Abstract
Background:
Acute kidney injury (AKI) is a common complication in patients, especially elderly patients, who undergo cardiac surgery with cardiopulmonary bypass. Studies have indicated a protective role of autophagy in AKI. However, the mechanisms underlying the regulatory effect of autophagy in AKI among patients undergoing cardiac surgeries are poorly understood. In this study, we aimed to test the hypothesis that exosomal microRNAs (miRNAs) regulate autophagy in tubular epithelial cells after AKI.
Methods:
Plasma exosomal RNA was extracted from young and elderly AKI patients undergoing cardiac surgery, and the miRNAs expression during the perioperative period were analyzed using next-generation sequencing. The screened miRNAs and their target genes were subjected to gene oncology function and Kyoto Encyclopedia of Genes and Genome enrichment analyses. Renal tubular epithelial cell line (HK-2 cells) was cultured and hypoxia/reoxygenation (H/R) model was established, which is an in vitro renal ischemia/reperfusion (I/R) model. We used Western blot analysis, cell viability assay, transfection, luciferase assay to investigate the mechanisms underlying the observed increases in the levels of renal I/R injury-mediated exosomal miRNAs and their roles in regulating HK-2 cells autophagy.
Results:
miR-590-3p was highly enriched in the plasma exosomes of young AKI patients after cardiac surgery. Increased levels of miR-590-3p led to the increases in the expression of autophagy marker proteins, including Beclin-1 and microtubule associated protein 1 light chain 3 beta (LC3II), and prolonged the autophagic response in HK-2 cells after H/R treatment. These effects were achieved mainly via increases in the exosomal miR-590-3p levels, and the tumor necrosis factor receptor-associated factor 6 protein was shown to play a key role in I/R injury-mediated autophagy induction.
Conclusion:
Exosomes released from HK-2 cells after renal I/R injury regulate autophagy by transferring miR-590-3p in a paracrine manner, which suggests that increasing the miR-590-3p levels in HK-2 cell-derived exosomes may increase autophagy and protect against kidney injury after renal I/R injury.
Keywords: Acute kidney injury, Autophagy, Exosome, Hypoxia, Ischemia, miR-590-3p, Reperfusion
Introduction
Acute kidney injury (AKI) is a syndrome characterized by a sudden decrease in glomerular filtration function and is classified into grades 1 to 3 according to severity.[1] AKI is a common complication observed in patients, particularly elderly patients, who undergo cardiac surgery with cardiopulmonary bypass (CPB).[2,3] The rapid expansion of the aging population has led to a concomitant increase in the number of older adults undergoing cardiac surgery. Therefore, AKI in elderly patients after cardiac surgery has become an urgent clinical problem.
The use of the CPB procedure during cardiac surgery often leads to a unique low-flow, low-pressure, and non-circulating perfusion state. After CPB, perfusion is restored in the kidneys of the patients, and renal ischemia/reperfusion (I/R) injury may occur. AKI caused by I/R injury mainly manifests as sublethal damage to the renal tubules, and incomplete repair of renal tubular damage can lead to the progression of renal fibrosis.[4]
After kidney damage occurs, damage and repair mechanisms are usually activated simultaneously. Residual renal tubular epithelial cells are the main source of cells for replacing lost tubular epithelial cells.[5] Studies have shown that young patients with AKI are often able to restore their kidneys to a healthy state in terms of both structure and function, but older patients have difficulty in recovering from kidney damage.[6] The poor prognosis of elderly patients with AKI after cardiac surgery with CPB may be associated with decreased autophagic capacity. Moderate levels of autophagy are currently thought to play an important role in maintaining the structure and function of tubular epithelial cells.[7–9] Recent research has suggested that microRNAs (miRNAs) are closely related to autophagy.[10] miRNAs are single-stranded non-coding RNAs of 19–23 nucleotides that regulate gene expression through the posttranscriptional repression of their target mRNAs. Recently, substantial evidence has implicated miRNAs in kidney diseases, particularly AKI.[11] miR-590-3p inhibits autophagy and promotes apoptosis by downregulating Beclin1 expression, and these effects are involved in renal cell injury caused by the contrast agent urografin.[12] In addition, miR-590-3p expression is upregulated by the combination of endothelial-monocyte-activating polypeptide-II and temozolomide, and upregulated miR-590-3p expression inhibits metastasis-associated in colon cancer 1 (MACC1)-induced glioblastoma stem cell autophagy by inhibiting the PI3K/AKT/mTOR pathway.[13]
Exosomes may contain miRNAs, which are released into the peripheral circulatory system or extracellular matrix to mediate intercellular communication.[14] Exosomes, extracellular vesicles with a diameter of 40 to 160 nm, are formed by the cellular process of exocytosis.[15]
In patients who undergo cardiac surgery with CPB, tubular epithelial cells are thought to enhance autophagy, facilitate adaptative responses to renal I/R injury, and decrease renal damage soon after surgery. We herein tested the hypothesis that exosomal miR-590-3p regulates autophagy in tubular epithelial cells after renal I/R injury.
Methods
Ethics approval
The experimental procedures involving human samples were approved by the Ethics Committee of Beijing Tongren Hospital Affiliated to Captical Medical University (TRECKY2019-143 V1.0). All the study samples were anonymized during the analysis. Written informed consent was obtained from each participant. The study was conducted in accordance with the Declaration of Helsinki.
Patient identification and selection
The study was conducted in patients who underwent coronary artery bypass surgery at Beijing Tongren Hospital between December 2019 and August 2020. Evaluations regarding the inclusion and exclusion criteria were performed during the preoperative visit by study personnel after informed consent was obtained. The exclusion criteria were the following: (1) lack of consent; (2) age ≤18 years; (3) pregnancy; (4) severe organ dysfunction (liver or renal); (5) preoperative acute or chronic renal insufficiency or previous use of renal replacement therapy; (6) mental illness or psychotropic drug use; and (7) secondary surgery. The patients from the young AKI group were aged 18 to 44 years, and the patients from the old AKI group were at least 60 years of age.[16]
We evaluated the clinical history and laboratory examination results of all patients. Plasma samples were collected from patients with AKI after cardiac surgery, and exosomes were extracted. Plasma exosome RNA was extracted from young and elderly patients with AKI, and miRNAs that were significantly changed before and after surgery were identified using high-throughput sequencing. After excluding patients with <750,000 labeled miRNA reads at each time point, the final study population included 12 young and 13 elderly AKI patients.
Anesthesia process and sample collection
All the subjects were anesthetized and monitored in a standardized manner. Electrocardiogram (ECG), pulse oximetry, and invasive blood pressure measurements were obtained from all the patients; and these measurements were initiated before anesthesia induction. Detailed descriptions of the anesthesia process are provided in the Supplemental Materials.
Before and after the surgery, approximately 10 mL of venous blood was collected from each patient under aseptic conditions, stored in a vacuum tube containing ethylenediaminetetraacetic acid (EDTA), gently inverted, and mixed several times; the supernatant was promptly harvested by centrifugation at 1500 × g and 4°C for 15 min. The plasma samples were stored at −80°C.
Exosome isolation, identification, and trafficking assay
To separate plasma exosomes, plasma samples were further filtered into new clean Eppendorf tubes using a syringe and a 0.45 μm filter (Millipore-Sigma, Massachusetts, USA), and the exosomes were then isolated from the plasma samples using a ExoQuick ULTRA EV Isolation Kit (System Biosciences, Bay area, USA). The cell-derived exosomes were separated by ultracentrifugation. Detailed descriptions of the process are provided in the Supplemental Materials. The sizes and numbers of the exosomes were determined by nanoparticle tracking analysis (NTA) with a ZetaView PMX 110 (ZetaView, Particle Metrix, Germany). The exosomes were identified by transmission electron microscope (TEM) at Shanghai Umibio Biotechnology Co., Ltd (China).
Exosomes were isolated from the culture medium of HK-2 cells subjected to H/R for 6 h. To monitor exosome trafficking, the isolated exosomes were labeled with the red fluorescent dye PKH26 (Sigma-Aldrich, MO, USA), and then ultracentrifuged again for 120 min at 140,000 × g at 4°C, and excessive staining was stopped by the addition of complete medium. An additional ultracentrifugation step was then performed at the same high speed for 70 min. After washing with phosphate buffered saline (PBS), exosomes were added to HK-2 cells, and 12 h later, the cell cytoskeletons were stained with Actin-Tracker Green (C1033, Beyotime, Shanghai, China). The human renal proximal tubular cell line HK-2 cells were obtained from the Cell Data Center, Shanghai Institute of Biological Sciences, Chinese Academy of Sciences. The exosomal trafficking was visualized with a fluorescence microscope (Leica, Germany).
RNA extraction, library preparation, and next-generation sequencing (NGS)
Total RNA was extracted from plasma exosomes using TRIzol reagent (Thermo Fisher Scientific, Waltham, USA). The concentration and quality of RNA were evaluated using a nanodrop spectrophotometer (ND-2000; Thermo Fisher Scientific). In addition, complementary DNA (cDNA) synthesis and real-time quantitative polymerase chain reaction (RT–PCR) were performed using the Hairpin-it miR-590-3p/mRNA RT–PCR Quantitation Kit (Gene Pharma, Shanghai, China) with corresponding primers according to the manufacturer's protocol. U6 was used as the internal control for miR-590-3p. RT–PCR was performed with the Q5 system (Thermo Fisher) to determine the Ct values, and the relative expression of target genes was determined using the 2−ΔΔCt method. The following primers were synthesized by Gene Pharma Co. (Shanghai, China): miR-590-3p, forward 5′CGGGGGTAATTTTATGTATAAGCTAGT-3′ and reverse 5′-CTCAACTGGTGTCGTGGA-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACG CTTCACGAATTTGCGT-3′.
Plasma exosomal RNA was extracted from young and elderly AKI patients, and miRNAs that showed significant change before and after surgery were analyzed using high-throughput sequencing. After harvesting the total RNA, 18–30 nt fragments were separated using agarose gel electrophoresis (AGE). The 3′ and 5′ junctions were ligated separately, and the small RNAs from both sides of the ligated junctions were amplified by reverse transcription and PCR. After recovery and purification using AGE, 140 bp bands were obtained and used to construct the library, and the resulting library was subjected to quality control using an Agilent 2100 system (Agilent Technologies, Palo Alto, USA) and Real-time Quantitative PCR Detecting System, then sequenced with the instrument.
Western blot analysis
Protein extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (0.22 μm, Millipore, Massachusetts, USA). The membranes were then blocked with 5% skimmed milk for 1.5 hours. Secondary antibodies were then added for cultivation. An electrochemiluminescence system was subsequently used for the detection of the immunoreactive bands.
The antibodies included were anti-CD63 (Abcam, Cambridge, UK), anti-Alix (Abcam, USA), anti-CD 81 antibody (Abcam, UK), microtubule associated protein 1 light chain 3 beta (anti-LC3B) (Abcam, UK), anti-p62 (Abcam, UK), anti-Caspase-3 (Abcam, UK), anti-Bcl-2 (Abcam, UK), tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) (Abcam, UK), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam, UK) horseradish peroxidase-conjugated goat anti-rabbit IgG (Proteintech, Wuhan, China), and horseradish peroxidase-conjugated goat anti-mouse immunoglobin (IgG) (Proteintech, Wuhan, China).
Bioinformatic analysis
Among the miRNAs that were significantly changed in young and elderly AKI patients undergoing surgery, those with direct or inverse changes in expression as well as their target genes were subjected to gene ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathway enrichment analyses using Gene-Sifter software (PerkinElmer, Inc., Shanghai, China) and DAVID online tool (ncifcrf.go; https://david.ncifcrf.gov/tools.jsp).
Cell culture, hypoxia/reoxygenation (H/R) model, and transfection
The human renal proximal tubular cell line HK-2 cells were cultured in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) supplemented with 10% fetal bovine serum (FBS) and penicillin–streptomycin (100 U/mL) in 5% CO2 at 37°C. The medium was changed every day. The cells were exposed to hypoxia (5% CO2, 1% O2, and 94% N2) without nutrients (glucose-free, serum-free) for 12 h, and then subjected to 0.5 h, 3 h, 6 h, 12 h, and 24 h of reoxygenation (5% CO2, 21% O2, and 74% N2) to construct the H/R model, which is an in vitro renal I/R model.[10,17] The control group maintained adequate nutrition and normoxic environment as described above.
The miR-590-3p mimic (5′-UAAUUUUAUGUAUAAGCUAGU-3′) and its corresponding negative control (NC, 5′-UUCUCCGAACGUGUCACGUTT-3′), as well as the miR-590-3p inhibitor (5′-ACUAGCUUAUACAUAAAAUUA-3′) and its corresponding NC (5′-CAGUACUUUUGUGUAGUACAA-3′) were purchased from Gene Pharma Co. (Shanghai, China). HK-2 cells were transfected with lipofectamine 3000 transfection reagent at a final concentration of 50 nmol/L. The transfected cells were cultured in an incubator, and the medium was replaced with complete medium supplemented with antibiotics 4–6 h later. After 48 h of transfection, the cells were prepared for subsequent experiments. The changes in the expression of miR-590-3p in HK-2 cells and their exosomes were measured using RT–PCR to evaluate the effect of the transfection.
Cell viability assay
Cells were counted and seeded in 96-well plates at a density of 5000 cells per well, and cell viability was measured using the cell counting kit-8 (CCK-8) cell viability assay (10 μL/well, Tongren, Tokyo, Japan). At a specified time point, HK-2 cells were treated with CCK-8 reagent for 2 h according to the manufacturer's instructions, and the absorbance was recorded at 450 nm using a microplate absorbance meter. Empty wells served as blank controls, and the experiment was performed three times under the same operating conditions.
Luciferase assay
The potential binding sites between miR-590-3p and TRAF6 were identified using Target Scan (https://www.targetscan.org/). The TRAF6-3′ untranslated region sequence containing the miR-590-3p-binding site and a corresponding mutated sequence were synthesized using PCR amplification. Briefly, 293T cells were seeded in 12-well plates, and 24 h later, the cells were transfected with 50 nmol/L miR-590-3p mimic or NC using lipofectamine 3000. The cells were then cotransfected with 2 μg of the wildtype (WT) or mutant TRAF6-3′ untranslated region (UTR) sequence, and 48 h after transfection, the luciferase activities were measured using the Luciferase Reporter Detection Kit (C0037, Promega, Madison, USA) in accordance with the manufacturer's instructions. Each sample was assessed at least three times.
Statistical analysis
The DESeq2.0 algorithm was used to analyze differentially expressed miRNAs, and a fold change (FC) >2 indicated statistically significant difference in miRNA expression. The data were expressed as the mean ± standard deviation. The statistical analyses were performed using SPSS 22.0 (SPSS, Inc., Chicago, IL, USA). The parameters were analyzed using one-way analysis of variance (ANOVA) with Tukey's post hoc test. P < 0.05 was considered statistically significant.
Results
Basic clinical characteristics of patients
The 25 patients included in this study consisted of 12 young and 13 elderly AKI patients. The differences in baseline data and perioperative indicators other than age were not statistically significant (P > 0.05), as shown in Table 1 and Supplementary Table 1.
Table 1.
Comparison of intraoperative characteristics between young AKI and elderly AKI groups.
| Parameters | Young AKI (n = 12) | Elderly AKI (n = 13) | Statistics | P value |
| Anesthetics for maintenance during surgery | ||||
| Propofol, without volatile | 3 | 2 | – | 0.645 |
| Volatile, without propofol | 1 | 2 | – | >0.999 |
| Volatile, with propofol | 8 | 9 | – | >0.999 |
| Operative details | ||||
| Aortic cross-clamp time (min) | 63.83 ± 9.30 | 65.31 ± 10.77 | −0.365 | 0.718 |
| Circulatory arrest time (min) | 98.58 ± 12.21 | 101.54 ± 12.56 | −0.596 | 0.557 |
| AKI grade | ||||
| 1 | 8 | 6 | – | 0.529 |
| 2 | 2 | 5 | – | 0.378 |
| 3 | 2 | 2 | – | >0.999 |
Data are shown as n or mean ± standard deviation. AKI: Acute kidney injury; –: Not available.
Isolation of plasma exosomes from patients and identification of exosomes
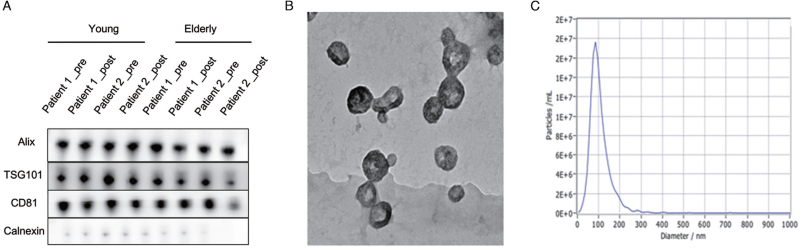
The levels of exosome-enriched and exosome-non-enriched proteins were analyzed using immunoblotting [Figure 1A]. The exosome markers Alix, TSG101, and CD81 were detected in samples from both groups, and the signal intensities of these proteins did not differ between vesicles collected before and after the surgery. The TEM images showed “cup-shaped” membrane vesicles in the field of vision [Figure 1B]. The peak diameter of the particles was further determined to be 40–160 nm [Figure 1C].
Figure 1.
Characterization of plasma exosomes. (A) Pre- and post-operative exosomes in the young AKI (left) and elderly AKI (right) groups were positive for the protein markers Alix, TSG101, and CD81. (B) The plasma exosomes were identified using TEM. (C) The size of exosomes analyzed by NTA. AKI: Acute kidney injury; NTA: Nanoparticle tracking analysis; TEM: Transmission electron microscope.
Characterization of the RNA in plasma exosomes from patients
The sequencing libraries derived from patients in both groups were consistent in size and composition [Supplementary Figure 1]. The presurgical baseline concentrations (young group: 7.36E6 ± 2.50E6; old group: 6.95E6 ± 2.25E6) were similar between groups (P = 0.675). Compared to the results before surgery, postsurgical mean library sizes were lower in patients from young group (7.16E6 ± 2.53E6 reads) and elderly group (6.85E6 ± 2.23E6 reads), reaching no statistical significance (P = 0.856) and (P = 0.906). [Supplementary Figure 1A]. The mean library sizes were 7.36E6 ± 2.50E6 reads (preoperative/young group), 6.95E6 ± 2.25E6 (preoperative/old group) and 7.16E6 ± 2.53E6 reads (postoperative/young group), 6.85E6 ± 2.23E6 reads (postoperative/old group). Analysis of the relative mapping frequencies showed an enrichment of miRNA reads in both groups [Supplementary Figure 1B]. On average, 37 ± 9% (preoperative/young group), 40 ± 10% (postoperative/young group), 44 ± 11% (preoperative/old group), and 40 ± 8% (postoperative/old group) of all the reads mapped to miRNAs. Four young and two elderly patients were excluded from the original cohort because of insufficient miRNA reads (<750,000).
After applying stringent filtering criteria for miRNA expression and magnitude of changes in expression, a total of 56 miRNAs were found to be significantly differentially expressed in the young AKI group (17 upregulated, log2-FC range: 2.87–9.63; 38 downregulated, log2-FC range: −8.97 to −2.74). The expression levels of 24 miRNAs were significantly altered in the elderly AKI group vs. the young AKI group (8 upregulated, log2-FC range: 4.63–7.50; 16 downregulated, log2-FC range: −6.96 to −5.89). In addition, we assessed potentially overlapping changes in miRNA expression between the groups. As demonstrated in Supplementary Figure 2A the postoperatively upregulated miRNAs showed no overlap between the groups. One miRNA showed significant downregulation after surgery in both groups. Information about all the differentially expressed miRNAs in the preoperative samples from young and elderly AKI patients is provided in Supplementary Tables 2 and 3.
Among the miRNAs showing significantly differential expression in the plasma exosomes of young and elderly AKI patients before and after surgery, miRNAs showing consistent and opposite changes in expression were identified. By comparing the differences in exosomal miRNA expression between the young and elderly AKI groups, the expression of hsa-miR-590-3p was found to be significantly increased during surgery in young AKI patients (P = 0.049); however, hsa-miR-590-3p expression was significantly downregulated in exosomal miRNAs in elderly AKI patients perioperatively (P = 0.041; Supplementary Table 4). Functional analysis of the target genes of the differentially expressed miRNAs was performed using GO [Supplementary Figure 2B], and biological pathway enrichment was analyzed using the KEGG Biological Pathway Database (http://www.genome.jp/) [Supplementary Figure 2C]. In addition, hsa-miR-590-3p expression was shown to be significantly different between 10 young and 10 elderly AKI patients. In addition, 10 young and 10 elderly AKI patients with were selected to verify the expression of hsa-miR-590-3p in the plasma exosomes of the patients by RT-PCR. A statistically significant difference in hsa-miR-590-3p expression was detected between the two groups of patients [Supplementary Figure 2D].
H/R-induced autophagy in vitro
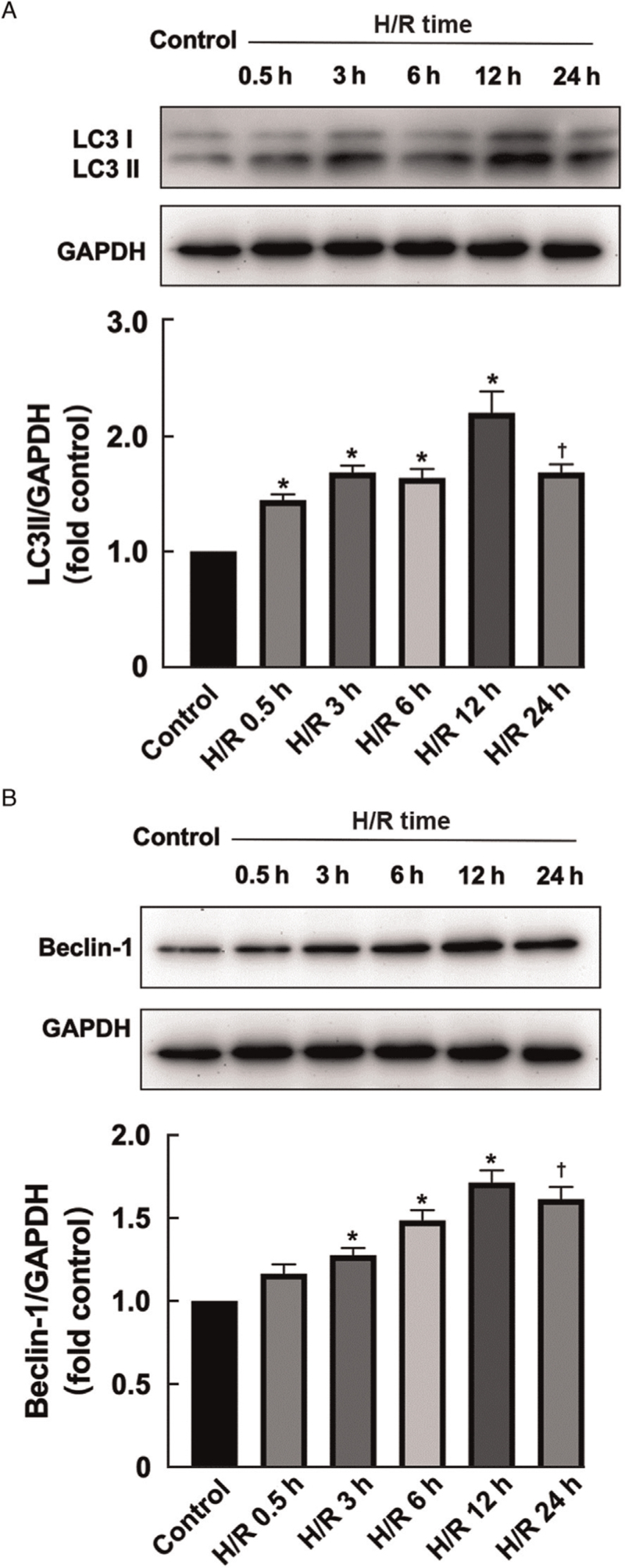
After H/R treatment, the expression of microtubule associated protein 1 light chain 3 beta (LC3II) and Beclin-1 in HK-2 cells was increased at 0.5 h, peaked at 6 h, and remained higher than that in the normoxic group at 24 h after reperfusion [Supplementary Figure 3A, 3B]. The induction of autophagy was also morphologically confirmed by analyzing LC3 fluorescent spots. Immunofluorescence studies have shown that at 6 h after H/R exposure in vitro, many clear green LC3 spots indicate the induction of autophagy, and autophagy continues until 24 h after H/R treatment. In the control group, only weak background immunofluorescence was observed [Supplementary Figure 3C]. The results showed that after H/R treatment, the rate of autophagy in HK-2 cells peaked at 6 h and then gradually declined.
The levels of cleaved caspase 3, Bcl-2, and LC3 were higher in the chloroquine (CQ) treatment group than in the H/R group, and the cell viability of the H/R + CQ group was significantly lower than that of the H/R group, as determined by the CCK-8 assay [Supplementary Figure 4]. These results indicate that H/R treatment significantly induces autophagy. The inhibition of autophagy after H/R in vitro may be the cause of HK-2 cell damage, and autophagy may exert a protective effect on renal I/R injury.
miR-590-3p is transferred between HK-2 cells via exosomes after H/R treatment
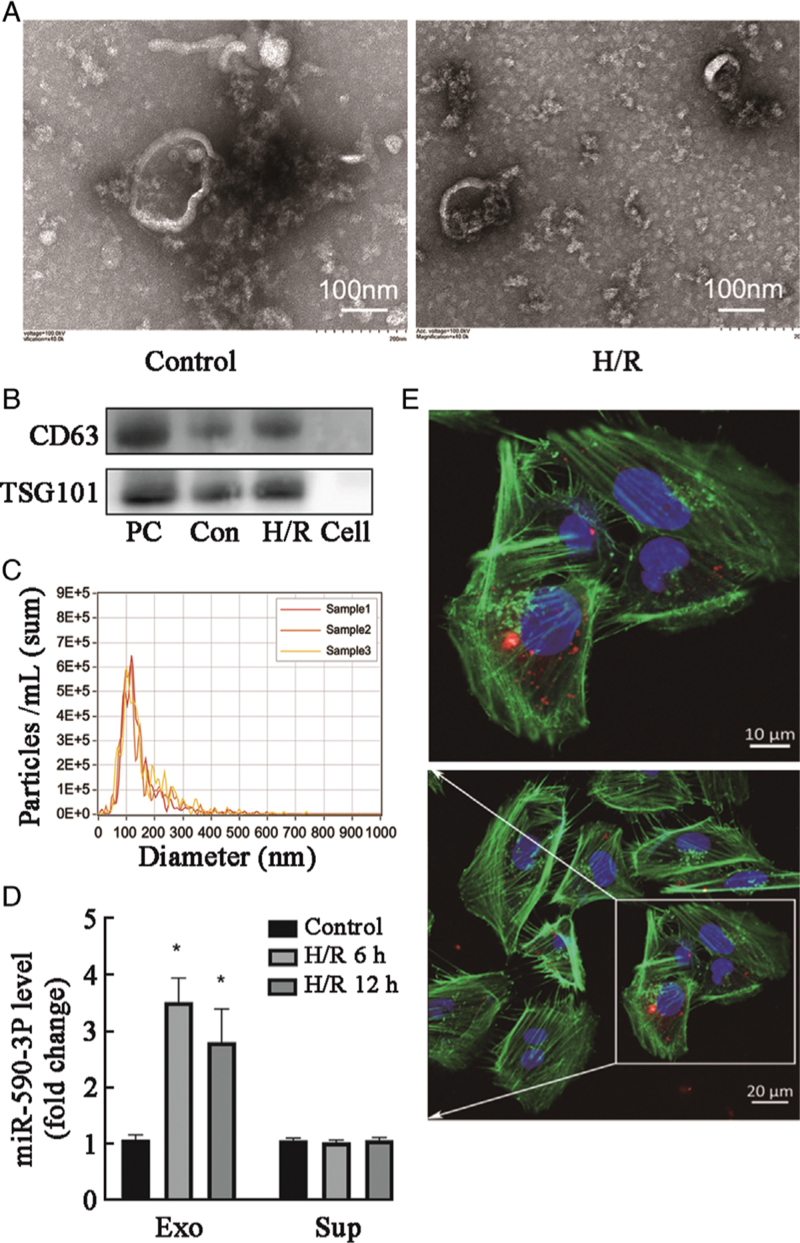
Under normoxic and H/R conditions, HK-2 cells released circular structures with diameters of 40–160 nm using TEM [Figure 2A], which indicated that they secreted exosomes under both conditions. The exosomes were characterized using Western blot and NTA [Figure 2B,C].
Figure 2.
miR-590-3p is transferred between HK-2 cells via exosomes after H/R treatment. (A) Secretory vesicles isolated from HK-2 cell culture medium were identified using transmission electron microscopy. (B, C) Exosomes were quantified by Western blot and NTA. Cell: Cell supernatant; Con: Control. (D) Levels of miR-590-3p in the exosome (Exo) and supernatant (Sup) fractions of medium from HK-2 cells after 12 h of H/R as detected by real-time reverse transcription-polymerase chain reaction (RT–PCR). Data are expressed as the mean ± standard deviation; N = 3 for NTA, N = 4–5 for Western blot. ∗P < 0.01, vs. the control group. (E) Representative confocal microscopy images of HK-2 cells exposed to PKH26-labeled exosomes from HK-2 cells subjected to 12 h of H/R. Nuclei were stained with DAPI. Red: PKH26; Green: Actin tracker; Blue: DAPI (nuclei). DAPI: 4′,6-diamidino-2-phenylindole; H/R: Hypoxia/reoxygenation; I/R: Ischemia/reperfusion; NTA: Nanoparticle tracking analysis; NC: Negative control; PC: Positive control.
Red fluorescence was observed after 12 h of incubation on the membranes of normoxic HK-2 cells, suggesting the efficient uptake and incorporation of exosomes from H/R-treated HK-2 cells to normoxic HK-2 cells [Figure 2D, E]. We separated the culture media of HK-2 cells exposed to H/R into exosome and supernatant fractions. Then, we quantified the miR-590-3p levels in these fractions at different time points. After H/R treatment, the miR-590-3p level in exosomes increased in a time-dependent manner, whereas that in supernatants did not increase, which indicated that the exosomes in H/R-treated HK-2 cell culture medium were highly enriched for miR-590-3p [Figure 2D].
HK-2 cell-derived exosomes with high miR-590-3p levels enhanced autophagy in adjacent cells after H/R treatment
HK-2 cells were transfected with miR-590-3p mimics or the NC using lipofectamine 3000 reagent. The cells were later collected, and the exosomes were isolated. Exosomes with high expression of miR-590-3p and normal expression of miR-590-3p were obtained. Then, these exosomes with differential expression levels of miR-590-3p levels were then incubated with normoxic HK-2 cells exposed to H/R, and the effects of exosomes with high miR-590-3p expression on autophagy in HK-2 cells after H/R were assessed by measuring the changes in the expression of autophagy-related proteins in the normoxic HK-2 cells.
First, the RT–PCR results showed that the transfection of HK-2 cells with miR-590-3p mimics after exposure to H/R significantly increased the expression of miR-590-3p in exosomes (P = 0.004) [Figure 3A]. This experiment was divided into a control group, H/R group, H/R + miR-590-3p exosome group, and H/R + control exosome group. Compared with that in the control group, the expression of miR-590-3p in HK-2 cells of the H/R group was significantly increased as determined by RT–PCR (P = 0.0027) . Compared with that in the H/R group, the expression of miR-590-3p in cells of the H/R + miR-590-3p exosome group was significantly increased (P = 0.0004) [Figure 3B].
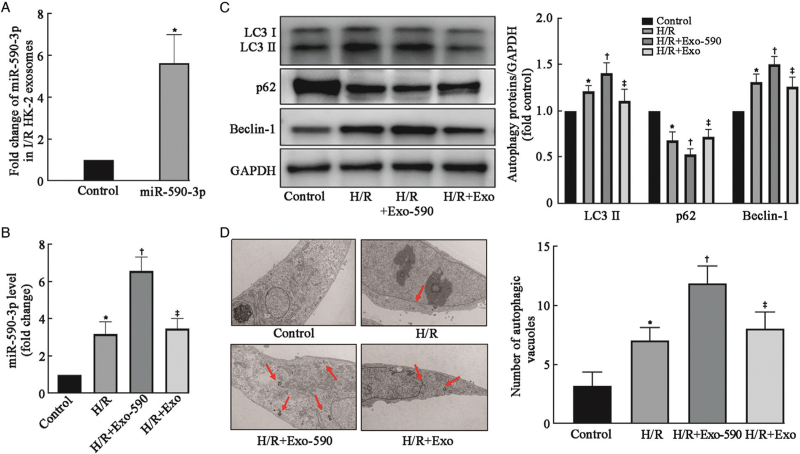
Figure 3.
Exosomal miR-590-3p derived from HK-2 cells after H/R enhanced the activation of autophagy. (A) miR-590-3p expression was significantly upregulated in HK-2 cell exosomes after transfection with a miR-590-3p mimic as determined by RT–PCR. (B) miR-590-3p expression was upregulated in HK-2 cells after H/R and further increased significantly after treatment with miR-590-3p-overexpressing exosomes as determined by RT–PCR. (C) Western blot analysis of autophagy-related proteins (LC3, p62, and Beclin-1) revealed that autophagy was induced in cultured normoxic HK-2 cells after H/R. The activation of autophagy was increased by exosomes overexpressing miR-590-3p from HK-2 cells. (D) Representative images and quantification of autophagic vacuoles in these groups. Autophagic vacuoles (red arrows) were detected by transmission electron microscopy at a magnification of ×10,000. Scale bars represent 2 μm. The data are expressed as the mean ± SD; N = 3 for RT–PCR, N = 4–5 for Western blot. ∗P < 0.05 vs. the control group; †P < 0.05, vs. the H/R group; ‡P < 0.05, vs. the H/R + Exo-590 group. Exo: Exosome; H/R: Hypoxia/reoxygenation; I/R: Ischemia/reperfusion; LC3: Microtubule associated protein 1 light chain 3; SD: Standard deviation.
The Western blot analysis showed that the H/R + miR-590-3p exosome group exhibited significantly higher in LC3 II and Beclin-1 protein expression and a significantly lower p62 protein expression compared with the H/R group, and these differences were statistically significant [Figure 3C]. Exosomes with high miR-590-3p expression presented increased autophagy levels in HK-2 cells. The H/R + miR-590-3p exosome group exhibited lower LC3 II (P = 0.0004) and Beclin-1 protein expression levels (P = 0.0005) and higher p62 protein expression levels than the H/R + miR-590-3p exosome group (P = 0.0082), and these were significant. The autophagic vacuoles in these four groups were observed and quantified using TEM, and the results regarding the number of autophagic vacuoles were similar to those obtained by Western blot [Figure 3D]. The number of autophagic vacuoles was increased in the H/R + miR-590-3p exosome group compared with the H/R group and the H/R + control exosome group. These results indicate that high levels of miR-590-3p in exosomes from HK-2 cells after H/R may enhance the autophagy of normoxic HK-2 cells.
HK-2 cells transfected with miR-590-3p mimics were subjected to H/R treatment under the same conditions. The Western blot was then performed to measure the expression levels of the autophagy-specific proteins LC3 II/I and Beclin-1 in the cells after H/R treatment. The LC II (H/R 6 h vs. H/R 12 h; P = 0.0001) and Beclin-1 (H/R 6 h vs. H/R 12 h; P = 0.0001) expression in HK-2 cells transfected with miR-590-3p mimics increased to the highest level at 12 h after H/R, and the high expression level was maintained for up to 12 h after H/R treatment (P = 0.0001) [Figure 4].
Figure 4.
High expression of miR-590-3p and intracellular autophagy-related proteins after H/R. (A) Expression levels of the HK-2 autophagy-related protein LC3II in HK-2 cells transfected with miR-590-3p mimics at different time points of H/R culture. (B) Expression levels of the autophagy-related protein Beclin-1 in HK-2 cells transfected with miR-590-3p mimics at different time points of H/R culture. The data are expressed as the mean ± standard deviation; N = 4–5 for Western blot. ∗P < 0.05 vs. the control group. †P < 0.05 vs. the H/R 12 h group. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; H/R: Hypoxia/reoxygenation; LC3: Microtubule associated protein 1 light chain 3.
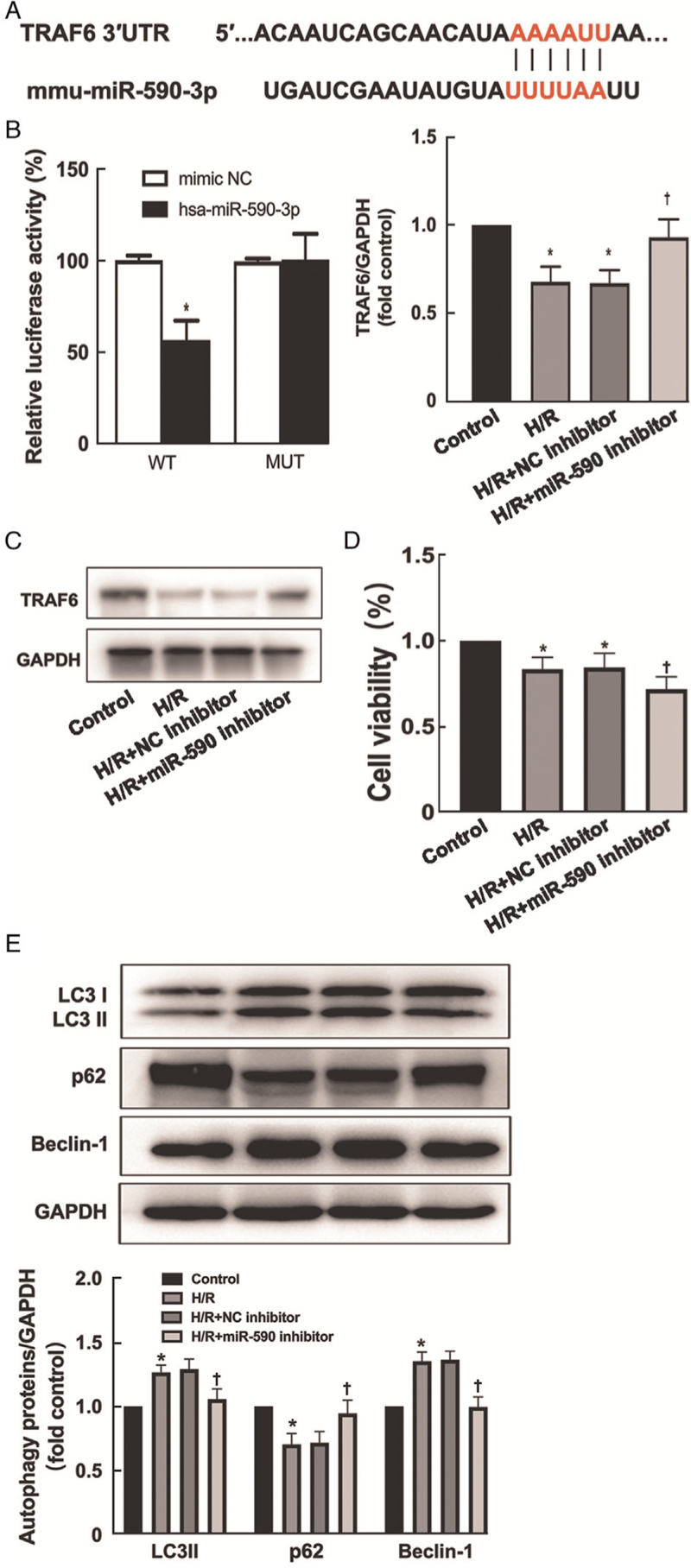
TRAF6 is a target gene of miR-590-3p
TRAF6, a member of the TRAF protein family, plays a pivotal role in the activation of autophagy induced by Toll-like receptor 4 (TLR4) signaling.[18] Analysis of the TargetScan online database revealed potential binding sites between miR-590-3p and TRAF6 [Figure 5A]. To confirm this finding, a pGL3 promoter-based TRAF6 3′-UTR was cotransfected with mimics or control sequences of miR-590-3p into HK-2 cells. The luciferase activity of the TRAF6 3′-UTR was significantly inhibited by the miR-590-3p mimics compared with the NC; however, the luciferase activity of the mutated TRAF6 3′-UTR was not changed by transfection with the mimics or control sequences of miR-590-3p (P = 0.005) [Figure 5B]. To elucidate the relationship between miR-590-3p and TRAF6, we further verified the presence of TRAF6 and the changes in its expression in H/R-treated HK-2 cells transfected with miR-590-3p inhibitor using Western blot (H/R vs. H/R+miR-590 inhibitor; P = 0.0004) [Figure 5C]. Transfection of the miR-590-3p inhibitor into HK-2 cells exposed to H/R increased the expression of TRAF6. The cell viability of the H/R + miR-590 inhibitor group was significantly lower than that of the H/R + NC inhibitor group, as determined by the CCK-8 assay (H/R vs. H/R +miR-590 inhibitor; P = 0.0362) [Figure 5D]. Western blot analysis of autophagy-related proteins (LC3, p62, Beclin-1) (H/R vs. H/R +miR-590 inhibitor; LC3: P = 0.0296; p62: P = 0.0032; Beclin-1: P = 0.0018) revealed that H/R treatment-induced autophagy was inhibited in the H/R + miR-590 inhibitor group [Figure 5E].
Figure 5.
TRAF6 is a direct target of miR-590-3p. (A) Schematic representation of the potential miR-590-3p binding sites in the 3′ UTR of TRAF6 as predicted by the online database. (B) Relative luciferase activity of the wild type (WT) and mutated (MUT) reporter constructs cotransfected with either the miR-590-3p mimic or scrambled oligonucleotides. (C) After the transfection of miR-590-3p inhibitors into HK-2 cells subjected to H/R, the expression of TRAF6 was assessed using Western blot. (D) The cell viability of the H/R + miR-590 inhibitor group was significantly lower than of the H/R + NC inhibitor group by the CCK-8 assay. (E) Western blot analysis of autophagy-related proteins (LC3, p62, Beclin-1) revealed that H/R-induced autophagy was inhibited in the H/R + miR-590 inhibitor group. The data are expressed as the mean ± standard deviation; N = 4–5 for Western blot. ∗P < 0.05 vs. the control group; †P < 0.05 vs. the NC inhibitor group. CCK-8: Cell Counting Kit-8; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; H/R: Hypoxia/reoxygenation; LC3: Microtubule associated protein 1 light chain 3; NC: Negative control; SD: Standard deviation; TRAF6: Tumor necrosis factor receptor-associated factor 6.
Discussion
Kidney tissues are susceptible to age-related damage, and AKI is more common but not restricted to the elderly population.[19,20] The increasing size of the population of individuals aged >60 years has led to a further increase in the incidence of AKI. The elderly population of patients with AKI after cardiac surgery involving CPB is of particular concern. miRNAs have been shown to play important roles in regulating gene expression in the pathophysiology of various renal diseases. miRNAs are stably expressed in clinical plasma samples, and their stability is derived from endogenous RNase activity.[21] Studies have shown the stable presence of extracellular miRNAs in vesicles derived from membrane structures, such as exosomes.[22,23] In the present work, miR-590-3p was highly enriched in the plasma exosomes of young AKI patients.
The KEGG analysis showed that miR-590-3p was closely related to the PI3K-Akt signaling pathway and autophagy pathway. Previous studies have shown that the PI3K-Akt signaling pathway plays a key regulatory role in autophagy.[24,25] Many studies have strongly demonstrated that the activation of the PI3K-Akt signaling pathway is associated with AKI.[26,27] Therefore, although the pathway was not explicitly explored in the present study, miR-590-3p may regulate autophagy through the PI3K-Akt pathway.
The involvement of renal I/R injury in AKI and the pathophysiology of renal I/R injury are closely related to renal tubular epithelial cell injury.[28,29] HK-2 cells are commonly used as model cells in studies of AKI in vitro and, in the present study, we used HK-2 cells H/R-induced human kidney proximal tubular epithelial cells as an in vitro renal I/R injury model.[30] The pathophysiology of AKI after cardiac surgery with CPB is complex and includes renal I/R injury, inflammation, and oxidative stress.[2] Renal I/R injury,[31] one of the most important mechanisms, was selected for the establishment of a cellular model.
Recent studies have shown that cells can communicate with neighboring or distant cells through exosomes.[32,33] As important vehicles for intercellular communication, exosomes can transmit genetic information that performs regulatory functions, such as miRNAs.[34] In this study, miR-590-3p was enriched in the plasma exosomes of young patients with AKI after cardiac surgery, which suggested that miR-590-3p is delivered to distant locations via exosomes. In fact, our experiments provide direct evidence showing that intercellular communication between HK-2 cells is mediated by exosomes. We confirmed that exosomes released from H/R-treated HK-2 cells could be taken up by other normoxic HK-2 cells. The uniformly distributed red fluorescence in the recipient cells indicated that exosomes were efficiently phagocytosed by normoxic HK-2 cells. Further experiments are needed to elucidate the mechanism underlying exosomal transfer between HK-2 cells and to determine how miR-590-3p is sorted into exosomes.
Previous studies have suggested that autophagy protects against cellular injury and plays a key role in delaying and preventing the progression of AKI to chronic kidney disease (CKD). Westhoff et al[35] found that knockdown of the telomerase gene affects the mTOR-mediated autophagic pathway in renal tubular epithelial cells and delays the repair process in the kidney after I/R injury. Several studies have established renal proximal tubule-specific ATG-knockout mouse models. As observed using these mouse models, renal tubular injury and deterioration of renal insufficiency are more pronounced in ATG-knockout mice after I/R than in control mice, confirming that autophagy exerts a protective effect on mouse renal I/R injury.[36,37] Moreover, the inactivation of autophagy promotes apoptosis, which suggests that increased autophagy inhibits apoptosis and plays a protective role.[8,38]
We obtained plasma exosomes from young and elderly AKI patients after cardiac surgery with CPB and found that the expression of miR-590-3p was significantly higher in plasma-derived exosomes from young AKI patients than in those derived from elderly patients. Therefore, we hypothesized that miR-590-3p in exosomes derived from renal tubular epithelial cells plays an important role in the repair of renal tubular epithelial cell damage after AKI. Numerous studies have shown that miRNAs can regulate autophagy by targeting autophagy-related genes. Therefore, we hypothesize that renal tubular epithelial cell-derived exosomes with high miR-590-3p expression enhance renal I/R injury-activated autophagy through paracrine secretion.
Autophagic flux is an objective indicator of autophagy activation and a dynamic indicator of successful autophagosome formation and degradation. LC3 and Beclin-1 are two key proteins in the autophagy process. The p62 protein, also known as SQSTM1, has substrate specificity, regulates the composition of autophagosomes, and is degraded in the middle and late stages of autophagy, and its expression level is inversely correlated with autophagic activity.[39] Autophagic flux is a dynamic process that can be evaluated by the simultaneous observation of p62 expression and LC3 II/I conversion. Autophagic flux is activated by an increase in the conversion of LC3 I to LC3 II and a decrease in p62 protein expression. Based on previous studies, we successfully established an in vitro H/R model by employing glucose-free medium and hypoxia-reoxygenation.[40] In this H/R model, the LC3 II and Beclin-1 expression levels were significantly upregulated in a time-dependent manner and peaked at 6 h post-H/R model. Consistent with this observation, the analysis of LC3 puncta revealed the same trend, which could explain the time-dependent increase in the autophagy in HK-2 cells after H/R treatment and suggests a potential mechanism through which the autophagy pathway is regulated.
Several studies have shown that these miRNAs can regulate autophagy by targeting various signaling pathways related to autophagy cascades.[41–43] miR-590-3p is a recently identified miRNA that is encoded by a gene located in intron 4H of the eukaryotic translation initiation factor.[44] miR-590-3p promotes cardiomyocyte proliferation and inhibits apoptosis by targeting RIPK1 to reduce oxidative stress in mice exposed to I/R.[45] Using an in vitro model of sepsis-induced cardiac dysfunction, Liu et al[46] demonstrated that miR-590-3p regulates the AMP-activated protein kinase (AMPK)/mTOR signaling pathway, inhibits cardiomyocyte pyroptosis, promotes autophagy by inhibiting NOD-like receptor family pyrin domain containing 3 (NLRP3) expression, and regulates AKI and podocyte apoptosis. In addition, some findings suggest that HMGB2 is a target gene of miR-590-3p and that HMGB2 is closely associated with the severity of IgA nephropathy.[29]
We cultured exosomes secreted by HK-2 cells with differentially expression levels of miR-590-3p with H/R-treated HK-2 cells. HK-2 cell-derived exosomes expressing high miR-590-3p levels enhanced H/R-induced cellular autophagy, which further suggested that exosomes from H/R-treated HK-2 cells enhance the autophagy of adjacent cells through miR-590-3p transport. The high expression levels of Beclin-1 and LC3 in HK-2 cells transfected with the miR-590-3p mimics after H/R were maintained longer than those in untransfected cells (until 12 h after H/R), thus confirming that the high expression of miR-590-3p prolonged the autophagic response of HK-2 cells.
Numerous studies have shown that miRNAs perform their biological functions mainly by regulating the expression of downstream target genes, and several studies have shown that TRAF6 is involved in apoptosis and tumor suppression.[47,48] A previous study found that miR-590-3p suppresses LPS-induced AKI and podocyte apoptosis by targeting TRAF6.[49] In this study, we identified TRAF6 as a potential target gene of miR-590-3p through bioinformatics analysis. Our results showed that HK-2 cell-derived exosomes enhance autophagy by transporting miR-590-3p in a paracrine manner after H/R to directly inhibit TRAF6 expression and thereby participate in the pathogenesis of AKI.
This study has several limitations. First, this study did not provide any evidence showing that exosomes extracted from plasma originate from tubular epithelial cells. Lots of exosome-related studies have reported that markers in peripheral blood are closely related to patients’ complications. Further experiments are needed to explain how miR-590-3p is sorted into exosomes and why exosomes extracted from plasma originate from tubular epithelial cells. Second, although we performed sequencing using clinical plasma samples and cell experiments to verify the mechanism, there remains a lack of animal experiments. In our future work, we will establish a renal I/R injury animal model and will identify whether autophagy is increased by abnormal serum creatinine in vivo. Finally, the effect of the miR-590-3p-mediated inhibition of TRAF6 on autophagy in HK-2 cells has not been studied in-depth, and the next step is to refine the relevant research.
In summary, this study showed that exosomes released from HK-2 cells may enhance autophagy by transferring miR-590-3p in a paracrine manner after renal I/R injury. Furthermore, our findings indicate that exosomes enriched in miR-590-3p can induce autophagic signaling in H/R-treated HK-2 cells by targeting TRAF6. These findings provide an attractive new therapeutic approach for AKI after cardiac surgery, especially in elderly patients.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81970344).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Chen Y, Zhang C, Du Y, Yang X, Liu M, Yang W, Lei G, Wang G. Exosomal transfer of microRNA-590-3p between renal tubular epithelial cells after renal ischemia-reperfusion injury regulates autophagy by targeting TRAF6. Chin Med J 2022;135:2467–2477. doi: 10.1097/CM9.0000000000002377
Supplemental digital content is available for this article.
References
- 1.Hu J, Chen R, Liu S, Yu X, Zou J, Ding X. Global incidence and outcomes of adult patients with acute kidney injury after cardiac surgery: a systematic review and meta-analysis. J Cardiothorac Vasc Anesth 2016; 30:82–89. doi: 10.1053/j.jvca.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Bellomo R. Cardiac surgery-associated acute kidney injury: risk factors, pathophysiology and treatment. Nat Rev Nephrol 2017; 13:697–711. doi: 10.1038/nrneph.2017.119. [DOI] [PubMed] [Google Scholar]
- 3.Schurle A, Koyner JL. CSA-AKI: incidence, epidemiology, clinical outcomes, and economic impact. J Clin Med 2021; 10:5746.doi: 10.3390/jcm10245746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. J Clin Invest 2011; 121:4210–4221. doi: 10.1172/jci45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lameire N, Van Biesen W, Vanholder R. Acute renal failure. Lancet 2005; 365:417–430. doi: 10.1016/s0140-6736(05)17831-3. [DOI] [PubMed] [Google Scholar]
- 6.Santin Y, Lluel P, Rischmann P, Gamé X, Mialet-Perez J, Parini A. Cellular senescence in renal and urinary tract disorders. Cells 2020; 9:2420.doi: 10.3390/cells9112420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura T, Takahashi A, Takabatake Y, Namba T, Yamamoto T, Kaimori JY, et al. Autophagy protects kidney proximal tubule epithelial cells from mitochondrial metabolic stress. Autophagy 2013; 9:1876–1886. doi: 10.4161/auto.25418. [DOI] [PubMed] [Google Scholar]
- 8.Park SH, Choi HI, Ahn J, Jang YJ, Ha TY, Seo HD, et al. Autophagy functions to prevent methylglyoxal-induced apoptosis in HK-2 cells. Oxid Med Cell Longev 2020; 2020:8340695.doi: 10.1155/2020/8340695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan L, Chen J, Fang W. Exosomes derived from calcium oxalate-treated macrophages promote apoptosis of HK-2 cells by promoting autophagy. Bioengineered 2022; 13:2442–2450. doi: 10.1080/21655979.2021.2012622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghafouri-Fard S, Shoorei H, Mohaqiq M, Majidpoor J, Moosavi MA, Taheri M. Exploring the role of non-coding RNAs in autophagy. Autophagy 2022; 18:949–970. doi: 10.1080/15548627.2021.1883881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou YF, Zhang W. Role of microRNA in the detection, progression, and intervention of acute kidney injury. Exp Biol Med (Maywood) 2018; 243:129–136. doi: 10.1177/1535370217749472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Li Q, Sun L, Chen L, Li Y, Huang B, et al. miR-30e-5p regulates autophagy and apoptosis by targeting beclin1 involved in contrast-induced acute kidney injury. Curr Med Chem 2021; 28:7974–7984. doi: 10.2174/0929867328666210526125023. [DOI] [PubMed] [Google Scholar]
- 13.Zhou W, Liu L, Xue Y, Zheng J, Liu X, Ma J, et al. Combination of endothelial-monocyte-activating polypeptide-II with temozolomide suppress malignant biological behaviors of human glioblastoma stem cells via miR-590-3p/MACC1 inhibiting PI3K/AKT/mTOR signal pathway. Front Mol Neurosci 2017; 10:68.doi: 10.3389/fnmol.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018; 19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 15.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020; 367:eaau6977.doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye P, Er Y, Wang H, Fang L, Li B, Ivers R, et al. Burden of falls among people aged 60 years and older in mainland China, 1990-2019: findings from the Global Burden of Disease Study 2019. Lancet Public Health 2021; 6:e907–e918. doi: 10.1016/s2468-2667(21)00231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang TT, Lv LL, Pan MM, Wen Y, Wang B, Li ZL, et al. Hydroxychloroquine attenuates renal ischemia/reperfusion injury by inhibiting cathepsin mediated NLRP3 inflammasome activation. Cell Death Dis 2018; 9:351.doi: 10.1038/s41419-018-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2001; 2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 19.Bellomo R, Kellum JA, Ronco C. Acute kidney injury. Lancet 2012; 380:756–766. doi: 10.1016/s0140-6736(11)61454-2. [DOI] [PubMed] [Google Scholar]
- 20.GBD Chronic Kidney Disease Collaboration. Global, regional, and national burden of chronic kidney disease, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020; 395:709–733. doi: 10.1016/s0140-6736(20)30045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittelbrunn M, Sánchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol 2012; 13:328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turturici G, Tinnirello R, Sconzo G, Geraci F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. Am J Physiol Cell Physiol 2014; 306:C621–C633. doi: 10.1152/ajpcell.00228.2013. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Jiang YY, Chen H, Wu YC, Zhang L. Tanshinone I attenuates the malignant biological properties of ovarian cancer by inducing apoptosis and autophagy via the inactivation of PI3K/AKT/mTOR pathway. Cell Prolif 2020; 53:e12739.doi: 10.1111/cpr.12739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Cai C, Min S, Yan B, Liu W, Yang X, Li L, et al. MiR-27a promotes the autophagy and apoptosis of IL-1(treated-articular chondrocytes in osteoarthritis through PI3K/AKT/mTOR signaling. Aging (Albany NY) 2019; 11:6371–6384. doi: 10.18632/aging.102194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Tian X, Ji Y, Liang Y, Zhang J, Guan L, Wang C. LINC00520 targeting miR-27b-3p regulates OSMR expression level to promote acute kidney injury development through the PI3K/AKT signaling pathway. J Cell Physiol 2019; 234:14221–14233. doi: 10.1002/jcp.28118. [DOI] [PubMed] [Google Scholar]
- 27.Tongqiang L, Shaopeng L, Xiaofang Y, Nana S, Xialian X, Jiachang H, et al. Salvianolic acid B prevents iodinated contrast media-induced acute renal injury in rats via the PI3K/Akt/Nrf2 pathway. Oxid Med Cell Longev 2016; 2016:7079487.doi: 10.1155/2016/7079487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira RC, Brito MV, Ribeiro RF, Jr, Oliveira LO, Monteiro AM, Brandão FM, et al. Influence of remote ischemic conditioning and tramadol hydrochloride on oxidative stress in kidney ischemia/reperfusion injury in rats. Acta Cir Bras 2017; 32:229–235. doi: 10.1590/s0102-865020170030000007. [DOI] [PubMed] [Google Scholar]
- 29.Zhai Y, Qi Y, Long X, Dou Y, Liu D, Cheng G, et al. Elevated hsa-miR-590-3p expression down-regulates HMGB2 expression and contributes to the severity of IgA nephropathy. J Cell Mol Med 2019; 23:7299–7309. doi: 10.1111/jcmm.14582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayek SS, Leaf DE, Samman Tahhan A, Raad M, Sharma S, Waikar SS, et al. Soluble urokinase receptor and acute kidney injury. N Engl J Med 2020; 382:416–426. doi: 10.1056/NEJMoa1911481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malek M, Nematbakhsh M. Renal ischemia/reperfusion injury; from pathophysiology to treatment. J Renal Inj Prev 2015; 4:20–27. doi: 10.12861/jrip.2015.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simons M, Raposo G. Exosomes - Vesicular carriers for intercellular communication. Curr Opin Cell Biol 2009; 21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell 2016; 164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 34.Pitt JM, Kroemer G, Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J Clin Invest 2016; 126:1139–1143. doi: 10.1172/jci87316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westhoff JH, Schildhorn C, Jacobi C, Hömme M, Hartner A, Braun H, et al. Telomere shortening reduces regenerative capacity after acute kidney injury. J Am Soc Nephrol 2010; 21:327–336. doi: 10.1681/asn.2009010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S, Hartleben B, Kretz O, Wiech T, Igarashi P, Mizushima N, et al. Autophagy plays a critical role in kidney tubule maintenance, aging and ischemia-reperfusion injury. Autophagy 2012; 8:826–837. doi: 10.4161/auto.19419. [DOI] [PubMed] [Google Scholar]
- 37.Jiang M, Wei Q, Dong G, Komatsu M, Su Y, Dong Z. Autophagy in proximal tubules protects against acute kidney injury. Kidney Int 2012; 82:1271–1283. doi: 10.1038/ki.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang ZS, Yang DY, Fu YB, Zhang L, Zhao QP, Li G. Knockdown of CkrL by shRNA deteriorates hypoxia/reoxygenation-induced H9C2 cardiomyocyte apoptosis and survival inhibition Via Bax and downregulation of P-Erk1/2. Cell Biochem Funct 2015; 33:80–88. doi: 10.1002/cbf.3093. [DOI] [PubMed] [Google Scholar]
- 39.Bjørkøy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2006; 2:138–139. doi: 10.4161/auto.2.2.2405. [DOI] [PubMed] [Google Scholar]
- 40.Pu T, Liao XH, Sun H, Guo H, Jiang X, Peng JB, et al. Augmenter of liver regeneration regulates autophagy in renal ischemia-reperfusion injury via the AMPK/mTOR pathway. Apoptosis 2017; 22:955–969. doi: 10.1007/s10495-017-1370-6. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Zhou D, Ren Y, Zhang Z, Guo X, Ma M, et al. Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy 2019; 15:478–492. doi: 10.1080/15548627.2018.1522467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu H, Liu C, Yang Q, Xin C, Du J, Sun F, et al. MIR145-3p promotes autophagy and enhances bortezomib sensitivity in multiple myeloma by targeting HDAC4. Autophagy 2020; 16:683–697. doi: 10.1080/15548627.2019.1635380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lan T, Shiyu H, Shen Z, Yan B, Chen J. New insights into the interplay between miRNAs and autophagy in the aging of intervertebral discs. Ageing Res Rev 2021; 65:101227.doi: 10.1016/j.arr.2020.101227. [DOI] [PubMed] [Google Scholar]
- 44.Salem M, O’Brien JA, Bernaudo S, Shawer H, Ye G, Brkić J, et al. miR-590-3p promotes ovarian cancer growth and metastasis via a novel FOXA2-versican pathway. Cancer Res 2018; 78:4175–4190. doi: 10.1158/0008-5472.Can-17-3014. [DOI] [PubMed] [Google Scholar]
- 45.Zhao C, Jiang J, Wang YL, Wu YQ. Overexpression of microRNA-590-3p promotes the proliferation of and inhibits the apoptosis of myocardial cells through inhibition of the NF-(B signaling pathway by binding to RIPK1. J Cell Biochem 2019; 120:3559–3573. doi: 10.1002/jcb.27633. [DOI] [PubMed] [Google Scholar]
- 46.Liu JJ, Li Y, Yang MS, Chen R, Cen CQ. SP1-induced ZFAS1 aggravates sepsis-induced cardiac dysfunction via miR-590-3p/NLRP3-mediated autophagy and pyroptosis. Arch Biochem Biophys 2020; 695:108611.doi: 10.1016/j.abb.2020.108611. [DOI] [PubMed] [Google Scholar]
- 47.Huang H, Li X, Yu L, Liu L, Zhu H, Cao W, et al. Wogonoside inhibits TNF receptor-associated factor 6 (TRAF6) mediated-tumor microenvironment and prognosis of pancreatic cancer. Ann Transl Med 2021; 9:1460.doi: 10.21037/atm-21-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Wen H, Fu J, Cai L, Li PL, Zhao CL, et al. Hepatocyte TNF receptor-associated factor 6 aggravates hepatic inflammation and fibrosis by promoting lysine 6-linked polyubiquitination of apoptosis signal-regulating kinase 1. Hepatology 2020; 71:93–111. doi: 10.1002/hep.30822. [DOI] [PubMed] [Google Scholar]
- 49.Ma J, Li YT, Zhang SX, Fu SZ, Ye XZ. MiR-590-3p attenuates acute kidney injury by inhibiting tumor necrosis factor receptor-associated factor 6 in septic mice. Inflammation 2019; 42:637–649. doi: 10.1007/s10753-018-0921-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.