Abstract
Subcellular compartmentalization is a defining feature of all cells. In prokaryotes, compartmentalization is generally achieved via protein-based strategies. The two main classes of microbial protein compartments are bacterial microcompartments and encapsulin nanocompartments. Encapsulins self-assemble into proteinaceous shells with diameters between 24 and 42 nm and are defined by the viral HK97-fold of their shell protein. Encapsulins have the ability to encapsulate dedicated cargo proteins, including ferritin-like proteins, peroxidases, and desulfurases. Encapsulation is mediated by targeting sequences present in all cargo proteins. Encapsulins are found in many bacterial and archaeal phyla and have been suggested to play roles in iron storage, stress resistance, sulfur metabolism, and natural product biosynthesis. Phylogenetic analyses indicate that they share a common ancestor with viral capsid proteins. Many pathogens encode encapsulins, and recent evidence suggests that theymaycontribute toward pathogenicity. The existing information on encapsulin structure, biochemistry, biological function, and biomedical relevance is reviewed here.
Keywords: encapsulin, nanocompartment, protein organelle, self-assembly, natural products, stress resistance
INTRODUCTION
It is increasingly evident that the interior of prokaryotic cells is highly organized (1, 2). In contrast to eukaryotes, prokaryotes do not generally possess lipid-based organelles but instead rely on protein-based compartmentalization strategies to achieve spatial control and subcellular organization. Historically, bacterial microcompartments (BMCs) were the main class of protein-based organelles recognized in bacteria (3). BMCs consist of sets of enzymes sequestered in large protein shells, often with diameters larger than 100 nm, consisting of multiple kinds of shell protomers (4, 5). BMCs can either function in an anabolic capacity, exemplified by the carbon-fixing carboxysome found in cyanobacteria and some chemoautotrophs (6, 7), or fulfill a catabolic role as metabolosomes in the utilization of carbon and nitrogen sources (8-10). More recently, a second widespread class of protein compartments called encapsulins has been discovered and represents an emerging paradigm for intracellular spatial control and organization. Encapsulins are proteinaceous nanoscale compartments also able to specifically encapsulate dedicated enzymatic components (11, 12). Intracellular compartments with a proteomically defined interior and a discrete boundary fulfilling distinct physiological functions are generally referred to as organelles (13). This includes lipid-based organelles, phase-separated structures, and protein-based compartments, all of which have been described in prokaryotic cells in recent years. Compartmentalization generally serves four distinct functions: the creation of distinct reaction spaces and environments, storage, transport, and regulation (13). Often, it serves several of these functions at the same time.
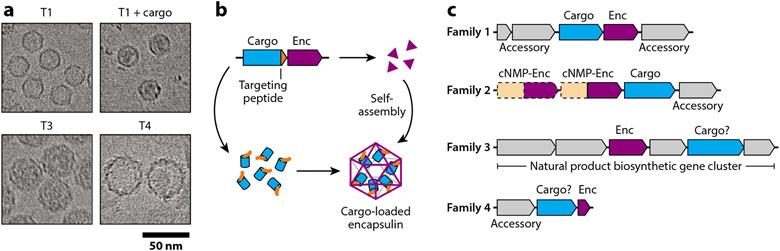
All encapsulins studied so far self-assemble from a single shell protomer into icosahedral compartments with diameters between 24 and 42 nm and with triangulation numbers of T1, T3, or T4 (12, 14) (Figure 1a). Their defining feature is the ability to encapsulate dedicated cargo proteins, including ferritin-like proteins (Flps), peroxidases, hemerythrins, and desulfurases (Figure 1b). Cargo encapsulation is mediated either by short, conserved peptide sequences at the termini of cargo proteins—referred to as targeting peptides (TPs) or cargo-loading peptides—or by more extended N-terminal encapsulation-mediating domains (15-17). TPs can be used to identify cargo proteins at the sequence level. Encapsulin systems can be found in many bacterial and archaeal phyla and have so far been suggested to play roles in iron storage, oxidative stress resistance, anaerobic ammonium oxidation, and sulfur metabolism (12, 14). Based on recent genome-mining studies, encapsulins have been grouped into four distinct families reflecting differences in sequence, structure, and operon organization (18) (Figure 1c). In contrast to all other known microbial protein compartments or organelles, the encapsulin shell protein shares the HK97 (Hong Kong 97) phage-like fold, pointing to an evolutionary connection with the world of bacteriophages and viruses (18).
Figure 1.
Encapsulin assembly and classification. (a) Representative cryogenic electron micrographs of differently sized encapsulin shells: T1, Thermotoga maritima; T1 + cargo, Acinetobacter sp. 1289694; T3, Kuenenia stuttgartiensis, and T4, Quasibacillus thermotolerans. (b) Schematic outlining concurrent Family 1 encapsulin cargo loading and shell self-assembly. (c) Four-family classification scheme for encapsulins based on sequence similarity and genome neighborhood analysis. Question marks indicate experimentally unconfirmed cargo proteins or enzymatic components. Abbreviations: cNMP, cyclic nucleotide; Enc, encapsulin.
A common theme for all encapsulin families is their historic misannotation and mischaracterization, which initially stifled progress in understanding their molecular and physiological functions. A contributing factor to this was likely their unusual HK97-fold, which does not show any homology to known cellular proteins. The specific histories of the four encapsulin families are discussed in more detail in the section titled Evolutionary Origin and Comparative Analysis. In 2008, encapsulins were finally recognized as protein-based compartmentalization systems with dedicated functions in cellular organisms (11).
Due to their robust self-assembly and engineerability, encapsulins have found widespread use in biomolecular and protein engineering applications. A number of recent in-depth reviews (14, 19-22) have discussed the engineering of encapsulins and their use in biomedical and biotechnological applications, and the reader is referred to them for more information on this topic.
DISTRIBUTION, DIVERSITY, AND CLASSIFICATION
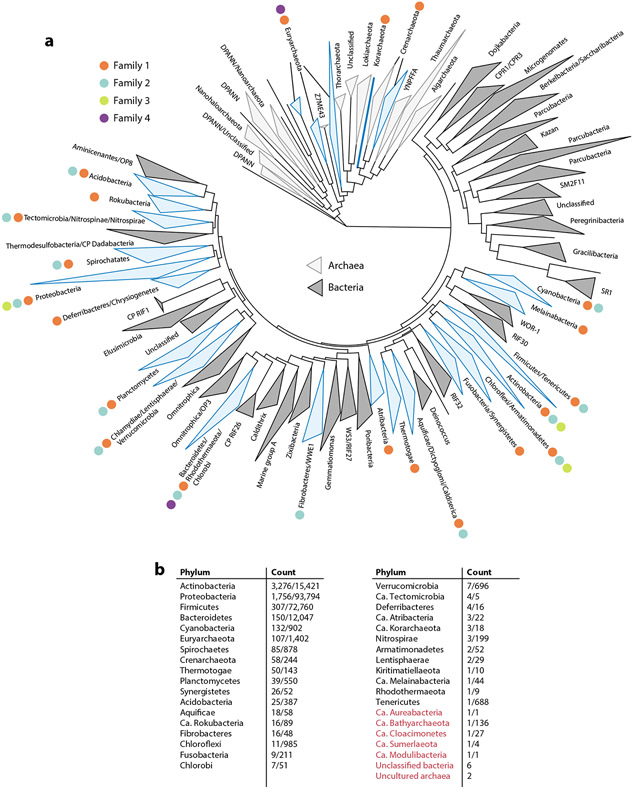
Recent large-scale computational searches for encapsulin-containing operons encoded in prokaryotic genomes have resulted in a curated list of over 6,000 encapsulin-like systems (18, 23) (Figure 2). These operons can be found in 31 bacterial and 4 archaeal phyla. Based on these recent data sets, a novel classification scheme for encapsulin systems—establishing four distinct encapsulin families—has been proposed based on sequence similarity, Pfam (protein family) membership, and genome-neighborhood composition (Figure 1c).
Figure 2.
Phylogenetic distribution of encapsulins. (a) Phylogenetic tree based on 108 of the major bacterial and archaeal phyla (147). Encapsulin-containing phyla are highlighted in blue. Different encapsulin families present in any given phylum are shown with differently colored dots, as detailed in the key. (b) A list of encapsulin-containing phyla. Values in the Count column give the number of identified encapsulin operons and the total number of proteomes available in UniProt for any given phylum (number of identified systems/number of UniProt proteomes). Phyla colored red indicate new, uncultured, or unclassified organisms not shown in the phylogenetic tree. [Ca. Modulibacteria has been proposed as a CP but is not annotated as such in UniProt (148).] Abbreviations: CP, candidate phylum; Ca., Candidatus; UniProt, Universal Protein Resource.
Family 1
Family 1 encapsulins (PF04454) represent the first recognized, most well-studied, and most widespread family of encapsulin-like systems. They were initially detected as high-molecular-weight aggregates via transmission electron microscopy in selectively bacteriostatic culture supernatants of Brevibacterium linens M18 (24). Based on this observation, the authors concluded that this high-molecular-weight protein component must be a homomultimeric bacteriocin and called it linocin M18. This led to the initial misannotation of Family 1 encapsulins as bacteriocins that still persists today. Further homologs were discovered in Mycobacterium tuberculosis (Cfp29) (25) and Thermotoga maritima (maritimacin) (26, 27) and were thought to exhibit proteolytic activity. However, neither bacteriostatic nor proteolytic activity could be validated in later studies using more purified protein preparations (11). Because of these early studies, Family 1 encapsulins are almost always misannotated within bacterial and archaeal genomes as bacteriocin, maritimacin, or linocin M18. Even very recently, reports can be found in the literature of partially purified encapsulins being falsely characterized as bacteriocins or proteases (28-32). Further confusion arises from the fact that in these early studies, encapsulins were isolated from culture supernatants and filtrates, leading to the misguided idea that they represent secreted proteins (24, 25, 33). The presence of encapsulins in culture supernatants is likely due to their release upon cell lysis and slow accumulation over time facilitated by their often-observed resistance toward proteolysis (15, 34, 35).
Family 1 encapsulins can be found in 31 out of 35 encapsulin-encoding prokaryotic phyla (18) (Figure 2). The majority of Family 1 operons are encoded by the phyla Proteobacteria, Actinobacteria, and Firmicutes and can be classified into at least seven main operon types based on co-encoded cargo proteins (Table 1). The generalized operon organization of Family 1 encapsulin systems consists of the encapsulin shell protein and a single primary cargo protein almost always encoded directly upstream of the encapsulin gene (18, 23) (Figure 1c). Many operons may also encode other noncargo but conserved accessory components that are coregulated and likely important for operon function (18, 23, 36).
Table 1.
Overview of all so-far characterized and proposed encapsulin systems organized by family, cargo type, proposed function, number of computationally identified operons, and status (confirmed as encapsulin or not)
| Family | Cargo/enzyme component | Proposed function | Number of putative operons (18) |
Status |
|---|---|---|---|---|
| 1 | DyP | Oxidative stress resistance | 1,505 | Confirmed (35, 63, 81, 150, 151) |
| Flp | Oxidative stress resistance/iron storage | 549 | Confirmed (11, 15, 16, 58, 59, 65-67, 75, 151-154) | |
| IMEF | Iron storage/oxidative stress resistance | 101 | Confirmed (23, 36) | |
| Fusion-Flp | Oxidative stress resistance/iron storage | 69 | Confirmed (68, 69) | |
| Hemerythrin | Oxidative/nitrosative stress resistance | 45 | Confirmed (23) | |
| Bacterioferritin | Iron storage/oxidative stress resistance | 15 | Putative | |
| Fusion-cytochrome c | Anaerobic ammonium oxidation/stress resistance | 9 | Confirmed (23, 84) | |
| 2 | Cysteine desulfurase | Sulfur metabolism | 1,480 | Confirmed (17) |
| Terpene cyclase | Isoprenoid biosynthesis | 912 | Putative | |
| Polyprenyl transferase | Isoprenoid biosynthesis | 900 | Putative | |
| Terpene cyclase + polyprenyl transferase | Isoprenoid biosynthesis | |||
| Xylulose kinase | Xylose utilization | 124 | Putative | |
| 3 | Variable biosynthetic enzymes | Peptide/polyketide natural product biosynthesis | 132 | Putative |
| 4 | DeoC | Nucleotide utilization | 32 | Putative |
| OsmC | Oxidative stress resistance | 29 | Putative | |
| GAPDH | Gluconeogenesis | 10 | Putative | |
| [NiFe] sulfhydrogenase | Hydrogen metabolism | 9 | Putative |
Abbreviations: DeoC, deoxyribose-phosphate aldolase; DyP, dye-decolorizing peroxidase; Flp, ferritin-like protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IMEF, iron-mineralizing encapsulin-associated firmicute cargo; OsmC, osmotically inducible protein C.
Family 2
Family 2 encapsulins can be distinguished from Family 1 due to their differing operon organization and lack of assigned Pfam family. They are now further divided into two subfamilies based on the absence (Family 2A) or presence (Family 2B) of a putative cyclic nucleotide (cNMP)-binding domain (PF00027) fused to the HK97-fold shell component (18) (Figure 1c). Family 2 encapsulins have only recently been recognized as such (17) and were historically misannotated and mischaracterized in various ways. Family 2A encapsulins were first reported in the membrane fraction of the intracellular parasite Mycobacterium leprae as the immunodominant major membrane protein I (MMPI) and were subsequently falsely considered to be surface-exposed membrane proteins (37). MMPI was studied in the context of different mycobacterial pathogens and is discussed in more detail in the section titled Biomedical and Agricultural Relevance. Family 2B encapsulins were also originally misannotated and mischaracterized as the multimeric transcription factor EshA in Streptomyces griseus, due to the presence of the aforementioned cNMP-binding domain often found in transcriptional regulators (38). EshA was proposed to be important for the formation of sporogenic hyphae in S. griseus and Streptomyces coelicolor A3(2) (38-40) and the regulation of antibiotic production in S. coelicolor A3(2) (41, 42), Streptomyces avermitilis (43), and Streptomyces albus (44), while a close homolog of EshA, dubbed EshB—a second Family 2B encapsulin system in the S. coelicolor A3(2) genome—was found to have no effect on antibiotic production (41). All these studies were carried out before Family 2 encapsulins were recognized as cargo-loaded protein compartments.
Family 2 encapsulins represent the most numerous encapsulin systems known to date and can be found in 14 bacterial phyla (18) (Figure 2). The majority of Family 2 encapsulins are found in the phyla Actinobacteria, Proteobacteria, Bacteroidetes, and Cyanobacteria. Family 2 consists of at least five distinct operon types based on cargo protein identity and co-occurrence (Table 1). Compared to Family 1, Family 2 operon organization is more varied due to the presence of cNMP-binding domains in Family 2B encapsulins and the variable occurrence of two distinct shell components within 2B operons (18). Noncargo accessory components may also be present, likely related to the biological function of a given operon (45) (Figure 1c).
Family 3
Recently, a third family of encapsulins (Family 3) has been proposed based on computational genome mining (18). Family 3 encapsulins are misannotated as phage major capsid proteins and have been classified together with the majority of HK97-fold viruses into Pfam family PF05065. However, close inspection of their genome neighborhoods has revealed that they are in fact not part of virus genomes but rather are often integrated into large peptide and polyketide biosynthetic gene clusters (Figure 1c). They can mostly be found in the phyla Actinobacteria and Proteobacteria, primarily in Streptomyces and Myxococcus species, as well as some other closely related genera (Figure 2). Streptomyces and Myxococcus species are widely known as being among the most prolific producers of bioactive natural products (46, 47). Due to the variability of co-occurring biosynthetic genes in Family 3 operons, a simple classification of these systems is difficult (Table 1). So far, no Family 3 encapsulin system has been experimentally characterized.
Family 4
Similar to Family 3, Family 4 encapsulins (PF08967) have only recently been identified through in silico searches and so far have not been experimentally characterized (18). They are the most distinct family of encapsulins discovered to date and are restricted to the archaeal phylum Euryarchaeota and bacteria of the phylum Bacteroidetes (Figure 2). All so-far sequenced Pyrococcus and Thermococcus genomes encode two distinct Family 4 operons. Family 4 encapsulin proteins are truncated and thus only one third the length of a standard HK97-fold protein (~100 residues) (48, 49). The reasons Family 4 has been suggested to represent a divergent type of encapsulin systems are that they are structurally similar to one of the core domains of the HK97-fold [A-domain (axial domain)], they are found outside viral genomes encoded in operons with enzymatic components, and they are part of the same Pfam clan as the other encapsulin families. All archaeal Family 4 operons consist of an enzyme component and a Family 4 encapsulin protein located directly downstream (Figure 1c). Four distinct and conserved archaeal operon types can be distinguished based on the type of enzymatic component (Table 1). Bacterial Family 4 encapsulins are not encoded in obvious operon-like structures, making their classification and function prediction more difficult.
Co-Occurrence of Multiple Encapsulin Operons
Many organisms possess more than one encapsulin system. About 15–20% of all encapsulin-containing prokaryotic genomes encode multiple encapsulin operons (18). The number of co-occurring systems can range from two operons of the same or different family—common combinations are Family 1 + 2B, 2× Family 2B, and 2× Family 4—up to seven distinct operons in Nocardia terpenica (1× Family 1, 1× Family 2A, and 5× Family 2B) or five distinct Family 2B operons in Sorangium cellulosum. Four of the S. cellulosum operons represent systems with two shell components each, resulting in a total of nine distinct shell proteins encoded in a single genome. The greatest family-level diversity of encapsulin systems can be found in a number of Actinomadura and Corallococcus spp., which encode Family 1, 2B, and 3 systems.
The widespread nature and diversity of encapsulin systems highlights the importance of protein-based compartmentalization systems for regulating and optimizing bacterial and archaeal metabolism. With the increasing number of available prokaryotic genomes and the sampling of previously unexplored environmental niches, more encapsulin families and operon types are likely to be discovered in the future.
EVOLUTIONARY ORIGIN AND COMPARATIVE ANALYSIS
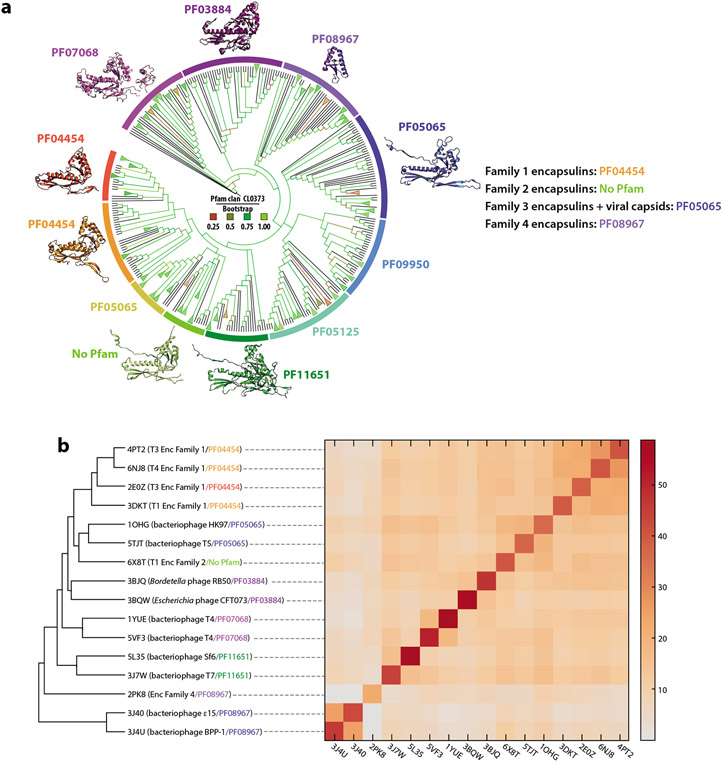
All encapsulin families with a recognized Pfam association belong to the Pfam clan CL0373, which contains the majority of Pfam-classified HK97-fold proteins (18). Pfam classification is based on a combination of structural and sequence similarity (50, 51). Pfam clans are meant to encompass only Pfam families that share a common evolutionary origin, thus indicating that by Pfam metrics all encapsulins are evolutionarily related to the other HK97-fold proteins—representing bacteriophage and virus capsid proteins—contained within CL0373. Sequence-based phylogenetic analysis indicates that all encapsulin families except Family 4 are more closely related to one another than they are to other HK97-fold proteins contained within CL0373 (18) (Figure 3a). The P22 bacteriophage coat protein family (PF11651) was identified as being most closely related to Family 1, 2, and 3 encapsulins. Bacterial and archaeal Family 1 encapsulins were found to be more closely related to one another than to other HK97-fold proteins, suggesting interdomain transfer of Family 1 encapsulins—likely from Bacteria to Archaea—which represents a well-documented phenomenon (52, 53). Family 4 encapsulins were suggested to be the most evolutionarily distinct family, more closely related to other HK97-fold capsid proteins than to Family 1, 2, or 3 encapsulins.
Figure 3.
Phylogenetic analysis and structural comparison of HK97-fold proteins. (a) Phylogenetic tree of Pfam clan CL0373. Branches are colored based on Bootstrap values. Representative protomer structures are shown with their respective Pfam family designations. (b) Structural comparison of CL0373 members using the DALI server (149). (Left) Dendrogram and (right) heat map based on pairwise Z score comparisons. Pfam families are colored as in panel a. Abbreviations: Enc, encapsulin; Pfam, protein family.
Complementary analyses using structure-based clustering of representatives of Pfam clan CL0373 showed that Family 1 encapsulins form an apparent monophyletic clade, while the single available Family 2A encapsulin structure (6X8T) was more similar to virus capsids of family PF05065 (18, 45) (Figure 3b). No Family 3 encapsulin structure is currently available; however, some Family 3 members have been automatically grouped within Pfam family PF05065, potentially indicating that they are structurally more similar to Family 2 than to Family 1 encapsulins. Even though Family 4 encapsulins are heavily truncated compared with the canonical HK97-fold, structure-based clustering was successfully used to confirm their membership in Pfam clan CL0373 (18). Family 4 was found to most closely resemble the A-domain of PF05065 capsid proteins.
Both sequence- and structure-based analyses suggest that encapsulins share a common ancestor with the HK97-fold proteins contained within Pfam clan CL0373, representing the ubiquitous viral order Caudovirales (54). The factors just discussed may point to a viral origin of encapsulin systems, potentially via molecular domestication of prophage HK97-type capsid proteins by their cellular hosts. This scenario is in agreement with the fact that HK97-fold viruses are widespread and often found as proviruses and prophages in the genomes of members of all domains of life, whereas encapsulins show a narrower phylogenetic distribution (18, 23, 54).
SHELL STRUCTURE AND FUNCTION
The defining feature of encapsulin systems is the formation of heteromeric complexes by HK97-fold structural proteins with enzymatic components. In the so-far characterized Family 1 and 2 systems, complex formation takes the form of selective sequestration of enzymes, referred to as cargo proteins, inside self-assembling icosahedral protein shells formed by the HK97-fold encapsulin shell protein (12). The HK97-fold is named after the gp5 protein of bacteriophage Hong Kong 97 (55, 56) and is predominantly found in the capsid proteins of viruses of the order Caudovirales infecting bacteria and archaea (48, 49, 54). It is further present in the floor domain of eukaryotic Herpesvirales capsid proteins (57). Outside of viruses, the HK97-fold is only found in bacterial and archaeal encapsulins (18).
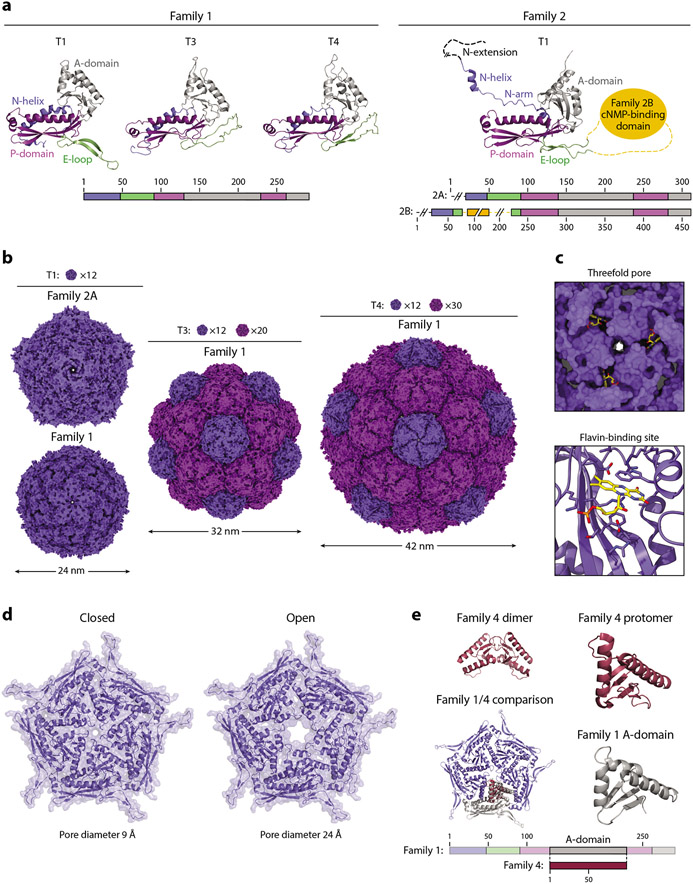
The HK97-fold protomer is roughly triangular in shape and consists of three conserved domains, the axial domain (A-domain), peripheral domain (P-domain), and extended loop (E-loop) (48, 49) (Figure 4a). Various other loops, extensions, and insertions have been reported in the HK97-fold capsid proteins of different viruses and phages. Family 1 encapsulins contain a unique N-terminal helix (N-helix) that directly interacts with the P-domain and forms part of the TP binding site and is thus important for anchoring encapsulated cargo proteins to the shell interior (11, 36). E-loop conformation has been observed to be a key assembly mode determinant that dictates the triangulation number of the resulting encapsulin shell, with T3 and T4 shells resulting from a more compact protomer while a more extended E-loop results in T1 assemblies. Instead of a simple N-helix, the single structurally characterized Family 2 encapsulin from Synechococcus elongatus (Family 2A) exhibits an extended N-arm followed by a short N-helix and a disordered N-extension that is more similar to the N termini found in many viral capsid proteins of the Pfam family PF05065 (17). The extended N-arm motif is likely also found in Family 3 encapsulins, as they are often automatically grouped with PF05065 as well, but no Family 3 structure is currently available to confirm this hypothesis. So far, no structure of a Family 2B system has been reported, but based on sequence alignments and annotations, these systems appear to contain an ~120-residue putative cNMP-binding insertion domain inside the E-loop. The function of this domain has so far not been elucidated; however, it has been suggested that binding cyclic nucleotides might allow the encapsulin shell to change conformation, disassemble, or relay a signal to the shell interior (18).
Figure 4.
Structure and function of the encapsulin shell. (a, left) Family 1 and (right) Family 2 encapsulin protomers colored by their conserved HK97-fold domains. Protomers of Family 1 T1 (3DKT), T3 (4PT2), and T4 (6NJ8), and Family 2 T1 (6X8M) shells are shown. (b) Exterior view down the fivefold symmetry axis of T1, T3, and T4 shells. Pentameric and hexameric facets are colored dark and light purple, respectively. The number of facets needed to tile a closed shell of a given triangulation number is shown. The same structures were used as in panel a. (c) Flavin-binding site in the Thermotoga maritima T1 shell (7KQ5). (Top) View down the threefold axis and (bottom) a zoomed-in view of the binding site. Residues within 5 Å of the flavin moiety are shown as sticks. (d) The closed and open states of the pentameric pores of the Haliangium ochraceum T1 shell are shown. (e) Comparison of a Family 4 encapsulin (2PK8) with a Family 1 pentameric facet (3DKT). Abbreviations: A-domain, axial domain; cNMP, cyclic nucleotide; E-loop, extended loop; N-helix, N-terminal helix; P-domain, peripheral domain.
Encapsulin protomers have been reported to self-assemble into three differently sized closed shells with icosahedral symmetry, roughly 24 nm (T1, 60 protomers), 32 nm (T3, 180 protomers), and 42 nm (T4, 240 protomers) in diameter (12) (Figure 4b). Analogous to viral capsids, encapsulin shells consist of pentameric and hexameric facets occupying the icosahedral fivefold and threefold symmetry axes, respectively. Even though the eponymous HK97 bacteriophage possesses a T7 capsid (~65 nm, 420 protomers) (55), encapsulin shell proteins by themselves seem unlikely to be able to form defined and stable T7 assemblies because, so far, all known HK97-fold T7 capsids require scaffolding components for the assembly and maturation of a stable T7 structure (48, 49). However, some encapsulin systems may utilize currently unknown scaffolding components to achieve an expanded T7 architecture. Encapsulins generally possess symmetrical pores at their five-, three-, or twofold symmetry axes, which can differ in size and charge and are hypothesized to facilitate and control the transit of small molecules relevant for the functioning of the respective encapsulin system while excluding unwanted proteins and compounds (12).
Even though the shell of Family 1 encapsulins was thought to fulfill a purely structural function by forming a diffusion barrier that allows cargo sequestration and control over molecular flux, recent structural studies have identified a flavin cofactor directly bound to the HK97-fold encapsulin protomer in the Thermotoga maritima T1 Flp system (58-60) (Figure 4c). The binding site for this redox cofactor is located on the outside of the shell around the threefold symmetry axis and pore. The precise function of the flavin moiety is currently unknown; however, it has been suggested to play a role in the redox chemistry necessary for dynamic iron storage inside the encapsulin shell (60). The flavin moiety could achieve this by relaying electrons through the compartment shell, thus enabling either oxidation and storage or reduction and release of iron. Based on sequence comparisons, it has been suggested that ~30% of bacterial Family 1 Flp systems may incorporate a flavin cofactor into their shell (60). Most of these are found in anaerobes, potentially indicating that a flavin cofactor is specifically necessary in anaerobic environments.
Another recent study of the Haliangium ochraceum T1 Flp encapsulin showed that encapsulin shells are not static but are in fact capable of substantial conformational change (61). Specifically, two distinct states of the fivefold pore—open and closed—could be resolved in cryogenic electron microscopy (cryo-EM) structures with a substantial difference in pore diameter of 15 Å (Figure 4d). This indicates that, at least for some T1 Flp systems, dynamic pores likely play a major role in controlling molecular flux of substrates, in this case iron, across the shell, thus adding another layer of complexity to encapsulin function.
The newly proposed Family 4 encapsulins represent the most structurally distinct encapsulin family. As mentioned in the section titled Distribution, Diversity, and Classification, they are heavily truncated and structurally similar to the A-domain of the HK97-fold. A crystal structure of a Family 4 encapsulin from Pyrococcus furiosus is available and crystallized as a dimer (62) (Figure 4e). Similar to the A-domain of the HK97-fold (48, 49), the Family 4 protomer consist of two α-helices surrounding a central four-stranded β-sheet (Figure 4e). Based on structural alignments, it has been suggested that the Family 4 protomer is the result of a loss of the HK97-fold N- and C-terminal domains, resulting in a contiguous stretch of about 100 amino acids representing most of the HK97-fold A-domain. The dimeric form of the Family 4 protomer may represent its native oligomeric state, which interacts with the co-encoded enzymatic component. Alternatively, based on the structural similarity with HK97-fold A-domains, Family 4 encapsulins may also be able to oligomerize into higher-order facets or complexes, potentially together with their partner enzymes.
It is currently unclear if a generalizable reason for encapsulating certain enzymes inside a protein shell exists. Given the diversity of so-far identified encapsulin operons, the protein shells formed by encapsulin systems seem likely to enclose certain cargo proteins and processes for different reasons, including dynamic storage, intermediate sequestration, and regulation, all of which likely benefit from compartmentalization for related but different molecular reasons.
CARGO PROTEINS, BIOCHEMISTRY, AND BIOLOGICAL FUNCTION
The specific function of each encapsulin system is primarily determined by the type of coregulated enzymatic component usually colocalized within a given encapsulin operon. In Family 1 and 2 systems, enzymatic components have been shown to represent dedicated cargo proteins selectively encapsulated within the self-assembling encapsulin shell (17, 36, 63). Family 3 and 4 systems have not been experimentally characterized, and variations on the mode of encapsulin-enzyme interaction are possible, specifically for Family 4 systems, in which an alternative dimeric oligomerization state of the HK97-fold protomer has been reported (62).
So far, experiments have revealed the general biochemical function of three types of encapsulin systems, two from Family 1 and one from Family 2. For Family 1, some information is available for ferroxidase- and peroxidase-containing operons, while for Family 2, a desulfurase-encoding operon has been studied. Here, these systems have been classified by their core catalytic capabilities and not necessarily by their physiological function or phylogenetic affiliation. Only limited data exist about the roles these systems play in prokaryotic cell biology and physiology.
Family 1 Ferroxidase Cargos: Flps and IMEF Proteins
The ferroxidases found in Family 1 systems are all part of the large protein superfamily of ferritin-like proteins characterized by helical bundle structures that harbor dinuclear iron active sites (64). Ferroxidase activity can variably rely on molecular oxygen, hydrogen peroxide, or other compounds as oxidants. The two so-far experimentally studied ferroxidase operon types contain Flp and iron-mineralizing encapsulin-associated firmicute (IMEF) cargo proteins (11, 36, 60, 61, 65). Due to their ferroxidase activities, both Flp and IMEF cargo proteins are able to oxidize ferrous to ferric iron inside the encapsulin shell, leading to the formation of encapsulated ferric iron precipitates, often referred to as ferrihydrites (36) (Figure 5a). Static and/or dynamic pores likely play a key role in controlling the flux of iron across the protein shell (61). It is currently unknown if or how stored iron precipitates are remobilized, but for a subset of Flp systems, a flavin cofactor coordinated to the shell has been suggested to play a role in iron reduction and release (58-60). Different mechanisms of iron storage and mobilization might be used under aerobic and anaerobic conditions.
Figure 5.
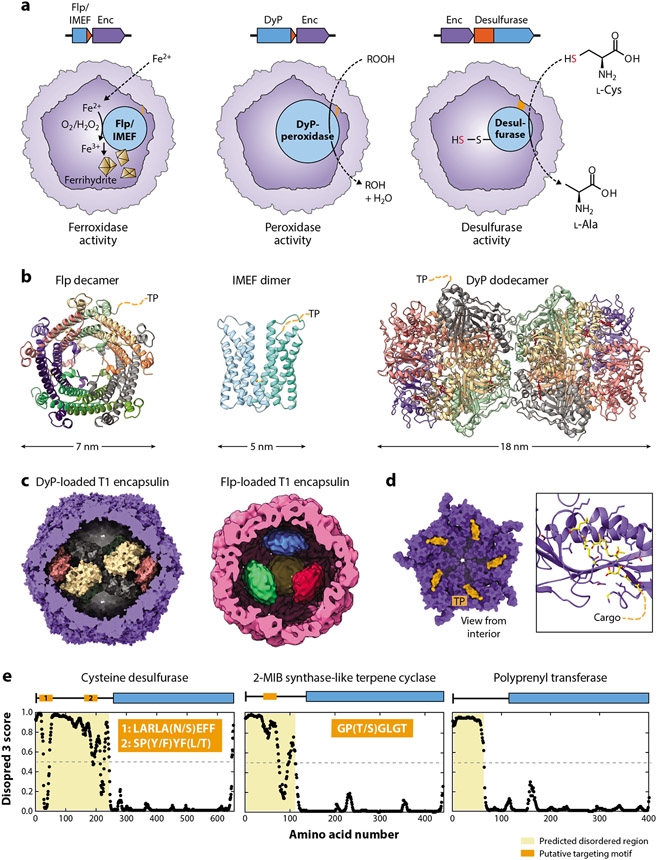
Cargo proteins and encapsulin biochemistry. (a) Overview of the three so-far partially characterized biochemical functions of encapsulins, namely, ferroxidase, peroxidase, and desulfurase activity. TPs/domains are highlighted in orange. (b) Structures of cargo proteins highlighting their oligomerization state. For clarity, only one TP per oligomer is shown. DyP-bound heme groups are shown in red. (c) DyP (Mycobacterium smegmatis) and Flp (Haliangium ochraceum) cargo proteins shown in their native assembly state inside cargo-loaded T1 encapsulins. (d) Family 1 cargo-loading mechanism mediated by TP binding to the interior of the encapsulin shell. (Left) A pentameric facet and (right) a zoomed-in view on a single binding site are shown (Thermotoga maritima; 3DKT). TPs are shown in orange/yellow. Residues within 5 Å of the TP are shown as sticks. (e) Proposed cargo-loading mechanism for Family 2 encapsulins. Shown are disorder plots generated by Disopred 3 highlighting the large, disordered N-terminal regions/domains in many putative Family 2 cargo proteins. Three examples (desulfurase, terpene cyclase, and polyprenyl transferase) are shown with proposed targeting motifs indicated in orange. Abbreviations: DyP, dye-decolorizing peroxidase; Enc, encapsulin; Flp, ferritin-like protein; IMEF, iron-mineralizing encapsulin-associated firmicute; MIB, methylisoborneol; TP, targeting peptide.
Crystal structures of Flp cargo from H. ochraceum and Rhodospirillum rubrum suggest that they form decameric assemblies with D5 symmetry (66, 67) (Figure 5b). Recent cryo-EM studies on cargo-loaded encapsulins strongly indicate that decameric Flp assemblies represent the native state of Flp inside the encapsulin shell as well (60, 61). It has been suggested that a tetrahedral arrangement of four Flp decamers may represent the native loading state of the H. ochraceum system (61) (Figure 5c). Interestingly, this assembly state leads to a symmetry mismatch between encapsulated Flp and the encapsulin shell. In all identified archaeal Flp systems, an Flp domain is directly fused to the N terminus of the encapsulin shell protein, resulting in a Fusion-Flp encapsulin and leading to the internalization of Flp domains upon shell assembly. Studies of P. furiosus and Sulfolobus solfataricus Fusion-Flp systems have shown that they form T3 shells and contain internalized Flp assemblies (68, 69). The excised P. furiosus Flp domain has been shown to form a D5 symmetrical decamer similar to the other characterized Flp cargos; however, the structural arrangement of fused Flp inside the shell remains unknown (66). Based on the number of protomers in T1 (60 subunits), T3 (180 subunits), and T4 (240 subunits) encapsulin shells, it is hard to imagine how the corresponding 6, 18, or 24 Flp decamers could be arranged in a symmetrical way in the encapsulin interior. Further studies are needed to elucidate the Flp assembly state in Fusion-Flp encapsulins, which might be more heterogeneous than that for nonfused Flp cargo proteins.
A recent study of the Quasibacillus thermotolerans IMEF system revealed it to be the so-far only confirmed and characterized T4 encapsulin (36). It appears that up to 42 IMEF dimers can be encapsulated within the T4 shell, one each for the 12 pentameric and 30 hexameric facets. IMEF is a member of the ferritin-like protein superfamily and forms an unusual dimer of four helix bundles harboring a ferroxidase active site at the dimer interface. Due to the large size of the T4 shell, large amounts of iron can be stored as ferric iron precipitates inside the compartment. Iron-rich cores 30 nm in diameter have been observed containing close to 25,000 iron atoms—an order of magnitude more than can be stored in standard (bacterio)ferritins. Intriguingly, many IMEF operons encode a conserved 2Fe–2S ferredoxin, homologous to bacterioferritin-associated ferredoxins, which have been shown to be responsible for iron reduction and release from bacterioferritin cages (70). This 2Fe–2S ferredoxin carries an N-terminal motif very similar to the conserved usually C-terminal TPs found in confirmed Family 1 cargo proteins and was shown to copurify with the T4 shell under heterologous expression conditions (36). Further studies are needed to conclusively determine if this ferredoxin might represent a low-abundance cargo protein or a component able to specifically interact with the shell exterior. In both scenarios, it may be involved in relaying electrons across the protein shell to allow the reduction and remobilization of stored iron.
Flp and IMEF encapsulins are reminiscent of the long-studied and ubiquitous (bacterio)ferritins that represent the major iron storage system for most prokaryotic and eukaryotic cells (71). Unlike (bacterio)ferritin cages, Flp and IMEF cargo proteins by themselves cannot store precipitated iron in a soluble and inert form and have to rely on the encapsulin shell to achieve iron precipitate sequestration. The vast majority of organisms containing Flp encapsulins encode standard (bacterio)ferritins in their genomes, suggesting that the primary role of Flp encapsulins might not be iron storage but rather combating oxidative stress by safely storing excess reactive iron under certain conditions. This hypothesis is in agreement with the upregulation of the T. maritima Flp encapsulin in oxidative stress–induced biofilms (72) and a recent study reporting the upregulation of an Flp encapsulin system in Phascolarctobacterium faecium as part of the coculture-induced stress response (73). Further, a Fusion-Flp encapsulin in the archaeon Palaeococcus pacificus has been proposed to play a role in oxygen stress resistance (74). Deletion of the encapsulin shell protein in the Myxococcus xanthus Flp operon resulted in tan-phase-locked mutants with abolished extracellular polysaccharide production, reduced swarming behavior, and defective sporulation, indicating a broader role of Flp encapsulins potentially related to iron or redox homeostasis and myxobacterial development (75). In contrast to Flp systems, almost all IMEF encapsulins are encoded in spore-forming Firmicutes genomes lacking standard (bacterio)ferritins (36). This lack of canonical iron storage proteins suggests that IMEF encapsulins may represent the primary iron storage system of these organisms. The reason why this subset of Firmicutes relies on encapsulins instead of standard (bacterio)ferritins for iron storage will require further study.
Family 1 Hemerythrin and Bacterioferritin Cargos
Besides Flp and IMEF, two other ferritin-like superfamily cargo proteins have been reported, namely hemerythrins and bacterioferritins (18, 23). Generally, hemerythrins are known to bind to oxygen, nitric oxide, and potentially other volatile or reactive small molecules (76, 77). Hemerythrin-containing encapsulins have been shown to assemble into T1 shells, while hemerythrin cargo proteins alone were reported to form dimers in solution (23). Hemerythrin encapsulins have further been shown to offer oxidative and nitrosative stress protection when overexpressed in a heterologous host (23). Hemerythrin systems are likely involved in the sequestration or detoxification of harmful low-molecular-weight compounds. No experimental information is currently available regarding the recently computationally identified bacterioferritin cargo proteins. Bacterioferritin monomers are composed of two four-helix bundles and are thus structurally distinct from the other identified ferritin-like superfamily cargos. They generally assemble into 12-nm cages and are used by themselves as the main iron storage system in many bacteria (78). Further studies are needed to elucidate the function and underlying logic of a putative shell-within-a-shell arrangement in the context of iron storage.
Family 1 Peroxidase Cargos
All Family 1 peroxidase-containing systems encode dye-decolorizing peroxidases (DyPs) (18). DyP-type peroxidases are heme proteins and are named for their ability to oxidize a broad range of anthraquinone dyes (79). They have further been shown to be able to break down lignin and other typical peroxidase substrates (80, 81). DyP from B. linens forms an overall hexameric assembly inside the B. linens T1 shell consisting of a trimer of dimers with D3 symmetry (82). In contrast, a recent cryo-EM study of the natively isolated Mycobacterium smegmatis DyP encapsulin revealed that the majority of isolated particles contained two DyP hexamers forming an overall twofold symmetrical dodecameric complex within the T1 shell (63) (Figure 5c). The encapsulin shell seems to stabilize this DyP dodecamer, because in its unencapsulated form, M. smegmatis DyP mainly forms hexamers. Encapsulin-associated DyPs have been shown to hydrolyze a number of peroxides, but their native substrate range is currently unknown. The general biological function of DyP encapsulin systems is also still speculative; however, a recent study showed that a DyP Family 1 system in Mycobacterium tuberculosis plays a direct role in oxidative stress resistance (83).
Family 1 Fusion Cytochrome Cargos
The final type of Family 1 encapsulin for which some experimental data are available is unusual fusion systems exclusively found in anaerobic ammonium oxidation (anammox) bacteria of the phylum Planctomycetes (23). They contain an N-terminal diheme cytochrome c fusion domain and have been shown to form T3 shells with the cytochrome c domains located on the shell interior (23). Their biological function has not been studied in detail; however, a role in detoxifying harmful intermediates produced during anammox like nitric oxide, hydroxylamine, or hydrazine has been proposed, as well as a role in iron storage inside the anammoxosome, the membrane-bound compartment enclosing all the components needed for anammox metabolism (23, 84).
Family 1 Targeting Peptides and Cargo Loading
In a number of instances, it was possible to obtain structural information for the TP–shell interaction that mediates cargo encapsulation in all nonfusion Family 1 encapsulin systems (Figure 5d). So far, shell-bound TPs of two T1 Flp systems (T. maritima: GGDLGIRK and H. ochraceum: GSLGIGSLR) and one T4 IMEF system (Q. thermotolerans: TVGSLIQ) could be resolved (11, 36, 61). Surprisingly, the TP in the high-resolution structure of the cargo-loaded M. smegmatis DyP encapsulin could not be observed, potentially hinting at increased flexibility or low occupancy (63). TPs are usually located at the C-terminus of any cargo protein and often contain a conserved single or double GSL motif followed by a positively charged residue (18). TPs are responsible for binding to a conserved binding site formed by the N-terminal helix and P-domain of Family 1 protomers during shell self-assembly. Protomers in pentameric (T1 and T4) and hexameric (T4) facets have been shown to bind TPs. The binding motifs are connected to the cargo protein via flexible linkers of variable length (~5–20 amino acids), often containing many glycine, alanine, and proline residues (18).
Family 2 Desulfurase Cargos
The third type of biochemically distinct encapsulin system for which experimental data are available is Family 2A encapsulins with SufS-like cysteine desulfurases as cargo (17). Cysteine desulfurase activity generally entails transfer of the side chain sulfur atom of l-cysteine to a conserved thiol on the surface of the desulfurase itself, thus forming l-alanine and a persulfide intermediate as the reaction products (Figure 5a). The sulfur atom intermittently stored as a persulfide is then transferred to downstream acceptors, often containing rhodanese domains, for further distribution to various metabolic processes, including Fe–S cluster formation and cofactor biosynthesis (85, 86). The presence of rhodaneses in many Family 2A desulfurase operons may indicate that a mechanism exists to transfer sulfur stored as persulfides on encapsulated desulfurases across the protein shell for downstream sulfur utilization. In S. elongatus, the desulfurase encapsulin operon is upregulated upon sulfate starvation. It was experimentally confirmed that the cysteine desulfurase is encapsulated inside a T1 shell, which enhances catalytic activity compared to the unencapsulated form of the enzyme. No concrete data regarding the physiological role of this encapsulin system is currently available; however, roles in sulfur utilization, sulfur storage, or redox homeostasis have been proposed (17). Experiments with truncation mutants showed that the large unannotated and disordered N-terminal domain found in all desulfurase cargo proteins is crucial for mediating cargo encapsulation. Two potential targeting motifs located in this N-terminal domain have been proposed (Figure 5e). This represents a novel cargo-loading mechanism for encapsulin systems, distinct from the C-terminal TP-based cargo loading observed for Family 1 encapsulins.
Family 2 Terpene Cyclase, Polyprenyl Transferase, and Xylulose Kinase Cargos
In addition to Family 2 desulfurase operons, a variety of other putative Family 2 cargo types have recently been computationally identified (18). This includes many different terpene cyclase, polyprenyl transferase, and xylulose kinase systems (Table 1). Terpene cyclase and polyprenyl transferase operons have been suggested to play a role in the biosynthesis and regulation of cyclic and linear isoprenoid natural products—including 2-methylisoborneol and geosmin—while xylulose kinase systems may be involved in xylose utilization. Similar to desulfurase cargos, Family 2–associated terpene cyclases and polyprenyl transferases contain large unannotated and disordered N termini, potentially harboring conserved motifs important for mediating cargo encapsulation (Figure 5e). In contrast, putative xylulose kinase cargo proteins do not contain any obviously conserved targeting domains or motifs. Further studies will be needed to explore the biochemical and physiological functions of Family 2 encapsulins.
Family 3
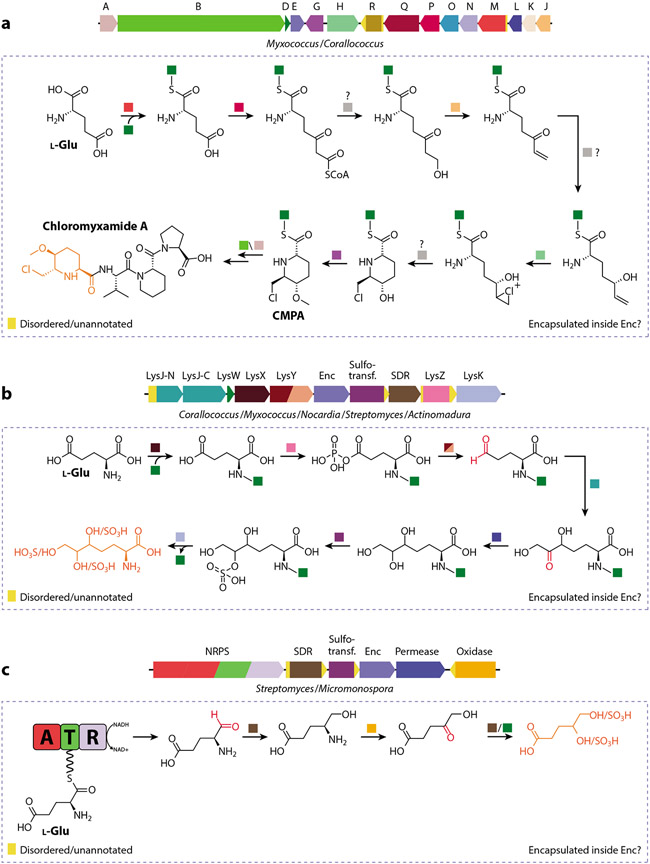
Family 3 encapsulin systems were recently identified as part of a large-scale computational study and are characterized by a putative HK97-fold shell protein located within different types of small-molecule biosynthetic gene clusters (18). No experimental data on the structure or biochemistry of Family 3 encapsulins are currently available. However, a Myxococcus gene cluster encoding a Family 3 encapsulin has recently been shown to produce a variety of chlorinated 6-chloromethyl-5-methoxypipecolic acid–containing peptide natural products dubbed chloromyxamides (87) (Figure 6a). One theme of many of the identified Family 3 systems is that they often contain genes for sulfotransferases and short-chain dehydrogenases, potentially indicating that the resulting natural products may contain sulfated hydroxyl groups generated through the successive action of these enzymes (88) (Figure 6b). Other Family 3 operons encode amino group carrier proteins or large genes coding for nonribosomal peptide synthetases and polyketide synthases (89) (Figure 6c). The diversity of genome neighborhoods surrounding Family 3 encapsulin genes suggests that Family 3 operons are capable of producing a structurally diverse set of natural products. Some of the enzymes encoded in Family 3 operons contain extended unannotated and possibly disordered N- or C-terminal regions, which may indicate that some of them do represent cargo proteins with a cargo-loading mechanism reminiscent of Family 2 systems. Active encapsulation of certain biosynthetic enzymes may allow Family 3 encapsulins to sequester reactive aldehyde or ketone intermediates—potentially generated by short-chain dehydrogenases/reductases—thus preventing aldehyde toxicity and unwanted side reactions. Similar molecular logic has been reported in bacterial microcompartments (90).
Figure 6.
Select Family 3 encapsulin operons and proposed biosynthetic pathways. (a) Chloromyxamide biosynthetic gene cluster and its proposed AmCP-dependent biosynthesis. Some of the depicted reactions may happen inside a Family 3 encapsulin, indicated by the dotted box. The chemical logic behind encapsulation may be the sequestration of reactive or toxic aldehyde/ketone or chlorination intermediates. The chlorinated CMPA building block is highlighted (orange). Question marks indicate uncertainty about steps as depicted. Abbreviations: A, ornithine cyclodeaminase; B, hybrid nonribosomal peptide synthetase/type I polyketide synthase; CMPA, 6-chloromethyl-5-methoxypipecolic acid; D, LysW; E, Enc, Family 3 encapsulin; G, SAM-dependent methyltransferase; H, rubber oxygenase A; J, enoyl-CoA hydratase; K, TetR/AcrR family transcriptional regulator; L, NADP-dependent oxidoreductase; M, AMP-dependent synthetase; N, acyl-CoA dehydrogenase; O, acyl-CoA dehydrogenase; P, acetyl-CoA acetyltransferase; Q, acyl-CoA dehydrogenase; R, aldehyde dehydrogenase. (b) Uncharacterized Family 3 gene cluster and proposed AmCP-dependent biosynthesis. Potentially reactive carbonyl intermediates are shown in red, while a putative product is shown in orange. Abbreviations: AmCP, amino group carrier protein; Enc, Family 3 encapsulin; LysJ-N/C, transketolase; LysK, AmCP-amino acid carboxypeptidase; LysW, AmCP; LysY, AmCP–amino acid phosphate reductase; LysX, amino acid–AmCP ligase; LysZ, AmCP–amino acid kinase; SDR, short-chain dehydrogenase/reductase; Sulfotrans., sulfotransferase. (c) Family 3 encapsulin embedded in an uncharacterized NRPS-dependent biosynthetic gene cluster. Potentially reactive carbonyl intermediates are shown in red, and a putative product is shown in orange. Abbreviations: A, adenylation domain; AmCP, amino group carrier protein; CMPA, 6-chloromethyl-5-methoxypipecolic acid; Enc, Family 3 encapsulin; NRPS, nonribosomal peptide synthetase; R, reduction domain; SDR, short-chain dehydrogenase/reductase; Sulfotrans., sulfotransferase; T, thiolation domain.
Family 4
In archaea, Family 4 encapsulin systems consist of a truncated HK97-fold protein and one of four conserved enzymatic components encoded in the same operon (18). No Family 4 system has been experimentally characterized. The four types of enzymes are: [NiFe] sulfhydrogenase (four subunits: α, β, γ, and δ), osmotically inducible protein C (OsmC), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and deoxyribose-phosphate aldolase (DeoC). These types of enzymes are known from various contexts to be involved in a broad range of biochemistries and physiological processes in archaea, including hydrogen utilization and energy metabolism ([NiFe] sulfhydrogenase) (91-96), organic hydroperoxide detoxification (OsmC) (97, 98), glycolysis and gluconeogenesis (GAPDH) (99-108), and nucleoside/nucleotide utilization (DeoC) (109-115). None of the Family 4–associated enzymatic components contain any obvious unannotated or disordered regions, suggesting a different mode of complex formation with the HK97 component compared with those in Family 1 and 2 systems. By analogy to all other known HK97-fold proteins, Family 4 encapsulins likely act as structural proteins and form heteromeric complexes with the respective coregulated enzymatic components. This is supported by a proteomics study in P. furiosus showing that GAPDH and the Family 4 encapsulin encoded in the same operon can form a stable complex (92). The form of this interaction and the general assembly state of Family 4 encapsulins is currently unknown. Family 4 encapsulins might stabilize the respective enzymatic components through close association, acting as specialized molecular chaperones (116, 117) and leading to increased thermal stability, increased resistance against oxidative stress, and a prolonged lifetime of the associated enzyme.
To summarize, proposed encapsulin functions include roles as reaction spaces for various anabolic (Family 2 and 3) and catabolic (Family 1 and 2) processes, storage compartments (Family 1 and 2), and enzyme regulatory systems (Family 2 and 4) as well as chaperones (Family 4). This remarkable breadth of function arises through the combination of a self-assembling protein shell—resulting in compartmentalized spaces with defined microenvironments and control over molecular flux—and a dedicated and modular cargo encapsulation mechanism. Future studies aimed at further elucidating structural and mechanistic details as well as the physiological roles of encapsulins will be needed to truly capture their functional diversity.
BIOMEDICAL AND AGRICULTURAL RELEVANCE
Encapsulins are found in the genomes of a wide variety of important Gram-negative and Gram-positive pathogens as well as many commensals of the human microbiota (18). For instance, Family 1 and 2 encapsulin systems encoding Flp, peroxidase, and desulfurase cargos are found in pathogenic Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii, all part of the highly virulent and antibiotic-resistant ESKAPE group of pathogens that is responsible for the majority of severe hospital-acquired infections worldwide (118). No specific information about the physiological functions of these systems within the respective pathogens or their roles during infection is currently available.
Encapsulins are also widely distributed in Mycobacteria, including various M. tuberculosis, M. leprae, and M. avium strains, the causative agents of tuberculosis, leprosy, and a variety of nontuberculous diseases, respectively (119, 120). In particular, peroxidase-containing Family 1 systems are found in many M. tuberculosis strains, while nontuberculous Mycobacteria often encode Family 2A operons with desulfurase cargos. Originally, the Family 1 encapsulin in M. tuberculosis was identified as an immunodominant 29-kDa culture filtrate protein (Cfp29) acting as a T cell antigen in both mice and human patients (121). A transposon screen has further shown that Cfp29 is necessary for M. tuberculosis growth in mice (122). Recently, a direct link between the oxidative stress resistance of M. tuberculosis during infection and the Cfp29 system has been reported, which represents the first direct evidence for the involvement of encapsulins in pathogenicity and virulence (83). It was shown both that peroxidase-loaded Cfp29 increases the resistance of M. tuberculosis to hydrogen peroxide treatment and that Cfp29 mutants show attenuated survival in macrophages while also being more susceptible to antibiotic exposure (83). The M. leprae Family 2A encapsulin system was initially misannotated as a membrane protein (MMPI). MMPI was shown to elicit a gamma interferon–secreting T cell proliferative response in leprosy patients (123). Because MMPI represents a dominant M. leprae antigen, its potential application in diagnostic tests or vaccines for leprosy was proposed (123). MMPI was also studied in the context of the pathogenic M. avium complex, specifically M. avium subsp. paratuberculosis (MAP), a nontuberculous mycobacterium that can cause severe lung disease in immunocompromised patients. MAP is also the causative agent of Johne’s disease (paratuberculosis) in ruminants and the suspected causative agent in human Crohn’s disease and rheumatoid arthritis (124, 125). MMPI was found to play a role in the invasion of bovine epithelial cells (126), to induce apoptosis in murine macrophages by targeting mitochondria (127), and to represent one of the immunodominant antigens in ruminant infections (128-131), which led to efforts aimed at using MMPI for the diagnosis and control of paratuberculosis in cattle (132, 133). Further, MMPI has shown promise as an attenuated ruminant paratuberculosis vaccine candidate (134, 135). All but one of these studies were carried out without knowing that encapsulins represent protein nanocompartments loaded with enzymatic cargo. Consequently, almost no details about the molecular mechanisms and precise roles different encapsulins play during infections are currently available.
Many Family 1 Flp and Family 2 desulfurase systems are found in Burkholderia species, including the pathogens Burkholderia cepacia (pulmonary infections and cystic fibrosis) and Burkholderia pseudomallei (melioidosis) (136), while Nocardia spp. (nocardiosis), Clostridium spp. (colitis, botulism, and gangrene), and Bordetella spp. (whooping cough) encode Family 1 peroxidase and Flp systems (137, 138). None of these systems have so far been studied.
Pathogenencoded encapsulins are likely involved in stress resistance—as has been confirmed for M. tuberculosis—or nutrient utilization and storage. Both of these broad functional categories are often important for host invasion and proliferation in the hostile environments encountered by pathogens during infections (139-141). Specialized encapsulin-based nutrient utilization systems—specifically for the scarce and essential elements iron (Family 1 Flp systems) and sulfur (Family 2 desulfurase systems)—may be able to increase pathogen fitness and survival, similar to the importance of bacterial microcompartment-based nutrient utilization systems for the virulence and adaptability of Salmonella typhimurium (food poisoning), Enterococcus faecalis (nosocomial infections), and Clostridium difficile (colitis) (142-144).
Future efforts to characterize pathogen-associated encapsulin systems may yield novel targets for therapeutic intervention. The fact that many encapsulin systems seem to be nonessential but beneficial under certain environmental conditions could mean that targeting them may lead to an attenuation of virulence while minimizing selection pressure for resistance development. In the agricultural context, increased focus on elucidating the roles of encapsulins in livestock infections may result in novel or improved diagnostics, treatments, or vaccines for major ruminant diseases that cause substantial economic losses every year (145, 146).
ACKNOWLEDGMENTS
I thank Jon Marles-Wright for providing structure files of the open and closed states of the Haliangium ochraceum pentameric facet. This work was supported by National Institutes of Health grant R35GM133325.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Cornejo E, Abreu N, Komeili A. 2014. Compartmentalization and organelle formation in bacteria. Curr. Opin. Cell Biol 26:132–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diekmann Y, Pereira-Leal JB. 2013. Evolution of intracellular compartmentalization. Biochem. J 449:319–31 [DOI] [PubMed] [Google Scholar]
- 3.Kerfeld CA, Heinhorst S, Cannon GC. 2010. Bacterial microcompartments. Annu. Rev. Microbiol 64:391–408 [DOI] [PubMed] [Google Scholar]
- 4.Melnicki MR, Sutter M, Kerfeld CA. 2021. Evolutionary relationships among shell proteins of carboxysomes and metabolosomes. Curr. Opin. Microbiol 63:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochoa JM, Yeates TO. 2021. Recent structural insights into bacterial microcompartment shells. Curr. Opin. Microbiol 62:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turmo A, Gonzalez-Esquer CR, Kerfeld CA. 2017. Carboxysomes: metabolic modules for CO2 fixation. FEMS Microbiol. Lett 364:fnx176. [DOI] [PubMed] [Google Scholar]
- 7.Liu LN. 2021. Advances in the bacterial organelles for CO2 fixation. Trends Microbiol. In press. 10.1016/j.tim.2021.10.004 [DOI] [PubMed] [Google Scholar]
- 8.Bobik TA, Lehman BP, Yeates TO. 2015. Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol. Microbiol 98:193–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferlez B, Sutter M, Kerfeld CA. 2019. Glycyl radical enzyme-associated microcompartments: redox-replete bacterial organelles. mBio 10:e02327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prentice MB. 2021. Bacterial microcompartments and their role in pathogenicity. Curr. Opin. Microbiol 63:19–28 [DOI] [PubMed] [Google Scholar]
- 11.Sutter M, Boehringer D, Gutmann S, Gunther S, Prangishvili D, et al. 2008. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol 15:939–47 [DOI] [PubMed] [Google Scholar]
- 12.Nichols RJ, Cassidy-Amstutz C, Chaijarasphong T, Savage DF. 2017. Encapsulins: molecular biology of the shell. Crit. Rev. Biochem. Mol. Biol 52:583–94 [DOI] [PubMed] [Google Scholar]
- 13.Greening C, Lithgow T. 2020. Formation and function of bacterial organelles. Nat. Rev. Microbiol 18:677–89 [DOI] [PubMed] [Google Scholar]
- 14.Jones JA, Giessen TW. 2021. Advances in encapsulin nanocompartment biology and engineering. Biotechnol. Bioeng 118:491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassidy-Amstutz C, Oltrogge L, Going CC, Lee A, Teng P, et al. 2016. Identification of a minimal peptide tag for in vivo and in vitro loading of encapsulin. Biochemistry 55:3461–68 [DOI] [PubMed] [Google Scholar]
- 16.Altenburg WJ, Rollins N, Silver PA, Giessen TW. 2021. Exploring targeting peptide-shell interactions in encapsulin nanocompartments. Sci. Rep 11:4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols RJ, LaFrance B, Phillips NR, Radford DR, Oltrogge LM, et al. 2021. Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism. eLife 10:e59288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreas MP, Giessen TW. 2021. Large-scale computational discovery and analysis of virus-derived microbial nanocompartments. Nat. Commun 12:4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demchuk AM, Patel TR. 2020. The biomedical and bioengineering potential of protein nanocompartments. Biotechnol. Adv 41:107547. [DOI] [PubMed] [Google Scholar]
- 20.Gabashvili AN, Chmelyuk NS, Efremova MV, Malinovskaya JA, Semkina AS, Abakumov MA. 2020. Encapsulins—bacterial protein nanocompartments: structure, properties, and application. Biomolecules 10:966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giessen TW. 2016. Encapsulins: microbial nanocompartments with applications in biomedicine, nanobiotechnology and materials science. Curr. Opin. Chem. Biol 34:1–10 [DOI] [PubMed] [Google Scholar]
- 22.Giessen TW, Silver PA. 2017. Engineering carbon fixation with artificial protein organelles. Curr. Opin. Biotechnol 46:42–50 [DOI] [PubMed] [Google Scholar]
- 23.Giessen TW, Silver PA. 2017. Widespread distribution of encapsulin nanocompartments reveals functional diversity. Nat. Microbiol 2:17029. [DOI] [PubMed] [Google Scholar]
- 24.Valdés-Stauber N, Scherer S. 1994. Isolation and characterization of Linocin M18, a bacteriocin produced by Brevibacterium linens. Appl. Environ. Microbiol 60:3809–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenkrands I, Rasmussen PB, Carnio M, Jacobsen S, Theisen M, Andersen P. 1998. Identification and characterization of a 29-kilodalton protein from Mycobacterium tuberculosis culture filtrate recognized by mouse memory effector cells. Infect. Immun 66:2728–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hicks PM, Chang LS, Kelly RM. 2001. Homomultimeric protease and putative bacteriocin homolog from Thermotoga maritima. Methods Enzymol. 330:455–60 [DOI] [PubMed] [Google Scholar]
- 27.Hicks PM, Rinker KD, Baker JR, Kelly RM. 1998. Homomultimeric protease in the hyperthermophilic bacterium Thermotoga maritima has structural and amino acid sequence homology to bacteriocins in mesophilic bacteria. FEBS Lett. 440:393–98 [DOI] [PubMed] [Google Scholar]
- 28.Anast JM, Schmitz-Esser S. 2020. The transcriptome of Listeria monocytogenes during co-cultivation with cheese rind bacteria suggests adaptation by induction of ethanolamine and 1,2-propanediol catabolism pathway genes. PLOS ONE 15:e0233945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira MM, Ramos ETA, Drechsel MM, Vidal MS, Schwab S, Baldani JI. 2018. Gluconacin from Gluconacetobacter diazotrophicus PAL5 is an active bacteriocin against phytopathogenic and beneficial sugarcane bacteria. J. Appl. Microbiol 125:1812–26 [DOI] [PubMed] [Google Scholar]
- 30.Ward DE, Shockley KR, Chang LS, Levy RD, Michel JK, et al. 2002. Proteolysis in hyperthermophilic microorganisms. Archaea 1:63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olano C, Garcia I, González A, Rodriguez M, Rozas D, et al. 2014. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol 7:242–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teran LC, Distefano M, Bellich B, Petrosino S, Bertoncin P, et al. 2020. Proteomic studies of the biofilm matrix including outer membrane vesicles of Burkholderia multivorans C1576, a strain of clinical importance for cystic fibrosis. Microorganisms 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, He Y, Zhang L, Xu Z, Ben H, et al. 2019. Discovery of potential pathways for biological conversion of poplar wood into lipids by co-fermentation of Rhodococci strains. Biotechnol. Biofuels 12:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loncar N, Rozeboom HJ, Franken LE, Stuart MCA, Fraaije MW. 2020. Structure of a robust bacterial protein cage and its application as a versatile biocatalytic platform through enzyme encapsulation. Biochem. Biophys. Res. Commun 529:548–53 [DOI] [PubMed] [Google Scholar]
- 35.Tamura A, Fukutani Y, Takami T, Fujii M, Nakaguchi Y, et al. 2015. Packaging guest proteins into the encapsulin nanocompartment from Rhodococcus erythropolis N771. Biotechnol. Bioeng 112:13–20 [DOI] [PubMed] [Google Scholar]
- 36.Giessen TW, Orlando BJ, Verdegaal AA, Chambers MG, Gardener J, et al. 2019. Large protein organelles form a new iron sequestration system with high storage capacity. eLife 8:e46070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter N, Triccas JA, Rivoire B, Pessolani MC, Eiglmeier K, et al. 1995. Characterization of the gene encoding the immunodominant 35 kDa protein ofMycobacterium leprae. Mol. Microbiol 16:865–76 [DOI] [PubMed] [Google Scholar]
- 38.Kwak J, McCue LA, Trczianka K, Kendrick KE. 2001. Identification and characterization of a developmentally regulated protein, EshA, required for sporogenic hyphal branches in Streptomyces griseus. J. Bacteriol 183:3004–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saito N, Matsubara K, Watanabe M, Kato F, Ochi K. 2003. Genetic and biochemical characterization of EshA, a protein that forms large multimers and affects developmental processes in Streptomyces griseus. J. Biol. Chem 278:5902–11 [DOI] [PubMed] [Google Scholar]
- 40.Salerno P, Persson J, Bucca G, Laing E, Ausmees N, et al. 2013. Identification of new developmentally regulated genes involved in Streptomyces coelicolor sporulation. BMC Microbiol. 13:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito N, Xu J, Hosaka T, Okamoto S, Aoki H, et al. 2006. EshA accentuates ppGpp accumulation and is conditionally required for antibiotic production in Streptomyces coelicolor A3(2). J. Bacteriol 188:4952–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawamoto S, Watanabe M, Saito N, Hesketh A, Vachalova K, et al. 2001. Molecular and functional analyses of the gene (eshA) encoding the 52-kilodalton protein of Streptomyces coelicolor A3(2) required for antibiotic production. J. Bacteriol 183:6009–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin P, Li Y-Y, Zhou J, Wang Y-H, Zhang S-L, et al. 2013. Direct proteomic mapping of Streptomyces avermitilis wild and industrial strain and insights into avermectin production. J. Proteom 79:1–12 [DOI] [PubMed] [Google Scholar]
- 44.Kuhl M, Gläser L, Rebets Y, Rückert C, Sarkar N, et al. 2020. Microparticles globally reprogram Streptomyces albus toward accelerated morphogenesis, streamlined carbon core metabolism, and enhanced production of the antituberculosis polyketide pamamycin. Biotechnol. Bioeng 117:3858–75 [DOI] [PubMed] [Google Scholar]
- 45.Nichols RJ, LaFrance B, Phillips NR, Oltrogge LM, Valentin-Alvarado LE, et al. 2021. Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism. eLife 10:e59288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward AC, Allenby NE. 2018. Genome mining for the search and discovery of bioactive compounds: the Streptomyces paradigm. FEMS Microbiol. Lett 365:fny240. [DOI] [PubMed] [Google Scholar]
- 47.Bader CD, Panter F, Müller R. 2020. In depth natural product discovery—Myxobacterial strains that provided multiple secondary metabolites. Biotechnol. Adv 39:107480. [DOI] [PubMed] [Google Scholar]
- 48.Suhanovsky MM, Teschke CM. 2015. Nature’s favorite building block: deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 479–480:487–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duda RL, Teschke CM. 2019. The amazing HK97 fold: versatile results of modest differences. Curr. Opin. Virol 36:9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, et al. 2006. Pfam: clans, web tools and services. Nucleic Acids. Res 34:D247–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soding J. 2005. Protein homology detection by HMM–HMM comparison. Bioinformatics 21:951–60 [DOI] [PubMed] [Google Scholar]
- 52.Boto L. 2010. Horizontal gene transfer in evolution: facts and challenges. Proc. Biol. Sci 277:819–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanhere A, Vingron M. 2009. Horizontal Gene Transfers in prokaryotes show differential preferences for metabolic and translational genes. BMC Evol. Biol 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krupovic M, Koonin EV. 2017. Multiple origins ofviral capsid proteins from cellular ancestors. PNAS 114:E2401–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helgstrand C, Wikoff WR, Duda RL, Hendrix RW, Johnson JE, Liljas L. 2003. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 Å resolution. J. Mol. Biol 334:885–99 [DOI] [PubMed] [Google Scholar]
- 56.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129–33 [DOI] [PubMed] [Google Scholar]
- 57.Baker ML, Jiang W, Rixon FJ, Chiu W. 2005. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol 79:14967–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caston JR. 2021. Cryo-EM structure of a thermophilic encapsulin offers clues to its functions. IUCrJ 8:333–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiryaman T, Toor N. 2021. Cryo-EM structure of a thermostable bacterial nanocompartment. IUCrJ 8:342–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LaFrance B, Cassidy-Amstutz C, Nichols RJ, Oltrogge L, Nogales E, Savage DF. 2021. The encapsulin from Thermatoga maritima is a flavoprotein with a symmetry matched ferritin-like cargo protein. bioRxiv 441214. 10.1101/2021.04.26.441214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ross J, McIver Z, Lambert T, Piergentili C, Gallagher KJ, et al. 2021. Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment. bioRxiv 439977. 10.1101/2021.04.15.439977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clancy Kelley L-L, Dillard BD, Tempel W, Chen L, Shaw N, et al. 2007. Structure of the hypothetical protein PF0899 from Pyrococcus furiosus at 1.85 Å resolution. Acta Crystallogr. F 63:549–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang Y, Mu A, Zhang Y, Zhou S, Wang W, et al. 2021. Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin. PNAS 118:e2025658118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andrews SC. 2010. The Ferritin-like superfamily: evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Biophys. Acta Gen. Subj 1800:691–705 [DOI] [PubMed] [Google Scholar]
- 65.McHugh CA, Fontana J, Nemecek D, Cheng N, Aksyuk AA, et al. 2014. A virus capsid-like nanocompartment that stores iron and protects bacteria from oxidative stress. EMBO J. 33:1896–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He D, Piergentili C, Ross J, Tarrant E, Tuck LR, et al. 2019. Conservation of the structural and functional architecture of encapsulated ferritins in bacteria and archaea. Biochem. J 476:975–89 [DOI] [PubMed] [Google Scholar]
- 67.He D, Hughes S, Vanden-Hehir S, Georgiev A, Altenbach K, et al. 2016. Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments. eLife 5:e18972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akita F, Chong KT, Tanaka H, Yamashita E, Miyazaki N, et al. 2007. The crystal structure of a virus-like particle from the hyperthermophilic archaeon Pyrococcus furiosus provides insight into the evolution of viruses. J. Mol. Biol 368:1469–83 [DOI] [PubMed] [Google Scholar]
- 69.Heinemann J, Maaty WS, Gauss GH, Akkaladevi N, Brumfield SK, et al. 2011. Fossil record of an archaeal HK97-like provirus. Virology 417:362–68 [DOI] [PubMed] [Google Scholar]
- 70.Yao H, Wang Y, Lovell S, Kumar R, Ruvinsky AM, et al. 2012. The structure of the BfrB–Bfd complex reveals protein–protein interactions enabling iron release from bacterioferritin. J. Am. Chem. Soc 134:13470–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arosio P, Elia L, Poli M. 2017. Ferritin, cellular iron storage and regulation. IUBMB Life 69:414–22 [DOI] [PubMed] [Google Scholar]
- 72.Pysz MA, Conners SB, Montero CI, Shockley KR, Johnson MR, et al. 2004. Transcriptional analysis of biofilm formation processes in the anaerobic, hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol 70:6098–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ikeyama N, Murakami T, Toyoda A, Mori H, Iino T, et al. 2020. Microbial interaction between the succinate-utilizing bacterium Phascolarctobacterium faecium and the gut commensal Bacteroides thetaiotaomicron. MicrobiologyOpen 9:e1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeng X, Zhang X, Shao Z. 2020. Metabolic adaptation to sulfur of hyperthermophilic Palaeococcus pacificus DY20341T from deep-sea hydrothermal sediments. Int. J. Mol. Sci 21:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim D, Choi J, Lee S, Hyun H, Lee K, Cho K. 2019. Mutants defective in the production of encapsulin show a tan-phase-locked phenotype in Myxococcus xanthus. J. Microbiol 57:795–802 [DOI] [PubMed] [Google Scholar]
- 76.Okamoto Y, Onoda A, Sugimoto H, Takano Y, Hirota S, et al. 2014. H2O2-dependent substrate oxidation by an engineered diiron site in a bacterial hemerythrin. Chem. Commun 50:3421–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alvarez-Carreno C, Alva V, Becerra A, Lazcano A. 2018. Structure, function and evolution of the hemerythrin-like domain superfamily. Protein Sci. 27:848–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rivera M. 2017. Bacterioferritin: structure, dynamics, and protein–protein interactions at play in iron storage and mobilization. Acc. Chem. Res 50:331–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim SJ, Shoda M. 1999. Purification and characterization of a novel peroxidase from Geotrichum candidum Dec 1 involved in decolorization of dyes. Appl. Environ. Microbiol 65:1029–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahmad M, Roberts JN, Hardiman EM, Singh R, Eltis LD, Bugg TD. 2011. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50:5096–107 [DOI] [PubMed] [Google Scholar]
- 81.Rahmanpour R, Bugg TD. 2013. Assembly in vitro of Rhodococcus jostii RHA1 encapsulin and peroxidase DypB to form a nanocompartment. FEBS J. 280:2097–104 [DOI] [PubMed] [Google Scholar]
- 82.Putri RM, Allende-Ballestero C, Luque D, Klem R, Rousou KA, et al. 2017. Structural characterization of native and modified encapsulins as nanoplatforms for in vitro catalysis and cellular uptake. ACS Nano 11:12796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lien KA, Dinshaw K, Nichols RJ, Cassidy-Amstutz C, Knight M, et al. 2020. A nanocompartment system contributes to defense against oxidative stress in Mycobacterium tuberculosis. eLife 10:e74358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tracey JC, Coronado M, Giessen TW, Lau MCY, Silver PA, Ward BB. 2019. The discovery of twenty-eight new encapsulin sequences, including three in anammox bacteria. Sci. Rep 9:20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cipollone R, Ascenzi P, Visca P. 2007. Common themes and variations in the rhodanese superfamily. IUBMB Life 59:51–59 [DOI] [PubMed] [Google Scholar]
- 86.Black KA, Dos Santos PC. 2015. Shared-intermediates in the biosynthesis of thio-cofactors: mechanism and functions of cysteine desulfurases and sulfur acceptors. Biochim. Biophys. Acta Mol. Cell. Res 1853:1470–80 [DOI] [PubMed] [Google Scholar]
- 87.Gorges J, Panter F, Kjaerulff L, Hoffmann T, Kazmaier U, Muller R. 2018. Structure, total synthesis, and biosynthesis of chloromyxamides: myxobacterial tetrapeptides featuring an uncommon 6-chloromethyl-5-methoxypipecolic acid building block. Angew. Chem. Int. Ed 57:14270–75 [DOI] [PubMed] [Google Scholar]
- 88.Kavanagh KL, Jornvall H, Persson B, Oppermann U. 2008. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci 65:3895–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ouchi T, Tomita T, Horie A, Yoshida A, Takahashi K, et al. 2013. Lysine and arginine biosyntheses mediated by a common carrier protein in Sulfolobus. Nat. Chem. Biol 9:277–83 [DOI] [PubMed] [Google Scholar]
- 90.Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, Sutter M. 2018. Bacterial microcompartments. Nat. Rev. Microbiol 16:277–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chandrayan SK, McTernan PM, Hopkins RC, Sun J, Jenney FE Jr., Adams MW. 2012. Engineering hyperthermophilic archaeon Pyrococcus furiosus to overproduce its cytoplasmic [NiFe]-hydrogenase. J. Biol. Chem 287:3257–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Menon AL, Poole FL II, Cvetkovic A, Trauger SA, Kalisiak E, et al. 2009. Novel multiprotein complexes identified in the hyperthermophilic archaeon Pyrococcus furiosus by non-denaturing fractionation of the native proteome. Mol. Cell. Proteom 8:735–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ash PA, Kendall-Price SET, Vincent KA. 2019. Unifying activity, structure, and spectroscopy of [NiFe] hydrogenases: combining techniques to clarify mechanistic understanding. Acc. Chem. Res 52:3120–31 [DOI] [PubMed] [Google Scholar]
- 94.Chou C-J, Shockley KR, Conners SB, Lewis DL, Comfort DA, et al. 2007. Impact of substrate glycoside linkage and elemental sulfur on bioenergetics of and hydrogen production by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol 73:6842–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fiala G, Stetter KO. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol 145:56–61 [Google Scholar]
- 96.Silva PJ, van den Ban EC, Wassink H, Haaker H, de Castro B, et al. 2000. Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem 267:6541–51 [DOI] [PubMed] [Google Scholar]
- 97.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol 180:2636–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alegria TG, Meireles DA, Cussiol JR, Hugo M, Trujillo M, et al. 2017. Ohr plays a central role in bacterial responses against fatty acid hydroperoxides and peroxynitrite. PNAS 114:E132–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seidler NW. 2013. Basic biology of GAPDH. Adv. Exp. Med. Biol 985:1–36 [DOI] [PubMed] [Google Scholar]
- 100.Seidler NW. 2013. GAPDH and intermediary metabolism. Adv. Exp. Med. Biol 985:37–59 [DOI] [PubMed] [Google Scholar]
- 101.Barber RD, Harmer DW, Coleman RA, Clark BJ. 2005. GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genom 21:389–95 [DOI] [PubMed] [Google Scholar]
- 102.Brasen C, Esser D, Rauch B, Siebers B. 2014. Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev 78:89–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siebers B, Schonheit P. 2005. Unusual pathways and enzymes of central carbohydrate metabolism in Archaea. Curr. Opin. Microbiol 8:695–705 [DOI] [PubMed] [Google Scholar]
- 104.Charron C, Talfournier F, Isupov MN, Branlant G, Littlechild JA, et al. 1999. Crystallization and preliminary X-ray diffraction studies of d-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Methanothermus fervidus. Acta. Crystallogr. D 55:1353–55 [DOI] [PubMed] [Google Scholar]
- 105.van der Oost J, Schut G, Kengen SW, Hagen WR, Thomm M, de Vos WM. 1998. The ferredoxin-dependent conversion of glyceraldehyde-3-phosphate in the hyperthermophilic archaeon Pyrococcus furiosus represents a novel site of glycolytic regulation. J. Biol. Chem 273:28149–54 [DOI] [PubMed] [Google Scholar]
- 106.Brunner NA, Brinkmann H, Siebers B, Hensel R. 1998. NAD+-dependentglyceraldehyde-3-phosphate dehydrogenase from Thermoproteus tenax. The first identified archaeal member of the aldehyde dehydrogenase superfamily is a glycolytic enzyme with unusual regulatory properties. J. Biol. Chem 273:6149–56 [DOI] [PubMed] [Google Scholar]
- 107.Zwickl P, Fabry S, Bogedain C, Haas A, Hensel R. 1990. Glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaebacterium Pyrococcus woesei: characterization of the enzyme, cloning and sequencing of the gene, and expression in Escherichia coli. J. Bacteriol 172:4329–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schut GJ, Brehm SD, Datta S, Adams MW. 2003. Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol 185:3935–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sakuraba H, Yoneda K, Yoshihara K, Satoh K, Kawakami R, et al. 2007. Sequential aldol condensation catalyzed by hyperthermophilic 2-deoxy-d-ribose-5-phosphate aldolase. Appl. Environ. Microbiol 73:7427–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sakuraba H, Tsuge H, Shimoya I, Kawakami R, Goda S, et al. 2003. The first crystal structure of archaeal aldolase. Unique tetrameric structure of 2-deoxy-d-ribose-5-phosphate aldolase from the hyperthermophilic archaea Aeropyrum pernix. J. Biol. Chem 278:10799–806 [DOI] [PubMed] [Google Scholar]
- 111.Rashid N, Imanaka H, Fukui T, Atomi H, Imanaka T. 2004. Presence of a novel phosphopentomutase and a 2-deoxyribose 5-phosphate aldolase reveals a metabolic link between pentoses and central carbon metabolism in the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Bacteriol 186:4185–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lomax MS, Greenberg GR. 1968. Characteristics of the deo operon: role in thymine utilization and sensitivity to deoxyribonucleosides. J. Bacteriol 96:501–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jia B, Liu J, Van Duyet L, Sun Y, Xuan YH, Cheong G-W. 2015. Proteome profiling of heat, oxidative, and salt stress responses in Thermococcus kodakarensis KOD1. Front. Microbiol 6:605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Orita I, Sato T, Yurimoto H, Kato N, Atomi H, et al. 2006. The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. J. Bacteriol 188:4698–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Salleron L, Magistrelli G, Mary C, Fischer N, Bairoch A, Lane L. 2014. DERA is the human deoxyribose phosphate aldolase and is involved in stress response. Biochim. Biophys. Acta Mol. Cell. Res 1843:2913–25 [DOI] [PubMed] [Google Scholar]
- 116.Niforou K, Cheimonidou C, Trougakos IP. 2014. Molecular chaperones and proteostasis regulation during redox imbalance. Redox Biol. 2:323–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Burston SG, Clarke AR. 1995. Molecular chaperones: physical and mechanistic properties. Essays Biochem. 29:125–36 [PubMed] [Google Scholar]
- 118.De Oliveira DMP, Forde BM, Kidd TJ, Harris PNA, Schembri MA, et al. 2020. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev 33:00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saxena S, Spaink HP, Forn-Cuni G. 2021. Drug resistance in nontuberculous mycobacteria: mechanisms and models. Biology 10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kanabalan RD, Lee LJ, Lee TY, Chong PP, Hassan L, et al. 2021. Human tuberculosis and Mycobacterium tuberculosis complex: a review on genetic diversity, pathogenesis and omics approaches in host biomarkers discovery. Microbiol. Res 246:126674. [DOI] [PubMed] [Google Scholar]
- 121.Weldingh K, Andersen P. 1999. Immunological evaluation of novel Mycobacterium tuberculosis culture filtrate proteins. FEMS Immunol. Med. Microbiol 23:159–64 [DOI] [PubMed] [Google Scholar]
- 122.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, et al. 2013. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155:1296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Triccas JA, Roche PW, Winter N, Feng CG, Butlin CR, Britton WJ. 1996. A 35-kilodalton protein is a major target of the human immune response to Mycobacterium leprae. Infect. Immun 64:5171–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bo M, Jasemi S, Uras G, Erre GL, Passiu G, Sechi LA. 2020. Role of infections in the pathogenesis of rheumatoid arthritis: focus on mycobacteria. Microorganisms 8:1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aitken JM, Phan K, Bodman SE, Sharma S, Watt A, et al. 2021. A Mycobacterium species for Crohn’s disease? Pathology 53:818–23 [DOI] [PubMed] [Google Scholar]
- 126.Bannantine JP, Huntley JFJ, Miltner E, Stabel JR, Bermudez LE. 2003. The Mycobacterium avium subsp. paratuberculosis 35 kDa protein plays a role in invasion of bovine epithelial cells. Microbiology 149:2061–69 [DOI] [PubMed] [Google Scholar]
- 127.Lee K-I, Whang J, Choi H-G, Son Y-J, Jeon HS, et al. 2016. Mycobacterium avium MAV2054 protein induces macrophage apoptosis by targeting mitochondria and reduces intracellular bacterial growth. Sci. Rep 6:37804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bannantine JP, Radosevich TJ, Stabel JR, Berger S, Griffin JF, Paustian ML. 2007. Production and characterization of monoclonal antibodies against a major membrane protein of Mycobacterium avium subsp. paratuberculosis. Clin. Vaccine Immunol 14:312–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bannantine JP, Bayles DO, Waters WR, Palmer MV, Stabel JR, Paustian ML. 2008. Early antibody response against Mycobacterium avium subspecies paratuberculosis antigens in subclinical cattle. Proteome Sci. 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li L, Munir S, Bannantine JP, Sreevatsan S, Kanjilal S, Kapur V. 2007. Rapid expression of Mycobacterium avium subsp. paratuberculosis recombinant proteins for antigen discovery. Clin. Vaccine Immunol 14:102–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shin SJ, Yoo HS, McDonough SP, Chang YF. 2004. Comparative antibody response of five recombinant antigens in related to bacterial shedding levels and development of serological diagnosis based on 35 kDa antigen for Mycobacterium avium subsp. paratuberculosis. J. Vet. Sci 5:111–17 [PubMed] [Google Scholar]
- 132.Tilocca B, Soggiu A, Greco V, Piras C, Arrigoni N, et al. 2020. Immunoinformatic-based prediction of candidate epitopes for the diagnosis and control of paratuberculosis (Johne’s disease). Pathogens 9:705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li L, Wagner B, Freer H, Schilling M, Bannantine JP, et al. 2017. Early detection of Mycobactenum avium subsp. paratuberculosis infection in cattle with multiplex-bead based immunoassays. PLOS ONE 12:e0189783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Abdellrazeq GS, Fry LM, Elnaggar MM, Bannantine JP, Schneider DA, et al. 2020. Simultaneous cognate epitope recognition by bovine CD4 and CD8 T cells is essential for primary expansion of antigen-specific cytotoxic T-cells following ex vivo stimulation with a candidate Mycobacterium avium subsp. paratuberculosis peptide vaccine. Vaccine 38:2016–25 [DOI] [PubMed] [Google Scholar]
- 135.Franceschi V, Mahmoud AH, Abdellrazeq GS, Tebaldi G, Macchi F, et al. 2019. Capacity to elicit cytotoxic CD8 T cell activity against Mycobacterium avium subsp. paratuberculosis is retained in a vaccine candidate 35 kDa peptide modified for expression in mammalian cells. Front. Immunol 10:2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chomkatekaew C, Boonklang P, Sangphukieo A, Chewapreecha C. 2020. An evolutionary arms race between Burkholderia pseudomallei and host immune system: What do we know? Front. Microbiol 11:612568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.José RJ, Periselneris JN, Brown JS. 2020. Opportunistic bacterial, viral and fungal infections of the lung. Medicine 48:366–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bowman JA, Utter GH. 2020. Evolving strategies to manage Clostridium difficile colitis. J. Gastrointest. Surg 24:484–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Harvey PC, Watson M, Hulme S, Jones MA, Lovell M, et al. 2011. Salmonella enterica serovar typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect. Immun 79:4105–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Klumpp J, Fuchs TM. 2007. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology 153:1207–20 [DOI] [PubMed] [Google Scholar]
- 141.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, et al. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. PNAS 108:17480–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Srikumar S, Fuchs TM. 2011. Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl. Environ. Microbiol 77:281–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pitts AC, Tuck LR, Faulds-Pain A, Lewis RJ, Marles-Wright J. 2012. Structural insight into the Clostridium difficile ethanolamine utilisation microcompartment. PLOS ONE 7:e48360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maadani A, Fox KA, Mylonakis E, Garsin DA. 2007. Enterococcus faecalis mutations affecting virulence in the Caenorhabditis elegans model host. Infect. Immun 75:2634–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ott SL, Wells SJ, Wagner BA. 1999. Herd-level economic losses associated with Johne’s disease on US dairy operations. Prev. Vet. Med 40:179–92 [DOI] [PubMed] [Google Scholar]
- 146.Rasmussen P, Barkema HW, Mason S, Beaulieu E, Hall DC. 2021. Economic losses due to Johne’s disease (paratuberculosis) in dairy cattle. J. Dairy Sci 104:3123–43 [DOI] [PubMed] [Google Scholar]
- 147.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, et al. 2016. A new view of the tree of life. Nat. Microbiol 1:16048. [DOI] [PubMed] [Google Scholar]
- 148.Sekiguchi Y, Ohashi A, Parks DH, Yamauchi T, Tyson GW, Hugenholtz P. 2015. First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking. PeerJ. 3:e740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Holm L. 2020. DALI and the persistence of protein shape. Protein Sci. 29:128–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Contreras H, Joens MS, McMath LM, Le VP, Tullius MV, et al. 2014. Characterization of a Mycobacterium tuberculosis nanocompartment and its potential cargo proteins. J. Biol. Chem 289:18279–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Snijder J, Kononova O, Barbu IM, Uetrecht C, Rurup WF, et al. 2016. Assembly and mechanical properties of the cargo-free and cargo-loaded bacterial nanocompartment encapsulin. Biomacromolecules 17:2522–29 [DOI] [PubMed] [Google Scholar]
- 152.Rurup WF, Cornelissen JJ, Koay MS. 2015. Recombinant expression and purification of “virus-like” bacterial encapsulin protein cages. Methods Mol. Biol 1252:61–67 [DOI] [PubMed] [Google Scholar]
- 153.Ross J, Lambert T, Piergentili C, He D, Waldron KJ, et al. 2020. Mass spectrometry reveals the assembly pathway of encapsulated ferritins and highlights a dynamic ferroxidase interface. Chem. Commun 56:3417–20 [DOI] [PubMed] [Google Scholar]
- 154.Piergentili C, Ross J, He D, Gallagher KJ, Stanley WA, et al. 2020. Dissecting the structural and functional roles of a putative metal entry site in encapsulated ferritins. J. Biol. Chem 295:15511–26 [DOI] [PMC free article] [PubMed] [Google Scholar]