Summary
Here, we present a protocol to identify and quantify phosphopeptides during the dynamic formation of an immunological synapse. We describe steps for mixing isotope-labeled immune and target cells, the stabilization of cell-to-cell conjugates by cross-linking, and their isolation by fluorescence-activated cell sorting. We detail the isolation of phosphopeptides by phosphopeptide enrichment and their subsequent measurement by mass spectrometry. Finally, we describe the analysis of the resulting data to separate cell-specific phosphopeptides using the isotope label and label-free quantification.
Subject areas: Cell Biology, Flow Cytometry/Mass Cytometry, Immunology, Molecular/Chemical Probes, Protein Biochemistry, Proteomics, Mass Spectrometry
Graphical abstract

Highlights
-
•
Isolation of NK cells forming an immune synapse
-
•
Cell-type-specific phosphoproteomics
-
•
SILAC-based quantification of the phosphopeptides
-
•
Mass-spectrometry-based quantification of the phosphopeptides
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we present a protocol to identify and quantify phosphopeptides during the dynamic formation of an immunological synapse. We describe steps for mixing isotope-labeled immune and target cells, the stabilization of cell-to-cell conjugates by cross-linking, and their isolation by fluorescence-activated cell sorting. We detail the isolation of phosphopeptides by phosphopeptide enrichment and their subsequent measurement by mass spectrometry. Finally, we describe the analysis of the resulting data to separate cell-specific phosphopeptides using the isotope label and label-free quantification.
Before you begin
Rapid killing of diseased cells, such as cancer cells, by cytotoxic lymphocytes relies on a highly specialized cell-to-cell conjugation termed the immunological synapse. Formation and activity of the immunological synapse involve multiple sequential steps, including but not limited to, recognition of and adhesion to the target cell, integration of signals at the target cell surface driving activation of effector functions, polarization of the lytic machinery toward the target cell and secretion of effector molecules, such as granzyme B and perforin, into the synaptic cleft.1,2,3
Cancer cells have evolved defense responses to evade lymphocyte cytotoxicity, such as rapid polarization of their actin cytoskeleton and vesicles toward the immunological synapse.4,5,6 Characterization of the signaling pathways controlling the key cellular processes at both pre- and post-synaptic sides of the immunological synapse is essential to develop novel strategies aimed at improving cytotoxic lymphocyte immune response.
In this protocol we describe the steps for the formation of immunological synapses between natural killer (NK) cells and target cancer cells, the stabilization and isolation of cell-to-cell conjugates, the enrichment and analysis of phospho-peptides by mass spectrometry, and the specific data analysis allowing to unravel phospho-signaling in each cell type. Although the protocol is detailed it is required that the experimenter is familiar with the main techniques used in the protocol, mass spectrometry of peptides, the analysis of proteomics data, and FACS sorting. The protocol can be used with less than 25 μg starting material because of the high efficiency of the preparation and the accuracy of the analysis.
Protocol overview
Although exemplified for the analysis of the lytic synapse between NK cells and tumor cells, our protocol could, in principle, apply to other types of immunological synapses, such as those involving T cells, dendritic cells, or B cells.1,7,8,9 The main principle of this protocol is that the two populations of interacting cells are labeled with different SILAC amino acids,10,11 allowing to trace back the origin of the phosphopeptides identified by mass spectrometry. Light-labeled (L) NK cells and heavy-labeled (H) breast cancer MDA-MB-231 cells are cocultured for a short period of time to allow immunological synapse formation. Cell-to-cell conjugates are then stabilized by chemical cross-linking and isolated using FACS sorting. Proteins are extracted and separated on an SDS-PAGE, and proteins are digested in situ. Phosphopeptides are enriched by Fe-chelate-chromatography and analyzed by mass spectrometry. Recorded spectra are analyzed using the MaxQuant software package,12,13 followed by bioinformatic data analysis (Figure 1).
Figure 1.
Overview of the experimental setup
SILAC-labeled NK92-MI cells are incubated with MDA-MB-231 (target cells) and establish an immunological synapse. NK92-MI -target cell conjugates are isolated based on fluorescence (red + green), and the proteins from these conjugates are extracted. After enrichment of the phosphopeptides the samples are analyzed by mass spectrometry and the resulting data is processed in silico.
Prepare buffers and reagents
Timing: 0.5–2 h
-
1.
Prepare buffers following the recipes in the materials and equipment.
-
2.All reagents can be stored at room temperature for 2 months, except the ones indicated by the manufacturer:
-
a.Trypsin should be prepared fresh at 0.25 μg/μL in 50 mM ammonium bicarbonate buffer.
-
b.Reduction buffer and Alkylation buffer should be prepared fresh.
-
a.
Note: Alkylation buffer should be kept in the dark.
Note: The protocol below describes the sample preparation for approximately 25 samples. For a larger number of samples, the volumes need to be adjusted.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE/Cyanine7 anti-human CD56 (NCAM) antibody | BioLegend | Cat# 318318, RRID: AB_604107 |
| Chemicals, peptides, and recombinant proteins | ||
| H2O, LC-MS grade | Supelco | 7732-18-5 |
| Precision Plus Protein™ Dual Color Standards | Bio-Rad | 1610374 |
| 10× Tris/Glycine/SDS | Bio-Rad | 1610732 |
| 2× Laemmli sample buffer | Bio-Rad | 1610737 |
| InstantBlue | Abcam | ISB1L-1L |
| Dithiothreitol (DTT) | Fisher Scientific | 3483-12-3 |
| 2-Cloroacetamide (CAA) | Aldrich | 22790-250G-F |
| Trypsin | Promega | V5111 |
| Formic acid LC-MS grade | ThermoScientific | 85178 |
| Ammonium bicarbonate | Fluka | 09830-500G |
| Acetonitrile | Supelco | EM-AX0145P-6 |
| Urea | Sigma-Aldrich | U5128 |
| Tris-HCl | Merck | 1185-53-1 |
| NaCl | Merck | 7647-14-5 |
| EDTA | Merck | 60-00-4 |
| 16% formaldehyde, methanol-free (PFA) | Thermo Fisher Scientific | Cat #28908 |
| Methanol HPLC grade | Sigma-Aldrich | 900688 |
| RPMI 1640 Medium for SILAC | Thermo Fisher Scientific | Cat# 88365 |
| DMEM:F-12 for SILAC | Thermo Fisher Scientific | Cat# 88370 |
| Fetal bovine serum | Sigma-Aldrich | Cat# F0392 |
| L-Arginine monohydrochloride | Sigma-Aldrich | Cat# A6969 |
| L-Arginine · HCl (13C6, 99%; 15N4, 99%) | Cambridge Isotope Laboratories | Cat# CNLM-539-H-0.05 |
| L-Lysine monohydrochloride | Sigma-Aldrich | Cat# L8662 |
| L-Lysine · 2HCl (13C6, 99%; 15N2, 99%) | Cambridge Isotope Laboratories | Cat# CNLM-291-H-0.05 |
| L-Leucine | Sigma-Aldrich | Cat# L8912 |
| Penicillin/Streptomycin | Lonza | Cat# DE17-602E |
| Puromycin dihydrochloride | Santa Cruz | Cat# CAS 58-58-2 |
| Dulbecco’s phosphate-buffered saline (DPBS), 1×, no calcium, no magnesium | Thermo Fisher Scientific | Cat# 10010015 |
| AutoMACS Running Buffer | Miltenyi Biotec | Cat #130-091-221 |
| Dulbecco’s phosphate-buffered saline (DPBS), 1× with Ca2+ & Mg2+ | Thermo Fisher Scientific | Cat# 14040-091 |
| PBS with Mg2+ and Ca2+ (PBS/Mg/Ca) | Thermo Fisher Scientific | Cat# 14040133 |
| Trypsin-EDTA | Thermo Fisher Scientific | Cat# 25300062 |
| Critical commercial assays | ||
| MycoAlert Mycoplasma Detection kit | Lonza | Cat# LT07-318 |
| Live-or-Dye NucFix Red Staining kit | Biotium | Cat# 32010 |
| High-Select Fe-NTA Phosphopeptide Enrichment Kit | Thermo Fisher Scientific | A32992 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat# 23225 |
| Trypan Blue Solution 0.4% | Thermo Fisher Scientific | Cat# 15250-061 |
| Deposited data | ||
| Proteomics dataset | MASSIVE proteomeXchange | ftp://massive.ucsd.edu/MSV000090645/ |
| Experimental models: Cell lines | ||
| NK-92MI | ATCC | Cat# CRL-2408, RRID:CVCL_3755 |
| MDA-MB-231 | ATCC | Cat# HTB-26, RRID:CVCL_0062 |
| Software and algorithms | ||
| FlowJo v10.8.1 | BD Bioscience https://www.flowjo.com/solutions/flowjo/downloads | |
| MaxQuant | https://maxquant.org | version 1.6.17.0 |
| Perseus | https://maxquant.org | Version 1.6.15.0 |
| R | http://r-project.org | Version 4.1.3 |
| Other | ||
| Thermomixer | Eppendorf | 5355 000.011 |
| SDS-PAGE Vertical Electrophoresis Cell | Bio-Rad | 1658005 |
| Microcentrifuge tubes, 2 mL | Eppendorf | 88379 |
| Vortex | Heidolph | 541-10000-00 |
| Centrifuge | Beckman Coulter | A46472 |
| Power basic power supply | Bio-Rad | 1645050 |
| 12% Mini-PROTEAN® TGX™ Precast Protein Gels | Bio-Rad | 4561045 |
| Razor blade | ||
| pH indicator paper | Merck | 1.09565.0001 |
| 0.2 mL Skirted 96-well Robotic Plate | Thermo Scientific | AB-1300 |
| Silicone sealing mat. For 96 well PCR plates | Nerbeplus | 04-090-0000 |
| Acclaim PepMap RSLC 75μm × 2 cm nanoViper | Thermo Fisher | PN164534 |
| Acclaim PepMap RSLC 75μm × 15 cm nanoViper | Thermo Fisher | PN164535 |
| Q Exactive™ HF hybrid quadrupole-Orbitrap mass spectrometer | Thermo Fisher | P/N 1372090 |
| Dionex Ultimate 3000 HPLC NCS-3500RS | Thermo Fisher | 5041.0010 |
| Autosampler WPS-3000TPL RS | Thermo Fisher | Autosampler WPS-3000TPL RS |
| GPS200 pipette tip | Eppendorf | 0030000889 |
| Octadecyl (C18)-bonded silica | 3M | 2215 |
| Tube with round bottom for culture 5 mL | Falcon | Cat# 352054 |
| Protein LoBind tubes, 5 mL, with snap cap | Eppendorf | Cat# 0030108302 |
| Tube, 15 mL, pp, 17/120 mm, conical bottom | Greiner | Cat# 188271 |
| Cell culture flask, 550 mL, 175cm2, PS | Greiner | Cat# 660175 |
| Cell culture flask, 250 mL, 75cm2, PS | Greiner | Cat# 658175 |
| BD FACS Aria flow cytometer | Becton Dickinson Biosciences | |
Note: MDA-MB-231 were purchased from ATCC and subsequently genetically modified to stably express a fluorescent actin reporter for our specific research interest (Al Absi et al.4).
Materials and equipment
Buffers resource table
NK-92MI medium
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 medium for SILAC | 400 mL | |
| FBS dialyzed by ultracentrifugation 0.15 M NaCl | 20% | 100 mL |
| L-Arginine monohydrochloride | 1% 100 μg/mL |
5 mL |
| L-Lysine monohydrochloride | 1% 100 μg/mL |
5 mL |
| L-Leucine | 1% 100 μg/mL | 5 mL |
| Penicillin/Streptomycin | 1% | 5 mL |
| Total | N/A | 520 mL |
Note: All reagents are mixed and stored at 4°C for up to 1 month, except for amino acids that are stored at −20°C and added just before using the medium.
MDA-MB-231 medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM:F-12 for SILAC | 450 mL | |
| FBS dialyzed by ultracentrifugation 0.15 M NaCl | 10% | 50 mL |
| L-Arginine · 2HCl (13C6, 99%; 15N4, 99%) | 1% 100 μg/mL | 5 mL |
| L-Lysine · 2HCl (13C6, 99%; 15N2, 99%) | 1% 100 μg/mL | 5 mL |
| L-Leucine | 1% 100 μg/mL | 5 mL |
| Penicillin/Streptomycin | 1% | 5 mL |
| Puromycin dihydrochloride | 0.5 μg/mL | 250 μL |
| Total | N/A | 520.25 mL |
Note: All reagents are mixed and stored at 4°C for up to 1 month except for amino acids that are stored at −20°C and added just before using the medium.
Lysis Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Urea | 8 M | 9.6 g solid urea in 9.6 mL of H20 |
| NaCl | 75 mM | 1.5 mL 1 M NaCl |
| Tris-HCl pH 8.0 | 50 mM | 1 mL 1 M Tris HCl pH 8.0 |
| EDTA | 1 mM | 40 μL of 500 mM EDTA |
| H2O, LC-MS grade | N/A | 7.86 mL |
| Total | N/A | 20 mL |
-
•
Dilution Buffer: 50 mM Tris-HCl pH 8.0.
-
•
Washing Buffer A: dissolve 59.3 mg ammonium bicarbonate in 15 mL MS-grade water, pH 8.5.
-
•
Washing Buffer B: take 5 mL of acetonitrile and 39.5 mg ammonium bicarbonate and fill up to 10 mL with MS-grade water, pH 8.5.
-
•
Reduction buffer: Add 19.8 mg ammonium bicarbonate and 77.1 mg DTT and fill up to 5 mL with MS-grade water, pH 8.5.
-
•
Alkylation buffer: Add 19.8 mg ammonium bicarbonate and 46.8 chloroacetamide and fill up to 5 mL with MS-grade water, pH 8.5.
-
•
Ammonium bicarbonate buffer: Dissolve 39.5 g ammonium bicarbonate in 100 mL MS-grade water, pH 8.5.
-
•
Digestion Buffer: Add 1 μg Trypsin in 150 μL ammonium bicarbonate buffer (150 μL per sample), pH 8.5.
-
•
Mobile phase A: Dilute 100 μL formic acid in 100 mL MS-grade water (0.1%).
-
•
Mobile phase B: Dilute 100 μL formic acid in 100 mL HPLC-grade acetonitrile.
-
•
ST buffer A: 2 mL 100 mL HPLC-grade methanol.
-
•
ST buffer B: Take 5 μL formic acid, 250 μL acetonitrile and fill up to 5 mL with MS-grade water.
-
•
ST buffer C: Take 5 μL formic acid, 4 mL acetonitrile and fill up to 5 mL with MS-grade water.
-
•
Stop buffer: Take 20 μL formic acid and fill up to 2 mL with MS-grade water.
-
•
Fixation buffer: Dissolve 200 mg of paraformaldehyde in 10 mL double distilled water.
Note: All reagents are mixed and stored at 4°C for up to 1 month.
Mass spectrometry parameters
This protocol is specifically set up for phosphopeptide analysis in low concentration. The conditions are optimized for Ultimate 3000 UHPLC system (DIONEX) coupled to Q Exactive HF mass spectrometer (ThermoFisher Scientific), using the Nanospray Flex ion source at 275°C (ThermoFisher Scientific). The MS was calibrated following the manufacturer instructions. The MS was operated in data-dependent mode with the following parameters:
| General | |
| Runtime | 0–149 min |
| Polarity | Positive |
| Default Charge State | 2 |
| Full MS | |
| Microscans | 1 |
| Resolution | 60000 |
| AGC target | 3e6 |
| Maximum IT | 120 MS |
| Number of scan ranges | 1 |
| Scan range | 375–1,700 m/z |
| Spectrum data type | Profile |
| dd-MS2 | |
| Microscans | 1 |
| Resolution | 60000 |
| AGC target | 2e5 |
| Maximum IT | 90 ms |
| Loop count | 7 |
| MSX count | 1 |
| Isolation Window | 1.6 m/z |
| Isolation offset | 0.0 m/z |
| Fixed first mass | 100.0 m/z |
| NCE | 28 |
| Spectrum data type | Profile |
| dd Settings | |
| Maximum AGC target | 4.50e3 |
| Charge exclusion | Unassigned, 1, 6–8, >8 |
| Exclude isotopes | On |
| Dynamic exclusion | 20.0 s |
UHPLC parameters
The UHPLC setup was injecting in two-column mode: the loading pump injects the sample in the loop in “microliter pickup mode”, and sample is transferred to the Acclaim PepMap RSLC 75μm × 2 cm nanoViper (pepTrap) for 8 min at 5 μL/min. After 8 min, the column-oven valve switches to put both pepTrap and the Acclaim PepMap RSLC 75μm × 15 cm nanoViper online to start the gradient. The parameters are described below:
| Time (min) | Flow (μL/min) | % Mobile phase B |
|---|---|---|
| 0 | 0.300 | 2 |
| 8 | 0.300 | 2 |
| 134 | 0.300 | 35 |
| 134.1 | 0.300 | 90 |
| 138 | 0.300 | 90 |
| 138.1 | 0.300 | 2 |
| 149 | 0.300 | 2 |
CRITICAL: Formic acid is corrosive and irritant. Handle inside a chemical hood and use appropriate safety gear.
CRITICAL: Methanol is a flammable liquid and poisonous by inhalation, prepare aliquots and handle inside a chemical hood.
Alternatives: Other LC-MS/MS setups can be used for the MS analysis, as long as they can analyze SILAC-labeled samples.10,14 The conditions must be adjusted according to the instrumentation, and the final concentrations and volumes of the samples have to be adjusted. Other SDS-PAGE systems can be used for sample cleaning and fractionation, but it is important to keep the protein amount at 25 μg for further phosphopeptide enrichment and mass spectrometry analysis.
Step-by-step method details
Production of SILAC-labeled NK-target cell conjugates
Cell labeling in SILAC medium
Timing: 3 weeks (for steps 1 to 2)
This section describes the labeling of the cells in SILAC media. NK-92MI cells are cultured in light-labeled and the MDA-MB-231 cells in heavy-labeled SILAC media.10 Duration of cell culture in SILAC media depends on two parameters: the labeling efficiency of proteins and the quantity of cells needed to form a sufficient number of cell-to-cell conjugates. The labeling efficiency for each cell type is evaluated after five serial cell passages and should exceed 95% labeling. If the labeling efficiency is not high enough, the cells should be cultured over an extended period of time until the threshold of 95% is reached.
To optimize cell culture, the size of cell culture flasks is adjusted to the number of cells necessary for coculture.
-
1.NK-92MI cell culture.
-
a.NK-92MI are cultured in 30 mL RPMI SILAC medium in 75 cm2 flasks and used after 5 passages if the labeling efficiency of the cells exceeds 95%.Note: add the isotope-labeled amino acids freshly.
-
b.Every 2–3 days, cells are counted to adjust the concentration at 3∗105 cells/mL (1 passage).
-
c.To proceed with step 2: 105–106 cells are sufficient for a preliminary test.
-
d.To proceed with step 3: 3∗106 are sufficient.
-
e.To proceed with step 4: 35∗106 NK-92MI cells are required.
-
a.
-
2.MDA-MB-231 cell culture.
-
a.MDA-MB-231 are cultured in 20 mL DMEM SILAC medium in 175 cm2 flasks and used after 7 passages.Note: add the isotope-labeled amino acids freshly.
-
b.At the confluence, cells are washed with PBS to remove the excess medium.
-
c.Add Trypsin-EDTA for 2 min.
-
d.Collect the detached cells by adding MDA-MB-231 SILAC-cell culture medium.
-
e.Seed the cells at a 1/5 dilution (1 passage).
-
f.To proceed with step 2: 105–106 cells are sufficient for a preliminary test.
-
g.To proceed with step 3: 106 cells are sufficient.
-
h.To proceed with step 4: 15∗106 MDA-MB-231 cells are required.
 CRITICAL: It is necessary to perform a labeling efficiency test on heavy K and R to confirm that >95% of the proteins are heavy-labeled before proceeding to the next step. In case the labeling efficiency is < 95% go to problem 1.
CRITICAL: It is necessary to perform a labeling efficiency test on heavy K and R to confirm that >95% of the proteins are heavy-labeled before proceeding to the next step. In case the labeling efficiency is < 95% go to problem 1. CRITICAL: It is necessary to cultivate both cell lines in SILAC media, even if light amino acids are used, to ensure that no excess amino acids are carried over into the next steps.
CRITICAL: It is necessary to cultivate both cell lines in SILAC media, even if light amino acids are used, to ensure that no excess amino acids are carried over into the next steps.
-
a.
Determination of the labeling efficiency
Timing: 2 days (for steps 3 to 22)
In this section, the degree of incorporation of the isotope-labeled amino acids is determined by a mass spectrometric measurement.
-
3.
Take an aliquot of the heavy labeled cells (cell culture section). A total of 105–106 cells are sufficient for the preliminary test.
-
4.
Lyse cells with 100 μL of lysis buffer on ice.
Note: prepare Lysis buffer freshly before use.
-
5.
Vortex for 10 s at maximum setting.
-
6.
Incubate cells at 4°C for 15 min to allow the mixture to sit.
-
7.
Vortex for 10 s at maximum setting.
-
8.
Centrifuge in a tabletop bench centrifuge at 4°C at maximum speed (16,000 g) to remove cell debris.
-
9.
Transfer supernatant to a 2 mL Eppendorf tube.
-
10.
Estimate protein concentration with BCA assay. The protein concentration should be between 0.5–2 mg/mL.
-
11.
Add 5 μL of reduction buffer and incubate for 30 min at RT.
-
12.
Add 5 μL of alkylation buffer and incubate for 30 min at RT in the dark.
-
13.
Dilute sample 1:4 with dilution buffer to decrease urea concentration below 2 M.
-
14.
Add trypsin buffer (substrate ratio of 1:50) for overnight at RT.
-
15.
Add 10 μL of stop buffer.
-
16.
Perform step 5 subsection, “digest clean up” to clean the peptides.
-
17.
Reconstitute the samples in ST buffer B to obtain approximately 1 μg/μL.
-
18.
Inject 1 μg of material into the LC-MS/MS with the parameters described in the mass spectrometry resource parameters section.
-
19.
Analyze the data with MaxQuant following the protocol described in section: automated interpretation of the mass spectra using MaxQuant, with exceptions:
-
20.
Under “variable modifications”, add Pro6 (determine arginine-to-proline conversion).
-
21.
Select multiplicity 2 and mark the labels as Lys8 and Arg10. Do not select the “re-quantify” option (described in10).
-
22.
The labeling efficiency should be calculated based on the non-normalized SILAC ratios and calculated for K and R separately.
Note: Determine the incorporation rate as:
Complete labeling is considered when the incorporation rate is higher than 95%. Heavy proline (a side-product of arginine labeling) should not exceed 1%.
Cell-to-cell conjugation efficiency test
Timing: 3 h (for step 23)
The following section describes the formation and subsequent chemical cross-linking of cell-to-cell conjugates. For the isolation of the conjugates, natural killer cells and target cells are marked by two different fluorescent labels. Labeling can be achieved using immunofluorescence staining or expression of recombinant fluorescent proteins. Here, NK-92MI cells are labeled using a PE-Cy7 conjugated anti-CD56 antibody prior to conjugate formation, while MDA-MB-231 cells are identified by mEmerald-actin-fusion.4 Both cell types are co-incubated for 10 min to allow the formation of cell-to-cell conjugates and fixed with paraformaldehyde (PFA). Double-labeled conjugates are identified and isolated using FACS.
-
23.
Calculate the number of cells required to reach the critical amount of cell-to-cell conjugates necessary for subsequent mass spectrometry analysis.
Note: An example is given below (Figure 2) following the method described in step 4 and step 5 with a 1:10 cell amount. In our experiment, we use the BD FACS Aria (Becton Dickinson Bioscience) for FACS analysis and sorting. Other instruments can be used, but the parameters have to be adjusted accordingly.
Figure 2.
Example of dot plot of PE-Cy7 and mEmerald fluorescence intensity following NK92-MI cell-MDA-MB-231 cell conjugation formation
Double positive (PE-Cy7+ mEmerald+) NK92-MI cell-MDA-MB-231 cell conjugates are selected. The evaluated percentage of cell-to-cell conjugates is 6,43%. Note: In our experimental conditions and using a ratio of NK92-MI cells:MDA-MB-231 cells of 3:1, we usually observed rate of 5%–10%.
Conjugate formation and chemical cross-linking
Timing: 3 h (for steps 24 to 28)
A minimum of 25 μg of cell-to-cell conjugates (approximately 106 conjugated cells) is necessary for mass spectrometry analysis. To calculate the number of NK-92MI cells and MDA-MB-231 cells that should be mixed in order to reach the critical number of cell-to-cell conjugates, both the NK-92MI:MDA-MB-231 cell ratio (i.e., 3:1) and the conjugation rate at this given ratio (i.e., 5%–10% of the total cell number) should be taken into account. It is important to test the cell-to-cell conjugation efficiency test first (conditions described in step 3) for further mass spectrometry experiments.
-
24.Staining of the NK-92 MI cells.
-
a.Count NK-92MI cells using a Neubauer chamber with trypan blue solution (follow the manufacturer’s instructions).
-
b.Transfer 35∗106 NK-92MI cells in a 15 mL falcon tube.
-
c.Centrifuge at 300 × g for 5 min.
-
d.Discard the supernatant and resuspend the cells in 10 mL of AutoMACS Running Buffer.
-
e.Centrifuge at 300 × g for 5 min.
-
f.Discard the supernatant and resuspend the cells at 10∗106 cells per mL.
-
g.Distribute 1 mL per 15 mL falcon tube.
-
h.Add 2.5 μL anti-human CD56-PECy7 antibody per million of cells and vortex briefly (2.5 μg/mL).
-
i.Incubate at 4°C for 30 min in the dark.
-
j.Add 10 mL of AutoMACS Running Buffer.
-
k.Centrifuge at 300 × g for 5 min.
-
l.Discard the supernatant and resuspend the cells at 9∗105 cells in 200 μL of RPMI light medium.
-
m.Incubate the cells for 15 min at 37°C in a cell culture incubator.
-
a.
Note: The required quantities should be adjusted according to the number of cells.
-
25.Detaching of the MDA-MB-231 cells.
-
a.Remove the cell culture medium.
-
b.Wash with 5 mL of DPBS.
-
c.Add 3 mL of Trypsin-EDTA and incubate for 2 min at 37°C in a cell culture incubator.
-
d.Stop trypsinization of the MDA-MB-231 cells by adding complete DMEM medium.
-
e.Count MDA-MB-231 cells and resuspend at 3∗105 cells in 200 μL in RPMI light culture medium.
-
a.
Note: The required quantities should be adjusted according to the number of cells.
-
26.NK-92MI cells and MDA-MB-231 cells co-culture, formation of the cell-to-cell conjugates.
-
a.Conjugates and control cells.
-
i.Cell-to-cell Conjugates: Mix 9∗105 NK92-MI (200 μL) and 3∗105 MDA-MB-231 (200 μL) in a round bottom tube (cell ratio 3:1). In total, 30∗106 NK92-MI cells are conjugated with 10∗106 MDA-MB-231 cells in 33 round bottom tubes.
-
ii.Control cells: As a control, keep unmixed cells distributed in the same conditions as the mixed cells (200μL in round-bottom tubes).
-
iii.Incubate 10 min at 37°C in a cell culture incubator.
-
i.
-
a.
Note: In our example, 5∗106 of MDA-MB-231 cells were distributed in 16 round bottom tubes at 3∗105 cells per tube, and 5∗106 of NK-92MI cells were distributed in 6 round bottom tubes at 9∗105 cells per tube.
Note: Excessive or ungentle centrifugation reduces the number of conjugated cells. See problem 2.
-
27.Stain for viability.
-
a.Add 1 mL of PBS/Mg/Ca per tube.
-
b.Centrifuge at 300 × g for 5 min.
-
c.Discard the supernatant.
-
d.Resuspend in PBS/Mg/Ca with 0.1× Live-or-Dye NucFix Red staining kit.
-
e.Incubate for 15 min at 4°C.
-
f.Add 1 mL of PBS/Mg/Ca per tube.
-
g.Centrifuge at 300 × g for 5 min.
-
a.
Note: The required quantities should be recalculated according to the number of cells.
Note: To reduce the risk of interfering with the subsequent steps, in particular mass spectrometric analysis, and recover unlabeled living cells, we recommend using (fixable) cell membrane impermeant viability dyes that specifically stain the nucleus of dead cells, such as Live-or-Dye NucFix™ Red.
Note: Live-or-Dye NucFix Red must be diluted in PBS/Mg/Ca to keep conjugates maintained.
Note: Resuspend by gently pipetting to avoid alteration of formed cell-to-cell conjugates.
-
28.Fix the cells.
-
a.Add 200 μL of fixation buffer per tube.
-
b.Incubate for 15 min at room temperature.
-
c.Add 1 mL of PBS/Mg/Ca per tube.
-
d.Centrifuge at 400 × g for 5 min.
-
e.Add 200 μL PBS/Mg/Ca per tube.
-
a.
Note: PFA must be diluted in PBS/Mg/Ca to keep conjugates maintained.
Note: The required quantities should be recalculated according to the number of cells.
Cell sorting
Timing: 1 h (for step 29)
Timing: 1 day (for step 30)
Step 5 describes the key steps for data acquisition and/or cell sorting. For further details on flow cytometry, the reader is referred to Cossarizza et al.15
- 29.
-
30.Isolation of control cells and cell-to-cell conjugates.
-
a.Pipette gently to resuspend the cell suspension.
-
b.Filter cell suspensions with 100μm cell strainers.
-
c.Sort the Cell-to-Cell conjugates: sort NK92-MI cell-MDA-MB-231 cell conjugates by selecting the double positive PE-Cy7+ mEmerald+ cell population (Figure 3).
-
d.Sort the Control cells: sort unmixed NK92-MI cells or unmixed MDA-MB-231 cells by selecting the single positive PE-Cy7+ cell population or single positive mEmerald+ cell population, respectively (Figure 4).
-
e.Centrifuge at 500 × g for 10 min.
-
f.Remove carefully as much supernatant as possible by pipette aspiration.
-
a.
Pause point: The pellets can be stored at -80°C.
Note: Recover at least 0.35 × 106 cells (cell-to-cell conjugates or control cells) from cell sorting.
Note: Pipet gently when resuspending cells to avoid dissociating cell-to-cell conjugates. If the sorting efficiency is below 30%, see problem 3.
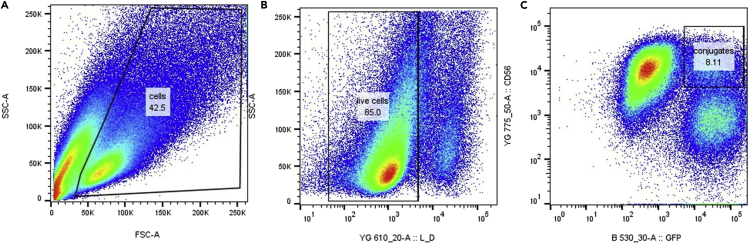
Figure 3.
Gating strategy for identification and isolation of NK92-MI cell-MDA-MB-231 conjugates
(A) Flow cytometry scatter FSC/SSC profile after co-incubation of effector and target cells. Cells are discriminated from cell debris based on size and granularity. The gate is intentionally extended (upper right corner) to maximize the number of cell-to-cell conjugates.
(B) Live cells are discriminated from dead cells based on Live-or-Dye NucFix™ Red staining.
(C) Double positive (PE-Cy7+ mEmerald+) NK92-MI -MDA-MB-231 cell conjugates are selected and sorted for subsequent mass spectrometry analysis.
Figure 4.
Gating strategy for identification and isolation of control cells
(A) Live NK92-MI cells are discriminated from dead NK92-MI cells based on Live-or-Dye NucFix™ Red staining (left panel). PE-Cy7+ NK cells are selected and sorted for subsequent mass spectrometry analysis (right panel).
(B) Live MDA-MB-231 cells are discriminated from dead MDA-MB-231 cells based on Live-or-Dye NucFix™ Red staining (left panel). mEmerald+ MDA-MB-231 cells are selected and sorted for subsequent mass spectrometry analysis (right panel).
CAUTION: This strategy allows selecting only NK92-MI cell-MDA-MB-231 cell conjugates, but the number of NK92-MI cells or MDA-MB-231 cells in the conjugate is uncertain. Several MDA-MB-231 and NK92-MI could be conjugated in the same cell complex. Filtering the cell suspension before the sorting will avoid larger aggregates of several cells.
Production of protein extracts and mass spectrometry analysis
Protein extraction, digestion, and cleaning
Timing: 2 days (for steps 31 to 34)
Proteins are separated on a pre-casted SDS gel into three fractions to decrease the complexity of the sample. A lower complexity will increase the number of identified phosphopeptides. The SDS-gel functions two-fold: first, the heating of the samples reverses the PFA cross-links, and second, it removes substances that are interfering with the mass spectrometric analysis. Proteins are digested into peptides in the SDS-gel, and the resulting peptides are extracted. The contained phospho-peptides are enriched using metallo-chelate chromatography before being analyzed by mass spectrometry.
Note: The starting material should not be below 0.35∗106 cell-to-cell conjugates, which approximately corresponds to 25 μg per sample.
-
31.SDS-Page setup.
-
a.Place the Bis-Tris gel in the tank.
-
b.Fill the tank with running buffer.
-
a.
-
32.Sample cleaning and fractionation by SDS-PAGE.
-
a.Resuspend the PFA-treated samples in 25 μL 2× Laemmli buffer and add 5 μL 10% SDS.
-
b.Vortex vigorous for 20 s.
-
c.Incubate 10 min at 95°C.
-
d.Put the samples on ice for 5 min.
-
e.Centrifuge for 10 min at maximum speed in a tabletop centrifuge (16,000 × g).
-
f.Load the samples and protein size standard in the different wells.
-
g.Program the power supply to 120 V constant per gel.
-
h.Run the gel until the running front reaches approximately half of the gel.
-
i.Stain the gel with Coomassie brilliant blue staining solution.
-
j.Cut each lane with a razor blade in three equal slices. Every slice should be cut into cubes of approximately 1 mm2 size to optimize further cleaning and increase penetration of the different reagents.
-
k.Place all cubes from every slice in a different 2 mL Eppendorf tube.
-
a.
Note: Proteins should be clearly fractionated into different bands. See problem 4.
Note: The whole sample should run through the gel with no remaining sample at the top of the well. See problem 5.
-
33.Protein digestion.
-
a.Wash the gel pieces with 400 μL of washing buffer A for 10 min in a thermomixer, 800 g, at RT.
-
b.Discard the supernatant.
-
c.Wash the gel pieces with 400 μL of washing buffer B for 10 min in a thermomixer, 800 × g, at RT.
-
d.Discard the supernatant.
-
e.Repeat the steps (33a to 33c) four times.
-
f.After discarding the supernatant, incubate the sample with 200 μL of reduction buffer for 20 min in a thermomixer at 800 × g.
-
g.Discard the supernatant.
-
h.Add 200 μL of alkylation buffer for 30 min in a thermomixer at 800 × g (in the dark) at RT.
-
i.Discard the supernatant.
-
j.Add 200 μL washing buffer B for 10 min in a thermomixer at 800 × g.
-
k.Discard the supernatant.
-
l.Add 150 μL digestion buffer.
-
m.Incubate the samples for 16 h at 37°C.
-
n.Stop the digestion by adding 15 μL of stop buffer (final concentration 0.1% formic acid).
-
o.Place the supernatant in a fresh 2 mL Eppendorf tube.
-
p.Add 150 μL of ST buffer C to the gel pieces and incubate the sample for 30 min at 800 g at RT.
-
q.Pool the supernatant from the gel pieces with the previous extraction in the Eppendorf tubes.
-
r.Dry the samples in a speed-vac for 2 h at 45°C.
-
a.
CAUTION: The coomassie brilliant blue dye from the gel pieces should vanish for optimal digestion. If the coomassie brilliant blue dye remains, add 3 more washes (step 33c).
CAUTION: The gel pieces should be wet in all the procedures; otherwise, the reproducibility of digestion will be compromised.
CRITICAL: If the gel pieces absorb all the buffer, add 50 μL digestion buffer. In case the digestion buffer is increased, the stop buffer should also be adjusted to a final concentration of 0.1% formic acid.
Pause point: The dried peptides can be stored for 1 week at -20°C.
-
34.Digest clean-up.
-
a.Acidify digest (∼50 μL) by adding ST buffer A (check pH < 3).
-
b.Punch out 2 discs of Octadecyl (C18)-bonded silica material with a flat tip gauge 16 needle.
-
c.Push the 2 discs into a GPS200 pipette tip using PEEK tubing.
-
d.Add 20 μL ST buffer A to the disc.
-
e.Centrifuge the tip at 1,000 × g for 1 min.
-
f.Add 20μL ST buffer C to the disc.
-
g.Centrifuge the tip at 1,000 × g for 1 min.
-
h.Add 20 μL ST buffer B to the disc.
-
i.Centrifuge the tip at 1,000 × g for 1 min.
-
j.Repeat steps 34f to 34i twice.
-
k.Load acidified sample into the tip.
-
l.Centrifuge the tip at 1,000 × g for 3 min.
-
m.Wash the sample that is retained in the tip with 20 μL ST buffer B.
-
n.Centrifuge the tip at 1,000 × g for 1 min.
-
o.Repeat steps 34m and 34n 3 times.
-
p.Elute bound peptides with 50 μL of ST buffer C.
-
q.Transfer eluate in measuring plate.
-
r.Snap freeze and lyophilize in speed vac.
-
a.
Pause point: The purified peptides can be stored one month at -80°C.
Phosphopeptide enrichment
Timing: 1 h (for steps 35 to 36)
Phosphopeptides are usually low abundant and are difficult to be detected in the mass spectrometer. Therefore, the phosphopeptides need to be enriched using a specific metallo-chelate chromatography.
-
35.
Reconstitute the samples in the starting buffer and perform the phosphopeptide enrichment as specified in High-Select Fe-NTA Phosphopeptide Enrichment Kit.
Note: this step enables the enrichment of phosphorylated peptides from protein extracts using iron-chelate resin in spin columns. The capacity of the columns ranges from 0.5 to 5 mg of total digested protein sample and can enrich up to 150 μg phosphopeptides with high selectivity.
-
36.
In the last step of the kit protocol, resuspend the material in 12 μL of 0.1% formic acid to inject into the mass spectrometer.
CRITICAL: check the color of the Fe3+-NTA beads. The color should be black. If the beads turn yellow, the beads are oxidized and will not work.
Note: if the analysis of the sample does not show enough phosphopeptides, See the troubleshooting of the High-Select Fe-NTA Phosphopeptide Enrichment Kit.
Pause point: The eluted peptides can be stored at -80°C for one month.
Mass spectrometry analysis (MS)
Timing: 1 day (dependent on the computer hardware available) (for step 37)
The recorded mass spectrometric data has to be matched to peptides and proteins. For this, we use the MaxQuant software package. MaxQuant demands significant computing resources. In our case, we used a 72-core computer to analyze the data. Using a smaller computer will prolong the time needed for the analysis.
-
37.
Inject 0.2–0.5 μg of material into an LC-MS/MS. It is very important to inject exactly the same amount in all the samples (peptide abundance measured by Pierce™ BCA Protein Assay Kit).
Note: the parameters that are described below are a guide for a specific mass spectrometer, but others could be used to perform MS analysis.
Optional: The amount of phosphopeptides to inject could differ depending on the peptide abundance, the mass spectrometer, and the nanoHPLC system. Those conditions have to be adjusted injecting the corresponding volume into LC-MS/MS. For a Q Exactive HF (Thermo Scientific) 0.1 μg of material is enough to generate a robust dataset (around 2,500 peptides per measurement, which corresponds to approximately 2 × 109 of maximum total ion count (TIC)).
To interpret the mass spectrometry data, use the MaxQuant software package (here, we use version 1.6.17.0) with the following search parameters. The rest of the MaxQuant parameters should be maintained as default for optimal data processing.12,16,17
| Parameter | Value |
|---|---|
| Version | 2.0.1.0 |
| Include contaminants | TRUE |
| PSM FDR | 0.01 |
| PSM FDR Crosslink | 0.01 |
| Protein FDR | 0.01 |
| Site FDR | 0.01 |
| Use Normalized Ratios For Occupancy | TRUE |
| Min. peptide Length | 7 |
| Min. score for unmodified peptides | 0 |
| Min. score for modified peptides | 40 |
| Min. delta score for unmodified peptides | 0 |
| Min. delta score for modified peptides | 6 |
| Min. unique peptides | 0 |
| Min. razor peptides | 1 |
| Min. peptides | 1 |
| Fixed modification | |
| Use only unmodified peptides and | TRUE |
| Modifications included in protein quantification | Oxidation (M);Acetyl (Protein N-term) |
| Peptides used for protein quantification | Razor |
| Discard unmodified counterpart peptides | TRUE |
| Label min. ratio count | 2 |
| Use delta score | FALSE |
| iBAQ | FALSE |
| iBAQ log fit | FALSE |
| Match between runs | TRUE |
| Matching time window [min] | 0.7 |
| Match ion mobility window [indices] | 0.05 |
| Alignment time window [min] | 20 |
| Alignment ion mobility window [indices] | 1 |
| Find dependent peptides | TRUE |
| Dependent peptide FDR | 0.01 |
| Mass bin size | 0.0065 |
| Decoy mode | revert |
| Include contaminants | TRUE |
| Advanced ratios | TRUE |
| Second peptides | TRUE |
| Stabilize large LFQ ratios | TRUE |
| Separate LFQ in parameter groups | FALSE |
| Require MS/MS for LFQ comparisons | TRUE |
| Calculate peak properties | FALSE |
| Main search max. combinations | 200 |
| Advanced site intensities | TRUE |
| Write msScans table | FALSE |
| Write msmsScans table | TRUE |
| Write ms3Scans table | TRUE |
| Write allPeptides table | TRUE |
| Write mzRange table | TRUE |
| Write DIA fragments table | FALSE |
| Write DIA fragments quant table | FALSE |
| Write pasefMsmsScans table | TRUE |
| Write accumulatedMsmsScans table | TRUE |
| Max. peptide mass [Da] | 4600 |
| Min. peptide length for unspecific search | 8 |
| Max. peptide length for unspecific search | 25 |
| Razor protein FDR | TRUE |
| Disable MD5 | FALSE |
| Max mods in site table | 3 |
| Match unidentified features | FALSE |
| Evaluate variant peptides separately | TRUE |
| Variation mode | None |
| MS/MS tol. (FTMS) | 20 ppm |
| Top MS/MS peaks per Da interval. (FTMS) | 12 |
| Da interval. (FTMS) | 100 |
| MS/MS deisotoping (FTMS) | TRUE |
| MS/MS deisotoping tolerance (FTMS) | 7 |
| MS/MS deisotoping tolerance unit (FTMS) | ppm |
| MS/MS higher charges (FTMS) | TRUE |
| MS/MS water loss (FTMS) | TRUE |
| MS/MS ammonia loss (FTMS) | TRUE |
| MS/MS dependent losses (FTMS) | TRUE |
| MS/MS recalibration (FTMS) | FALSE |
| MS/MS tol. (Unknown) | 20 ppm |
| Top MS/MS peaks per Da interval. (Unknown) | 12 |
| Da interval. (Unknown) | 100 |
| MS/MS deisotoping (Unknown) | TRUE |
| MS/MS deisotoping tolerance (Unknown) | 7 |
| MS/MS deisotoping tolerance unit (Unknown) | ppm |
| MS/MS higher charges (Unknown) | TRUE |
| MS/MS water loss (Unknown) | TRUE |
| MS/MS ammonia loss (Unknown) | TRUE |
| MS/MS dependent losses (Unknown) | TRUE |
| MS/MS recalibration (Unknown) | FALSE |
| Site tables | Deamidation (NQ)Sites.txt;Oxidation (M)Sites.txt;Phospho (STY)Sites.txt |
| Enzyme | Trypsin/P |
| Enzyme mode | Specific |
| Use enzyme first search | FALSE |
| Use variable modifications first search | FALSE |
| Requantify | TRUE |
| Multiplicity | 2 |
| Max. missed cleavages | 2 |
| Max. labeled AAs | 3 |
| Labels0 | |
| Labels1 | Arg10;Lys8 |
| LC-MS run type | Standard |
Note: The MaxQuant software has high demands on computer hardware. The runs may take a long time on a normal computer.
Bioinformatic analysis and data interpretation
Timing: 2 days (for steps 38 to 46)
In this step, the data is analyzed using statistical tools (Figure 5).
Figure 5.
Data analysis scheme
Cell types are SILAC labeled. Both the single cells and the cell-to-cell conjugates are measured by mass spectrometry. The two different SILAC isotope markers are used as different channels and compared to the single cells using LFQ quantification to identify cell type-specific events.
The MaxQuant search engine generates several files while processing the raw data from ThermoFisher instruments. To extract the files needed for the analysis: go to the combined folder, and then to the txt folder. The samples are analyzed by R-based programming or Perseus.18,19 Perseus is convenient for filtering data (114–116). For the quantification (117–121) we recommend R-based scripting to analyze the data. An example of the data analysis is shown in Table S1.
-
38.
Filter out the phosphopeptides belonging to proteins that are contaminants, reverse or identified by site in proteinGroups.txt file and Phospho (STY)Sites.txt file.
-
39.In Phospho (STY)Sites.txt file, Phosphopeptides with at least two individual SILAC ratios in each type of sample can be selected with the following threshold parameters:
-
a.score > 40.
-
b.score for localization > 40.
-
c.PEP <0.015.
-
a.
-
40.
Separate the filtered phosphopeptides by the two SILAC channels. In this case, the Light (L) channel corresponds to Natural Killer cells (NK92-MI), and the Heavy channel (H) corresponds to MDA-MB-231 cells.
Note: a minimum of two biological replicates have to be considered to calculate the abundance of every single phosphosite.
-
41.
The phosphosites are quantified using the Intensity that is differentially labeled by SILAC. To validate a significant change of phosphorylation, compare the Intensity H from the MDA-MB-231 cells (Intensity H MDA-MB-231) to Intensity H from the conjugates (Intensity H MDA-MB-231 - NK92-MI), or the Intensity L from NK92-MI cells (Intensity L NK92-MI) to the Intensity L from the conjugate (Intensity L MDA-MB-231 - NK92-MI).
-
42.
ONLY differences of the absolute value of
are considered as valid phosphorylation (positive) or dephosphorylation (negative). The rest of the phosphosites will be discarded as the signal is not sufficient to distinguish from background signals.
-
43.
The abundance changes in the phosphosites also depend on the differences in protein abundance between those two samples. We monitor this difference by Z-scoring of the LFQ Intensity H in proteingroups.txt file.
-
44.
will not be considered as protein abundance change.
-
45.
Proteins that include phosphosites changing in the same direction will be discarded.
-
46.
The SILAC label compares the protein abundance between MDA-MB-231 and NK92-MI in conjugates.
Note: the SILAC pairs in NK92-MI will be considered as an artifact (only L label in the sample), but the SILAC pair in MDAs (H label) will be considered to perform a quality control for labeling efficiency.
Expected outcomes
The provided datasets originate from an analysis using the described method. The data analysis of the dataset leads to the identification of 1197 phosphoproteins and 2800 phosphopeptides, which can be assigned to different cell types of cell-to-cell conjugates. Experiments using other cell types should identify phosphoproteins and -peptides in the same range.
Limitations
The analysis is based on SILAC-labeling to distinguish where the phosphorylated peptides originate. Thus, the method can only be used on cells that can be SILAC-labeled. The number of cell-to-cell conjugates formed by the two cell populations can vary depending on the cell type and might be a limiting factor for some cell type combinations. If one cell type can form homotypic cell-to-cell conjugates, the strategy for FACS needs to be adjusted and fitted to select conjugated cells from single cells. A minimal number of phosphorylated peptides are necessary for the mass spectrometric analysis to get sufficient identifications. Depending on the conjugation efficiency, the amounts have to be adjusted. In general, more replicates will lead to a more robust coverage of the phospho-proteome.
Troubleshooting
Problem 1
The labeling efficiency of the cells is <95% due to problems with the dialyzed serum or the number of passages is not enough to incorporate all the labeled amino acids (steps 1 and 2).
Potential solution
Wash cells with PBS and perform 4 more passages with a fresh dialyzed serum before a new test. Check if the SILAC media does contain the right amount of SILAC amino acids for labeling.
Problem 2
Centrifugation and pipetting before the fixation step could alter cell-to-cell conjugates (steps 26–28). The sheer forces within the pipette can separate cells from each other.
Potential solution
Pipet gently and limit washing steps by pre-labeling cells for viability before NK92-MI cells and MDA-MB-231 conjugation. The 10 min co-culture time is insufficient to induce strong NK92-MI cell cytotoxic activity, specifically with MDA-MB-231 characterized by their resistance to NK92-MI cell attack.
Problem 3
The sorting efficiency (for both NK92-MI cell-MDA-MB-231 cell conjugates and unmixed cells), as defined by the number of positive events sorted divided by the number of positive events detected, has to be larger than 30%.
Potential solution
The sorting efficiency depends on several parameters that could be modified to improve it:
-
•
Aggregates formed by cell clustering can be limited by a filtration step with 100 cell strainers before the sorting.
-
•
The cell concentration per mL is important to limit the exclusion of positive events because of too close proximity to a negative event. The cell suspension could be diluted to limit this phenomenon.
-
•
A first sorting can improve the percentage of positive events in the cell suspension based on yield and not purity. The machine’s settings on yield mode allow the recovery of all positive events, despite the proximity to negative events that could be included in the sorted drop. The recovered cell suspension is enriched in positive events and can be sorted again on purity mode. The machine will exclude all drops that could contain any non-target events.
Problem 4
There is no clean separation between the different bands in the SDS-PAGE.
Potential solution
Generally, this is due to the high concentration of PFA or high amounts of DNA in the sample. Add 8 μL of 10% SDS instead of 5 μL and boil for 15 min at 95°C. Use sonication to break down the DNA or add 5 μL of Benzonase for 20 min at RT.
Problem 5
The sample is not homogeneous, and some of the samples is not passing through the gel or is trapped in the gel pocket.
Potential solution
The sample was not soluble because of the cell debris. Perform an ultra-centrifugation at 100,000 g for 20 min after boiling the sample, recovering the non-solid phase (upper phase) before loading it onto the SDS-PAGE.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gunnar Dittmar (gunnar.dittmar@lih.lu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
C.T. and L.F. are supported by the Luxembourg National Research Fund (ACTIVASION, C19/BM/13579644) and La Fondation Cancer (Luxembourg, ACTIMMUNE, FC/2019/02). We acknowledge the National Cytometry Platform (NCP) for assistance with the single-cell sorting. The NCP is supported by funding from Luxembourg’s Ministry of Higher Education and Research (MESR).
Author contributions
D.P.H. and L.F. performed the experiments and developed the protocols. C.T. and G.D. conceptualized and supervised the project. D.P.H. and L.F. wrote the manuscript draft. D.P.H., L.F., C.T., and G.D. wrote the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102104.
Contributor Information
Daniel Perez-Hernandez, Email: daniel.perezhernandez@lih.lu.
Gunnar Dittmar, Email: gunnar.dittmar@lih.lu.
Supplemental information
Data and code availability
The mass spectrometric dataset generated during this study are available at MASSIVE proteomeXchange: MSV000090645. ftp://massive.ucsd.edu/MSV000090645/
References
- 1.Dustin M.L. The immunological synapse. Cancer Immunol. Res. 2014;2:1023–1033. doi: 10.1158/2326-6066.CIR-14-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orange J.S. Formation and function of the lytic NK-cell immunological synapse. Nat. Rev. Immunol. 2008;8:713–725. doi: 10.1038/nri2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wurzer H., Hoffmann C., Al Absi A., Thomas C. Actin cytoskeleton straddling the immunological synapse between cytotoxic lymphocytes and cancer cells. Cells. 2019;8 doi: 10.3390/cells8050463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al Absi A., Wurzer H., Guerin C., Hoffmann C., Moreau F., Mao X., Brown-Clay J., Petrolli R., Casellas C.P., Dieterle M., et al. Actin cytoskeleton remodeling drives breast cancer cell escape from natural killer-mediated cytotoxicity. Cancer Res. 2018;78:5631–5643. doi: 10.1158/0008-5472.CAN-18-0441. [DOI] [PubMed] [Google Scholar]
- 5.Biolato A.M., Filali L., Wurzer H., Hoffmann C., Gargiulo E., Valitutti S., Thomas C. Actin remodeling and vesicular trafficking at the tumor cell side of the immunological synapse direct evasion from cytotoxic lymphocytes. Int. Rev. Cell Mol. Biol. 2020;356:99–130. doi: 10.1016/bs.ircmb.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Wurzer H., Filali L., Hoffmann C., Krecke M., Biolato A.M., Mastio J., De Wilde S., François J.H., Largeot A., Berchem G., et al. Intrinsic resistance of chronic lymphocytic leukemia cells to NK cell-mediated lysis can Be overcome in vitro by pharmacological inhibition of Cdc42-induced actin cytoskeleton remodeling. Front. Immunol. 2021;12:619069. doi: 10.3389/fimmu.2021.619069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diella F., Haslam N., Chica C., Budd A., Michael S., Brown N.P., Trave G., Gibson T.J. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 8.Friedl P., den Boer A.T., Gunzer M. Tuning immune responses: diversity and adaptation of the immunological synapse. Nat. Rev. Immunol. 2005;5:532–545. doi: 10.1038/nri1647. [DOI] [PubMed] [Google Scholar]
- 9.Lanzavecchia A. Antigen-specific interaction between T and B cells. Nature. 1985;314:537–539. doi: 10.1038/314537a0. [DOI] [PubMed] [Google Scholar]
- 10.Ong S.-E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Deng J., Erdjument-Bromage H., Neubert T.A. Quantitative comparison of proteomes using SILAC. Curr. Protoc. Protein Sci. 2019;95:e74. doi: 10.1002/cpps.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 13.Tyanova S., Mann M., Cox J. MaxQuant for in-depth analysis of large SILAC datasets. Methods Mol. Biol. 2014;1188:351–364. doi: 10.1007/978-1-4939-1142-4_24. [DOI] [PubMed] [Google Scholar]
- 14.Ong S.E., Foster L.J., Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/S1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 15.Cossarizza A., Chang H., Radbruch A., Acs A., Adam D., Adam-Klages S., Agace W.W., Aghaeepour N., Akdis M., Allez M., et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition) Eur. J. Immunol. 2019;49:1457–1973. doi: 10.1002/eji.201970107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J.V., Mann M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 17.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 18.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 19.Tyanova S., Cox J. In: Cancer Systems Biology Methods in Molecular Biology. von Stechow L., editor. Springer New York; 2018. Perseus: a bioinformatics platform for integrative analysis of proteomics data in cancer research; pp. 133–148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometric dataset generated during this study are available at MASSIVE proteomeXchange: MSV000090645. ftp://massive.ucsd.edu/MSV000090645/