Abstract
The seminal discovery of ribonuclease P (RNase P) and its catalytic RNA by Sidney Altman has not only revolutionized our understanding of life, but also opened new fields for scientific exploration and investigation. This review focuses on human RNase P and its use as a gene-targeting tool, two topics initiated in Altman's laboratory. We outline early works on human RNase P as a tRNA processing enzyme and comment on its expanding nonconventional functions in molecular networks of transcription, chromatin remodeling, homology-directed repair, and innate immunity. The important implications and insights from these discoveries on the potential use of RNase P as a gene-targeting tool are presented. This multifunctionality calls to a modified structure–function partitioning of domains in human RNase P, as well as its relative ribonucleoprotein, RNase MRP. The role of these two catalysts in innate immunity is of particular interest in molecular evolution, as this dynamic molecular network could have originated and evolved from primordial enzymes and sensors of RNA, including predecessors of these two ribonucleoproteins.
Keywords: RNase P, RNA polymerase III, tRNA, innate immunity, antisense, gene targeting
EARLY STUDIES OF HUMAN RNase P
Initial biochemical studies of human RNase P, which were conducted in parallel to those of bacterial RNase P by Sidney Altman, revealed that this endoribonuclease had enzymatic properties similar to those of its bacterial counterpart, particularly in the use of divalent cations for phosphodiester bond hydrolysis and generation of tRNA with 5′-phosphoryl termini (Bothwell and Altman 1975; Koski et al. 1976). These founding works were followed by those of others who used autoimmune antibodies for immunoprecipitation of human RNase P, leading to the identification of H1 RNA and cloning its corresponding gene (Gold et al. 1988; Bartkiewicz et al. 1989; Baer et al. 1990). The gene, designated RPPH1, is transcribed by two polymerases, Pol II and Pol III (James Faresse et al. 2012), possibly as part of alternative defense strategies of the cell against pathogens (see below).
Extensive biochemical purification of human RNase P showed that numerous proteins copurified with the activity of RNase P, which were each designated Rpp, for RNase P protein, followed by their molecular mass in kilodaltons (Eder et al. 1997). Rpp30 and Rpp38 were the first two subunits to be characterized (Eder et al. 1997) and those were followed by the characterization of Rpp14, Rpp20, Rpp21, Rpp29, Rpp40 (Fig. 1; Jarrous et al. 1998, 1999a, 2001) and Rpp25 (Guerrier-Takada et al. 2002). The subunits Pop1, Pop5, and Pop4 (i.e., Rpp29) were identified by using phylogenetic comparative sequence analyses with yeast homologs (Lygerou et al. 1994, 1996; van Eenennaam et al. 1999, 2000, 2001). In the case of the yeast Rpp20 and Rpp30, they were identified based on their homology with human counterparts and characterized by genetic and functional studies (Stolc and Altman 1997; Stolc et al. 1998). The biochemical purification of the Saccharomyces cerevisiae nuclear RNase P and subsequent genetic analyses of its nine protein subunits were thoroughly carried out by David Engelke and his team (Chamberlain et al. 1998), who also characterized the RNA component several years before (Lee et al. 1991). Except for Rpp21, and its yeast homolog Rpr2p, the remaining protein subunits are shared by RNase MRP, a mitochondrial and nucleolar rRNA processing ribonucleoprotein endoribonuclease (Chang and Clayton 1987; Lygerou et al. 1994, 1996; Chu et al. 1997; Dichtl and Tollervey 1997; Lee and Clayton 1997; Chamberlain et al. 1998; van Eenennaam et al. 1999, 2000).
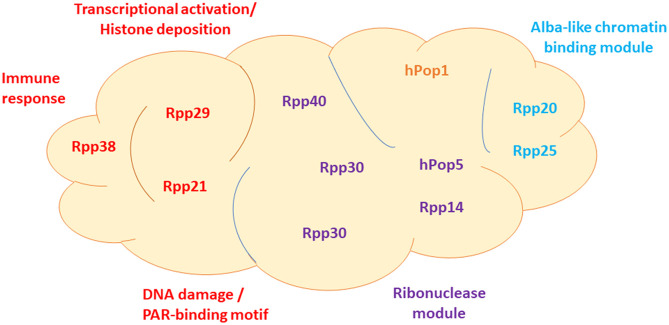
FIGURE 1.
Functional modules of human RNase P. Human RNase P has 10 protein subunits that associate with H1 RNA. A new functional arrangement of these protein subunits in the ribonucleoprotein complex is shown. The approximate positions of the subunits are based on the solved Cryo-EM structure of human RNase P. Traditionally, RNase P RNA is divided into two functional units, the specificity domain and catalytic domain, which refer to binding and cleavage of precursor tRNA by bacterial RNase P. However, following new roles of human RNase P and its subunits in myriad biological processes, other than 5′ end cleavage of precursor tRNA (see text), a new rough arrangement of the subunits in defined functional modules is represented. The functional domains cover transcription, chromatin remodeling, innate immunity, DNA damage repair, and ribonuclease module. Since molecular processes are interconnected, such as chromatin remodeling and transcription, the cooperation of these conjoining domains and sharing of subunits is expected. The Alba-like domain is composed of Rpp20 and Rpp25 and shared by RNase MRP and MRP-TERT. In yeast, the homologs of these two proteins are shared by the telomerase (Lemieux et al. 2016; Garcia et al. 2020).
HUMAN RNase P HAS A CATALYTIC RNA SUBUNIT
The cloning of the cDNAs coding for the protein subunits of human RNase P enabled the preparation of recombinant polypeptides for reconstitution of the endoribonucleolytic cleavage of precursor tRNA by RNase P in vitro (N Orlovetskie and N Jarrous, in prep.; Mann et al. 2003; Reiner et al. 2011). It has been shown that the combination of an in vitro transcribed H1 RNA with recombinant Rpp21 and Rpp29 was sufficient to excise the 5′ leader of precursor tRNA in the presence of ≥10 mM MgCl2. The cleavage was site-specific, dependent on properly folded H1 RNA, particularly its universally conserved P4 pseudoknot, and produced a tRNA with 5′-phosphoryl terminus (Mann et al. 2003). The role of Rpp29 as a cofactor in RNA-based catalysis has been corroborated by demonstrating that it activates M1 RNA, the RNA subunit of Escherichia coli, in a heterologous reconstitution system containing low concentrations of Mg2+ ions (Mann et al. 2003; Sharin et al. 2005). A well-designed heterologous reconstitution system has been established by combining M1 RNA with recombinant Rpp29 homolog of the soil-dwelling amoeba Dictyostelium discoideum using Schizosaccharomyces pombe pSupS1 as substrate (Stamatopoulou et al. 2010). These reconstitution systems underscore the essential role of the eukaryotic RNA subunits of RNase P in substrate recognition and site-specific cleavage and that the protein subunits serve as cofactors in activating the RNA at low concentrations of divalent ions. Direct evidence that H1 RNA is catalytic was provided by demonstrating that this RNA alone excises the 5′ leader of precursor tRNA under reaction conditions of high concentrations of Mg2+ ions (Kikovska et al. 2007). A similar result was obtained in the same study using the RNase P RNA from the lower eukaryote Giardia lamblia (Kikovska et al. 2007). The findings exclude the requirement for any protein cofactor, including Pop1, in hydrolyzing the phosphodiester bond or recognizing the precursor tRNA as substrate. In fact, the formation of binary complexes of H1 RNA-precursor tRNA (pSupS1) was illustrated in vitro (Mann et al. 2003). Thus, the 10 protein subunits of human RNase P serve auxiliary roles in enzyme function, such as proper folding of the RNA or enhancing and diversifying recognition of substrates, such as tRNA and tRNA-like containing substrates (Lyons and Robertson 2003; Wilusz 2016; Jarrous 2017). The diversification of substrates for a catalytic RNase P RNA in the presence and absence of its protein cofactor has been exemplified in vitro (Liu and Altman 1994).
HUMAN RNase P IS PART OF THE TRANSCRIPTION COMPLEXES OF POL III
The availability of recombinant Rpp polypeptides described above allowed the preparation of rabbit polyclonal antibodies, which were used for cell biology studies. Indirect immunofluorescent analysis and expression of fusion Rpp-reporter proteins have revealed that these proteins are primarily localized in distinct nuclear compartments in the cell (Jarrous et al. 1999b, 2001; Abu-Zhayia et al. 2017). This implies that the catalytic ribonucleoprotein form is not confined to a single locale in the nucleus. Importantly, some Rpp subunits rapidly mobilize to induced DNA double-strand breaks (DSBs) sites (Abu-Zhayia et al. 2017). Rpp21 and Rpp29 are promptly recruited to laser-induced and γ-irradiated DSBs and are vital to the subsequent homology-directed repair, but not non-homologous end joining. H1 RNA is implicated in the recruitment of Rpp21 and Rpp29 to the damaged sites (Abu-Zhayia et al. 2017). In contrast, Rpp14, Rpp25, and Rpp38 do not respond to induced DSBs. Hence, an RNase P with protein composition different from the conventional ribonucleoprotein form is used for genome maintenance and preservation. The role of protein subunits of human RNase P, for example, Rpp29, Rpp21, and Pop1, in chromatin assembly and transcriptional regulation has been demonstrated (Newhart et al. 2016; Shastrula et al. 2018). Additionally, two protein subunits, Rpp20 and Rpp25, of human RNase P belong to the Alba-like chromatin binding proteins that regulate transcription (Aravind et al. 2003) and are shared by the telomerase in yeast (Lemieux et al. 2016; Garcia et al. 2020). Knockdown of Rpp25 inhibits 5S rRNA gene transcription by Pol III, but not the tRNA processing activity of RNase P in extracts (Serruya et al. 2015). Thereby, protein subunits of human RNase P are not all dedicated to the endonucleolytic cleavage of precursor tRNA, but rather serve other linked functions. Together, these results dictate a modified structure–function partitioning of human RNase P (Fig. 1), broadening the classical division to specificity and catalytic domains first described for the bacterial RNA counterpart (Haas et al. 1991; Westhof and Altman 1994; Kazantsev et al. 2005; Marquez et al. 2006; Mondragón 2013).
The above differential distribution of human RNase P subunits in the nucleus suggested that the assembly of this ribonucleoprotein is dynamic and related to gene transcription (Jarrous 2002,2017). RNase P could be immunoprecipitated by monoclonal antibodies directed against specific protein subunits of Pol III in coimmunoprecipitation experiments (Reiner et al. 2006, 2008; Serruya et al. 2015). Knockdown of Rpp subunit expression or targeted digestion of the H1 RNA leads to a severe inhibition of transcription of small noncoding RNA genes, including those coding for tRNA, 5S rRNA, 7SL RNA and U6 snRNA. Moreover, subunits of RNase P bind to genetic loci of transcriptionally active tRNA and 5S rRNA genes in a cell cycle-dependent manner (Reiner et al. 2006,2008; Jarrous et al. 2010). These observations support a link between RNase P and Pol III at these loci. Proficient initiation complexes assembled on 5S rRNA and tRNA genes in S100 cell extracts and then purified by gel filtration chromatography and velocity sedimentation analyses contain RNase P (Fig. 2; Serruya et al. 2015; Ramanathan et al. 2020). These purified transcription complexes have molecular weights of 1.5–3 × 106 and carry out transcription and subsequent processing of the nascent precursor tRNA to mature forms. Hence, RNase P and other tRNA processing enzymes coexist with the polymerase in multifunctional transcription complexes (Fig. 2; Jarrous et al. 2022). Knockdown of RNase P expression leads to a severe inhibition of transcription by Pol III (Reiner et al. 2006, 2008; Serruya et al. 2015). Hence, destruction of RNase P leads to inhibition of formation of initiation complexes of this polymerase in cells and extracts (Serruya et al. 2015). In the budding yeast, however, transcription and processing of tRNA are not linked (Chamberlain et al. 1998; Turowski and Tollervey 2015). Moreover, splicing of intron-containing precursor tRNA occurs on the surface of mitochondria (Yoshihisa et al. 2007; Wan and Hopper 2018), whereas knockout of protein subunits of nuclear RNase P showed no inhibitory effect on the synthesis of precursor tRNAs (Lee et al. 1991; Chamberlain et al. 1998). Nonetheless, more localization studies may be needed to determine the precise locales of tRNA splicing, following the evidence for physical and functional contacts of the nucleus to mitochondria (Eisenberg-Bord et al. 2021).
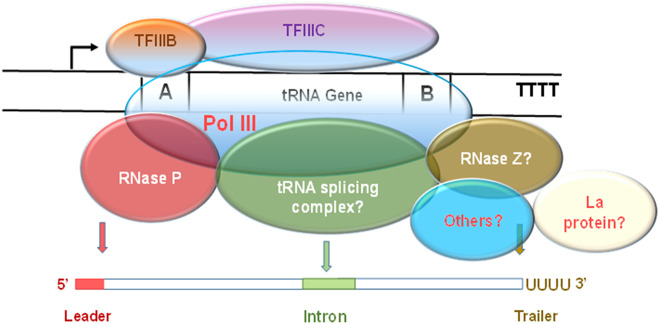
FIGURE 2.
A model of human transcription complex of Pol III assembled on intron-containing tRNA gene. The depicted tRNA gene has two main regulatory elements, Box A and Box B. These elements are bound by the transcription factors TFIIIB and TFIIIC that recruit Pol III. The polymerase initiates transcription from a start point (arrow) and terminates at a short poly-T sequence (TTTT), the terminator. Pol III reinitiates via a facilitated recycling mechanism by which it performs rounds of transcription without leaving the gene. Depending on the tRNA gene, a nascent precursor tRNA can have 5′ leader (red), short intron (green), and 3′ trailer of uridines. The positions of the polymerase, TFIIIB, TFIIIC, RNase P, tRNA splicing complex, RNase Z, La protein, and others (e.g., nucleotide modifiers, helicases, transport factors) are arbitrary.
RNase P FUNCTIONS IN THE INNATE IMMUNE SYSTEM: A PRIMORDIAL RNA-BASED SYSTEM?
Recent genetic and biochemical studies revealed that human Pol III and RNase P are implicated in innate immune responses against DNA and RNA viruses (Ramanathan et al. 2020; Jarrous and Rouvinski 2021). It has been shown that a recessive mutation in the POLR3E gene, which codes for a protein subunit of Pol III, in a child patient impairs production of interferon in response to infection of derived cells with cytomegalovirus and herpes simplex virus type 1 (Ramanathan et al. 2020). The impairment of the antiviral response is linked to compromised activity of mutated Pol III owing to assembly of structurally aberrant transcription complexes. Unexpectedly, expression of protein subunits of Pol III and RNase P also responds to infection of the cell with an RNA virus, the Sindbis virus (Ramanathan et al. 2020). The latter virus has a positive single-stranded RNA genome and belongs to the prevalent genus of alphaviruses. In fact, cells having defects in the biosynthesis of tRNA display high sensitivity to infection with the Sindbis virus (D Mani and N Jarrous, in prep.). The cell responds to an invading RNA virus via a signal transduction pathway that engages the Pol III/RNase P axis as a part of the innate immunity system (D Mani and N Jarrous, in prep.; Jarrous and Rouvinski 2021). The role of RNase P and its subunits in innate immunity appears to be wide-ranging. Thus, it has been shown that the homolog of the Rpp30 subunit of RNase P in rice confers resistance to fungal and bacterial pathogens (Li et al. 2021). Additionally, a role of the evolutionary related RNase MRP in viral infection has also been studied (Jaag et al. 2011; Mattijssen et al. 2011). Notably, transcription of the RPPH1 gene coding for the H1 RNA by two polymerases, Pol II and Pol III (James Faresse et al. 2012), emphasizes the importance of this universally conserved catalytic RNA for the cell defense against invading pathogens (D Mani and N Jarrous, in prep.; Jarrous and Rouvinski 2021). In a hypothetical RNA World rich with protocells and viruses with genomic RNAs, an RNase P RNA ancestor (Evans et al. 2006) might have been crucial for the development of primitive innate immune systems, possibly made of communicating RNAs.
HUMAN RNase P AND INACTIVATION OF GENE EXPRESSION BY EGS
Recognition of precursor tRNAs by RNase P RNA is essentially based on common structural features. This structural basis of substrate recognition was used in the development of a targeting system for inactivation of gene expression by RNase P (Fig. 3; Forster and Altman 1990). Thus, RNase P is directed to recognize and cleave any target RNA, pathogenic or otherwise, by use of a custom-designed external guide sequence (EGS) that forms a tRNA-like structure with the target (Fig. 3B–D). An EGS can be separate or attached to an RNase P RNA, such as the M1 RNA, for generating M1GS ribozymes. Both EGS forms were used first in Sidney Altman's laboratory to demonstrate their potential use for gene-targeting applications (Yuan et al. 1992; Yuan and Altman 1994; Guerrier-Takada et al. 1995; Liu and Altman 1995). Subsequently, results in many laboratories have shown that RNase P and the EGSs effectively inhibit gene expression in gene-targeting applications, such as antiviral, antibacterial, antiparasitic, and anticancer applications.
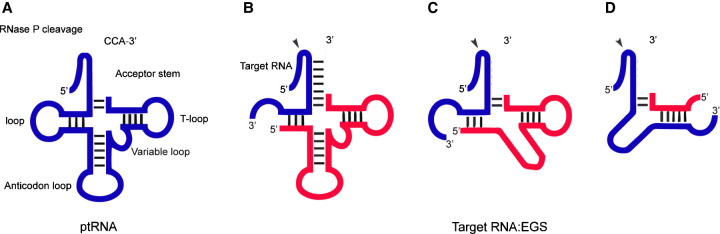
FIGURE 3.
RNase P substrates and EGS technology. (A) Representation of a natural substrate (ptRNA). (B–D) A hybridized complex of a target RNA (e.g., mRNA) and an EGS that resembles part of the structure of a tRNA and can be cleaved by RNase P. (C) results from B by deleting the anticodon domain of the EGS, which is dispensable for EGS targeting activity. (D) represents an alternative design based on C. The arrow shows the site of the cleavage by RNase P and RNase P catalytic RNA.
The EGS technology has been applied to inhibit expression of essential genes of several bacteria, including Salmonella typhimurium, Yersinia pestis, Mycobacterium marinum, Staphylococcus aureus, and Francisella tularensis, and achieve an antimicrobial effect for the infections of several pathogenic bacteria (Altman 2014). Moreover, it was demonstrated that EGSs were effective in inhibiting gene expression and development of Plasmodium falciparum, the causative agent of malaria (Augagneur et al. 2012). This technology can be used in antimicrobial therapy, its ability to achieve species-specific inhibition of bacterial viability could become very useful in circumventing the current limitation of narrow spectrum antimicrobials in inhibiting commensal nonpathogenic bacteria. These results clearly demonstrate the feasibility of using EGS technology in antibacterial strategies (Altman 2014).
The aforementioned EGS forms were tested in cultured human cells and mice for modulating gene expression and blocking viral infection (Bai et al. 2010, 2011; Li et al. 2017; Deng et al. 2019). In human cells, RNase P RNA coupled with designed external guide sequences effectively inhibits the gene expression and replication of many human viruses such as influenza virus, human immunodeficiency virus, human herpes simplex 1, human cytomegalovirus, and Kaposi sarcoma associated herpesvirus (Liu and Altman 1996; Kim and Liu 2007; Jiang et al. 2011). When delivered in mice using a hydrodynamic transfection procedure, a M1GS ribozyme was expressed in the spleen and liver, and effectively blocked cytomegalovirus gene expression and infection in vivo (Li et al. 2018). In an additional study, a M1GS ribozyme was delivered using a Salmonella-based vector and was shown to effectively block cytomegalovirus infection and improve animal survival. Furthermore, when EGSs capable of directing mouse RNase P for gene targeting were delivered in the animal, they were found to be expressed in various tissues, and effectively inhibit cytomegalovirus gene expression and block viral replication and pathogenesis (Li et al. 2017).
The RNase P-EGS technology has also been applied to anti-cancer applications. Sánchez-García and colleagues constructed M1GS ribozymes to specifically cleave chimeric RNA molecules originating from chromosomal abnormalities (Cobaleda and Sánchez-García 2000). They used a well-characterized model of BCR and ABL genes where aberrant translocation results in BCR–ABL oncogenes that cause chronic myelogenous leukemia and acute lymphoblastic leukemias. M1GS RNAs specifically and efficiently cleaved the target mRNA in vitro and inhibited the effect of BCR–ABL function in cultured cells (Cobaleda and Sánchez-García 2000). In another study, Stein and colleagues constructed EGSs to induce RNase P-mediated cleavage of the mRNA that encodes protein kinase C-α (PKC-α) (Ma et al. 2000). Chemically synthesized 2′-O-methyl modified EGSs were administered into T24 bladder carcinoma cells and were shown to be effective in reducing PKC-α expression. These experiments provided direct evidence that RNase P-mediated cleavage directed by EGS is highly specific in targeting its mRNA (Ma et al. 2000).
FUTURE DIRECTIONS AND CHALLENGES OF THE RNase P-EGS TECHNOLOGY
Compared to other nucleic acid-based gene interference approaches, the RNase P-EGS technology exhibits several unique and attractive features as a gene-targeting tool. First, the mechanism of the EGS technology for degradation of a specific mRNA is different from other RNA- or DNA-based gene-targeting approaches. It uses the endogenous RNase P, which is one of the most ubiquitous, abundant, stable and efficient enzymes in all types of cells. RNase P is also highly expressed (5 × 104 copies per cell) and is responsible for the processing of all tRNA precursors in the cell. Second, the sequence specificity of the EGS technology is governed by two different types of interactions between the EGS and the target mRNA: (a) the base-pairing interactions in which the sequence of 10–13 nt in the EGS hybridizes with the target mRNA, and (b) the interactions between the target mRNA and the other part of the EGS sequence (equivalent to the T-stem and T-loop, and variable regions of a tRNA) which are required for folding of the RNase P-recognizable tertiary structure. Thus, the EGS-based technology is specific and does not generate nonspecific “irrelevant cleavage” that is observed in RNase H-mediated cleavage induced by conventional antisense phosphorothioate molecules (Yuan et al. 1992; Ma et al. 2000).
Tremendous progress has recently been made in using nucleic acid gene-targeting tools, such as the CRISPR/Cas9 system, for clinical applications. RNase P-associated EGS and M1GS ribozyme represent promising gene-targeting agents for both basic and clinical applications. Future studies may be needed to address several challenges and develop these agents with the following considerations. The cleavage efficiency and specificity of the RNase P guide sequence technology in vivo will be further improved by better design of EGSs, such as in vitro selection of EGSs and modeling of EGSs with its specific tertiary interactions with the target mRNA and RNase P (Kim and Liu 2007). Moreover, the delivery and expression of the EGSs can be optimized with the recently developed novel vectors and lipid carrier methods that have been shown to be successful for clinical applications.
Further understanding of human RNase P should provide new exciting insight and challenge for the development of this ribonuclease as a gene-targeting tool. For example, recent studies suggest that human RNase P is a part of the network for innate immune responses. Is the expression of RNase P or its activity modulated during innate immune responses such as those induced by viral infections? If this is the case, how are the expressions of different protein subunits and the H1 RNA coordinately regulated during normal and induced conditions for the innate immune responses? Little is currently known about the effects of overloading of RNase P with the EGSs for gene-targeting applications. Will the overloading stimulate the expression of RNase P and modulate innate immune responses? These issues are highly relevant to the RNase P-EGS technology. Further studies on these issues will facilitate the development of the RNase P guide sequence technology in clinic for treatment of various human diseases including infections and cancers.
ACKNOWLEDGMENTS
We thank Paul S. Eder (NIH) for valuable comments and editing of the manuscript, and Isadora Zhang and Eduardo Lujan for critical reading and comments. This research was supported by the Israel Science Foundation (#538/21 and #1205/17) and the United States-Israel Binational Science Foundation (#2015/157) to N.J. and by a University of California Start-Up Fund to F.L. This essay is dedicated to the memory of Sidney Altman, a supreme scientist and caring mentor.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079475.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Abu-Zhayia ER, Khoury-Haddad H, Guttmann-Raviv N, Serruya R, Jarrous N, Ayoub N. 2017. A role of human RNase P subunits, Rpp29 and Rpp21, in homology directed-repair of double-strand breaks. Sci Rep 7: 1002. 10.1038/s41598-017-01185-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman S. 2014. Antibiotics present and future. FEBS Lett 588: 1–2. 10.1016/j.febslet.2013.10.048 [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Anantharaman V. 2003. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol 4: R64. 10.1186/gb-2003-4-10-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augagneur Y, Wesolowski D, Tae HS, Altman S, Ben Mamoun C. 2012. Gene selective mRNA cleavage inhibits the development of Plasmodium falciparum. Proc Natl Acad Sci 109: 6235–6240. 10.1073/pnas.1203516109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer M, Nilsen TW, Costigan C, Altman S. 1990. Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res 18: 97–103. 10.1093/nar/18.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Li H, Vu G-P, Gong H, Umamoto S, Zhou T, Lu S, Liu F. 2010. Salmonella-mediated delivery of RNase P-based ribozymes for inhibition of viral gene expression and replication in human cells. Proc Natl Acad Sci 107: 7269–7274. 10.1073/pnas.0912813107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Gong H, Li H, Vu G-P, Lu S, Liu F. 2011. Oral delivery of RNase P ribozymes by Salmonella inhibits viral infection in mice. Proc Natl Acad Sci 108: 3222–3227. 10.1073/pnas.1014975108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkiewicz M, Gold H, Altman S. 1989. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev 3: 488–499. 10.1101/gad.3.4.488 [DOI] [PubMed] [Google Scholar]
- Bothwell ALM, Altman S. 1975. Partial purification and properties of an endoribonuclease isolated from human KB cells. J Biol Chem 250: 1451–1459. 10.1016/S0021-9258(19)41834-6 [DOI] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR. 1998. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev 12: 1678–1690. 10.1101/gad.12.11.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. 1987. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J 6: 409–417. 10.1002/j.1460-2075.1987.tb04770.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Zengel JM, Lindahl L. 1997. A novel protein shared by RNase MRP and RNase P. RNA 3: 382–391. [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Sánchez-García I. 2000. In vivo inhibition by a site-specific catalytic RNA subunit of RNase P designed against the BCR-ABL oncogenic products: a novel approach for cancer treatment. Blood 95: 731–737. 10.1182/blood.V95.3.731.003k28_731_737 [DOI] [PubMed] [Google Scholar]
- Deng Q, Liu Y, Li X, Yan B, Sun X, Tang W, Trang P, Yang Z, Gong H, Wang Y, et al. 2019. Inhibition of human cytomegalovirus major capsid protein expression and replication by ribonuclease P-associated external guide sequences. RNA 25: 645–655. 10.1261/rna.069682.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Tollervey D. 1997. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J 16: 417–429. 10.1093/emboj/16.2.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder PS, Kekuda R, Stolc V, Altman S. 1997. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc Natl Acad Sci 94: 1101–1106. 10.1073/pnas.94.4.1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord M, Zung N, Collado J, Drwesh L, Fenech EJ, Fadel A, Dezorella N, Bykov YS, Rapaport D, Fernandez-Busnadiego R, et al. 2021. CNM1 mediates nucleus–mitochondria contact site formation in response to phospholipid levels. J Cell Biol 220: e202104100. 10.1083/jcb.202104100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D, Marquez SM, Pace NR. 2006. RNase P: interface of the RNA and protein worlds. Trends Biochem Sci 31: 333–341. 10.1016/j.tibs.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Forster AC, Altman S. 1990. External guide sequences for an RNA enzyme. Science 249: 783–786. 10.1126/science.1697102 [DOI] [PubMed] [Google Scholar]
- Garcia PD, Leach RW, Wadsworth GM, Choudhary K, Li H, Aviran S, Kim HD, Zakian VA. 2020. Stability and nuclear localization of yeast telomerase depend on protein components of RNase P/MRP. Nat Commun 11: 2173. 10.1038/s41467-020-15875-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold HA, Craft J, Hardin JA, Bartkiewicz M, Altman S. 1988. Antibodies in human serum that precipitate ribonuclease P. Proc Natl Acad Sci 85: 5483–5487. 10.1073/pnas.85.15.5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Li Y, Altman S. 1995. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc Natl Acad Sci 92: 11115–11119. 10.1073/pnas.92.24.11115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada C, Eder PS, Gopalan V, Altman S. 2002. Purification and characterization of Rpp25, an RNA-binding protein subunit of human ribonuclease P. RNA 8: 290–295. 10.1017/S1355838202027954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas ES, Morse DP, Brown JW, Schmidt FJ, Pace NR. 1991. Long-range structure in ribonuclease P RNA. Science 254: 853–856. 10.1126/science.1719634 [DOI] [PubMed] [Google Scholar]
- Jaag HM, Lu Q, Schmitt ME, Nagy PD. 2011. Role of RNase MRP in viral RNA degradation and RNA recombination. J Virol 85: 243–253. 10.1128/JVI.01749-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James Faresse N, Canella D, Praz V, Michaud J, Romascano D, Hernandez N. 2012. Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators. PLoS Genet 8: e1003028. 10.1371/journal.pgen.1003028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N. 2002. Human ribonuclease P: subunits, function, and intranuclear localization. RNA 8: 1–7. 10.1017/S1355838202011184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N. 2017. Roles of RNase P and its subunits. Trends Genet 33: 594–603. 10.1016/j.tig.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Jarrous N, Rouvinski A. 2021. RNA polymerase III and antiviral innate immune response. Transcription 12: 1–11. 10.1080/21541264.2021.1890915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Eder PS, Guerrier-Takada C, Hoog C, Altman S. 1998. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA 4: 407–417. [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Eder PS, Wesolowski D, Altman S. 1999a. Rpp14 and Rpp29, two protein subunits of human ribonuclease P. RNA 5: 153–157. 10.1017/S135583829800185X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Wolenski JS, Wesolowski D, Lee C, Altman S. 1999b. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J Cell Biol 146: 559–572. 10.1083/jcb.146.3.559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Reiner R, Wesolowski D, Mann H, Guerrier-Takada C, Altman S. 2001. Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA 7: 1153–1164. 10.1017/S1355838201010469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Reiner R, Dehtiar Y. 2010. Human RNase P and transcription. Protein Rev 10: 223–234. 10.1007/978-1-4419-1142-1_12 [DOI] [Google Scholar]
- Jarrous N, Mani D, Ramanathan A. 2022. Coordination of transcription and processing of tRNA. FEBS J 289: 3630–3641. 10.1111/febs.15904 [DOI] [PubMed] [Google Scholar]
- Jiang X, Bai Y, Rider P, Kim K, Zhang C-Y, Lu S, Liu F. 2011. Engineered external guide sequences effectively block viral gene expression and replication in cultured cells. J Biol Chem 286: 322–330. 10.1074/jbc.M110.158857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. 2005. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci 102: 13392–13397. 10.1073/pnas.0506662102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikovska E, Svärd SG, Kirsebom LA. 2007. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci 104: 2062–2067. 10.1073/pnas.0607326104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Liu F. 2007. Inhibition of gene expression in human cells using RNase P-derived ribozymes and external guide sequences. Biochim Biophys Acta 1769: 603–612. 10.1016/j.bbaexp.2007.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski RA, Bothwell ALM, Altman S. 1976. Identification of a ribonuclease P-like activity from human KB cells. Cell 9: 101–116. 10.1016/0092-8674(76)90056-8 [DOI] [PubMed] [Google Scholar]
- Lee DY, Clayton DA. 1997. RNase mitochondrial RNA processing correctly cleaves a novel R loop at the mitochondrial DNA leading-strand origin of replication. Genes Dev 11: 582–592. 10.1101/gad.11.5.582 [DOI] [PubMed] [Google Scholar]
- Lee JY, Rohlman CE, Molony LA, Engelke DR. 1991. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol 11: 721–730. 10.1128/MCB.11.2.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux B, Laterreur N, Perederina A, Noël J-F, Dubois M-L, Krasilnikov AS, Wellinger RJ. 2016. Active yeast telomerase shares subunits with ribonucleoproteins RNase P and RNase MRP. Cell 165: 1171–1181. 10.1016/j.cell.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Sheng J, Xu M, Vu G-P, Yang Z, Liu Y, Sun X, Trang P, Lu S, Liu F. 2017. Inhibition of murine cytomegalovirus infection in animals by RNase P-associated external guide sequences. Mol Ther Nucleic Acids 9: 322–332. 10.1016/j.omtn.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Liu Y, Wang Y, Li R, Trang P, Tang W, Yang Z, Wang Y, Sun X, Xing X, et al. 2018. Engineered RNase P ribozymes effectively inhibit the infection of murine cytomegalovirus in animals. Theranostics 8: 5634–5644. 10.7150/thno.27776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xiong Y, Lai LB, Zhang K, Li Z, Kang H, Dai L, Gopalan V, Wang GL, Liu W. 2021. The rice RNase P protein subunit Rpp30 confers broad-spectrum resistance to fungal and bacterial pathogens. Plant Biotechnol J 19: 1988–1999. 10.1111/pbi.13612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Altman S. 1994. Differential evolution of substrates for an RNA enzyme in the presence and absence of its protein cofactor. Cell 77: 1093–1100. 10.1016/0092-8674(94)90448-0 [DOI] [PubMed] [Google Scholar]
- Liu F, Altman S. 1995. Inhibition of viral gene expression by the catalytic RNA subunit of RNase P from Escherichia coli. Genes Dev 9: 471–480. 10.1101/gad.9.4.471 [DOI] [PubMed] [Google Scholar]
- Liu F, Altman S. 1996. Requirements for cleavage by a modified RNase P of a small model substrate. Nucleic Acids Res 24: 2690–2696. 10.1093/nar/24.14.2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Séraphin B, Tollervey D. 1994. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev 8: 1423–1433. 10.1101/gad.8.12.1423 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Pluk H, van Venrooij WJ, Séraphin B. 1996. hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J 15: 5936–5948. 10.1002/j.1460-2075.1996.tb00980.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons AJ, Robertson HD. 2003. Detection of tRNA-like structure through RNase P cleavage of viral internal ribosome entry site RNAs near the AUG start triplet. J Biol Chem 278: 26844–26850. 10.1074/jbc.M304052200 [DOI] [PubMed] [Google Scholar]
- Ma M, Benimetskaya L, Lebedeva I, Dignam J, Takle G, Stein CA. 2000. Intracellular mRNA cleavage induced through activation of RNase P by nuclease-resistant external guide sequences. Nat Biotechnol 18: 58–61. 10.1038/71924 [DOI] [PubMed] [Google Scholar]
- Mann H, Ben-Asouli Y, Schein A, Moussa S, Jarrous N. 2003. Eukaryotic RNase P: role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol Cell 12: 925–935. 10.1016/S1097-2765(03)00357-5 [DOI] [PubMed] [Google Scholar]
- Marquez SM, Chen JL, Evans D, Pace NR. 2006. Structure and function of eukaryotic ribonuclease P RNA. Mol Cell 24: 445–456. 10.1016/j.molcel.2006.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattijssen S, Hinson ER, Onnekink C, Hermanns P, Zabel B, Cresswell P, Pruijn GJM. 2011. Viperin mRNA is a novel target for the human RNase MRP/RNase P endoribonuclease. Cell Mol Life Sci 68: 2469–2480. 10.1007/s00018-010-0568-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón A. 2013. Structural studies of RNase P. Annu Rev Biophys 42: 537–557. 10.1146/annurev-biophys-083012-130406 [DOI] [PubMed] [Google Scholar]
- Newhart A, Powers SL, Shastrula PK, Sierra I, Joo LM, Hayden JE, Cohen AR, Janicki SM. 2016. RNase P protein subunit Rpp29 represses histone H3.3 nucleosome deposition. Mol Biol Cell 27: 1154–1169. 10.1091/mbc.E15-02-0099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Weintraub M, Orlovetskie N, Serruya R, Mani D, Marcu O, Stepensky P, Weisblum Y, Djian E, Shaag A, et al. 2020. A mutation in POLR3E impairs antiviral immune response and RNA polymerase III. Proc Natl Acad Sci 117: 22113–22121. 10.1073/pnas.2009947117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner R, Ben-Asouli Y, Krilovetzky I, Jarrous N. 2006. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev 20: 1621–1635. 10.1101/gad.386706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner R, Krasnov-Yoeli N, Dehtiar Y, Jarrous N. 2008. Function and assembly of a chromatin-associated RNase P that is required for efficient transcription by RNA polymerase I. PLoS ONE 3: e4072. 10.1371/journal.pone.0004072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner R, Alfiya-Mor N, Berrebi-Demma M, Wesolowski D, Altman S, Jarrous N. 2011. RNA binding properties of conserved protein subunits of human RNase P. Nucleic Acids Res 39: 5704–5714. 10.1093/nar/gkr126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serruya R, Orlovetskie N, Reiner R, Dehtiar-Zilber Y, Wesolowski D, Altman S, Jarrous N. 2015. Human RNase P ribonucleoprotein is required for formation of initiation complexes of RNA polymerase III. Nucleic Acids Res 43: 5442–5450. 10.1093/nar/gkv447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharin E, Schein A, Mann H, Ben-Asouli Y, Jarrous N. 2005. RNase P: role of distinct protein cofactors in tRNA substrate recognition and RNA-based catalysis. Nucleic Acids Res 33: 5120–5132. 10.1093/nar/gki828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastrula PK, Lund PJ, Garcia BA, Janicki SM. 2018. Rpp29 regulates histone H3.3 chromatin assembly through transcriptional mechanisms. J Biol Chem 293: 12360–12377. 10.1074/jbc.RA118.001845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatopoulou V, Toumpeki C, Tzakos A, Vourekas A, Drainas D. 2010. Domain architecture of the DRpp29 protein and its interaction with the RNA subunit of Dictyostelium discoideum RNase P. Biochemistry 49: 10714–10727. 10.1021/bi101297z [DOI] [PubMed] [Google Scholar]
- Stolc V, Altman S. 1997. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev 11: 2926–2937. 10.1101/gad.11.21.2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Katz A, Altman S. 1998. Rpp2, an essential protein subunit of nuclear RNase P, is required for processing of precursor tRNAs and 35S precursor rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci 95: 6716–6721. 10.1073/pnas.95.12.6716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski TW, Tollervey D. 2015. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev RNA 6: 129–139. 10.1002/wrna.1263 [DOI] [PubMed] [Google Scholar]
- van Eenennaam H, Pruijn GJM, Van Venrooij WJ. 1999. hPop4: a new protein subunit of the human RNase MRP and RNase P ribonucleoprotein complexes. Nucleic Acids Res 27: 2465–2472. 10.1093/nar/27.12.2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eenennaam H, Jarrous N, Van Venrooij WJ, Pruijn GJM. 2000. Architecture and function of the human endonucleases RNase P and RNase MRP. IUBMB Life 49: 265–272. 10.1080/15216540050033113 [DOI] [PubMed] [Google Scholar]
- van Eenennaam H, Lugtenberg D, Vogelzangs JHP, van Venrooij WJ, Pruijn GJM. 2001. hPop5, a protein subunit of the human RNase MRP and RNase P endoribonucleases. J Biol Chem 276: 31635–31641. 10.1074/jbc.M103399200 [DOI] [PubMed] [Google Scholar]
- Wan Y, Hopper AK. 2018. From powerhouse to processing plant: conserved roles of mitochondrial outer membrane proteins in tRNA splicing. Genes Dev 32: 1309–1314. 10.1101/gad.316257.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Altman S. 1994. Three-dimensional working model of M1 RNA, the catalytic RNA subunit of ribonuclease P from Escherichia coli. Proc Natl Acad Sci 91: 5133–5137. 10.1073/pnas.91.11.5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. 2016. Long noncoding RNAs: re-writing dogmas of RNA processing and stability. Biochim Biophys Acta 1859: 128–138. 10.1016/j.bbagrm.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T. 2007. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells 12: 285–297. 10.1111/j.1365-2443.2007.01056.x [DOI] [PubMed] [Google Scholar]
- Yuan Y, Altman S. 1994. Selection of guide sequences that direct efficient cleavage of mRNA by human ribonuclease P. Science 263: 1269–1273. 10.1126/science.8122108 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Hwang ES, Altman S. 1992. Targeted cleavage of mRNA by human RNase P. Proc Natl Acad Sci 89: 8006–8010. 10.1073/pnas.89.17.8006 [DOI] [PMC free article] [PubMed] [Google Scholar]