Abstract
In recent years, there have been calls for implementation of “epitope matching” in deceased-donor organ allocation policies (later changed to “eplet matching”). Emerging data indeed support the use of molecular mismatch load analysis in specific patient groups, with the objective of posttransplant stratification into different treatment arms. For this purpose, the expectation is to statistically categorize patients as low- or high-immune-risk. Importantly, these patients will continue to be monitored‚ and their risk category, as well as their management, can be adjusted according to on-going findings. However, when discussing deceased donor organ allocation and matching algorithms, where the decision is not modifiable and has lasting impact on outcomes, the situation is fundamentally different. The goal of changing allocation schemes is to achieve the best possible HLA compatibility between donor and recipient. Immunologically speaking, this is a very different objective. For this purpose, the specific interplay of immunogenicity between the donor and any potential recipient must be understood. In seeking compatibility, the aim is not to redefine matching but to identify those mismatches that are “permissible” or‚ in other words, less immunogenic. In our eagerness to improve transplant outcome, unfortunately, we have conflated the hype with the hope. Terminology is used improperly, and new terms are created in the process with no sufficient support. Here, we call for a cautious evaluation of baseline assumptions and a critical review of the evidence to minimize unintended consequences.
Transplantation networks around the world have implemented specific deceased-donor organ allocation policies to safeguard distribution of this scarce resource. The complexity of these policies stems from the need to achieve a delicate balance between competing needs to accomplish the best utilization of these organs while maintaining equity, practicality, efficiency, and quality of posttransplant results. A critical component in determining prolonged graft survival is the degree of HLA matching between the transplant recipient and their donor. However, the weight given for HLA matching in the final allocation algorithm was empirically compromised (to a different degree in different allocation schemes) because of competing priorities overall. Moreover, the introduction of molecular typing techniques emphasized the vast polymorphism of the HLA system and underscored the lower likelihood of finding a fully HLA-matched donor. This was the landscape into which HLAMatchmaker,1 an algorithm developed by Rene Duquesnoy, was conceived. Importantly, the initial goal of this software was to overcome the extensive workload required to accurately identify the full range of HLA antibodies exhibited by highly sensitized patients‚ in other words, to provide a tool that can predict those handful of crossmatch-negative potential donors without the need to perform multiple futile physical crossmatches.2 This is, in fact, the basis for the Eurotransplant Acceptable Mismatch Program.3 The excitement about potential use of this software to support donor/recipient matching for all waitlist patients came about only later.
Early studies using the HLAMatchmaker software demonstrated statistical correlation between the number of amino acid “triplet” differences (the initial output of the Matchmaker) and the likelihood of patients to develop HLA donor specific antibodies (DSAs)4 but not cytotoxic T-cell reactivity.5 Soon afterward‚ HLAMatchmaker was used to retrospectively explain generation of de novo HLA DSAs in patients that lost their graft because of rejection.6 In response to critical review, demonstrating that antibody recognition sites do not necessarily include amino acids adjacent linearly and to account for other observations not explained by the original “triplet” concept, HLAMatchmaker software was modified to output “eplets” rather than triplets.7 That led the road for Duquesnoy to describe the HLAMatchmaker software as a structurally based matching program.8 The baseline assumption was that polymorphic amino acids present within the patient’s HLA antigens cannot contribute to immune reactivity. With emphasis on advancement in HLA molecular typing, the field was ripe to explore new approaches to explain HLA antibody reactivity, and additional software was introduced soon after including the Electrostatic Mismatch Score, PIERCHE, and EMMA (a summary and comparison between the different methods can be found in Saleem et al9).
In a leap of faith, supported by the landmark publication by Wiebe and colleagues10 in which the term “epitope matching” was used, our field embraced the application of HLAMatchmaker and other molecular mismatch load (MML) analysis software as an approach to assess HLA compatibility between transplant recipients and their donors. At this point‚ there are 2 schools of thoughts proposing how to utilize MML analysis:
use for “epitope matching,” thus use in deceased-donor allocation (and kidney paired donation) algorithms;
use for risk stratification posttransplantation.
The information and requirements to support either of these approaches are different. Our goal is to provide a critical review of the HLAMatchmaker software. We further provide an in-depth scrutiny of its utilization, especially in publications advocating for its use in allocation matching algorithms. Most of the comments are applicable to other MML analysis tools. We believe these issues must be rigorously considered before implementation of changes to current practices.
HLAMATCHMAKER, THE SOFTWARE
Conflating Epitopes and Eplets
The very brief chronicle provided above details the transition from using the term triplet, in describing groups of 3 amino acid sequences, to the assumption we can relate to these few amino acids as epitopes. However, there are no experimental data to support that an eplet indeed equals an epitope and that the 2 concepts are interchangeable. This is a problematic misconception with serious potential ramifications.
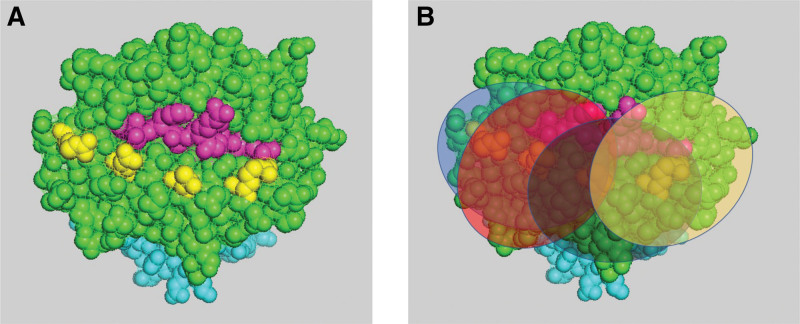
Studies to define epitopes originally flourished in investigating T-cell epitopes, especially as it relates to vaccine development and drug design. The size of a T-cell epitope (the area recognized by the T-cell receptor [TCR]) is similar in size to that of the B-cell epitope, recognized by the B-cell receptor/antibody. Multiple estimations were reported based on crystallography studies, mostly stating an average surface area of 600 to 900Å‚2,11 much larger than the 3 to 4 amino acids represented by an eplet. Indeed, a TCR specificity is determined based on a small peptide (roughly 9–15 amino acid long), that is, presented in the context of self-HLA molecules. Because the background of self-HLA molecules is constant within the individual, the field took to using a shorthand in referring to the polymorphic peptide as the epitope, although the footprint of the TCR is much larger. Considering B-cell epitopes, if the target is a viral or bacterial protein that differs from self, focusing on a small portion of the epitope may be inconsequential. However, when the target is another HLA molecule that bears over 90% sequence homology with self but likely has multiple mismatches in a small area, assuming each eplet represents a full epitope can lead to misinterpretation. Furthermore, Figure 1 demonstrates the significant overlap that will happen for some “epitopes” if indeed each eplet represents a different epitope. Consequently, access of the cognate antibodies to some of these epitopes will be blocked. Thus, whereas the assumption that each eplet is a small representative unit of an epitope is very appealing, it does not make immunologic sense in the context of HLA molecules.
FIGURE 1.
The difference between an eplet and an epitope: (A) 4 eplets (yellow) are depicted on the background of an HLA class I molecule (α chain—green, β2 microglobulin—light blue, peptide—pink); (B) a rough depiction of the antibody footprint/epitope, assuming each eplet is a “core” for an epitope, is represented by a circle of a different color (purple, orange, blue, and yellow). This schematic representation clearly demonstrates that the footprint of the antibody may include additional eplets, beyond the “core” eplet. It further demonstrates that there is an overlap between the area of these epitopes such that‚ if 1 antibody binds to its target, other antibodies will be blocked from accessing their cognate epitope and therefore will likely not bind. Simple counting such as the molecular mismatch load approach will result in 4 eplets; however, only 1 or 2 antibodies can bind to this HLA molecule. Thus, counting the number of eplet mismatches does not make immunologic sense (although it does make statistical sense).
To address these issues, new terminology—structural epitopes versus functional epitopes—was introduced.12 The term functional epitope had been in use previously to highlight the portion of the full/structural epitope that likely affects binding affinity or rate of association/dissociation between an epitope and an antibody. Importantly, though, determination of which amino acids are more critical, and therefore potentially more functional, was experimentally determined. For example, combinatorial alanine-scanning strategy was used to determine relative contribution of amino acids in multiple epitopes, as summarized in Weiss et al.13 Other studies used single, multiple, and combinatorial mutagenesis to allow comprehensive scanning of target antigens.14 There are no empirical data to substantiate the claim that eplets, as defined by HLAMatchmaker‚ are functional epitopes.
An additional modification to the HLAMatchmaker software was the “self/nonself-eplets phenomenon,”15 specifically that “epitopes can be defined by eplet pairs, involving 1 nonself-eplet and a self-eplet between the immunizing antigen and the antibody producer.” It was stated that “the nonself-self paradigm provides a new insight of HLA epitope immunogenicity and may explain why sensitized patients have antibodies to a restricted number of mismatched epitopes.” The concept of self-nonself–recognition was previously introduced in studies of molecular mimicry, in attempts to explain nonimmunogenicity of critical regulatory foreign proteins,16 for example, studies of immune responses to malignant melanoma. Although the original nonimmunogenicity hypothesis was based on analyzing sequence homology, the concept was not accepted before presentation of experimental evidence.17 There are no empirical data to support this nonimmunogenicity hypothesis in HLA alloimmune responses.
Antibody-verified and Antibody-nonverified Eplets
As has been alluded to above, comparison between any 2 alleles within an HLA locus is likely to show >90% homology. The polymorphic amino acids tend to cluster in specific areas of the molecule, although some are scattered throughout the gene.18 The original list of triplets defined in HLAMatchmaker was compiled by aligning the sequences of all HLA alleles and identifying the polymorphic amino acids, thus providing an exhaustive list of all “theoretically defined triplets” (later referred to as eplets). With the utilization of HLAMatchmaker by multiple investigators, however, it became evident that not all eplets, mismatches between 2 individuals are necessarily associated with antibody production/recognition. As a result, a new database was created in conjunction with the 16th International Histocompatibility and Immunogenicity workshop.19 This new compilation, the Epitope Registry,20 made a distinction between antibody verified (AbVer) eplets and non-antibody verified (non-AbVer) eplets. What is less transparent is the wide definition used for “antibody verification.” In a small fraction of cases, verification is based on extensive adsorption elution studies. In many other cases, however, “verification” relies on serological reactivity patterns observed 20 plus y ago, when serology testing was common (but the breadth of the polymorphism in the HLA system, and molecular typing were not). Some “verification” used murine monoclonal antibodies that do not necessarily represent human immunogenicity or are based on analyzing single antigen bead (SAB) reactivity pattern from polyclonal sera. These reactivity patterns were often obtained before the implementation of measures to remove inhibition (such that under-representation of antibody presence may have affected the interpretation of these results, per STAR recommendations).21 Many of the AbVer eplets were recorded using sera from multiparous women rather than from transplant recipients who lost their graft due to AMR. Importantly, a substantial portion of the verification evidence has not been published in peer-reviewed literature and thus did not undergo critical evaluation.
Review of the literature demonstrates that some studies used all potential eplets for their analysis, whereas other publications used only the AbVer eplets, with the assumption that those provide more rigorous analysis. To review the evidence of alloreactivity and immunogenicity of those AbVers, a comprehensive evaluation was put forth by the Leiden group.22,23 The investigators used HLA-specific human monoclonal antibodies and lymphocytotoxicity assays, as well as a newer version of an in silico analysis approach (HLA-EMMA).24 These studies demonstrate that not all AbVer eplets are in fact associated with immune responses, documenting that there is a lack of empirical data to identify immunogenic epitopes.
Determining the Donor and the Recipient HLA Milieu
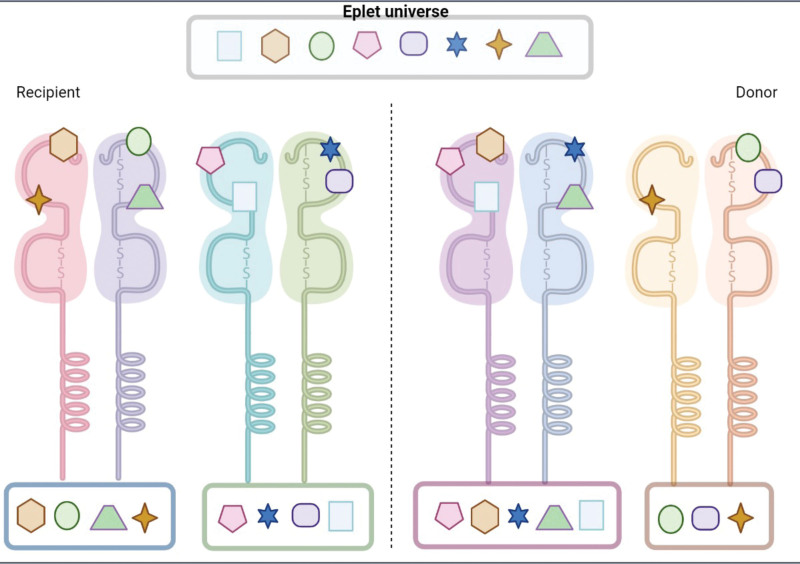
HLAMatchmaker considers the patient as the “antibody producer” and the donor as the “stimulator.” As such, the software combines the 2 alleles of each locus into 1 “eplet universe.” The software then compares the patient’s “eplet universe” with the donor’s “eplet universe” and outputs a calculation of the eplet mismatch load. For example, consider a patient typed as HLA-DQ5 and HLA-DQ7 and a donor typed as HLA-DQ6 and HLA-DQ8. HLAMatchmaker inputs all eplets found in the amino acid sequences of HLA-DQ5 and -DQ7, combined, into 1 bucket. Similarly, the donor’s eplets found in the HLA-DQ6 and -DQ8 are combined into a second bucket. Finally, the eplets from the donor bucket are subtracted from those of the patient’s bucket, outputting the final eplet mismatch load. Creating these eplet universes leads to the assumption that a donor and a recipient can be fully mismatched at the antigen level but matched at the eplet level (Figure 2).
FIGURE 2.
The eplet universe: The HLAMatchmaker software identifies eplets from both alleles of the recipient or the donor (within each locus), and combines them into 1 bucket representing the “universe of eplets” of either the recipient or the donor. Each eplet is represented by a different symbol. For ease of reference, eplets from each allele are presented within a box below the allele from which they are derived. Thus, the recipient has 2 alleles with 4 eplets for each of them (blue and green boxes)‚ and the donor has 1 allele with 5 eplets (purple box) and the other allele with 3 eplets (brown box). HLAMatchmaker combines the recipient or the donor “boxes” into 1 universe; that summation is presented within the gray box on top. Although the recipient and donor have completely different alleles, this manipulation generates the illusion that their eplet universes are identical.
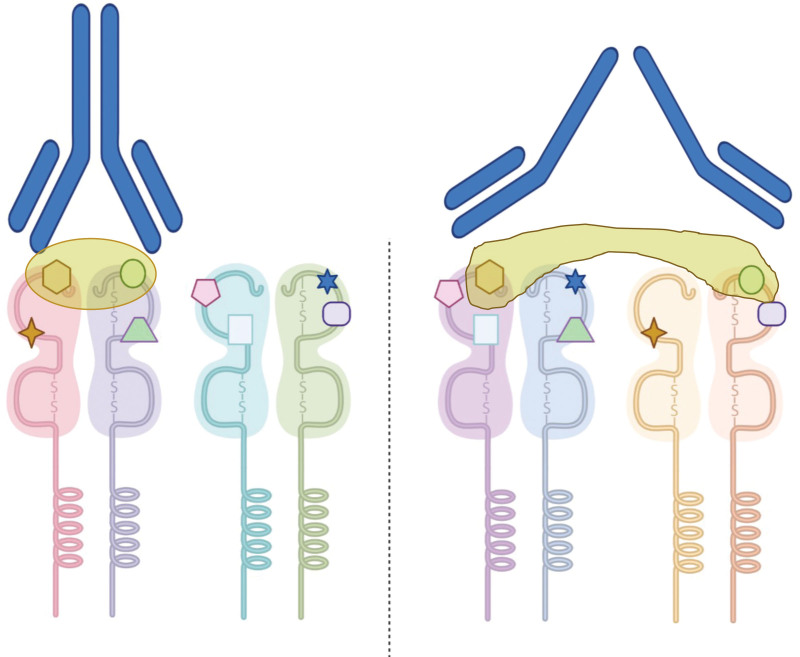
Immunologically speaking, however, there is a profound mistake in this concept. There are no immunologic systems, neither the TCR, nor the B-cell receptor (antibody), that combine 2 potentially foreign antigens into 1 universe. This statement stands true when discussing viral or bacterial immunogens and similarly when discussing HLA alloantigens (Figure 3). Furthermore, untangling the 3-dimensional polymorphic structure of the HLA molecule into small independent building blocks and combining building blocks from 2 separate alleles into 1 “universe” carry the risk of facing cis/trans ambiguities. In fact, this is a well-known difficulty in resolving some molecular HLA typing, such as sequence-specific oligo nucleotide probe and sequence-based typing. Both methods amplify DNA from both alleles of each locus simultaneously, virtually generating a “universe” of genetic amplicons. In HLA-sequence–specific oligo nucleotide probe, this universe is probed for recognition of small, independent, (epletlike) nucleotide sequences, leading to a unique pattern of recognition. Unfortunately, it is not uncommon to receive 2 or more potential patterns identifying different pairs of alleles (eg, DRB1*11:01/DRB1*13:01 or DRB1*11:04/DRB1*13:02). These ambiguities must be resolved by additional testing to determine the correct typing.25 By not considering the individual alleles, HLAMatchmaker’s “eplet universe” creates a similar environment of potential ambiguities.
FIGURE 3.
Can an antibody recognize similar eplets in different allele configurations? Similar to Figure 2, the recipient and the donor have the same “eplet universe.” The brown circle on the left represents the footprint of the antibody = epitope. To support the assumption that an eplet universe can immunologically represent an individual regardless of the context in which the eplets are organized, one must assume that an antibody can recognize disparate HLA molecules at the same time. This abnormality is schematically depicted in the “broken antibody” on the right.
To overcome this particular hurdle, MML analysis must be performed against each of the donor alleles (rather than a donor eplet universe), as we have previously demonstrated26 and as was recently published by the Manitoba group.27 Most other studies neglected to resolve or even acknowledge the problematic use of the “eplet universe” concept, jeopardizing the validity of their reported results. It is yet to be determined whether combining the patients’ alleles into 1 universe has immunologic validity, or whether a more sophisticated analysis must be used.
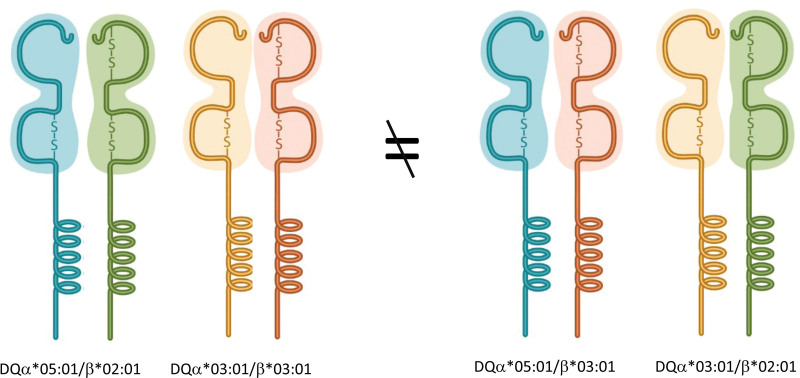
When considering HLA-DQ alleles, there is an additional layer of complexity. HLA-DQ antigens are composed of 2 chains (α and β) that are each encoded by a separate gene. First, most studies include eplet information only for the DQβ chain. Since the DQα chain is also polymorphic, this means that the analysis presented in these studies lacks information about half of the HLA-DQ polymorphism. Importantly, because HLA-DQ DSAs are the most frequent and the most detrimental to transplant outcome,28-30 this omission may lead to major flaws in data interpretation. Furthermore, analyzing the DQα information independent of its DQβ chain counterpart can mask real differences between patient and donor. For example, a patient typed as HLA-DQα*05:01/β*02:01 and DQα*03:01/β*03:01 will show no eplet mismatches when compared with a donor typed as HLA-DQα*05:01/β*03:01 and DQα*03:01/β*02:01, using the “eplet universe” approach. We have previously shown that these molecules have different physiochemical and electrostatic properties31 (Figure 4 is a schematic representation).
FIGURE 4.
HLA-DQ molecules are composed of 2 polymorphic chains, an α and a β chain. The specific pairing between these chains determines the specificity of the molecule. In this example, the 2 α and the 2 βchains are identical in both individuals. However, they are combined to generate different products that will have different physiochemical properties. Therefore, they are likely to elicit recognition of different antibodies (or T-cell receptors).
STUDIES USING HLAMATCHMAKER
Different Versions Use Somewhat Different Definitions and Provide Different Output
Since the introduction of the HLAMatchmaker software into transplantation literature in 2002, there have been over 1700 publications using the keywords HLA epitope, eplet, mismatch, and transplantation. During these 20 y, there have been significant changes to the software output. Those include (1) a change from using triplets to eplets; (2) a change from including only contiguous/linear sequences to including also noncontiguous sequences; (3) the introduction of the self/nonself theory and so on. Different versions of HLAMatchmaker were also introduced as an analysis companion for commercial solid phase antibody testing kits, leading to discrepancies depending on the version used for analysis. For example, the number of unique eplets in version 2.1 of the software is larger than the number of unique eplets in the current version.32 Thus, investigation of the same patient cohort, using 2 versions of the software, may yield different results. This raises the question of how these eplets were determined and what criteria must be fulfilled in order to assign “eplet” designation. This issue must be resolved, and MML computational algorithm software must undergo curation and validation, before implementation into any clinical application.
An example of the potential impact of such changes is seen in 2 publications from Wiebe et al. In their earlier publication,10 the investigators determined that an eplet mismatch load of 10 for DR and 17 for DQ defines the threshold for patients at higher risk to develop de novo HLA-DR or HLA-DQ DSAs. In a follow-up publication, however, where the next version of the HLA Matchmaker was used, the threshold was changed to 11 eplets for both HLA-DR and HLA-DQ.33 Although the authors of these publications promote the use of eplet mismatch load for risk stratification posttransplantation, this observation is applicable, and even more problematic, for utilization of MML analysis in organ allocation algorithms.
Large Dataset Cohorts With Long-term Follow-up Suffer From Significant Missing Information
Missing HLA Typing Information and the Use of Imputation
Assignment of eplets, or any other MML approach, requires HLA typing at the high resolution (2-field typing). Unfortunately, this level of typing information is currently available only in some small, single-center cohorts. These studies often suffer from lack of ethnic diversity and‚ per definition, from small numbers. Large cohort datasets suffer from lack of high-resolution typing data and‚ even more so, complete lack of information regarding typing at certain loci. Engen et al34 and Senev et al35 demonstrated the pitfalls of using imputation, especially for the non-Caucasian (a priori disadvantaged) populations. Indeed, data from the National Marrow Donor Program showed that having low-resolution information for all loci (HLA-A, -B, -C, -DR, -DQ) in combination with reliable information of the individual’s ethnic background may aid in predicting high-resolution typing information with reasonable accuracy (especially for Caucasian patients). However, data obtained, for example, from the Scientific Registry of Transplant Recipients (SRTR) have neither of these qualifiers—typing is available at low resolution, and many patients and donors completely lack HLA-DQ (and often HLA-C) typing. Information entered before 2010 is based on serological typing, which is significantly less accurate. Moreover, the ethnic background definition in the SRTR is self-declared and is restricted to the 4 major ethnic groups. The consequences of inaccuracies introduced via imputation are even more alarming in countries where low- resolution typing remains the standard method of testing and the population is mostly non-Caucasian. Not only the conversion to MML is affected; interpretation of routine DSA testing using the Luminex SAB assay can be impacted. In fact, it was noted that some of the common alleles in non-Caucasian populations are not even represented as part of the SAB panel.
Imputation of high-resolution HLA typing is presented by some investigators as an unavoidable necessity when collating a dataset of sufficient size and with adequate duration of follow-up to address the question of the clinical relevance of “epitope matching.” This is a puzzling statement. Is recognition of a methodological limitation sufficient to avoid or eliminate potential detrimental implications of misanalysing data? In a recent report from the Genome Canada group, the investigators used a large dataset from the SRTR including over 118 000 patients transplanted between the years 2000 and 2015.36 Using the maximum-likelihood estimation method for imputation, the investigators were not able to yield imputed allele level typing for 15% of the cohort. This is a substantial portion of the cohort, raising the question of why this sophisticated imputation method failed to generate information for so many individuals. Was a particular ethnic group affected? And knowing this level of failure, how does that support reliability of the data? What level of error can be considered acceptable? Interestingly, some of the investigators of this study published an article the following year describing multiple limitations of HLAMatchmaker and the imputation approaches.37
Other Missing Data
Studies using large cohort data often do so to benefit from long-term follow-up data on transplant outcome. An inclusion criterion, in many of these studies, is absence of any HLA antibodies (cPRA of 0%) at the time of transplant. The goal is to avoid inclusion of patients that may had preformed antibodies at the time of transplantation. However, achieving the desired length of follow-up means that many of those patients were tested using cytotoxic, less sensitive antibody evaluation and crossmatch assays. In other words, it is conceivable that many of these patients would have tested positive using solid phase assays. The STAR working group21 recommends the utilization MML tools specifically for patients that are alloimmune naive to their donors. The likelihood of having low levels of preformed DSAs in these long-term cohorts is yet another significant limitation that may affect reliability of the results. Large cohort studies also suffer from limitations such as granular definition of graft loss, adequacy of immunosuppression, additional immune assaults, and so on. Anecdotally, it will be fascinating to see what correlations are found if the investigators simply use the imputed high-resolution HLA typing information (alongside the other presumed missing data) instead of MML approaches.
Statistical Analysis—the Use of Bounded Rationality
To demonstrate a utility for “epitope matching” to support improved transplant outcome, investigators compared between patients with and without the tested outcome—for example, generation of de novo DSAs. This appears to be a simple but solid statistical evaluation. The problem lies in the inclusion criteria for the control group. For example, should a patient who received an HLA-matched organ be included in these studies? Is it reasonable to assume that a patient with an HLA-DR– or an HLA-DQ– matched transplant is capable of generating antibodies to the donor’s HLA-DR or -DQ, respectively? Reviewing the literature, most studies include patients with zero MML in certain loci as part of their control groups. Indeed, it is desirable to have a patient cohort composed of consecutive cases with minimal exclusions. However, a simple analysis performed at our center demonstrates a significant reduction of the P depending on the characteristics of the control cohort. If the control cohort included all patients who did not develop DSAs, the P was highly significant. If the control cohort included only those patients who were mismatched with their donor, the P was much lower (data not shown). This limitation is well described in the field of data and decision science and is known as the use of bounded rationality.38 We believe that including patients who a priori are not expected to develop DSAs as part of the control cohort can lead to misinterpretation of the data.
Additional statistical creativity is observed in other studies where MML analysis from multiple loci is summed into a combined score. This score is then compared with DSAs generated at a particular locus or at any of the loci‚ for example, adding eplet mismatches in all HLA loci, correlating with generation of de novo DSAs against the donor HLA-DQ alleles.
ASSUMPTIONS MADE IN DATA INTERPRETATION TO SUPPORT THE USE OF MML IN ORGAN ALLOCATION POLICIES
Epitope Matching Will Provide Easier Matching Than Antigen/Allele Level Matching
A common argument for the use for epitope or eplet “matching” claims that the frequency of individual alleles in the population is smaller than the frequency of the different eplets, and therefore, it will be easier to find a match based on a polymorphism that is more prevalent in the population. For example, Tran et al39 studied 2000 subjects from the BC transplant program who had high-resolution typing for all HLA loci. Of those, 154 were excluded because their alleles are not present as part of the HLAMatchmaker database. The remaining subjects included 1049 patients with kidney failure and 797 kidney donors. Within this population, the investigators identified a total of 361 unique HLA alleles and 150 unique eplets. As a result, they reported that “eplets are more common and uniformly distributed between donors and recipients than the respective HLA isoforms.” Although these are indeed the facts, one must assume that each eplet is recognized as an independent unit. We have demonstrated above that this is not the case. Each unique HLA allele is composed of several eplets, which are presented as a combined unit for antibodies or TCR scrutiny (refer to Figure 2).
In fact, the authors continued to provide data demonstrating the futile exercise of performing “epitope matching.” Specifically, analyzing genotype distribution, they found 1017 unique genotypes among patients and 756 unique genotypes among donors and only 1.5% of the genotypes occurred in both groups. They further calculated “epitypes” (a new terminology, probably referring to the identity of eplets within a genotype = 2 per subject).39 Similar to the genotype data, they found 1010 unique epitypes among patients, 751 unique epitypes donors with 1.8% of epitypes occurring in both groups. In conclusion, the authors recognized that identical matching at the epitype, or at each gene region, is improbable in a diverse transplant population. Collectively, this information indicates that using eplet analysis cannot provide the claimed reduction in complexity of the matching,40 and therefore, it cannot support matching algorithms for organ allocation.
Practical Considerations—What Additional Assumptions Will Need to Be Made?
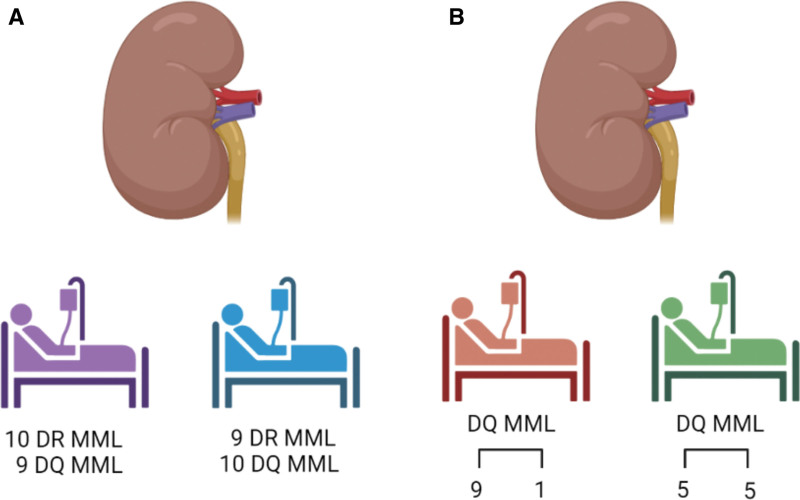
Separate from the concerns raised above, it is not clear how “epitope matching” can practically be implemented in allocation algorithms. Currently, patients receive X points for matching (depending on the specific organ allocation system around the world). In a hypothetical scenario, we have 2 potential recipients with similar priorities due to non-HLA factors. The 1 patient has 10 mismatches at the DR locus and 9 mismatches at the DQ locus. The other patient has 9 mismatches at the DR locus but 10 mismatches at the DQ locus. Which of them should get higher priority? The one with less mismatches at the DR locus or the one with less mismatches at the DQ locus? Or maybe they should both receive the same priority‚ as both have a total of 19 mismatches? In a different scenario, looking only at the DQ locus to simplify the case, both potential recipients have 10 mismatches with the donor DQ alleles, but one has 9 mismatches to allele-one and 1 mismatch with allele-two, compared with the second patient that has 5 mismatches with each of the 2 donor alleles. Which of these potential recipients should get priority? (Figure 5). This example highlights the required depth of understanding that is currently sorely missing.
FIGURE 5.
Utilization of “epitope matching” or eplet mismatch load in organ allocation algorithms: 2 scenarios are presented. A, A deceased-donor kidney is offered to patient Purple and patient Blue. The first has 10 HLA-DR mismatches and 9 HLA-DQ mismatches‚ and the second patient has 9 HLA-DR mismatches and 10 HLA-DQ mismatches. Using the current HLAMatchmaker eplet universe approach‚who should get priority? B, A deceased-donor kidney is offered to patient Brown and patient Green. Using the individual donor-allele analysis—the first patient has 9 mismatches to the first HLA-DQ allele and 1 mismatch with the other allele‚ and the second patient has 5 mismatches to either of the donor alleles—who should get priority? MML, molecular mismatch load.
Can We Use MML in Patients With Prior Sensitization?
An interesting approach was taken by Wen et al41 performing a retrospective evaluation of high-risk patients with 10 y follow-up posttransplantation and detailed maintenance immunosuppression and medication adherence information. Patients with preformed or early development (3 mo posttransplant) of DSAs were excluded. Generation of de novo DSAs was compared with eplet mismatch load, with biopsy-proven diagnosis of AMR, and death-censored graft survival. The investigators concluded that the level of eplet mismatch load was not associated with AMR or allograft loss; rather, poor outcome was associated with nonadherence and high levels of DSAs. This observation is particularly of interest, as the STAR working group recommended the use of MML as risk stratification for patients that are alloimmune naive (no history of pregnancies, transfusions, or prior transplants). Additional studies are needed to determine if there is a role for MML risk stratification in patients with potential latent memory, as those comprise roughly half of the patients on the kidney waitlist.
Words of Caution
With the hope to improve organ allocation, we witness a flurry of rapid publications based on multiple misconceptions and misinformation. Building on those, new bioinformatic tools are being developed, for example, HLA-Epi.42 New terms are introduced, for example, “epitope typing,”43 “antibody-binding eplets‚” or “epitypes.”39 The use of such tools and the associated evocative terminology continues to build on precarious infrastructure and supports a false sense of familiarity and confidence. The use of a computer program, as well as terms that sound sophisticated and advanced, may help promote and market tools that have poor scientific basis. It is important to note that a recommendation from the International Histocompatibility Workshop and Conference, Epitope component was to refrain from using the term “epitope” when referring to eplets. In fact, the recommendation was to rename the “epitope registry” as the Eplet Registry. Furthermore, the term “epitope matching” should be eliminated. Rather, the use of Acceptable Mismatching should be used when referring to organ allocation and analysis of histocompatibility between recipient and donor.
INCORPORATION OF MML INFORMATION INTO CLINICAL USE—CURRENT STATUS
The Acceptable Mismatch (AMM) program had utilized HLAMatchmaker from its inception as an approach to predict the range of unacceptable antigens in highly sensitized patients, based on reactivity patterns of a relatively small panel of cells.2 In other words, the initial purpose of using the HLAMatchmaker was to decipher all potential antibody reactivity and to better forecast a negative crossmatch. This was a significant advancement from the Eurotransplant operation that was based, at the time, on antibody assignment determined by serologic testing of a relatively small panel of cells. Importantly, the AMM program still does not attempt to predict which patients will generate de novo DSAs or will have new alloimmune responses. Clearly‚ the long-term transplant outcome of the AMM program is exceptionally high‚ as demonstrated by Heidt et al.44 Notwithstanding, though, performing thorough analysis of HLA antibody reactivity, using the SAB panel, and employing strict concerted effort not to cross any DSA barriers at the time of transplantation yield similarly good results. This was nicely demonstrated by Bray et al using the “Emory algorithm” in 2006.45 Specifically, in their cohort of 492 patients, including unsensitized, sensitized, and highly sensitized patients, all showed similar 5-y deceased-donor graft survival if unacceptable antigens were avoided and flow cytometry crossmatch was negative.
To the best of our knowledge, thus far‚ only 1 group utilized eplet mismatch load analysis for prospective decision making in determining organ allocation. In this publication, Kausman et al46 reports on a group of 7 pediatric patients who received renal transplantation through kidney paired exchange program. The patients were followed up for 8 to 54 mo with graft survival comparable to the 1- and 5-y graft survival in the general pediatric transplant population in Australia. Nonetheless, 3 of 6 patients developed DSAs (1 patient was not tested). The authors concluded that “it remains unclear whether improved epitope matching will reduce the risk of de novo HLA antibodies and contribute to improved graft outcomes and reduced rates of sensitization.” Although other groups examined the eplet mismatch load at the time of organ offers (eg, Bryan et al47), no other transplant centers made prospective decisions based on these results. It does appear, though, that the National Kidney Registry is promoting “eplet matching” in their algorithms‚48 although no outcome data, nor information on how these decisions are being validated, are available.
Risk Stratification Posttransplantation
Notwithstanding the limitations of HLAMatchmaker and other MML analysis tools, a large body of literature suggests a statistical worse graft outcome for patients transplanted across a high MML (although definition of high MML differs between studies). The most comprehensive and rigorous series of studies was published by the Manitoba group.10,27,33,49 The STAR working group21,50 provided clear recommendations and highlighted gaps that must be addressed before implementation of MML analysis for risk stratification for the purpose of posttransplant management decisions. Specifically, these recommendations include the need for 2-field typing resolution for HLA alleles (rather than imputation) and incorporation of the DQα/β (and DPα/β) as heterodimers in the analysis. The STAR working group strongly supports the need to perform MML assessment at the single donor-allele level (rather than the donor “universe”) and to consider the relative contribution of the individual alleles to the observed outcome. There must be accurate and stringent definition for generation of de novo DSAs (with prospective and continuous monitoring) and stringent definition of biopsy results. For statistical analysis, multivariable statistics should be performed, with adjustment for potential confounders, and external validity of the final model.
MOVING FORWARD—UNDERSTANDING IMMUNOGENICITY
Immunogenicity is a complex term that aims to describe and define the ability of the donor organ (in the specific case of transplantation) to provoke an immune response. Other than multiple host immune factors, there are additional modifiers such as adequacy of immunosuppression, additional immunologic assaults, and so on. It is indeed affected by the dissimilarity between donor and recipient, but probably not by the degree of dissimilarity but rather by qualitative differences. Although the puzzle of immunogenicity may not be solved soon, we believe that determining permissible from nonpermissible mismatches is a reasonable goal.
In acknowledgement that MML does not equate immunogenicity, several investigators utilized the same MML tools, applying somewhat different statistics. For example, McCaughan et al51 studied heart and lung transplant recipients using eplet analysis and electrostatic potential modeling. They identified HLA-DQA1*05/DQB1*02:01 and HLA-DQA1*05/DQB1*03:01 as 2 donor alleles inducing high rates of de novo DSA generation. Schawalder et al52 analyzed mother-child pairs with sera obtained at days 1 to 4 postdelivery. This group concentrated their analysis of HLA-DQ mismatches, identifying those eplets that were more frequent using “reacting” and “nonreacting” eplets to propose a concept for calculating the immunogenicity score. Finally, the Genome Canada Transplant Consortium focused their latest efforts on trying to identify a hierarchy of eplets associated with transplant loss. Looking at eplet mismatches in all HLA loci, the group endeavored to distinguish a smaller subset of mismatched eplets as those that are more likely to be associated with death-censored graft failure.53
The most illustrative data demonstrating that MML does not equate immunogenicity was published by Lucas et al54 Studying >700 donor/recipient pairs, the investigators showed that there is a directionality in responses. Specifically, donor/recipient pairs with reverse mismatches (eg, in pair 1, the donor was typed as A2 and recipient A68, and in pair 2, the donor was typed as A68 and recipient as A2) did not show the same degree of generating de novo DSAs. In other words, not only did these 2 pairs have the same MML, but the mismatches were identical, although in different direction. The investigators concluded that the directional of mismatch, and hence the larger background on which they are present, is at least as important as the MML.
Currently, there is very little research to decipher immunogenicity that is not fully dependent on an MML tool. The Leiden group has been diligently engaged in producing human monoclonal class II antibodies from multiparous women or transplant recipients. These monoclonal antibodies are then undergoing thorough investigation using multiple approaches.23 We have developed a unique approach to study immunogenicity in a cohort of transplant recipients who received a kidney from a donor with 2 HLA-DQ mismatches but have developed antibodies only against one of the donor’s mismatched DQ antibodies26 (2MM1DSA). Sera from these patients is then interrogated using adsorption elution studies and is evaluated using in silico mutagenesis. Several groups have initiated site-directed mutagenesis experiments with the goal to better characterize the antibody binding footprint. Crystallography studies will likely shed more light onto the exact dimension and orientation of this binding. We believe that progress requires the use of multiprong approaches. It is important to mention that‚ to fully comprehend immunogenicity, one must also study the role and relevance of T-cell epitopes in the context of solid organ transplantation.
IN SUMMARY
HLAMatchmaker and the work pioneered by Rene Duquesnoy redirected discourse in the field of transplantation from a stagnated look at HLA matching as a necessary burden, to the meaningful contribution of HLA compatibility in avoiding transplant rejection. This work complemented the humoral theory promoted by Paul Terasaki,55,56 leading to attempts to curtail the generation of de novo DSAs. In an editorial from 2015‚ Glotz and Tambur57 cautioned that “we are yet to comprehend what determines immunogenicity and pathogenicity of HLA mismatches…and…it is likely to assume that there is more to an epitope than just the sheer number of eplet mismatches.” Although our community is changing the discussion from epitope matching to the understanding that some eplets may carry higher immunologic risk than others, many misconceptions remain.
This review highlights the multiple reasons why MML analysis is not suitable to guide organ allocation algorithms. Currently, the Food and Drug Administration is evaluating the use of MML analysis as a tool in support of pharmacologic clinical trials, with the goal to enrich populations at risk of developing allograft rejection and DSAs. Although MML is likely to support risk stratification posttransplantation, we strongly recommend combining its implementation only as part of a holistic, 1-y posttransplant patient evaluation before changes in immunosuppression management decisions.
There is much to be done to decipher permissible from nonpermissible mismatches. Rather than adopting an approach of “done beats perfect,” our community has the responsibility to rigorously study and understand immunogenicity such that we can find the best approach to implement lessons learned.
ACKNOWLEDGMENTS
Anat R. Tambur is a Paul I Terasaki Scholar.
Footnotes
The authors declare no funding or conflicts of interest.
A.R.T. and R.S. participated in the writing of the article.
Supplemental Visual Abstract; http://links.lww.com/TP/C539
Supplemental digital content (SDC) is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.transplantjournal.com).
REFERENCES
- 1.Duquesnoy RJ. HLAMatchmaker: a molecularly based algorithm for histocompatibility determination. I. Description of the algorithm. Hum Immunol. 2002;63:339–352. [DOI] [PubMed] [Google Scholar]
- 2.Duquesnoy RJ, Witvliet M, Doxiadis II, et al. HLAMatchmaker-based strategy to identify acceptable HLA class I mismatches for highly sensitized kidney transplant candidates. Transpl Int. 2004;17:22–30. [DOI] [PubMed] [Google Scholar]
- 3.Claas FH, Witvliet MD, Duquesnoy RJ, et al. The acceptable mismatch program as a fast tool for highly sensitized patients awaiting a cadaveric kidney transplantation: short waiting time and excellent graft outcome. Transplantation. 2004;78:190–193. [DOI] [PubMed] [Google Scholar]
- 4.Dankers MK, Witvliet MD, Roelen DL, et al. The number of amino acid triplet differences between patient and donor is predictive for the antibody reactivity against mismatched human leukocyte antigens. Transplantation. 2004;77:1236–1239. [DOI] [PubMed] [Google Scholar]
- 5.Dankers MK, Heemskerk MB, Duquesnoy RJ, et al. HLAMatchmaker algorithm is not a suitable tool to predict the alloreactive cytotoxic T-lymphocyte response in vitro. Transplantation. 2004;78:165–167. [DOI] [PubMed] [Google Scholar]
- 6.Adeyi OA, Girnita AL, Howe J, et al. Serum analysis after transplant nephrectomy reveals restricted antibody specificity patterns against structurally defined HLA class I mismatches. Transpl Immunol. 2005;14:53–62. [DOI] [PubMed] [Google Scholar]
- 7.Duquesnoy RJ. A structurally based approach to determine HLA compatibility at the humoral immune level. Hum Immunol. 2006;67:847–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duquesnoy RJ. Clinical usefulness of HLAMatchmaker in HLA epitope matching for organ transplantation. Curr Opin Immunol. 2008;20:594–601. [DOI] [PubMed] [Google Scholar]
- 9.Saleem N, Das R, Tambur AR. Molecular histocompatibility beyond Tears: the next generation version. Hum Immunol. 2022;83:233–240. [DOI] [PubMed] [Google Scholar]
- 10.Wiebe C, Pochinco D, Blydt-Hansen TD, et al. Class II HLA epitope matching-A strategy to minimize de novo donor-specific antibody development and improve outcomes. Am J Transplant. 2013;13:3114–3122. [DOI] [PubMed] [Google Scholar]
- 11.Stave JW, Lindpaintner K. Antibody and antigen contact residues define epitope and paratope size and structure. J Immunol. 2013;191:1428–1435. [DOI] [PubMed] [Google Scholar]
- 12.Duquesnoy RJ, Marrari M. HLAMatchmaker-based definition of structural human leukocyte antigen epitopes detected by alloantibodies. Curr Opin Organ Transplant. 2009;14:403–409. [DOI] [PubMed] [Google Scholar]
- 13.Weiss GA, Watanabe CK, Zhong A, et al. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 2000;97:8950–8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojas G, Tundidor Y, Infante YC. High throughput functional epitope mapping: revisiting phage display platform to scan target antigen surface. MAbs. 2014;6:1368–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duquesnoy RJ. The antibody response to an HLA mismatch: a model for nonself-self discrimination in relation to HLA epitope immunogenicity. Int J Immunogenet. 2012;39:1–9. [DOI] [PubMed] [Google Scholar]
- 16.Willers J, Lucchese A, Kanduc D, et al. Molecular mimicry of phage displayed peptides mimicking GD3 ganglioside. Peptides. 1999;20:1021–1026. [DOI] [PubMed] [Google Scholar]
- 17.Kanduc D. “Self-nonself” peptides in the design of vaccines. Curr Pharm Des. 2009;15:3283–3289. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Sanchez C, Usenko CY, Herrera ND, et al. The shared epitope phenomenon-A potential impediment to virtual crossmatch accuracy. Clin Transplant. 2020;34:e13906. [DOI] [PubMed] [Google Scholar]
- 19.Duquesnoy RJ, Marrari M, da M Sousa LC, et al. 16th IHIW: a website for antibody-defined HLA epitope Registry. Int J Immunogenet. 2013;40:54–59. [DOI] [PubMed] [Google Scholar]
- 20.HLA Epitope Registry. Available at http://epregistry.ufpi.br/index/index. Accessed March 17, 2022.
- 21.Tambur AR, Campbell P, Claas FH, et al. Sensitization in transplantation: assessment of risk (STAR) 2017 Working Group Meeting Report. Am J Transplant. 2018;18:1604–1614. [DOI] [PubMed] [Google Scholar]
- 22.Kramer CSM, Roelen DL, Heidt S, et al. Defining the immunogenicity and antigenicity of HLA epitopes is crucial for optimal epitope matching in clinical renal transplantation. HLA. 2017;90:5–16. [DOI] [PubMed] [Google Scholar]
- 23.Bezstarosti S, Bakker KH, Kramer CSM, et al. A comprehensive evaluation of the antibody-verified status of eplets listed in the HLA epitope registry. Front Immunol. 2021;12:800946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer CSM, Koster J, Haasnoot GW, et al. HLA-EMMA: a user-friendly tool to analyse HLA class I and class II compatibility on the amino acid level. HLA. 2020;96:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paunić V, Gragert L, Madbouly A, et al. Measuring ambiguity in HLA typing methods. PLoS One. 2012;7:e43585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tambur AR, McDowell H, Hod-Dvorai R, et al. The quest to decipher HLA immunogenicity: telling friend from foe. Am J Transplant. 2019;19:2910–2925. [DOI] [PubMed] [Google Scholar]
- 27.Wiebe C, Kosmoliaptsis V, Pochinco D, et al. HLA-DR/DQ molecular mismatch: a prognostic biomarker for primary alloimmunity. Am J Transplant. 2019;19:1708–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeVos JM, Gaber AO, Knight RJ, et al. Donor-specific HLA-DQ antibodies may contribute to poor graft outcome after renal transplantation. Kidney Int. 2012;82:598–604. [DOI] [PubMed] [Google Scholar]
- 29.Willicombe M, Brookes P, Sergeant R, et al. De novo DQ donor-specific antibodies are associated with a significant risk of antibody-mediated rejection and transplant glomerulopathy. Transplantation. 2012;94:172–177. [DOI] [PubMed] [Google Scholar]
- 30.Senev A, Coemans M, Lerut E, et al. Eplet mismatch load and de novo occurrence of donor-specific anti-HLA antibodies, rejection, and graft failure after kidney transplantation: an observational cohort study. J Am Soc Nephrol. 2020;31:2193–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tambur AR, Kosmoliaptsis V, Claas FHJ, et al. Significance of HLA-DQ in kidney transplantation: time to reevaluate human leukocyte antigen-matching priorities to improve transplant outcomes? An expert review and recommendations. Kidney Int. 2021;100:1012–1022. [DOI] [PubMed] [Google Scholar]
- 32.Tassone G, De Santis D, Vukovic I, et al. Different eplet software programs give discordant and incorrect results: an analysis of HLAMatchmaker vs Fusion Matchmaker eplet calling software. HLA. 2020;96:52–63. [DOI] [PubMed] [Google Scholar]
- 33.Wiebe C, Rush DN, Nevins TE, et al. Class II Eplet mismatch modulates tacrolimus trough levels required to prevent donor-specific antibody development. J Am Soc Nephrol. 2017;28:3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engen RM, Jedraszko AM, Conciatori MA, et al. Substituting imputation of HLA antigens for high-resolution HLA typing: evaluation of a multiethnic population and implications for clinical decision making in transplantation. Am J Transplant. 2021;21:344–352. [DOI] [PubMed] [Google Scholar]
- 35.Senev A, Emonds MP, Van Sandt V, et al. Clinical importance of extended second field high-resolution HLA genotyping for kidney transplantation. Am J Transplant. 2020;20:3367–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sapir-Pichhadze R, Zhang X, Ferradji A, et al. Epitopes as characterized by antibody-verified eplet mismatches determine risk of kidney transplant loss. Kidney Int. 2020;97:778–785. [DOI] [PubMed] [Google Scholar]
- 37.Lemieux W, Mohammadhassanzadeh H, Klement W, et al. Matchmaker, matchmaker make me a match: opportunities and challenges in optimizing compatibility of HLA eplets in transplantation. Int J Immunogenet. 2021;48:135–144. [DOI] [PubMed] [Google Scholar]
- 38.Simon HA. Theories of bounded rationality. McGuire CB, Radner R, Arrow KJ, eds. In: Decision and Organization. North-Holland Publishing Company; 1972. [Google Scholar]
- 39.Tran JN, Günther OP, Sherwood KR, et al. ; Genome Canada Transplant Consortium. High-throughput sequencing defines donor and recipient HLA B-cell epitope frequencies for prospective matching in transplantation. Commun Biol. 2021;4:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran J. Human Leukocyte Antigen B Cell Epitope Matching in Kidney Transplantation. Dissertation. The University of British Columbia; 2022. [Google Scholar]
- 41.Wen J, Basu A, Bentall A, et al. Is the level of HLA eplet mismatch a risk factor for graft loss among kidney transplant recipients who have already formed de novo donor specific antibody? Hum Immunol. 2021;82:240–246. [DOI] [PubMed] [Google Scholar]
- 42.Geffard E, Boussamet L, Walencik A, et al. HLA-EPI: a new EPIsode in exploring donor/recipient epitopic compatibilities. HLA. 2022;99:79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherwood KR, Tran J, Günther OP, et al. ; Genome Canada Transplant Consortium. Genome Canada precision medicine strategy for structured national implementation of epitope matching in renal transplantation. Hum Immunol. 2022;83:264–269. [DOI] [PubMed] [Google Scholar]
- 44.Heidt S, Haasnoot GW, Claas FHJ. How the definition of acceptable antigens and epitope analysis can facilitate transplantation of highly sensitized patients with excellent long-term graft survival. Curr Opin Organ Transplant. 2018;23:493–499. [DOI] [PubMed] [Google Scholar]
- 45.Bray RA, Nolen JD, Larsen C, et al. Transplanting the highly sensitized patient: the emory algorithm. Am J Transplant. 2006;6:2307–2315. [DOI] [PubMed] [Google Scholar]
- 46.Kausman JY, Walker AM, Cantwell LS, et al. Application of an epitope-based allocation system in pediatric kidney transplantation. Pediatr Transplant. 2016;20:931–938. [DOI] [PubMed] [Google Scholar]
- 47.Bryan CF, Chadha V, Warady BA. Donor selection in pediatric kidney transplantation using DR and DQ eplet mismatching: a new histocompatibility paradigm. Pediatr Transplant. 2016;20:926–930. [DOI] [PubMed] [Google Scholar]
- 48.National Kidney Registry. Kidney compatibility: finding the best match. Available at https://www.kidneyregistry.org/for-patients/finding-the-best-match/. Accessed March 17, 2022.
- 49.Wiebe C, Nickerson P. Strategic use of epitope matching to Improve outcomes. Transplantation. 2016;100:2048–2052. [DOI] [PubMed] [Google Scholar]
- 50.Tambur AR, Campbell P, Chong AS, et al. Sensitization in transplantation: assessment of risk (STAR) 2019 Working Group Meeting Report. Am J Transplant. 2020;20:2652–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCaughan JA, Battle RK, Singh SKS, et al. Identification of risk epitope mismatches associated with de novo donor-specific HLA antibody development in cardiothoracic transplantation. Am J Transplant. 2018;18:2924–2933. [DOI] [PubMed] [Google Scholar]
- 52.Schawalder L, Hönger G, Kleiser M, et al. Development of an immunogenicity score for HLA-DQ eplets: a conceptual study. HLA. 2021;97:30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohammadhassanzadeh H, Oualkacha K, Zhang W, et al. ; Genome Canada Transplant Consortium. On path to informing hierarchy of eplet mismatches as determinants of kidney transplant loss. Kidney Int Rep. 2021;6:1567–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucas DP, Leffell MS, Zachary AA. Differences in immunogenicity of HLA antigens and the impact of cross-reactivity on the humoral response. Transplantation. 2015;99:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terasaki PI. Humoral theory of transplantation. Am J Transplant. 2003;3:665–673. [DOI] [PubMed] [Google Scholar]
- 56.Terasaki PI. A personal perspective: 100-year history of the humoral theory of transplantation. Transplantation. 2012;93:751–756. [DOI] [PubMed] [Google Scholar]
- 57.Glotz D, Tambur A. Stratifying patients based on epitope mismatching: ready for primetime? Am J Transplant. 2015;15:2021–2022. [DOI] [PubMed] [Google Scholar]