Abstract
Introduction:
Purine-rich element-binding protein A (PURA) encodes Pur-alpha, a transcriptional activator protein is crucial for normal brain development. Pathogenic variants in PURA are known to cause mental retardation, autosomal dominant 31, characterized by psychomotor delay, absent or poor speech, hypotonia, feeding difficulties, seizures or ‘seizure-like’ movements, and dysmorphism. PURA-related neurodevelopmental disorder result either from heterozygous pathogenic sequence variants in PURA or microdeletions spanning PURA.
Methods:
Singleton exome sequencing was performed for index patient. The pathogenic variant identified was validated and segregation analysis was performed by Sanger sequencing.
Results:
We report on a patient of PURA-related neurodevelopmental disorder identified with a novel de novo stop-gain variant. In addition to typical phenotype, patient also had hypersensitivity to various stimuli which was not reported in PURA-related disorder. However, patient shares phenotype with ‘infantile hypotonia with psychomotor retardation and characteristic facies 2’ (IHPRF2) caused by biallelic pathogenic variants in UNC80.
Conclusion:
This study expands the phenotypic spectrum of PURA-related neurodevelopmental disorder and adds on to genetic heterogeneity of IHPRF phenotype. We propose PURA-related neurodevelopmental disorder to be considered as one of the subtypes of IHPRF.
Keywords: Dysmorphism, hypersensitivity, hypotonia, neurodevelopmental disorder, psychomotor retardation, purine-rich element-binding protein A, UNC80, whole exome sequencing
Introduction
Neurodevelopmental disorders (NDD) are genetically heterogeneous disorders characterized by various intellectual, behavioral and motor disabilities. Cumulatively, it affects about 2-5% of the population with varying degrees of severity (Boyle et al., 2011). The advent of next-generation sequencing technique has led to the rapid discovery of new NDD genes. The application of trio whole exome sequencing (WES) has enabled the identification of pathogenic de novo variants in a significant proportion of patients with NDD (Hamdan et al., 2014).
Mental retardation, autosomal dominant 31 (MIM# 616158) is a neurodevelopmental disorder caused by heterozygous pathogenic variants in the PURA gene. It is characterized by moderate to severe psychomotor delay, absent or poor speech development, hypotonia, feeding difficulties, respiratory problems, seizures or ‘seizure-like’ movements, dyskinesia, and dysmorphism. PURA-related neurodevelopmental disorders result either from heterozygous pathogenic sequence variants in PURA or microdeletions spanning PURA (Reijnders et al., 2017). To date, including the present patient, a total of 80 patients of PURA-related NDD have been reported (Shimojima et al., 2011; Hosoki et al., 2012; Brown et al., 2013; Hunt et al., 2014; Lalani et al., 2014; Bonaglia et al., 2015; Tanaka et al., 2015; Reijnders et al., 2017; Okamoto et al., 2017).
We describe a case of PURA-related NDD identified through WES with de novo pathogenic variant. Furthermore, we compare the phenotype of PURA-related NDD with a clinically indistinguishable phenotype of disorders with ‘infantile hypotonia with psychomotor retardation and characteristic facies’ (IHPRF) resulting from biallelic pathogenic variants in either of these genes, viz., UNC80, NALCN, or TBCK and propose that PURA-related NDD should be classified as an additional subtype of IHPRF.
Methods
Clinical report
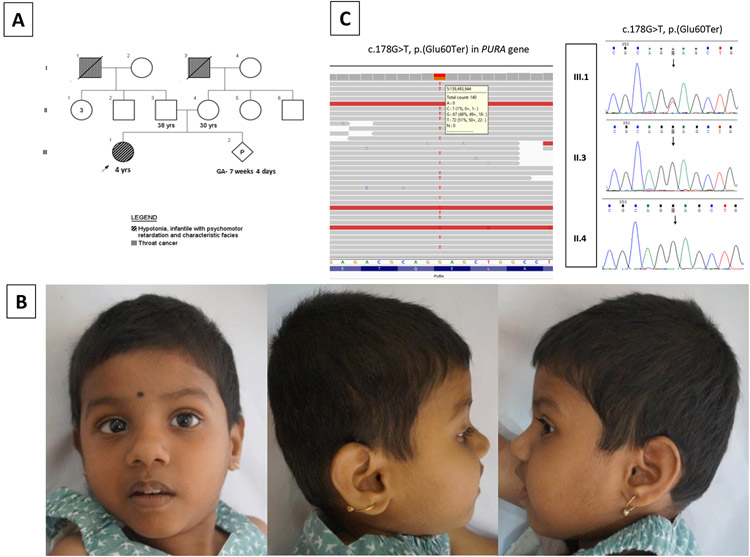
We ascertained a 4-year-old girl with a complaint of developmental delay. She was the first child of non-consanguineous healthy couple, born at term following uncomplicated pregnancy with a birth weight of 3.2 kg (0 SD) (Fig. 1A). She was transferred to neonatal intensive care unit for generalized hypotonia, lethargy, and abnormal ‘seizure-like’ movements on day 2 of life. Early-onset feeding difficulties including poor sucking, dysphagia, and drooling were present. She attained social smile at 6 months, neck holding at 14 months, unsupported sitting at 3 years, and bisyllables at 2 years. No further milestones were achieved. She had poor speech with only 3-4 words vocabulary. There was no history of regression of milestones. On clinical examination, her occipitofrontal circumference was 47 cm (−3.5 SD), length 93 cm (−2 SD) and weight 10.6 kg (−3.5 SD). Dysmorphic features including brachycephaly, hypotonic facies, telecanthus, downslanting palpebral fissures, strabismus, wide nasal bridge, bulbous nasal tip, thick vermilion of lips, and open mouth with drooling of saliva were noted (Fig. 1B). She had profound truncal as well as peripheral hypotonia, poor coordination, and dyskinetic face and limb movements. Hypersensitivity to various stimuli (sound, light, and on eliciting deep tendon reflexes) were observed. Rest of the systemic examination was unremarkable. Ophthalmological evaluation revealed nystagmus with alternating esotropia. Hearing assessment, echocardiography and magnetic resonance imaging (MRI) of the brain were normal.
Figure 1.
(A) Pedigree of affected family
(B) Clinical photographs of proband: subtle dysmorphic features with brachycephaly, hypotonic facies, telecanthus, downslanting palpebral fissures, strabismus, wide nasal bridge, bulbous nasal tip, thick vermilion of lips, open mouth and profound hypotonia
(C) IGV of proband showing sequence variant c.178G>T, p.(Glu60Ter) in PURA in heterozygous state and sequence chromatograms of affected family showing heterozygous variant c.178G>T, p.(Glu60Ter) in PURA in the proband. The variant was not observed in her parents
Molecular testing
The EDTA blood samples were collected from the patient and her parents after obtaining written informed consent from the parents. WES (Illumina, Inc., San Diego, California, USA) was performed for the patient as described earlier (Girisha et al., 2016; Shukla et al., 2017). For WES data analysis, the raw data was processed and converted to a FASTQ format using an in-house pipeline based on GATK Best Practices. This data was annotated by ANNOVAR and in-house scripts. The called variants were analyzed based on variant prioritization and filtering strategy outlined in Supplementary Table S1. Validation of the identified variant and segregation analysis were done by Sanger sequencing.
Results
On analysis of the WES data, a novel stop-gain variant c.178G>T, p.(Glu60Ter) was observed in exon 1 of PURA (NM_005859.4) in heterozygous state in the patient (Fig. 1C). This variant is likely to result in premature termination of translation leading to either nonsense-mediated mRNA decay or a truncated protein. The variant was not observed in population databases like gnomAD, 1000 Genomes Project, Exome Aggregate Consortium, and in our in-house data of 700 exomes in either heterozygous or homozygous state. Multiple lines of in-silico prediction tools (MutationTaster, FATHMM-MKL and LRT) were predicting the variant to be disease causing. Sanger validation of the identified variant was done in the patient. On segregation analysis, the variant was not observed in her parents (Fig. 1C). As per ACMG guidelines, variant was classified as pathogenic (PVS1, PS2, PM2, PP3) and submitted in the ClinVar database ().
Discussion
The proband was clinically diagnosed as IHPRF in view of developmental delay, significant hypotonia, characteristic facies, and hypersensitivity to sensory stimuli. However, WES revealed a novel pathogenic variant in PURA known to cause mental retardation, autosomal dominant 31. Individuals with pathogenic variants in PURA present with consistent phenotype of developmental delay with lack of independent walking and poor speech, hypotonia, early-onset feeding difficulties, abnormal ‘seizure-like’ movements, and dyskinesia (Hunt et al., 2014; Lalani et al., 2014; Tanaka et al., 2015). Visual abnormalities like strabismus, nystagmus, and esotropia are frequently observed. Subtle dysmorphic features noted in most individuals including the present proband, includes hypotonic facies, telecanthus, downslanting palpebral fissures, bulbous nasal tip, and open mouth (Hunt et al., 2014; Lalani et al., 2014; Tanaka et al., 2015). Growth abnormalities like microcephaly, short stature and poor weight gain are additional findings in this disorder (Hunt et al., 2014; Reijnders et al., 2017). Excessive hiccups, hypersomnolence, congenital apnea, stereotypic hand movements, regression and scoliosis have been identified in significant number of patients earlier, but not seen in present proband (Reijnders et al., 2017). On brain MRI, white matter abnormalities or delayed myelination were reported in 23/79 patients (Hunt et al., 2014; Lalani et al., 2014; Tanaka et al., 2015; Reijnders et al., 2017). However, brain MRI of present proband was normal at the age of 4 years.
PURA-related NDD and IHPRF (Table 1) share a common findings of psychomotor retardation, profound hypotonia, seizures or ‘seizure-like’ movements, dyskinesia, visual abnormalities, and characteristic facial dysmorphism. Growth delay and musculoskeletal abnormalities including scoliosis and joint contractures were also reported in all. In literature, exaggerated startle reflex to sound has been reported in PURA-related NDD. The proband demonstrated hypersensitivity to various stimuli like sound, light, and on eliciting deep tendon reflexes, reported frequently in IHPRF2 (MIM# 616801) caused by biallelic pathogenic variants in UNC80 (Stray-Pedersen et al., 2016). Other subtypes of IHPRF i.e. IHPRF1 (MIM# 615419) and IHPRF3 (MIM# 616900) share similar phenotype to UNC80-related IHPRF, except hypersensitivity to stimuli. Non-specific white matter abnormalities have been reported in PURA-related NDD as well as all subtypes of IHPRF (Al-Sayed et al., 2013; Köroğlu et al., 2013; Hunt et al., 2014; Lalani et al., 2014; Tanaka et al., 2015; Bhoj et al., 2016; Perez et al., 2016; Shamseldin et al., 2016; Stray-Pedersen et al., 2016; Reijnders et al., 2017).
Table 1:
Comparison of phenotypes of our patient, PURA-related neurodevelopmental disorder, IHPRF1, 2, and 3
| Phenotypic characteristics |
Proband |
PURA-related
neurodevelopmental disorder |
IHPRF2 (UNC80) | IHPRF1 (NALCN) | IHPRF3 (TBCK) |
|---|---|---|---|---|---|
| Neurodevelopmental abnormality | |||||
| Motor delay | + | + | + | + | + |
| Speech delay | + | + | + | + | + |
| Cognitive delay | + | + | + | + | + |
| Hypotonia | + | + | + | + | + |
| Seizures or ‘seizure-like’ activity | + | + | + | + | + |
| Feeding difficulties | + | + | + | + | + |
| Oral motor dysfunction | + | + | + | + | + |
| Dyskinesia | + | + | + | + | − |
| Sensory hypersensitivity$ | + | Only startle reflex | + | − | − |
| Facial features | |||||
| Brachycephaly | + | + | + | + | + |
| Hypotonic facies | + | + | + | + | + |
| Broad forehead | + | + | + | + | + |
| Downslant palpebral fissures | + | + | + | + | + |
| Broad nasal bridge | + | + | + | + | + |
| Bulbous nasal tip | + | + | + | − | + |
| Open mouth | + | + | + | + | + |
| Thick vermilion of upper lip | + | − | − | − | + |
| Tented upper lip | − | + | + | + | + |
| Ocular abnormality | |||||
| Strabismus | + | + | + | + | + |
| Nystagmus | + | + | + | + | + |
| Esotropia | + | + | + | + | + |
| Musculoskeletal abnormality | |||||
| Scoliosis | − | + | + | + | + |
| Joint contractures | − | + | + | + | + |
| Growth | |||||
| Microcephaly | + | + | + | + | + |
| Short stature | + | + | + | + | + |
| Postnatal weight < 3rd centile | + | + | + | + | + |
| Brain imaging | |||||
| MRI brain findings | Normal | Delayed myelination; nonspecific white matter hyperintensities; thin corpus callosum; enlargement of extra-axial spaces; mild cerebral atrophy | Nonspecific white matter hyperintensities; thin corpus callosum; enlargement of ventricles and extra-axial spaces; mild cerebral atrophy | Cerebral and cerebellar atrophy | Nonspecific white matter abnormalities; thin corpus callosum; enlargement of ventricles; cerebral atrophy |
(+): present; (−): absent
IHPRF: Infantile hypotonia with psychomotor retardation and characteristic facies
Sensory hypersensitivity: hypersensitivity to various stimuli (like sound, light, and on eliciting deep tendon reflexes)
The human purine-rich element-binding protein A (PURA) is located on chromosome 5q31.2. It encodes a single-stranded nucleic acid binding protein, Pur-alpha. It is a highly conserved multifunctional protein with unwindase activity, which plays important role in DNA replication, DNA transcription, RNA trafficking, and translation (Weber et al 2016). The functionality of Pur-alpha relies on the three conserved PUR repeat motifs: PUR I, PUR II, and PUR III (Graebsch et al., 2009, Weber et al., 2016). Functional studies in mice suggest that PURA is required for normal brain development, neuronal proliferation, dendritic maturation, and synapse formation. PURA knockout mice have demonstrated ataxic gait, seizures, tremor, lethargy, and early death with white matter abnormalities, reduced number of neurons, and altered synapse morphology recapitulating the phenotype observed in human beings (Khalili et al., 2003; Hokkanen et al. 2012).
Of the 80 reported PURA-related NDD patients, including the present patient, 72 patients had PURA intragenic sequence variants and eight patients had nonrecurrent 5q31.3 microdeletions spanning the PURA gene (Reijnders et al., 2017). The missense pathogenic variants occur in one of the three PUR repeats, while truncating pathogenic variants span across the entire gene (Hunt et al., 2014; Lalani et al., 2014; Tanaka et al., 2015; Okamoto et al., 2017). These variants probably result in functional haploinsufficiency of the protein (Hunt et al., 2014). Almost all patients, including the present patient, had de novo variants except for one patient where gonosomal mosaicism was observed in one of the clinically unaffected parents (Reijnders et al., 2017). The neurodevelopmental features of patients with PURA pathogenic variants and those carrying microdeletions are similar. However, phenotype is more severe in patients with a larger deletion spanning multiple genes in addition to PURA (Hosoki et al., 2012; Brown et al., 2013; Lalani et al., 2014; Bonaglia et al., 2015).
Hence, we conclude that PURA-related NDD should be considered in the evaluation of children with profound hypotonia, psychomotor retardation, dyskinesia, sensory hypersensitivity, and characteristic dysmorphic facies. We propose PURA-related NDD to be considered as one of the subtypes of IHPRF due to striking similarity of their phenotypes.
Supplementary Material
Acknowledgements
We thank the family who cooperated with the evaluation of the child and consented for participation in this study. The Department of Health Research, Ministry of Health and Family Welfare, Government of India funded the project titled “Clinical and molecular characterization of leukodystrophies in Indian children” (V.25011/379/2015-GIA/HR).
Footnotes
Conflicts of interest
The enclosed manuscript has been seen and approved by all the authors and there is no conflict of interest concerning this work.
References
- Al-Sayed MD, Al-Zaidan H, Albakheet A, Hakami H, Kenana R, Al-Yafee Y, et al. (2013). Mutations in NALCN cause an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay. Am J Hum Genet 93:721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoj EJ, Li D, Harr M, Edvardson S, Elpeleg O, Chisholm E, et al. (2016). Mutations in TBCK, Encoding TBC1-Domain-Containing Kinase, Lead to a Recognizable Syndrome of Intellectual Disability and Hypotonia. Am J Hum Genet 98:782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Zanotta N, Giorda R, D'Angelo G, Zucca C (2015). Long-term follow-up of a patient with 5q31.3 microdeletion syndrome and the smallest de novo 5q31.2q31.3 deletion involving PURA. Mol Cytogenet 8:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle CA, Boulet S, Schieve LA, Cohen RA, Blumberg SJ, Yeargin-Allsopp M, et al. (2011). Trends in the prevalence of developmental disabilities in US children, 1997-2008. Pediatrics 127:1034–1042. [DOI] [PubMed] [Google Scholar]
- Brown N, Burgess T, Forbes R, McGillivray G, Kornberg A, Mandelstam S, Stark Z (2013) 5q31.3 Microdeletion syndrome: clinical and molecular characterization of two further cases. Am J Med Genet A 161A:2604–2608. [DOI] [PubMed] [Google Scholar]
- Girisha KM, Shukla A, Trujillano D, Bhavani GS, Hebbar M, Kadavigere R, Rolfs A (2016). A homozygous nonsense variant in IFT52 is associated with a human skeletal ciliopathy. Clin Genet 90:536–539. [DOI] [PubMed] [Google Scholar]
- Graebsch A, Roche S, Niessing D (2009). X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface. Proc Natl Acad Sci U S A 106:18521–18526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Srour M, Capo-Chichi JM, Daoud H, Nassif C, Patry L, et al. (2014). De novo mutations in moderate or severe intellectual disability. PLoS Genet 10:e1004772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokkanen S, Feldmann HM, Ding H, Jung CK, Bojarski L, Renner-Müller I, et al. (2012). Lack of Pur-alpha alters postnatal brain development and causes megalencephaly. Hum Mol Genet 21:473–484. [DOI] [PubMed] [Google Scholar]
- Hosoki K, Ohta T, Natsume J, Imai S, Okumura A, Matsui T, et al. (2012). Clinical phenotype and candidate genes for the 5q31.3 microdeletion syndrome. Am J Med Genet A 158A:1891–1896. [DOI] [PubMed] [Google Scholar]
- Hunt D, Leventer RJ, Simons C, Taft R, Swoboda KJ, Gawne-Cain M, et al. (2014). Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J Med Genet 51:806–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, et al. (2003). Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol 23:6857–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köroğlu Ç, Seven M, Tolun A (2013). Recessive truncating NALCN mutation in infantile neuroaxonal dystrophy with facial dysmorphism. J Med Genet 50:515–520. [DOI] [PubMed] [Google Scholar]
- Lalani SR, Zhang J, Schaaf CP, Brown CW, Magoulas P, Tsai AC, et al. (2014). Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am J Hum Genet 95:579–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Nakao H, Niihori T, Aoki Y (2017). Patient with a novel purine-rich element binding protein A mutation. Congenit Anom (Kyoto) 57:201–204. [DOI] [PubMed] [Google Scholar]
- Perez Y, Kadir R, Volodarsky M, Noyman I, Flusser H, Shorer Z, et al. (2016). UNC80 mutation causes a syndrome of hypotonia, severe intellectual disability, dyskinesia and dysmorphism, similar to that caused by mutations in its interacting cation channel NALCN. J Med Genet 53:397–402. [DOI] [PubMed] [Google Scholar]
- Reijnders MRF, Leventer RJ, Lee BH, Baralle D, Selber P, Paciorkowski AR, Hunt D (2017). PURA-Related Neurodevelopmental Disorders. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A (editors). GeneReviews®. Seattle (WA): University of Washington, Seattle; 1993–2020. [PubMed] [Google Scholar]
- Shamseldin HE, Faqeih E, Alasmari A, Zaki MS, Gleeson JG, Alkuraya FS (2016). Mutations in UNC80, Encoding Part of the UNC79-UNC80-NALCN Channel Complex, Cause Autosomal-Recessive Severe Infantile Encephalopathy. Am J Hum Genet 98:210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima K, Isidor B, Le Caignec C, Kondo A, Sakata S, Ohno K, Yamamoto T (2011). A new microdeletion syndrome of 5q31.3 characterized by severe developmental delays, distinctive facial features, and delayed myelination. Am J Med Genet A 155A:732–736. [DOI] [PubMed] [Google Scholar]
- Shukla A, Hebbar M, Srivastava A, Kadavigere R, Upadhyai P, Kanthi A, et al. (2017). Homozygous p.(glu87lys) variant in ISCA1 is associated with a multiple mitochondrial dysfunctions syndrome. J Hum Genet 62:723–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray-Pedersen A, Cobben JM, Prescott TE, Lee S, Cang C, Aranda K, et al. (2016). Biallelic Mutations in UNC80 Cause Persistent Hypotonia, Encephalopathy, Growth Retardation, and Severe Intellectual Disability. Am J Hum Genet 98:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka AJ, Bai R, Cho MT, Anyane-Yeboa K, Ahimaz P, Wilson AL, et al. (2015). De novo mutations in PURA are associated with hypotonia and developmental delay. Cold Spring Harb Mol Case Stud 1:a000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J, Bao H, Hartlmüller C, Wang Z, Windhager A, Janowski R, et al. (2016). Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha. Elife 5:e11297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.