Abstract
The occurrence and transmission of carbapenemase-producing-Enterobacterales (CPE) on a global scale has become a major issue. Clinical reports are rarely providing information on the genomic and plasmid features of carbapenem-resistant Serratia marcescens. Our objective was to investigate the resistance and transmission dynamics of two carbapenem-resistant S. marcescens that are resistant to carbapenem and have caused bacteremia in China. Blood specimens were taken from two individuals with bacteremia. Multiplex PCR was employed to identify genes that code for carbapenemase. Antimicrobial susceptibility tests and plasmid analysis were conducted on S. marcescens isolates SM768 and SM4145. The genome of SM768 and SM4145 were completely sequenced using NovaSeq 6000-PE150 and PacBio RS II platforms. Antimicrobial resistance genes (ARGs) were predicted using the ResFinder tool. S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and southern blotting were employed to analyze plasmids. Two S. marcescens that produced KPC-2 were identified from bloodstream infections. The antimicrobial susceptibility testing demonstrated that both of the isolates had a resistance to various antibiotics. The whole-genome sequence (WGS) and plasmid analysis revealed the presence of bla KPC-2-bearing IncR plasmids and multiple plasmid-borne antimicrobial resistance genes in the isolates. Our comparative plasmid analysis suggested that the two IncR plasmids identified in this study could be derived from a common ancestor. Our findings revealed the emergence of bla KPC-2-bearing IncR plasmid in China, which could be a hindrance to the transmission of KPC-2-producing S. marcescens in clinical settings.
Keywords: carbapenem-resistant S. marcescens , KPC-2, bacteremia, CTX-M-14, IncR
Introduction
Serratia marcescens is a type of Gram-negative bacteria belonging to the order Enterobacterales, family Enterobacteriaceae (Park et al., 2007). S. marcescens is frequently encountered in a range of habitats, such as moist areas, prosthetic material, and within the respiratory tract and gastrointestinal flora (Casolari et al., 2013; Rosenthal et al., 2020). Its capacity to cause opportunistic infections in medical facilities is enhanced by these factors. This opportunistic pathogen frequently responsible for hospital-acquired infections, including urinary tract infections (UTIs), respiratory tract infections, conjunctivitis, tear duct infections, and keratitis (Elsherbiny et al., 2018; Konecka et al., 2019; Millan-Lou et al., 2022; Prado et al., 2022; Anderson et al., 2022). S. marcescens is a significant hospital-acquired pathogen, and its invasive infections have resulted in high mortality rates (Yeo et al., 2020; do Prado et al., 2021; Bes et al., 2021).
The emergence of acquired antimicrobial resistance (AMR) in S. marcescens has become a major health risk to the general public (Prado et al., 2022). There are now several reports of multi-drug resistant (MDR) S. marcescens outbreaks carrying either extended-spectrum β-lactamases (ESBLs) or carbapenemases, which confer extended spectrum cephalosporin and carbapenem resistance, respectively (Elsherbiny et al., 2018; Phan et al., 2018; Firmo et al., 2020; Bes et al., 2021; Tamma et al., 2021; Loqman et al., 2021).
In hospital-acquired infections, the prevalence rate of carbapenem-resistant S. marcescens has increased recently (Prado et al., 2022). The production of carbapenemase is the primary cause for the rapid and widespread proliferation of S. marcescens drug resistance (Firmo et al., 2020; Jimenez et al., 2020). Despite its clinical relevance and the increasing concerns about AMR, little is known about the epidemiology and genetic diversity of carbapenem-resistant S. marcescens within healthcare institutions.
Here, we present two cases of bloodstream infections caused by S. marcescens isolates that produce KPC-2. We further sequenced two isolates to characterize their genetic diversity, assess for evidence of nosocomial transmission, and determine the genetic context of acquired AMR determinants. These results help the timely implementation of infection control measures.
Materials and methods
Bacterial isolation
Identification of the bacterial species was performed by MALDI-TOF MS and 16S rRNA sequence analysis, as described previously (Wang et al., 2019). The isolates were further subjected to PCR to detect the carbapenemase gene as previously described (Zheng et al., 2015).
Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MICs) of ten antimicrobial agents (imipenem, meropenem, piperacillin-tazobactam, cefotaxime, ceftazidime, aztreonam, ciprofloxacin, gentamicin, amikacin, tobramycin) were determined by agar dilution method, except for colistin and tigecycline, which were determined by the broth microdilution method. Susceptibility was interpreted according to CLSI (2022) and European Committee on Antimicrobial Susceptibility Testing (EUCAST 2022) guidelines (https://www.eucast.org/). Escherichia coli ATCC25922 was used as quality control (Wang et al., 2019).
Whole-genome sequencing and in silico analysis
Genomic DNA was extracted using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) and sequenced using NovaSeq 6000-PE150 (Illumina, San Diego, CA, USA) and PacBio RS II platform (Pacific Biosciences, California, USA). De novo assembly was generated by using SPAdes 3.11.0 (Hu et al., 2011). Plasmid replicons and antimicrobial resistance genes were predicted through the PlasmidFinder (https://bitbucket.org/genomicepidemiology/plasmidfinder/src/master/) and ResFinder website (https://cge.cbs.dtu.dk/services/ResFinder/), respectively. The genetic environment surrounding bla KPC-2 was annotated using RAST3 and Easyfig 2.2.3 (Chi et al., 2019). The complete genome sequences of S. marcescens SM768 and SM4145 were uploaded to NCBI with the following project number: PRJNA841282.
Plasmid characterization
SM768 and SM4145 were tested by S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and southern blotting to validate the location of bla KPC-2. Whole-cell DNA of two isolates was extracted and embedded in gold agarose gel plugs (SeaKem® Gold Agarose, Lonza, Atlanta, GA, USA). The plugs were digested with S1 nuclease (TaKaRa, Dalian, China) and separated by PFGE. Plasmids obtained by PFGE were transferred horizontally to a nylon membrane (Millipore, USA) and hybridized with digoxin-labelled bla KPC-2-specific probes obtained and the Dig High Prime DNA Labeling and Detection Starter Kit (Roche Diagnostics) (Zheng et al., 2019). The Salmonella enterica serotype Braenderup strain H9812 was used as the DNA marker.
Conjugation assays
Transfer of bla KPC-2 was investigated using conjugation for isolates SM768 and SM4145. The recipient strains for this experiment were E. coli J53 (azide-resistant) and E. coli EC600 (rifampicin-resistant), while SM768 and SM4145 were selected as donor strains (Wang et al., 2019).
Results
KPC-2-producing S. marcescens isolations and patients
Samples of blood from a 36-year-old female and a 49-year-old male yielded two isolates of S. marcescens, SM768 and SM4145, both of which were found to be resistant to carbapenems. PCR test for carbapenemase-encoding genes confirmed that both isolates carried the bla KPC-2 gene.
Antimicrobial resistance profiles and antimicrobial resistance genes
Isolates SM768 and SM4145 showed resistance to imipenem, piperacillin-tazobactam, cefotaxime, ceftazidime, and aztreonam ( Table S1 ). In addition, two isolates were susceptible to amikacin and tobramycin. It is expected that SM768 and SM4145 both exhibited high-level resistance to colistin, as Serratia has an intrinsic resistance to this antibiotic. Whole-genome sequencing showed that SM4145 carried multiple antimicrobial resistance genes (ARGs). These include ESBLs resistance gene bla CTX-M-14, the plasmid-encoded quinolone resistance gene qnrS1, and the aminoglycoside resistance gene aac6’-Ic.
Determination of the bla KPC-2 gene location and transferability
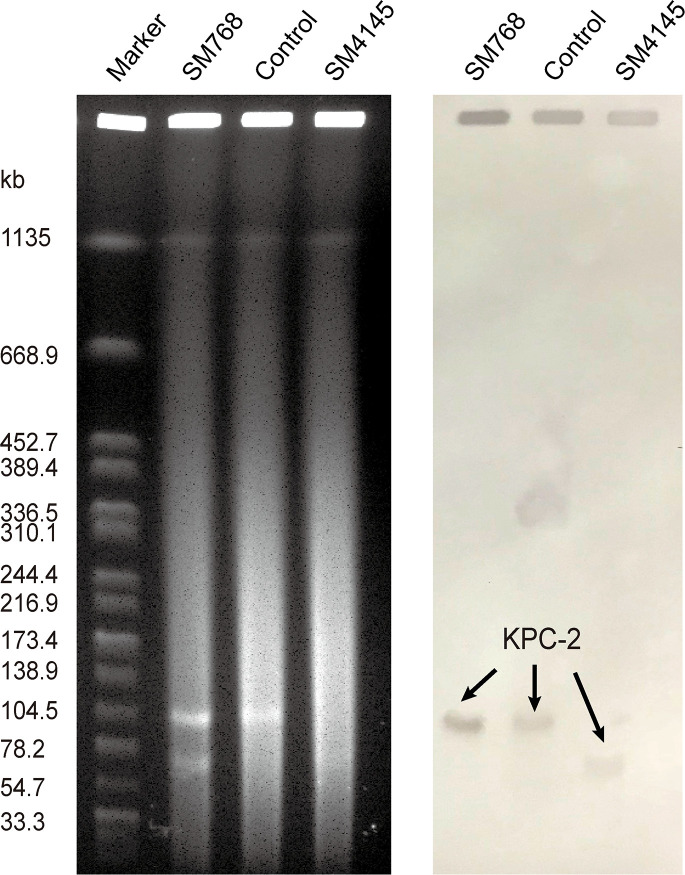
S1-PFGE and Southern blot revealed that SM768 isolate carried a ~105 kb plasmid harbouring bla KPC-2 gene. In contrast, SM4145 isolate harbouring a ~70 kb plasmid encoding bla KPC-2 gene ( Figure 1 ). Furthermore, the bla KPC-2 genes could be transferred from SM768 into the recipient E. coli strains via conjugation, confirmed by PCR. In contrast, SM4145 was negative for the transferability test (data not shown).
Figure 1.
S1-PFGE analysis of KPC-2-producing S. marcescens isolates and southern hybridization using a bla KPC-2 probe. Lane M, molecular weight marker Salmonella Braenderup H9812; Lane SM768, isolate S. marcescens SM768; Lane control, KPC-2-producing Klebsiella pneumoniae isolate 1095 served as the control; Lane SM4145, isolate S. marcescens SM4145.
Genetic context of bla KPC-2 gene
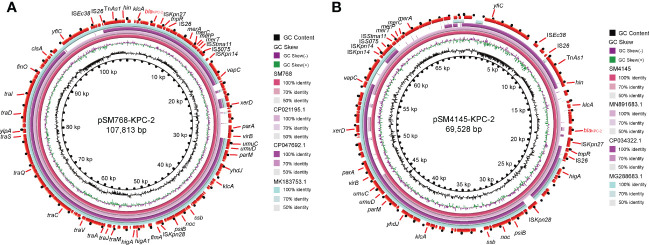
The WGS results demonstrated that two bla KPC-2-encoding plasmids, pSM768-KPC-2, were IncR-type plasmids with 107,813 bp, and pSM4145-KPC-2 was also an IncR-type plasmid with the size of 69,528 bp, respectively ( Figure 2 ). Plasmid pSM768-KPC-2 contained a collection of genes involved in segregation, fertility inhibition, stability, and conjugal transfer of the plasmid (parA, parM, finO, umuCD, and tra regulon), which together constructed the essential backbone of pSM768-KPC-2. In contrast, the tra regulon was absent in plasmid pSM4145-KPC-2.
Figure 2.
Complete sequences of two bla KPC-2 -harbouring IncR plasmids were recovered in this work. (A) Alignment of plasmid sequence of pSM768-KPC-2 with other bla KPC-2 -bearing plasmids pH17-2 (CP021195), pC110-KPC (CP047692), and p17-15-KPC (MK183753). (B) Comparison of pSM4145-KPC-2 with plasmids pK033_1 (CP034322), p314013-KPC (MN891683), and pE20-NR (MG288683). Circles inside to outside denote the GC content, GC screw, and the ORFs in both DNA strands. Block arrows represent coding sequences and indicate the direction of transcription. bla KPC-2 gene is highlighted in red. Arrow size is proportional to gene length. The circular image of multiple plasmids comparisons was generated with the BLAST Ring Image Generator.
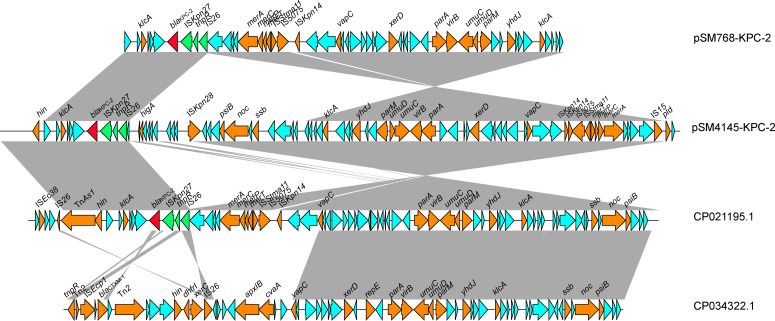
The bla KPC-2 gene surrounding pSM768-KPC-2 and pSM4145-KPC-2 showed the same genetic background. The bla KPC-2 gene was flanked by an ISKpn27, tnpR, and IS26 elements downstream and a klcA gene upstream ( Figure 3 ). Comparative plasmid analysis revealed that pSM768-KPC-2 shares a high identity with pSM4145-KPC-2, except a ~38 kb sequence was inserted in pSM768-KPC-2 plasmid. This indicated that pSM768-KPC-2 and pSM4145-KPC-2 might be derived from a common ancestor. In addition, in silico analysis also found that pSM768-KPC-2 exhibits high relatedness with a ~107 kb KPC-2-carrying IncR plasmid pH17-2 (CP021195) from Escherichia coli and a ~150 kb plasmid pK033_1 (CP034322) from K. pneunomiae.
Figure 3.
Colinear genome alignment of pSM768-KPC-2, pSM4145-KPC-2, pH17-2, and pK033_1. Arrows represent the direction of transcription. Red open reading frames (ORFs) indicate KPC-2.
Discussion
Serratia marcescens is a well-known opportunistic pathogen that is often seen to be responsible for infections in intensive care, surgical and dialysis facilities (Ferreira et al., 2020). Antimicrobial resistance of S. marcescens has not been widely explored in China (Lin et al., 2016; Su et al., 2017; Huang et al., 2021; Zhong et al., 2022; Hou et al., 2022). Herein, we found two carbapenem-resistant S. marcescens from patients with bloodstream infections in China, and the bla KPC-2 gene, which is native to the area, is likely to be disseminated through the transmission of IncR plasmids.
The global spread of the resistance of Enterobacterales to many antibiotics, seriously affecting the treatment of infections, has become a major public issue (Hu et al., 2018; Dong et al., 2018; Dong et al., 2018; Zheng et al., 2020). The major plasmid types carrying bla KPC-2 published included IncFII, IncF, IncP-6, IncN, ColRNAI, and IncI2 (Xu et al., 2018; Hu et al., 2019; Xiaoliang et al., 2019; Abril et al., 2021; Peng et al., 2022). To the best of our knowledge, this is the first time that bla KPC-2 genes have been found to be carried by IncR plasmid in S. marcescens isolates from bacteremia.
It is now acknowledged that S. marcescens has the potential to cause disease in vulnerable individuals. Although S. marcescens displayed relatively low virulence, the emergence of carbapenem-resistant S. marcescens has become a real threat to patients (Zhang et al., 2007). In China, the identification of KPC-2-producing S. marcescens was first described in 2007 (Zhang et al., 2007). Since then, related reports mainly originated from eastern China (Miao et al., 2018; Li et al., 2018; Xie et al., 2020; Xu et al., 2020), which indicates that regional dissemination of such pathogens might be due to the bla KPC-2-harbouring plasmids.
The global distribution of CTX-M variants showed that the CTX-M-1 group (especially CTX-M-15) was the dominant genotype in most regions, while the CTX-M-9 group (especially CTX-M-14) has been reported to be the most common genotypes in China in recent years (Zhang et al., 2016; Shen et al., 2017; Huang et al., 2018; Feng et al., 2019; Yuan et al., 2019; Zheng et al., 2021). A previous study found that CTX-M-14 was the most common gene type in S. marcescens isolates in China (Yang et al., 2012). Our data is consistent with these findings.
Previous studies have indicated that S. marcescens often presents carbapenem resistance caused by plasmids with differing replicon types that contain the bla KPC gene (Xu et al., 2020). In this work, we identified and characterized two IncR plasmids from S. marcescens. We further employed NovaSeq and PacBio RS II platforms to clarify the genetic context of IncR plasmids, which are acutely lacking for bla KPC-bearing IncR plasmids. However, the small size of bla KPC-harbouring IncR plasmids identified in this study is the limitation merit mentioning. A further comprehensive investigation is warranted on the spread of bla KPC-harbouring IncR plasmid in other Enterobacterales.
To summarize, we characterized KPC-2-producing S. marcescens isolates from bloodstream infections in terms of their antimicrobial susceptibility, antimicrobial resistance genes, and plasmid transfer mechanism. Both bla KPC genes were located on the IncR plasmid, which presents a potential challenge for interrupting the transmission of KPC-2-producing S. marcescens in clinical settings. Our insights may have implications in the clinical care and monitoring of KPC-2-producing bacteria.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .
Author contributions
AX and JJ designed and executed the study, performed the results analyses, and drafted the manuscript. JJ, LH, LZ, and YS handled the molecular experiments. WC and HX established and performed the whole genome sequencing. All authors contributed to the article and approved the submitted version.
Funding Statement
This work was supported by the Major Medical Science and Technology Projects in Henan Province (SBGJ202001006).
Conflict of interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1075255/full#supplementary-material
References
- Abril D., Vergara E., Palacios D., Leal A. L., Marquez-Ortiz R. A., Madronero J., et al. (2021). Within patient genetic diversity of blaKPC harboring klebsiella pneumoniae in a Colombian hospital and identification of a new NTEKPC platform. Sci. Rep. 11 (1), 21409. doi: 10.1038/s41598-021-00887-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. T., Himpsl S. D., Mitchell L. A., Kingsley L. G., Snider E. P., Mobley H. L. T. (2022). Identification of distinct capsule types associated with serratia marcescens infection isolates. PloS Pathog. 18 (3), e1010423. doi: 10.1371/journal.ppat.1010423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bes T., Nagano D., Martins R., Marchi A. P., Perdigao-Neto L., Higashino H., et al. (2021). Bloodstream infections caused by klebsiella pneumoniae and serratia marcescens isolates co-harboring NDM-1 and KPC-2. Ann. Clin. Microbiol. Antimicrob. 20 (1), 57. doi: 10.1186/s12941-021-00464-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari C., Pecorari M., Della Casa E., Cattani S., Venturelli C., Fabio G., et al. (2013). Serratia marcescens in a neonatal intensive care unit: Two long-term multiclone outbreaks in a 10-year observational study. New Microbiol. 36 (4), 373–383. [PubMed] [Google Scholar]
- Chi X., Berglund B., Zou H., Zheng B., Borjesson S., Ji X., et al. (2019). Characterization of clinically relevant strains of extended-spectrum beta-Lactamase-Producing klebsiella pneumoniae occurring in environmental sources in a rural area of China by using whole-genome sequencing. Front. Microbiol. 10, 211. doi: 10.3389/fmicb.2019.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI . (2022). M100–performance standards for antimicrobial susceptibility testing, 32nd edition. Ed. Wayne P. A. (Malvern, PA: Clinical and Laboratory Standards Institute; ). [Google Scholar]
- Dong N., Lin D., Zhang R., Chan E. W., Chen S. (2018). Carriage of blaKPC-2 by a virulence plasmid in hypervirulent klebsiella pneumoniae. J. Antimicrob. Chemother. 73 (12), 3317–3321. doi: 10.1093/jac/dky358 [DOI] [PubMed] [Google Scholar]
- Dong N., Yang X., Zhang R., Chan E. W., Chen S. (2018). Tracking microevolution events among ST11 carbapenemase-producing hypervirulent klebsiella pneumoniae outbreak strains. Emerg. Microbes Infect. 7 (1), 146. doi: 10.1038/s41426-018-0146-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Prado G. V. B., Mendes E. T., Martins R. C. R., Perdigao-Neto L. V., Freire M. P., Spadao F., et al. (2021). Carbapenem-resistant serratia marcescens bloodstream infection in hematopoietic stem cell transplantation patients: Will it be the next challenge? Transpl Infect. Dis. 23 (4), e13630 doi: 10.1111/tid.13630. [DOI] [PubMed] [Google Scholar]
- Elsherbiny N. M., Ali I. M., Hassanein K. M., Ahmed M. T. (2018). Extended-spectrum beta-lactamase (ESBL)-producing serratia marcescens causing healthcare-associated infections in assiut university hospitals, Egypt. J. Glob Antimicrob. Resist. 13, 96–97. doi: 10.1016/j.jgar.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Feng C., Wen P., Xu H., Chi X., Li S., Yu X., et al. (2019). Emergence and comparative genomics analysis of extended-Spectrum-beta-Lactamase-Producing escherichia coli carrying mcr-1 in fennec fox imported from Sudan to China. mSphere 4 (6), e00732-19. doi: 10.1128/mSphere.00732-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R. L., Rezende G. S., Damas M. S. F., Oliveira-Silva M., Pitondo-Silva A., Brito M. C. A., et al. (2020). Characterization of KPC-producing serratia marcescens in an intensive care unit of a Brazilian tertiary hospital. Front. Microbiol. 11, 956. doi: 10.3389/fmicb.2020.00956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firmo E. F., Beltrao E. M. B., Silva F., Alves L. C., Brayner F. A., Veras D. L., et al. (2020). Association of blaNDM-1 with blaKPC-2 and aminoglycoside-modifying enzyme genes among klebsiella pneumoniae, Proteus mirabilis and serratia marcescens clinical isolates in Brazil. J. Glob Antimicrob. Resist. 21, 255–261. doi: 10.1016/j.jgar.2019.08.026 [DOI] [PubMed] [Google Scholar]
- Hou J., Mao D., Zhang Y., Huang R., Li L., Wang X., et al. (2022). Long-term spatiotemporal variation of antimicrobial resistance genes within the serratia marcescens population and transmission of s. marcescens revealed by public whole-genome datasets. J. Hazard Mater 423 (Pt B), 127220. doi: 10.1016/j.jhazmat.2021.127220 [DOI] [PubMed] [Google Scholar]
- Huang X., Shen S., Shi Q., Ding L., Wu S., Han R., et al. (2021). First report of bla (IMP-4) and bla (SRT-2) coproducing serratia marcescens clinical isolate in China. Front. Microbiol. 12, 743312. doi: 10.3389/fmicb.2021.743312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Zheng B., Yu W., Niu T., Xiao T., Zhang J., et al. (2018). Antibacterial effect evaluation of moxalactam against extended-spectrum beta-lactamase-producing escherichia coli and klebsiella pneumoniae with in vitro pharmacokinetics/pharmacodynamics simulation. Infect. Drug Resist. 11, 103–112. doi: 10.2147/IDR.S150431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Xu H., Shang Y., Guo L., Song L., Zhang H., et al. (2018). First genome sequence of a blaKPC-2-carrying citrobacter koseri isolate collected from a patient with diarrhoea. J. Glob Antimicrob. Resist. 15, 166–168. doi: 10.1016/j.jgar.2018.09.016 [DOI] [PubMed] [Google Scholar]
- Hu X., Yu X., Shang Y., Xu H., Guo L., Liang Y., et al. (2019). Emergence and characterization of a novel IncP-6 plasmid harboring bla KPC-2 and qnrS2 genes in aeromonas taiwanensis isolates. Front. Microbiol. 10, 2132. doi: 10.3389/fmicb.2019.02132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Zhang W., Liang H., Liu L., Peng G., Pan Y., et al. (2011). Whole-genome sequence of a multidrug-resistant clinical isolate of acinetobacter lwoffii. J. Bacteriol. 193 (19), 5549–5550. doi: 10.1128/JB.05617-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A., Abbo L. M., Martinez O., Shukla B., Sposato K., Iovleva A., et al. (2020). KPC-3-Producing serratia marcescens outbreak between acute and long-term care facilities, Florida, USA. Emerg. Infect. Dis. 26 (11), 2746–2750. doi: 10.3201/eid2611.202203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecka E., Mokracka J., Krzyminska S., Kaznowski A. (2019). Evaluation of the pathogenic potential of insecticidal serratia marcescens strains to humans. Pol. J. Microbiol. 68 (2), 185–191. doi: 10.33073/pjm-2019-018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Feng J., Zhan Z., Yin Z., Jiang Q., Wei P., et al. (2018). Dissemination of KPC-2-Encoding IncX6 plasmids among multiple enterobacteriaceae species in a single Chinese hospital. Front. Microbiol. 9, 478. doi: 10.3389/fmicb.2018.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Hu Q., Zhang R., Hu Y., Xu X., Lv H. (2016). Emergence of serratia marcescens isolates possessing carbapenem-hydrolysing beta-lactamase KPC-2 from China. J. Hosp Infect. 94 (1), 65–67. doi: 10.1016/j.jhin.2016.04.006 [DOI] [PubMed] [Google Scholar]
- Loqman S., Soraa N., Diene S. M., Rolain J. M. (2021). Dissemination of carbapenemases (OXA-48, NDM and VIM) producing enterobacteriaceae isolated from the Mohamed VI university hospital in marrakech, Morocco. Antibiotics (Basel) 10 (5), 492. doi: 10.3390/antibiotics10050492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M., Wen H., Xu P., Niu S., Lv J., Xie X., et al. (2018). Genetic diversity of carbapenem-resistant enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in Eastern China. Front. Microbiol. 9, 3341. doi: 10.3389/fmicb.2018.03341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan-Lou M. I., Lopez C., Bueno J., Perez-Laguna V., Lapresta C., Fuertes M. E., et al. (2022). Successful control of serratia marcescens outbreak in a neonatal unit of a tertiary-care hospital in Spain. Enferm Infecc Microbiol. Clin. 40 (5), 248–254. doi: 10.1016/j.eimc.2021.05.003 [DOI] [PubMed] [Google Scholar]
- Park Y. J., Yu J. K., Lee S., Oh E. J., Woo G. J. (2007). Prevalence and diversity of qnr alleles in AmpC-producing enterobacter cloacae, enterobacter aerogenes, citrobacter freundii and serratia marcescens: A multicentre study from Korea. J. Antimicrob. Chemother. 60 (4), 868–871. doi: 10.1093/jac/dkm266 [DOI] [PubMed] [Google Scholar]
- Peng C., Feng D. H., Zhan Y., Wang Q., Chen D. Q., Xu Z., et al. (2022). Molecular epidemiology, microbial virulence, and resistance of carbapenem-resistant enterobacterales isolates in a teaching hospital in guangzhou, China. Microb. Drug Resist 28(6), 698-709. doi: 10.1089/mdr.2021.0156 [DOI] [PubMed] [Google Scholar]
- Phan H. T. T., Stoesser N., Maciuca I. E., Toma F., Szekely E., Flonta M., et al. (2018). Illumina short-read and MinION long-read WGS to characterize the molecular epidemiology of an NDM-1 serratia marcescens outbreak in Romania. J. Antimicrob. Chemother. 73 (3), 672–679. doi: 10.1093/jac/dkx456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado G., Mendes E. T., Martins R. C. R., Perdigao-Neto L. V., Freire M. P., Marchi A. P., et al. (2022). Phenotypic and genotypic characteristics of a carbapenem-resistant serratia marcescens cohort and outbreak: Describing an opportunistic pathogen. Int. J. Antimicrob. Agents 59 (1), 106463. doi: 10.1016/j.ijantimicag.2021.106463 [DOI] [PubMed] [Google Scholar]
- Rosenthal V. D., Belkebir S., Zand F., Afeef M., Tanzi V. L., Al-Abdely H. M., et al. (2020). Six-year multicenter study on short-term peripheral venous catheters-related bloodstream infection rates in 246 intensive units of 83 hospitals in 52 cities of 14 countries of middle East: Bahrain, Egypt, Iran, Jordan, kingdom of Saudi Arabia, Kuwait, Lebanon, Morocco, Pakistan, Palestine, Sudan, Tunisia, Turkey, and united Arab Emirates-international nosocomial infection control consortium (INICC) findings. J. Infect. Public Health 13 (8), 1134–1141. doi: 10.1016/j.jiph.2020.03.012 [DOI] [PubMed] [Google Scholar]
- Shen P., Fan J., Guo L., Li J., Li A., Zhang J., et al. (2017). Genome sequence of shigella flexneri strain SP1, a diarrheal isolate that encodes an extended-spectrum beta-lactamase (ESBL). Ann. Clin. Microbiol. Antimicrob. 16 (1), 37. doi: 10.1186/s12941-017-0212-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W. Q., Zhu Y. Q., Deng N. M., Li L. (2017). Imipenem-resistance in serratia marcescens is mediated by plasmid expression of KPC-2. Eur. Rev. Med. Pharmacol. Sci. 21 (7), 1690–1694. [PubMed] [Google Scholar]
- Tamma P. D., Smith T. T., Adebayo A., Karaba S. M., Jacobs E., Wakefield T., et al. (2021). Prevalence of bla CTX-m genes in gram-negative bloodstream isolates across 66 hospitals in the united states. J. Clin. Microbiol. 59 (6), e00127-21. doi: 10.1128/JCM.00127-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xu L., Chi X., Li Y., Kou Z., Hou P., et al. (2019). Emergence of NDM-1- and CTX-M-3-Producing raoultella ornithinolytica in human gut microbiota. Front. Microbiol. 10, 2678. doi: 10.3389/fmicb.2019.02678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaoliang W., Huiming H., Chunlei C., Beiwen Z. (2019). Genomic characterisation of a colistin-resistant klebsiella pneumoniae ST11 strain co-producing KPC-2, FloR, CTX-M-55, SHV-12, FosA and RmtB causing a lethal infection. J. Glob Antimicrob. Resist. 19, 78–80. doi: 10.1016/j.jgar.2019.08.023 [DOI] [PubMed] [Google Scholar]
- Xie S., Fu S., Li M., Guo Z., Zhu X., Ren J., et al. (2020). Microbiological characteristics of carbapenem-resistant enterobacteriaceae clinical isolates collected from county hospitals. Infect. Drug Resist. 13, 1163–1169. doi: 10.2147/IDR.S248147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Fu Y., Zhao F., Jiang Y., Yu Y. (2020). Molecular characterization of carbapenem-resistant serratia marcescens clinical isolates in a tertiary hospital in hangzhou, China. Infect. Drug Resist. 13, 999–1008. doi: 10.2147/IDR.S243197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Wang X., Yu X., Zhang J., Guo L., Huang C., et al. (2018). First detection and genomics analysis of KPC-2-producing citrobacter isolates from river sediments. Environ. pollut. 235, 931–937. doi: 10.1016/j.envpol.2017.12.084 [DOI] [PubMed] [Google Scholar]
- Yang H. F., Cheng J., Hu L. F., Ye Y., Li J. B. (2012). Plasmid-mediated quinolone resistance in extended-spectrum-beta-lactamase- and AmpC beta-lactamase-producing serratia marcescens in China. Antimicrob. Agents Chemother. 56 (8), 4529–4531. doi: 10.1128/AAC.00493-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo K. T., Octavia S., Lim K., Lin C., Lin R., Thoon K. C., et al. (2020). Serratia marcescens in the neonatal intensive care unit: A cluster investigation using molecular methods. J. Infect. Public Health 13 (7), 1006–1011. doi: 10.1016/j.jiph.2019.12.003 [DOI] [PubMed] [Google Scholar]
- Yuan S., Wu G., Zheng B. (2019). Complete genome sequence of an IMP-8, CTX-M-14, CTX-M-3 and QnrS1 co-producing enterobacter asburiae isolate from a patient with wound infection. J. Glob Antimicrob. Resist. 18, 52–54. doi: 10.1016/j.jgar.2019.05.029 [DOI] [PubMed] [Google Scholar]
- Zhang R., Zhou H. W., Cai J. C., Chen G. X. (2007). Plasmid-mediated carbapenem-hydrolysing beta-lactamase KPC-2 in carbapenem-resistant serratia marcescens isolates from hangzhou, China. J. Antimicrob. Chemother. 59 (3), 574–576. doi: 10.1093/jac/dkl541 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhou K., Zheng B., Zhao L., Shen P., Ji J., et al. (2016). High prevalence of ESBL-producing klebsiella pneumoniae causing community-onset infections in China. Front. Microbiol. 7, 1830. doi: 10.3389/fmicb.2016.01830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Feng C., Xu H., Yu X., Guo L., Jiang X., et al. (2019). Detection and characterization of ESBL-producing escherichia coli expressing mcr-1 from dairy cows in China. J. Antimicrob. Chemother. 74 (2), 321–325. doi: 10.1093/jac/dky446 [DOI] [PubMed] [Google Scholar]
- Zheng B., Xu H., Lv T., Guo L., Xiao Y., Huang C., et al. (2020). Stool samples of acute diarrhea inpatients as a reservoir of ST11 hypervirulent KPC-2-Producing klebsiella pneumoniae. mSystems 5 (3), e00498-20. doi: 10.1128/mSystems.00498-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Yue M., Zhang J., Ruan Z. (2021). Coexistence of two bla(CTX-M-14) genes in a bla(NDM-5)-carrying multidrug-resistant escherichia coli strain recovered from a bloodstream infection in China. J. Glob Antimicrob. Resist. 26, 11–14. doi: 10.1016/j.jgar.2021.05.002 [DOI] [PubMed] [Google Scholar]
- Zheng B., Zhang J., Ji J., Fang Y., Shen P., Ying C., et al. (2015). Emergence of raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob Agents Chemother 59 (11), 7086–7089. doi: 10.1128/AAC.01363-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y., Liu W., Yuan P., Yang L., Xu Z., Chen D. (2022). Occurrence of serratia marcescens carrying bla(IMP-26) and mcr-9 in southern China: New insights in the evolution of megaplasmid IMP-26. Antibiotics 11 (7), 869. doi: 10.3390/antibiotics11070869 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material .