Abstract
Cannabis-based products have experienced notable increases in co-usage alongside tobacco products. Several cannabinoids exhibit inhibition of a number of cytochrome P450 (CYP) and UDP glucuronosyltransferase (UGT) enzymes, but few studies have examined their inhibition of enzymes involved in nicotine metabolism. The goal of the present study was to examine potential drug–drug interactions occurring in the nicotine metabolism pathway perpetrated by cannabidiol (CBD) and its active metabolite, 7-hydroxy-CBD (7-OH-CBD). The inhibitory effects of CBD and 7-OH-CBD were tested in microsomes from HEK293 cells overexpressing individual metabolizing enzymes and from human liver tissue. Assays with overexpressing microsomes demonstrated that CBD and 7-OH-CBD inhibited CYP-mediated nicotine metabolism. Binding-corrected IC50,u values for CBD inhibition of nicotine metabolism to cotinine and nornicotine, and cotinine metabolism to trans-3′-hydroxycotinine (3HC), were 0.27 ± 0.060, 0.23 ± 0.14, and 0.21 ± 0.14 μM, respectively, for CYP2A6; and 0.26 ± 0.17 and 0.029 ± 0.0050 μM for cotinine and nornicotine formation, respectively, for CYP2B6. 7-OH-CBD IC50,u values were 0.45 ± 0.18, 0.16 ± 0.08, and 0.78 ± 0.23 μM for cotinine, nornicotine, and 3HC formation, respectively, for CYP2A6, and 1.2 ± 0.44 and 0.11 ± 0.030 μM for cotinine and nornicotine formation, respectively, for CYP2B6. Similar IC50,u values were observed in HLM. Inhibition (IC50,u = 0.37 ± 0.06 μM) of 3HC to 3HC-glucuronide formation by UGT1A9 was demonstrated by CBD. Significant inhibition of nicotine metabolism pathways by CBD and 7-OH-CBD suggests that cannabinoids may inhibit nicotine metabolism, potentially impacting tobacco addiction and cessation.
Introduction
The co-use of cannabis and nicotine-based products has been steadily increasing among adults in the United States, coinciding with increased availability of cannabis and tobacco vaping products.1 In the National Survey on Drug Use and Health, nearly all daily cannabis users are cigarette smokers.2 It is thought that this reflects shared psychosocial and environmental risk factors for the use of these products.3 While some research has focused on adverse health outcomes of long-term cannabis use and how co-use with tobacco may affect cannabis outcomes, there is no data on the impact of cannabis use on tobacco use behaviors. Specifically, the pharmacologic effect of combined exposure to nicotine and cannabis compounds is virtually unknown, with few studies focusing on how co-use affects nicotine intake and metabolism. These patterns of co-usage may result in unexpected drug–drug interactions (DDI) as many of the compounds in tobacco and cannabis utilize the same enzymatic systems for metabolism and detoxification.
Of the major cannabinoids in cannabis, cannabidiol (CBD) interacts with the CB1 and CB2 receptors in the brain with a much lower affinity as compared to (−)-trans-Δ9-tetrahydrocannabinol (THC; the main psychoactive component of cannabis), resulting in extremely low psychoactive effects.4 However, CBD displays a broad range of potential therapeutic applications, including anti-inflammatory properties, antipsychotic and antiepileptic effects, as well as modulation of the immune system and the central nervous system.5 CBD usage is rapidly expanding among many patient populations due in part to its good safety profile, and recent industry statistics put CBD usage higher than recreational marijuana usage among the general population.6 A 2019 Gallup poll found that 12% of Americans consider themselves active users of recreational marijuana, compared to 14% who reported regular CBD usage.7
CBD metabolism to its major active metabolite, 7-hydroxy-CBD (7-OH-CBD), and its subsequent inactive metabolite, 7-carboxy-CBD (CBD-COOH), is mediated by cytochromes P450 (CYPs) 2C9, 2C19, and 3A4.8 While the pharmacology of 7-OH-CBD is not well studied, 7-OH-CBD exhibits a t1/2 that is longer than that of CBD and is found in the plasma of CBD users. The plasma Cmax (maximum concentration) of CBD following a single dose regimen of Epidiolex (6000 mg) is 2.5 μM9 with a calculated unbound plasma Cmax of 0.075 μM.10 These values are similar for 7-OH-CBD, with the total and calculated unbound plasma Cmax following this same dosing regimen at 1.569 and 0.0936 μM,10 respectively. 7-OH-CBD exhibits anticonvulsant properties comparable to CBD and is currently a patented medication for NAFLD, used to lower blood triglyceride levels.11,12
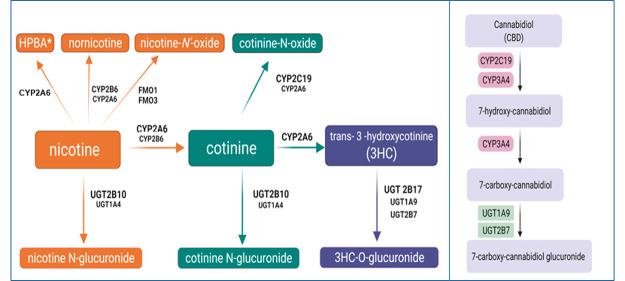
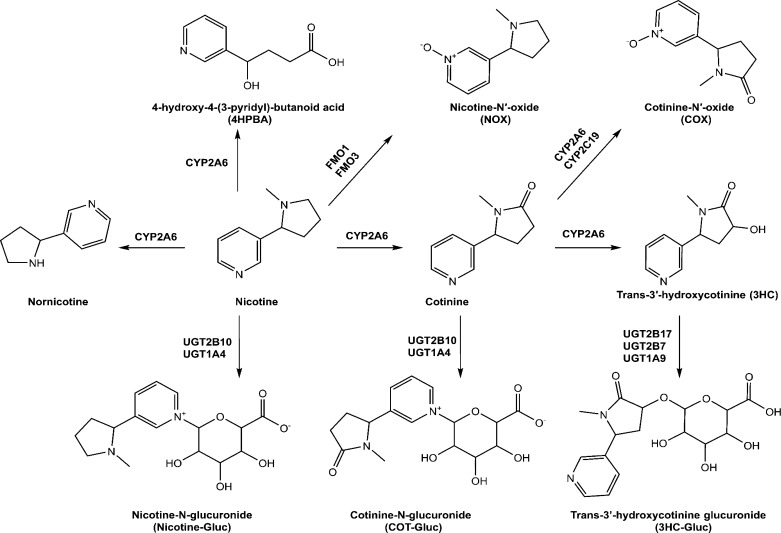
Nicotine is the major pharmacologically active substance in tobacco products and e-cigarettes and is responsible for mediating dependence. Nicotine is rapidly metabolized in vivo, with a t1/2 of 1–2 h,13 and smokers modulate their tobacco consumption to achieve a brain nicotine concentration that maintains the desired effects. The major metabolic pathway for nicotine is oxidation by CYP2A6 to form the inactive metabolite, cotinine (Figure 1), with some studies suggesting that CYP2B6 plays a minor role in cotinine formation.14 Nicotine undergoes additional metabolism through glucuronidation, catalyzed primarily by UGT2B1015 to form nicotine glucuronide (Nic-Gluc) and oxygenation by flavin monooxygenases (FMOs) including FMO1 and FMO316 to form nicotine-N′-oxide (NOX). Minor metabolites include nornicotine, formed primarily by CYPs 2A6 and 2B6, and 4-hydroxy-4-(3-pyridyl)-butanoic acid (HPBA), possibly formed by CYP2A6.17−21 Cotinine is also metabolized by CYP2A6 to form trans-3′-hydroxycotinine (3HC) and, like nicotine, can undergo conversion by UGT2B10-mediated glucuronidation to form cotinine glucuronide (Cot-Gluc)22 or oxidation by CYPs 2C19 and 2A6 to form cotinine N-oxide (COX).16 3HC is glucuronidated in vivo on the 3′-hydroxyl group, a reaction mediated by UDP glucuronosyltransferases (UGTs) 1A9, 2B7, and 2B17.23
Figure 1.
Schematic of nicotine metabolic pathways.
Functional variants of several of these metabolizing enzymes were linked to altered nicotine metabolism in smokers,15,16 and individuals with reduced or nonfunctional CYP2A6 activity were shown to be significantly less likely to become smokers, and for those who do become smokers, they consumed fewer cigarettes and scored lower for nicotine dependence.24 Decreased expression or inhibition of CYP2A6 activity reduces nicotine metabolism and has been suggested as a primary target for tobacco cessation strategies, resulting in a reduction of the number of cigarettes necessary for a smoker to maintain a given nicotine level leading to diminished smoking overall.
Previous studies suggest that CBD may be an effective treatment for tobacco addiction.25 In one study, the number of cigarettes smoked was decreased by as much as 40% over the course of treatment with a CBD inhaler.26 Previous studies have shown that cannabinoids and several major THC metabolites can inhibit several important hepatic CYPs27,28 as well as UGT enzymes.29 The goal of the present study was to examine the inhibitory potential of CBD and its major metabolite, 7-OH-CBD, on nicotine metabolic pathways, with the goal of better understanding the potential effect of these cannabinoids on tobacco addiction.
Experimental Procedures
Chemicals and Reagents
CBD and 7-OH-CBD were purchased from Cayman Chemicals (Ann Arbor, MI) or Sigma-Aldrich (St. Louis, MO). Pooled human liver microsomes (HLM) [n = 50, mixed-gender (21 female and 29 male), race (42 Caucasian, 4 Hispanic, 2 African American, and 2 Asian), and age (5–77 years old)] were obtained from Sekisui Xenotech, LLC (Lenexa, Kansas). NADPH-regenerating system (1.3 mM NADP, 3.3 mM glucose 6-phosphate, and 0.4 U/mL glucose 6-phosphate dehydrogenase) was obtained from Corning (Bedford, MA). Nicotine, nornicotine, HPBA, NOX, Nic-Gluc, cotinine, COX, Cot-Gluc, 3HC, and 3HC glucuronide (3HC-Gluc) standards were purchased from Sigma-Aldrich. Fluconazole, benzydamine hydrochloride, clopidogrel, trifluoperazine, diclofenac, amitriptyline, and tranylcypromine were also purchased from Sigma-Aldrich. Optima grade methanol, acetonitrile, and formic acid were obtained from Fisher Scientific (Waltham, MA). Ultra-low-binding microcentrifuge tubes, Dulbecco’s modified Eagle’s medium, Dulbecco’s phosphate-buffered saline, UDP glucuronic acid (UDPGA), alamethicin, MgCl2, and Geneticin (G418) were purchased from VWR (Radnor, PA). BCA protein assays were purchased from Pierce (Rockford, IL); premium-grade fetal bovine serum (FBS) was purchased from Seradigm (Radnor, PA), and ChromatoPur bovine serum albumin (BSA) was purchased from MB Biomedicals (Santa Ana, CA).
Inhibition Assays
HEK293 cells individually overexpressing V5-tagged CYP enzymes (CYP2A6, CYP2B6, and CYP2C19) and V5-tagged FMO enzymes (FMO1 and FMO3) were developed and described previously.16 Microsomal membrane fractions of CYP- and FMO-overexpressing cell lines were prepared by differential centrifugation as previously described, with protein concentrations estimated using the BCA assay, as per the manufacturer’s recommendations.16 An initial screening of the inhibition potential of CBD and 7-OH-CBD against individual CYP and FMO enzymes was determined using microsomes (30–100 μg) from CYP- or FMO-overexpressing HEK293 cell lines in reactions containing either 1 or 10 μM CBD or 7-OH-CBD as potential inhibitors, nicotine or cotinine as substrates, 100 mM potassium phosphate buffer (pH 7.4), and 3 mM MgCl2 in a final reaction volume of 30 μL. The nicotine and cotinine concentrations used in these reactions are described in Table 1 and were at or near their known Michaelis–Menten constant values (KM) for a given enzyme (e.g., nicotine to cotinine, 100 μM; cotinine to 3HC, 100 μM; nicotine to nornicotine, 50 μM; nicotine to NOX, 1 mM; and cotinine to COX, 1 mM14,16,19,30−33). As CBD exhibits extensive nonspecific binding (70–90%) to protein and labware, low-binding 0.6 mL microcentrifuge tubes were used for all reactions. Assays were preincubated for 5 min at 37 °C, initiated by the addition of 1.8 μL of NADPH-regenerating system, and incubated for 30 min at 37 °C. Reactions were terminated by the addition of 30 μL of ice-cold stop solution (acetonitrile/methanol; 1:1). Samples were mixed on a vortex mixer and centrifuged at 17 000g for 15 min at 4 °C. The supernatant (∼50 μL) was transferred to an ultra-high-performance liquid chromatography (UPLC) sample vial for subsequent UPLC-MS/MS analysis. Reactions with HLM were performed as described above for CYP- or FMO-overexpressing cell microsomes except with a 15 min incubation time. As positive controls for inhibition assays, tranylcypromine was added as an inhibitor for CYP2A6,34−36 fluconazole was added as an inhibitor for CYP2C19,37 and clopidogrel was added as an inhibitor of CYP2B6.38,39 As there are no known probe inhibitors of FMOs, a probe substrate, the FMO3 substrate benzydamine, was added as a potential inhibitor of FMOs 1 and 3.40−43 Substrate (nicotine/cotinine) without any inhibitor was used as the reference assay for 100% activity. All analyses were performed in triplicate.
Table 1. IC50 Valuesa of CBD and 7-OH-CBD against in Microsomes from Recombinant CYP and UGT-Overexpressing Cells and in HLM.
| CBD |
7-OH-CBD |
||||||
|---|---|---|---|---|---|---|---|
| metabolite | substrate | substrate concentrationb (μM) | microsomes | IC50 (μM) | IC50,uc (μM) | IC50 (μM) | IC50,uc (μM) |
| cotinine | nicotine | 100 μM | recd CYP2A6 | 4.4 ± 1.0 | 0.27 ± 0.06 | 5.8 ± 2.3 | 0.45 ± 0.18 |
| 500 μM | rec CYP2B6 | 4.2 ± 2.7 | 0.26 ± 0.17 | 15 ± 5.7 | 1.2 ± 0.44 | ||
| 500 μM | HLM | 10 ± 2.9 | 0.98 ± 0.28 | 21 ± 5.2 | 2.0 ± 0.49 | ||
| 3HC | cotinine | 100 μM | rec CYP2A6 | 3.4 ± 2.3 | 0.21 ± 0.14 | 10 ± 3.0 | 0.78 ± 0.23 |
| 900 μM | HLM | 8.8 ± 1.3 | 0.86 ± 0.13 | 9.1 ± 3.4 | 0.86 ± 0.32 | ||
| nornicotine | nicotine | 50 μM | rec CYP2A6 | 3.7 ± 2.2 | 0.23 ± 0.14 | 2.1 ± 0.97 | 0.16 ± 0.08 |
| 100 μM | rec CYP2B6 | 0.47 ± 0.077 | 0.029 ± 0.005 | 1.4 ± 0.40 | 0.11 ± 0.03 | ||
| 500 μM | HLM | 5.1 ± 1.1 | 0.50 ± 0.11 | 7.9 ± 2.5 | 0.74 ± 0.24 | ||
| 3HC-Gluc | 3HC | 30 mM | rec UGT1A9 | 9.7 ± 1.5 | 0.37 ± 0.06 | NAe | NAe |
IC50 values are presented as mean ± SD of three independent experiments.
Shown are the concentrations of substrate used in the respective assays for activity screenings and IC50 determinations.
IC50,u, binding-corrected IC50 values.
rec, recombinant.
NA, not analyzed.
Since purchased nicotine stocks had small amounts of nornicotine as a contaminant, nornicotine peak areas were calculated after subtracting out the “background” nornicotine peak observed within the stock nicotine prep, as determined by UPLC.
HEK293 cells individually overexpressing UGT isoforms 1A4, 1A9, 2B7, 2B10, and 2B17 were developed and described previously.44 Initial screenings of the inhibition potential of CBD and 7-OH-CBD against UGTs 1A4, 1A9, 2B7, 2B10, and 2B17 were performed similarly to that described above for CYP/FMO assays, using microsomes from UGT-overexpressing HEK293 cell lines or HLM (50–100 μg for both) in reactions containing either 10 or 100 μM CBD or 7-OH-CBD, substrate at concentrations approaching their known KM’s for a given enzyme (5 mM nicotine, 5 mM cotinine, or 30 mM 3HC22,23,45), 50 mM Tris–HCl buffer (pH 7.4), and 5 mM MgCl2 in a final reaction volume of 25 μL. As higher concentrations of cannabinoids were required for the UGT experiments, 2% BSA was added to increase the solubility of cannabinoids as well as to sequester inhibitory long-chain unsaturated fatty acids. Microsomes were preincubated with alamethicin (50 μg/mg of microsomal protein) on ice for 20 min prior to the addition of reaction components. Reactions were then initiated by the addition of 4 mM UDPGA and incubated for 60–120 min at 37 °C. Assays containing substrate without any inhibitor were used as the reference assay for 100% activity. All analyses were performed in triplicate. Reactions were terminated by the addition of 25 μL of ice-cold stop solution (acetonitrile/methanol; 1:1), mixed on a vortex mixer, and centrifuged at 17 000g for 15 min at 4 °C. The supernatant (∼40 μL) was transferred to a UPLC sample vial for subsequent UPLC-MS/MS analysis. As positive controls for inhibition assays, trifluoperazine was added as a potential inhibitor of UGT1A4,46,47 diclofenac as a potential inhibitor of UGTs 1A9 and 2B7,48,49 amitriptyline as a potential inhibitor of UGT2B10,50 and imatinib as a potential inhibitor of UGT2B17.51
Incubation conditions were optimized for HLM and overexpressing cell lines for amount of microsomal protein and reaction time, with optimal conditions chosen based on the following criteria: (1) Metabolite formation was linear with time and protein concentration. (2) Substrate consumption was no more than 20% of the initial amount. (3) Metabolite formation was reliably and reproducibly detected by the UPLC–MS/MS method utilized.
Metabolite Detection
For nicotine metabolite detection, we used a method identical to that described and validated in previous studies.33 Briefly, nicotine, NOX, Nic-Gluc, HPBA, cotinine, COX, Cot-Gluc, 3HC, and 3HC-Gluc were detected using a UPLC (Waters Acquity; Waters Corp, Milford, MA) coupled to a triple-quadrupole mass spectrometer equipped with a Zspray electrospray ionization interface (Waters Xevo TQD; Waters Corp) by multiple reaction monitoring (MRM). While a nornicotine standard was not available for the present studies, nornicotine was identified using both parent and daughter scans to develop and confirm an MRM method for nornicotine detection. Except for the detection of NOX, all samples were injected (2–5 μL) onto an Acquity UPLC BEH C18 column (1.7 μM, 2.1 × 100 mm). A 9 min gradient elution was used with mobile phases A (0.1% formic acid in water) and B (100% methanol) as follows: 1 min at 95% A; 5% B followed by a linear gradient for 7 min to 5% A; 95% B, 1 min at 5% A; 95% B and re-equilibration for 1 min at 95% A; 5% B. The flow rate was 0.4 mL/min, and the column temperature was 40 °C. A UPLC-BEH-HILIC column (2.1 × 100 mm, 1.7 μm) was used for the detection of NOX, with 5 mmol/L NH4AC in 50% acetonitrile as buffer A and 100% acetonitrile as buffer B, with elution as follows: 20% buffer A for 1.5 min, a linear gradient to 100% buffer A from 1.5 to 2.5 min, maintenance of 100% buffer A for 3 min, and a re-equilibrium step to the initial 20% buffer A conditions from 5.5 to 7 min (flow rate = 0.4 mL/min). The injection volume was 2 μL using a column temperature of 30 °C. Analytes were detected with the mass spectrometer operated in the positive ion mode for all metabolites tested in this study using dwell times of 10–53 ms and collision energy and cone voltages of 15–28 and 20–40 V, respectively. Ultrapure argon was used for collision-induced dissociation. The mass transitions (Q1/Q3), collision energies, and cone voltages for each substrate and metabolite are listed in Table S1. The desolvation temperature was 500 °C, with 600 L/h of nitrogen gas for desolvation and 30 L/h for the cone, while the temperature of the source was 120 °C. Observed metabolite retention times (see Table S1) were compared with the retention times of corresponding substrate or metabolite standards.
Assay accuracy and precision accuracy were validated on LC-MS/MS by repeated sample quantification by preparing five individual samples of reaction matrix containing 1 ppm cotinine, COX, or 3HC standard, processing them for LC-MS/MS, and immediately measuring cotinine, COX, or 3HC concentrations for each standard sample as compared to individually prepared standard curves all measured by LC-MS/MS as previously described.52 The mean recovery for the five samples was >88% while the coefficient of variation (CV) was 3.1% for these samples, suggesting high accuracy and precision for these studies.
Determination of IC50 Values
CBD or 7-OH-CBD concentrations that inhibited relative enzyme activity by ≥50% at 10 μM for CYPs and FMOs and 100 μM for UGTs were further investigated by the determination of IC50 values for each inhibitor–enzyme combination. IC50 determinations were performed in HLM and microsomes from HEK293-overexpressing cell lines using multiple concentrations of CBD and 7-OH-CBD ranging between 0.5 and 120 μM, with all determinations performed in three independent experiments. Nonspecific binding constants (fu,inc) for CBD in HEK293 microsomes and HLM were determined previously.29,53 For the fu,inc for 7-OH-CBD, the previously determined fu,inc for the structurally similar 11-OH-THC53 was used as a surrogate. IC50 values were determined using the following equation:
where “bottom” is the maximum observed inhibition (lowest percent activity), and “top” is the minimum observed inhibition (highest percent activity) for a given cannabinoid.
Statistical Analysis
Data were exported and analyzed using an Excel spreadsheet (Microsoft). The amount of metabolite formed at each concentration of inhibitor relative to the control (% relative activity) was calculated as
IC50 values were calculated by plotting the relative activity of each enzyme versus the log concentration of the test inhibitors using GraphPad Prism 7.04 software (GraphPad Software Inc., San Diego, CA). IC50 for unbound drug (IC50,u) values were calculated using the following equation: IC50,u = IC50 × fu,inc.
Results
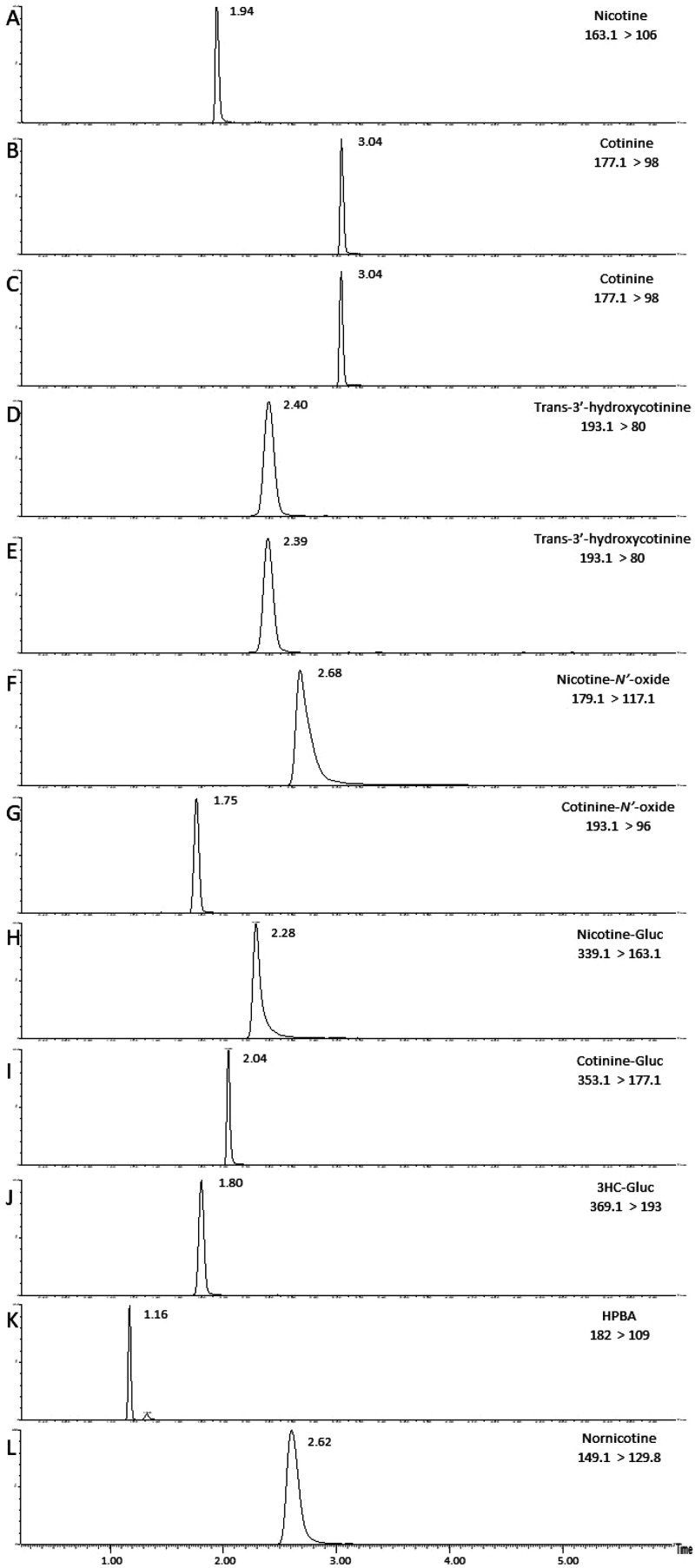
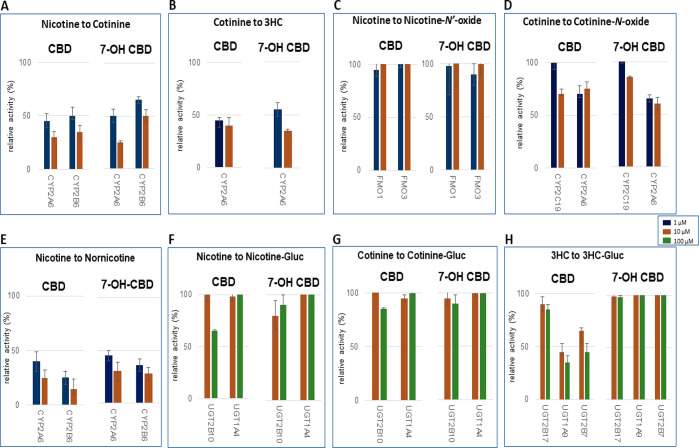
Metabolite peaks were detected by UPLC-MS/MS (Figure 2) in assays utilizing substrates from the nicotine metabolic pathway, as described above. Using nicotine as the substrate, preliminary screening studies demonstrated that, in assays containing CYP2A6-overexpressing cell microsomes, CBD and 7-OH-CBD inhibited cotinine formation by 55% and 50% at 1 μM, and 70% and 75% at 10 μM, respectively, as compared to 100% activity control reactions without added cannabinoid (Figure 3A). A similar pattern was observed when using cotinine as the substrate in CYP2A6-overexpressing cell microsomes, with CBD and 7-OH-CBD decreasing 3HC formation by 55% and 45% at 1 μM, and 60% and 65% at 10 μM, respectively, as compared to 100% activity control reactions (Figure 3B). A similar pattern was also observed for microsomes from CYP2B6-overexpressing cells, with 1 and 10 μM CBD exhibiting 50% and 65% inhibition, respectively, and 1 and 10 μM 7-OH-CBD exhibiting 35% and 50% inhibition, respectively, against cotinine formation using nicotine as the substrate (Figure 3A). No inhibitory effect on NOX formation was observed with either cannabinoid in FMO1- or FMO3-overexpressing cell microsomes in assays where nicotine was the substrate (Figure 3C), while only marginal inhibition was observed for 10 μM CBD and 7-OH-CBD against COX formation in microsomes from cells overexpressing either CYP2C19 (30% and 15% inhibition, respectively) or CYP2A6 (25% and 40%, respectively) with cotinine as the substrate (Figure 3D).
Figure 2.
Chromatograms of nicotine and its metabolites formed in assays containing microsomes from CYP-overexpressing HEK293 cells and standards. The metabolites examined are described with the mass transition shown in brackets. (A) Nicotine standard; (B) cotinine standard; (C) cotinine formation in CYP2A6 microsomes; (D) 3HC standard; (E) 3HC formation in CYP2A6 microsomes; (F) nicotine-N′-oxide standard; (G) cotinine-N′-oxide standard; (H) nicotine-Gluc standard; (I) cotinine-Gluc standard; (J) 3HC-Gluc standard; (K) HPBA standard; and (L) nornicotine formation in CYP2A6 microsomes.
Figure 3.
Screening of CBD and 7-OH-CBD inhibition of major nicotine metabolic pathways in microsomes from CYP-, FMO-, and UGT-overexpressing HEK293 cells. Incubations were performed using 1 or 10 μM cannabinoid, with nicotine, cotinine, and 3HC concentrations at or close to their known KM for their corresponding enzyme: 100 μM, 500 μM, 1 mM, 50 μM, 100 μM, and 5 mM nicotine for cotinine (CYP2A6), cotinine (CYP2B6), NOX (FMOs 1 and 3), nornicotine (CYP2A6), nornicotine (CYP2B6), and nicotine-Gluc (UGT2B10 and UGT1A4) formation reactions, respectively; 100 μM (CYP2A6), 1 mM (CYP2C19 and CYP2A6), and 5 mM (UGT2B17) cotinine for 3HC, COX, and cotinine-Gluc formation reactions, respectively; and 30 mM 3HC for 3HC-Gluc formation reactions (UGT1A9 and UGT2B7). (A) Nicotine to cotinine formation; (B) cotinine to 3HC formation; (C) nicotine to NOX formation; (D) cotinine to COX formation; (E) nicotine to nornicotine formation; (F) nicotine to nicotine-Gluc formation; (G) cotinine to cotinine-Gluc formation; and (H) 3HC to 3HC-Gluc formation. Shown are the mean inhibitions of three individual experiments performed for each cannabinoid against nicotine, cotinine, or 3HC as substrate. Data are expressed as a percentage of metabolite formation formed in assays with cannabinoid compared to assays without cannabinoid.
As controls for the inhibition assays, potential inhibitors exhibited the following levels of inhibition of nicotine metabolism using microsomes from recombinant enzymes: 70% and 95% inhibition of cotinine and nornicotine formation, respectively, in CYP2A6 microsomes by 10 μM tranylcypromine, 68% inhibition of Nic-Gluc formation in UGT1A4 microsomes by 100 μM trifluoperazine, and 50 μM clopidogrel exhibited 60% inhibition of cotinine formation by CYP2B6. Similar levels of inhibition were observed when using cotinine as the substrate (results not shown), with 55% of COX formation inhibited by 10 μM fluconazole in CYP2C19 microsomes. In addition, up to 65% inhibition was observed for 3HC-Gluc formation in UGT2B7, UGT1A9, or UGT2B17 microsomes containing 200 μM imatinib, and nicotine-Gluc or cotinine-Gluc formation in UGT2B10 microsomes containing 100 μM amitriptyline. Known inhibitors of FMO activity have not been previously described, and no significant inhibition was observed when adding up to 50 μM benzydamine as a potential inhibitor of FMO activities in the present studies.
Screening for inhibition by CBD and 7-OH-CBD was also performed for two additional nicotine metabolites (nornicotine and HPBA) in both CYP2A6- and CYP2B6-overexpressing microsomes. Both CBD and 7-OH-CBD exhibited inhibition of nornicotine formation in CYP2A6-overexpressing cell microsomes, with 60% and 55% inhibition observed with 1 μM CBD and 7-OH-CBD, respectively, and 75% and 70% inhibition observed with 10 μM CBD and 7-OH-CBD, respectively (Figure 3E). Similarly strong inhibition was also observed for nornicotine formation in CYP2B6-overexpressing cell microsomes, with 1 and 10 μM CBD exhibiting 75% and 85% inhibition, respectively, and 1 and 10 μM 7-OH-CBD exhibiting 64% and 72% inhibition, respectively (Figure 3E). No HPBA formation was observed with either CYP2A6- or CYP2B6-overexpressing microsomes when nicotine was used as substrate in the present studies, a pattern consistent with that observed in previous studies demonstrating a lack of HPBA formation activity by CYP2A6 with nicotine as the substrate.
Previous studies have shown that CBD does not inhibit nicotine-Gluc formation by UGT2B10.29 In the present study, no significant inhibition was observed for either CBD or 7-OH-CBD for nicotine-Gluc or cotinine-Gluc formation in microsomes from UGT2B10- or UGT1A4-overexpressing cells using nicotine or cotinine as the substrate (Figure 3F,G). No effect on 3HC-Gluc formation was observed for either cannabinoid using 3HC as the substrate in microsomes from cells overexpressing UGTs 2B17, 1A9, or 2B7 for 7-OH-CBD. In contrast, 1 and 10 μM CBD exhibited 55% and 65% decreases, respectively, in 3HC-Gluc formation for UGT1A9 microsomes, and 35% and 55% decreases, respectively, in 3HC-Gluc formation for UGT2B7 microsomes (Figure 3H).
The inhibitory effects of CBD and 7-OH-CBD were extended to establish IC50 and binding-corrected IC50 values (IC50,u) for CBD and 7-OH-CBD against both CYP and UGT enzymes shown to be inhibited in the screening assays (described above). As described above, the known fu,inc for CBD in HLM or microsomes containing CYP- or UGT-overexpressed enzymes29,53 and the fu,inc for 11-OH-THC53 as a surrogate for the unbound fraction of 7-OH-CBD in HLM or microsomes containing CYP-overexpressing enzymes were used to calculate IC50,u values. Since IC50 values were not calculated for 7-OH-CBD in UGT microsomal incubations (it exhibited no significant inhibition of UGT enzymes in the screening studies described above), determinations of unbound fractions for UGT assays including 7-OH-CBD were not performed.
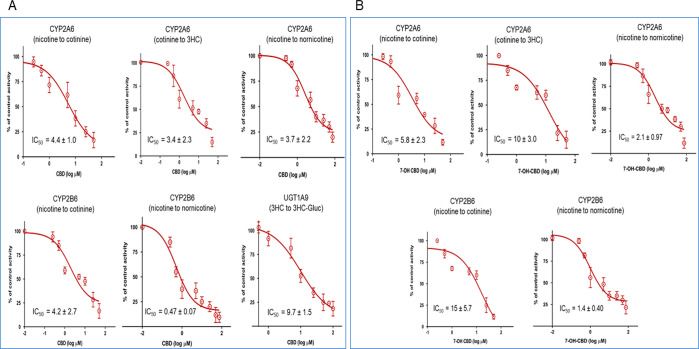
Inhibition curves for enzymes shown to be inhibited by CBD and 7-OH-CBD are shown in Figure 4. CBD showed strong inhibition of CYP2A6 activity in CYP2A6-overexpressing microsomes using either nicotine or cotinine as substrate, with IC50,u values of 0.27 ± 0.06 and 0.21 ± 0.14 μM for cotinine and 3HC formation, respectively (Table 1). Similarly, CBD inhibited CYP2B6-mediated metabolism of nicotine to cotinine with an IC50,u value of 0.26 ± 0.17 μM. Moreover, CBD inhibited CYP2A6- and CYP2B6-mediated nicotine to nornicotine formation with an IC50,u value of 0.23 ± 0.14 and 0.030 ± 0.0050 μM, respectively. To confirm that the effects observed by CBD in CYP-overexpressing microsomes are also observed in human liver, inhibition assays were performed in HLM, with similar IC50,u values observed for CBD in HLM (0.98 ± 0.28 μM for cotinine formation and 0.50 ± 0.11 μM for nornicotine formation) as compared to that observed in incubations with microsomes from CYP2A6- or CYP2B6-overexpressing cells. CBD also exhibited strong inhibition of glucuronidation activity in UGT1A9-overexpressing microsomes against 3HC, with an IC50,u value of 0.37 ± 0.06 μM. While 100 μM CBD exhibited >50% inhibition of UGT2B7 activity in the activity screenings described above, IC50 values could not be calculated for UGT2B7 due to inconsistent inhibition at higher CBD concentrations with this enzyme (results not shown). IC50 values could also not be calculated for 3HC-Gluc formation for CBD in HLM (inhibition reached <45% with up to 100 μM CBD), likely due to the presence of other UGTs (i.e., UGT2B17) which also glucuronidate 3HC but which are not inhibited by CBD.
Figure 4.
Inhibitory effects of CBD and 7-OH-CBD in microsomes from CYP- and UGT-overexpressing HEK293 cells. (A) Incubations performed using CBD as the inhibitor; (B) incubations performed using 7-OH-CBD as the inhibitor. Shown are averages of triplicate plots comparing CBD or 7OH-CBD concentration (X-axis) with the percent of control activity (Y-axis) against substrates in CYP- or UGT-overexpressing microsomes. Incubations were performed for 30–120 min at 37 °C using 30–100 μg of microsomal protein, 0.5–120 μM of either CBD or 7-OH-CBD, and 100 μM (CYP2A6) and 500 μM (CYP2B6) nicotine for cotinine formation reactions, 50 μM (CYP2A6) and 100 μM (CYP2B6) nicotine for nornicotine formation reactions, 100 μM cotinine for 3HC formation reactions, and 30 mM 3HC for 3HC-Gluc formation reactions. Individual metabolites were analyzed by UPLC-MS/MS as described in the Experimental Procedures section.
7-OH-CBD also exhibited significant inhibition against both cotinine and 3HC formation in reactions containing nicotine or cotinine, with IC50,u values of 0.45 ± 0.18 and 0.78 ± 0.23 μM, respectively, in incubations containing microsomes from CYP2A6-overexpressing cells (Figure 4B and Table 1). 7-OH-CBD also showed strong inhibition of cotinine formation (IC50,u = 1.2 ± 0.44 μM) in incubations containing microsomes from CYP2B6-overexpressing cells and showed strong inhibition of nornicotine formation in incubations containing nicotine and microsomes overexpressing either CYP2A6 (IC50,u = 0.16 ± 0.08 μM) or CYP2B6 (IC50,u = 0.11 ± 0.03 μM). Comparable IC50,u values of 2.0 ± 0.49, 0.86 ± 0.32, and 0.74 ± 0.24 μM were also observed for 7-OH-CBD inhibition of cotinine, 3HC, and nornicotine formation, respectively, in incubations containing HLM.
Discussion
The present study is the first to conduct a comprehensive examination of the inhibitory effects of CBD and its active metabolite 7-OH-CBD on nicotine metabolism, with the enzymatic activities of each of the major enzymes individually assessed for potential drug–drug interactions with these cannabinoids. The results indicate that CBD and 7-OH-CBD strongly inhibit CYP2A6 activity, as both cannabinoids inhibited the formation of cotinine and 3HC in incubations containing CYP2A6-overexpressing microsomes, with nonspecific binding-corrected IC50,u values <0.30 μM for CBD and <0.80 μM for 7-OH-CBD. Interestingly, the IC50,u values for the metabolism of nicotine and cotinine by CYP2A6 and CYP2B6 are similar. In addition, both CBD and 7-OH-CBD inhibited the formation of the secondary nicotine metabolite, nornicotine, in CYP2A6- or CYP2B6-overexpressing microsomal reactions, suggesting that the inhibition observed for these cannabinoids was not metabolite-specific for nicotine metabolites. This pattern of inhibition of nicotine metabolism by CYP2A6 and CYP2B6 by CBD and 7-OH-CBD was also observed in HLM. While the strong inhibition observed for CBD and 7-OH-CBD for both enzymes is similar to that observed for CBD in previous studies of several major hepatic CYP450 enzymes including CYP2A6 and CYP2B6 using probe substrates,28,53 differences in IC50,u values were observed for the substrates used in the present study for both enzymes as compared to the probe substrates used in those studies. This may be due to differences in the substrates used between studies as inhibition can be substrate specific.29,53 While IC50,u values collected from in vitro experiments are not necessarily good predictors of DDIs, the IC50,u values observed for both CBD and 7-OH-CBD in both recombinant enzymes systems and in HLM approach the physiological levels of CBD and 7-OH-CBD in the plasma corrected for plasma protein binding after a single clinical dose of CBD (0.06–0.15 μM CBD; 0.04–0.09 μM 7-OH-CBD9,10). With additional chronic or frequent dosing of CBD, the physiologic unbound fractions of plasma CBD and 7-OH-CBD are likely higher than the levels necessary for an in vivo interaction. Together, this suggests that these cannabinoids may be inhibiting overall nicotine metabolism in smokers.
Significant inhibition of several other major nicotine metabolism pathways was not observed by CBD or 7-OH-CBD in the present studies. This includes the oxidation of nicotine by FMOs 1 and 3 and the glucuronidation of nicotine or cotinine by UGT2B10. This is consistent with previous observations of FMOs not being shown to be subject to induction or inhibition by most xenobiotics.54 Additionally, previous studies have also observed a lack of cannabinoid inhibition of the UGT2B10-mediated glucuronidation of nicotine.29 The inhibition of UGT1A9-mediated 3HC-Gluc formation by CBD observed in this study could potentially cause increases in plasma 3HC in smokers. This could affect the inherent ratio of plasma 3HC:cotinine, which is often used as a biomarker of CYP2A6 activity.55
A limitation of the present study was that the inhibition by CBD or 7-OH-CBD of cotinine formation directly from the nicotine Δ1′,5′-iminium ion was not tested in the present study. While CYP2A6 has been previously shown to be active against the iminium ion, aldehyde oxidase has also been shown to be active.56,57 However, it is likely that the inhibition of cotinine formation from nicotine by CBD and 7-OH-CBD observed in the present study was not due to an effect on aldehyde oxidase since HLM and microsomal preparations of CYP2A6-overexpressing HEK293 cells were used, which should have minimal contamination by cytosolic proteins including aldehyde oxidase. The results presented in this study are also consistent with previous studies of both HLM and recombinant CYP2A6 demonstrating cotinine formation from nicotine.19,58,59
Tobacco users are known to precisely titrate nicotine levels throughout the day, finding the perfect balance between reward and aversion.60 While some nicotinic receptor agonists and partial agonists help tobacco users quit,61 these therapies have low success rates and significant side effects. Since greater than 70% of nicotine is metabolized to cotinine by CYP2A6 in the majority of tobacco users,62 reducing CYP2A6-mediated nicotine metabolism has been suggested as a primary target to reduce the number of cigarettes necessary for a smoker to maintain a given nicotine level, leading to diminished smoking overall.21,63 Functional variants of CYP2A6 (and consequent variations in nicotine metabolism across individuals) are known to affect human smoking behavior,64 with individuals carrying reduced-activity or nonfunctional CYP2A6 variants (i.e., slow metabolizers) less likely to become smokers,65−67 and those who do become smokers consuming significantly fewer cigarettes65,67,68 and scoring lower for nicotine dependence.65,67−69 The major endogenous function of CYP2A6 is still unknown, and subjects homozygous for CYP2A6 low-activity or deletion alleles (1:100 to 1:1000 people) have no discernible biological or physical complications.70 In addition, there are few major drugs [selegiline (Ki = 4.6 μM) is one of the few71] that are metabolized primarily by CYP2A6 and have the potential for drug–drug interactions. While there are few randomized trials of cessation stratified by CYP2A6 genotype, the meta-relative risk of 6-month abstinence was 0.54 (95% CI 0.37–0.78) in normal vs slow metabolizers.72 A systematic review of the literature found that faster nicotine metabolizers smoke more cigarettes per day (cpd) due to a decreased half-life of nicotine.73 Several studies focusing on CYP2A6 support this, with CYP2A6 activity correlated with total cigarette consumption in smokers of both Japanese and European ancestry.74 Faster CYP2A6-mediated nicotine metabolism was shown to be associated with greater total daily puffs and puff volume.75 In addition, a predictive nicotine metabolism model based on CYP2A6 haplotypes showed a similar association between nicotine oxidation to cotinine and cpd in African American smokers,76 and recent clinical studies recommend the targeting of CYP2A6 activity for tobacco use disorder treatment.77 Therefore, with the vast majority of the population having active CYP2A6, pharmacological reduction of CYP2A6 activity is expected to mimic the effects of naturally occurring CYP2A6 variants in the human population, reducing both smoking behavior and dependence.
In a previous randomized, double-blind placebo-controlled study, the ad-hoc use of CBD by inhaler in cigarette smokers resulted in a significant ∼40% reduction of the number of cigarettes smoked during treatment.26 In addition, Hindocha et al. conducted a series of studies in which they discovered that vaping cannabis was associated with decreased tobacco consumption, and a single 800 mg oral dose of CBD reduced the salience and pleasantness of cigarette cues, compared with a placebo group.78 These results are consistent with the inhibitory effects by CBD and 7-OH-CBD on CYP2A6-mediated nicotine metabolism described in the present study, with decreases in nicotine metabolism potentially leading to increased plasma nicotine levels per cigarette smoked and a reduction in the number of cigarettes smoked, thus diminishing the adverse health effects of smoking. Further investigations will be required to determine the potential for CBD and potentially other cannabinoids as agents for tobacco cessation therapy.
Acknowledgments
The authors would like to thank Gang Chen for his helpful contributions to the study. Figure 1 was created with BioRender.com.
Glossary
Abbreviations
- AUC
area under the curve
- BCA
bicinchoninic acid
- BSA
bovine serum albumin
- CBD
cannabidiol
- CBN
cannabinol
- CBD-COOH
7-carboxy-cannabidiol
- 7-OH-CBD
7-hydroxy-cannabidiol
- CYP
cytochrome P450
- COX
cotinine-N-oxide
- Cot-Gluc
cotinine glucuronide
- DDI
drug–drug interaction
- FBS
fetal bovine serum
- FMO
flavin monooxygenase
- HEK
human embryonic kidney
- HLM
human liver microsomes
- IC50
half-maximal inhibitory concentration
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MRM
multiple reaction monitoring
- NADP
β-nicotinamide adenine dinucleotide phosphate
- NOX
nicotine-N′-oxide
- Nic-Gluc
nicotine glucuronide
- THC
(−)-trans-Δ9-tetrahydrocannabinol
- 3HC
trans-3′-hydroxycotinine
- UDPGA
UDP glucuronic acid
- UPLC
ultraperformance liquid chromatography
- HPBA
4-hydroxy-4-(3-pyridyl)-butanoic acid
- 3HC-Gluc
3HC glucuronide
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.2c00259.
Mass spectrometry conditions used to analyze individual nicotine metabolites (PDF)
Author Contributions
§ S.N., S.C., and K.B. contributed equally to this work. CRediT: Shamema Nasrin conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing-original draft, writing-review & editing; Shelby Coates data curation, formal analysis, validation, visualization, writing-review & editing; Keti Bardhi data curation, formal analysis, validation, visualization, writing-review & editing; Christy Watson methodology, writing-original draft; Joshua E. Muscat writing-original draft, writing-review & editing; Philip Lazarus conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing-original draft, writing-review & editing.
This work was supported by the National Institutes of Health National Institutes of Environmental Health Sciences [Grants R01-ES025460] to P.L.
The authors declare no competing financial interest.
Notes
Significance Statement. The present study is the first to comprehensively examine the effects of CBD and its major active metabolite, 7-OH-CBD, on the metabolism of nicotine. Results from this study demonstrate that both CBD and 7-OH-CBD inhibit nicotine metabolism by inhibiting several enzymes important in nicotine metabolism, including the major nicotine metabolizing enzyme, CYP2A6. This inhibition has implications for co-users of tobacco and CBD, with decreases in overall nicotine metabolism potentially influencing tobacco addiction and cessation strategies.
Supplementary Material
References
- McClure E. A.; Tomko R. L.; Salazar C. A.; Akbar S. A.; Squeglia L. M.; Herrmann E.; Carpenter M. J.; Peters E. N. Tobacco and cannabis co-use: Drug substitution, quit interest, and cessation preferences. Exp Clin Psychopharmacol 2019, 27, 265–275. 10.1037/pha0000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin R. D.; Pacek L. R.; Copeland J.; Moeller S. J.; Dierker L.; Weinberger A.; Gbedemah M.; Zvolensky M. J.; Wall M. M.; Hasin D. S. Trends in Daily Cannabis Use Among Cigarette Smokers: United States, 2002–2014. Am. J. Public Health. 2018, 108, 137–142. 10.2105/AJPH.2017.304050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook J. S.; Lee J. Y.; Finch S. J.; Brown E. N. Course of comorbidity of tobacco and marijuana use: Psychosocial risk factors. Nicotine Tob Res. 2010, 12, 474–482. 10.1093/ntr/ntq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee R. G. The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: delta9-tetrahydrocannabinol, cannabidiol and delta9-tetrahydrocannabivarin. Br. J. Pharmacol. 2008, 153, 199–215. 10.1038/sj.bjp.0707442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S.; Maharao N.; Paine M. F.; Unadkat J. D. Predicting the Potential for Cannabinoids to Precipitate Pharmacokinetic Drug Interactions via Reversible Inhibition or Inactivation of Major Cytochromes P450. Drug Metab. Dispos. 2020, 48, 1008–1017. 10.1124/dmd.120.000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen C.; Shahinas J. Dosage, Efficacy and Safety of Cannabidiol Administration in Adults: A Systematic Review of Human Trials. J. Clin Med. Res. 2020, 12, 129–141. 10.14740/jocmr4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14% of Americans Say They Use CBD Products, 2019. https://news.gallup.com/poll/263147/americans-say-cbd-products.aspx.
- Beers J. L.; Fu D.; Jackson K. D. Cytochrome P450-Catalyzed Metabolism of Cannabidiol to the Active Metabolite 7-Hydroxy-Cannabidiol. Drug Metab. Dispos. 2021, 49, 882–891. 10.1124/dmd.120.000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L.; Gidal B.; Blakey G.; Tayo B.; Morrison G. A Phase I, Randomized, Double-Blind, Placebo-Controlled, Single Ascending Dose, Multiple Dose, and Food Effect Trial of the Safety, Tolerability and Pharmacokinetics of Highly Purified Cannabidiol in Healthy Subjects. CNS Drugs 2018, 32, 1053–1067. 10.1007/s40263-018-0578-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highlights of prescribing information: Epidiolex (cannabidiol) oral solution; US Food and Drug Administration; Greenwich Biosciences Inc.https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/210365lbl.pdf.
- Vlad R. A.; Hancu G.; Ciurba A.; Antonoaea P.; Rédai E. M.; Todoran N.; Silasi O.; Muntean D. L. Cannabidiol - therapeutic and legal aspects. Die Pharmazie 2020, 75, 463–469. [DOI] [PubMed] [Google Scholar]
- Stott C.; Jones N.; Whalley B.; Stephens G.; Williams C.. 7-oh-cannabidiol (7-oh-cbd) and/or 7-oh-cannabidivarin (7-oh-cbdv) for use in the treatment of epilepsy; GW Pharma Limited. US Patent US20220062197A1, 2015.
- Benowitz N. L.; Jacob P. Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther 1994, 56, 483–493. 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- Yamazaki H.; Inoue K.; Hashimoto M.; Shimada T. Roles of CYP2A6 and CYP2B6 in nicotine C-oxidation by human liver microsomes. Arch. Toxicol. 1999, 73, 65–70. 10.1007/s002040050588. [DOI] [PubMed] [Google Scholar]
- Chen G.; Giambrone N. E.; Dluzen D. F.; Muscat J. E.; Berg A.; Gallagher C. J.; Lazarus P. Glucuronidation Genotypes and Nicotine Metabolic Phenotypes: Importance of Functional UGT2B10 and UGT2B17 Polymorphisms. Cancer Res. 2010, 70, 7543–7552. 10.1158/0008-5472.CAN-09-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Paramo Y. X.; Chen G.; Ashmore J. H.; Watson C. J. W.; Nasrin S.; Adams-Haduch J.; Wang R.; Gao Y.-T.; Koh W.-P.; Yuan J.-M.; Lazarus P. Nicotine-N′-Oxidation by Flavin Monooxygenase Enzymes. Cancer Epidemiol Biomarkers Prev 2019, 28, 311–320. 10.1158/1055-9965.EPI-18-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. M.; von Weymarn Lb Fau - Murphy S. E.; Murphy S. E. Identification of N-(hydroxymethyl) norcotinine as a major product of cytochrome P450 2A6, but not cytochrome P450 2A13-catalyzed cotinine metabolism. Chem. Res. Toxicol. 2005, 18 (12), 1792–1798. 10.1021/tx0501381. [DOI] [PubMed] [Google Scholar]
- Wade E.; Bowman E. R.; Turnbull L. B.; McKennis H. Norcotinine (desmethylcotinine) as a urinary metabolite of nornicotine. J. Med. Pharm. Chem. 1961, 4, 21–30. 10.1021/jm50017a002. [DOI] [PubMed] [Google Scholar]
- Murphy S. E.; Raulinaitis V Fau - Brown K. M.; Brown K. M. Nicotine 5′-oxidation and methyl oxidation by P450 2A enzymes. Drug Metab. Dispos. 2005, 33 (8), 1166–1173. 10.1124/dmd.105.004549. [DOI] [PubMed] [Google Scholar]
- Hecht S. S.; Hochalter Jb Fau - Villalta P. W.; Villalta Pw Fau - Murphy S. E.; Murphy S. E. 2′-Hydroxylation of nicotine by cytochrome P450 2A6 and human liver microsomes: formation of a lung carcinogen precursor. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (23), 12493–12497. 10.1073/pnas.220207697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz N. L. Pharmacology of Nicotine: Addiction, Smoking-Induced Disease, and Therapeutics. Annu. Rev. Pharmacol Toxicol 2009, 49, 57–71. 10.1146/annurev.pharmtox.48.113006.094742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Blevins-Primeau A. S.; Dellinger R. W.; Muscat J. E.; Lazarus P. Glucuronidation of Nicotine and Cotinine by UGT2B10: Loss of Function by the UGT2B10 Codon 67 (Asp > Tyr) Polymorphism. Cancer Res. 2007, 67, 9024–9029. 10.1158/0008-5472.CAN-07-2245. [DOI] [PubMed] [Google Scholar]
- Chen G.; Giambrone N. E.; Lazarus P. Glucuronidation of trans-3′-hydroxycotinine by UGT2B17 and UGT2B10. Pharmacogen and Genomics 2012, 22, 183–190. 10.1097/FPC.0b013e32834ff3a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianezza M. L.; Sellers E. M.; Tyndale R. F. Nicotine metabolism defect reduces smoking. Nature 1998, 393 (6687), 750. 10.1038/31623. [DOI] [PubMed] [Google Scholar]
- Lekhram Changoer G. A.Chewing gum composition comprising cannabinoids and nicotine; Apirx Pharmaceutical Usa LLC, 2020. [Google Scholar]
- Morgan C. J. A.; Das R. K.; Joye A.; Curran H. V.; Kamboj S. K. Cannabidiol reduces cigarette consumption in tobacco smokers: Preliminary findings. Addictive Behav 2013, 38 (9), 2433–6. 10.1016/j.addbeh.2013.03.011. [DOI] [PubMed] [Google Scholar]
- Cox E. J.; Maharao N.; Patilea-Vrana G.; Unadkat J. D.; Rettie A. E.; McCune J. S.; Paine M. F. A marijuana-drug interaction primer: Precipitants, pharmacology, and pharmacokinetics. Pharmacol Ther 2019, 201, 25–38. 10.1016/j.pharmthera.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S.; Paine M. F.; Unadkat J. D. Comprehensive Predictions of Cytochrome P450 (P450)-Mediated In Vivo Cannabinoid-Drug Interactions Based on Reversible and Time-Dependent P450 Inhibition in Human Liver Microsomes. Drug Metab. Dispos. 2022, 50, 351–360. 10.1124/dmd.121.000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin S.; Watson C. J. W.; Bardhi K.; Fort G.; Chen G.; Lazarus P. Inhibition of UDP-Glucuronosyltransferase Enzymes by Major Cannabinoids and Their Metabolites. Drug Metab. Dispos. 2021, 49, 1081–1089. 10.1124/dmd.121.000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M.; Yamamoto T.; Nunoya K.; Yokoi T.; Nagashima K.; Inoue K.; Funae Y.; Shimada N.; Kamataki T.; Kuroiwa Y. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab. Dispos. 1996, 24 (11), 1212. [PubMed] [Google Scholar]
- Nakajima M.; Yamamoto T.; Nunoya K.; Yokoi T.; Nagashima K.; Inoue K.; Funae Y.; Shimada N.; Kamataki T.; Kuroiwa Y. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J. Pharmacol Exp Ther 1996, 277 (2), 1010–1015. [PubMed] [Google Scholar]
- Yamanaka H.; Nakajima M.; Fukami T.; Sakai H.; Nakamura A.; Katoh M.; Takamiya M.; Aoki Y.; Yokoi T. CYP2A6 AND CYP2B6 are involved in nornicotine formation from nicotine in humans: interindividual differences in these contributions. Drug Metab. Dispos. 2005, 33 (12), 1811–1818. 10.1124/dmd.105.006254. [DOI] [PubMed] [Google Scholar]
- Perez-Paramo Y. X.; Watson C. J. W.; Chen G.; Thomas C. E.; Adams-Haduch J.; Wang R.; Khor C. C.; Koh W.-P.; Nelson H. H.; Yuan J.-M.; Lazarus P. Impact of Genetic Variants in the Nicotine Metabolism Pathway on Nicotine Metabolite Levels in Smokers. Cancer Epidemiol Biomarkers Prev EPI-22-0868 2022, OF1. 10.1158/1055-9965.EPI-22-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper A. J.; Madan A.; Fau; Parkinson A.; Parkinson A. Inhibition of coumarin 7-hydroxylase activity in human liver microsomes. Arch. Biochem. Biophys. 1997, 341 (1), 47–61. 10.1006/abbi.1997.9964. [DOI] [PubMed] [Google Scholar]
- Zhang W.; Kilicarslan T.; Tyndale R.; Sellers E. Evaluation of methoxsalen, tranylcypromine, and tryptamine as specific and selective CYP2A6 inhibitors in vitro. Drug Metab. Dispos. 2001, 29 (6), 897–902. [PubMed] [Google Scholar]
- Taavitsainen P.; Juvonen R.; Fau; Pelkonen O.; Pelkonen O. In vitro inhibition of cytochrome P450 enzymes in human liver microsomes by a potent CYP2A6 inhibitor, trans-2-phenylcyclopropylamine (tranylcypromine), and its nonamine analog, cyclopropylbenzene. Drug Metab. Dispos. 2001, 29 (3), 217–222. [PubMed] [Google Scholar]
- Wienkers L. C.; Wurden C.J.; Storch E.; Kunze K. L.; Rettie A. E.; Trager W. F. Formation of (R)-8-hydroxywarfarin in human liver microsomes. A new metabolic marker for the (S)-mephenytoin hydroxylase, P4502C19. Drug Metab. Dispos. 1996, 24 (5), 610–614. [PubMed] [Google Scholar]
- Richter T.; Mürdter T. E.; Heinkele G.; Pleiss J.; Tatzel S.; Schwab M.; Eichelbaum M.; Zanger U. M. Potent mechanism-based inhibition of human CYP2B6 by clopidogrel and ticlopidine. J. Pharmacol Exp Ther 2004, 308 (1), 189–197. 10.1124/jpet.103.056127. [DOI] [PubMed] [Google Scholar]
- Hagihara K.; Nishiya Y.; Kurihara A.; Kazui M.; Farid N. A.; Ikeda T. Comparison of human cytochrome P450 inhibition by the thienopyridines prasugrel, clopidogrel, and ticlopidine. Drug Metab Pharmacokinet. 2008, 23 (6), 412–420. 10.2133/dmpk.23.412. [DOI] [PubMed] [Google Scholar]
- Capolongo F.; Santi A.; Anfossi P.; Montesissa C. Benzydamine as a useful substrate of hepatic flavin-containing monooxygenase activity in veterinary species. J. Vet Pharmacol Ther 2010, 33, 341–346. 10.1111/j.1365-2885.2009.01145.x. [DOI] [PubMed] [Google Scholar]
- Taniguchi-Takizawa T.; Shimizu M.; Kume T.; Yamazaki H. Benzydamine N-oxygenation as an index for flavin-containing monooxygenase activity and benzydamine N-demethylation by cytochrome P450 enzymes in liver microsomes from rats, dogs, monkeys, and humans. Drug Metab Pharmacokinet. 2015, 30 (1), 64–9. 10.1016/j.dmpk.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Störmer E.; Roots I.; Brockmöller J. Benzydamine N-oxidation as an index reaction reflecting FMO activity in human liver microsomes and impact of FMO3 polymorphisms on enzyme activity. Br. J. Clin. Pharmacol. 2000, 50 (6), 553–561. 10.1046/j.1365-2125.2000.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D. H.; Rettie A. E. In vitro evaluation of potential in vivo probes for human flavin-containing monooxygenase (FMO): metabolism of benzydamine and caffeine by FMO and P450 isoforms. Br. J. Clin. Pharmacol. 2000, 50, 311–314. 10.1046/j.1365-2125.2000.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger R. W.; Fang J.-L.; Chen G.; Weinberg R.; Lazarus P. Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab. Dispos. 2006, 34, 943. 10.1124/dmd.105.009100. [DOI] [PubMed] [Google Scholar]
- Kaivosaari S.; Toivonen P.; Hesse L. M.; Koskinen M.; Court M. H.; Finel M. Nicotine glucuronidation and the human UDP-glucuronosyltransferase UGT2B10. Mol. Pharmacol. 2007, 72 (3), 761. 10.1124/mol.107.037093. [DOI] [PubMed] [Google Scholar]
- Uchaipichat V.; Mackenzie P. I.; Elliot D. J.; Miners J. O. Selectivity of substrate (trifluoperazine) and inhibitor (amitriptyline, androsterone, canrenoic acid, hecogenin, phenylbutazone, quinidine, quinine, and sulfinpyrazone) ″probes″ for human udp-glucuronosyltransferases. Drug Metab. Dispos. 2006, 34 (3), 449–56. 10.1124/dmd.105.007369. [DOI] [PubMed] [Google Scholar]
- Jiang L.; Liang S.-C.; Wang C.; Ge G.-B.; Huo X.-K.; Qi X.-Y.; Deng S.; Liu K.-X.; Ma X.-C. Identifying and applying a highly selective probe to simultaneously determine the O-glucuronidation activity of human UGT1A3 and UGT1A4. Sci. Rep 2015, 5, 9627. 10.1038/srep09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano Y.; Usui T.; Kamimura H. In vitro inhibitory effects of non-steroidal anti-inflammatory drugs on 4-methylumbelliferone glucuronidation in recombinant human UDP-glucuronosyltransferase 1A9--potent inhibition by niflumic acid. Biopharm Drug Dispos 2006, 27 (1), 1–6. 10.1002/bdd.475. [DOI] [PubMed] [Google Scholar]
- Ammon S.; von Richter O.; Hofmann U.; Thon K. P.; Eichelbaum M.; Mikus G. In vitro interaction of codeine and diclofenac. Drug Metab. Dispos. 2000, 28 (10), 1149–1152. [PubMed] [Google Scholar]
- Pattanawongsa A.; Nair P. C.; Rowland A.; Miners J. O. Human UDP-Glucuronosyltransferase (UGT) 2B10: Validation of Cotinine as a Selective Probe Substrate, Inhibition by UGT Enzyme-Selective Inhibitors and Antidepressant and Antipsychotic Drugs, and Structural Determinants of Enzyme Inhibition. Drug Metab. Dispos. 2016, 44 (3), 378–88. 10.1124/dmd.115.068213. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Basit A.; Busch D.; Yabut K.; Bhatt D. K.; Drozdzik M.; Ostrowski M.; Li A.; Collins C.; Oswald S.; Prasad B. Quantitative characterization of UDP-glucuronosyltransferase 2B17 in human liver and intestine and its role in testosterone first-pass metabolism. Biochem. Pharmacol. 2018, 156, 32–42. 10.1016/j.bcp.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Paramo Y. X.; Watson C. J. W.; Chen G.; Lazarus P. CYP2C19 plays a major role in the hepatic N-oxidation of cotinine. Drug Metab Dispos DMD-AR-2021-000624 2022, DMD-AR-2021-000624. 10.1124/dmd.121.000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrin S.; Watson C. J. W.; Perez-Paramo Y. X.; Lazarus P. Cannabinoid Metabolites as Inhibitors of Major Hepatic CYP450 Enzymes, with Implications for Cannabis-Drug Interactions. Drug Metab. Dispos. 2021, 49, 1070–1080. 10.1124/dmd.121.000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson E.Introduction to Biotransformation (Metabolism). In Hayes’ Handbook of Pesticide Toxicology; Elsevier, 2010; pp 865–875. [Google Scholar]
- Perez-Paramo Y. X.; Lazarus P. Pharmacogenetics factors influencing smoking cessation success; the importance of nicotine metabolism. Expert Opin Drug Metab Toxicol 2021, 17, 333–349. 10.1080/17425255.2021.1863948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandänge S.; Lindblom L. The enzyme “aldehyde oxidase” is an iminium oxidase. Reaction with nicotine Δ1′(5′) iminium ion. Biochem. Biophys. Res. Commun. 1979, 91, 991–996. 10.1016/0006-291X(79)91977-6. [DOI] [PubMed] [Google Scholar]
- Gorrod J. W.; Hibberd A. R. The metabolism of nicotine-delta 1′(5′)-iminium ion, in vivo and in vitro. Eur. J. Drug Metab Pharmacokinet 1982, 7 (4), 293–298. 10.1007/BF03189632. [DOI] [PubMed] [Google Scholar]
- Dicke K. E.; Skrlin S. M.; Murphy S. E. Nicotine and 4-(methylnitrosamino)-1-(3-pyridyl)-butanone metabolism by cytochrome P450 2B6. Drug Metab. Dispos. 2005, 33 (12), 1760–1764. 10.1124/dmd.105.006718. [DOI] [PubMed] [Google Scholar]
- Von Weymarn L. B.; Retzlaff C.; Murphy S. E. CYP2A6- and CYP2A13-Catalyzed Metabolism of the Nicotine Δ5′(1′)Iminium Ion. J. Pharmacol Exp Ther 2012, 343 (2), 307–15. 10.1124/jpet.112.195255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler C. D.; Kenny P. J. Nicotine aversion: Neurobiological mechanisms and relevance to tobacco dependence vulnerability. Neuropharmacology 2014, 76, 533–544. 10.1016/j.neuropharm.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningfield J. E.; Shiffman S.; Ferguson S. G.; Gritz E. R. Tobacco dependence and withdrawal: Science base, challenges and opportunities for pharmacotherapy. Pharmacol Ther 2009, 123, 1–16. 10.1016/j.pharmthera.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. L.; Tiirikainen M. I.; Patel Y. M.; Wilkens L. R.; Stram D. O.; Le Marchand L.; Murphy S. E. Genetic determinants of CYP2A6 activity across racial/ethnic groups with different risks of lung cancer and effect on their smoking intensity. Carcinogenesis 2016, 37, 269–279. 10.1093/carcin/bgw012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndale R. F.; Sellers E. M. Variable CYP2A6-mediated nicotine metabolism alters smoking behavior and risk. Drug Metab. Dispos. 2001, 29, 548–552. [PubMed] [Google Scholar]
- Murphy S. E.; Wickham K. M.; Lindgren B. R.; Spector L. G.; Joseph A. Cotinine and trans 3′-hydroxycotinine in dried blood spots as biomarkers of tobacco exposure and nicotine metabolism. J. Expo Sci. Environ. Epidemiol 2013, 23, 513–518. 10.1038/jes.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T.; Nakajima-Taniguchi C.; Fukuda T.; Funamoto M.; Maeda M.; Tange E.; Ueki R.; Kawashima K.; Hara H.; Fujio Y.; Azuma J. CYP2A6 polymorphisms are associated with nicotine dependence and influence withdrawal symptoms in smoking cessation. Pharmacogenomics J. 2006, 6, 115–119. 10.1038/sj.tpj.6500348. [DOI] [PubMed] [Google Scholar]
- Mroziewicz M.; Tyndale R. F. Pharmacogenetics: a tool for identifying genetic factors in drug dependence and response to treatment. Addict Sci. Clin Pract 2010, 5 (2), 17–29. [PMC free article] [PubMed] [Google Scholar]
- Jarvik M. E.; Madsen D. C.; Olmstead R. E.; Iwamoto-Schaap P. N.; Elins J. L.; Benowitz N. L. Nicotine Blood Levels and Subjective Craving for Cigarettes. Pharmacol., Biochem. Behav. 2000, 66, 553–558. 10.1016/S0091-3057(00)00261-6. [DOI] [PubMed] [Google Scholar]
- Fujieda M. Evaluation of CYP2A6 genetic polymorphisms as determinants of smoking behavior and tobacco-related lung cancer risk in male Japanese smokers. Carcinogenesis 2004, 25, 2451–2458. 10.1093/carcin/bgh258. [DOI] [PubMed] [Google Scholar]
- Kortmann G. L.; Dobler C. J.; Bizarro L.; Bau C. H. Pharmacogenetics of smoking cessation therapy. Am. J. Med. Genet B Neuropsychiatr Genet 2009, 9999B, 17–28. 10.1002/ajmg.b.30978. [DOI] [PubMed] [Google Scholar]
- Tanner J.-A.; Tyndale R. Variation in CYP2A6 Activity and Personalized Medicine. J. Pers Med. 2017, 7, 18. 10.3390/jpm7040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu E. C. K.; Tyndale R. F. Selegiline Is a Mechanism-Based Inactivator of CYP2A6 Inhibiting Nicotine Metabolism in Humans and Mice. J. Pharmacol Exp Ther 2008, 324, 992–999. 10.1124/jpet.107.133900. [DOI] [PubMed] [Google Scholar]
- Schuit E.; Panagiotou O. A.; Munafò M. R.; Bennett D. A.; Bergen A. W.; David S. P. Pharmacotherapy for smoking cessation: effects by subgroup defined by genetically informed biomarkers. Cochrane Database of Sys Rev. 2017, 9 (9), CD011823. 10.1002/14651858.CD011823.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- West O.; Hajek P.; McRobbie H. Systematic review of the relationship between the 3-hydroxycotinine/cotinine ratio and cigarette dependence. Psychopharmacology 2011, 218, 313–322. 10.1007/s00213-011-2341-1. [DOI] [PubMed] [Google Scholar]
- Buchwald J.; Chenoweth M. J.; Palviainen T.; Zhu G.; Benner C.; Gordon S.; Korhonen T.; Ripatti S.; Madden P. A. F.; Lehtimäki T.; Raitakari O. T.; Salomaa V.; Rose R. J.; George T. P.; Lerman C.; Pirinen M.; Martin N. G.; Kaprio J.; Loukola A.; Tyndale R. F. Genome-wide association meta-analysis of nicotine metabolism and cigarette consumption measures in smokers of European descent. Mol. Psychiatry 2021, 26, 2212–2223. 10.1038/s41380-020-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.; Krebs N. M.; Zhu J.; Muscat J. E. Nicotine metabolite ratio predicts smoking topography: The Pennsylvania Adult Smoking Study. Drug Alcohol Depend. 2018, 190, 89–93. 10.1016/j.drugalcdep.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom A. J.; Harari O.; Martinez M.; Madden P. A. F.; Martin N. G.; Montgomery G. W.; Rice J. P.; Murphy S. E.; Bierut L. J.; Goate A. Use of a predictive model derived from in vivo endophenotype measurements to demonstrate associations with a complex locus, CYP2A6. Hum. Mol. Genet. 2012, 21, 3050–3062. 10.1093/hmg/dds114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler K.; Chenoweth M. J.; El-Boraie A.; Giratallah H.; Kowalczyk W. J.; Heishman S. J.; Tyndale R. F.; Le Foll B. Impact of CYP2A6 Activity on Nicotine Reinforcement and Cue-Reactivity in Daily Smokers. Nicotine Tob Res. 2021, 23, 1735–1743. 10.1093/ntr/ntab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindocha C.; Freeman T. P.; Grabski M.; Stroud J. B.; Crudgington H.; Davies A. C.; Das R. K.; Lawn W.; Morgan C. J. A.; Curran H. V. Cannabidiol reverses attentional bias to cigarette cues in a human experimental model of tobacco withdrawal. Addiction 2018, 113, 1696–1705. 10.1111/add.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.