INTRODUCTION:
In the United States, the effectiveness of anal cancer screening programs has been limited by a lack of trained professionals proficient in high-resolution anoscopy (HRA) and a high patient lost-to-follow-up rate between diagnosis and treatment. Simplifying anal intraepithelial neoplasia grade 2 or more severe (AIN 2+) detection could radically improve the access and efficiency of anal cancer prevention. Novel optical imaging providing point-of-care diagnoses could substantially improve existing HRA and histology-based diagnosis. This work aims to demonstrate the potential of high-resolution microendoscopy (HRME) coupled with a novel machine learning algorithm for the automated, in vivo diagnosis of anal precancer.
METHODS:
The HRME, a fiber-optic fluorescence microscope, was used to capture real-time images of anal squamous epithelial nuclei. Nuclear staining is achieved using 0.01% wt/vol proflavine, a topical contrast agent. HRME images were analyzed by a multitask deep learning network (MTN) that computed the probability of AIN 2+ for each HRME image.
RESULTS:
The study accrued data from 77 people living with HIV. The MTN achieved an area under the receiver operating curve of 0.84 for detection of AIN 2+. At the AIN 2+ probability cutoff of 0.212, the MTN achieved comparable performance to expert HRA impression with a sensitivity of 0.92 (P = 0.68) and specificity of 0.60 (P = 0.48) when using histopathology as the gold standard.
DISCUSSION:
When used in combination with HRA, this system could facilitate more selective biopsies and promote same-day AIN2+ treatment options by enabling real-time diagnosis.
KEY WORDS: anal cancer, artificial intelligence, diagnostic imaging, human papillomavirus, human immunodeficiency virus
INTRODUCTION
In the United States, the incidence of anal cancer is over 30 times more common in persons living with HIV than in the general population (1,2). A compromised immune system makes people living with HIV (PLWH) susceptible to coinfection with the human papillomavirus (HPV) in the anal canal, linked to 90% of anal cancer cases (3).
Although HPV vaccination is the most effective method to prevent HPV-associated cancers, the administration is limited to individuals aged 9 to 45 (4). For PLWH, clinical experts are increasingly advising regular screening with cytology (5). Cytology screen-positive patients (atypical squamous cells of undetermined significance [ASCUS] or more severe) are asked to return for a second visit to undergo high-resolution anoscopy (HRA) (6). Suspicious areas are biopsied and evaluated by a pathologist. HRA-guided biopsy requires a high degree of expertise; new HRA practitioners take around 200 cases to begin consistently identifying all precancerous lesions (high-grade squamous intraepithelial lesions [HSILs] or anal intraepithelial neoplasia grade 2 or more severe [AIN 2+]) in a patient (7). Patients diagnosed with AIN 2+ return for a third visit to receive treatment through the topical application of therapeutic creams, photodynamic therapy, thermal destruction, or lesion excision. Although the treatment of anal precancer has been shown to reduce the risk of progression to cancer significantly, the multiple visit approach to care has led to high patient lost-to-follow-up rates (8). A study on the outcomes of an anal cancer-screening program from 2009 to 2019 found that only 58% of patients diagnosed with AIN 2+ returned for treatment (9).
Simplifying AIN 2+ detection could radically improve the access and efficiency of anal cancer prevention. Novel optical imaging to identify histologic AIN 2+ could substantially improve existing HRA and histology-based diagnostic strategies. By offering an in vivo diagnosis, more selective biopsies can be performed. In addition, the ability to delineate normal from neoplastic mucosa in real-time may reduce the number of patients lost to follow-up and facilitate “see and treat” approaches.
One promising technology is high-resolution microendoscopy (HRME), where a fluorescence microscope coupled to a fiber-optic probe is used to capture real-time images of squamous epithelial nuclei after topical application of proflavine. HRME images of precancerous anal lesions show characteristic changes in nuclear morphology including nuclear enlargement, crowding, and pleomorphism. In a pilot study of 41 patients, Varela et al. (10) trained a group of 8 clinicians to visually interpret HRME images of anal epithelium and identify anal precancers. The group achieved a sensitivity of 0.93 and specificity of 0.87. Automated algorithms have been developed to interpret HRME images and provide real-time feedback in other organ sites (11,12).
In this work, we describe the use of a novel automated machine learning method to analyze HRME images of anal epithelium (13). The novel deep learning algorithm was initially developed to detect cervical precancer in HRME images and achieved a performance comparable to expert colposcopy with a sensitivity of 0.94 (P = 0.3) and specificity of 0.58 (P = 1.0) (13). In this work, we collected HRME images from 77 patients living with HIV and analyzed them with the proposed algorithm. We found that the performance of this model was comparable to expert HRA impression with a sensitivity of 0.92 (P = 0.68) and specificity of 0.60 (P = 0.48) in detection of anal precancer. To the best of our knowledge, this is the first evaluation of an automated, in vivo imaging-based system for the detection of anal precancer.
METHODS
High-resolution microendoscopy
Instrumentation
The HRME is a fiber-optic fluorescence microscope that captures real-time images of proflavine-stained squamous epithelial nuclei (12,14). Nuclear staining is achieved using 0.01% wt/vol proflavine, a topical antiseptic with a long history of safe clinical use (15). Illumination is provided by a 460-nm LED directed through a narrow bandpass filter. Proflavine emission (peak wavelength at 515 nm) travels back through the probe and a long-pass filter before being focused on a monochrome CCD camera (16). The HRME has a lateral spatial resolution of 4.4 μm and a field of view of 790 μm. As shown in Figure 1, a live feed from the HRME was displayed on a tablet and a foot pedal was used by the clinician to capture images.
Figure 1.
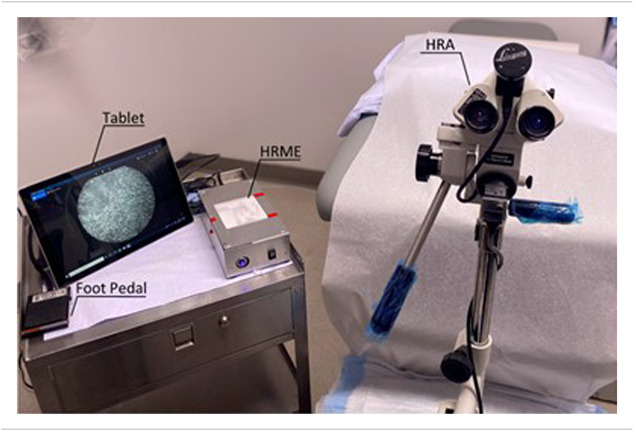
High-resolution microendoscopy (HRME) and high-resolution anoscopy (HRA) device at the point of care.
Development of image analysis algorithm using cervical HRME data
Images acquired with the HRME are analyzed using a multitask deep learning network (MTN) for automated image interpretation. In this study, we applied the MTN optimized for detection of cervical precancer without modification to analyze HRME images of anal tissue. The MTN was trained and validated for detection of cervical precancer using histologically correlated cervical HRME images from over 1,600 screen-positive women (13). External validation in an additional set of 508 cervical patients confirmed that the model's performance was on par with expert coloscopic impression (13).
Study participants
HRME images were collected from patients scheduled to undergo routine surveillance with HRA at the Icahn School of Medicine at Mount Sinai or affiliated clinics. Patient medical records were reviewed to identify potential subjects. PLWH were eligible for this study if they were 18 years or older and had either a history of biopsy-proven anal precancer (AIN 2+) or an abnormal cytology result of ASCUS or more severe within the past 2 years. Exclusion criteria encompassed individuals who could not provide written informed consent or if they had a platelet count less than 75,000 cells/mm3 and an absolute neutrophil count less than 1,000 cells/mm3, a known permanent or irreversible bleeding disorder, or allergy or previous reaction to proflavine.
This study was reviewed and approved by the Institutional Review Board (IRB) of Baylor College of Medicine and Affiliated Hospitals (Houston, Texas), which served as the IRB of record for the study (ID: H-44616). The study was registered at ClinicalTrials.gov (NCT04563754). All subjects provided written informed consent.
Study procedures
All diagnostic examinations and study procedures were performed by 1 expert anoscopist with more than 10 years of clinical experience. Participants underwent anal swab testing for HPV DNA (Roche Cobas), gonorrhea (Labcorp), and chlamydia (Labcorp) using an aliquot of the anal sample. Standard of care HRA was performed using a high-resolution colposcope (Leisegang OptiK Model 1 Colposcope). During the procedure, the clinician inserted an anoscope to open the anal cavity and used the colposcope to inspect for abnormalities indicative of disease such as abnormal vascular patterns, ulceration, mass effect, and mucosal friability (17). At the clinician's discretion, 5% acetic acid and Lugol’s iodine were used as contrast agents. Clinical impression of abnormal areas was documented as either HSIL or non-HSIL. The corresponding location of clinically abnormal areas was also documented (level and quadrant). After HRA imaging, the colposcope was set aside and the HRME imaging procedure begun. Proflavine was applied and HRME imaging was performed before diagnostic biopsy. HRME images were acquired from all clinically documented lesions and 1 clinically normal-appearing site. Subsequently, biopsies of all clinically abnormal areas were obtained and 1 biopsy from a normal area at the clinician's discretion. Histopathology was reported as benign, anal intraepithelial neoplasia (AIN) 1, condyloma acuminatum, AIN 2, AIN 3, or cancer. p16 immunohistochemistry was used to validate cases, with strong and diffuse positive p16 staining supporting AIN 2+ diagnoses.
Image analysis
An HRME image quality assessment was performed by 4 raters blinded to pathology. If a majority of raters agreed that more than 50% of the field of view was out of focus in a given image, it was deemed to fail quality control and was excluded from the analysis.
The MTN was used to analyze HRME images passing quality control (13). In brief, the MTN consists of a nuclear segmentation component and a diagnostic branch. The nuclear segmentation architecture resembles a U-Net, with an encoder and decoder joined through skip connections (18). The diagnostic branch originates from the last encoding block in the segmentation network and is composed of sequential encoding blocks that focus on diagnostic feature extraction. Two fully connected layers perform the final prediction, and a softmax function normalizes the output to a probability distribution of the likelihood of HSIL. To restrict analysis to the portion of the image containing the fiber-optic probe, HRME images were cropped and divided into 4 nonoverlapping quadrants; each quadrant was processed independently by the MTN. Processing the image as quadrants retains the full-image resolution, while minimizing the number of learnable parameters necessary. The final image score was computed by averaging the probability of anal HSIL given to each quadrant.
Evaluation metrics
Results were reported for each imaged site. Consensus diagnoses among 3 pathologists were used as the gold standard for this study. The sensitivity and specificity of HRA with respect to histopathology were calculated using clinical impression of HSIL as the threshold for a positive HRA. The receiver operating characteristic curve of the MTN prediction with respect to histopathology was computed. A high-sensitivity operating point with sensitivity equivalent to that of HRA impression was selected for the MTN. The sensitivity and specificity of the MTN and HRA impression were statistically compared using the McNemar test (19). Finally, the agreement between HRA impression and MTN was evaluated using Cohen’s κ (20).
RESULTS
A total of 77 PLWH were enrolled and underwent diagnostic HRA, HRME, and tissue biopsy. None of the patients approached for the study met the exclusion criteria. Of the 77 patients enrolled, 2 lacked biopsy-correlated HRME images and were excluded. An additional 22 sites were eliminated from the analysis: 1 site lacked an HRA impression and 21 sites failed HRME image quality control. Throughout the study, the fraction of images that passed quality control for each patient increased. In total, 17% of HRME images were removed from analysis because of poor image quality. However, in the last third of the study, 95% of HRME images passed quality control. No correlations were identified between clinical characteristics and HRME image quality. As a result, the final analysis set included data from 67 patients and 99 imaging/biopsy sites (see Supplementary Figure 1, Supplementary Digital Content 1, http://links.lww.com/CTG/A900). Table 1 summarizes the demographic characteristics of the patients in the analysis cohort. The prevalence of abnormal anal cytology (ASCUS or more severe) was 81%; 72% of participants tested positive for at least 1 type of high-risk HPV (Table 1). Table 2 details the histopathologic diagnoses for sites in the analysis set. No adverse events occurred throughout the study.
Table 1.
Demographic characteristics of patients in the analysis cohort
| Demographic characteristics | N = 67 n (%) |
| Race/ethnicity | |
| White | 19 (28) |
| Hispanic | 26 (39) |
| Black | 17 (25) |
| Other | 5 (7) |
| Age, yr | |
| <40 | 19 (28) |
| 40–49 | 19 (28) |
| ≥50 | 29 (43) |
| Sex | |
| Female | 3 (4) |
| Male | 64 (96) |
| Sexually transmitted diseases | |
| Negative | 59 (88) |
| Gonorrhea | 2 (3) |
| Chlamydia | 3 (4) |
| Not performed | 3 (4) |
| Cytology | |
| Benign | 4 (6) |
| ASCUS/LSIL | 44 (66) |
| LSIL-H | 2 (3) |
| HSIL | 8 (12) |
| Indeterminate | 8 (12) |
| Not performed | 1 (1) |
| Anal high-risk HPV | |
| Negative | 11 (16) |
| Positive | 48 (72) |
| Indeterminate | 1 (1) |
| Not performed | 7 (10) |
ASCUS, atypical squamous cells of undetermined significance; HPV, human papillomavirus; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion.
Table 2.
Histopathologic diagnoses for sites in the analysis set imaged with high-resolution microendoscopy
| Histopathologic diagnosis | No. of sites (%) |
| Benign | 29 (29) |
| AIN1 | 31 (31) |
| AIN2+ | 39 (39) |
| Total | 99 (100) |
AIN1, anal intraepithelial neoplasia grade 1; AIN2+, anal intraepithelial neoplasia grade 2 or more severe.
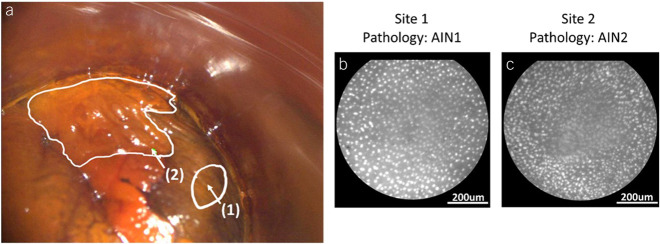
Figure 2 shows an example HRA image; sites where HRME images were obtained are labeled and corresponding HRME images are shown. The HRA image was captured after application of Lugol’s iodine and shows (i) normal-appearing anal mucosa with adequate Lugol’s uptake and (ii) a distinct posterior anal canal lesion with negative Lugol’s uptake (Figure 2a). White outlines indicate lesion boundaries based on clinical impression and arrows denote the sites imaged with HRME and biopsied. HRME images captured at sites 1 and 2 are shown in Figure 2b and Figure 2c, respectively. Visually, nuclei in site 1 appear more circular and evenly distributed than nuclei in site 2. Histopathologic diagnosis determined that site 1 was AIN 1, whereas site 2 was AIN 2. Both HRA impression and the MTN correctly classified the 2 sites. Site 1 received an HRA impression of non-HSIL and an AIN 2+ probability of 0.03 by the MTN (Figure 2b). Site 2 received an HRA impression of HSIL and an AIN 2+ probability of 0.33 by the MTN (Figure 2c). The AIN 2+ probability cutoff that best approximated the sensitivity of HRA impression was 0.212.
Figure 2.
(a) Clinical high-resolution anoscopy (HRA) image of the right lateral squamocolumnar junction in the anal canal with (1) normal-appearing anal mucosa and (2) a distinct posterior anal canal lesion. Outlines indicate lesion boundaries and arrows denote the sites imaged with high-resolution microendoscopy (HRME) and biopsied. (b) HRME image of site 1, which was classified as negative by the multitask deep learning network (MTN) and HRA impression, and was determined to be anal intraepithelial neoplasia grade 1 (AIN 1) by histopathology. (c) HRME image of site 2, which was classified as positive by the MTN and HRA impression, and was determined to be anal intraepithelial neoplasia grade 2 (AIN 2) by histopathology. The contrast of the HRA image was improved through dynamic range adjustment.
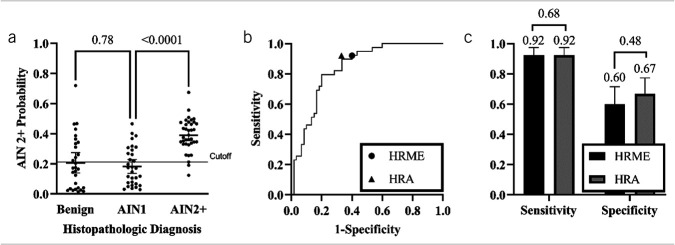
Figure 3a shows the AIN 2+ probability predicted by the MTN stratified by histopathologic diagnosis. The mean AIN 2+ probability for sites with a histologic diagnosis of benign, AIN 1, and AIN 2+ was 0.207, 0.182, and 0.380, respectively. We found no statistically significant difference between the scores of sites with a histologic diagnosis of benign or AIN 1 (P = 0.78). However, a significant difference was identified between sites with a histologic diagnosis of AIN 1 and AIN 2+ (P < 0.0001) and between sites diagnosed as benign/AIN 1 and AIN 2+ (P < 0.0001). Figure 3b depicts the receiver operating curve and operating point of the MTN alongside the sensitivity and specificity of HRA impression. The area under the receiver operating curve was 0.84. At the AIN 2+ probability cutoff of 0.212, the MTN achieved comparable performance to expert HRA impression, with a sensitivity of 0.92 (P = 0.68, 95% confidence interval [CI] 0.80–0.97) and specificity of 0.60 (P = 0.48, 95% CI 0.47–0.71) when using histopathology as the gold standard (Figure 3c). The agreement between the MTN and HRA impression stratified by histopathologic diagnosis can be seen in Table 3. The overall agreement between the MTN and HRA impression was 75.8% (κ = 0.501). The MTN and HRA impression disagreed in 24 sites of which 7 were benign (29%), 11 were AIN 1 (46%), and 6 were AIN 2+ (25%). Among these discordant sites, the MTN correctly classified 10 sites, including 3 AIN 2+ sites missed by HRA impression, while HRA impression correctly classified 14 sites, including 3 AIN 2+ sites missed by the MTN. The MTN and HRA agreed on the remaining 75 sites.
Figure 3.
Diagnostic performance of multitask deep learning network (MTN) and high-resolution anoscopy (HRA) impression using histopathology as the gold standard. (a) Per site anal intraepithelial neoplasia grade 2 or more severe (AIN 2+) probability stratified by histopathologic diagnosis. Histopathologic diagnosis of AIN 2+ was considered positive. Error bars indicate the mean and 95% confidence intervals (CIs), while the solid line across all classes denotes a retrospective cutoff to discriminate AIN 2+ lesions. (b) Receiver operating characteristic curve for MTN; operating points for MTN and HRA impression are indicated with symbols. (c) Sensitivity and specificity of MTN and HRA impression at corresponding operating points. Error bars indicate 95% CI.
Table 3.
Agreement between MTN and HRA impression stratified by histopathologic diagnosis
| HRA | MTN | Histopathologic diagnosis | ||
| Benign (n = 29) | AIN1 (n = 31) | AIN2+ (n = 39) | ||
| − | − | 15 | 14 | 0 |
| + | − | 2 | 5 | 3 |
| − | + | 5 | 6 | 3 |
| + | + | 7 | 6 | 33 |
Histopathologic diagnosis of AIN 2+ was considered positive.
AIN1, anal intraepithelial neoplasia grade 1; AIN2+, anal intraepithelial neoplasia grade 2 or more severe; HRA, high-resolution anoscopy; MTN, multitask deep learning network.
DISCUSSION
This study demonstrates that an automated, in vivo imaging-based system for anal precancer detection has comparable performance to expert HRA impression for diagnosis of histologic anal HSIL. There is an urgent need to improve the effectiveness of anal cancer screening and prevention for high-risk groups including PLWH. Current anal cancer screening capacity remains limited by the number of trained HRA practitioners and a high patient lost-to-follow-up rate between diagnosis and treatment. When used in combination with HRA, novel optical imaging systems to identify histologic AIN 2+, like the one presented in this work, have the potential to facilitate more selective biopsies and promote same-day treatment options by enabling real-time diagnosis.
In contrast to the in vivo diagnostic system described here, previous studies of machine learning diagnosis of anal precancer have focused on the analysis of ex vivo samples. Wentzensen et al. (21) developed a deep learning model for AIN 2+ detection from cytology slides that achieved an AUC of 0.77 in a test set with 299 patients. Similar to our approach to training the MTN, Wentzensen et al. used cervical cytology imaging data to assist in training their model. Using cervical data is a powerful strategy to overcome the limited availability of anal data, given the limited availability of anal cytology/biopsy results. Although the structure of the anal cavity presents unique challenges for in vivo imaging, the similarities between the progression of anal and cervical precancers are well documented (22). Future work could benefit from continued use of cervical data to aid the development of computer-aided diagnostic systems for anal precancer detection.
One limitation of this system is that placement of the HRME probe depends on HRA guidance and therefore on appropriate identification of a possible anal precancerous lesion by the clinician. It will consequently not alleviate the need for procedural acumen, and proficiency in HRA techniques will remain a prerequisite for successful HRME or tissue diagnosis. Advancements in the development of algorithms to localize suspicious lesions in HRA images, like those in the field of colposcopy image analysis, may aid HRME probe placement (23,24). Furthermore, the HRME could be used as a triaging tool which may allow for a potential “see and treat” approach, eliminating the need for additional treatment-related visits. In addition, because of the HRME's small field of view and its susceptibility to motion artifacts, only a small area of the anal anatomy can be surveyed within the limited time frame of a clinical visit. Thus, the clinician operating the instrument must carefully decide which areas to prioritize for HRME imaging. This decision will affect the performance of the HRME in anal precancer detection. To overcome these limitations, recent developments in HRME instrumentation have sought to reduce motion blur by using a higher frame rate camera and increasing the HRME field of view by mosaicking sequentially acquired frames (25). The new instrumentation coupled with rapid real-time scoring would allow clinicians to survey more extensive areas of interest at a higher speed.
This study does not assess the tradeoff of training and validating in our model using cervical instead of anal HRME images. Based on our previous assessment of the number of samples required to train the MTN for cervical precancer detection, we can infer that we currently do not have sufficient samples to train and validate the MTN exclusively on anal HRME data (13). However, future work could explore using anal HRME data to refine the MTN and improve its diagnostic performance once more data become available.
This pilot study shows that high-resolution microscopy combined with deep learning image interpretation can achieve a sensitivity and specificity equivalent to expert HRA impression which could decrease the need for pathology-based diagnoses by providing point-of-care diagnostic capabilities. These results encourage a follow-up study with higher statistical power to compare the HRME performance with the expert impression of multiple anoscopists.
CONFLICTS OF INTEREST
Guarantor of the article: Elizabeth Chiao, MD, MPH.
Specific author contributions: D.B.: formal analysis, software, data curation, methodology, visualization, writing—original draft, writing—review & editing. A.K.: investigation, resources, data curation, writing—original draft, visualization, writing—review & editing. J.C.: supervision, writing—review & editing, project administration. T.M.: data curation, visualization, project administration, writing—review & editing. R.S.: supervision, writing—review & editing. Y.L.: investigation, resources, writing—review & editing, funding. K.S.: supervision, project administration, writing—review & editing, funding acqusition. R.R.-K. and S.A.: supervision, conceptualization, writing—review & editing, funding acqusition. M.G.: supervision, conceptualization, investigation, resources, writing—review & editing, funding acqusition. E.C.: supervision, conceptualization, writing—review & editing, funding acquisition.
Financial support: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers: R01CA232890, R01CA251911. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Potential competing interests: S.A. is a gastric cancer screening consultant for Roche. All other authors declare no competing interests.
Citation diversity: Recent work in several fields of science has identified a bias in citation practices such that papers from women and other minority scholars are undercited relative to the number of papers in the field (26–29). We recognize this bias and have worked diligently to ensure that we are referencing appropriate papers with fair gender and racial author inclusion.
Data availability statement: The data that support the findings of this study are available from the corresponding author on reasonable request through a data-sharing agreement that provides for (i) a commitment to securing the data only for research purposes and not to identify any individual participant; (ii) a commitment to securing the data using appropriate computer technology; and (iii) a commitment to destroying or returning the data after analyses are completed.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank all the participants who volunteered to participate in the study and Courtney Chan for her contributions to the study.
Study Highlights.
WHAT IS KNOWN
✓ People living with HIV are susceptible to coinfection with the human papillomavirus, linked to 90% of anal cancer cases.
✓ Early detection and treatment of anal precancer reduces the risk of progression to cancer.
✓ There is a scarcity of professionals proficient in anal precancer diagnosis.
✓ There is a high patient lost-to-follow-up rate after high-resolution anoscopy and biopsy.
WHAT IS NEW HERE
✓ In vivo imaging with high-resolution microendoscopy reveals changes in nuclear morphology associated with high-grade anal precancer.
✓ A deep learning model trained to detect cervical precancer was used unmodified to interpret images of anal tissue.
✓ The deep learning–enabled image interpretation had a similar performance for detection of anal intraepithelial neoplasia grade 2 or more severe (AIN 2+) as expert anoscopy which could support a “see and treat” approach for AIN 2+ by providing a point-of-care diagnosis at the time of anoscopy.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A900
Contributor Information
Alex Kortum, Email: ask5@rice.edu.
Jennifer Carns, Email: jcarns@rice.edu.
Tinaye Mutetwa, Email: tinaye.mutetwa@icahn.mssm.edu.
Richard Schwarz, Email: schwarzr@rice.edu.
Yuxin Liu, Email: Yuxin.liu@mountsinai.org.
Keith Sigel, Email: keith.sigel@mssm.edu.
Rebecca Richards-Kortum, Email: rkortum@rice.edu.
Sharmila Anandasabapathy, Email: Anandasa@bcm.edu.
Michael Gaisa, Email: michael.gaisa@mssm.edu.
Elizabeth Chiao, Email: eychiao@mdanderson.org.
REFERENCES
- 1.Colón-López V, Shiels MS, Machin M, et al. Anal cancer risk among people with HIV infection in the United States. J Clin Oncol 2018;36(1):68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Souza G, Wiley DJ, Li X, et al. Incidence and epidemiology of anal cancer in the multicenter AIDS cohort study. J Acquir Immune Defic Syndr 2008;48(4):491–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Martel C, Plummer M, Vignat J, et al. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer 2017;141(4):664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palefsky JM, Giuliano AR, Goldstone S, et al. HPV vaccine against anal HPV infection and anal intraepithelial neoplasia. N Engl J Med 2011;365(17):1576–85. [DOI] [PubMed] [Google Scholar]
- 5.Albuquerque A, Rios E, Schmitt F. Recommendations favoring anal cytology as a method for anal cancer screening: A systematic review. Cancers (Basel) 2019;11(12):1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke MA, Wentzensen N. Strategies for screening and early detection of anal cancers: A narrative and systematic review and meta-analysis of cytology, HPV testing, and other biomarkers. Cancer Cytopathol 2018;126(7):447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richel O, Prins JM, de Vries HJ. Screening for anal cancer precursors: What is the learning curve for high-resolution anoscopy? AIDS 2014;28(9):1376–7. [DOI] [PubMed] [Google Scholar]
- 8.Lee JY, Lensing SY, Berry‐Lawhorn JM, et al. Design of the ANal Cancer/HSIL Outcomes Research study (ANCHOR study): A randomized study to prevent anal cancer among persons living with HIV. Contemporary Clinical Trials 2022;113:106679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silvera R, Martinson T, Gaisa MM, et al. The other side of screening: Predictors of treatment and follow-up for anal precancers in a large health system. AIDS 2021;35(13):2157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varela BR, Perl D, Zhou E, et al. Mo1645 High-resolution microendoscopy (HRME) in the detection of anal intraepithelial neoplasia: Assessment of accuracy and interobserver variability. Gastrointest Endosc 2013;77(5):AB456–AB457. [Google Scholar]
- 11.Grant BD, Schwarz RA, Quang T, et al. High-resolution microendoscope for the detection of cervical neoplasia. Methods Mol Biol 2015;1256:421–34. [DOI] [PubMed] [Google Scholar]
- 12.Yang EC, Vohra IS, Badaoui H, et al. Development of an integrated multimodal optical imaging system with real-time image analysis for the evaluation of oral premalignant lesions. J Biomed Opt 2019;24(2):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenes D, Barberan CJ, Hunt B, et al. Multi-task network for automated analysis of high-resolution endomicroscopy images to detect cervical precancer and cancer. Comput Med Imaging Graph 2022;97:102052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quang T, Schwarz RA, Dawsey SM, et al. A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointest Endosc 2016;84(5):834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pantano N, Hunt B, Schwarz RA, et al. Is proflavine exposure associated with disease progression in women with cervical dysplasia? A brief report. Photochem Photobiol 2018;94(6):1308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prieto SP, Powless AJ, Boice JW, et al. Proflavine hemisulfate as a fluorescent contrast agent for point-of-care cytology. PLoS One 2015;10(5):e0125598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jay N, Michael Berry J, Miaskowski C, et al. Colposcopic characteristics and Lugol's staining differentiate anal high-grade and low-grade squamous intraepithelial lesions during high resolution anoscopy. Papillomavirus Res 2015;1:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, 2015, pp 234–41. [Google Scholar]
- 19.McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947;12(2):153–7. [DOI] [PubMed] [Google Scholar]
- 20.Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas 1960;20(1):37–46. [Google Scholar]
- 21.Wentzensen N, Lahrmann B, Clarke MA, et al. Accuracy and efficiency of deep-learning-based automation of dual stain cytology in cervical cancer screening. J Natl Cancer Inst 2021;113(1):72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darragh TM, Winkler B. Anal cancer and cervical cancer screening: Key differences. Cancer Cytopathol 2011;119(1):5–19. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Chen J, Xue P, et al. Computer-aided cervical cancer diagnosis using time-lapsed colposcopic images. IEEE Trans Med Imaging 2020;39(11):3403–15. [DOI] [PubMed] [Google Scholar]
- 24.Hu L, Bell D, Antani S, et al. An observational study of deep learning and automated evaluation of cervical images for cancer screening. J Natl Cancer Inst 2019;111(9):923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt B, Coole J, Brenes D, et al. High frame rate video mosaicking microendoscope to image large regions of intact tissue with subcellular resolution. Biomed Opt Express 2021;12(5):2800–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caplar N, Tacchella S, Birrer S. Quantitative evaluation of gender bias in astronomical publications from citation counts. Nat Astron 2017;1(6):141. [Google Scholar]
- 27.Dworkin JD, Linn KA, Teich EG, et al. The extent and drivers of gender imbalance in neuroscience reference lists. Nat Neurosci 2020;23(8):918–26. [DOI] [PubMed] [Google Scholar]
- 28.Maliniak D, Powers R, Walter BF. The gender citation gap in international relations. Int Organ 2013;67(4):889–922. [Google Scholar]
- 29.Dion ML, Sumner JL, Mitchell SM. Gendered citation patterns across political science and social science methodology fields. Polit Anal 2018;26(3):312–27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.