Abstract
Acetylcholinesterase (AChE) is an important enzyme and target for human therapeutics, environmental safety, and global food supply. Inhibitors of this enzyme are also used for pest elimination and can be misused for suicide or chemical warfare. Adverse effects of AChE pesticides on nontarget organisms, such as fish, amphibians, and humans, have also occurred as a result of biomagnifications of these toxic compounds. We have exhaustively curated the public data for AChE inhibition data and developed machine learning classification models for seven different species. Each set of models were built using up to nine different algorithms for each species and Morgan fingerprints (ECFP6) with an activity cutoff of 1 μM. The human (4075 compounds) and eel (5459 compounds) consensus models predicted AChE inhibition activity using external test sets from literature data with 81% and 82% accuracy, respectively, while the reciprocal cross (76% and 82% percent accuracy) was not species-specific. In addition, we also created machine learning regression models for human and eel AChE inhibition to return a predicted IC50 value for a queried molecule. We did observe an improved species specificity in the regression models, where a human support vector regression model of human AChE inhibition (3652 compounds) predicted the IC50s of the human test set to a better extent than the eel regression model (4930 compounds) on the same test set, based on mean absolute percentage error (MAPE = 9.73% vs 13.4%). The predictive power of these models certainly benefits from increasing the chemical diversity of the training set, as evidenced by expanding our human classification model by incorporating data from the Tox21 library of compounds. Of the 10 compounds we tested that were predicted active by this expanded model, two showed >80% inhibition at 100 μM. This machine learning approach therefore offers the ability to rapidly score massive libraries of molecules against the models for AChE inhibition that can then be selected for future in vitro testing to identify potential toxins. It also enabled us to create a public website, MegaAChE, for single-molecule predictions of AChE inhibition using these models at megaache.collaborationspharma.com.
Introduction
Acetylcholinesterase (AChE) is the primary cholinesterase in the body and is an enzyme that catalyzes the breakdown of acetylcholine (ACh) and of some other choline esters that function as neurotransmitters. AChE is found mainly at neuromuscular junctions and in chemical synapses of the cholinergic type, where its activity serves to terminate synaptic transmission. AChE belongs to the carboxylesterase family of enzymes and is the primary inhibition target of organophosphorus (OP) pesticides and nerve agents.1 AChE hydrolyzes choline esters and has a very high catalytic activity, with each molecule of AChE degrading ∼25,000 molecules of ACh per second, approaching the limit allowed by diffusion of the substrate.2,3 OPs irreversibly and noncompetitively inhibit AChE, causing poisoning by phosphorylating the serine hydroxyl residue on AChE, which inactivates AChE.4 AChE has been implicated in a multitude of biological functions, including, but not limited to, neurite outgrowth, bone development, and regulation of proliferation and apoptosis in hematopoietic stem cells and of amyloid fibril formation.5−9 AChE is also critical for nerve function; hence, the inhibition of this enzyme causes ACh accumulation and results in muscle overstimulation. Inactivation of AChE in nerve synapses can therefore be lethal while OP-inhibited AChE can be reactivated with oximes, provided the OP has not aged.10 Two classes of ACh receptors are activated differently in the body, namely, nicotinic receptors and muscarinic receptors. Overstimulation of nicotinic acetylcholine receptors in the central nervous system, due to accumulation of ACh, results in anxiety, headache, convulsions, ataxia, depression of respiration and circulation, tremor, general weakness, and potentially coma.11,12 When there is expression of muscarinic overstimulation due to excess ACh at muscarinic acetylcholine receptors there can be symptoms of visual disturbances, tightness in the chest, wheezing due to bronchoconstriction, increased bronchial secretions, increased salivation, lacrimation, sweating, and peristalsis, and urination can occur.13
AChE is also a validated therapeutic target in the symptomatic treatment of Alzheimer’s disease to treat cognitive deficiency.14 A number of FDA approved AChE inhibitor drugs including tacrine, rivastigmine, donepezil, and galantamine are widely prescribed for this disease. Our understanding of AChE inhibition has been enhanced by various crystal structures and the computational design of drugs addressing this target at both the catalytic and peripheral anionic site.15 For example, donepezil occupies an anionic subsite in the active site gorge by engaging in π–π stacking and cation−π interactions.16 Synthetic efforts employing medicinal chemistry have been a fertile source of multiple classes of AChE inhibitors,15 which in turn have been employed to build a wide array of quantitative structure activity relationship (QSAR) machine learning models.17−24 Additional modeling efforts have employed different methodologies such as molecular docking,25−28 structure-based pharmacophore modeling,29 machine learning,30 2D and 3D similarity searches,31 MIA-QSAR modeling,32 or even combinations of these strategies.33−35 Based on these and other previously published studies, such computational methods can be used to help identify AChE inhibitors that may have deleterious effects on human or other species health with ultimate impacts on the environment.
Inhibition of recombinant or purified AChE is often used as a predictor of toxicity in vitro and appears in off-target safety panels used by commercial companies for drug discovery.36 The EPA Office of Pesticide Programs uses cholinesterase inhibition in risk assessments of carbamate and organophosphorus pesticides. The majority of the original 309 compounds compiled by the EPA in the ToxCast Phase 1 chemical library were pesticides or the active ingredients thereof,37 and a recent screen of the ten-thousand-compound Tox21 library used three different in vitro HTS assays to identify AChE inhibitors.38 Despite divergence in primary sequences, and several duplication events in invertebrates, the 3D structure of the enzyme is mostly shared across species.39−43 Although the electric eel AChE is regularly used in the search for human drugs,44−53 some studies have documented interspecies differences in inhibition for certain classes of compounds.54−56
In this current study, we have compiled AChE inhibition data from seven different species using publicly available information and then used our Assay Central machine learning software57,58 to build classification and regression models using 15 different algorithms with ECFP6 fingerprints. We have also explored additional more recently described machine learning algorithms. These models were validated in most cases using nested, 5-fold cross-validation, an external data set from the literature, as well as prospective testing and have resulted in the production of a public website for the use of these models.
Experimental Procedures
Data Sets
Data Curation
Data for AChE inhibition (IC50) were downloaded from ChEMBL 3059 for human (CHEMBL220), eel (CHEMBL4078), cow (CHEMBL4768), rat (CHEMBL3199), ray (CHEMBL4780), mosquito (CHEMBL2046266), and mouse (CHEMBL3198). Additional entries for human AChE inhibition were obtained from BindingDB,60 but these were limited to those that did not list ChEMBL as their source. Entries without a numerical value in the “IC50” value column were removed. Entries for which the assay description listed an organism other than the one listed under “Target Organism” were removed. Entries without a species of origin listed in the assay description were further investigated in the primary literature, under the listed Document ID. If the species of origin for the AChE inhibition data in the citation differed from the Target Organism, or if there was no species of origin given in the primary literature, these entries were removed. We did not distinguish between method of assay listed but retained all entries that described AChE inhibition. Relational IC50 values (i.e., IC50 > 1000 nM) were retained for the classification but not for regression models. Due to this, the data set sizes for the classification and regression models were variant. We next put each data set through our proprietary software “E-Clean”, which uses open-source RDKit tools, to remove duplicate compounds and salt as well as neutralize charges. For regression models, the IC50 values were converted to −log M, and then averaged for duplicate compounds. For classification models, −log M values were binarized on a threshold of 1 μM (−log M = 6) and assigned a binary activity. The binary activities of duplicate compounds were averaged, and those with an average activity ≥0.5 were assigned an activity of 1. Both continuous (regression) and binarized (classification) data sets were then further standardized within the Assay Central software using the Indigo Toolkit as described previously,58 and final data sets were created for model training (See Supplemental Datafiles). The “uncurated” human data set is composed of all entries in ChEMBL 30 with pChEMBL values listed under human AChE (CHEMBL220), excluding any entries with text in the “Data Validity Comment” column of the spreadsheet. This includes anything flagged as “Non standard unit for type”, “Outside typical range”, “Potential missing data”, or “Potential transcription error”. The entries were then cleaned using E-Clean and the Indigo Toolkit in Assay Central, as per the curated data sets. The classification data set was binarized as per the specifications above.
Data Curation for Human and Eel Test Sets
A search of PubMed Central was performed searching for novel AChE inhibitors published between 2017 and 2021 that had not previously been recorded in ChEMBL. These human and eel test sets were composed of 208 and 203 compounds, respectively, from >20 different papers each (Supporting Information). The compounds in both test sets were absent from their respective training data sets based on INCHI key comparisons.
Incorporating Tox21 Screen Data
Three simultaneous high-throughput screens of Tox21 compounds were performed recently in a search to find AChE inhibitors, investigating the inhibition of AChE in assays using either cells, human liver microsomes, or enzyme-only conditions.38 We obtained these data from these screens from PubChem bioassays (1347395, 1347399, and 1347397, respectively) and also compiled a list of compounds that were scored “inactive” in all three screens [i.e., assigned them IC50 values of “> 100 μM” (−log M = 4)]. We then processed this list using “E-Clean” and added these compounds (6455 postprocessing) to our human ChEMBL and BindingDB training data. We also included 177 IC50 values from the enzyme-only follow-up assay in this study. The resulting classification model consisted of 10,382 unique compounds.
Assay Central
Computing
All computing was conducted with the same setup as previously described.58
Model Building
The Assay Central software builds classification models using eight different machine learning algorithms, as described previously.58 For model internal validation we used nested, 5-fold internal cross-validation for the AdaBoosted decision trees (ADA), Bernoulli naïve Bayes (BNB), K-nearest neighbors (kNN), LogisticRegression (LREG), random forest (RF), support vector classification (SVC), and XGBoost (XGB) algorithms, and a 20% leave-out set for the deep learning 3 layers (DL) algorithm.
Model Validation
Applicability Domains
Applicability domain (AD) scores for classification models were calculated using a reliability-density neighborhood (RDD) algorithm described previously.61 Compounds that received an AD score of 1 were in the model and were retroactively removed from the test set.
t-SNE Visualization
This was performed as previously described.58
Statistical Analysis
Statistical analyses for the internal cross-validation were performed in Assay Central, and statistical analysis for external validation was performed in Microsoft Excel. Statistical analysis for linear regression, nonlinear regression [log(inhibitor) vs response–variable slope (four parameters), least-squares regression], Pearson correlation, Welch’s t test, and Kruskal–Wallis ANOVAs were performed in GraphPad Prism 9.4.1.
Library Scoring
We downloaded several libraries from the EPA CompTox Chemicals Dashboard for scoring with our AChE inhibition models including the KEMI Market List (30,418 compounds), the HBM4 EU CEC Screen (70,000 compounds and 300,000 predicted metabolites), and the 62,000 compound NORMAN SusDat list. We also scored 11,000 PFAS compounds compiled from 37 individual chemical lists on the CompTox dashboard as well as 12 additional chemical lists.62 We did not screen the predicted metabolites of the CEC list due to the inability to purchase compounds for verification.
In Vitro Validation
We performed AChE inhibition studies using the Abcam Acetylcholinesterase Inhibitor Screening Kit [ab283363, previously BioVision K197-100 (Colorimetric)] with modifications, as described previously.57 Briefly, compounds were evaluated for their ability to inhibit the AChE hydrolysis of acetylthiocholine to thiocholine in the presence of 5,5′-dithiobis-2-nitrobenzoic acid (DNTB), which forms the colorimetric 5-thio-2-nitrobenzoic acid (TNB) anion.63 Kinetic assays were performed, measuring the change in absorbance at 412 nM over time (slope), and percent inhibition was calculated in regard to the solvent control (SC) of 1% DMSO. %Inhibition = (slope of SC – slope of (C))/slope of SC × 100. Reported percent inhibition is the average of at least two technical replicates ± standard deviation. Dose–response curves were performed in technical triplicate.
Results
Data Curation
After significant pruning of the AChE data sets for each species, we had curated training sets which varied substantially in size by species (Table 1). Human and eel data sets were the largest with the final sets having 4075 and 5459, respectively. With the exception of a subset of compounds for human AChE inhibition found in BindingDB, all of the compound IC50 data for the initial models were downloaded from ChEMBL. Close to two thousand entries were removed from the human data set based on filter criteria, with the overwhelming majority being removed due to them having an undisclosed species in their primary source material. During this curation step we also found instances where the species was mislabeled in the ChEMBL data set, and at multiple stages during the model building and validation stages we returned to the source data to find that data was mislabeled or mistranscribed from the source material. Due to a transposition error in the human data set which we discovered, this heavily skewed the range of the data set; we therefore carefully investigated the original source material for every entry flagged by ChEMBL “outside normal values”, and we corrected or discarded those that were incorrect. We supplemented the ChEMBL data for the human data set with unique entries for human AChE inhibition from BindingDB.
Table 1. Curation of ChEMBL IC50 Data for Classification Models of Species-Specific Inhibition of AChE.
| organism | number of IC50 entries downloaded from ChEMBL | number remaining after entries—manual curation | number of remaining compounds after cleaning | number remaining of actives/inactives at 1 μM threshold |
|---|---|---|---|---|
| Human (Homo sapiens) | 8205 (plus 706 from BindingDB) | 4749 (plus 200 from BindingDB) | 4075 | 1813/2262 |
| Eel (Electrophorus electricus) | 6534 | 6265 | 5459 | 2084/3375 |
| Rat (Rattus norvegicus) | 1764 | 1653 | 1406 | 687/719 |
| Mouse (Mus musculus) | 529 | 429 | 368 | 145/223 |
| Cow (Bos taurus) | 616 | 599 | 457 | 239/218 |
| Ray (Torpedo californica) | 430 | 385 | 307 | 156/151 |
| Mosquito (Anopheles gambiae) | 139 | 102 | 72 | 27/45 |
Machine Learning Model Building: Classification Models
We built classification models for each of the seven species with AChE data listed in Table 2 using 8 different machine learning algorithms ADA, BNB, kNN, LREG, RF, SVC, XGB, and DL. We also performed a comparison of our descriptor-based models against a more recent machine learning algorithm called AttentiveFP64 using the larger eel and human data sets. AttentiveFP is a graph-based model which has showed improved results over descriptor-based models in a number of tasks. We used the DeepChem65 Library for the AttentiveFP implementation, keeping the hyperparameters default except for dropout, which we set to 0.2 based on model performance in the original paper. Internal cross-validation of the models was performed using nested, 5-fold cross-validation with the exception of DL, which used a 20% leave-out set, and AttentiveFP, which used 5-fold cross-validation (Table 2). These 5-fold cross-validation statistics looked very good overall, with most AUC and F1 scores >0.9 across algorithms for each organism except mosquito, which were created from the data set with the fewest number of compounds The AttentiveFP models did not outperform our descriptor-based models. This is not surprising, as more complex models often require larger amounts of data than what we typically have, which is often <10,000 data points, in order to outperform descriptor-based models.66
Table 2. Nested, 5-Fold Cross-Validation Statistics of Classification Models of AChE Inhibition at 1 μM Threshold for Seven Speciesa.
| Human AChE Training Set: 1813 Active/4075 Total Compounds | ||||||||
|---|---|---|---|---|---|---|---|---|
| model | AUC | F1 score | precision | recall | accuracy | specificity | Cohen’s kappa | MCC |
| DLb | 0.93 | 0.84 | 0.84 | 0.83 | 0.85 | 0.87 | 0.70 | 0.70 |
| ADA | 0.91 | 0.80 | 0.81 | 0.80 | 0.82 | 0.85 | 0.64 | 0.64 |
| BNB | 0.84 | 0.73 | 0.71 | 0.75 | 0.75 | 0.76 | 0.51 | 0.51 |
| kNN | 0.92 | 0.84 | 0.81 | 0.87 | 0.85 | 0.84 | 0.71 | 0.71 |
| LREG | 0.91 | 0.82 | 0.82 | 0.81 | 0.84 | 0.86 | 0.67 | 0.67 |
| RF | 0.94 | 0.84 | 0.86 | 0.81 | 0.86 | 0.90 | 0.71 | 0.72 |
| SVC | 0.94 | 0.86 | 0.86 | 0.85 | 0.87 | 0.89 | 0.74 | 0.74 |
| XGB | 0.93 | 0.85 | 0.85 | 0.85 | 0.87 | 0.88 | 0.73 | 0.73 |
| AttentiveFPc | 0.75 | 0.76 | 0.72 | 0.83 | 0.75 | 0.74 | 0.43 | 0.43 |
| Eel AChE Training Set: 2084 Active/5459 Total Compounds | ||||||||
|---|---|---|---|---|---|---|---|---|
| model | AUC | F1 score | precision | recall | accuracy | specificity | Cohen’s kappa | MCC |
| DLb | 0.90 | 0.79 | 0.76 | 0.82 | 0.83 | 0.84 | 0.65 | 0.66 |
| ADA | 0.91 | 0.78 | 0.81 | 0.76 | 0.84 | 0.89 | 0.66 | 0.66 |
| BNB | 0.85 | 0.71 | 0.74 | 0.69 | 0.79 | 0.85 | 0.55 | 0.55 |
| kNN | 0.93 | 0.84 | 0.82 | 0.85 | 0.87 | 0.89 | 0.73 | 0.73 |
| LREG | 0.91 | 0.80 | 0.81 | 0.80 | 0.85 | 0.88 | 0.68 | 0.68 |
| RF | 0.94 | 0.84 | 0.86 | 0.81 | 0.88 | 0.92 | 0.74 | 0.74 |
| SVC | 0.93 | 0.84 | 0.83 | 0.84 | 0.87 | 0.89 | 0.73 | 0.73 |
| XGB | 0.93 | 0.83 | 0.84 | 0.82 | 0.87 | 0.90 | 0.73 | 0.73 |
| AttentiveFPc | 0.86 | 0.82 | 0.83 | 0.82 | 0.87 | 0.76 | 0.53 | 0.53 |
| Rat AChE Training Set: 687 Active/1406 Total Compounds | ||||||||
|---|---|---|---|---|---|---|---|---|
| model | AUC | F1 score | precision | recall | accuracy | specificity | Cohen’s kappa | MCC |
| DLb | 0.95 | 0.87 | 0.85 | 0.90 | 0.87 | 0.85 | 0.74 | 0.75 |
| ADA | 0.92 | 0.84 | 0.86 | 0.82 | 0.85 | 0.87 | 0.69 | 0.69 |
| BNB | 0.87 | 0.80 | 0.82 | 0.78 | 0.81 | 0.84 | 0.62 | 0.62 |
| kNN | 0.93 | 0.86 | 0.83 | 0.89 | 0.86 | 0.83 | 0.72 | 0.72 |
| LREG | 0.94 | 0.86 | 0.86 | 0.86 | 0.86 | 0.87 | 0.72 | 0.72 |
| RF | 0.94 | 0.87 | 0.87 | 0.87 | 0.87 | 0.87 | 0.74 | 0.74 |
| SVC | 0.93 | 0.87 | 0.85 | 0.88 | 0.87 | 0.86 | 0.73 | 0.73 |
| XGB | 0.94 | 0.87 | 0.87 | 0.87 | 0.87 | 0.87 | 0.74 | 0.74 |
| Mouse AChE Training Set: 145 Active/368 Total Compounds | ||||||||
|---|---|---|---|---|---|---|---|---|
| model | AUC | F1 score | precision | recall | accuracy | specificity | Cohen’s kappa | MCC |
| DLb | 0.95 | 0.89 | 0.96 | 0.83 | 0.92 | 0.98 | 0.83 | 0.83 |
| ADA | 0.93 | 0.84 | 0.86 | 0.83 | 0.88 | 0.91 | 0.74 | 0.75 |
| BNB | 0.89 | 0.79 | 0.82 | 0.78 | 0.84 | 0.88 | 0.66 | 0.67 |
| kNN | 0.90 | 0.85 | 0.82 | 0.90 | 0.88 | 0.86 | 0.75 | 0.76 |
| LREG | 0.94 | 0.86 | 0.84 | 0.88 | 0.88 | 0.89 | 0.76 | 0.76 |
| RF | 0.95 | 0.86 | 0.87 | 0.86 | 0.89 | 0.91 | 0.77 | 0.77 |
| SVC | 0.93 | 0.87 | 0.84 | 0.91 | 0.89 | 0.88 | 0.78 | 0.78 |
| XGB | 0.93 | 0.87 | 0.87 | 0.87 | 0.89 | 0.91 | 0.78 | 0.78 |
| Cow AChE Training Set: 239 Active/457 Total Compounds | ||||||||
|---|---|---|---|---|---|---|---|---|
| model | AUC | F1 score | precision | recall | accuracy | specificity | Cohen’s kappa | MCC |
| DLb | 0.94 | 0.85 | 0.89 | 0.81 | 0.85 | 0.89 | 0.70 | 0.70 |
| ADA | 0.92 | 0.87 | 0.88 | 0.86 | 0.87 | 0.87 | 0.73 | 0.73 |
| BNB | 0.93 | 0.87 | 0.89 | 0.85 | 0.87 | 0.89 | 0.73 | 0.74 |
| kNN | 0.92 | 0.88 | 0.86 | 0.90 | 0.87 | 0.84 | 0.74 | 0.74 |
| LREG | 0.94 | 0.89 | 0.90 | 0.88 | 0.89 | 0.89 | 0.77 | 0.77 |
| RF | 0.94 | 0.88 | 0.91 | 0.85 | 0.88 | 0.90 | 0.76 | 0.76 |
| SVC | 0.94 | 0.89 | 0.90 | 0.88 | 0.89 | 0.89 | 0.77 | 0.77 |
| XGB | 0.93 | 0.88 | 0.89 | 0.88 | 0.88 | 0.88 | 0.75 | 0.76 |
| Ray AChE Training Set: 156 Active/307 Total Compounds | ||||||||
|---|---|---|---|---|---|---|---|---|
| model | AUC | F1 score | precision | recall | accuracy | specificity | Cohen’s kappa | MCC |
| DLb | 0.95 | 0.89 | 0.88 | 0.91 | 0.89 | 0.87 | 0.77 | 0.77 |
| ADA | 0.93 | 0.87 | 0.92 | 0.83 | 0.88 | 0.93 | 0.75 | 0.76 |
| BNB | 0.89 | 0.83 | 0.81 | 0.85 | 0.82 | 0.80 | 0.65 | 0.65 |
| kNN | 0.92 | 0.88 | 0.92 | 0.85 | 0.89 | 0.92 | 0.77 | 0.78 |
| LREG | 0.94 | 0.89 | 0.91 | 0.87 | 0.89 | 0.91 | 0.78 | 0.78 |
| RF | 0.94 | 0.87 | 0.94 | 0.82 | 0.88 | 0.95 | 0.77 | 0.78 |
| SVC | 0.95 | 0.89 | 0.92 | 0.86 | 0.89 | 0.93 | 0.79 | 0.79 |
| XGB | 0.94 | 0.90 | 0.91 | 0.88 | 0.90 | 0.91 | 0.79 | 0.79 |
| Mosquito AChE Training Set: 27 Active/72 Total Compounds | ||||||||
|---|---|---|---|---|---|---|---|---|
| model | AUC | F1 score | precision | recall | accuracy | specificity | Cohen’s kappa | MCC |
| DLb | 1 | 0.67 | 1 | 0.50 | 0.80 | 1 | 0.55 | 0.61 |
| ADA | 0.83 | 0.73 | 0.79 | 0.71 | 0.81 | 0.87 | 0.59 | 0.60 |
| BNB | 0.85 | 0.56 | 0.61 | 0.55 | 0.71 | 0.78 | 0.33 | 0.35 |
| kNN | 0.88 | 0.80 | 0.85 | 0.82 | 0.85 | 0.87 | 0.69 | 0.71 |
| LREG | 0.86 | 0.77 | 0.86 | 0.75 | 0.82 | 0.89 | 0.65 | 0.67 |
| RF | 0.88 | 0.81 | 0.88 | 0.78 | 0.85 | 0.91 | 0.70 | 0.72 |
| SVC | 0.87 | 0.75 | 0.71 | 0.82 | 0.81 | 0.80 | 0.60 | 0.61 |
| XGB | 0.82 | 0.70 | 0.83 | 0.64 | 0.79 | 0.89 | 0.55 | 0.58 |
DL = deep learning; ADA = AdaBoosted decision trees; BNB = Bernoulli naïve Bayes; kNN = K-nearest neighbors; LREG = LogisticRegression; RF = random forest; SVC = support vector classification; XGB = XGBoost.
Statistics for a 20% leave-out set.
Statistics for 5-fold cross-validation.
External Model Validation of Classification Models
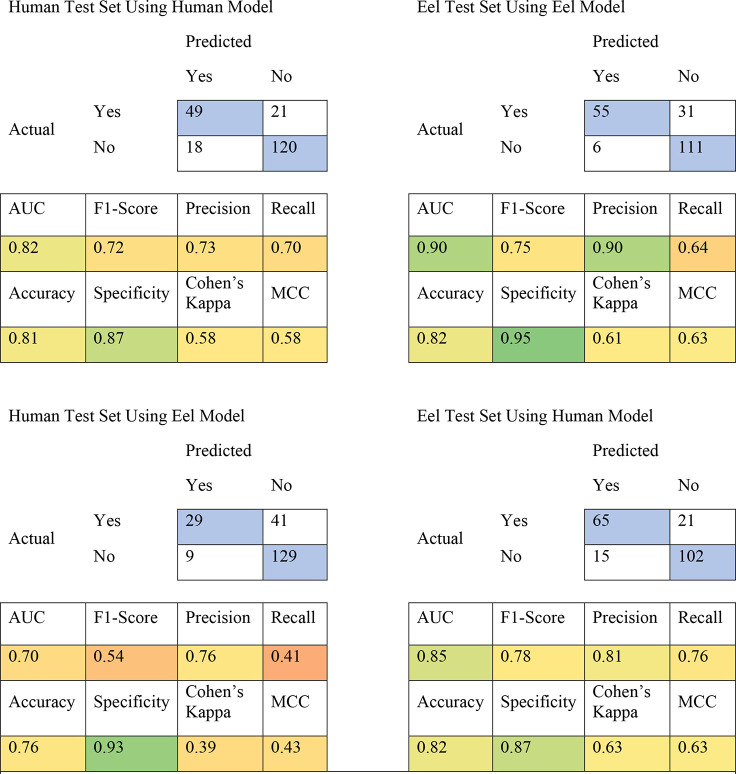
External validation for the human and eel models was performed using test sets manually curated from the literature for inhibitors of human and eel AChE. We predicted these test sets against consensus models composed of all eight classification models for each species, and we saw that the human and eel consensus models were able to predict activity in the human and eel test sets with 81% and 82% accuracy, respectively (Table 3). While this indicated an excellent predictive power of these models, the reciprocal crosses (e.g., eel model vs human test set and vice versa) showed accuracies just as high. This demonstrates that while these models are good at predicting general AChE inhibition accurately, there is no appreciable species specificity between these models. This would suggest that either species could be used to predict the classification of a molecule for the other species.
Table 3. Truth Tables for Human and Eel AChE Test Sets Predicted against Human and Eel Consensus Modelsa.
Blue squares in the truth tables indicate correct predictions. Colors in the statistic squares represent a color range from worse (red) to better (green).
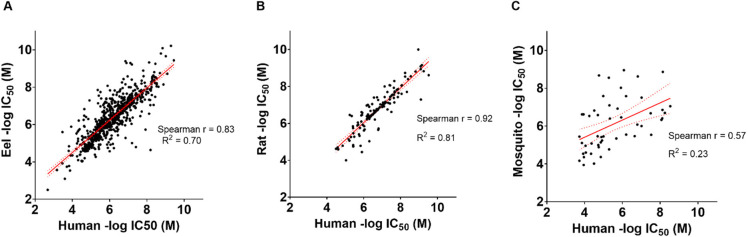
These results are unsurprising, given what we know about AChE; the enzyme is highly conserved in vertebrates, and while some differences in IC50 values between eel and human have been reported,39,54−56 it has usually been a matter of degree instead of a stark activity difference. A comparison of the −log M IC50 values of the 664 compounds found in common between the human and eel continuous data sets shows a strong correlation between the activity of the compounds in both species (Spearman r = 0.83, Figure 1A). An even stronger correlation was seen between human and rat, although only 127 compounds were shared between the two data sets (Figure 1B), so this difference may be based on sample size. This contrasts sharply with the mosquito data, which showed the weakest correlation between the two species (Figure 1C). This is not entirely surprising, as the mosquito is the furthest away evolutionarily, and this difference is the basis for the validity of mosquito AChE being a viable druggable target for OP pesticides.
Figure 1.
Comparison between IC50 values of human against three other species (eel, rat, and mosquito). Each point represents the −log IC50 (M) value of a single molecule tested in human AChE, plotted against the value of the same molecule tested in a different species. Red lines represent a simple linear regression. Dotted lines represent 95% confidence bands. P value <0.0001. (A) Eel, (B) rat, and (C) mosquito.
Comparison of Data Set Overlap for Classification Models
In our literature search for AChE inhibitors that were not yet deposited into ChEMBL, less than ten percent of the papers we encountered were focused on organisms other than human or eel. Therefore, finding external test sets for those species-specific models was challenging. Lacking external test sets and given the results of the human and eel test sets, we were keen to see if there was anything chemically distinguishable to be found in the data sets compiled for the other, underrepresented species. We undertook this by comparing the chemical property space, using ECFP6 descriptors of the data sets for each species-specific data set using a t-SNE plot. A t-SNE plot is a method that has been used previously to visualize library differences. The ECFP6 fingerprints of these molecules are condensed into a two-dimensional vector that can be plotted, and the resulting plot is a two-dimensional representation of a given chemical space.58Figure S1 demonstrates that the chemical property spaces of the rat, mouse, cow, ray, and mosquito training sets are, with a few exceptions, largely encapsulated by the chemical property space of the eel and or human training sets, and these models are likely useful surrogates for the individual models built for each vertebrate species.
Given the often-small variation between species-specific activity, any differences are unlikely to be easily captured by a classification model. However, given the modest difference in correlation between the human and eel log IC50’s (Figure 1A), we were keen to see if we could model the degree of separation between human and eel activity. We therefore also investigated the ability of regression models to distinguish between species-specific differences in the test sets.
Machine Learning Model Building: Regression Models
We created regression models for the human and eel data sets using ADAR, BayesianRidge (BR), ElasticNet regression (ENR), kNN regression (kNNR), random forest regression (RFR), support vector regression (SVR), and XGB regression (XGBR). We also implemented AttentiveFP, as described in the classification models section above, to compare against descriptor-based models. The data sets for the regression models were smaller than those for the classification models (human = 3652, eel = 4930) as we excluded compounds with relational IC50 values. The SVR algorithm outperformed all the other algorithms for both human and eel with R2 values of 0.81 and 0.75, respectively (Table 4). While not the best-performing algorithm, AttentiveFP outperformed some of the descriptor-based models, suggesting it is a competitive model and may outperform some descriptor-based models for regression with larger data sets.
Table 4. Nested, 5-Fold Cross-Validation Statistics for Human and Eel Regression Models for AChE Inhibitiona.
| Human: 3652 Compounds (1.06–11.22 −log M) | |||||
|---|---|---|---|---|---|
| model | MAE | RMSE | R2 | MPD | MGD |
| ADAR | 0.96 | 1.17 | 0.42 | 0.24 | 0.04 |
| BR | 0.68 | 0.88 | 0.67 | 0.14 | 0.03 |
| ENR | 0.88 | 1.13 | 0.46 | 0.22 | 0.04 |
| kNNR | 0.53 | 0.75 | 0.76 | 0.10 | 0.02 |
| RFR | 0.55 | 0.75 | 0.76 | 0.10 | 0.02 |
| SVR | 0.50 | 0.67 | 0.81 | 0.08 | 0.02 |
| XGBR | 0.62 | 0.86 | 0.68 | 0.13 | 0.03 |
| AttentiveFPb | 0.55 | 0.58 | 0.74 | 0.10 | 0.03 |
| Eel: 4930 Compounds (0.62–10.42 −log M) | |||||
|---|---|---|---|---|---|
| model | MAE | RMSE | R2 | MPD | MGD |
| ADAR | 0.79 | 0.98 | 0.34 | 0.17 | 0.03 |
| BR | 0.57 | 0.75 | 0.61 | 0.10 | 0.02 |
| ENR | 0.73 | 0.92 | 0.42 | 0.15 | 0.03 |
| kNNR | 0.47 | 0.66 | 0.70 | 0.08 | 0.01 |
| RFR | 0.46 | 0.64 | 0.72 | 0.07 | 0.01 |
| SVR | 0.43 | 0.60 | 0.75 | 0.06 | 0.01 |
| XGBR | 0.53 | 0.75 | 0.61 | 0.10 | 0.02 |
| AttentiveFPb | 0.50 | 0.46 | 0.69 | 0.09 | 0.02 |
ADAR = AdaBoosted decision trees; BR = BayesianRidge; ENR = ElasticNet regression; kNNR = K-nearest neighbors regression; RFR = random forest regression; SVR = support vector regression; XBGR = XGBoost regression; R2 = coefficient of determination; MAE = mean of absolute value of errors; RMSE = root-mean-squared error; MPD = mean Poisson deviance; MGD = mean gamma deviance.
Statistics from 5-fold cross-validation
External Model Validation of Regression Models
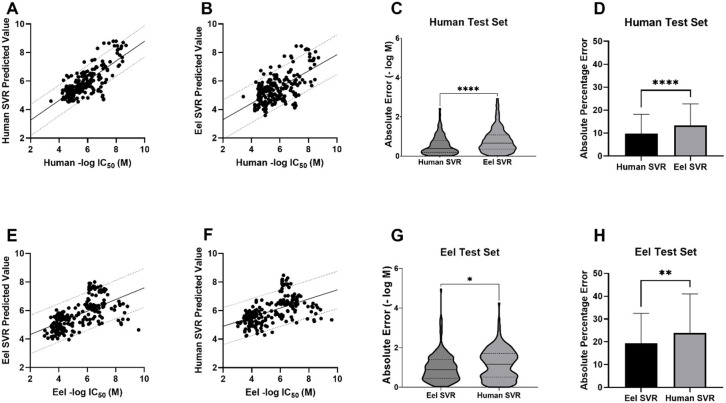
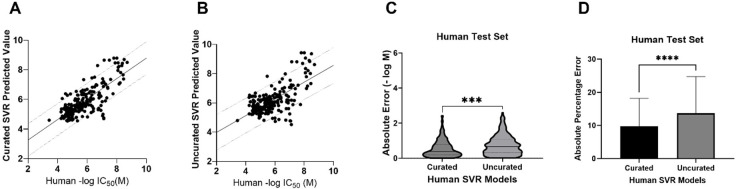
We also used these regression models to predict the potency of the compounds in the human and eel external test sets. The best agreement between measured and predicted values was demonstrated with the human test set predicted with the human regression SVR model (Figure 2A). The predicted −log(M) IC50 values had the highest correlation (Pearson’s r = 0.76) and the lowest mean absolute percentage error (MAPE = 9.73%) as compared to the measured values of any of the four tested model/test set pairings (Figure 2B–H). The differences in absolute error and MAPE between the human and eel models for the human test set were statistically significant (Figure 2C,D). Despite the less accurate predictions for the eel test set on both the eel and human SVR models (Figure 2E,F), the MAPE for the eel test set predicted by the eel model was statistically significantly lower than for the human model (Figure 2F), suggesting some species specificity. This outcome was surprising, given the lack of species specificity found with our classification models (Table 3). This discrepancy illustrates one of the differences in regression and classification models as information is likely lost in assigning classification based on IC50 values to very closely related compounds that lie on the activity threshold cutoff.
Figure 2.
SVR models for the prediction of AChE inhibition in human and eel test sets. (A) The human test set predicted with the human SVR model. MAPE = 9.73%, MAE = 0.55 ± 0.48 (SD), RMSE = 0.73, Pearson’s r = 0.76, R2 = 0.58, two-tailed P value <0.0001. The line shows a simple linear regression with 90% prediction bands. (B) The human test set predicted with the eel SVR model. MAPE = 13.4%, MAE = 0.79 ± 0.60, RMSE = 0.99, Pearson’s r = 0.61, R2 = 0.38, two-tailed P value <0.0001. (C) Comparison of absolute error for real vs predicted value of the human test set using the human or the eel SVR model (−log M). Unpaired t test with Welch’s correction, P < 0.0001. (D) Comparison of absolute percentage errors for real vs predicted value of the human test set using the human or the eel SVR model. Unpaired t test with Welch’s correction, P < 0.0001. (E) The eel test set predicted with the eel SVR model. MAPE = 19.4%, MAE = 0.99 ± 0.71, RMSE = 1.22, Pearson’s r = 0.60, R2 = 0.36, two-tailed P value <0.0001 F) The eel test set predicted with the human SVR model. MAPE = 23.9%, MAE = 1.16 ± 0.77, RMSE = 1.39, Pearson’s r = 0.52, R2 = 0.27, two-tailed P value <0.0001. (G) Comparison of absolute error for real vs predicted value of the eel test set using the human or the eel SVR model, in −log(M). Unpaired t test with Welch’s correction, P = 0.0226. (H) Comparison of absolute percentage errors for real vs predicted value of the eel test set using the human or the eel SVR model. Unpaired t test with Welch’s correction, P = 0.0033. (MAPE = mean absolute percentage error, MAE = mean absolute error, R2 = coefficient of determination; RMSE = root-mean-squared error.)
Following further analysis, we also found that the eel test set showed two distinct clusters of molecules that were either over- or underpredicted in potency by both models (Figure S2A). These molecules are from three different publications exploring AChE inhibition of hybrid molecules. Conjugates of the well-known AChE inhibitor tacrine67 (IC50 = 159 nM) were predicted by the model to be 4–30 times more potent than their measured IC50 values, while conjugates of coumarin68 (IC50 = 38,500 nM) and ligustrazine69 (not active) were predicted by the model to be 4–90,000 times less potent than their measured submicromolar, or subnanomolar, IC50 values. It is not surprising that hybrid molecules would not easily be distinguishable from their origin molecules using ECFP6 fingerprints. Interestingly, if these molecules are removed from the test set, the correlation and R2 of the remaining set improve dramatically and are on par with the human test set on the human model (Figure S2B), although the MAPE improves only slightly (Figure S2C).
Exploration of Data Curation on Results
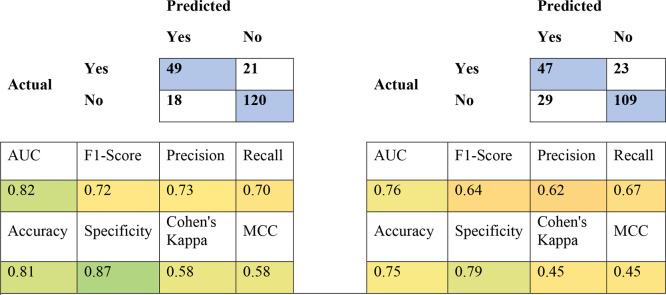
Due to the expense of manual curation that went into building these data sets, we were also interested to learn if our extensive pruning strategies produced a measurable result. The human data set required extensive correction. Retrospectively, we were interested to see how models built from “uncurated” ChEMBL data performed with internal and external validation. The uncurated human data set is composed of all entries under human AChE in ChEMBL30, minus those entries with a warning under the “Data Validity Comment” section in ChEMBL. The internal cross-validation statistics for the uncurated human classification models were still good (Table 5), as was the predictive power of the uncurated consensus model against our external test set (Table 6).
Table 5. Internal 5-Fold Cross-Validation Statistics of Uncurated Human Classification Models.
| Uncurated Human AChE Training Set: 2315 Active/5035 Total Compounds | ||||||||
|---|---|---|---|---|---|---|---|---|
| modela | AUC | F1 score | precision | recall | accuracy | specificity | Cohen’s kappa | MCC |
| DL | 0.91 | 0.82 | 0.81 | 0.84 | 0.83 | 0.83 | 0.67 | 0.67 |
| ADA | 0.88 | 0.77 | 0.79 | 0.75 | 0.79 | 0.83 | 0.58 | 0.58 |
| BNB | 0.82 | 0.72 | 0.72 | 0.73 | 0.74 | 0.75 | 0.48 | 0.48 |
| kNN | 0.91 | 0.84 | 0.81 | 0.86 | 0.85 | 0.83 | 0.69 | 0.69 |
| LREG | 0.89 | 0.79 | 0.80 | 0.78 | 0.81 | 0.83 | 0.62 | 0.62 |
| RF | 0.92 | 0.82 | 0.86 | 0.78 | 0.84 | 0.89 | 0.68 | 0.69 |
| SVC | 0.92 | 0.84 | 0.84 | 0.85 | 0.86 | 0.86 | 0.71 | 0.71 |
| XGB | 0.92 | 0.83 | 0.83 | 0.83 | 0.84 | 0.86 | 0.68 | 0.68 |
DL = Deep learning; ADA = AdaBoosted decision trees; BNB = Bernoulli naïve Bayes; kNN = K-nearest neighbors; LREG = LogisticRegression; RF = random forest; SVC = support vector classification; XGB = XGBoost.
Table 6. Truth Tables for Human AChE Test Set Predicted against Human Curated (Left) and Human Uncurated Consensus (Right) Modelsa.
Blue squares in the truth tables indicate correct predictions. Colors in the statistic squares represent a color range from worse (red) to better (green).
Similar to the regression models created with the curated human data set, the uncurated human SVR had the best internal cross-validation statistics (Table 7). However, unlike the classification models, the accuracy of the curated SVR model (Figure 3A) surpassed the accuracy of the uncurated SVR model (Figure 3B), and this difference was statistically significant (Figure 3C,D). This illustrates how data-centric curation improves regression model results, suggesting that a stronger focus on data curation is likely required for accurate regression models as compared to classification models.
Table 7. Nested, 5-Fold Cross-Validation Statistics for Uncurated Human Regression Models for AChE Inhibitiona.
| Uncurated Human: 4511 Compounds (3.76–10.96 −log M) | |||||
|---|---|---|---|---|---|
| model | MAE | RMSE | R2 | MPD | MGD |
| ADAR | 0.93 | 1.11 | 0.27 | 0.20 | 0.03 |
| BR | 0.65 | 0.85 | 0.57 | 0.12 | 0.02 |
| ENR | 0.81 | 1.02 | 0.38 | 0.17 | 0.03 |
| kNNR | 0.54 | 0.77 | 0.65 | 0.09 | 0.02 |
| RFR | 0.55 | 0.74 | 0.68 | 0.09 | 0.01 |
| SVR | 0.49 | 0.68 | 0.73 | 0.07 | 0.01 |
| XGBR | 0.61 | 0.83 | 0.59 | 0.11 | 0.02 |
ADAR = AdaBoosted decision trees; BR = BayesianRidge; ENR = ElasticNet regression; kNNR = K-nearest neighbors regression; RFR = random forest regression; SVR = support vector regression; XBGR = XGBoost regression; R2 = coefficient of determination; MAE = mean of absolute value of errors; RMSE = root-mean-squared error; MPD = mean Poisson deviance; MGD = mean gamma deviance.
Figure 3.
Comparison of the predictive power of the curated and uncurated human SVR models. (A) The human test set predicted with the human SVR model. MAPE = 9.73%, MAE = 0.55 ± 0.48 (SD), RMSE = 0.73, Pearson’s r = 0.76, R2 = 0.58, two-tailed P value <0.0001. The line shows a simple linear regression with 90% prediction bands. (B) The human test set predicted with the uncurated human SVR model. MAPE = 13.7%, MAE = 0.73 ± 0.55, RMSE = 0.93, Pearson’s r = 0.66, R2 = 0.44, two-tailed P value <0.0001. (C) Comparison of absolute error for real vs predicted values (−log M) of the human test set using the human curated or uncurated SVR models. Unpaired t test with Welch’s correction, P value = 0.0002. (D) Comparison of absolute percentage errors for the real vs predicted value of the human test set using the human curated or uncurated SVR models. Unpaired t test with Welch’s correction, P value <0.0001.
Machine Learning Model Building: Expanded Human Classification Model
While our models were initially assessed by predicting activity of test sets extracted from a literature search for new AChE inhibitors, we also predicted the activity of additional compounds. The data sets that were used to train the human and eel classification models are mostly nonoverlapping (they share only 771 compounds in common), and both occupy a similar chemical property space that encapsulates the test sets (Figure S3). This is likely because the compounds found in searches of PubChem, BindingDB, and PubMed largely represent AChE inhibitor-like molecules, or compounds derived from known inhibitors. The KEMI Market List (Stellan Fischer, KEMI) is a list of ∼30,000 compounds compiled from different regulatory agencies that are expected to appear in the EU market. Unlike the data in the aforementioned public databases, the market list is not drug-centric, nor has it been selected for AChE inhibition. Predictably, it occupies a very different chemical property space from our predictive models (Figure S4A). We were able to increase the chemical property space of our human classification models using data from a recent screen for inhibition of human AChE in three different assays against the Tox21 10,000 compound library.38 Most of the IC50 values reported from the follow-up studies from this screen qualified as inactive in our original human classification models based on the 1 μM activity cutoff. We therefore rebuilt these models with an activity cutoff of 100 μM and after incorporation of the Tox21 compounds added 177 additional active compounds (enzyme-only assay). We chose the enzyme-only assay for this as it represents most of our in vitro data from ChEMBL and BindingDB. We then added any compounds that scored “inactive” for all three AChE screens (enzyme-only, enzyme with human liver microsomes, and cell-based; ∼6400 compounds) to our classification models as inactive. Between the combined inactives of three AChE screens, the 177 new active compounds, and our reclassified ChEMBL and BindingDB data, the resulting human AChE classification models are composed of 10,382 compounds and now overlap much more chemical property space when compared with the KEMI library (Figure S4B).
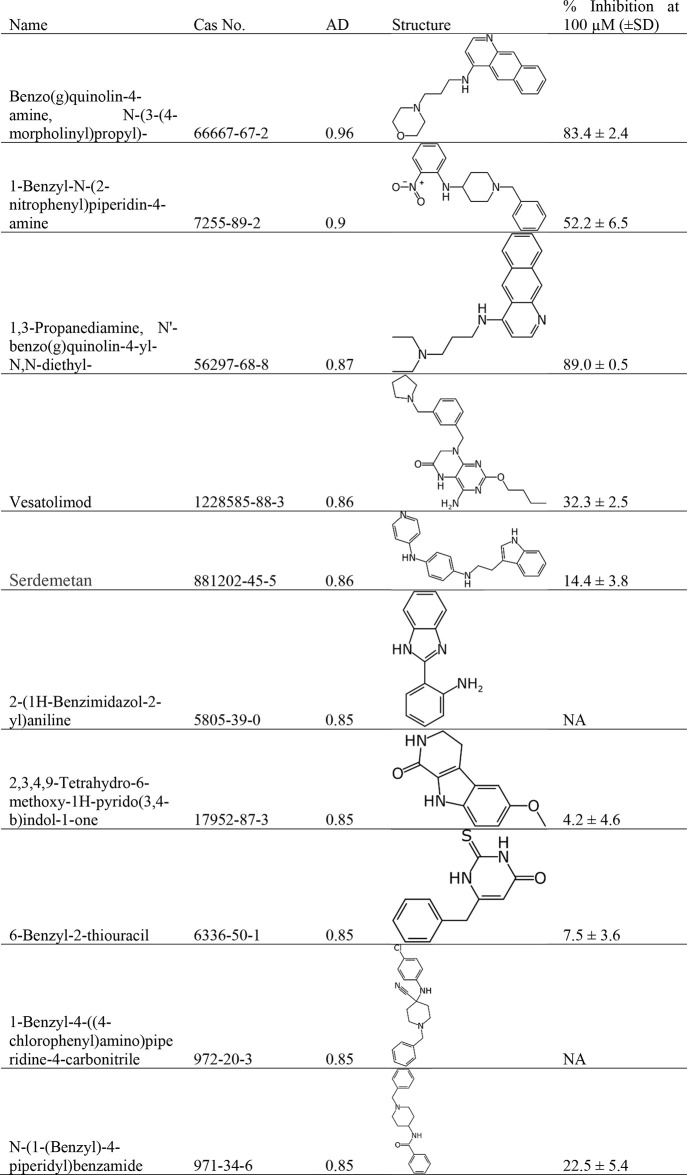
These new models performed well with internal cross-validation (Table S1), so we used them to score the KEMI market list, the 70,000-compound HBM4 EU CEC Screen list, as well as 50 other additional chemical lists from the CompTox Dashboard. Of the >195,000 compounds scored by our 100 μM human AChE model, there were 111 compounds predicted to be active with an applicability domain score ≥0.8 (Table S2). The applicability domain score was used as a metric as it is an indirect measure of the confidence of the prediction score, calculated using the RDD algorithm.61 While many of the top scoring compounds were unavailable (prohibitively expensive, or already known AChE inhibitors not in the model), we tested ten compounds for potential AChE inhibitory activity, picking those with the highest AD score that were readily available for purchase (Table 8, Figure S5).
Table 8. Inhibition of AChE by Compounds Predicted Active by the Expanded Human Classification Model.
Eight of the ten selected compounds displayed modest inhibition of AChE at 100 μM, with six of the ten displaying >10% activity at this concentration and two of those IC50s of between 7 and 14 μM (Figure S5). While this is below the predicted ≥50% activity at 100 μM, as defined by the model, the fact that most display at least partial dose–response curves (Figure S5) suggests that this model is able to identify some newly described AChE inhibitors.
Discussion
To date there have been several examples of other research groups using machine learning to create models for AChE inhibition,30,70−78 including recent papers using ChEMBL or BindingDB as sources for their training sets.72,76,79 Many of the studies used 3D-QSAR and a docking step for scoring. Our models in contrast use only ECFP6 fingerprints, which do not require 3D information on the target. Additionally, these prior examples tend to focus on a single organism to model, while in contrast we have carefully curated data sets and created machine learning models for AChE inhibition in seven different species. Given the disparity between numbers of unique compounds from public databases in some of these recent papers, and the number we arrived at after our extensive pruning and cleaning of data from these same databases, it suggests that this curation process was more selective in ensuring the human AChE models were composed of human-only data. This extra curation step also improved the predictive power of our regression models.
We have generated and validated our AChE inhibition classification models using internal and additional external methodologies. The excellent internal validation statistics in all of these models also led us to use consensus models for predictions of external test sets. We have shown that the consensus classification models for human and eel AChE inhibition can predict activity in external test sets with 81% and 82% accuracy, respectively (Table 3). We have also generated an SVR model for human AChE inhibition that predicted −log M IC50 values of a test set well (MAE = 0.55, RMSE = 0.73, Figure 2). External test sets created from the literature may also avoid bias that may arise from the partitioning of the data set in leave-out cross-validation, in the case that similar compounds from the same paper might appear in both the model and the external test set. When comparing the accuracy of the eel and human models against the species-specific test sets, we did not observe any species selectivity in our classification models. This reinforces our assertion that either the human or eel consensus classification model could be a good proxy for vertebrate AChE, as the structural similarities are higher among the vertebrates.39 Our human SVR model demonstrated species specificity and was the best model/test set pair for predicting the potency of the compounds against AChE. This represents a useful tool for evaluating which compounds may be worth pursuing when looking for new AChE inhibitors as therapeutics (for humans or as pesticides) or by scoring molecules that may have an impact on the environment via nontarget species. Our further exploration of the value of curation efforts on the human AChE inhibition data sets showed a marked improvement in the predictive power of these human regression models.
There are many potential machine learning algorithms that could be utilized, and as we have shown previously there is minimal difference between their performance.80 More recently described approaches such as transformer-based models are thought to improve over more classical machine learning models for tasks such as lipophilicity predictions; however, they tend to perform worse at bioactivity/IC50 prediction tasks. For example, it has been shown that Chemformer performs substantially worse than SVM models trained on 2048-bit ECFP4 feature inputs.81 Another recently described method, AttentiveFP, was also outperformed on the Beta-Secretase 1 (BACE) bioactivity IC50 data set by a random forest model.64 These results can be explained as more complex models require more training data in order to outperform their classical algorithm counterparts, and bioactivity data sets are often small, with our data sets having under 10,000 data points.66 To show whether these results hold true with our largest data sets, we trained an AttentiveFP model to compare against our other machine learning methods using the implementation from the DeepChem library. We found that AttentiveFP did not outperform our best models for regression or classification for either the eel or human AChE data sets using cross-validation. As additional algorithms come to our attention they can be benchmarked with these data sets.
The expanded human classification model has potential uses outside of the therapeutic space. Compounds that inhibit AChE may also pose potential health risks. Over 50 chemical lists from the CompTox dashboard62 that have implications for human and environmental safety have been evaluated. We have shown that there are over 100 compounds predicted to be active against AChE, with a high degree of confidence based on our predictions and applicability domain score. Future investigations of additional libraries of such compounds for potential AChE inhibition would help to further diversify and strengthen this model. However, these do not necessarily reflect available physical libraries, and we have encountered challenges acquiring several of our top-scoring compounds, either because the compounds are restricted, or the compounds are simply not available from chemical vendors.
Machine learning models were also created for other species but consisted of much smaller data sets. The mosquito AChE model being the smallest did not perform as well on internal cross-validation as the other species. It is also the only invertebrate data set that we modeled, as well as the only organism that is classified as a pest. As such, it potentially deserves more attention, as the majority of AChE inhibitors found in the environment are in the form of OP or carbamate pesticides. Using machine learning models to predict compounds that could target the invertebrate forms of AChE, without targeting the vertebrate forms, is a compelling option for future investigation, as well as exploring compounds specific to butyrylcholinesterase without inhibiting the closely related AChE.82,83
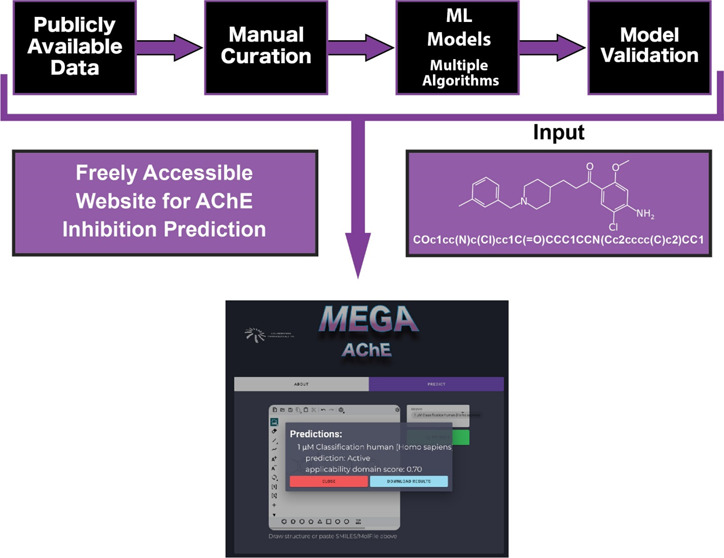
Finally, to make all our AChE models more widely available to the scientific community we have created MegaAChE: a website that can be used for predictions of a limited number of molecules from a molecule structure (megaache.collaborationspharma.com, Figure S6). We have also recently pointed out that such machine learning methods have a potential for dual use which we need to guard against and ensure valid use of the data sets.84,85
Glossary
Abbreviations
- AChE
acetylcholinesterase
- Ach
acetylcholine
- OP
organophosphorus
- QSAR
quantitative structure activity relationship
- ECFP6
Extended Connectivity Fingerprints Diameter 6
- AD
applicability domain
- RDD
reliability-density neighborhood
- SVR
support vector regression
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.2c00283.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval of the final version of the manuscript. CRediT: Patricia A Vignaux data curation, formal analysis, investigation, validation, writing-review & editing; Thomas R Lane formal analysis, supervision, writing-review & editing; Fabio Urbina methodology, software, writing-review & editing; Jacob Gerlach software; Ana C Puhl conceptualization, funding acquisition, writing-original draft; Scott H Snyder software, visualization; Sean Ekins conceptualization, funding acquisition, project administration, supervision, writing-original draft, writing-review & editing.
We kindly acknowledge NIH funding: R44GM122196-02A1 from NIGMS (PI: S.E.), 1R43ES033855-01 from NIEHS. Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number 1R43ES033855-01. We kindly acknowledge a matching grant award under the FY 21-22 One North Carolina SBIR/STTR Matching Funds Program Solicitation.
The authors declare the following competing financial interest(s): S.E. is founder and owner of Collaborations Pharmaceuticals, Inc. P.A.V., T.R.L., F.U., J.G., A.C.P., and S.H.S. are employees of Collaborations Pharmaceuticals, Inc.
Notes
Statement on Dual-Use. The AChE machine learning models described in this study have potential dual-use capabilities, and we therefore propose to implement restrictions to control who has access to these models and limit the number of molecules predicted when used on the website. We believe such precautions are necessary, and these will evolve over time as we integrate further software features to control this dual-use potential.
Supplementary Material
References
- King A. M.; Aaron C. K. Organophosphate and carbamate poisoning. Emerg Med. Clin North Am. 2015, 33, 133–151. 10.1016/j.emc.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Taylor P.; Radic Z. The cholinesterases: from genes to proteins. Annu. Rev. Pharmacol Toxicol 1994, 34, 281–320. 10.1146/annurev.pa.34.040194.001433. [DOI] [PubMed] [Google Scholar]
- Quinn D. M. Acetylcholinesterase: enzyme structure, reaction dynamics, and virtual transition states. Chem. Rev. 1987, 87, 955–979. 10.1021/cr00081a005. [DOI] [Google Scholar]
- Hsieh B. H.; Deng J. F.; Ger J.; Tsai W. J. Acetylcholinesterase inhibition and the extrapyramidal syndrome: a review of the neurotoxicity of organophosphate. Neurotoxicology 2001, 22, 423–427. 10.1016/S0161-813X(01)00044-4. [DOI] [PubMed] [Google Scholar]
- Day T.; Greenfield S. A. A non-cholinergic, trophic action of acetylcholinesterase on hippocampal neurones in vitro: molecular mechanisms. Neuroscience 2002, 111, 649–656. 10.1016/S0306-4522(02)00031-3. [DOI] [PubMed] [Google Scholar]
- Luo X.; Lauwers M.; Layer P. G.; Wen C. Non-neuronal Role of Acetylcholinesterase in Bone Development and Degeneration. Frontiers in Cell and Developmental Biology 2021, 8, 620543. 10.3389/fcell.2020.620543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq H.; Patinkin D.; Lev-Lehman E.; Grifman M.; Ginzberg D.; Eckstein F.; Zakut H. Antisense oligonucleotide inhibition of acetylcholinesterase gene expression induces progenitor cell expansion and suppresses hematopoietic apoptosis ex vivo. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 7907–7911. 10.1073/pnas.91.17.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq H.; Seidman S. Acetylcholinesterase--new roles for an old actor. Nat. Rev. Neurosci 2001, 2, 294–302. 10.1038/35067589. [DOI] [PubMed] [Google Scholar]
- Nalivaeva N. N.; Turner A. J. AChE and the amyloid precursor protein (APP) - Cross-talk in Alzheimer’s disease. Chem. Biol. Interact 2016, 259, 301–306. 10.1016/j.cbi.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Zhuang Q.; Young A.; Callam C. S.; McElroy C. A.; Ekici Ö. D.; Yoder R. J.; Hadad C. M. Efforts toward treatments against aging of organophosphorus-inhibited acetylcholinesterase. Ann. N.Y. Acad. Sci. 2016, 1374, 94–104. 10.1111/nyas.13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karalliedde L.; Senanayake N. Organophosphorus insecticide poisoning. Br J. Anaesth 1989, 63, 736–750. 10.1093/bja/63.6.736. [DOI] [PubMed] [Google Scholar]
- Naik K.; Aralikatte S.; Hesarur N.; Patil R. Prospective hospital-based clinical and electrophysiological evaluation of acute organophosphate poisoning. Annals of Indian Academy of Neurology 2019, 22, 91. 10.4103/aian.AIAN_137_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waseem M.; Perry C.; Bomann S.; Pai M.; Gernsheimer J. Cholinergic crisis after rodenticide poisoning. West J. Emerg Med. 2010, 11, 524–527. [PMC free article] [PubMed] [Google Scholar]
- Wang T.; Liu X. H.; Guan J.; Ge S.; Wu M. B.; Lin J. P.; Yang L. R. Advancement of multi-target drug discoveries and promising applications in the field of Alzheimer’s disease. Eur. J. Med. Chem. 2019, 169, 200–223. 10.1016/j.ejmech.2019.02.076. [DOI] [PubMed] [Google Scholar]
- Castro A.; Martinez A. Peripheral and dual binding site acetylcholinesterase inhibitors: implications in treatment of Alzheimer’s disease. Mini Rev. Med. Chem. 2001, 1, 267–272. 10.2174/1389557013406864. [DOI] [PubMed] [Google Scholar]
- Camps P.; Gomez E.; Munoz-Torrero D.; Badia A.; Vivas N. M.; Barril X.; Orozco M.; Luque F. J. Synthesis, in vitro pharmacology, and molecular modeling of syn-huprines as acetylcholinesterase inhibitors. J. Med. Chem. 2001, 44, 4733–4736. 10.1021/jm010949b. [DOI] [PubMed] [Google Scholar]
- Kaur J.; Zhang M. Q. Molecular modelling and QSAR of reversible acetylcholines-terase inhibitors. Curr. Med. Chem. 2000, 7, 273–294. 10.2174/0929867003375254. [DOI] [PubMed] [Google Scholar]
- Tong W.; Collantes E. R.; Chen Y.; Welsh W. J. A comparative molecular field analysis study of N-benzylpiperidines as acetylcholinesterase inhibitors. J. Med. Chem. 1996, 39, 380–387. 10.1021/jm950704x. [DOI] [PubMed] [Google Scholar]
- Golbraikh A.; Bernard P.; Chretien J. R. Validation of protein-based alignment in 3D quantitative structure-activity relationships with CoMFA models. Eur. J. Med. Chem. 2000, 35, 123–136. 10.1016/S0223-5234(00)00108-2. [DOI] [PubMed] [Google Scholar]
- El Yazal J.; Rao S. N.; Mehl A.; Slikker W. Jr. Prediction of organophosphorus acetylcholinesterase inhibition using three-dimensional quantitative structure-activity relationship (3D-QSAR) methods. Toxicol. Sci. 2001, 63, 223–232. 10.1093/toxsci/63.2.223. [DOI] [PubMed] [Google Scholar]
- Sutherland J. J.; O’Brien L. A.; Weaver D. F. A comparison of methods for modeling quantitative structure-activity relationships. J. Med. Chem. 2004, 47, 5541–5554. 10.1021/jm0497141. [DOI] [PubMed] [Google Scholar]
- Fernandez M.; Caballero J. Ensembles of Bayesian-regularized genetic neural networks for modeling of acetylcholinesterase inhibition by huprines. Chem. Biol. Drug Des 2006, 68, 201–212. 10.1111/j.1747-0285.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- Jung M.; Tak J.; Lee Y.; Jung Y. Quantitative structure-activity relationship (QSAR) of tacrine derivatives against acetylcholinesterase (AChE) activity using variable selections. Bioorg. Med. Chem. Lett. 2007, 17, 1082–1090. 10.1016/j.bmcl.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Manchester J.; Czermiński R. SAMFA: Simplifying Molecular Description for 3D-QSAR. J. Chem. Inf. Model. 2008, 48, 1167–1173. 10.1021/ci800009u. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Li D.; Cao F.; Xiao W.; Zhao L.; Ding G.; Wang Z. Z. Identification of Human Acetylcholinesterase Inhibitors from the Constituents of EGb761 by Modeling Docking and Molecular Dynamics Simulations. Comb Chem. High Throughput Screen 2018, 21, 41–49. 10.2174/1386207320666171123201910. [DOI] [PubMed] [Google Scholar]
- Iman K.; Mirza M. U.; Mazhar N.; Vanmeert M.; Irshad I.; Kamal M. A. In silico Structure-based Identification of Novel Acetylcholinesterase Inhibitors Against Alzheimer’s Disease. CNS Neurol Disord Drug Targets 2018, 17, 54–68. 10.2174/1871527317666180115162422. [DOI] [PubMed] [Google Scholar]
- Telpoukhovskaia M. A.; Patrick B. O.; Rodriguez-Rodriguez C.; Orvig C. In silico to in vitro screening of hydroxypyridinones as acetylcholinesterase inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 1624–1628. 10.1016/j.bmcl.2016.01.080. [DOI] [PubMed] [Google Scholar]
- Akula N.; Lecanu L.; Greeson J.; Papadopoulos V. 3D QSAR studies of AChE inhibitors based on molecular docking scores and CoMFA. Bioorg. Med. Chem. Lett. 2006, 16, 6277–6280. 10.1016/j.bmcl.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Shiri F.; Pirhadi S.; Ghasemi J. B. Dynamic structure based pharmacophore modeling of the Acetylcholinesterase reveals several potential inhibitors. J. Biomol Struct Dyn 2019, 37, 1800–1812. 10.1080/07391102.2018.1468281. [DOI] [PubMed] [Google Scholar]
- Chekmarev D.; Kholodovych V.; Kortagere S.; Welsh W. J.; Ekins S. Predicting Inhibitors of Acetylcholinesterase by Regression and Classification Machine Learning Approaches with Combinations of Molecular Descriptors. Pharm. Res. 2009, 26, 2216–2224. 10.1007/s11095-009-9937-8. [DOI] [PubMed] [Google Scholar]
- Cui L.; Wang Y.; Liu Z.; Chen H.; Wang H.; Zhou X.; Xu J. Discovering New Acetylcholinesterase Inhibitors by Mining the Buzhongyiqi Decoction Recipe Data. J. Chem. Inf Model 2015, 55, 2455–2463. 10.1021/acs.jcim.5b00449. [DOI] [PubMed] [Google Scholar]
- Muthukumaran P.; Rajiniraja M. MIA-QSAR based model for bioactivity prediction of flavonoid derivatives as acetylcholinesterase inhibitors. J. Theor. Biol. 2018, 459, 103–110. 10.1016/j.jtbi.2018.09.030. [DOI] [PubMed] [Google Scholar]
- Jang C.; Yadav D. K.; Subedi L.; Venkatesan R.; Venkanna A.; Afzal S.; Lee E.; Yoo J.; Ji E.; Kim S. Y.; Kim M. H. Identification of novel acetylcholinesterase inhibitors designed by pharmacophore-based virtual screening, molecular docking and bioassay. Sci. Rep 2018, 8, 14921. 10.1038/s41598-018-33354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida J. R.; Figueiro M.; Almeida W. P.; de Paula da Silva C. H. T. Discovery of novel dual acetylcholinesterase inhibitors with antifibrillogenic activity related to Alzheimer’s disease. Future Med. Chem. 2018, 10, 1037–1053. 10.4155/fmc-2017-0201. [DOI] [PubMed] [Google Scholar]
- Jiang C. S.; Ge Y. X.; Cheng Z. Q.; Song J. L.; Wang Y. Y.; Zhu K.; Zhang H. Discovery of new multifunctional selective acetylcholinesterase inhibitors: structure-based virtual screening and biological evaluation. J. Comput. Aided Mol. Des 2019, 33, 521–530. 10.1007/s10822-019-00202-2. [DOI] [PubMed] [Google Scholar]
- Bowes J.; Brown A. J.; Hamon J.; Jarolimek W.; Sridhar A.; Waldron G.; Whitebread S. Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nat. Rev. Drug Discovery 2012, 11, 909–922. 10.1038/nrd3845. [DOI] [PubMed] [Google Scholar]
- Kavlock R.; Chandler K.; Houck K.; Hunter S.; Judson R.; Kleinstreuer N.; Knudsen T.; Martin M.; Padilla S.; Reif D.; Richard A.; Rotroff D.; Sipes N.; Dix D. Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 2012, 25, 1287–1302. 10.1021/tx3000939. [DOI] [PubMed] [Google Scholar]
- Li S.; Zhao J.; Huang R.; Travers J.; Klumpp-Thomas C.; Yu W.; MacKerell A. D. Jr.; Sakamuru S.; Ooka M.; Xue F.; Sipes N. S.; Hsieh J. H.; Ryan K.; Simeonov A.; Santillo M. F.; Xia M. Profiling the Tox21 Chemical Collection for Acetylcholinesterase Inhibition. Environ. Health Perspect 2021, 129, 47008. 10.1289/EHP6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner J.; Kriz Z.; Kuca K.; Jun D.; Koca J. Acetylcholinesterases--the structural similarities and differences. J. Enzyme Inhib Med. Chem. 2007, 22, 417–424. 10.1080/14756360701421294. [DOI] [PubMed] [Google Scholar]
- Arabshahi H. J.; Trobec T.; Foulon V.; Hellio C.; Frangež R.; Sepčić K.; Cahill P.; Svenson J. Using Virtual AChE Homology Screening to Identify Small Molecules With the Ability to Inhibit Marine Biofouling. Frontiers in Marine Science 2021, 8, 762287. 10.3389/fmars.2021.762287. [DOI] [Google Scholar]
- Kim Y. H.; Lee S. H. Which acetylcholinesterase functions as the main catalytic enzyme in the Class Insecta?. Insect Biochem. Mol. Biol. 2013, 43, 47–53. 10.1016/j.ibmb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Combes D.; Fedon Y.; Grauso M.; Toutant J.-P.; Arpagaus M. Four Genes Encode Acetylcholinesterases in the Nematodes Caenorhabditis elegans and Caenorhabditis briggsae. cDNA Sequences, Genomic Structures, Mutations and in vivo Expression. J. Mol. Biol. 2000, 300, 727–742. 10.1006/jmbi.2000.3917. [DOI] [PubMed] [Google Scholar]
- Chatonnet A.; Lenfant N.; Marchot P.; Selkirk M. E. Natural genomic amplification of cholinesterase genes in animals. J. Neurochem 2017, 142, 73–81. 10.1111/jnc.13990. [DOI] [PubMed] [Google Scholar]
- Duarte Y.; Gutierrez M.; Alvarez R.; Alzate-Morales J. H.; Soto-Delgado J. Experimental and Theoretical Approaches in the Study of Phenanthroline-Tetrahydroquinolines for Alzheimer’s Disease. ChemistryOpen 2019, 8, 627–636. 10.1002/open.201900073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves S.; Resta S.; Rinaldo F.; Costa M.; Josselin R.; Gwizdala K.; Piemontese L.; Capriati V.; Pereira-Santos A. R.; Cardoso S. M.; Santos M. A. Design, Synthesis, and In Vitro Evaluation of Hydroxybenzimidazole-Donepezil Analogues as Multitarget-Directed Ligands for the Treatment of Alzheimer’s Disease. Molecules 2020, 25, 985. 10.3390/molecules25040985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.; Yun Y.; Miao Y.; Sun J.; Wang X. Synthesis and biological evaluation of 3-arylbenzofuranone derivatives as potential anti-Alzheimer’s disease agents. J. Enzyme Inhib Med. Chem. 2020, 35, 805–814. 10.1080/14756366.2020.1740694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takomthong P.; Waiwut P.; Yenjai C.; Sripanidkulchai B.; Reubroycharoen P.; Lai R.; Kamau P.; Boonyarat C. Structure-Activity Analysis and Molecular Docking Studies of Coumarins from Toddalia asiatica as Multifunctional Agents for Alzheimer’s Disease. Biomedicines 2020, 8, 107. 10.3390/biomedicines8050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.; Yang H.; Huang Z.; Tian S.; Li Q.; Du C.; Chen T.; Liu Y.; Sun H.; Liu Z. Design, Synthesis, and Evaluation of Acetylcholinesterase and Butyrylcholinesterase Dual-Target Inhibitors against Alzheimer’s Diseases. Molecules 2020, 25, 489. 10.3390/molecules25030489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okello E. J.; Mather J. Comparative Kinetics of Acetyl- and Butyryl-Cholinesterase Inhibition by Green Tea Catechins|Relevance to the Symptomatic Treatment of Alzheimer’s Disease. Nutrients 2020, 12, 1090. 10.3390/nu12041090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourshojaei Y.; Abiri A.; Eskandari K.; Haghighijoo Z.; Edraki N.; Asadipour A. Phenoxyethyl Piperidine/Morpholine Derivatives as PAS and CAS Inhibitors of Cholinesterases: Insights for Future Drug Design. Sci. Rep 2019, 9, 19855. 10.1038/s41598-019-56463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudel P.; Seong S. H.; Zhou Y.; Ha M. T.; Min B. S.; Jung H. A.; Choi J. S. Arylbenzofurans from the Root Bark of Morus alba as Triple Inhibitors of Cholinesterase, beta-Site Amyloid Precursor Protein Cleaving Enzyme 1, and Glycogen Synthase Kinase-3beta: Relevance to Alzheimer’s Disease. ACS Omega 2019, 4, 6283–6294. 10.1021/acsomega.9b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira C. Brum J.; Neto D. C. F.; de Almeida J.; Lima J. A.; Kuca K.; Franca T. C. C.; Figueroa-Villar J. D. Synthesis of New Quinoline-Piperonal Hybrids as Potential Drugs against Alzheimer’s Disease. Int. J. Mol. Sci. 2019, 20, 3944. 10.3390/ijms20163944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scipioni M.; Kay G.; Megson I. L.; Kong Thoo Lin P. Synthesis of novel vanillin derivatives: novel multi-targeted scaffold ligands against Alzheimer’s disease. Medchemcomm 2019, 10, 764–777. 10.1039/C9MD00048H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasteel E. E. J.; Nijmeijer S. M.; Darney K.; Lautz L. S.; Dorne J.; Kramer N. I.; Westerink R. H. S. Acetylcholinesterase inhibition in electric eel and human donor blood: an in vitro approach to investigate interspecies differences and human variability in toxicodynamics. Arch. Toxicol. 2020, 94, 4055–4065. 10.1007/s00204-020-02927-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajda M.; Jonczyk J.; Malawska B.; Czarnecka K.; Girek M.; Olszewska P.; Sikora J.; Mikiciuk-Olasik E.; Skibinski R.; Gumieniczek A.; Szymanski P. Synthesis, biological evaluation and molecular modeling of new tetrahydroacridine derivatives as potential multifunctional agents for the treatment of Alzheimer’s disease. Bioorg. Med. Chem. 2015, 23, 5610–5618. 10.1016/j.bmc.2015.07.029. [DOI] [PubMed] [Google Scholar]
- Russom C. L.; LaLone C. A.; Villeneuve D. L.; Ankley G. T. Development of an adverse outcome pathway for acetylcholinesterase inhibition leading to acute mortality. Environ. Toxicol. Chem. 2014, 33, 2157–2169. 10.1002/etc.2662. [DOI] [PubMed] [Google Scholar]
- Vignaux P. A.; Minerali E.; Lane T. R.; Foil D. H.; Madrid P. B.; Puhl A. C.; Ekins S. The Antiviral Drug Tilorone Is a Potent and Selective Inhibitor of Acetylcholinesterase. Chem. Res. Toxicol. 2021, 34, 1296–1307. 10.1021/acs.chemrestox.0c00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T. R.; Urbina F.; Rank L.; Gerlach J.; Riabova O.; Lepioshkin A.; Kazakova E.; Vocat A.; Tkachenko V.; Cole S.; Makarov V.; Ekins S. Machine Learning Models for Mycobacterium tuberculosis In Vitro Activity: Prediction and Target Visualization. Mol. Pharmaceutics 2022, 19, 674–689. 10.1021/acs.molpharmaceut.1c00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez D.; Gaulton A.; Bento A. P.; Chambers J.; De Veij M.; Félix E.; Magariños M. P.; Mosquera J. F.; Mutowo P.; Nowotka M.; Gordillo-Marañón M.; Hunter F.; Junco L.; Mugumbate G.; Rodriguez-Lopez M.; Atkinson F.; Bosc N.; Radoux C. J.; Segura-Cabrera A.; Hersey A.; Leach A. R. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.; Lin Y.; Wen X.; Jorissen R. N.; Gilson M. K. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 2007, 35, D198–D201. 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniceto N.; Freitas A. A.; Bender A.; Ghafourian T. A novel applicability domain technique for mapping predictive reliability across the chemical space of a QSAR: reliability-density neighbourhood. Journal of Cheminformatics 2016, 8, 69. 10.1186/s13321-016-0182-y. [DOI] [Google Scholar]
- Williams A. J.; Grulke C. M.; Edwards J.; McEachran A. D.; Mansouri K.; Baker N. C.; Patlewicz G.; Shah I.; Wambaugh J. F.; Judson R. S.; Richard A. M. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. Journal of Cheminformatics 2017, 9, 61. 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman G. L.; Courtney K. D.; Andres V. Jr.; Feather-Stone R. M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Xiong Z.; Wang D.; Liu X.; Zhong F.; Wan X.; Li X.; Li Z.; Luo X.; Chen K.; Jiang H.; Zheng M. Pushing the Boundaries of Molecular Representation for Drug Discovery with the Graph Attention Mechanism. J. Med. Chem. 2020, 63, 8749–8760. 10.1021/acs.jmedchem.9b00959. [DOI] [PubMed] [Google Scholar]
- Ramsundar B.; Eastman P.; Walters P.; Pande V.; Leswing K.; Wu Z.. Deep Learning for the Life Sciences; O’Reilly media, 2019. [Google Scholar]
- Wu Z.; Ramsundar B.; Feinberg E. N.; Gomes J.; Geniesse C.; Pappu A. S.; Leswing K.; Pande V. MoleculeNet: a benchmark for molecular machine learning. Chemical Science 2018, 9, 513–530. 10.1039/C7SC02664A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z. Q.; Song J. L.; Zhu K.; Zhang J.; Jiang C. S.; Zhang H. Total Synthesis of Pulmonarin B and Design of Brominated Phenylacetic Acid/Tacrine Hybrids: Marine Pharmacophore Inspired Discovery of New ChE and Abeta Aggregation Inhibitors. Mar Drugs 2018, 16, 293. 10.3390/md16090293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khunnawutmanotham N.; Laongthipparos C.; Saparpakorn P.; Chimnoi N.; Techasakul S. Synthesis of 3-aminocoumarin-N-benzylpyridinium conjugates with nanomolar inhibitory activity against acetylcholinesterase. Beilstein J. Org. Chem. 2018, 14, 2545–2552. 10.3762/bjoc.14.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.; Liang X.; Xie G.; Chen L.; Liu W.; Luo G.; Zhang P.; Yu L.; Zheng X.; Ji H.; Zhang C.; Yi W. Synthesis and Evaluation of Novel Ligustrazine Derivatives as Multi-Targeted Inhibitors for the Treatment of Alzheimer’s Disease. Molecules 2018, 23, 2540. 10.3390/molecules23102540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.; Fallarero A.; Vainio M. J.; Saravanan P.; Santeri Puranen J.; Järvinen P.; Johnson M. S.; Vuorela P. M.; Mohan C. G. Molecular Docking Guided Comparative GFA, G/PLS, SVM and ANN Models of Structurally Diverse Dual Binding Site Acetylcholinesterase Inhibitors. Molecular Informatics 2011, 30, 689–706. 10.1002/minf.201100029. [DOI] [PubMed] [Google Scholar]
- A. Bermudez-Lugo J.; C. Rosales-Hernandez M.; Deeb O.; Trujillo-Ferrara J.; Correa-Basurto J. In Silico Methods to Assist Drug Developers in Acetylcholinesterase Inhibitor Design. Curr. Med. Chem. 2011, 18, 1122–1136. 10.2174/092986711795029681. [DOI] [PubMed] [Google Scholar]
- Simeon S.; Anuwongcharoen N.; Shoombuatong W.; Malik A. A.; Prachayasittikul V.; Wikberg J. E.S.; Nantasenamat C. Probing the origins of human acetylcholinesterase inhibition via QSAR modeling and molecular docking. PeerJ. 2016, 4, e2322. 10.7717/peerj.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B.; Zhao M.; Su Q.; Zhang M.; Lv W.; Chen Q.; Chen F.; Chu D.; Du D.; Zhang Y. 2D-SAR and 3D-QSAR analyses for acetylcholinesterase inhibitors. Molecular Diversity 2017, 21, 413–426. 10.1007/s11030-017-9732-0. [DOI] [PubMed] [Google Scholar]
- Adeshina Y. O.; Deeds E. J.; Karanicolas J. Machine learning classification can reduce false positives in structure-based virtual screening. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 18477–18488. 10.1073/pnas.2000585117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamodharan G.; Mohan C. G. Machine learning models for predicting the activity of AChE and BACE1 dual inhibitors for the treatment of Alzheimer’s disease. Mol. Divers. 2022, 26, 1501. 10.1007/s11030-021-10282-8. [DOI] [PubMed] [Google Scholar]
- Herrera-Acevedo C.; Perdomo-Madrigal C.; Herrera-Acevedo K.; Coy-Barrera E.; Scotti L.; Scotti M. T. Machine learning models to select potential inhibitors of acetylcholinesterase activity from SistematX: a natural products database. Mol. Divers 2021, 25, 1553–1568. 10.1007/s11030-021-10245-z. [DOI] [PubMed] [Google Scholar]
- Ganeshpurkar A.; Singh R.; Shivhare S.; Divya; Kumar D.; Gutti G.; Singh R.; Kumar A.; Singh S. K. Improved machine learning scoring functions for identification of Electrophorus electricus’s acetylcholinesterase inhibitors. Mol. Divers. 2022, 26, 1455. 10.1007/s11030-021-10280-w. [DOI] [PubMed] [Google Scholar]
- Nguyen T. H.; Tran P.-T.; Pham N. Q. A.; Hoang V.-H.; Hiep D. M.; Ngo S. T. Identifying Possible AChE Inhibitors from Drug-like Molecules via Machine Learning and Experimental Studies. ACS Omega 2022, 7, 20673. 10.1021/acsomega.2c00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu H.; Kumar R. N.; Garg P. Machine learning-based modeling to predict inhibitors of acetylcholinesterase. Mol. Divers 2022, 26, 331–340. 10.1007/s11030-021-10223-5. [DOI] [PubMed] [Google Scholar]
- Lane T. R.; Foil D. H.; Minerali E.; Urbina F.; Zorn K. M.; Ekins S. Bioactivity Comparison across Multiple Machine Learning Algorithms Using over 5000 Datasets for Drug Discovery. Mol. Pharmaceutics 2021, 18, 403–415. 10.1021/acs.molpharmaceut.0c01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin R.; Dimitriadis S.; He J.; Bjerrum E. J. Chemformer: a pre-trained transformer for computational chemistry. Machine Learning: Science and Technology 2022, 3, 015022 10.1088/2632-2153/ac3ffb. [DOI] [Google Scholar]
- Fang J.; Yang R.; Gao L.; Zhou D.; Yang S.; Liu A.-l.; Du G.-h. Predictions of BuChE Inhibitors Using Support Vector Machine and Naive Bayesian Classification Techniques in Drug Discovery. J. Chem. Inf. Model. 2013, 53, 3009–3020. 10.1021/ci400331p. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Lu X.; Yang H.; Chen Y.; Wang F.; Li J.; Tang Z.; Cheng X.; Yang Y.; Xu L.; Xia Q. Discovery of Selective Butyrylcholinesterase (BChE) Inhibitors through a Combination of Computational Studies and Biological Evaluations. Molecules 2019, 24, 4217. 10.3390/molecules24234217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina F.; Lentzos F.; Invernizzi C.; Ekins S. Dual use of artificial-intelligence-powered drug discovery. Nature Machine Intelligence 2022, 4, 189–191. 10.1038/s42256-022-00465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina F.; Lentzos F.; Invernizzi C.; Ekins S. A teachable moment for dual-use. Nature Machine Intelligence 2022, 4, 607–607. 10.1038/s42256-022-00511-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.