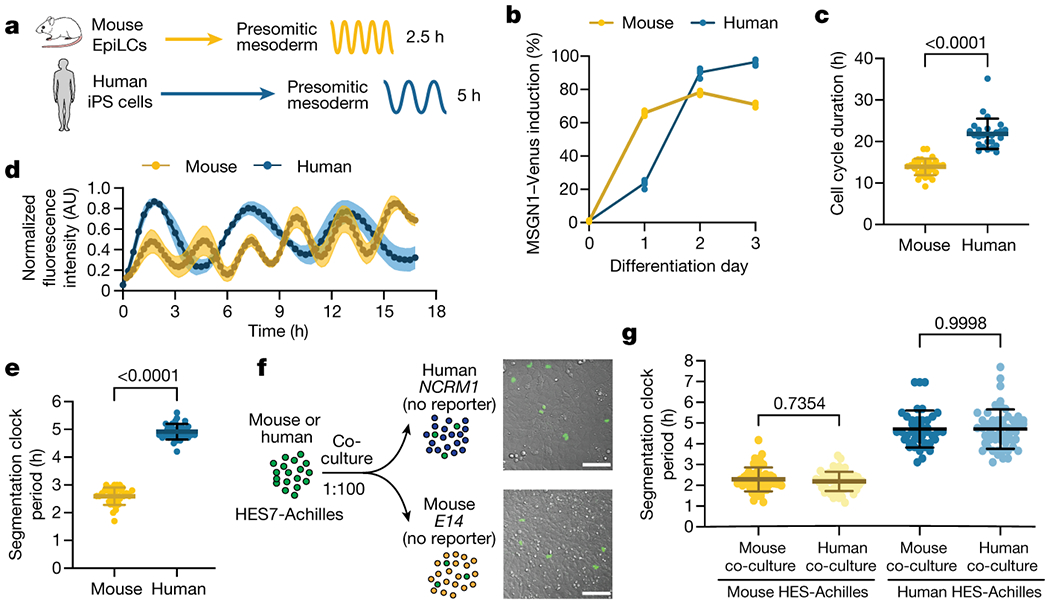
Cell-autonomous differences in developmental rate between differentiating mouse and human PSM cells
a. Schematic illustrating the differentiation of mouse and human PSCs towards PSM fate. The accelerated developmental pace of mouse cells is reflected in the reduced induction time and short oscillatory period relative to human cells. EpiLCs = Epiblast-Like Cells, iPSCs = induced Pluripotent Stem Cells.
b. PSM induction efficiency over the course of 3 days of differentiation for mouse and human PSCs. The percentage of cells expressing MSGN1-Venus was assessed by flow cytometry. n=5 independent experiments.
c. Duration of the cell cycle in hours for PSC-derived mouse and human PSM cells. Mean ±SD. n=33 (mouse); n=26 (human). Unpaired two-sided t-test: p=2.88x10−15
d. HES7-Achilles oscillation profiles for PSC-derived mouse and human PSM cells over the course of 18 hours. Mean ±SEM. n=5 independent experiments.
e. Period of HES7-Achilles oscillations in PSC-derived mouse and human PSM cells. Mean ±SD. n=25. Unpaired two-sided t-test: p=7.33x10−41
f. Left: Experimental strategy for the co-culture of CAG-H2B-mCherry; HES7-Achilles human or CAG-NLS-BFP; Hes7-Achilles mouse PSM cells with non-reporter mouse (E14) or human (NCRM1) PSM cells at a ratio of 1:100.
Right: Merged brightfield and human HES7-Achilles fluorescence images of human-human (top) and human-mouse (bottom) co-cultures. Scale bar = 100μm.
g. Period of HES7-Achilles oscillations in mouse (left) or human (right) HES7-Achilles PSM cells co-cultured with an excess of either mouse or human non-reporter PSM cells. Mean ±SD. n=56 (mouse-mouse), n=56 (mouse-human), n=41 (human-mouse), n=53 (human-human). One-way ANOVA with Šidák correction.
In vitro, the segmentation clock retains its species-specific period in isolated cells [1, 11, 15]. When we performed homo- or hetero-specific co-cultures of individual mouse Hes7-Achilles reporter PSM cells mixed with unlabeled PSM cells of either human or mouse origin at a ratio of 1:100 (Fig. 1f), the segmentation clock period remained unchanged (2.29 ± 0.57 hours vs. 2.19 ± 0.46 hours, p=0.73) (Fig. 1g, Extended Data Fig. 1d, f, h–I, Supplementary Video 3). This was also true for individual human reporter cells co-cultured with an excess of either human or mouse PSM cells (4.71 ± 0.94 hours vs. 4.71 ± 0. 88 hours, p=0.99) (Fig. 1g. Extended Data Fig. 1e–f, j–k, Supplementary Video 4). Although interspecies co-culture conditions gave rise to variable effects in oscillation amplitude (Extended Data Fig. 1g, h–k), these results indicated that the segmentation clock period is controlled cell autonomously.
The cell cycle has been proposed to function as a clock that controls developmental speed [2, 16]. However, treating human PSM cells with aphidicolin led to near-complete cell cycle arrest but did not impact the oscillatory period (4.93 ± 0.47 hours vs. 4.88 ± 0.31 hours, p=0.7378) (Extended Data Fig. 2a–b). Thus, the cell cycle does not contribute to the regulation of the segmentation clock period.
Species-specific metabolic rates
According to Kleiber’s law, mass-specific metabolic rates scale allometrically with adult body mass [17]. Gestation length also scales with adult body mass [18], suggesting that differences in basal metabolism could potentially explain the accelerated biochemical kinetics associated with faster development in mouse compared to human cells [1, 2]. When comparing similar numbers of PSM cells, basal glycolytic proton efflux rate (glycoPER) was not different between mouse and human cells (8.9x10−3 ± 6.3x10−4 vs. 8.8x10−3 ± 4.4x10−4 pmol/min/cell, p=0.783) while basal oxygen consumption rate (OCR) was slightly elevated in human cells (1.01x10−3 ± 4.3x10−5 vs. 1.09x10−3 ± 4.5x10−5 pmol/min/cell, p=0.0002) (Extended Data Fig. 2c–d).
However, we noted that human PSM cells were approximately twice as large as mouse cells (2060 ± 524 fL vs. 885.9 ± 157.9 fL, p<0.0001) (Fig. 2a; Extended Data Fig. 2e). When measured with the suspended microchannel resonator [31, 32], the total mass of human cells was also approximately twice that of mouse cells (2002 ± 71 pg vs. 1066 ± 56 pg. p<0.0001) (Fig. 2b; Supplementary Table 2), such that density was qualitatively similar for both species (1.057 ± 0.003 pg/fL vs. 1.061 ± 0.002 pg/fL, p=0.03) (Extended Data Fig. 2f, Supplementary Table 2). We also measured dry mass, dry volume and dry density of single cells by comparing the cell’s buoyant mass in media prepared with either normal water (H2O) or deuterium-based heavy water (D2O) [19, 20]. Dry mass of mouse PSM cells was less than half that of human cells (162 ± 5.7 pg vs. 380 ± 29.6 pg, p=0.0002), as was dry volume (113 ± 5 fL vs. 278 ± 24 fL, p=0.0003) (Extended Data Fig. 2g–h, Supplementary Table 2). Dry mass density, which depends on the molecular composition of the cell, was thus qualitatively similar between the two species (1.435 ± 0.015 pg/fL vs. 1.365 ± 0.012 pg/fL, p=0.004) (Extended Data Fig. 2i, Supplementary Table 2). Thus, human cells contained twice as much biological material as mouse cells. Consequently, normalization by either volume or mass is required to correct for cell size difference between the two species. Mass-specific OCR (1.013x10−6 ± 4.333x10−8 vs. 5.475x10−7 ± 2.272x10−8 pmol/min/pg, p<0.0001) and glycoPER (8.905x10−6 ± 6.372x10−7 vs. 4.425x10−6 ± 2.212x10−7 pmol/min/pg, p<0.0001) were twice as high in mouse than in human cells (Fig. 2c–d). The extracellular acidification rate (ECAR), which accounts for glycolytic and other acidification sources such as CO2 secretion, was also twice as high per unit mass in mouse compared to human cells (5.685x10−7 ± 3.493x10−8 vs. 2.805x10−7 ± 1.315x10−8 mpH/min/pg) (Extended Data Fig. 2j).
Figure 2.
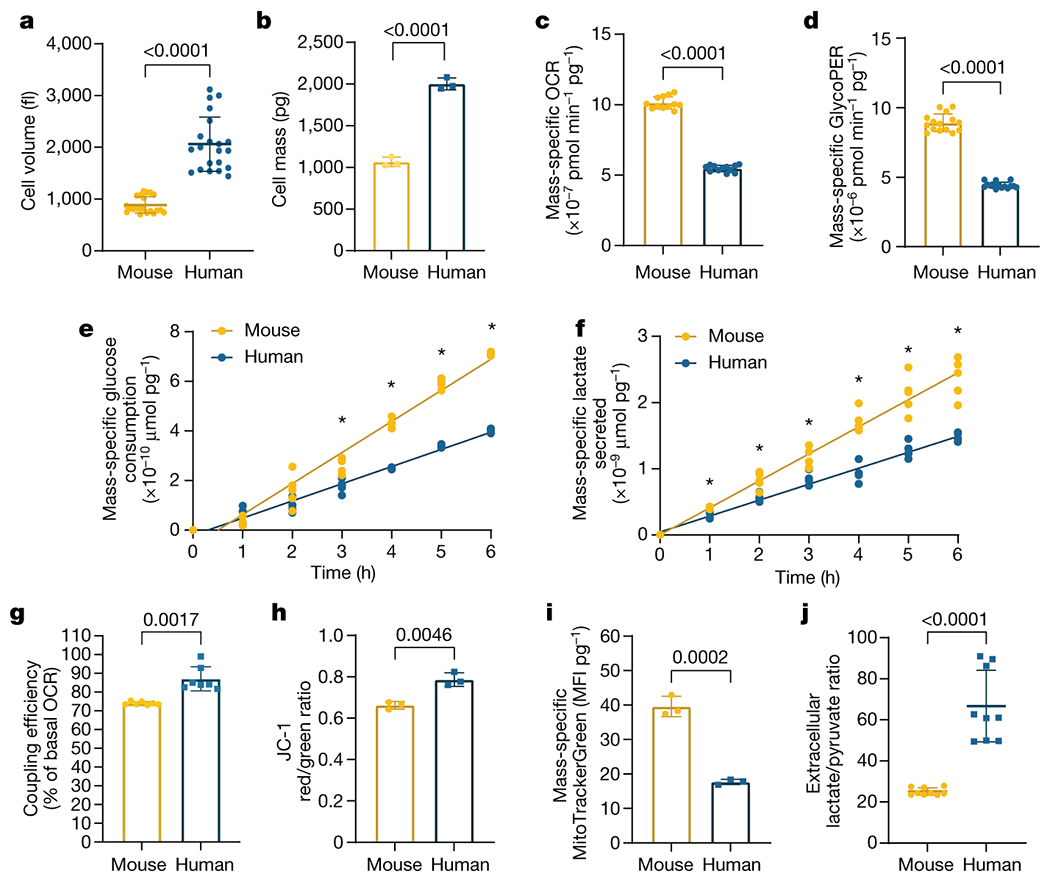
Elevated mass-specific metabolic rates in mouse PSM cells compared to human PSM cells
a. Volume of MSGN1-Venus+ PSC-derived mouse and human PSM cells as measured with a coulter counter. Mean ±SD. n=21. Unpaired two-sided t-test: p=3.25x10−12
b. Total cell mass of MSGN1-Venus+ PSC-derived mouse and human PSM cells as measured on a suspended microchannel resonator. Each datapoint represents the mean of >200 individual cells. Mean ±SD. n=3 independent experiments. Unpaired two-sided t-test: p=5.77x10−5
c. Mass-specific oxygen consumption rate for MSGN1-Venus+ PSC-derived mouse and human PSM cells. Mean ±SD. n=12. Unpaired two-sided t-test: p=3.19x10−20
d. Mass-specific glycolytic proton efflux rate for MSGN1-Venus+ PSC-derived mouse and human PSM cells. Mean ±SD. n=15. Unpaired two-sided t-test: p=5.05x10−21
e. Mass-specific cumulative glucose consumption for MSGN1-Venus+ PSC-derived mouse and human PSM cells. n=5. *denotes p<0.05, multiple unpaired two-sided t-tests with FDR=1%, p values: 1 hour=0.6074, 2 hours=0.0691, 3 hours=0.0013, 4 hours=7.44x10−8, 5 hours=2.29x10−9, 6 hours=3.58x10−12.
f. Mass-specific cumulative lactate secretion for MSGN1-Venus+ PSC-derived mouse and human PSM cells. n=5. *denotes p<0.05, multiple unpaired two-sided t-tests with FDR=1%, p values: 1 hour=0.00364, 2 hours=0.00246, 3 hours=0.0128, 4 hours=0.000039, 5 hours=0.000259, 6 hours=0.000172
g. Coupling efficiency shown as the percent of basal oxygen consumption linked to ATP production in MSGN1-Venus+ PSC-derived mouse and human PSM cells. Mean ±SD. n=7 biological replicates. Unpaired two-sided t-test.
h. Inner mitochondrial membrane potential (ΔΨm) in PSC-derived mouse and human PSM cells as measured by the ratiometric JC-1 dye. Mean ±SD. n=4 biological replicates. Unpaired two-sided t-test.
i. Mass-specific mitochondrial content (MitoTracker Green) in PSC-derived mouse and human PSM cells. Mean ±SD. n=6 biological replicates. Unpaired two-sided t-test
j. Extracellular lactate to pyruvate ratio in PSC-derived mouse and human PSM cells, which reflects the cytosolic NADH/NAD+ ratio. Mean ±SD. n=9. Unpaired two-sided t-test with Welch’s correction: p=9.02x10−5
Rates of glucose consumption (mouse: 1.252e-10, 95% CI: 1.172e-10 to 1.333e-10 vs. human: 6.922e-11, 95% CI: 6.524e-11 to 7.319e-11; p<0.0001 μmoles/pg/hour) and lactate secretion (mouse: 4.085e-010, 95% CI: 3.777e-10 to 4.394e-10 vs. human: 2.410e-10, 95% CI: 2.249e-10 to 2.571e-10, p<0.0001 μmoles/pg/hour) per unit mass were significantly higher in mouse cells (Fig. 2e–f). These data are consistent with the Warburg-like metabolism of PSM cells, wherein active glycolysis producing large amounts of lactate coexists with aerobic respiration [21, 22]. Mass-specific glutamine consumption was also significantly elevated in mouse PSM cells (0.00136 ± 0.00022 vs. 0.00078 ± 0.0002237 RLU/pg, p=0.0103) (Extended Data Fig. 2k).
We compared the metabolic rates of mouse and human neural progenitors differentiated in vitro from PSCs. PAX6+ neural progenitors were induced after 5 days for mouse and 7 days for human PSCs (Extended Data Fig. 2l–m). We did not detect significant volume differences between mouse and human neural progenitors (1255 ± 88 fL vs. 1203 ± 86 fL, p=0.5101) (Extended Data Fig. 2n). However, mass-specific OCR (8.437x10−7 ± 6.779x10−8 vs. 1.449x10−7 ± 1.859x10−8 pmol/min/pg, p<0.0001) and ECAR (2.99x10−7 ± 5.781x10−8 vs. 1.76x10−7 ± 2.521x10−8 mpH/min/pg, p<0.0001) were significantly higher in mouse (Extended Data Fig. 2o–p). Thus, mass-specific metabolic rates are higher in mouse PSM and neural progenitor cells compared to equivalent human cells.
We next performed stable isotope tracing with uniformly labeled [U-13C6]-Glucose and [U-13C5]-Glutamine in mouse and human PSM cells. Temporal labeling profiles were qualitatively similar between both species and labeling patterns for most metabolites were stabilized by 24 hours (Extended Data Fig. 3a–f; Supplementary Table 3). At isotopic steady state, glucose tracing led to high labeling levels for pyruvate and lactate, as well as partial labeling of TCA intermediates (Extended Data Fig. 3g–l). Glutamine tracing showed intermediate labeling levels for glutamate and TCA metabolites, but not pyruvate or lactate, supporting an anaplerotic role in the TCA cycle (Extended Data Fig. 3m–r). Importantly, stable isotope labeling patterns were almost identical between species, indicating that only the rate of glucose and glutamine utilization by downstream metabolic pathways differs between mouse and human.
We next performed mitochondrial stress tests using a Seahorse instrument. Mouse and human PSM cells differed in their maximal mass-specific respiration rate (2.117x10−6 ± 1.559x10−7 vs. 4.467x10−7 ± 5.737x10−8 pmol/min/pg, p<0.0001) and their spare respiratory capacity (192 ± 11.6 vs. 119.7 ± 4.2 percent of basal OCR, p<0.0001), which were significantly higher in mouse cells (Extended Data Fig. 4a–b). Respiration was less coupled to ATP production in mouse cells than in human cells (74 ± 0.9 vs. 87 ± 6.4 percent of basal OCR, p=0.0017), reflecting a higher proton leak (Fig. 2g) [23]. Measurement of the inner mitochondrial membrane potential (ΔΨm) with the ratiometric dye JC-1 revealed lower ΔΨm in mouse cells than in human cells (0.66 ± 0.018 vs. 0.78 ± 0.033 red/green fluorescence ratio, p=0.0046) (Fig. 2h). Staining differentiated cells with MitoTracker Green revealed that the mitochondrial content per unit mass is approximately twice higher in mouse than in human cells (39.5 ± 2.9 vs. 17.7 ± 0.7 MFI/pg, p=0.0002) (Fig. 2i). Moreover, when we isolated mitochondria from PSM cells and cultured them with Complex I substrates (i.e. pyruvate and malate), mouse mitochondria OCR was consistently elevated compared to human (Extended Data Fig. 4c). Thus, mitochondrial abundance and inherent differences in mitochondrial properties contribute to the elevated respiration in mouse PSM cells [24].
ETC impairment slows down the clock
In the chicken embryo, inhibition of respiration but not glycolysis alters segmentation clock oscillations [21]. We thus partially impaired the electron transport chain (ETC) in human PSM cells, using inhibitors at sub-lethal concentrations, and measured the effect on the segmentation clock (Extended Data Fig. 4f). Treatment with inhibitors of ETC complexes I (rotenone), II (atpenin A5), III (antimycin A) and IV (sodium azide) (Fig. 3a), led to basal respiration decrease by more than 50% (control: 100 ± 21.6%, 20nM rotenone: 41.5 ± 8.3%, 50nM atpenin: 44.9 ± 9.6%, 100nM antimycin A: 20.4 ± 6.4, 1mM sodium azide: 9.5 ± 3% of control, p<0.0001 in all cases) (Extended Data Fig. 5a). These inhibitors led to premature arrest of segmentation clock oscillations (control: 5.3 ± 0.6, 20nM rotenone: 3.5 ± 0.5, 50nM atpenin: 3.5 ± 0.8, 100nM antimycin A: 3.1 ± 0.3, 1mM sodium azide: 3.2 ± 0.6 oscillations in 25 hours, p<0.001 in all cases) with variable effects on oscillation amplitude (Extended Data Fig. 5b–c, e–h, q–t). We observed a significant lengthening of the segmentation clock period (control: 4.86 ± 0.36h, 20nM rotenone: 5.56 ± 0.31h, 50nM atpenin: 5.66 ± 0.48h, 100nM antimycin A: 6.05 ± 0.31h, 1mM sodium azide: 6.57 ± 0.42h, p<0.0001 in all cases) (Fig. 3b, Extended Data Fig. 5d, k–n, Supplementary Video 5). As ETC impairment leads to cell cycle arrest [25], we could not measure cell cycle length. Thus, decreasing metabolic rate can decrease the clock period.
Figure 3.
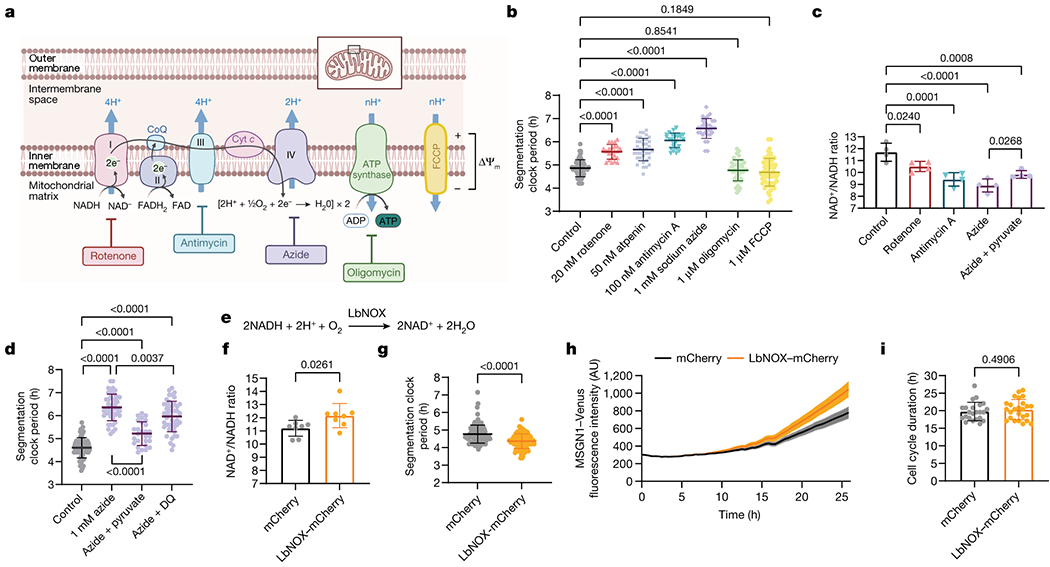
Regulation of the segmentation clock by the NAD+/NADH ratio
a. Electron transport chain and relevant small molecule inhibitors. Adapted from “Electron Transport Chain” by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates
b. HES7-Achilles oscillatory period in human PSM cells treated with DMSO control (n=53), 20nM rotenone (n=23), 50nM atpenin A5 (n=36), 100nM antimycin A (n=26), 1mM sodium azide (n=30), 1μM oligomycin (n=44), and 1μM FCCP (n=55) for 24 hours. Mean ±SD. One-way ANOVA with Šidák correction: rotenone p=1.1x10−8, atpenin p=6.4x10−14, antimycin p=1.85x10−23, azide p=1.88x10−42.
c. Whole-cell NAD+/NADH ratio in human PSM cells treated with DMSO control, 20nM rotenone, 100nM antimycin A, 1mM sodium azide, and 1mM sodium azide with 1mM sodium pyruvate for 24 hours. Mean ±SD. n=4 biological replicates. One-way ANOVA with Dunnett correction: control vs. azide p=1.1x10−5
d. HES7-Achilles oscillatory period in human PSM cells treated with DMSO control (n=67),1mM sodium azide alone (n=46), azide with 1mM sodium pyruvate (n=27), and azide with 5μM duroquinone (n=46). Mean ±SD. One-way ANOVA with Tukey’s correction: control vs. azide p=7.4x10−14, control vs. azide+pyr p=1.2x10−5, control vs. azide+DQ p=7.5x10−14, azide vs. azide+pyr p=1.4x10−13
e. NADH oxidation reaction catalyzed by LbNOX [28].
f. Whole-cell NAD+/NADH ratio in human PSM cells transduced with a lentivirus expressing either mCherry alone or LbNOX with mCherry. Mean ±SD. n=8 biological replicates. Unpaired two-sided t-test.
g. HES7-Achilles oscillatory period in human PSM cells transduced with a lentivirus expressing either mCherry alone (n=113) or LbNOX with mCherry (n=116). Mean ±SD. Unpaired two-sided t-test: p=3.7x10−10
h. MSGN1-Venus fluorescence during days 1–2 of human PSM differentiation, transduced with a lentivirus expressing either mCherry alone or LbNOX with mCherry. Mean ±SEM. n=7 biological replicates.
i. Cell cycle length in human PSM cells transduced with a lentivirus expressing either mCherry alone (n=23) or LbNOX with mCherry (n=25). Mean ±SD. Unpaired two-sided t-test.
ETC activity builds up a proton gradient across the inner mitochondrial membrane that powers oxidative phosphorylation. Inhibiting ATP synthase with oligomycin led to premature arrest of oscillations (5.3 ± 0.6 vs. 3.6 ± 1 oscillations in 25 hours, p<0.0001) and reduced oscillation amplitude (99.7 ± 29.4% vs. 42.9 ± 11.6% of control, p<0.0001) (Extended Data Fig. 5b–c, i, u). Surprisingly, this did not alter the period (4.86 ± 0.36h vs. 4.76 ±0.45h, p=0.8541) (Fig. 3b, Extended Data Fig. 5d, o), despite significant reduction of OCR (100 ± 21.6% vs. 56.7 ± 22.8% of control, p<0.001) (Extended Data Fig. 5a). Treatment with the ionophore FCCP decreased ΔΨm (0.99 ± 0.02 vs. 0.11 ± 0.01 fluorescence ratio, p<0.0001), uncoupling ETC activity from ATP production (Extended Data Fig. 5w). In cells treated with 1μM FCCP for 24 hours, oxygen consumption levels were indistinguishable from control cells (100 ± 21.6% vs. 99.5 ± 17.2% of control, p>0.999), representing entirely leak respiration (Extended Data Fig. 5a). FCCP-treated cells did not show premature arrest of oscillations (5.3 ± 0.6 vs. 5.1 ± 4 oscillations in 25 hours, p=0.968) but displayed decreased amplitude (99.7 ± 29.4% vs. 61.9 ± 15.% of control, p=0.0047) (Extended Data Fig. 5b–c, j, v). FCCP did not impact the clock period (4.86 ± 0.36h vs. 4.68 ± 0.59h, p=0.1849) (Fig. 3b; Extended Data Fig. 5d, p). Thus, ETC activity, rather than oxidative phosphorylation, is involved in controlling the segmentation clock period.
These results suggested that cellular ATP levels are not involved in regulating the clock period. In fact, ADP/ATP ratio was higher in mouse PSM cells than in human cells (0.42 ± 0.06 vs. 0.22 ± 0.03, p<0.0001) (Extended Data Fig. 4d), suggesting that an increased cellular energy charge does not mediate the accelerated developmental rate of mouse cells. To test this hypothesis, we cultured human PSM cells under conditions increasing cellular ATP concentration. First, we supplemented the media with succinate to directly feed the ETC at complex II. Second, we replaced glucose with galactose in the medium to increase reliance on oxidative phosphorylation for ATP production. Third, we treated the cells with the pyruvate dehydrogenase kinase (PDK) inhibitor dichloroacetate (DCA), to increases the conversion of pyruvate to acetyl-CoA for consumption by the TCA cycle (Extended Data Fig. 6a). Under all three conditions, ATP levels increased (control: 0.005826 ± 0.00027, 25mM succinate: 0.006776 ± 0.00027, 10mM galactose: 0.007315 ± 0.00009, 6.25mM DCA: 0.006778 ± 0.00036 μg ATP/well, p<0.0001 in all cases)(Extended Data Fig. 6b) along with OCR/ECAR ratio (control: 1.757 ± 0.24, 25mM succinate: 2.03 ± 0.14 p=0.0016, 10mM galactose: 10.08 ± 1.99 p<0.0001, 6.25mM DCA: 2.03 ± 0.21 p=0.0033 OCR/ECAR ratio)(Extended Data Fig. 6c–e). Replacement of glucose by galactose decreased ECAR compared to control and other treatments (control: 49.21 ± 4.07, 25mM succinate: 46.35 ± 1.91 p=0.0099, 10mM galactose: 7.92 ± 1.06 p<0.0001, 6.25mM DCA: 31.76 ± 2.66 p<0.0001 mpH/min/well) (Extended Data Fig. 6e), such that glycolytic ATP production was strongly downregulated (control: 72.04 ± 0.95, 25mM succinate: 65.96 ± 0.82, 10mM galactose: 16.82 ± 2.37, 6.25mM DCA: 67.02 ± 1.5 %glycoATP, p<0.0001 in all cases) (Extended Data Fig. 6f). The clock period in cells treated with succinate, galactose, or DCA was moderately lengthened (control: 4.73 ± 0.4h, 25mM succinate: 5.07 ± 0.41h p=0.0111, 10mM galactose: 5.28 ± 0.52h p<0.0001, 6.25mM DCA: 5.22 ± 0.38h p<0.0001) (Extended Data Fig. 6g–m, q). Although amplitude was decreased in galactose and DCA-treated cells (control: 99.59 ± 19.6, 25mM succinate: 81.67 ± 17.9 p=0.0715, 10mM galactose: 67.45 ± 18.5 p=0.0018, 6.25mM DCA: 79.29 ± 16.6 p=0.0347 percent of control) (Extended Data Fig. 6n–p, r), oscillations persisted normally (control: 5.26 ± 0.4, 25mM succinate: 5 ± 0, 10mM p=0.2647, galactose: 5 ± 0, 6.25mM p=0.3537, DCA: 5.25 ± 0.4 p=0.9999 oscillations in 25 hours) (Extended Data Fig. 6s). Cell cycle length was also increased (control: 20.84 ± 3.1h, 25mM succinate: 23.04 ± 3.8h p=0.0105, 10mM galactose: 28.66 ± 3.7h p<0.0001, 6.26mM DCA: 23.66 ± 2.9h p=0.0024) (Extended Data Fig. 6t). Together, these results confirmed that higher ATP concentrations do not promote faster oscillations or proliferation.
NAD+/NADH regulates the clock period
In cancer cells, PDK inhibition also decreases cellular proliferation [26]. This activates pyruvate dehydrogenase, shunting pyruvate away from lactate dehydrogenase (LDH), leading to a lower NAD+/NADH ratio (Extended Data Fig. 6a). Consequently, complex I of the ETC cannot sufficiently regenerate cellular NAD+ because the inner mitochondrial membrane becomes hyperpolarized and opposes the pumping of additional protons. Supplementing cells with pyruvate, which can be rapidly reduced by LDH to generate NAD+, rescues the proliferation rate of cells treated with a PDK inhibitor [26]. Also, restoring ETC activity by dissipating the elevated ΔΨm with uncoupling agents like FCCP rescues the NAD+/NADH ratio and proliferation rate [26]. A similar mechanism may be responsible for the increased segmentation clock period in PSM cells subjected to PDK inhibition. We observed significantly lower total NAD+/NADH ratio (11.59 ± 0.7 vs. 9.63 ± 0.5, p=0.0003) (Extended Data Fig. 7a) and increased ΔΨm (0.99 ± 0.02 vs. 1.12 ± 0.05, p=0.038) (Extended Data Fig. 7b) in PSM cells treated with 6.25mM DCA. The NAD+/NADH ratio could be restored to control levels by pyruvate supplementation or FCCP treatment (control: 11.59 ± 0.7, DCA+Pyr: 12.4 ± 0.09 p=0.084, DCA+10nMFCCP: 12.02 ± 0.3 p=0.4634) (Extended Data Fig. 7a).
These total NAD+/NADH ratio measurements included both mitochondrial and cytoplasmic NAD(H) pools. We generated a human PSC line carrying the fluorescent sensor Peredox, which displays higher fluorescence when cytoplasmic -but not mitochondrial- levels of NADH are increased [27] (Extended Data Fig. 7c–d). Using this reporter line, we confirmed the changes in cytosolic NAD+/NADH ratio upon DCA treatment and its restoration by pyruvate and FCCP (control: 1.053 ± 0.04, DCA: 1.114 ± 0.02 p=0.0074, DCA+Pyr: 1.08 ± 0.01 p=0.2425, DCA+10nM FCCP: 1.07 ± 0.02 p=0.6327 Peredox/mCherry ratio) (Extended Data Fig. 7e). Importantly, the clock period in DCA-treated cells was fully rescued by both pyruvate and FCCP (control: 4.63 ± 0.4h, DCA: 4.9 ± 0.4h p=0.0001, DCA+Pyr: 4.53 ± 0.3h p=0.5455, DCA+10nMFCCP: 4.77 ± 0.4h p=0.0927) (Extended Data Fig. 7f). Thus, the segmentation clock period depends on NAD+ availability rather than ATP supply. A similar mechanism may underlie the extended segmentation clock period when supplementing with succinate. This also increased ΔΨm (2.044 ± 0.11 vs. 2.465 ± 0.19, p=0.0341) (Extended Data Fig. 7g) and reduced the total NAD+/NADH ratio which could be rescued by pyruvate (control: 9.65 ± 0.25, succinate: 8.26 ± 0.68 p=0.024, succinate+pyr: 8.94 ± 0.44 p=0.2126) (Extended Data Fig. 7h) together with oscillatory period (control: 4.47 ± 0.5, succinate: 4.76 ± 0.5 p=0.017, succinate+pyr: 4.58 ± 0.6 p=0.5299) (Extended Data Fig. 7i).
Impaired ETC activity also decreases the NAD+/NADH ratio because NAD+ regeneration by complex I is altered. In cells treated with complex I, III and IV inhibitors, we observed a significant NAD+/NADH ratio decrease (control: 11.7 ± 0.7, 20nM rotenone: 10.5 ± 0.4 p=0.0240, 100nM antimycin A: 9.4 ± 0.5 p=0.0001, 1mM sodium azide: 8.9 ± 0.5 p<0.0001) (Fig. 3c) with increased Peredox fluorescence (control: 0.947 ± 0.05, 20nM rotenone: 1.18 ± 0.02, 100nM antimycin A: 1.22 ± 0.01, 1mM sodium azide: 1.35 ± 0.02 Peredox/mCherry ratio, p<0.0001 in all cases) (Extended Data Fig. 7j). Pyruvate supplementation of azide-treated cells partially restored the NAD+/NADH ratio [28] (total NAD+/NADH: 8.9 ± 0.5 vs. 9.8 ± 0.3, p=0.0268; Peredox/mCherry: 1.35 ± 0.02 vs. 1.20 ± 0.01, p<0.0001) (Fig. 3c, Extended Data Fig. 7j)) and the clock period (6.3 ± 0.6h vs. 5.2 ± 0.5h, p<0.0001) (Fig. 3d; Extended Data Fig. 7k, Supplementary Video 5). We also supplemented azide-treated cells with duroquinone, which can mediate NAD+ regeneration via the quinone oxidase NQO1 [29]. Duroquinone treatment led to modest recovery of the cytoplasmic NAD+/NADH ratio as measured by Peredox fluorescence (1.35 ± 0.02 vs. 1.27 ± 0.01, p<0.0001) (Extended Data Fig. 7j), but the total NAD+/NADH ratio did not increase (6.63 ± 0.2 vs. 6.60 ± 0.5, p=0.999) (Extended Data Fig. 7l). Nevertheless, clock oscillations were slightly but significantly accelerated in azide and duroquinone-treated cells relative to cells treated with azide alone (6.35 ± 0.6 vs. 5.96 ± 0.6, p=0.0037) (Fig. 3d; Extended Data Fig. 7m). This is consistent with ETC inhibition slowing the segmentation clock by impairing NAD(H) redox homeostasis.
Our results suggested that differences in NAD(H) redox balance may underlie the developmental rate differences between mouse and human cells. NAD+ is used as an electron acceptor in many metabolic reactions and is required for key steps in nucleotide synthesis and in central carbon, amino acid and lipid metabolism [30]. We compared the NAD+/NADH ratio in mouse vs. human PSM cells and found that, contrary to our expectations, the total NAD+/NADH ratio was significantly lower in mouse (6.81 ± 0.3 vs. 9.66 ± 0.4, p<0.0001) (Extended Data Fig. 4e). However, the dramatic difference in mitochondrial content between mouse and human PSM cells confounded this whole-cell NAD+/NADH measurement as mitochondria exhibit much lower NAD+/NADH values relative to cytoplasm [31, 32]. Our measurements of total NAD+/NADH ratio therefore most likely reflected the higher mitochondrial density of mouse cells compared to human cells. We therefore measured the extracellular lactate/pyruvate ratio, a proxy for cytoplasmic NADH/NAD+ [31]. This ratio was more than two-fold higher in human than mouse PSM cells (25.1 ± 1.8 vs. 66.7 ± 17.4, p<0.0001) (Fig. 2j), suggesting that the cytosolic NAD+/NADH redox balance scales with developmental rate.
We next inhibited LDH in human PSM cells with sodium oxamate to prevent NAD+ regeneration. This led to a decreased NAD+/NADH ratio (total NAD+/NADH: 11.19 ± 1.3 vs. 8.12 ± 0.3, p=0.0002; Peredox/mCherry: 0.6 ± 0.01 vs. 0.78 ± 0.01, p<0.0001) (Extended Data Fig. 8a–b) and concomitant increase in clock period (4.75 ± 0.5 vs. 5.04 ± 0.3, p=0.0404) (Extended Data Fig. 8c–f). Oscillations displayed normal amplitude (Extended Data Fig. 8g) but occasionally arrested early (5.2 ± 0.4 vs. 4.6 ± 0.5 oscillations in 25 hours, p=0.0406) (Extended Data Fig. 8h). Thus, lowering the cytosolic NAD+/NADH ratio could slow down the segmentation clock tempo.
The Lactobacillus brevis NADH oxidase LbNOX [28] can be expressed in mammalian cells to drive regeneration of NAD+ from NADH with concomitant reduction of oxygen to water (Fig. 3e). We used lentiviruses to express either mCherry or cytosolic LbNOX together with mCherry in human PSM cells (Extended Data Fig. 8i). LbNOX modestly increased the total NAD+/NADH ratio (11.20 ± 0.6 vs. 12.16 ± 0.9, p=0.0261) (Fig. 3f). We could not measure Peredox fluorescence ratios due to spectral overlap with mCherry. However, we confirmed LbNOX activity by measuring non-mitochondrial oxygen consumption, which was increased due to oxygen reduction by LbNOX (0.25 ± 0.04 vs. 0.36 ± 0.02 fraction of basal OCR, p<0.0001) (Extended Data. Fig. 8j). When we transduced HES7-Achilles reporter cells with LbNOX-mCherry, we observed a clock period decrease by approximately 25 minutes each cycle (4.76 ± 0.5 vs. 4.36 ± 0.4, p<0.0001) (Fig. 3g, Extended Data Fig. 8k–m, Supplementary Video 6). The oscillation amplitude was not affected (14.3 ± 2.9 vs. 12.5 ± 3.4, p=0.1196) (Extended Data Fig. 8n–o), and there was an increase in mean number of oscillations observed in 25 hours (5.6 ± 0.5 vs. 6 ± 0, p=0.0092) (Extended Data Fig. 8p). LbNOX-mCherry also accelerated the induction dynamics of the MSGN1-Venus reporter relative to mCherry alone (rate of growth (k): mCherry 0.048, 95% CI 0.04561 to 0.05064 vs. LbNOX-mCherry 0.06438, 95% CI 0.06140 to 0.06742, p<0.0001) (Fig. 3h). Cell cycle length was not different between cells transduced with LbNOX-mCherry or mCherry alone (19.78 ± 2.6 vs. 20.33 ± 2.8, p=0.4906) (Fig. 3i). Thus, modulation of the NAD+/NADH ratio can not only slow down but also accelerate the segmentation clock.
ETC effect is mediated by translation
Increased mitochondrial activity is correlated with faster transcription, faster translation, and accelerated growth in human cell lines [33–35]. We compared translation rates by pulsing PSM cells with puromycin for one hour. We then measured the amount of puromycilated peptides produced [36, 37] (Fig. 4a). The mass-specific translation rate is almost twice as high in mouse PSM cells compared to human cells (antibody: 28.95 ± 1.3 vs. 19.15 ± 1.4 MFI/pg, p=0.001; click-it: 13.01 ± 1.1 vs. 7.44 ± 0.3 MFI/pg, p=0.011) (Fig. 4b; Extended Data Fig. 9a). Slowing down translation by treating human PSM cells with low doses of cycloheximide (CHX) lengthened the clock period (control: 4.92 ± 0.3h, 40nM CHX: 5.43 ± 0.5h p=0.0001, 80nM CHX: 5.49 ± 0.3h p=0.0007, 160nM CHX: 5.58 ± 0.3h p<0.0001) with little effect on oscillation number or amplitude (Fig. 4c, Extended Data Fig. 9b–h, Supplementary Video 7). Cell cycle length was also increased by CHX (20.89 ± 3.1 vs. 25.17 ± 3.6, p<0.0001) (Fig. 4d). Thus, the global translation rate could potentially control developmental rate, although this effect could also be mediated by a subset of specific transcripts whose translation is slowed by CHX.
Figure 4.
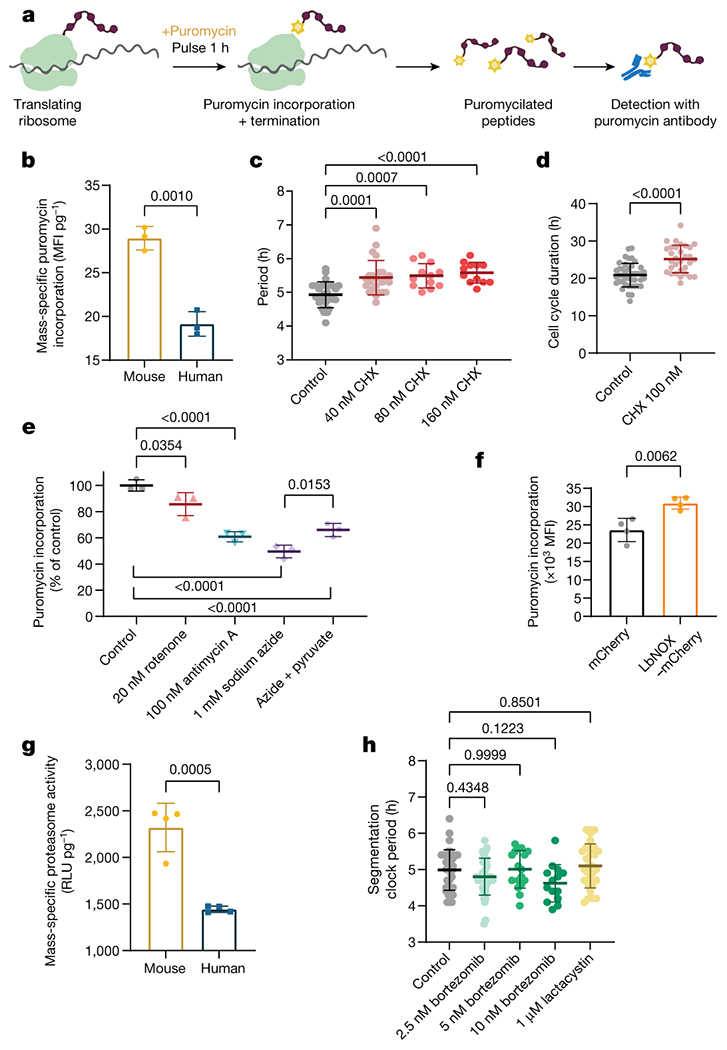
The global rate of protein synthesis acts downstream of the electron transport chain to regulate developmental speed
a. Experimental approach to measure global protein synthesis by detection of puromycilated peptides following a 1-hour pulse with puromycin (puro). Created withBioRender.com.
b. Mass-specific global translation rate as measured by puromycin incorporation in MSGN1-Venus+ PSC-derived mouse and human PSM cells immediately after a 1-hour puromycin pulse and detection by directly conjugated AlexaFluor647 anti-puromycin antibody. Mean ±SD. n=3 biological replicates. Unpaired two-sided t-test.
c. Period of HES7-Achilles oscillations in human PSM cells treated with vehicle control (DMSO, n=27) or increasing doses of cycloheximide (40nM, n=24; 80nM, n=12; 160nM, n=12). Mean ±SD. One-way ANOVA with Dunnett correction: control vs. 160nM CHX p=7.1x10−5
d. Duration of the cell cycle in control (DMSO-treated; n=42) human PSM cells and cells treated with 100nM cycloheximide (CHX; n=31). Mean ±SD. Unpaired two-sided t-test, p=1.1x10−6
e. Relative translation rate expressed as puromycin incorporation normalized to control (DMSO treatment) in human PSM cells treated with 20nM rotenone, 100nM antimycin A, 1mM sodium azide, and azide with 1mM sodium pyruvate for 24 hours. Mean ±SD. n=3 biological replicates. One-way ANOVA with Šidák correction: control vs. antimycin p=2.8x10−5, control vs. azide p=2.8x10−6, control vs. azide+pyr p=9.6x10−5
f. Global translation rate as measured by puromycin incorporation in human PSM cells transduced with a lentivirus expressing either mCherry alone or LbNOX with mCherry. Mean ±SD. n=4 biological replicates. Unpaired two-sided t-test.
g. Mass-specific proteasome activity in MSGN1-Venus+ PSC-derived mouse and human PSM cells as measured by cleavage of a luminogenic proteasome substrate. Mean ±SD. n=4 biological replicates. Unpaired two-sided t-test.
h. Period of HES7-Achilles oscillations in human PSM cells treated with DMSO control (n=35), 2.5nM (n=37), 5nM (n=17) or 10nM (n=14) bortezomib, or 1μM lactacystin (n=30). Mean ±SD. One-way ANOVA with Dunnett correction.
We observed that rotenone, antimycin A and sodium azide all decreased translation rate (control: 100.0 ± 4.2, 20nM rotenone: 85.7 ± 8.8 p=0.0354, 100nM antimycin A: 60.8 ± 3.8 p<0.0001, 1mM sodium azide: 49.5 ± 4.8 p<0.0001 percent of contol) (Fig. 4e). Supplementing azide-treated cultures with pyruvate partially rescued translation rate (49.5 ± 4.8 vs. 65.9 ± 5.0 p=0.0153) (Fig. 4e). Cultures treated with sodium azide for one hour displayed reduced translation, indicating rapid downregulation of protein synthesis (39851 ± 2298 vs. 30262 ± 5267 MFI, p=0.0446) (Extended Data Fig. 9i). These results suggested that ETC inhibition and NAD+ depletion can slow down the segmentation clock in part by reducing protein translation. Transduction with LbNOX-mCherry also significantly increased puromycin incorporation over mCherry (23601 ± 3167 vs. 30953 ± 1646 MFI, p=0.0062) (Fig. 4f). We performed Seahorse experiments in human PSM cells treated with cycloheximide. All aspects of respiration were indistinguishable between cycloheximide-treated and control cells (Extended Data Fig. 9j–l). Thus, mitochondrial activity acts upstream of translation rate to control the segmentation clock period.
Differences in global protein stability also correlate with species-specific developmental rates. Proteome half-life is two times shorter in mouse neural progenitors compared to human [2]. The half-life of puromycilated peptides was shorter in mouse compared to human PSM cells (2.23 ± 0.63h vs. 4.46 ± 1.47h, p<0.0001) (Extended Data Fig. 10a). Mass-specific proteasome activity was significantly elevated in mouse PSM cells (2323 ± 259 vs. 1446 ± 34 RLU/pg, p=0.0005) (Fig. 4g), suggesting that reduced protein stability is caused by higher proteasome activity.
Inhibiting proteasome function can lead to disruption of segmentation clock oscillations [38]. Treating human PSM cells with low doses of proteasome inhibitors bortezomib (BTZ) or lactacystin caused premature oscillations arrest (control: 5.17 ± 0.39, 2.5nM BTZ: 5.50 ± 0.54 p=0.459, 5nM BTZ: 2.69 ± 0.63, p=<0.0001, 10nM BTZ: 2.16 ± 0.38 p<0.0001, 1μM lactacystin: 4.77 ± 0.44 p=0.1635 oscillations in 25 hours) but did not change oscillatory period (control: 4.98 ± 0.5h, 2.5nM BTZ: 4.8 ± 0.5h p=0.4348, 5nM BTZ: 5.00 ± 0.5h p=0.9999, 10nM BTZ: 4.62 ± 0.5h p=0.1223, 1μM lactacystin: 5.10 ± 0.6h p=0.8501) despite significant inhibition of proteasome activity (control: 2257774 ± 122326, 2.5nM BTZ: 1707326 ± 63125, 5nM BTZ: 1590243 ± 122000, 10nM BTZ: 1309836 ± 102240, 1μM lactacystin: 1536581 ± 195052 RLU, p<0.0001 in all cases) (Fig. 4h; Extended Data Fig. 10b–I, Supplementary Video 8). Cell cycle duration could not be assessed under these conditions as proteasome inhibitors induce cell cycle arrest [39].
Azide treatment did not reduce proteasome activity (control: 2082679 ± 23055, 100nM antimycin A: 2452103 ± 30891, 1mM sodium azide: 2778603 ± 37391 RLU, p<0.0001 in both cases) or increase the stability of puromycilated peptides (Extended Data Fig. 10j–k). To assess the degradation profile of the full-length proteome, we performed pulse-chase experiments with the methionine analog L-azidohomoalanine (AHA). Unlike puromycin, AHA incorporation into growing peptides does not induce chain termination and thus labels full-length proteins. AHA labeling resulted in indistinguishable decay profiles between control and azide-treated cells over the timeframe relevant to segmentation clock oscillations (Extended Data Figure 10l). Together, these experiments suggest that the segmentation clock period is more sensitive to inhibition of protein production than degradation.
Discussion
In summary, we found that mass-specific respiration rates scale with and regulate the segmentation clock period by modulating NAD redox balance and, more downstream, translation rate. Given that the segmentation clock period can be used as a proxy for developmental rate [1], our results may explain at least in part the differences observed at early stages of mouse and human development. Such a mechanism may also be modulated locally in the embryo to generate heterochronic changes, as seen for instance in the acceleration of the segmentation clock period relative to growth rate in snakes [40]. Additional studies in other embryonic cell types and mammalian species are required to assess the generality of our findings. Our results also suggest that mass-specific metabolic rates for embryonic cell types may scale with adult body mass as predicted by Kleiber’s law, even under uniform culture conditions [23]. Our studies also revealed a striking parallel between the metabolic requirements of cancer cell proliferation and those of the segmentation clock [41]. In both cases, NAD+ redox homeostasis is more important than ATP availability to maintain normal growth and oscillation rates [26]. Given that PSM cells exhibit Warburg-like metabolism with high levels of aerobic glycolysis [21], these similarities further strengthen the notion that cancer cells resemble embryonic progenitors.
Moreover, the finding that the segmentation clock period is sensitive to NAD+ levels draws a parallel between developmental rate and aging. NAD+ decreases progressively with age and restoring NAD+ levels can ameliorate aging-associated phenotypes [42]. The aging process and developmental rate may share some regulatory mechanisms, especially given that lifespan and gestation period are positively correlated [43]. Future work should focus on the identification of factors that can modulate mass-specific respiration rates in vertebrates [23]. Ultimately, interspecific differences in developmental rate must be traceable to genetic causes. Lastly, the implication of translation rate downstream of mitochondrial activity strongly suggests that the integrated stress response may play a role in determining developmental rate [44, 45]. Continued research in this area will reveal how developmental time can be manipulated externally, with important applications in human stem cell therapy and in vitro disease modeling.
Materials and Methods
Pluripotent Stem Cell Culture
E14 and its derivative reporter lines pMsgn1-Venus [46] and Hes7-Achilles [11] mouse ESCs were maintained under feeder-free conditions on gelatin-coated dishes (StemCell Technologies cat. no. 07903) with 2i medium composed of high glucose DMEM (Gibco cat. no. 11965-118) supplemented with 1% GlutaMAX (Gibco cat. no. 35050061), 1% Non-Essential Amino Acids (Gibco cat. no. 11140-050), 1% Sodium Pyruvate (Gibco cat. no. 11360-070), 0.01% Bovine Serum Albumin (Gibco cat. no. 15260-037), 0.1% β-mercaptoethanol (Gibco cat. no. 21985-023), 15% Fetal Bovine Serum (EMD Millipore cat. no. ES009B), 1000 U/mL LIF (EMD Millipore cat. no. ESG1106), 3μM CHIR99021 (Tocris cat. no. 4423) and 1μM PD0325901 (Stemgent cat. no. 04-006). mESCs were passaged by TrypLE Express (Gibco cat. no. 12605010) dissociation every two days at a density of 1× 104 cells per cm2.
Human stem cell work was approved by Partners Human Research Committee (Protocol Number 2017P000438/PHS). We complied with all relevant ethical regulations. Written informed consent from the donor of the NCRM1 iPS cells was obtained by Rutgers University at the time of sample collection. NCRM1 iPS cells (RUCDR, Rutgers University) and lines carrying the MSGN1-Venus [47], HES7-Achilles [11], HES7-Achilles; pCAG-H2B-mCherry [11] reporters and the AAVS1-CAG-Peredox-mCherry-NLS sensor were maintained on Matrigel-coated plates (Corning, cat. no. 35277) in mTeSR1 medium (StemCell Technologies cat. no. 05851) as previously described [48]. hiPSCs were passaged every 4-5 days by Accutase (Corning cat. no. 25058CI) dissociation and seeded at a density of 5x 104 cells per cm2 in mTeSR1 supplemented with 10μM Y-27362 dihydrochloride (Rocki; Tocris Bioscience, cat. no. 1254).
Generation of NLS-BFP;Hes7-Achilles mESC line
To track individual mouse pPSM cells in homo- or hetero-specific co-culture conditions, we integrated a constitutive nuclear label into the Hes7-Achilles mouse ESC line using the Tol2 system. We used Lipofectamine 3000 (Invitrogen cat. no. L3000001) to co-transfect the following plasmids into Hes7-Achilles mouse ESCs: (i) pCAGGS-Tol2-NLS-BFP-Tol2, (ii) pCAGGS-Transposase, (iii) pMAX-GFP. We isolated GFP+ cells by FACS and plated them at low density on 35mm gelatin-coated dishes in 2i media. Note the GFP transfection was transient and only used for selection of successfully transfected cells. After clonal expansion, we visually inspected colonies, manually picked those expressing NLS-BFP and seeded them into individual wells of a 96-well plate. A set of 5 different clones were verified to stably transmit the CAG-NLS-BFP transgene and used for subsequent experiments.
Presomitic Mesoderm Differentiation
Mouse ESCs were pre-differentiated to an epiblast-like state as previously described [11] by seeding fibronectin-coated dishes (BD Biosciences cat. no. 356008) at a density of 0.8x 104 cells per cm2 in NDiff227 media (Takara cat. no. Y40002) supplemented with 1% KSR (Gibco cat. no. 10828028), 25 ng/ml Activin A (R&D systems cat. no. 338-AC-050) and 12 ng/ml bFGF (PeproTech cat. no. 450-33). The medium was refreshed after 24 hours. Human iPSCs were seeded on Matrigel-coated plates at a density of 3x 104 cells per cm2 in mTeSR1 with 10μM Rocki. After 24 hours, the medium was replaced by mTeSR1 without Rocki.
Presomitic mesoderm differentiation was initiated 48 hours after initial seeding for both mouse and human PSCs. Cultures were switched to DMEM/F12 GlutaMAX (Gibco cat. no. 10565042) supplemented with 1% Insulin-Transferrin-Selenium (ITS; Gibco cat. no. 41400045), 5% Fetal Bovine Serum (EMD Millipore cat. no. ES009B), 6 μM Chir 99021 (Tocris cat. no. 4423), 20ng/ml murine bFGF (PeproTech cat. no. 450-33) and 30 ng/ml Activin A (R&D systems cat. no. 338-AC-050). After 24 hours, the medium was replaced by DMEM/F12 GlutaMAX with 1% ITS, 5% FBS, 6 μM Chir 99021, 20ng/ml bFGF and 0.5 μM LDN193189 (Stemgent cat. no. 04-0074). If PSM cultures needed to be maintained for longer than 2 days, the medium was refreshed at 48 hours with DMEM/F12 GlutaMAX, 1% ITS, 5% FBS, 6 μM Chir 99021, 0.5 μM LDN193189, 50 ng/ml mFgf4 (R&D Systems cat. no. 5846-F4-025), 1 μg/ml Heparin (Sigma Aldrich cat. no. H3393-100KU), 2.5 μM BMS493 (Sigma Aldrich cat. no. B6688-5MG) and 10 μM Rocki to maintain the posterior PSM fate.
For live imaging experiments, cells were seeded on 24 well glass-bottom plates (In Vitro Scientific cat. no. P24-1.5H-N) on day 0 and cultured in DMEM/F12 without phenol red (Gibco cat. no. 31053028). Whenever the effects of chemical inhibitors or culture conditions on the segmentation clock were tested, human HES7-Achilles or HES7-Achilles; AAVS1-CAG-H2B-mCherry cells were differentiated under serum-free conditions as previously described [11] to avoid confounding factors from FBS composition.
Neural Progenitor Differentiation
Neural progenitor induction relied on dual Smad inhibition and was adapted from previously described protocols [49]. Mouse ESCs were seeded on fibronectin-coated dishes at a density of 1x 104 cells/cm2 and pre-differentiated to epiblast state as described above. Human iPSCs were seeded on matrigel-coated plates at a density of 3.5x 104 cells per cm2 in mTeSR1 with 10μM Rocki. At 24 hours, the medium was replaced by mTeSR1 without Rocki. Two days after initial seeding, both mouse and human cells were switched to NDiff227 supplemented with 1% FBS, 0.1 μM LDN193189, and 10 μM SB431542 (Selleck Chemicals cat. no. S1067). The media was refreshed daily. Mouse cells were cultured for 5 days and human cells for 7 days. Neural progenitor fate was confirmed by PAX6 immunofluorescence as described below.
Immunofluorescence
For immunostaining of 2D cultures, cells were grown on Matrigel-coated 24 well glass-bottom plates (In Vitro Scientific cat. no. P24-1.5H-N). Cells were rinsed in PBS and fixed in a 4% paraformaldehyde solution (Electron Microscopy Sciences cat. no. 15710) for 20 minutes at room temperature, then washed 3 times with phosphate buffered saline (PBS). Samples were permeabilized by washing three times for three minutes each in Tris buffered saline (TBS) with 0.1% Tween (TBST) and blocked for one hour at room temperature in TBS-0.1% Triton-3% FBS. The primary antibody (Rabbit α PAX6 Biolegend cat. no. 901301, lotB277104; Rabbit α pHistone H3 (Ser10) Santa Cruz cat. no. sc-8656, lot D1615; or Rabbit α Flag Cell Signaling Technologies cat. no. 14793S, lot 7) was diluted in blocking solution at 1:350 and incubated overnight at 4°C with gentle rocking. Following three TBST washes and a 10-minute block, cells were incubated with a Goat anti-Rabbit Alexa-Fluor 488 conjugated secondary antibody (ThermoFisher cat. no. A11034) or Goat anti-Rabbit Alexa Fluor 594 conjugated secondary antibody (ThermoFisher cat. no. A-11037)(1:500) and Hoechst33342 (1:1000) overnight at 4°C with gentle rocking. Three final TBST washes and a PBS rinse were performed, and cells were mounted in Fluoromount G (Southern Biotech cat. no. 0100-01). Images were acquired using a Zeiss LSM780 point scanning confocal microscope with a 20X objective.
PAX6 Intracellular staining
Samples were washed in PBS and dissociated with TrypLE. One million cells per sample were fixed with 4% formaldehyde and then permeabilized with 0.3% Triton, 0.5% BSA in PBS. Cells were washed once in 0.5% BSA. PAX6 primary antibody (Rabbit α PAX6 Biolegend cat. no. 901301, lotB277104) was diluted 1:100 in 0.5% BSA and samples were incubated for 1 hour. Following a wash in PBS, Goat anti-Rabbit Alexa-Fluor 488 conjugated secondary antibody (ThermoFisher cat. no. A11034) (1:500) was applied for 30 minutes. Samples were then washed and analyzed by flow cytometry.
Generation, Validation and Imaging of AAVS1-CAG-Peredox-mCherry-NLS
CAG-Peredox-mCherry-NLS was inserted into the AAVS1 safe harbor locus using the approach previously described by Oceguera-Yanez and colleagues [50]. For plasmid and cloning design, we used the following software: NEBuilder Assembly Tool (https://nebuilderv1.neb.com/), Geneious 9.1.5, In-Fusion cloning tools (https://www.takarabio.com/learning-centers/cloning/in-fusion-cloning-tools), and ApE v2.0.49.10. Briefly, we cloned the CAG-Peredox-mCherry-NLS sequence from pcDNA3.1-Peredox-mCherry-NLS (Addgene cat. no. 32384) into the pAAVS1-P-CAG-DEST vector (Addgene cat. no. 80490) by Gibson assembly and co-transfected it along with the pXAT2 vector (Addgene cat. no. 80494) into NCRM1 cells. Two days after transfection, we selected positive clones by supplementing mTeSR1 with puromycin (0.5 μg/mL, Sigma–Aldrich cat. no. P7255) for a total of 10 days. To enhance single cell survival, we added CloneR (StemCell Technologies cat. no. 05888) to the media during the first 2 days of selection. We obtained several positive clones and confirmed the homozygous insertion of the sensor by PCR as previously described [50].
To validate that the Peredox NADH/NAD+ sensor worked as expected in the newly generated AAVS1- CAG-Peredox-mCherry-NLS line, we first performed fluorescence lifetime imaging (FLIM) under different conditions (Extended Data Fig. 4f). We differentiated the sensor line to PSM fate under serum-free conditions on glass coverslips. Cells on coverslips were submerged in a recording chamber filled with a balanced salt solution (140mM NaCl, 2.5mM KCl, 10mM HEPES, 1mM MgCl2, 2mM CaCl2, and pH 7.4) at ~34°C, using a perfusion rate of 5 ml/min. Cells were sequentially perfused with the following solutions: 2mM glucose, 0.2mM, 0mM glucose, 0mM glucose with 10mM lactate, 0mM glucose with 10mM pyruvate, and finally 0mM glucose with a mixture of lactate to pyruvate at a ratio of 30:1. Cells were visualized with a Thorlabs Bergamo II microscope (Thorlabs Imaging Systems, Sterling, VA), with hybrid photodetectors R11322U-40 (Hamamatsu Photonics, Shizuoka, Japan). The objective lens used for cell visualization was an Olympus LUMPLFLN 60x/W (NA 1.0). Biosensor fluorescence was excited using light with a wavelength of 790 nm, delivered by a Chameleon Vision-S tunable Ti-Sapphire mode-locked laser (80 MHz,~75 fs; Coherent, Santa Clara, CA). Fluorescence emission light was split with an FF562-Di03 dichroic mirror and bandpass filtered for green (FF01-525/50) and red (FF01-641/75) channels (all filter optics from Semrock, Rochester, NY). Peredox emission was recorded in the green channel. The photodetector signals and laser sync signals were preamplified and then digitized at 1.25 gigasamples per second using a field programmable gate array board (PC720 with FMC125 and FMC122 modules, 4DSP, Austin, TX). Microscope control and image acquisition, as well as laboratory-built firmware and software for fluorescence lifetime determination, can be found elsewhere [51].
To further confirm that similar results could be obtained by ratiometric measurements, we imaged AAVS1- CAG-Peredox-mCherry-NLS cells with an LSM880 confocal microscope using a 20X/0.8 objective (Extended Data Fig. 4g) PSM cells were cultured in a balanced salt solution supplemented with 2mM, 0.2mM or 0mM glucose, 10mM lactate, 10mM pyruvate or a mixture of lactate to pyruvate at a ratio of 30:1 as described above. Images were thresholded based on mCherry-NLS fluorescence and nuclei were automatically segmented in Fiji [52]. A ratio of the mean Peredox: mCherry fluorescence intensity was calculated for each cell. All cells within an image were averaged to obtain the mean ratio for each sample. This ratiometric imaging approach was used in all experiments.
Mouse and Human PSM Co-Culture
Mouse E14 (no reporter), mouse NLS-BFP; Hes7-Achilles, human NCRM1 (no reporter), and human H2B-mCherry; HES7-Achilles PSCs were differentiated to PSM fate separately as described above. On day 2 of differentiation, PSM cells were dissociated with TrypLE. Mixed cell suspensions consisting of either mouse or human reporter cells combined with either human NCRM1 or mouse E14 cells at a ratio of 1:100 were prepared. This ratio allowed individual cells of the minority species to be completely surrounded by cells of the majority species and prevented the formation of cell clusters corresponding to the minority species. Cells were then reseeded on fibronectin-coated 24 well glass-bottom plates at a density of 2.5 x 106 cells per cm2 when human cells were in excess or 4 x 106 cells per cm2 when mouse cells were in excess. These densities allowed the cultures to be fully confluent upon attachment. Cells were then allowed to attach for one hour and subjected to timelapse imaging. Human H2B-mCherry+ cells were automatically segmented and tracked as previously described [11] to obtain single-cell HES7-Achilles fluorescence intensity profiles. Mouse NLS-BFP+ cells were manually tracked.
Primary Mouse PSM Explant culture
Explant culture was performed as described by Hubaud and colleagues [15]. LuVeLu [53] CD1 E9.5 mice (both male and female) were sacrificed according to local regulations in agreement with national and international guidelines. We complied with all relevant ethical regulations. Study protocol was approved by Brigham and Women’s Hospital IACUC/CCM (Protocol number N000478). Sample size was not estimated, nor were randomization or blinding performed. Tailbud was dissected with a tungsten needle and ectoderm was removed using Accutase. Explants were then cultured on fibronectin-coated plate (LabTek chamber). The medium consisted of DMEM, 4.5g/L Glucose, 2mM L-Glutamine, non-essential amino acids 1x, Penicillin 100U/mL, Streptomycin 100μg/mL, 15% fetal bovine serum (FBS), Chir-99021 3μM, LDN193189 200nM, BMS-493 2.5 μM, mFgf4 50ng/mL, heparin 1μg/mL, HEPES 10mM and Y-27632 10μM. Explants were incubated at 37°C, 7.5% CO2.
Timelapse Microscopy
Time lapse-imaging of mouse and human PSM cells and of mouse explants was performed on a Zeiss LSM 780 point-scanning confocal inverted microscope fitted with a large temperature incubation chamber and a CO2 module. An Argon laser at 514 nm and 7.5% or 2% power was used to excite Achilles or Venus fluorescent proteins, respectively. A DPSS 561 laser at 561nm and 2% laser power was used to excite mCherry. In all cases, a 20X Plan Apo (N.A. 0.8) objective was used to acquire images with an interval of 18 or 25 minutes in the case of human samples and 11 or 12 minutes for mouse samples, for a total of 16-48 hours. A 3x3 tile of 800x800 pixels per tile with a single z-slice of 18 μm thickness and 16-bit resolution was acquired per position. Multiple positions, with at least two positions per sample, were imaged simultaneously using a motorized stage. For mouse explants , a single section (~19.6μm thick) with tiling (3x3) of a 512x512 pixels field was acquired every 7.5 minutes at 8-bit resolution.
Oscillation analysis
We used HES7 knock-in fluorescent reporters to assess segmentation clock oscillations. Although dozens of genes display cyclic gene expression as part of the segmentation clock network [12, 54, 55], HES7 was chosen because it is considered the core oscillator for the mammalian segmentation clock. Hairy and enhancer of split (Her/Hes) genes represent the only gene family that is known to oscillate in all vertebrates [55]. Moreover, we had previously established pluripotent stem cell-based in vitro clock models based on HES7 reporters [11][56]. Time lapse movies of HES7-Achilles were first stitched and separated into subsets by position in the Zen program (Zeiss). Then, background subtraction and Gaussian blur filtering were performed in Fiji [52] to enhance image quality. A small region of interest (ROI) was drawn and the mean fluorescence intensity over time was calculated. For co-culture experiments, we instead tracked individual cells as described in the “Mouse and Human PSM Co-Culture section”. Intensity is presented in arbitrary units. For figure 1e, intensity profiles were smoothened in GraphPad Prism using 6 neighboring data points and a 2nd order polynomial. Profiles were then normalized between zero and one.
For each raw intensity profile, we manually identified peaks and calculated the time interval between each pair of consecutive peaks. All peaks in a given profile were measured. We did not distinguish peaks based on their order in the oscillation profile. The oscillatory period was defined as the average time between two peaks in HES7-Achilles profiles. This value is shown in figures as the segmentation clock period. Note that HES7-Achilles oscillatory profiles in figures are shown as the mean ± SEM for a set of individual profiles originating from the same experimental batch.
To measure how the oscillation parameters changed over time, we performed Hilbert analysis [57] in MATLAB. Oscillation profiles for a given condition were saved as CSV files where the first column represents timepoints (hours) and each subsequent column represents an individual profile. To smoothen the oscillation profiles, we applied the Savitzky-Golay filtering function with a polynomial order of 3 and a frame length of 11. At this point, we manually identified peaks and counted the number of peaks between 0 and 25 hours. Peaks were identified by visual inspection based on width and height (i.e. no narrower than 8 datapoints, not shallower than 50 a.u.). We then subtracted the moving mean with a time window of 10 timepoints. This normalized the data to oscillate about zero. An example of signal processing and peak calling can be found in Supplementary Figure 1. Next, we applied the Hilbert transformation to the data. The instantaneous amplitude was extracted as the complex magnitude (also called modulus) of the Hilbert transform. Figures showing mean amplitude or relative amplitude simply display the mean over the entire time course. The instantaneous phase was calculated by unwrapping the phase angle of the Hilbert transform. We then obtained the instantaneous frequency by differentiating the phase. We converted frequency to period and plotted this over time. As the Hilbert transformation is overly sensitive to drifts and changes in the shape of oscillations, we excluded non-physiological period values (e.g. >30 hours, <0) resulting from small blips in the profiles. The MATLAB code for this analysis can be found in github: https://github.com/md2981/Hilbert-Segmentation-Clock
Cell Cycle Length Measurements
To generate cultures with sparsely labeled cells, we mixed HES7-Achilles; AAVS1-CAG-H2B-mCherry human iPSCs or pMsgn1-Venus mouse ESCs with their parental line (NCRM1 or E14, respectively) at a ratio of 1:100 during the initial seeding. Cultures were then differentiated normally and subjected to timelapse imaging. Individual reporter cells were tracked manually on Fiji [52] and the time of cell division was recorded. The cell cycle length was defined as the time elapsed between the time a cell first divides and the time one of its daughter cells divides again.
To measure cell cycle length in primary mouse PSM explants, lentiviral infection was used to sparsely label cells with an SV40-mCherry reporter [15]. The plasmid E[beta]C (Addgene Plasmid #24312) was cut with BamHI and self-ligated to remove the original insert, thus generating a lentiviral transfer vector that expresses only mCherry. Lentivirus was produced in 293T cells, which were transfected using the CaCl2 method with the packaging plasmids psPAX2 (Addgene cat. no. 12260) and pVSVG (gift from M. Wernig lab). Supernatant was collected, filtered using a 0.45μM filter and concentrated by centrifugating 4 volumes of supernatant on 1 volume of TNE buffer (50mM Tris pH7.2, 100mM NaCl, 0.5mM EDTA, 15% sucrose) at 7197 relative centrifugal field (rcf) for 4 hours at 4°C. Explants were infected for ~4 hours and further incubated overnight before imaging. SV40-mCherry+ cells were manually tracked, and the cell cycle length was calculated as described above.
Flow Cytometry and Cell Sorting
To determine the fraction of PSM cells expressing MSGN1-Venus, cultures were dissociated with Accutase and analyzed by flow cytometry using an S3 cell sorter (Biorad). Undifferentiated ESCs or iPS cells, which do not express the fluorescent protein, were used as negative control for gating purposes. Samples were analyzed in biological triplicates. Results are presented as the percentage of Venus-positive cells among singlets. The same gating strategy was used to sort MSGN1-Venus+ for subsequent experiments. The gating strategy is illustrated in Supplementary Figure 2. All other flow cytometry analyses were performed on a 5-laser Fortessa analyzer (BD). Automatic compensation was set up whenever more than one dye or fluorescent protein was used at a time. Flow cytometry data was analyzed in FlowJo. The mean fluorescence intensity (MFI) for 10,000 cells is presented. In the case of mouse vs. human comparisons, only MSGN1-Venus+ cells were considered in the analysis and MFI was normalized to cell mass.
LbNOX Lentiviral Overexpression
A plasmid containing the LbNOX sequence was obtained from Addgene (Plasmid #75285) [28]. We cloned the LbNOX coding sequence into the BamHI-digested E[beta]C (Addgene Plasmid #24312) transfer plasmid. This plasmid was linearized by MluI digestion and LbNOX was inserted by Gibson assembly. The resulting transfer plasmid therefore expressed LbNOX under the control of the EF-1α promoter and mCherry under the control of the SV40 promoter. We produced lentivirus in Lenti-X 293T cells (Takara cat no. 632180) by co-transfecting the transfer plasmid along with psPAX2 (Addgene cat. no. 12260) and pVSVG (gift from M. Wernig lab). Transfection was performed with the jetPEI reagent (Polyplys cat. no. 101000053). Supernatants were collected at 24, 48, and 72 hours. Lentiviruses were concentrated using the Lenti-X Concentrator (Takara cat no. 631232), resuspended in DMEM and stored in single-use aliquots at −80°C. In parallel, we also produced the control BamHI-digested E[beta]C lentivirus, which only expresses mCherry. We did not determine the viral titer.
To transduce human iPSCs, we combined the lentiviruses with dissociated iPSCs immediately prior to seeding. We used a volume of 5μl lentivirus concentrated supernatant per 100,000 cells. Cultures were incubated overnight and re-infected when PSM differentiation was initiated. This achieved high efficiency of transduction as judged by mCherry expression. Due to issues with silencing, we did not maintain stable lines but rather transduced cells anew for each experiment. We verified that PSM induction was not affected by lentiviral transduction. We confirmed that LbNOX was expressed by immunofluorescence against the C-terminal flag tag.
Mitochondrial content and ΔΨm measurement
PSM cells were washed in PBS, dissociated in TryplE, washed in DMEM and then incubated for 30 minutes at 37°C 5% CO2 in DMEM supplemented with the appropriate dye. For mitochondrial content measurements, we used 25nM Mitotracker Green (Invitrogen cat. no. M7514). For comparison of ΔΨm between mouse and human PSM cells, cells were treated with 1μg/mL JC-1 (Invitrogen cat. no. T3168). For ΔΨm measurements in human PSM cells under different conditions, we used 20nM TMRM (Invitrogen, T668) in combination with 25nM Mitotracker Green for normalization. The cells were then washed in PBS, span down, and resuspended in PBS-1%FBS prior to analysis by flow cytometry. As a control, 1uM FCCP was used to depolarize the inner mitochondrial membrane.
Seahorse Assays
PSM cells were dissociated on day 2 of differentiation and reseeded onto fibronectin-coated Seahorse plates (Agilent cat. no. 101085-004) at a density of 7 x 105 cells per cm2 in Seahorse XF DMEM (Agilent cat. no. 103575-100) supplemented with 10mM glucose (Agilent cat. no. 103577-100), 1mM pyruvate (Agilent cat. no. 103578-100) and 2mM glutamine (Agilent cat. no. 103579-100). For mouse vs. human comparisons, MSGN1-Venus+ cells were pre-sorted. Cells were allowed to attach at room temperature for 20 minutes and then transferred to a 37°C incubator without CO2 for 1 hour. The Seahorse cartridge was hydrated and calibrated as per the manufacturer instructions. For the Mitochondrial Stress Test (Agilent cat. no. 103015-100), we used oligomycin at 1μM, FCCP at 1μM, rotenone at 0.5μM and antimycin A at 0.5μM. No glucose controls were used to calculate the CO2 contribution factor for glycolytic proton efflux rate determination. To quantify glycolytic vs. mitochondrial ATP production, we used the ATP Rate Assay kit (Agilent cat. no. 103592-100). All samples were run in six to ten replicates in either a Seahorse XF96 or XFe24 Analyzer and the data were analyzed in Wave and Microsoft Excel using macros provided by the manufacturer.
For the isolation of mitochondria from mouse and human PSM cells, we followed the protocol described by Bharadwaj and colleagues with some modifications [58]. To obtain sufficient starting material, mouse and human PSM cells from four confluent 10 cm dishes for each species were dissociated using TryplE. Following a PBS wash, the cell pellets were resuspended in 250μl ice-cold Chappel-Perry buffer I (see [58] for formulation) and transferred to 2mL glass-glass douncers. All further steps were performed on ice, including centrifugation at steps at 4°C. Cells were broken apart with 30 douncer strokes. An additional 250μl of Chappel-Perry buffer I were added, followed by 500μl of Chappel-Perry buffer II. Samples were transferred to 1.5mL microcentrifuge tubes and centrifuged at 900g for 10 minutes. Supernatants were transferred to new tubes and centrifuged at 10,000g for 10 minutes. Mitochondrial pellets were resuspended in 500μl Chappel-Perry buffer II, then centrifuged again at 10,000g for 10 minutes and resuspended in 500μl Chappel-Perry buffer I. Aliquots were taken for BCA protein quantification at this time. Samples were centrifuged one last time at 10,000g for 10 minutes and resuspended in 50μl Mitochondrial Assay Solution (see [58] for formulation). Mitochondria were diluted to the desired concentration in Mitochondrial Assay Solution supplemented with 10mM pyruvic acid and 2mM malic acid. A total amount of 10μg mitochondria in a volume of 20μl were seeded per well of an Agilent Seahorse XFe96 plate and attached by centrifugation at 2,000g for 20 minutes. The volume was then completed to 180μl per well with Mitochondrial Assay Solution containing pyruvic and malic acids, and the samples were loaded into the Seahorse XFe96 analyzer. Mix, wait and measurement intervals were followed as per [58]. Injections consisted of final concentrations 2mM ADP (Port A), 5μM Oligomycin (Port B), 6μM FCCP (Port C), and 1μM Rotenone with 1μM Antimycin A (Port D).
Extracellular Glucose, Lactate and Glutamine Quantification
Mouse and human MSGN1-Venus+ cells were pre-sorted and seeded onto a fibronectin-coated 96 well plate at a density of 4x105 cells/cm2. The media consisted of DMEM (Gibco cat. no. A1443001) containing 2mM glucose, 1mM glutamine, 1mM pyruvate, 0.1mM non-essential amino acids, 1% ITS, 5% dialyzed FBS supplemented (Cytiva cat. no. SH30079.01) with 6 μM Chir 99021, 0.5 μM LDN193189, 50 ng/ml mFgf4, 1 μg/ml Heparin, 2.5 μM BMS493 and 10 μM Rocki. Control samples with media only (no cells) were also included. For glucose and lactate detection, 5μl of media were collected every hour for a total of 6 hours, diluted in 195μl PBS and frozen at −20°C. The Promega Glucose-Glo (Promega cat. no. J6021) and Lactate-Glo (Promega cat. no. J5021) kits were used according to manufacturer protocols on white 384 well plates. Standard curves of glucose and lactate were used to calculate metabolite concentration in the media. For glutamine detection, media was collected at a single timepoint (12 hours) and the Promega Glutamate/Glutamine-Glo kit (Promega cat. no. J8021) was used. Luminescence was measured after the incubation time indicated by the manufacturer using a GloMax Promega plate reader with 1 second integration. Measurements were normalized to cell mass.
Stable Isotope Tracing
Sample preparation:
Mouse and human PSM cells were differentiated as described above in 6 well plates. On day 2 of differentiation, the plates were washed once with PBS and replaced with tracer medium. Tracer medium consisted of 25mM either unlabeled or [U-13C6]-glucose (Cambridge Isotope Laboratories cat. no. CLM-1396-0.5), 4mM either unlabeled or [U-13C5]-glutamine (Cambridge Isotope Laboratories cat. no. CLM-1822-H-0.1), 1mM Sodium Pyruvate, 0.1mM Non-Essential Amino Acids, 100u/ml Penicillin, 100ug/ml Streptomycin, 1% ITS, 5% dialyzed FBS supplemented with 6 μM Chir 99021, 0.5 μM LDN193189, 50 ng/ml mFgf4, 1 μg/ml Heparin, 2.5 μM BMS493 and 10 μM Rocki. For time-course experiments, samples were collected at the following timepoints: 0h, 3h, 6h, 9h, 12h, 18h, 24h, 36h, 48h. We could not extend the time-course further because PSM cells differentiate to somitic fate past this time window [11]. For steady-state measurements, cultures were incubated for 24 hours prior to metabolite extraction. Experiments were performed three times independently.
Metabolite extraction:
Intracellular metabolites were obtained after washing cells with 2 volumes of room temperature HPLC-grade water and floating the dry plates on liquid nitrogen to quench metabolism. Plates were stored at −80 °C until extraction. Metabolites were extracted with 1 mL 80% MeOH pre-cooled to −80 °C. Insoluble material was removed by centrifugation at 21,000 xg for 15 min at 4 °C. The supernatant was evaporated to dryness at 42 °C using a SpeedVac concentrator (Thermo Savant). Samples were resuspended in 35 μL LC-MS-grade water prior to analysis.
Acquisition parameters:
LC-MS analysis was performed on a Vanquish ultra-high-performance liquid chromatography system coupled to a Q Exactive orbitrap mass spectrometer by a HESI-II electrospray ionization probe (Thermo). External mass calibration was performed weekly. Metabolite samples (2.5 μL) were separated using a ZIC-pHILIC stationary phase (2.1 x 150 mm, 5 μm) (Merck). The autosampler temperature was 4 °C and the column compartment was maintained at 25 °C. Mobile phase A was 20 mM ammonium carbonate and 0.1% ammonium hydroxide. Mobile phase B was acetonitrile. The flow rate was 0.1 mL/min. Solvent was introduced to the mass spectrometer via electrospray ionization with the following source parameters: sheath gas 40, auxiliary gas 15, sweep gas 1, spray voltage +3.0 kV for positive mode and −3.1 kV for negative mode, capillary temperature 275 °C, S-lens RF level 40, and probe temperature 350 °C. Data were acquired and peaks integrated using TraceFinder 4.1 (Thermo).
Stable isotope quantification:
All metabolites were measured using the following mobile phase gradient: 0 min, 80% B; 5 min, 80% B; 30 min, 20% B; 31 min, 80% B; 42 min, 80% B. The mass spectrometer was operated in selected ion monitoring mode with an m/z window width of 9.0 centered 1.003355-times half the number of carbon atoms in the target metabolite. The resolution was set at 70,000 and AGC target was 1 x 105 ions. Peak areas were corrected for quadrupole bias as previously described [59]. Raw mass isotopomer distributions were corrected for natural isotope abundance using a custom R package (mzrtools, https://github.com/wmoldham/mzrtools) employing the method of Fernandez, et al. [60].
Cell Volume Measurements
Cells were dissociated in TrypLE, washed and resuspended in PBS. Volume was measured on a Moxi Go II Coulter-principle cell sizer and flow cytometer. When mouse vs. human PSM cells were compared, only MSGN1-Venus+ cells were considered. Data was analyzed in FlowJo.
Mass and Density Measurements – Suspended Microchannel Resonator
Mouse and human PSM cells were dissociated in TrypLE and MSGN1-Venus+ cells were sorted. Cells were then counted and resuspended in DMEM/F12, 1% ITS, 5% FBS with 6 μM Chir 99021, 20ng/ml bFGF and 0.5 μM LDN193189 at a concentration of 3x105 cells/ml. The cells were kept on ice and their total mass and density were measured using the suspended microchannel resonator (SMR) according to a previously developed fluid-switching method [20]. The SMR is a vibrating cantilever with a fluidic channel inside. In the absence of cells, the vibration frequency of the cantilever is proportional to the density of the fluid flowing through the cantilever. As a cell flows through the cantilever, the vibration frequency of the cantilever changes proportionally to the buoyant mass of the cell. Following the measurements of normal media density and the cell’s buoyant mass with this media, the cell is immersed in culture media that has been made denser by the addition of 35% OptiPrep (Sigma-Aldrich cat. no. D1556-250ML). The cell then flows back through the cantilever in the high-density media to obtain a second set of buoyant mass and media density measurements. The total mass and density of the cell is calculated by comparing these two sets of measurements according to the equation , where is buoyant mass, is volume, is density of the cell, and is the density of the media. After each cell is measured, the cell is flushed out of the SMR before fresh media and the next cell is loaded into the SMR. For measurements of cells’ dry mass, dry volume and the density of the dry mass, a similar fluid switching protocol was followed but, instead of using OptiPrep containing media, the second measurement was carried out in media where 50% of the water content was heavy water (D2O). Cellular water content exchanges rapidly [19, 20] causing the intracellular water content to be identical and neutrally buoyant to the extracellular water content. Therefore, the measurements in normal and heavy water can be used to calculate the dry mass, dry volume and the density of the dry mass according to the equation , where is buoyant mass, is dry mass of the cell, is density of the dry mass, and is the density of the extracellular fluid [19, 20]. All measurements were carried out at +4°C. Calibration of the SMR frequency response to a cell was done using NIST traceable 10.12 μm diameter polystyrene beads (Thermo Scientific, Duke Standard beads, cat. no. 4210A), and the calibration of SMR baseline frequency to fluid density was done using NaCl solutions of known density [61].
Small Molecule Inhibitor Treatments
Human PSM cells were differentiated using the serum-free protocol [11] and treated chronically with the relevant inhibitors or supplements as indicated on Supplementary Table 1 starting on day 2 of differentiation. Timelapse imaging started approximately 2 hours after the inhibitor addition. In the case of aphidicolin, cells were pre-treated for 24 hours before imaging. All other assays (Seahorse, proteasome activity, NAD+/NADH, Peredox fluorescence, etc.) were performed after 16-24 hours of treatment to observe chronic effects.
Whole-Cell NAD+/NADH Ratio
Cells were dissociated with TrypLE, washed and resuspended in PBS at a density of 8x105 cells/ml. For each sample, 50μl were distributed in triplicate wells of a 96 well plate. In the case of mouse vs. human comparisons, MSGN1-Venus+ cells were pre-sorted. Samples were lysed by adding 50μl 0.2N NaOH with 1% dodecyltrimethylammonium bromide (DTAB) to preserve the stability of dinucleotides and incubated for 10 minutes at room temperature with gentle shaking. Half of each sample was then transferred to an empty well within the same plate and 25μl 0.4N HCl were added. Samples were then incubated for 15 minutes at 65°C to selectively denature NAD+ in the basic solution and NADH in the acidic solution. Samples were then cooled down to room temperature for 5 minutes. pH was restored to neutral conditions by adding 25μl 0.5M Trizma® base to acid-treated samples and 50μl Trizma/HCl solution (1:1 mixture of 0.5M Trizma base and 0.4N HCl) to base-treated samples. 40μl of each sample were then transferred to a white 96 well plate. The detection reagent mixture was prepared according to the instructions in the Promega NAD/NADH (cat. no. G9071) kit and 40μl were added per sample. Luminescence was measured after an incubation of 45 minutes at room temperature using a GloMax Promega plate reader with 1 second integration. The NAD+/NADH ratio was calculated as the luminescence ratio of the acid-treated sample to the base-treated sample.
Extracellular Lactate/Pyruvate measurements
On day 2 of differentiation, the media was refreshed, and cells were incubated for 10 hours. Spent media was collected and centrifuged at 300g for 5 minutes to pellet cell debris. The supernatant was transferred to a new tube and flash frozen in liquid nitrogen. Samples were kept at −80°C until extraction. Media lactate and pyruvate quantitation was performed by using an LC/MS method with stable isotope dilution. 15 μL of spent media was extracted with 117 μL of acetonitrile and 45 μL of labeled lactate (D3-lactate, CDN Isotopes) and pyruvate (13C3-pyruvate, Sigma) diluted in water. The samples were then vortexed and left on ice for 5 minutes, vortexed again, and then spun at 21.1 x kg for 10 minutes. 100 μL of the supernatant was loaded into a glass vial for LC/MS analysis. 10 μL of each sample was analyzed using a Q Exactive Plus Orbitrap Mass Spectrometer with a DionexUltiMate 3000 UHPLC system (Thermo Fisher Scientific). Metabolites were separated on an Xbridge amide HILIC column (2.1 X100 mm, 2.5 μM particle size, Waters). Mobile phase A was 20mM ammonium acetate, 0.25 % ammonium hydroxide, 5% acetonitrile, pH 9.0. Mobile phase B was 100% acetonitrile. The gradient was: 85%B for 0-0.5 minutes, decreased to 35% B from 0.5-9 mins, decreased to 2% B from 9-11 minutes, held at 2% B from 11-12 minutes, increased to 85% B from 12-13.5 minutes, held at 85% B from 13.5-18 minutes. The flow rate was 220 μL/min from 0-14.6 minutes and 420 μL/min from 15-18 minutes. The MS data acquisition was polarity-switching full scan mode in a range of 70-1000 m/z, with resolution 70,000, AGC target of 3E6, and maximum injection time of 80 msec. All LC-MS data were collected with samples injected in a randomized order. Absolute quantitation was determined with a standard curve of known concentrations of unlabeled lactate and pyruvate extracted with the same labeled internal standards.
ADP/ATP Ratio and ATP content measurements
The BioVision ApoSENSOR™ ADP/ATP Ratio Bioluminescent Assay Kit (cat. no. K255-200) was used according to manufacturer’s instructions. Cells were dissociated with TrypLE, washed and resuspended in Nucleotide Releasing Buffer at a density of 3x105 cells/ml. In the case of mouse vs. human comparisons, MSGN1-Venus+ cells were pre-sorted. Background luminescence was measured first, followed by ATP-linked luminescence. To measure ADP, samples were treated with ADP-converting enzyme to generate ATP from ADP. Total (ATP+ADP) luminescence was then recorded. ADP-linked luminescence was calculated by subtracting the ATP-linked luminescence from the total luminescence. A GloMax Promega plate reader with 1 second integration was used.
Puromycin Incorporation and Pulse-Chase Experiments
To pulse cells with puromycin, samples were washed in PBS and incubated for 1 hour at 37°C 5% CO2 in DMEM containing 1μg/ml puromycin (Puro) or 20μM O-propargyl-puromycin (OPP-Puro). Samples were collected immediately following the incubation period for translation rate measurements. For degradation rate measurements, samples were washed with DMEM and chased for the indicated period of time. Specifically, samples were collected every 3 hours for a total of 12 hours. After collection, samples were washed again in PBS and dissociated with TrypLE. One million cells per sample were fixed with 4% formaldehyde and then permeabilized with 0.3% Triton in PBS. Puro-treated samples were then incubated for 1 hour with anti-puromycin Alexa Fluor 647 directly conjugated antibody (1:100; Millipore Sigma cat. no. MABE343). OPP-Puro samples were processed for Click-chemistry detection with Alexa Fluor 647 picolyl azide as per the instructions in the Click-iT® Plus OPP Protein Synthesis Assay Kit (Invitrogen cat. no. C10458). Samples were then analyzed by flow cytometry. In the case of mouse vs. human comparisons, only MSGN1-Venus+ cells were considered.
L-Azidohomoalanine (AHA) Pulse-Chase Experiments
The Click-it AHA Alexa Fluor 488 Protein Synthesis HCS Assay (Invitrogen cat. no. C10289) was used. Cells were washed in PBS and pre-incubated in DMEM lacking methionine (Gibco cat. no. 21013024 supplemented with 2mM glutamine, 1mM sodium pyruvate, 1% non-essential amino acids and 0.2mM cysteine) for one hour prior to the AHA pulse. The medium was then replaced by methionine-free DMEM containing 50μM AHA and cells were incubated for one hour. Samples were either processed immediately or after the indicated chase time as per the manufacturer instructions. Samples were then analyzed by flow cytometry.
Proteasome Activity Assays
The Promega Proteasome-Glo Chemotrypsin-like assay (Promega cat. no. G8660) was used following the manufacturer’s protocols. Cells were dissociated with TrypLE, washed several times and resuspended in DMEM at a density of 8x105 cells/ml. In the case of mouse vs. human comparisons, MSGN1-Venus+ cells were pre-sorted. Luminescence was recorded on a GloMax Promega plate reader with 1 second integration.
Cell Viability Assay
To document that inhibitor doses are sub-lethal, we assessed cell viability with trypan blue following 24 hours of incubation with the relevant inhibitor.
Statistical Analyses
Statistical analyses were performed with Prism 9 software (GraphPad). P values <0.05 were considered significant. Details of statistical analyses are indicated in figure legends. Unpaired t-tests or ordinary one-way ANOVA were performed with Tukey correction for multiple comparisons. All differentiation experiments were performed a minimum of three independent times (rounds of differentiation), each containing at least three technical replicates (wells) per condition.
Extended Data
Extended Data Figure 1.
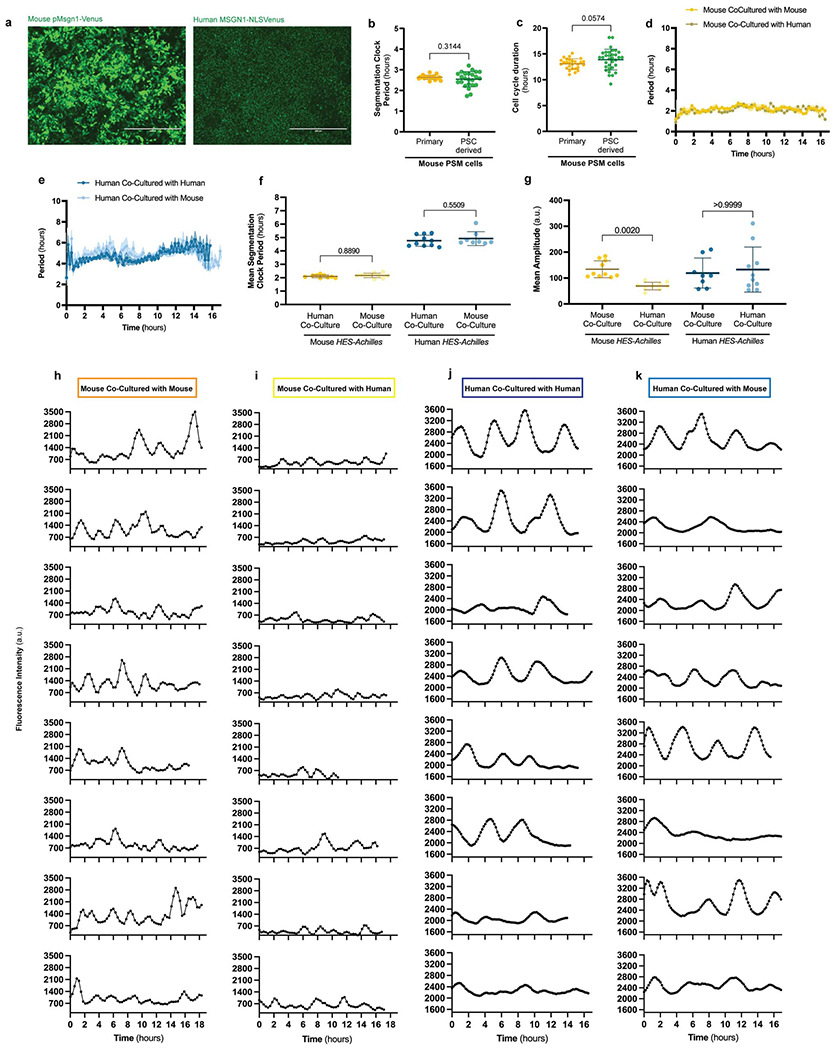
The segmentation clock-period is cell-autonomous even in chimeric conditions.
a. Representative micrographs of MSGN1-Venus fluorescence in mouse (left) and human (right) PSC-derived PSM cells on day 2 of differentiation. Note the reporter is cytoplasmic in mouse cells but nuclear in human cells. Similar results were obtained n=15 times. Scale bar = 400μm.
b. Period of segmentation clock oscillations in primary PSM tissue dissected from E9.5 mouse embryos carrying the LuVeLu reporter (n=18) and PSC-derived PSM expressing the Hes7-Achilles reporter (n=24). Mean ±SD. Unpaired two-sided t-test.
c. Cell cycle duration in primary PSM tissue dissected from E9.5 mouse embryos (n=27) and PSC-derived PSM (n=33). Mean ±SD. Unpaired two-sided t-test.
d. Instantaneous HES7-Achilles oscillatory period over the course of 16 hours as calculated by Hilbert transformation for mouse PSM cells co-cultured with mouse (n=10) or human (n=7) non-reporter PSM cells. Mean ±SEM.
e. Instantaneous HES7-Achilles oscillatory period over the course of 16 hours as calculated by Hilbert transformation for human PSM cells co-cultured with human (n=11) or mouse (n=8) non-reporter PSM cells. Mean ±SEM.
f. Mean segmentation clock period as calculated by Hilbert transformation for mouse (left) or human (right) PSM cells co-cultured with either mouse or human non-reporter PSM cells. Mean ±SD. n=10 (mouse-mouse), n=9 (mouse-human), n=8 (human-mouse), n=10 (human-human). One-way ANOVA with Šidák correction.
g. Mean amplitude of HES7-Achilles oscillations in mouse (left) or human (right) PSM cells co-cultured with either mouse or human non-reporter PSM cells. Mean ±SD. n=10 (mouse-mouse), n=9 (mouse-human), n=8 (human-mouse), n=10 (human-human). Kruskal-Wallis test with Dunn’s correction.
h. Representative single-cell tracks of HES7-Achilles fluorescence for mouse PSM cells cultured with non-reporter mouse PSM cells.
i. Representative single-cell tracks of HES7-Achilles fluorescence for mouse PSM cells cultured with non-reporter human PSM cells.
j. Representative single-cell tracks of HES7-Achilles fluorescence for human PSM cells cultured with non-reporter human PSM cells.
k. Representative single-cell tracks of HES7-Achilles fluorescence for human PSM cells cultured with non-reporter mouse PSM cells.
Extended Data Figure 2.
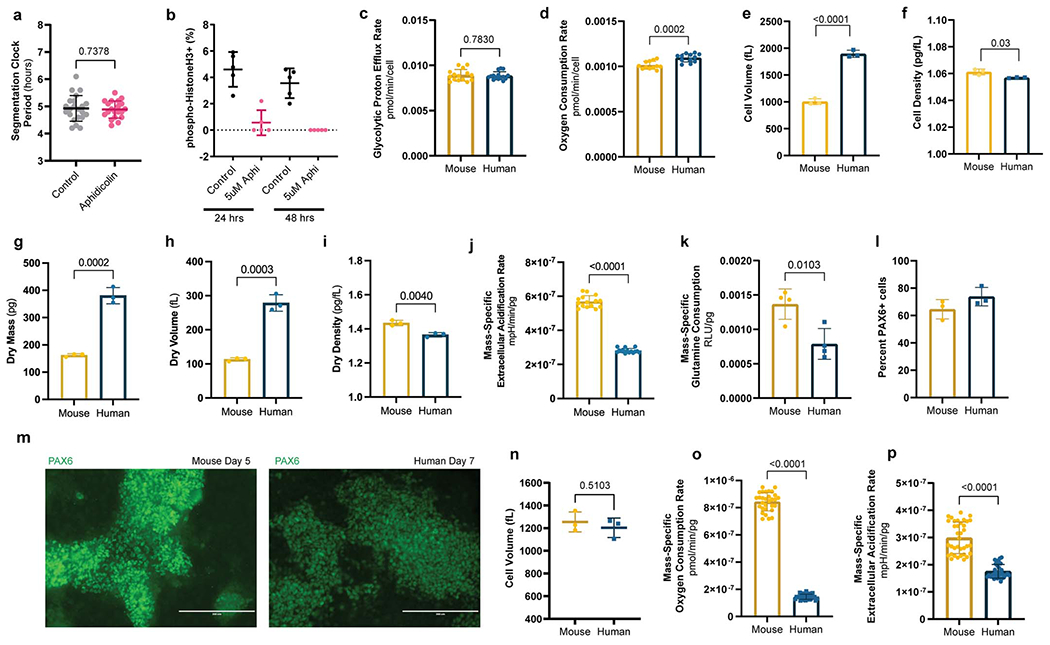
Comparison of metabolic and physical parameters in mouse vs. human PSM and neural progenitor cells
a. HES7-Achilles oscillatory period in human PSM cells under control (DMSO; n=22) or 5μM aphidicolin (n=20) conditions. Cultures were pre-treated with DMSO or aphidicolin for 24 hours to induce cell cycle arrest. Mean ±SD. Unpaired two-sided t-test.
b. Quantification of immunofluorescence staining for histone H3 phosphorylated at Ser10 in human PSM cells treated with vehicle control (DMSO) or 5 μM aphidicolin for 24 or 48 hours. Mean ±SD. n=5 biological replicates.
c. Glycolytic proton efflux rate per cell for MSGN1-Venus+ PSC-derived mouse and human PSM cells. Same data as Fig. 2d but normalized by cell. Mean ±SD. n=15. Unpaired two-sided t-test.
e. Oxygen consumption rate per cell for MSGN1-Venus+ PSC-derived mouse and human PSM cells. Same data as Fig. 2d but normalized by cell. Mean ±SD. n=12. Unpaired two-sided t-test.
f. Total cell volume as measured in a suspended microchannel resonator for MSGN1-Venus+ PSC-derived mouse and human PSM cells. Each datapoint represents the mean of >200 individual cells. Mean ±SD. n=3 independent experiments. Unpaired two-sided t-test: p=5.9x10−5
g. Total cell density of MSGN1-Venus+ PSC-derived mouse and human PSM cells as measured on a suspended microchannel resonator. Each datapoint represents the mean of >200 individual cells. Mean ±SD. n=3 independent experiments. Unpaired two-sided t-test
h. Dry mass as measured in a suspended microchannel resonator for MSGN1-Venus+ PSC-derived mouse and human PSM cells. Each datapoint represents the mean of >200 individual cells. Mean ±SD. n=3 independent experiments. Unpaired two-sided t-test.
i. Dry volume as measured in a suspended microchannel resonator for MSGN1-Venus+ PSC-derived mouse and human PSM cells. Each datapoint represents the mean of >200 individual cells. Mean ±SD. n=3 independent experiments. Unpaired two-sided t-test.
j. Dry density as measured in a suspended microchannel resonator for MSGN1-Venus+ PSC-derived mouse and human PSM cells. Each datapoint represents the mean of >200 individual cells. Mean ±SD. n=3 independent experiments. Unpaired two-sided t-test.
k. Mass-specific extracellular acidification rate (ECAR) in MSGN1-Venus+ PSC-derived mouse and human PSM cells. Mean ±SD. n=15. Unpaired two-sided t-test: p=8.7x10−23
l. Relative mass-specific glutamine consumption after 12 hours of culture for MSGN1-Venus+ PSC-derived mouse and human PSM cells. Mean ±SD. n=4 biological replicates. Unpaired two-sided t-test.
m. Percent PAX6+ cells in mouse (day 5) and human (day 7) neural progenitor cultures as measured by intracellular staining and flow cytometry. Mean ±SD. n=3 independent experiments.
n. Representative micrographs of immunofluorescence staining for PAX6 in PSC-derived mouse (left) and human (right) neural progenitor cells on days 5 and 7 of differentiation, respectively. Similar results were obtained n=3 times. Scale bar=200μm.
o. Volume of mouse (day 5) and human (day 7) neural progenitor cells as measured by a coulter counter. Mean ±SD. n=3 independent experiments. Unpaired two-sided t-test.
p. Mass-specific oxygen consumption rate for PSC-derived mouse and human neural progenitor cells. Mean ±SD. n=30. Unpaired two-sided t-test: p=1.69x10−51
q. Mass-specific extracellular acidification rate for PSC-derived mouse and human neural progenitor cells. Mean ±SD. n=36. Unpaired two-sided t-test: p=3.75x10−18
Extended Data Figure 3.
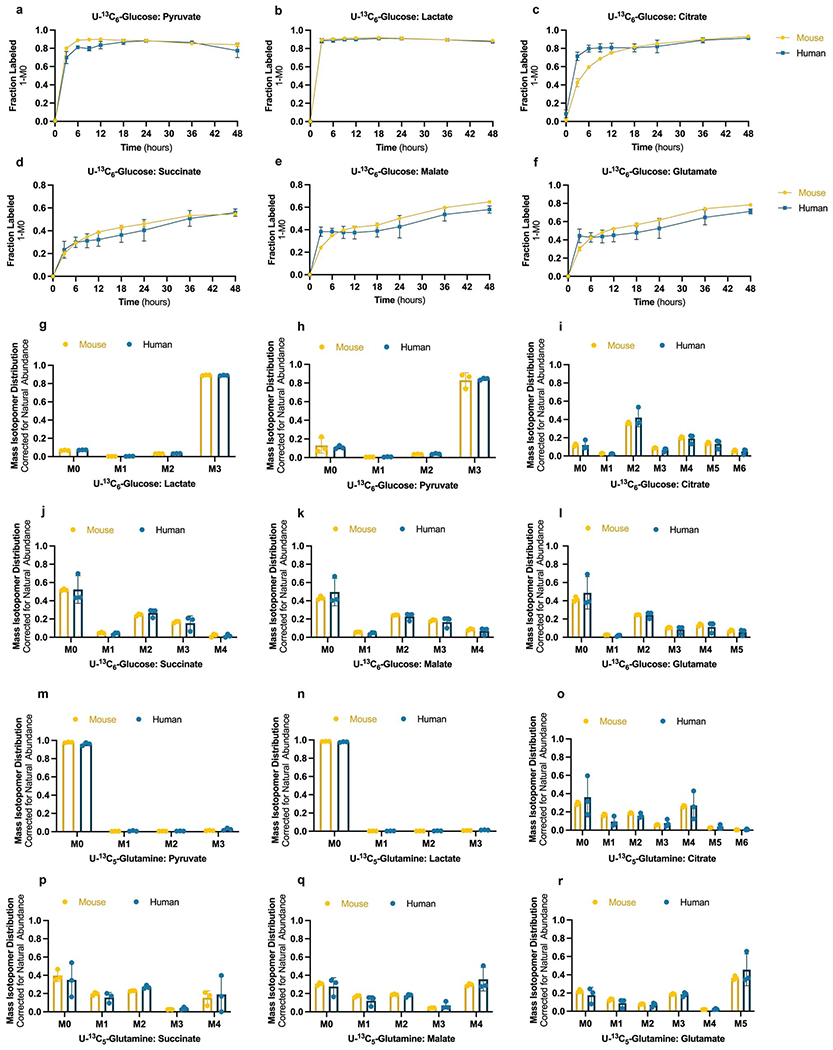
Stable isotope tracing of glucose and glutamine utilization patterns in mouse and human PSM cells
a-f. Stable isotope labeling with 25mM U13C6-Glucose over the course of 48 hours for PSC-derived mouse and human PSM cells. Total fraction labeled by any isotopomer is expressed as 1-M0 for pyruvate (a), lactate (b), citrate (c), succinate (d), malate (e), and glutamate (f). Mean ±SD. n=3 independent experiments.
g-l. Mass isotopomer distribution, adjusted for natural abundance, for pyruvate (e), lactate (f), citrate (g), succinate (h), malate (i), and glutamate (j) after 24 hours of labeling with 25mM U13C6-Glucose in mouse and human PSM cells. Labels in the x-axis correspond to distinct mass isotopomers with increasing number of heavy carbons. Mean ±SD. n=3 independent experiments.
m-r. Mass isotopomer distribution, adjusted for natural abundance, for pyruvate (k), lactate (l), citrate (m), succinate (n), malate (o), and glutamate (p) after 24 hours of labeling with 4mM U13C5-Glutamine in mouse and human PSM cells. Labels in the x-axis correspond to distinct mass isotopomers with increasing number of heavy carbons. Mean ±SD. n=3 independent experiments.
Extended Data Figure 4.
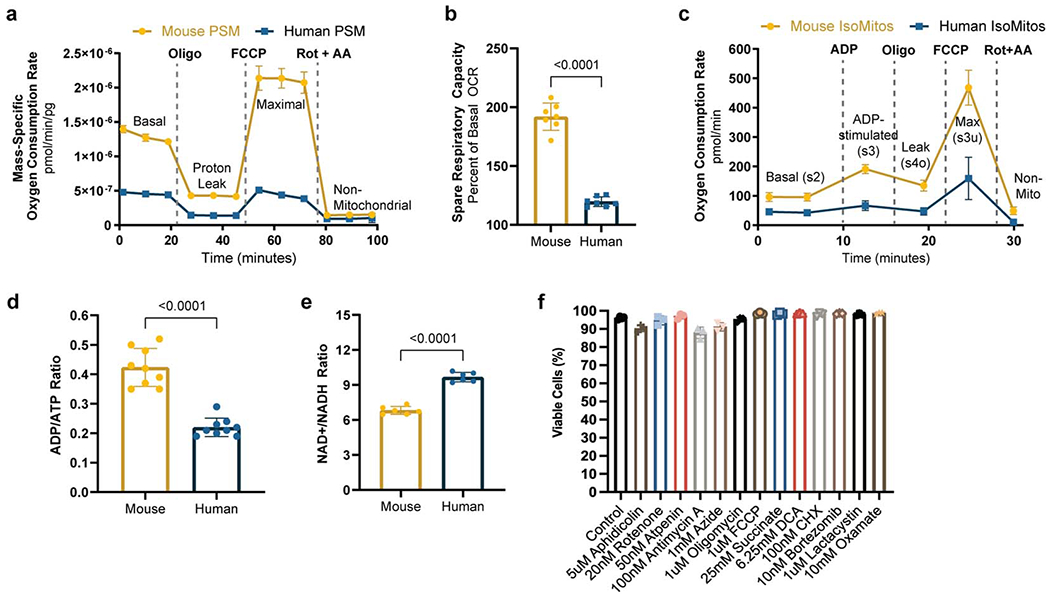
Mitochondrial properties of mouse and human PSM cells
a. Mass-specific oxygen consumption rate measured over the course of the mitochondrial stress test for MSGN1-Venus+ PSC-derived mouse (n=9) and human (n=7) PSM cells. 1μM oligomycin, 1μM FCCP, and 0.5μM Rotenone + 0.5μM Antimycin A were added at the timepoints marked by dotted lines. The first three timepoints denote basal respiration;, respiration after oligomycin addition corresponds to proton leak; FCCP induces maximal respiration; and rotenone/antimycin reveal non-mitochondrial respiration. Spare capacity refers to the difference between maximal and basal respiration rates. Mean ±SD.
b. Spare respiratory capacity in MSGN1-Venus+ PSC-derived mouse and human PSM cells. Mean ±SD. n=7 biological replicates. Unpaired two-sided t-test: p=2.7x10−9
c. Oxygen consumption rate profiles for mitochondria isolated from mouse and human PSC-derived PSM cells. Rates correspond to 10ug mitochondria seeded per assay well. ETC complex I substrates pyruvate and malate were provided to fuel respiration. 2mM ADP, 5μM Oligomycin, 6μM FCCP, and 1μM Rotenone with 1μM Antimycin A were injected at the timepoints marked by dotted lines. First two timepoints correspond to basal respiration (state 2), followed by ADP-stimulated respiration (state 3), then leak respiration (state 4o), followed by maximal FCCP-stimulated respiration (state 3u), and finally non-mitochondrial respiration. Mean ±SD. n=5 biological replicates.
d. Whole-cell NAD+/NADH ratio in MSGN1-Venus+ PSC-derived mouse and human PSM cells. Each datapoint represents the average of 3 technical replicates. Mean ±SD. n=6 biological replicates. Unpaired two-sided t-test: p=1.2x10−7
e. Whole-cell ADP/ATP ratio in MSGN1-Venus+ PSC-derived mouse and human PSM cells. Mean ±SD. n=9. Each datapoint represents the average of 3 technical replicates. Unpaired two-sided t-test: p=2.42x10−7
f. Percent viable cells as measured by trypan blue staining in human PSM cells treated with the indicated inhibitors for 24 hours. n=3 independent experiments.
Extended Data Figure 5.
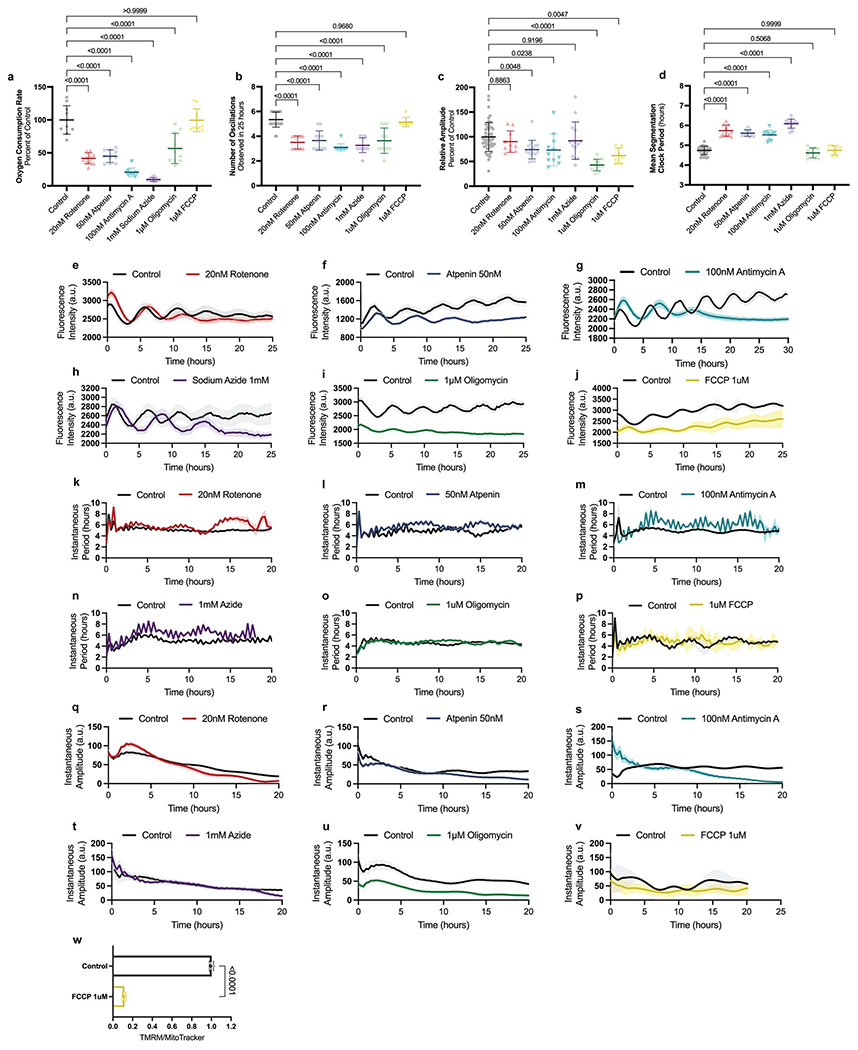
Effect of electron transport chain inhibitors on the segmentation clock
a. Basal oxygen consumption rate in human PSM cells treated with vehicle control (DMSO), 20nM rotenone, 50nM atpenin A5, 100nM antimycin A, 1mM sodium azide, 1μM oligomycin or 1μM FCCP. Mean ±SD. n=10 biological replicates. One-way ANOVA with Šidák correction: control vs. rotenone p=5.4x10−12, control vs. atpenin p=4.3x10−11, control vs. antimycin p=3.8x10−18, control vs. azide p=1.04x10−20, control vs. oligomycin p=5.9x10−8, control vs. FCCP p=0.999
b. Number of HES7-Achilles oscillations observed in 25 hours for human PSM cells treated with vehicle control (DMSO, n=29), 20nM rotenone (n=10), 50nM atpenin A5 (n=17), 100nM antimycin A (n=11), 1mM sodium azide (n=15), 1μM oligomycin (n=11), or 1μM FCCP (n=7). n denotes independent experiments. Mean ±SD. One-way ANOVA with Šidák correction: control vs. rotenone p=1.1x10−10, control vs. atpenin p=2.3x10−12, control vs. antimycin p=6.1x10−15, control vs. azide p=2.0x10−15, control vs. oligomycin p=4.7x10−10
c. Mean amplitude expressed as a percent of control for HES7-Achilles oscillations in human PSM cells treated with vehicle control (DMSO, n=53), 20nM rotenone (n=10), 50nM atpenin A5 (n=18), 100nM antimycin A (n=11), 1mM sodium azide (n=14), 1μM oligomycin (n=11), or 1μM FCCP (n=7). n denotes independent experiments. Mean ±SD. One-way ANOVA with Dunnett correction: control vs. oligomycin p=3.4x10−8
d. Mean segmentation clock period as calculated by Hilbert transformation for human PSM cells treated with vehicle control (DMSO, n=53), 20nM rotenone (n=10), 50nM atpenin A5 (n=18), 100nM antimycin A (n=11), 1mM sodium azide (n=14), 1μM oligomycin (n=11), or 1μM FCCP (n=7). n denotes independent experiments. Mean ±SD. One-way ANOVA with Šidák correction: control vs. rotenone p=8.9x10−21, control vs. atpenin p=1.8x10−18, control vs. antimycin p=2x10−15, control vs. azide p=1.4x10−33
e. HES7-Achilles oscillatory profile in human PSM cultures treated with DMSO control (n=10) or 20nM rotenone (n=11). n denotes independent experiments. Mean ±SEM.
f. HES7-Achilles oscillatory profile in human PSM cultures treated with DMSO control (n=12) or 50nM atpenin A5 (n=18). n denotes independent experiments. Mean ±SEM.
g. HES7-Achilles oscillatory profile in human PSM cultures treated with DMSO control (n=11) or 100nM antimycin A (n=13). n denotes independent experiments. Mean ±SEM.
h. HES7-Achilles oscillatory profile in human PSM cultures treated with DMSO control (n=9) or 1mM sodium azide (n=15). n denotes independent experiments. Mean ±SEM.
i. HES7-Achilles oscillatory profile in human PSM cultures treated with DMSO control (n=12) or 1μM oligomycin (n=17). n denotes independent experiments. Mean ±SEM.
j. HES7-Achilles oscillatory profile in human PSM cultures treated with DMSO control (n=3) or 1μM FCCP (n=7). n denotes independent experiments. Mean ±SEM.
k. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=10) or 20nM rotenone (n=11). n denotes independent experiments. Mean ±SEM.
l. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=12) or 50nM atpenin A5 (n=18). n denotes independent experiments. Mean ±SEM.
m. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=11) or 100nM antimycin A (n=13). n denotes independent experiments. Mean ±SEM.
n. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=9) or 1mM sodium azide (n=15). n denotes independent experiments. Mean ±SEM.
o. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=12) or 1μM oligomycin (n=17). n denotes independent experiments. Mean ±SEM.
p. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=3) or 1μM FCCP (n=7). n denotes independent experiments. Mean ±SEM.
q. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=10) or 20nM rotenone (n=11). n denotes independent experiments. Mean ±SEM.
r. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=12) or 50nM atpenin A5 (n=18). n denotes independent experiments. Mean ±SEM.
s. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=11) or 100nM antimycin A (n=13). n denotes independent experiments. Mean ±SEM.
t. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=9) or 1mM sodium azide (n=15). n denotes independent experiments. Mean ±SEM.
u. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=12) or 1μM oligomycin (n=17). n denotes independent experiments. Mean ±SEM.
v. Instantaneous HES7-Achilles oscillatory amplitude over the time as calculated by Hilbert transformation in human PSM cultures treated with DMSO control (n=3) or 1μM FCCP (n=7). n denotes independent experiments. Mean ±SEM.
w. Inner mitochondrial membrane potential (ΔΨm) in human PSM cells under control conditions or treated acutely with 1μM FCCP. TMRM fluorescence was normalized by mitochondrial content (MitoTracker Green) following flow cytometry. Mean ±SD. n=3 biological replicates. Unpaired two-sided t-test: p=6.4x10−7
Extended Data Figure 6.
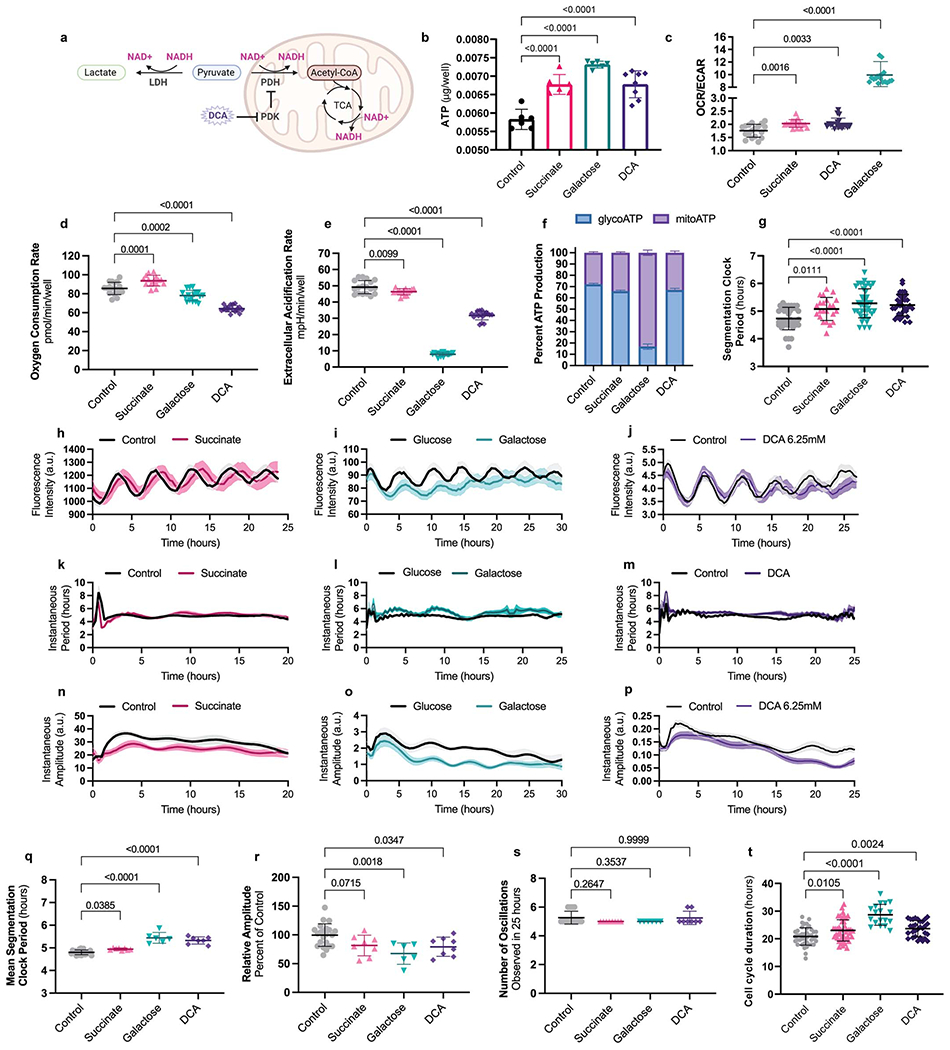
Increased ATP concentrations do not accelerate the segmentation clock
a. Illustration depicting the alternate fates of pyruvate and their regulation by metabolic enzymes. Lactate dehydrogenase (LDH) converts pyruvate to lactate and regenerates NAD+ from NADH. Pyruvate dehydrogenase (PDH) oxidizes pyruvate to acetyl coenzyme-A (acetyl-CoA) in the mitochondria and consumes NAD+. Acetyl -CoA then enters the tricarboxylic acid (TCA) cycle, which also consumes NAD+. Pyruvate dehydrogenase kinase (PDK) inhibits PDH by phosphorylating it. DCA is a PDK inhibitor that promotes the conversion of pyruvate to acetyl-CoA by relieving PDH inhibition. Created withBioRender.com.
b. ATP content per well for human PSM cells in control (water; n=6), 25mM succinate supplementation (n=6), 10mM galactose in the absence of glucose (n=6), and 6.25mM DCA (n=8) conditions after 24 hours of culture. In each case, 30,000 cells were seeded per assay well. Each datapoint represents the average of 3 technical replicates. Mean ±SD. One-way ANOVA with Šidák correction: control vs. succinate p=2x10−5, control vs. galactose p=1.6x10−8, control vs. DCA p=7.4x10−6
c. Ratio of Oxygen consumption rate (OCR) to extracellular acidification rate (ECAR) in human PSM cells treated with water control (n-18), 25mM succinate (n=15), 6.25mM DCA (n=18), or cultured with 10mM galactose instead of glucose (n=18) for 24 hours. Mean ±SD. One-way Brown-Forsythe and Welch ANOVA with Dunnett T3 correction: control vs. galactose p=2.5x10−12
d. Oxygen consumption rate (OCR) in human PSM cells treated with water control (n=18), 25mM succinate (n=15), 6.25mM DCA (n=18), or cultured with 10mM galactose instead of glucose (n=18) for 24 hours. Mean ±SD. One-way ANOVA with Dunnett correction: control vs. DCA p=3.8x10−18
e. Extracellular acidification rate (ECAR) in human PSM cells treated with water control (n=18), 25mM succinate (n=15), 6.25mM DCA (n=18), or cultured with 10mM galactose instead of glucose (n=18) for 24 hours. Mean ±SD. One-way ANOVA with Šidák correction: control vs. galactose p=3.05x10−51, control vs. DCA p=1x10−28
f. Percent of total ATP production corresponding to glycolysis (glycoATP) or mitochondrial respiration (mitoATP) in human PSM cells treated with water control (n=6), 25mM succinate (n=5), 6.25mM DCA (n=6), or cultured with 10mM galactose instead of glucose (n=6) for 24 hours. Mean ±SD.
g. HES7-Achilles oscillatory period in human PSM cells treated with vehicle control (water; n=36), 25mM succinate (n=23), 10mM galactose in the absence of glucose (n=43), and 6.25mM DCA (n=34). Mean ±SD. One-way ANOVA with Šidák correction: control vs. galactose p=5.9x10−7, control vs. DCA p=3.1x10−5
h. HES7-Achilles oscillatory profile in human PSM cells cultured under control conditions or supplemented with 25mM succinate. Mean ±SEM. n=8.
i. HES7-Achilles oscillatory profile in human PSM cells cultured with either 10mM glucose or 10mM galactose. Mean ±SEM. n=6.
j. HES7-Achilles oscillatory profile in human PSM cultures under control conditions (n=9) or 6.25mM DCA (n=8). Mean ±SEM.
k. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cells cultured under control conditions or supplemented with 25mM succinate. Mean ±SEM. n=8.
l. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cells cultured with either 10mM glucose or 10mM galactose. Mean ±SEM. n=6.
m. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cells cultured under control conditions (n=9) or supplemented with 6.25mM DCA (n=7). Mean ±SEM.
n. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cells cultured under control conditions or supplemented with 25mM succinate. Mean ±SEM. n=8.
o. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cells cultured with either 10mM glucose or 10mM galactose. Mean ±SEM. n=6.
p. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cells cultured under control conditions (n=9) or supplemented with 6.25mM DCA (n=8). Mean ±SEM.
q. Mean segmentation clock period as calculated by Hilbert transformation for human PSM cells treated with vehicle control (water; n=21), 25mM succinate (n=8), 10mM galactose in the absence of glucose (n=6), and 6.25mM DCA (n=8). n denotes independent experiments. Mean ±SD. One-way ANOVA with Šidák correction: contro vs. galactose p=3.3x10−12, control vs. DCA p=1.4x10−10.
r. Mean amplitude expressed as a percent of control for HES7-Achilles oscillations in human PSM cells treated with vehicle control (water; n=22), 25mM succinate (n=8), 10mM galactose in the absence of glucose (n=6), and 6.25mM DCA (n=7). n denotes independent experiments. Mean ±SD. One-way ANOVA with Dunnett correction.
s. Number of HES7-Achilles oscillations observed in 25 hours for human PSM cells treated with vehicle control (water; n=23), 25mM succinate (n=8), 10mM galactose in the absence of glucose (n=6), and 6.25mM DCA (n=8). n denotes independent experiments. Mean ±SD. One-way ANOVA with Dunnett correction.
t. Duration of the cell cycle in hours for human PSM cells treated with vehicle control (DMSO; n=42), 25mM succinate (n=44), 10mM galactose in the absence of glucose (n=18), and 6.25mM DCA (n=30). Mean ±SD. One-way ANOVA with Dunnett correction: control vs. galactose p=1.2x10−12
Extended Data Figure 7.
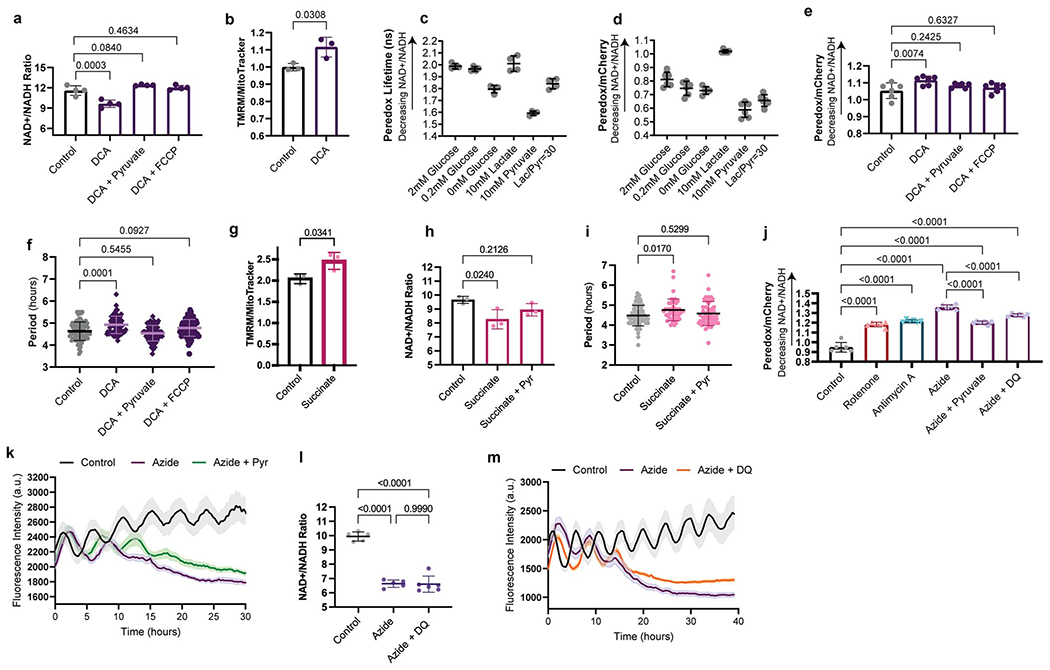
Rescue of the segmentation clock period by restoration of the NAD+/NADH ratio
a. Whole-cell NAD+/NADH ratio in vehicle-treated human PSM cells and cells treated with either 6.25mM DCA alone or DCA in combination with 1mM sodium pyruvate or 10nM FCCP for 24 hours. Each datapoint represents the average of 3 technical replicates. Mean ±SD. n=4. One-way ANOVA with Dunnett correction.
b. Inner mitochondrial membrane potential (ΔΨm) in human PSM cells under control conditions or treated with 6.25mM DCA for 24 hours. TMRM fluorescence was normalized by mitochondrial content (MitoTracker Green) following flow cytometry. Mean ±SD. n=3 biological replicates. Unpaired two-sided t-test.
c. Peredox-mCherryNLS fluorescence lifetime in human PSM cells cultured acutely in a balanced salt solution and supplemented with the indicated concentrations of glucose, lactate or pyruvate. Mean ±SD. n=4 biological replicates.
d. Ratiometric Peredox-to-mCherry fluorescence signal in human PSM cells cultured acutely in a balanced salt solution and supplemented with the indicated concentrations of glucose, lactate or pyruvate. Mean ±SD. n=6 biological replicates.
e. Ratiometric Peredox/mCherry signal in vehicle-treated human PSM cells and cells treated with either 6.25mM DCA alone or DCA in combination with 1mM sodium pyruvate or 10nM FCCP for 24 hours. Each datapoint represents the average of >200 individual cells analyzed within a biological replicate. Mean ±SD. n=6 biological replicates. One-way ANOVA with Dunnett correction.
f. HES7-Achilles oscillatory period in human PSM cells treated with vehicle control (water; n=78), 6.25mM DCA alone (n=68), DCA with 1mM sodium pyruvate (n=73), and DCA with 10nM FCCP (n=85). Mean ±SD. One-way ANOVA with Dunnett correction.
g. Inner mitochondrial membrane potential (ΔΨm) in human PSM cells under control conditions or treated with 25mM succinate for 24 hours. TMRM fluorescence was normalized by mitochondrial content (MitoTracker Green) following flow cytometry. Mean ±SD. n=3 biological replicates. Unpaired two-sided t-test.
h. Whole-cell NAD+/NADH ratio in vehicle-treated human PSM cells and cells treated with either 25mM succinate alone or succinate in combination with 1mM sodium pyruvate for 24 hours. Each datapoint represents the average of 3 technical replicates. Mean ±SD. n=3 biological replicates. One-way ANOVA with Dunnett correction.
i. HES7-Achilles oscillatory period in human PSM cells treated with vehicle control (water; n=62), 25mM succinate alone (n=46), or succinate with 1mM sodium pyruvate (n=46). Mean ±SD. One-way ANOVA with Dunnett correction.
j. Ratiometric Peredox/mCherry signal in DMSO-treated human PSM cells and cells treated with 20nM rotenone, 100nM antimycin A, 1mM sodium azide alone, azide with 1mM sodium pyruvate, and azide with 5μM duroquinone (DQ) for 24 hours. Each datapoint represents the average of >200 individual cells analyzed in a biological replicate. Mean ±SD. n=6. One-way ANOVA with Šidák correction: control vs. rotenone p= 2.9x10-15, control vs. antimycin p=7.1x10−17, control vs. azide p=4.4x10−22, control vs. azide+pyr p=3.3x10−16, control vs. azide+DQ p=2.2x10−19, azide vs. azide+pyr p=9.7x10−10, azide vs. azide+DQ p=5.3x10−6
k. HES7-Achilles oscillatory profile in human PSM cells cultures treated with DMSO control (n=10), 1mM sodium azide alone (n=13), and azide with 1mM sodium pyruvate (n=13). Mean ±SEM.
l. Whole-cell NAD+/NADH ratio in human PSM cells treated with vehicle-control (n=5), 1mM sodium azide alone (n=5), or azide with 5μM duroquinone (DQ) (n=6) for 24 hours. Each datapoint represents the average of 3 technical replicates. Mean ±SD. One-way ANOVA with Tukey correction: control vs. azide p=3.5x10−8, control vs. azide+DQ p=1.8x10−8
m. HES7-Achilles oscillatory profile in human PSM cells cultures treated with DMSO control (n=11), 1mM sodium azide alone (n=19), and azide with 5μM duroquinone (n=21). Mean ±SEM.
Extended Data Figure 8.
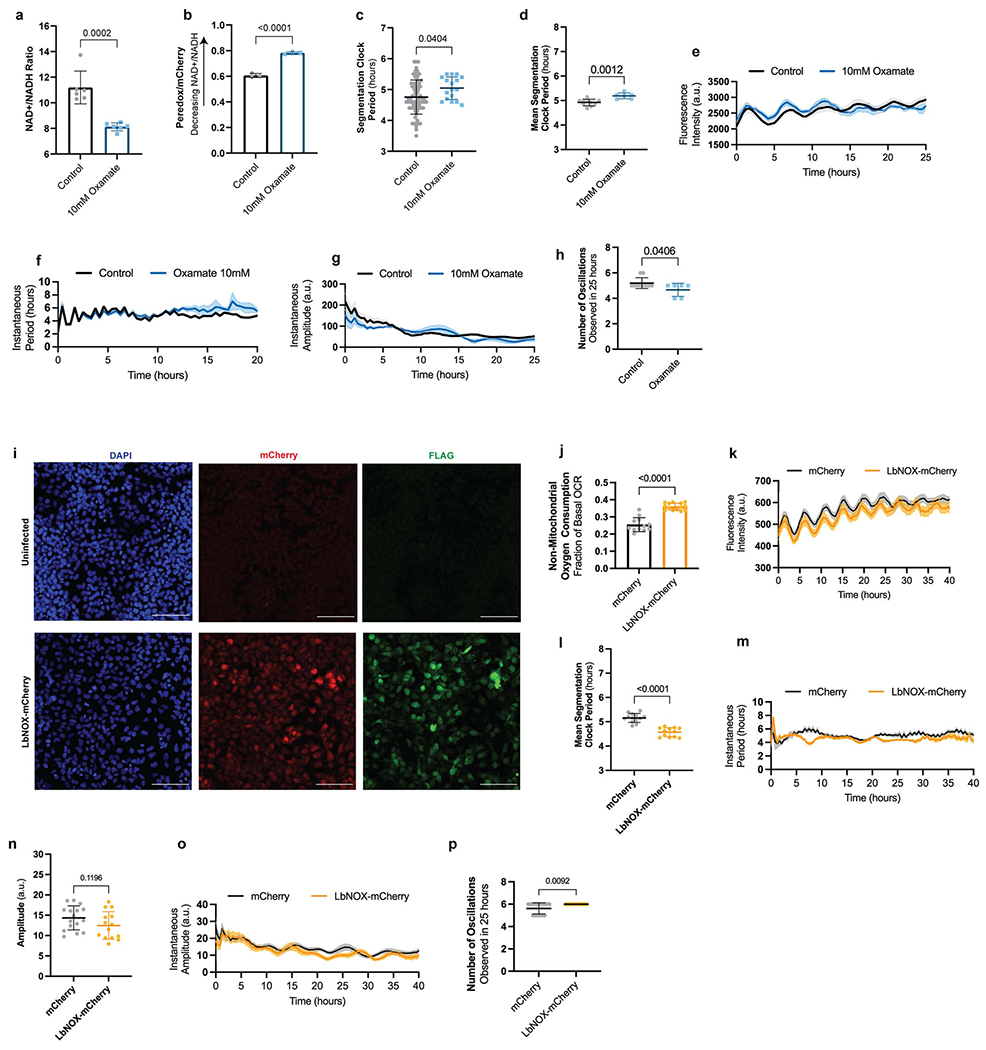
Modulation of the segmentation clock period by direct manipulation of the NAD+/NADH ratio
a. Whole-cell NAD+/NADH ratio human PSM cells under control conditions or treated acutely with 10mM oxamate. Each datapoint represents the average of 3 technical replicates. Mean ±SD. n=6. Unpaired two-sided t-test.
b. Ratiometric Peredox/mCherry signal in human PSM cells cultured under control conditions or treated acutely with 10mM oxamate. Each datapoint represents the average of >200 individual cells analyzed within a biological replicate. Mean ±SD. n=3. Unpaired two-sided t-test, p=8.3x10−5
c. Period of HES7-Achilles oscillations in human PSM cells cultures treated with water control (n=73) or 10mM sodium oxamate (n=17). Mean ±SEM. Unpaired two-sided t-test.
d. Mean segmentation clock period as calculated by Hilbert transformation for human PSM cells cultures treated with water control (n=10) or 10mM sodium oxamate (n=6). n denotes independent experiments. Mean ±SD. Unpaired two-sided t-test.
e. HES7-Achilles oscillatory profile in human PSM cells cultures treated with water control (n=10) or 10mM sodium oxamate (n=3). n denotes independent experiments. Mean ±SEM.
f. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cells cultures treated with water control (n=10) or 10mM sodium oxamate (n=3). n denotes independent experiments. Mean ±SEM.
g. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cells cultures treated with water control (n=10) or 10mM sodium oxamate (n=3). n denotes independent experiments. Mean ±SEM.
h. Number of HES7-Achilles oscillations observed in 25 hours in human PSM cells cultures treated with water control (n=10) or 10mM sodium oxamate (n=6). n denotes independent experiments. Mean ±SD. Unpaired two-sided t-test.
i. Representative micrographs of DAPI nuclear stain, mCherry endogenous fluorescence, and anti-FLAG immunofluorescence (LbNOX is flag-tagged in the C terminus [28]) in human PSM cells subjected to mock transduction (top) or transduced with LbNOX-mCherry (bottom). Similar results were obtained n=15 times. Scale bar = 100μm.
j. Non-mitochondrial oxygen consumption in human PSM cells transduced with a lentivirus expressing either mCherry alone (n=12) or LbNOX in combination with mCherry (n=14). OCR after addition of 0.5μM rotenone and 0.5μM antimycin A is expressed as fraction of basal OCR. Mean ±SD. Unpaired two-sided t-test: p=1.04x10−8
k. HES7-Achilles oscillatory profile over the course of 40 hours for human PSM cells transduced with a lentivirus expressing either mCherry alone (n=16) or LbNOX in combination with mCherry (n=14). Mean ±SEM.
l. Mean segmentation clock period as calculated by Hilbert transformation for human PSM cells transduced with a lentivirus expressing either mCherry alone (n=13) or LbNOX in combination with mCherry (n=12). Mean ±SD. Unpaired two-sided t-test: p=4.87x10−8
m Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation in human PSM cells transduced with a lentivirus expressing either mCherry alone (n=13) or LbNOX in combination with mCherry (n=12). Mean ±SD.
n. Mean HES7-Achilles oscillation amplitude in human PSM cells transduced with a lentivirus expressing either mCherry alone (n=16) or LbNOX in combination with mCherry (n=14). Mean ±SD. Unpaired two-sided t-test.
o. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation in human PSM cells transduced with a lentivirus expressing either mCherry alone (n=16) or LbNOX in combination with mCherry (n=14). Mean ±SD.
p. Number of oscillations (peaks) observed in 25 hours for human PSM cells transduced with a lentivirus expressing either mCherry alone (n=16) or LbNOX in combination with mCherry (n=14). Mean ±SD. Unpaired two-sided t-test.
Extended Data Figure 9.
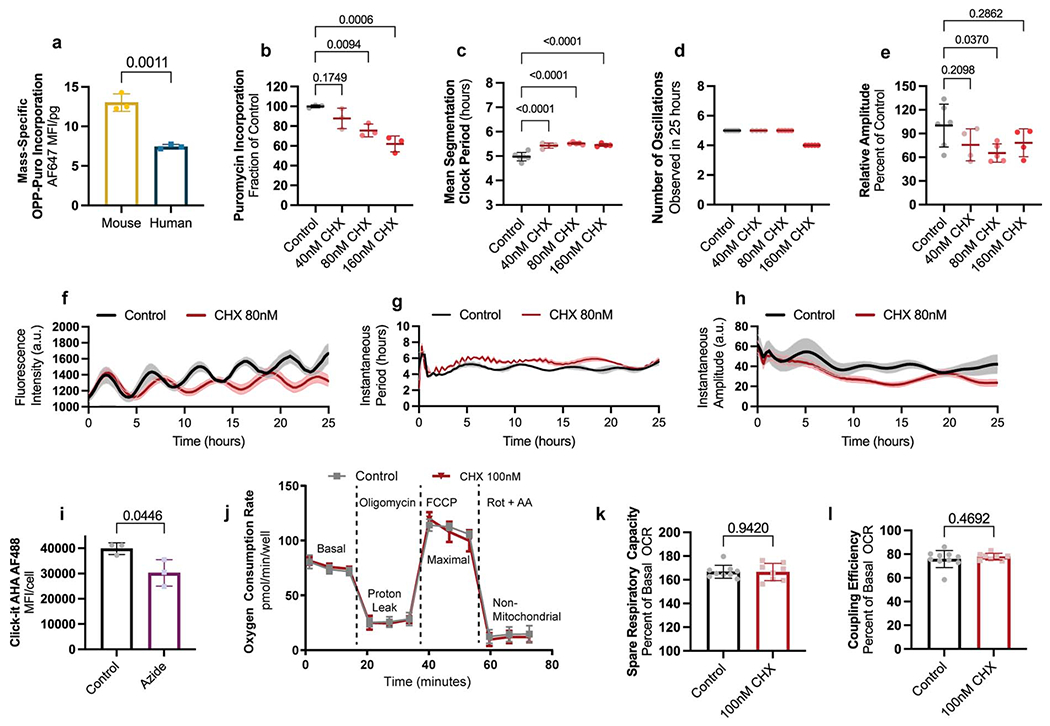
The segmentation clock is sensitive to the rate of translation
a. Mass-specific OPP-Puromycin incorporation as a measure of global translation rate in MSGN1-Venus+ PSC-derived mouse and human PSM cells. OPP-Puromycilated peptides were detected by click chemistry with AlexaFluor647-Picoyl Azide. Mean ±SD. n=3 biological replicates. Unpaired two-sided t-test.
b. Translation rate as measured by puromycin incorporation expressed as percent of control for human PSM cells treated with DMSO, 40nM, 80nM or 160nM cycloheximide (CHX) for 24 hours. Mean ±SD. n=3 biological replicates. One-way ANOVA with Dunnett correction.
c. Mean segmentation clock period as calculated by Hilbert transformation for human PSM cells treated with DMSO (n=6), 40nM (n=4), 80nM (n=5) or 160nM (n=4) cycloheximide (CHX) for 24 hours. n denotes independent experiments. Mean ±SD. One-way ANOVA with Šidák correction: control vs. 40nM p=7.6x10−5, control vs. 80nM p=4.5x10−6, control vs. 160nM p=4.2x10−5
d. Number of HES7-Achilles oscillations observed in 25 hours in human PSM cells treated with DMSO (n=6), 40nM (n=4), 80nM (n=5) or 160nM (n=5) cycloheximide (CHX) for 24 hours. n denotes independent experiments. Mean ±SD.
e. Mean amplitude expressed as a percent of control in human PSM cells treated with DMSO (n=6), 40nM (n=4), 80nM (n=5) or 160nM (n=4) cycloheximide (CHX) for 24 hours. n denotes independent experiments. Mean ±SD. One-way ANOVA with Dunnett correction
f. HES7-Achilles oscillatory profile for human PSM cells treated with DMSO-control (n=3) or 80nM cycloheximide (CHX; n=5). n denotes independent experiments. Mean ±SEM.
g. Instantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation for human PSM cells treated with DMSO-control (n=3) or 80nM cycloheximide (CHX, n=5). n denotes independent experiments. Mean ±SEM.
h. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation for human PSM cells treated with DMSO-control (n=3) or 80nM cycloheximide (CHX, n=5). n denotes independent experiments. Mean ±SEM.
k. Translation rate as measured by incorporation of the methionine analog AHA in human PSM cells treated with either DMSO control or 1mM sodium azide for one hour. Mean ±SD. n=3 biological replicates. Unpaired two-sided t-test.
j. Oxygen consumption rate measured over the course of the mitochondrial stress test for human PSM cells treated with DMSO control or 100nM cycloheximide for 24 hours. 1μM Oligomycin, 1μM FCCP, and 0.5μM Rotenone + 0.5μM Antimycin A were added at the timepoints marked by dotted lines. Mean ±SD. n=9 biological replicates.
k. Spare respiratory capacity in human PSM cells treated with vehicle control (DMSO) or 100nM cycloheximide (CHX) for 24 hours. Mean ±SD. n=8 biological replicates. Unpaired two-sided t-test.
l. Coupling efficiency shown as the percent of basal oxygen consumption that is linked to ATP production in human PSM cells treated with vehicle control (DMSO) or 100nM cycloheximide (CHX) for 24 hours. Mean ±SD. n=8 biological replicates. Unpaired two-sided t-test.
Extended Data Figure 10.
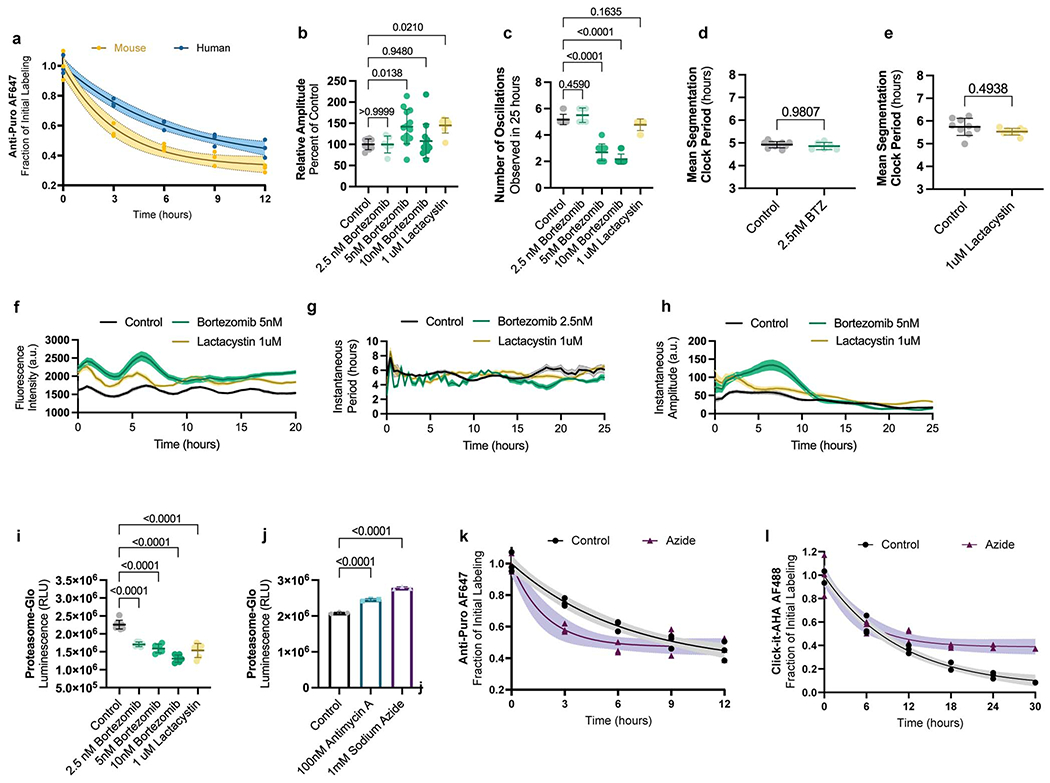
Protein stability differences between mouse and human PSM cells
a. Pulse-chase experiment tracking the degradation of puromycilated peptides over the course of 12 hours in MSGN1-Venus+ PSC-derived mouse and human PSM cells following a 1-hour pulse with puromycin. Solid line represents best one-phase decay fit with the 95% confidence intervals shown as shaded regions. n=3 independent experiments.
b. Mean amplitude expressed as a percent of control in human PSM treated with DMSO control (n=9), 2.5nM (n=6), 5nM (n=13) or 10nM (n=12) bortezomib, or 1μM lactacystin (n=8). n denotes independent experiments. Mean ±SD. One-way ANOVA with Dunnett correction.
c. Number of HES7-Achilles oscillations observed in 25 hours in human PSM treated with DMSO control (n=17), 2.5nM (n=6), 5nM (n=13) or 10nM (n=12) bortezomib, or 1μM lactacystin (n=9). n denotes independent experiments. Mean ±SD. One-way ANOVA with Šidák correction: control vs. 5nM bortezomib p=2.4x10−19, control vs. 10nM bortezomib p=1.8x10−22
d. Mean segmentation clock period as calculated by Hilbert transformation for human PSM cells treated with DMSO control (n=10) or 2.5nM bortezomib (BTZ, n=6). n denotes independent experiments. Mean ±SD. Unpaired two-sided t-test.
e. Mean segmentation clock period as calculated by Hilbert transformation for human PSM cells treated with DMSO control (n=9) or 1μM lactacystin (n=8). n denotes independent experiments. Mean ±SD. Unpaired two-sided t-test.
f. HES7-Achilles oscillatory profile for human PSM cells treated with DMSO-control (n=9), 5nM bortezomib (n=13), or 1μM lactacystin (n=8). Mean ±SEM.
g. stantaneous HES7-Achilles oscillatory period over time as calculated by Hilbert transformation for human PSM cells treated with DMSO-control (n=9), 2.5nM bortezomib (n=6), or 1μM lactacystin (n=8). Mean ±SEM.
h. Instantaneous HES7-Achilles oscillatory amplitude over time as calculated by Hilbert transformation for human PSM cells treated with DMSO-control (n=9), 5nM bortezomib (n=13), or 1μM lactacystin (n=8). Mean ±SEM.
i. Proteasome activity cells as measured by cleavage of a luminogenic proteasome substrate in human PSM treated with DMSO control, 2.5nM, 5nM or 10nM bortezomib, or 1μM lactacystin for 24 hours. Mean ±SD. n=6 biological replicates. One-way ANOVA with Šidák correction: control vs. 2.5nM bortezomib p=5.8x10−8, control vs. 5nM bortezomib p=1.2x10−9, control vs. 10nM bortezomib p=4.3x10−13, control vs. 1μM lactacystin p=2.2x10−10.
j. Proteasome activity cells as measured by cleavage of a luminogenic proteasome substrate in human PSM treated with DMSO control, 100nM antimycin A, or 1mM sodium azide for 24 hours. Mean ±SD. n=3 biological replicates. One-way ANOVA with Šidák correction: control vs. antimycin p=1.3x10−5, control vs. azide p=3.1x10−7
k. Pulse-chase experiment tracking the degradation of puromycilated peptides over the course of 12 hours in human PSM cells treated with DMSO control or 1mM sodium azide following a 1-hour pulse with puromycin. Solid line represents best one-phase decay fit with the 95% confidence intervals shown as shaded regions. n=3 independent experiments.
l. Pulse-chase experiment tracking the degradation of AHA-labeled proteins over the course of 30 hours in human PSM cells treated with DMSO control of 1mM sodium azide following a 1-hour pulse with AHA. Solid line represents best one-phase decay fit with the 95% confidence intervals shown as shaded regions. n=3 independent experiments.
Supplementary Material
Acknowledgements:
We thank members of the Pourquié laboratory and Dr. Jason Locasale, Dr. Gary Ruvkun, Dr. Clifford Tabin, Dr. Norbert Perrimon, Dr. Sneha Rath and Dr. Vamsi Mootha for critical reading of the manuscript and discussions. The Pourquié laboratory and M.D.C. were funded by grants from the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD) of the National Institutes of Health (NIH) under award numbers 2R01HD085121-06A and F31HD100033, respectively. MDC is a recipient of the NASEM Ford Foundation Dissertation fellowship. This work was supported by the Cancer Systems Biology Consortium funding (U54-CA217377) from the National Cancer Institute (S.R.M.) and the Virginia and D.K. Ludwig Fund for Cancer Research (S.R.M.). OSS was supported by the F32GM133047 from the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank the NeuroTechnology Studio at Brigham and Women’s Hospital for providing access to the Agilent Seahorse XF96 extracellular flux analyzer and Zeiss LSM880 confocal microscope, and for consultation on data acquisition and data analysis. We also thank the Center for Neurologic Diseases (ARCND) Flow Cytometry Core Facility at Brigham and Women’s Hospital for access to the BD Fortessa flow cytometer. In addition, we thank the Seahorse core facility at Brigham and Women’s hospital for training and access to the Agilent Seahorse XFe24 extracellular flux analyzer. We also express our gratitude towards Dr. Seungeun Oh and the Kirschner laboratory at Harvard Medical School for generously providing access to the Moxi Go II for cell volume measurements. We also thank the Olga Goldberger and Vamsi Mootha for providing the LbNOX plasmids. Finally, we acknowledge Manish Bharadwaj from Agilent Technologies for guidance in the implementation of Seahorse measurements on isolated mitochondria.
Footnotes
Competing interests: O.P. is scientific founder of Anagenesis Biotechnologies. S.R.M. is a co-founder of Travera and Affinity Biosensors, which develop technologies relevant to the research presented in this work. All other authors declare no competing interests.
Biological Materials: All materials used in this study, including stem cell lines carrying knock-in reporters, are available by request from the corresponding author.
Code Availability Statement: The MATLAB code for Hilbert analysis of oscillation properties can be found in github: https://github.com/md2981/Hilbert-Segmentation-Clock. The R custom package, mzrtools, for correction of natural isotope abundance in raw mass isotopomer distributions can also be found in github: https://github.com/wmoldham/mzrtools.
Data Availability Statement:
All data are available within the article, supplementary files and source data files. Source data are provided with this paper.
References
- 1.Matsuda M, et al. , Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science, 2020. 369(6510): p. 1450. [DOI] [PubMed] [Google Scholar]
- 2.Rayon T, et al. , Species-specific pace of development is associated with differences in protein stability. Science, 2020. 369(6510): p. eaba7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoyle NP and Ish-Horowicz D, Transcript processing and export kinetics are rate-limiting steps in expressing vertebrate segmentation clock genes. Proceedings of the National Academy of Sciences, 2013. 110(46): p. E4316–E4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stearns SC, The Evolution of Life History Traits: A Critique of the Theory and a Review of the Data. Annual Review of Ecology and Systematics, 1977. 8(1): p. 145–171. [Google Scholar]
- 5.Ricklefs RE, Embryo development and ageing in birds and mammals. Proceedings. Biological sciences, 2006. 273(1597): p. 2077–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blagosklonny MV, Big mice die young but large animals live longer. Aging, 2013. 5(4): p. 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otis EM and Brent R, Equivalent ages in mouse and human embryos. Anat Rec, 1954. 120(1): p. 33–63. [DOI] [PubMed] [Google Scholar]
- 8.Schröter C, et al. , Dynamics of zebrafish somitogenesis. Dev Dyn, 2008. 237(3): p. 545–53. [DOI] [PubMed] [Google Scholar]
- 9.Pourquié O, Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell, 2011. 145(5): p. 650–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubaud A and Pourquie O, Signalling dynamics in vertebrate segmentation. Nat Rev Mol Cell Biol, 2014. 15(11): p. 709–21. [DOI] [PubMed] [Google Scholar]
- 11.Diaz-Cuadros M, et al. , In vitro characterization of the human segmentation clock. Nature, 2020. 580(7801): p. 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda M, et al. , Recapitulating the human segmentation clock with pluripotent stem cells. Nature, 2020. 580(7801): p. 124–129. [DOI] [PubMed] [Google Scholar]
- 13.Chu LF, et al. , An In Vitro Human Segmentation Clock Model Derived from Embryonic Stem Cells. Cell Rep, 2019. 28(9): p. 2247–2255.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumiya M, et al. , ES cell-derived presomitic mesoderm-like tissues for analysis of synchronized oscillations in the segmentation clock. Development, 2018. 145(4): p. dev156836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubaud A, et al. , Excitable Dynamics and Yap-Dependent Mechanical Cues Drive the Segmentation Clock. Cell, 2017. 171(3): p. 668–682 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson MH and Day ML, Egg timers: how is developmental time measured in the early vertebrate embryo? Bioessays, 2000. 22(1): p. 57–63. [DOI] [PubMed] [Google Scholar]
- 17.Kleiber M, Body size and metabolic rate. Physiol Rev, 1947. 27(4): p. 511–41. [DOI] [PubMed] [Google Scholar]
- 18.Martin RD, Genoud M, and Hemelrijk CK, Problems of allometric scaling analysis: examples from mammalian reproductive biology. Journal of Experimental Biology, 2005. 208(9): p. 1731–1747. [DOI] [PubMed] [Google Scholar]
- 19.Feijó Delgado F, et al. , Intracellular Water Exchange for Measuring the Dry Mass, Water Mass and Changes in Chemical Composition of Living Cells. PLOS ONE, 2013. 8(7): p. e67590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miettinen TP, et al. , Single-cell monitoring of dry mass and dry mass density reveals exocytosis of cellular dry contents in mitosis. eLife, 2022. 11: p. e76664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oginuma M, et al. , A Gradient of Glycolytic Activity Coordinates FGF and Wnt Signaling during Elongation of the Body Axis in Amniote Embryos. Developmental cell, 2017. 40(4): p. 342–353.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulusu V, et al. , Spatiotemporal Analysis of a Glycolytic Activity Gradient Linked to Mouse Embryo Mesoderm Development. Dev Cell, 2017. 40(4): p. 331–341.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter RK, Allometry of mammalian cellular oxygen consumption. Cellular and Molecular Life Sciences CMLS, 2001. 58(5): p. 815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter RK, Hulbert AJ, and Brand MD, Allometry of mitochondrial proton leak: influence of membrane surface area and fatty acid composition. Am J Physiol, 1996. 271(6 Pt 2): p. R1550–60. [DOI] [PubMed] [Google Scholar]
- 25.Harris M, Pyruvate blocks expression of sensitivity to antimycin A and chloramphenicol. Somatic Cell Genet, 1980. 6(6): p. 699–708. [DOI] [PubMed] [Google Scholar]
- 26.Luengo A, et al. , Increased demand for NAD(+) relative to ATP drives aerobic glycolysis. Mol Cell, 2021. 81(4): p. 691–707.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung YP, et al. , Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell metabolism, 2011. 14(4): p. 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Titov DV, et al. , Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science (New York, N.Y.), 2016. 352(6282): p. 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merker MP, et al. , Influence of pulmonary arterial endothelial cells on quinone redox status: effect of hyperoxia-induced NAD(P)H:quinone oxidoreductase 1. Am J Physiol Lung Cell Mol Physiol, 2006. 290(3): p. L607–19. [DOI] [PubMed] [Google Scholar]
- 30.Fessel JP and Oldham WM, Pyridine Dinucleotides from Molecules to Man. Antioxidants & Redox Signaling, 2017. 28(3): p. 180–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williamson DH, Lund P, and Krebs HA, The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J, 1967. 103(2): p. 514–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veech RL, Guynn R, and Veloso D, The time-course of the effects of ethanol on the redox and phosphorylation states of rat liver. Biochem J, 1972. 127(2): p. 387–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.das Neves RP, et al. , Connecting Variability in Global Transcription Rate to Mitochondrial Variability. PLOS Biology, 2010. 8(12): p. e1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnston IG, et al. , Mitochondrial Variability as a Source of Extrinsic Cellular Noise. PLOS Computational Biology, 2012. 8(3): p. e1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guantes R, et al. , Global variability in gene expression and alternative splicing is modulated by mitochondrial content. Genome Research, 2015. 25(5): p. 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hidalgo San Jose L and Signer RAJ, Cell-type-specific quantification of protein synthesis in vivo. Nat Protoc, 2019. 14(2): p. 441–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt EK, et al. , SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods, 2009. 6(4): p. 275–7. [DOI] [PubMed] [Google Scholar]
- 38.Bessho Y, et al. , Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes & development, 2003. 17(12): p. 1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonvini P, et al. , Bortezomib-mediated 26S proteasome inhibition causes cell-cycle arrest and induces apoptosis in CD-30+ anaplastic large cell lymphoma. Leukemia, 2007. 21(4): p. 838–42. [DOI] [PubMed] [Google Scholar]
- 40.Gomez C, et al. , Control of segment number in vertebrate embryos. Nature, 2008. 454(7202): p. 335–9. [DOI] [PubMed] [Google Scholar]
- 41.Hosios AM and Vander Heiden MG, The redox requirements of proliferating mammalian cells. J Biol Chem, 2018. 293(20): p. 7490–7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Covarrubias AJ, et al. , NAD+ metabolism and its roles in cellular processes during ageing. Nature Reviews Molecular Cell Biology, 2021. 22(2): p. 119–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fushan AA, et al. , Gene expression defines natural changes in mammalian lifespan. Aging Cell, 2015. 14(3): p. 352–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bao XR, et al. , Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mick E, et al. , Distinct mitochondrial defects trigger the integrated stress response depending on the metabolic state of the cell. eLife, 2020. 9: p. e49178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Materials and Methods References
- 46.Chal J, et al. , Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat Biotechnol, 2015. 33(9): p. 962–9. [DOI] [PubMed] [Google Scholar]
- 47.Chal J, et al. , Recapitulating early development of mouse musculoskeletal precursors of the paraxial mesoderm in vitro. Development, 2018. 145(6). [DOI] [PubMed] [Google Scholar]
- 48.Chal J, et al. , Generation of human muscle fibers and satellite-like cells from human pluripotent stem cells in vitro. Nat Protoc, 2016. 11(10): p. 1833–50. [DOI] [PubMed] [Google Scholar]
- 49.Tomishima M, Neural induction – Dual SMAD inhibition, in StemBook. 2008, Harvard Stem Cell Institute. Copyright: © 2012 Mark Tomishima.: Cambridge (MA). [PubMed] [Google Scholar]
- 50.Oceguera-Yanez F, et al. , Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods, 2016. 101: p. 43–55. [DOI] [PubMed] [Google Scholar]
- 51.Yellen G and Mongeon R, Quantitative two-photon imaging of fluorescent biosensors. Curr Opin Chem Biol, 2015. 27: p. 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindelin J, et al. , Fiji: an open-source platform for biological-image analysis. Nature Methods, 2012. 9(7): p. 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aulehla A, et al. , A beta-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat Cell Biol, 2008. 10(2): p. 186–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dequéant ML, et al. , A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science, 2006. 314(5805): p. 1595–8. [DOI] [PubMed] [Google Scholar]
- 55.Krol AJ, et al. , Evolutionary plasticity of segmentation clock networks. Development (Cambridge, England), 2011. 138(13): p. 2783–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niwa Y, et al. , The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev Cell, 2007. 13(2): p. 298–304. [DOI] [PubMed] [Google Scholar]
- 57.Oppenheim AV, Schafer RW, and Buck JR., Discrete-Time Signal Processing. 2nd ed. 1999, Upper Saddle River, NJ: Prentice Hall. [Google Scholar]
- 58.Bharadwaj MS, et al. , Preparation and respirometric assessment of mitochondria isolated from skeletal muscle tissue obtained by percutaneous needle biopsy. Journal of visualized experiments : JoVE, 2015(96): p. 52350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim D, et al. , SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature, 2015. 520(7547): p. 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez CA, et al. , Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom, 1996. 31(3): p. 255–62. [DOI] [PubMed] [Google Scholar]
- 61.Mu L, et al. , Mass measurements during lymphocytic leukemia cell polyploidization decouple cell cycle- and cell size-dependent growth. Proc Natl Acad Sci U S A, 2020. 117(27): p. 15659–15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheaff R, Ilsley D, and Kuchta R, Mechanism of DNA polymerase alpha inhibition by aphidicolin. Biochemistry, 1991. 30(35): p. 8590–7. [DOI] [PubMed] [Google Scholar]
- 63.Palmer G, et al. , Studies on the respiratory chain-linked reduced nicotinamide adenine dinucleotide dehydrogenase. XIV. Location of the sites of inhibition of rotenone, barbiturates, and piericidin by means of electron paramagnetic resonance spectroscopy. J Biol Chem, 1968. 243(4): p. 844–7. [PubMed] [Google Scholar]
- 64.Miyadera H, et al. , Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proceedings of the National Academy of Sciences, 2003. 100(2): p. 473–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slater EC, The mechanism of action of the respiratory inhibitor, antimycin. Biochim Biophys Acta, 1973. 301(2): p. 129–54. [DOI] [PubMed] [Google Scholar]
- 66.Keilin D, The action of sodium azide on cellular respiration and on some catalytic oxidation reactions. Proceedings of the Royal Society of London. Series B - Biological Sciences, 1936. 121(822): p. 165–173. [Google Scholar]
- 67.Kankotia S and Stacpoole PW, Dichloroacetate and cancer: new home for an orphan drug? Biochim Biophys Acta, 2014. 1846(2): p. 617–29. [DOI] [PubMed] [Google Scholar]
- 68.Benz R and McLaughlin S, The molecular mechanism of action of the proton ionophore FCCP (carbonylcyanide p-trifluoromethoxyphenylhydrazone). Biophys J, 1983. 41(3): p. 381–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Racker E, A mitochondrial factor conferring oligomycin sensitivity on soluble mitochondrial ATPase. Biochem Biophys Res Commun, 1963. 10: p. 435–9. [DOI] [PubMed] [Google Scholar]
- 70.Baliga BS, Pronczuk AW, and Munro HN, Mechanism of Cycloheximide Inhibition of Protein Synthesis in a Cell-free System Prepared from Rat Liver. Journal of Biological Chemistry, 1969. 244(16): p. 4480–4489. [PubMed] [Google Scholar]
- 71.Fu Z, et al. , PSMA5 promotes the tumorigenic process of prostate cancer and is related to bortezomib resistance. Anticancer Drugs, 2019. 30(7): p. e0773. [DOI] [PubMed] [Google Scholar]
- 72.Dick LR, et al. , Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem, 1997. 272(1): p. 182–8. [DOI] [PubMed] [Google Scholar]
- 73.Zhou M, et al. , Warburg effect in chemosensitivity: targeting lactate dehydrogenase-A re-sensitizes taxol-resistant cancer cells to taxol. Mol Cancer, 2010. 9: p. 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Aguer C, et al. , Galactose Enhances Oxidative Metabolism and Reveals Mitochondrial Dysfunction in Human Primary Muscle Cells. PLOS ONE, 2011. 6(12): p. e28536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available within the article, supplementary files and source data files. Source data are provided with this paper.


