Abstract
The eukaryotic initiation factor 4G2 (eIF4G2, DAP5, Nat1, p97) was discovered in 1997. Over the past two decades, dozens of papers have presented contradictory data on eIF4G2 function. Since its identification, eIF4G2 has been assumed to participate in noncanonical translation initiation mechanisms, but recent results indicate that it can be involved in scanning as well. In particular, eIF4G2 provides leaky scanning through some upstream open reading frames (uORFs), which are typical for long 5′ UTRs of mRNAs from higher eukaryotes. It is likely the protein can also help the ribosome overcome other impediments during scanning of the 5′ UTRs of animal mRNAs. This may explain the need for eIF4G2 in higher eukaryotes, as many mRNAs that encode regulatory proteins have rather long and highly structured 5′ UTRs. Additionally, they often bind to various proteins, which also hamper the movement of scanning ribosomes. This review discusses the suggested mechanisms of eIF4G2 action, denotes obscure or inconsistent results, and proposes ways to uncover other fundamental mechanisms in which this important protein factor may be involved in higher eukaryotes.
Keywords: cellular IRES, cap-independent translation, cap-dependent translation, ribosome collision, eIF4G1, eIF4F
INTRODUCTION
Canonical mechanism of translation initiation in eukaryotes
Since the discovery of eIF4G2, it has been clear that the protein is involved in specific translation initiation mechanisms, as it is partially homologous to the initiation factor eIF4G1, which plays a key role in the canonical pathway of translation initiation in eukaryotes. Therefore, it makes sense to recall first how the majority of eukaryotic mRNAs initiate translation, discuss proposed alternative initiation mechanisms, and only then proceed to ideas about eIF4G2 functions.
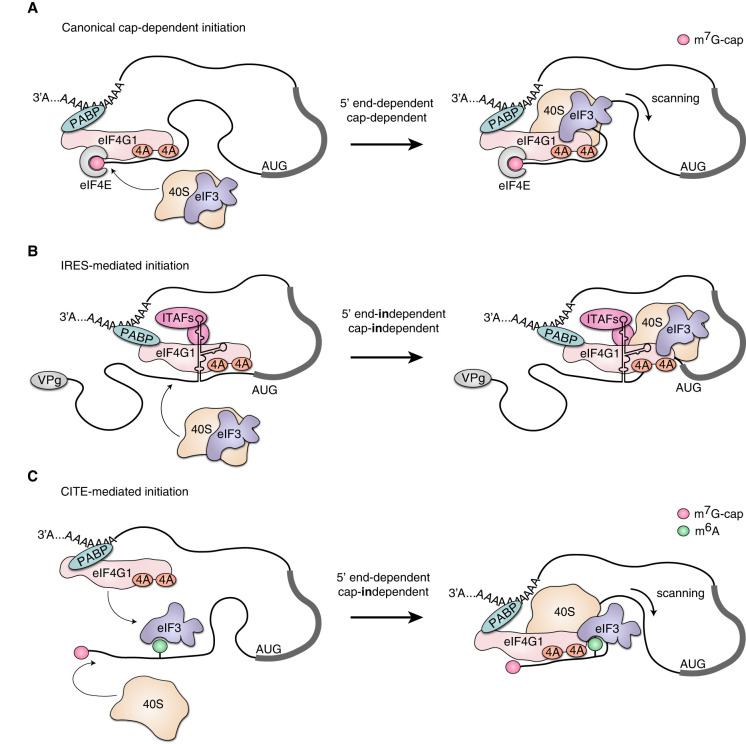
The main principles of the canonical translation initiation mechanism in eukaryotes are the recruitment of the small ribosomal subunit to the very 5′ end of an mRNA and its further sequential advancement along the 5′ UTR in the 3′ direction to locate a start codon (the scanning process) (Fig. 1A). After recognition of the start codon, the large subunit of the ribosome attaches and the synthesis of a polypeptide chain (translation elongation) begins. In eukaryotes, all cytoplasmic mRNAs possess a 7-methylguanosine (m7G) cap connected by a 5′,5′-triphosphate bond to a first nucleotide; the riboses of first two nucleotide residues can also be methylated. The ribosome is recruited to the 5′ end due to the ability of the eIF4F initiation complex to bind simultaneously with the m7G-cap and with the 43S complex that contains the small ribosomal subunit and other initiation factors; for recent detailed reviews, see (Hinnebusch 2014; Hinnebusch et al. 2016; Shirokikh and Preiss 2018). eIF4F consists of three eukaryotic initiation factors (eIFs): the cap-binding protein eIF4E, the helicase eIF4A, and the scaffold protein eIF4G1, which also stimulates eIF4A activity (Grifo et al. 1983). eIF4G1 interacts with eIF3, which in mammals consists of 13 subunits. In its turn, eIF3 binds to the 40S ribosomal subunit, forming the 43S scanning complex, which also includes eIF1, eIF1A, and a ternary complex consisting of eIF2, initiator methionyl-tRNA (Met-tRNAMeti), and GTP.
FIGURE 1.
Different modes of translation initiation. The eIF2 ternary complex, eIF1, eIF1A, eIF5, and eIF4B are not shown for simplicity. (A) Canonical cap-dependent translation. The small ribosomal subunit is recruited to the very 5′ end of mRNA by the eIF4E–eIF4G1–eIF3–40S interaction and then inspects the 5′ UTR of mRNA in the 3′ direction to locate the start codon (the scanning process). (B) IRES-mediated initiation. The small ribosomal subunit is recruited near the start codon of mRNA by the interaction of some translation initiation factors with specific secondary and tertiary structures. In the depicted case, the J-K domain of EMCV IRES attracts eIF4G1, forming a landing platform for the 40S ribosome. (C) CITE-mediated initiation. A tertiary structure or an RNA modification attracts the initiation factor (like the depicted N6-methyladenosine [m6A] attracts eIF3), then the 40S ribosomal complex is recruited and scanning starts from the very 5′ end of mRNA.
Thus, the 40S ribosome subunit is attracted to the capped 5′ end of mRNA through a chain of protein–protein interactions. The subsequent scanning requires the ATP-dependent activity of helicases (eIF4A and others) that unwind the secondary structure elements of 5′ UTRs in front of the scanning complexes (Voss et al. 2017; Shen and Pelletier 2020). Both eIF1 and eIF1A maintain an open conformation of the ribosomal complex competent for scanning and also influence start codon recognition. This is ensured by a constant check for complementarity of the scanned mRNA sequence to the anticodon of Met-tRNAMeti. The start codon must be in a proper nucleotide context for a greater probability of its recognition. For that, the two most important nucleotides around a potential start codon are purines at positions −3 and +4. For vertebrates, a highly efficient context, 5′-gccRccAUGG-3′, where R is purine, was found by Kozak (1987). Later high-throughput screenings confirmed this but also discovered efficient non-Kozak contexts (Noderer et al. 2014; Arce et al. 2017). The interplay of factors eIF2 (brings the initiator Met-tRNAMeti to the ribosome), eIF1 (destabilizes inappropriate codon-anticodon interactions, thereby promoting correct recognition), and eIF5 (activates the GTPase activity of eIF2) is critical for start codon recognition and its stringency. Successful recognition causes the formation of the 48S initiation complex, the rearrangement of Met-tRNAMeti, the release of eIF1, which lets eIF5 trigger eIF2-bound GTP hydrolysis, and the release of inorganic phosphate. The network of eIF1A–eIF5B–Met-tRNAMeti interactions ensures that the initiation complexes are competent for the large subunit to join. The 60S attachment triggers the hydrolysis of GTP bound to eIF5B, then the remaining initiating factors dissociate and polypeptide elongation begins.
At the moment, it is not entirely clear for how long the eIF4F complex remains bound to the m7G-cap during scanning. On longer 5′ UTRs, the connection is most likely broken at some point, probably due to a steric hindrance. Otherwise, only one scanning ribosome per the 5′ UTR could be present at a time. This would significantly reduce the efficiency of translation initiation in proportion to the 5′ UTR length (Bohlen et al. 2020). Futhermore, chains of several 40S ribosomal subunits followed by a single 80S ribosome were observed via electron microscopy of 80S elongating complexes arrested by cycloheximide. This supports the idea that a scanning complex dissociates from the m7G-cap as they move toward a start codon (Shirokikh et al. 2019).
In addition, eIF4G1 interacts with poly(A)-binding protein (PABP). As a result, the polyadenylated 3′ end of an mRNA is also connected to the initiation complex at the 5′ terminus, and thereby the mRNA forms a closed loop (Vicens et al. 2018; Biziaev et al. 2022). Cleavage of eIF4G1 by viral proteases leads to a loss of its scaffolding function. For instance, 2A and L proteases of some picornaviruses cleave off the binding sites of eIF4E and PABP from the eIF4G1 core (the remaining p100 fragment) (Fig. 2A; Lamphear et al. 1995), thereby disrupting cap-dependent translation initiation. The p100 fragment, which contains the binding sites for eIF4A and eIF3, is then used for cap-independent translation of viral mRNAs. In this way, the viral protease simultaneously suppresses translation of cellular mRNAs and stimulates that of the viral ones.
FIGURE 2.
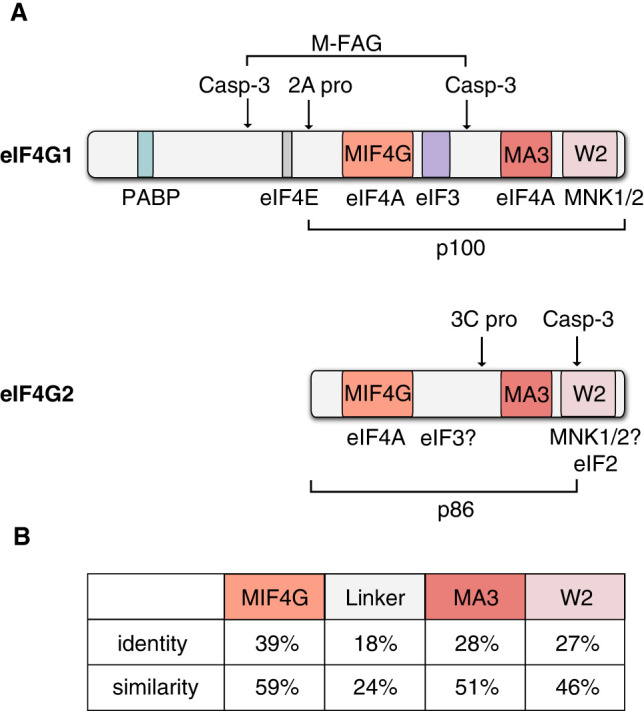
The comparison of mammalian eIF4G1 and eIF4G2. (A) The schematic structures of mammalian eIF4G1 and eIF4G2. Both eIF4G homologs possess three HEAT domains: MIF4G, MA3, and W2. The resolved structural domains are designated inside the boxes, and binding sites for the initiation factors and MNK1/2 are shown under the diagrams. The arrows denote cleavage sites for the picornaviral proteases and for caspase-3 (casp-3). The M-FAG (eIF4G1493–1136) and p86 fragment (eIF4G21–792) are produced by caspase-3. The picornaviral 2A protease (2A pro) separates a p100 fragment from eIF4G1, while eIF4G2 is cleaved by the picornaviral 3C protease (3C pro). (B) The comparison of eIF4G1 and eIF4G2 domains. The amino acid sequences of corresponding domains were pairwise aligned using the EMBL-EBI needle (EMBOSS) service with standard Needleman–Wunsch algorithm preset settings. The calculated percentages of identical and similar amino acids are indicated. The linker domain refers to the amino acid sequence between the MIF4G and MA3 domains.
Several structural homologs of eIF4G1, which are present in mammalian genomes, have been implicated in translation: eIF4G2, eIF4G3, PAIP1, PDCD4, CBP80, CTIF, and SLIP1. For a long time, eIF4G3 was thought to be functionally identical to eIF4G1, but a recent report suggests that it can specifically operate in a complex with eIF4E2 under hypoxic conditions (Ho et al. 2016). The other closest eIF4G1 homolog, eIF4G2, is the focus of this review.
Regulation of canonical translation initiation in animal cells
Regulation of translation often occurs at the stage of initiation. RNA-binding proteins or elements of RNA secondary structure inherent in many 5′ UTR sequences can interfere with scanning. Upstream open reading frames (uORFs) within the 5′ UTRs are another barrier to a scanning ribosome. To get to the main start codon, the scanning complex must either skip the uORF start codon (leaky scanning) or reinitiate scanning after its translation. Therefore, mRNA translation efficiency depends on the probability of these events (Hinnebusch et al. 2016). Clearly, influence of impediments on translation level differs for various mRNAs and depends on the length of their 5′ UTRs and their nucleotide sequences.
The regulation of general level of translation is carried out by changing the activities of two initiation factors: eIF2 and eIF4E. Inactivation of eIF2 due to phosphorylation by one of the four specific kinases reduces the efficiency of translation initiation (Kimball et al. 1998). However, in combination with the inhibitory effect of uORFs, phosphorylation of eIF2α can have the opposite effect: a decrease in the probability of initiation at the start codons of uORFs increases the number of scanning complexes that reach the main AUG. This mechanism, which has been called delayed reinitiation (Vattem and Wek 2004), was first described for the GCN4 mRNA in S. cerevisiae (Abastado et al. 1991) and, later, for the mRNAs of transcription factors ATF4 (Lu et al. 2004; Vattem and Wek 2004) and ATF5 (Zhou et al. 2008) in higher eukaryotes. There are also non-reinitiation-based cases of translational resistance to eIF2 inactivation associated with uORFs: UCP2, IFRD1 (Andreev et al. 2015), and GADD34 (Lee et al. 2009), but the mechanism of this resistance is not fully understood.
The other regulatable factor is the cap-binding protein eIF4E, whose activity is controlled by mTORC1 through phosphorylation of eIF4E-binding proteins (4E-BPs). The inactive kinase cannot phosphorylate 4E-BPs. Unphosphorylated 4E-BPs have a high affinity for eIF4E and thereby outcompete eIF4G1 in forming eIF4F and, consequently, hamper ribosome recruitment via the m7G-cap (Haghighat et al. 1995). However, although mTOR inhibitors have a significant negative effect on translation of cellular mRNAs, they do not block it completely (Gingras et al. 2001). This is associated with incomplete inhibition of eIF4E via 4E-BP or use of alternative translation initiation mechanisms inherent in some cellular mRNAs.
Noncanonical mechanisms to initiate translation in eukaryotic cells
Apart from the canonical cap-dependent mode of translation initiation, there is a mechanism that uses a specific ribosome landing on internal sections of mRNA, known as IRES (internal ribosome entry site) elements (Fig. 1B). In this case, the ribosome is recruited directly to a start codon or close to it instead of the 5′ end of mRNA. This mechanism was first discovered for genomic RNAs from poliomyelitis (PV) (Pelletier and Sonenberg 1988) and encephalomyocarditis (EMCV) viruses (Jang et al. 1988). The presence of IRES elements in viral genomic RNAs has been confirmed for many families of RNA viruses. IRES elements have highly specific secondary and tertiary structures that directly bind to initiation factors, for example, eIF4G1, eIF3, or even the ribosome itself (Yamamoto et al. 2017; Mailliot and Martin 2018; Sorokin et al. 2021). IRES elements from diverse families of RNA viruses have markedly different structures and modes of functioning, but all share the following specific features:
These are compact highly specific structures consisting of 200 to 400 nt residues.
Their function does not depend on a position within 5′ UTRs of mRNAs. They can be located at any distance from the 5′ end of mRNA, and their activity is not affected by upstream or downstream nucleotide sequences.
Depending on the type of IRES element, some (or even all) canonical initiation factors are not required for translation initiation, or, conversely, some additional specific proteins (IRES-transacting factors, ITAFs) may be involved.
The 40S ribosome can scan the 5′ UTR for a while or it can be placed directly at the start site. In the latter case, the start AUG codon is an integral part of the IRES element.
The study of viral IRES elements was especially prolific in the 1990s and early 2000s, using both gene engineering and chemical methods to determine the structures of various IRESs as well as the expression of model mRNAs in cell cultures or cell-free systems. In addition, the IRESs’ complexes with 40S ribosomes were isolated, and their protein compositions were determined. Finally, the assembly of functional initiating complexes from completely purified components was achieved for some IRESs. Since then, the fundamental conclusions drawn from these experiments have not undergone significant modifications (Pestova et al. 2001).
As expected, translation of viral mRNAs containing IRESs is cap-independent and therefore insensitive to inhibition of cap-dependent translation. Over the last three decades, many articles have reported a weak dependence of certain cytoplasmic mRNAs on m7G-cap or a persistent translation under stress conditions. This was observed in general for mRNAs that encode regulatory proteins for the cell cycle, embryogenesis, apoptosis, various stresses, and so on. In the absence of other explanations, such cap-independent translation was considered to be driven by internal initiation. In addition, artifact-prone approaches were used to prove the presence of cellular IRESs, which led to false positive results. Only recently has a reliable methodology for testing potential IRESs been developed (see Jackson 2013 and Terenin et al. 2017 for a recent overview of challenges in cellular IRES research).
Later, alternative mechanisms for cap-independent translation initiation emerged to explain the “atypical” behavior of certain cellular mRNAs. They are based on cap-independent translational enhancers (CITEs) (Fig. 1C). CITEs specifically bind to some initiation factors (eIF3, eIF4G1, eIF4E) or ribosomal subunits, just like IRESs. However, they direct initiation complexes to the very 5′ end of mRNA to scan the corresponding 5′ UTR. As such, CITEs promote translation initiation in a cap-independent but strictly 5′ end-dependent fashion (Terenin et al. 2013; Miras et al. 2017; Sorokin et al. 2021). For instance, eIF3 has been shown to drive an authentic CITE when it binds to methylated adenosines (m6A) and provides cap-independent but 5′ end-dependent translation of Hsp70 mRNA (Meyer et al. 2015; Zhou et al. 2015).
Since the discovery of the eIF4F complex, the initiation mechanism involving eIF4G1 has been regarded as the principal one. Although eIF4G1 is indeed vital for the development of organisms (Goyer et al. 1993; Contreras et al. 2008), its partial depletion in yeast or mammalian cells, contrary to expectations, does not have a critical effect on cell growth and viability under normal conditions (Ramírez-Valle et al. 2008; Park et al. 2011; Badura et al. 2012; Bryant et al. 2018). This indicates the ability of the cell to maintain the required level of protein synthesis at reduced concentrations of eIF4G1. Therefore, the role of eIF4G1 homologs in translation initiation should be determined.
STRUCTURE OF eIF4G2 AND ITS EXPRESSION
The eIF4G2 protein, also known as DAP5, Nat1, and p97, was discovered at the same time in 1997 by four research groups (Imataka et al. 1997; Levy-Strumpf et al. 1997; Shaughnessy et al. 1997; Yamanaka et al. 1997). In two cases, the protein was encountered during a search for apoptosis-associated factors (thus named death associated protein 5, DAP5) (Levy-Strumpf et al. 1997) or targets of the RNA-editing protein APOBEC1 (hence its name, novel APOBEC1 target 1, Nat1) (Yamanaka et al. 1997). Two other studies aimed to characterize an eIF4G1 homolog found by bioinformatic analysis of human cDNA (Imataka et al. 1997; Shaughnessy et al. 1997). These investigations showed that the gene encodes an ∼100 kDa protein homologous to the carboxy-terminal two-thirds of eIF4G1. Like eIF4G1, eIF4G2 binds to eIF4A and eIF3, but, unlike eIF4G1, it does not interact with the cap-binding factor eIF4E (Fig. 2A; Imataka et al. 1997; Yamanaka et al. 1997). Nevertheless, eIF4G2 is associated with polysomes (Lee and McCormick 2006; Nousch et al. 2007). The protein is ubiquitously expressed at a level comparable to that of eIF4G1, yet the exact eIF4G1:eIF4G2 ratio can vary from 2.4:1 (Schwanhäusser et al. 2011) to 1.4:1 (Itzhak et al. 2016) in NIH/3T3 or 1:4.5 (Nagaraj et al. 2011) in HeLa. These results have long suggested that eIF4G2 is an auxiliary protein that promotes elusive mechanisms of translation under rare conditions, and its specific role under normal conditions remained murky until very recently.
In retrospect, eIF4G1 was the first known eIF4G and was called just eIF4G or eIF4γ back then. Later, what is now called eIF4G3 was discovered and named eIF4GII, while eIF4G became eIF4GI, which mirrored the yeast eIF4G1 and eIF4G2 or plant eIF4G and eIF(iso)4G name pairs. In the same years, eIF4G2 was discovered and initially named DAP5, Nat1, and p97, and has retained these names until now. Collectively, all the data indicated that this protein is involved in translation initiation, so around 2005, reports started to emerge that referred to DAP5/Nat1/p97 and eIF4GII as eIF4G2 and eIF4G3, respectively. The problem is that eIF4G2 of higher eukaryotes is not an orthologue of yeast eIF4G2, and it is not obvious why eIF4GII is not the same as eIF4G2. Despite this apparent confusion, we tend to support the names eIF4G1-3 for the mammalian eIF4G orthologues.
Structure of the eIF4G2 protein
There is a recent detailed review of eIF4G1 and eIF4G2 structures (Friedrich et al. 2022), therefore we will just briefly outline some important points. Structural analysis of full-length eIF4G1 (the longest isoform consists of 1599 amino acids in humans) and eIF4G2 (907 amino acids in humans) has not yet been performed. However, crystal structures of individual domains have been obtained. Both homologs have three HEAT domains: MIF4G, MA3, and W2 (Fig. 2A). The X-ray diffraction data have been obtained for all three HEAT domains and the eIF4E-binding site of eIF4G1 (Friedrich et al. 2022), and for MIF4G (residues 61–323) (Virgili et al. 2013), MA3, and W2 domains (540–897) of eIF4G2 (Fig. 2A; Liberman et al. 2008; Fan et al. 2010). Maximum structural similarity is observed between the MIF4G domains of eIF4G1 and eIF4G2 (39% sequence identity, 59% similarity; see Fig. 2B; Virgili et al. 2013), which are both responsible for the interaction with eIF4A. The amino acid residues involved in eIF4A binding are especially conserved. However, the eIF4A binding to the MIF4G domain of eIF4G2 is characterized by a significantly higher dissociation constant compared to that of eIF4G1 (Virgili et al. 2013). The MA3 domain is the second eIF4A-binding site of eIF4G1. Despite the high overall similarity of the MA3 domains (Fig. 2B; Fan et al. 2010), that of eIF4G2 does not bind to eIF4A (Imataka and Sonenberg 1997). It is not known what this site is responsible for but, coincidentally, eIF4G2 stimulates eIF4A helicase activity two times less efficiently than eIF4G1 does (Virgili et al. 2013). The W2 domain of eIF4G2 resembles that of eIF5 and eIF2Bε and is involved in interaction with eIF2β. Although the eIF4G1 and eIF4G2 W2 domains share structural homology, eIF4G1 cannot bind eIF2β directly. A conserved negatively charged glutamate residue E862 of the eIF4G2 W2 domain is strictly required for binding to eIF2β, while the corresponding residue of the eIF4G1 W2 domain is a positively charged lysine (Liberman et al. 2008). Additionally, there is an eIF4G2-specific positively charged region in the MIF4G domain that was suggested to participate in RNA-binding (Virgili et al. 2013). The DETD792 caspase-3 cleavage site (marked with an arrow in Fig. 2A) is located in an unstructured loop of eIF4G2 and is absent in the W2 domains of other proteins (Liberman et al. 2008).
Translation of eIF4G2 mRNA
The eIF4G2 mRNA is characterized by an evolutionarily conserved non-AUG start codon: the GUG triplet is the start codon of human, mouse, zebrafish, and Xenopus eIF4G2 mRNA. Conservation among these species is 60%–80% for the mRNA nucleotide sequence and 75%–90% for the encoded amino acid sequence (Takahashi et al. 2005). The environment of the GUG codon is particularly conserved, with the codon located in a very favorable nucleotide context. The eIF4G2 homolog in Drosophila (dNAT1) is also translated from a non-AUG codon, but the translation start has not been precisely determined (Takahashi et al. 2005). Like other mRNAs with non-AUG start codons, the translation of eIF4G2 mRNA is regulated by eIF5 and 5MP (BZW): eIF5 overexpression enhances, and 5MP inhibits translation of the reporter containing eIF4G2 5′ UTR and a GUG codon as a start (in HEK293T cells) (Tang et al. 2017). Notably, eIF5, 5MP, and eIF1 have a significantly stronger effect on initiation on an AUG in a bad context or on a non-AUG codon compared to an AUG in a good context, allowing for more efficient regulation (Ivanov et al. 2010; Loughran et al. 2012). Translation of eIF4G2 and other mRNAs with non-AUG start codons was shown to be more resistant to a severe elongation-inhibiting stress. It is assumed that the scanning ribosomes are trapped just near the start codon due to the accumulation of elongating ribosomes, which increases the probability of initiation (Kearse et al. 2019). Whether such a phenomenon exists under physiological conditions is not yet known. In addition, the 5′ UTR of eIF4G2 mRNA contains an uORF that does not inhibit translation of the main ORF under normal conditions but may be involved in regulation (Tang et al. 2017). Thus, the synthesis of eIF4G2 protein can be regulated in the cell separately from proteins whose translation begins with the standard AUG codon. At the same time, eIF4G2 mRNA translation can be resistant to certain stresses that reduce the efficiency of protein synthesis on the vast majority of mRNAs. These properties of eIF4G2 mRNA suggest that the protein's function is associated with certain important events in the cell.
The 5′ UTR of eIF4G2 mRNA in all mammals and apparently in some other taxa bears an extended polypyrimidine sequence. Poly(rC)-binding protein 2 (PCBP2) binds to this part of 5′ UTR and thereby inhibits eIF4G2 translation (Smirnova et al. 2019). The estrogen receptor α (ERα) binds the 3′ UTR of eIF4G2 mRNA and promotes its translation via a yet undissected mechanism (Xu et al. 2021). Multiple miRNAs have also been reported to affect eIF4G2 abundance (Sanson et al. 2020; Tong et al. 2022); specifically, miR-16 modulates the expression of eIF4G2 in axons (Kar et al. 2013). Processing of the eIF4G2 mRNA can be accompanied by exon skipping that was observed in mouse lens development (Srivastava et al. 2017). RNA editing of eIF4G2 mRNA by the cytidine deaminase APOBEC1 that introduces multiple stop codons was observed in liver cells (Yamanaka et al. 1997), but its physiological consequences are still unknown.
eIF4G2 interaction with translation factors and other proteins
To determine the eIF4G2-mediated translation mechanism, it is important to understand which proteins it interacts with. Like eIF4G1, eIF4G2 binds both eIF4A isoforms (eIF4A1 and eIF4A2) (Imataka et al. 1997; Henis-Korenblit et al. 2000; Lee and McCormick 2006; Virgili et al. 2013; de la Parra et al. 2018), eIF3 (Imataka et al. 1997; Henis-Korenblit et al. 2000; Liberman et al. 2015; Sugiyama et al. 2017; de la Parra et al. 2018), well-known RNA-binding proteins FXR1/2 (Bukhari et al. 2016; Sugiyama et al. 2017; de la Parra et al. 2018), and FMR1 (Sugiyama et al. 2017; de la Parra et al. 2018), but unlike eIF4G1 (or significantly stronger than eIF4G1), it binds initiation factors eIF2β (Lee and McCormick 2006; Liberman et al. 2015; Bryant et al. 2018; de la Parra et al. 2018), and potential m6A readers PRRC2A-C and IGF2BP1 (Sugiyama et al. 2017; de la Parra et al. 2018; Wu et al. 2019). Whether eIF4G2 binds to the MAP kinase-interacting kinases MNK1 and MNK2 remains unclear: the eIF4G2–MNK1 complex could be detected in (Pyronnet et al. 1999), but not in (Brown et al. 2014; Bryant et al. 2018). The possible roles of some of these unique eIF4G2 partners are discussed below.
Binding to eIF2 is important for the mechanics of eIF4G2. Swapping the W2 domains with eIF4G1 results in eIF4G2 malfunction (Weber et al. 2022) and a point mutation (E862K) that prevents eIF2 binding inactivates eIF4G2 (Bryant et al. 2018). The eIF4G2–eIF2β interaction is correlated with MEK1/2 or CDK1 activation and eIF4G2 phosphorylation (T508), and treatment with MEK1/2 inhibitors completely abolishes both binding and phosphorylation. Yet, eIF4G2 mutants (T508A or T508E) form a complex with eIF2β in a MEK1/2-dependent fashion (Bryant et al. 2018). In the light of the recent structure of the 48S ribosomal complex (Querido et al. 2020), it might seem that the eIF4G2–eIF2β interaction does not fit the structural constraints of both proteins when bound to the small subunit. The resolved part of eIF4G1 (its MIF4G domain) is located rather far from the intersubunit surface-bound eIF2. Should eIF4G2 bind in a similar position, the amino-terminal part of eIF2β that interacts with eIF4G2 (Lee and McCormick 2006) and the linker between the MIF4G and MA3 domains of eIF4G2 are both predicted to be intrinsically disordered and therefore potentially span a considerable distance around the 40S subunit. The obtained 48S structure does not contradict the feasibility of an interaction between eIF4G2 and eIF2β.
The structural similarity of eIF4G1 and eIF4G2 implies their mutually exclusive interaction with eIF3. The decrease in number of eIF4G1–eIF3 complexes under mTOR inhibition (Harris et al. 2006; Thoreen et al. 2012) and the concomitant increase in eIF4G2 binding to eIF3 (Thoreen et al. 2012) indirectly support this notion. Besides, eIF4G1 binds the c, e, and d subunits of eIF3 (LeFebvre et al. 2006; Villa et al. 2013), and eIF4G2 also cross-links to eIF3d (if other eIF3 subunits cross-link to eIF4G2 has not been tested) (de la Parra et al. 2018). The position of the resolved part of eIF4G1 in the human 48S complex confirms previous biochemical findings (Querido et al. 2020). In particular, the amino-terminal domain of eIF3d is found near the MIF4G domain of eIF4G1, while the major part of eIF3d is located at a considerable distance from this position, that is, close to the mRNA exit site of the 40S subunit (Querido et al. 2020; Kratzat et al. 2021). In fact, all suggested mechanisms of eIF4G2 action imply mutually exclusive eIF4G1 and eIF4G2 interactions with ribosomal complexes, but this has never been shown directly and should not be taken for granted. For example, the unstructured linker between MIF4G and MA3 domains that interacts with eIF3e in the case of eIF4G1 (LeFebvre et al. 2006) is rather poorly conserved between eIF4G1 and eIF4G2 (Fig. 2B), and therefore eIF4G2 can interact with eIF3 in another manner. In addition, another MIF4G protein, PAIP1, binds to eIF3g instead (Martineau et al. 2008). Like eIF3d, eIF3g is stretched over the small ribosome subunit, and its parts are bound to it near the mRNA entry site (Simonetti et al. 2016; Querido et al. 2020) and to eIF3l near the eIF4G1 MIF4G position (Simonetti et al. 2016). PAIP1 can form ternary PAIP1–eIF3–eIF4G1 complexes (Martineau et al. 2008), and therefore, it is unclear how different MIF4G proteins coordinate their interactions with eIF3.
PROPOSED MECHANISMS OF eIF4G2 OPERATION IN TRANSLATION INITIATION
Four options for eIF4G2's function in translation initiation can be envisaged. The first two models suggest participation of eIF4G2 in cap-independent translation when eIF4G2 binds to its target cellular mRNAs and serves IRESs or CITEs, respectively. Two other models presume that eIF4G2 is involved in cap-dependent translation when eIF4G2 either uses another cap-binding protein instead of eIF4E or helps eIF4G1 in conventional cap-dependent initiation.
eIF4G2 and cap-independent translation initiation
The striking similarity between eIF4G2 and the carboxy-terminal fragment of eIF4G1 formed by picornaviral 2A proteases (p100) (Fig. 2A), as well as the inability to pull-down eIF4G2 using m7G-sepharose (Tcherkezian et al. 2014; Liberman et al. 2015), immediately implicated eIF4G2 in IRES-mediated translation translation of cellular mRNAs: c-MYC (Henis-Korenblit et al. 2002), XIAP (Henis-Korenblit et al. 2002; Hundsdoerfer et al. 2005), APAF1 (Henis-Korenblit et al. 2002; Nevins et al. 2003; Liberman et al. 2015), eIF4G2 itself (Nevins et al. 2003; Hundsdoerfer et al. 2005; Lewis et al. 2008; Liberman et al. 2015), c-IAP1 (Hundsdoerfer et al. 2005), HIAP2 (Lewis et al. 2008), BCL-2 (Marash et al. 2008; Liberman et al. 2015), CDK1 (Marash et al. 2008), p53 (Weingarten-Gabbay et al. 2014; Haizel et al. 2020), FGF-9 (Haizel et al. 2020), HMGN3 (Yoffe et al. 2016), C9ORF72 (uORF) (Spijker et al. 2021), DSCR1.4 (Seo et al. 2019). Most of these studies used a DNA bicistronic approach, which leads to artifacts and false positive results, and thus the conclusions drawn from such experiments are not persuasive (Kozak 2001; Andreev et al. 2009; Jackson 2013; Terenin et al. 2017). Indeed, the monocistronic reporters with CDK1 or eIF4G2 5′ UTRs failed to show any dependence on eIF4G2 (Smirnova et al. 2022). However, this does not necessarily mean that translation of all mentioned mRNAs does not require eIF4G2. For example, eIF4G2 is involved in translation of APAF1 and BCL2 mRNAs (Smirnova et al. 2022).
Some researchers used A-capped reporter mRNAs to show eIF4G2-dependence of translation, assuming these mRNAs could not be translated in a 5′ end-dependent fashion (Hundsdoerfer et al. 2005; Liberman et al. 2015). Whether this assumption was correct or not, A-capped reporters are not a palatable substitute because they require eIF4G2 for translation, even if their corresponding m7G-capped counterparts do not (Smirnova et al. 2022).
A possible eIF4G2 role in IRES-mediated translation of viral mRNAs is not well established. An isolated domain V of the poliovirus IRES binds to eIF4G1 with Kd = 75 nM (Avanzino et al. 2018), while the entire closely related coxsackie B3 virus (CBV3) IRES binds to eIF4G1 with Kd = 2 nM and eIF4G2 with Kd = 10 nM (Dave et al. 2019). In line with this, eIF4G2 depletion decreases the activity of CBV3 IRES and hampers virus propagation. Whether this reflects a difference in the operation of otherwise similar type 1 IRESs or in experimental setups is not known yet. Moreover, CBV3 2A protease cleaves not only eIF4G1 (Chau et al. 2007), but also eIF4G2 at G434 between the MIF4G and MA3 domains (Hanson et al. 2016) with yet unexplored consequences.
Interestingly, eIF4G2 and m6A mRNA modification are required for translation of at least some circular RNAs that apparently can only be translated through an internal initiation mechanism (Yang et al. 2017; Timoteo et al. 2020). Indeed, tethering either eIF4G1 (Gregorio et al. 1999; Terenin et al. 2013; Chen et al. 2022) or eIF4G2 (Nousch et al. 2007) to an mRNA internal position can drive low levels of internal initiation, and several eIF4G2-interacting proteins, such as FMR1, IGF2BP1, and PRRC2A, were all recently shown to be m6A-readers (Huang et al. 2018; Wu et al. 2019).
In a context of monocistronic mRNAs, the direct binding of eIF4G2 should probably drive CITE-mediated initiation. This may be the case for DSCR1.4 mRNA, which only marginally depends on m7G-cap and requires eIF4G2 for its expression (Seo et al. 2019). In fact, many mRNAs have been shown to interact with eIF4G2 (Marash et al. 2008; Weingarten-Gabbay et al. 2014; Yoffe et al. 2016; Seo et al. 2019; Haizel et al. 2020; Weber et al. 2022; David et al. 2022). In most cases, the binding was linked to eIF4G2-driven internal initiation, but it is not possible to unambiguously interpret the data as a result of the use of error-prone methodology. Moreover, not all eIF4G2-binding mRNA require the protein for their translation, and not every eIF4G2 mRNA target binds eIF4G2 directly (David et al. 2022).
Unconventional cap-dependent initiation modes that involve eIF4G2
Two alternative mechanisms of cap-dependent translation initiation that involve eIF4G2 have been proposed. The first one was discovered in the course of research on the unorthodox stimulation of translation by miRNA (Vasudevan et al. 2007; Mortensen et al. 2011). It only occurs in the G0-phase of the cell cycle (or in G0-like oocytes in stage IV), when cap-dependent translation initiation is inhibited due to mTOR inactivation, and the miRNA induces mRNA deadenylation by PARN. Later, it was found that eIF4G2 is a part of this miRNA-protein complex, together with FXR1 and AGO2 (a key RNA interference protein) (Bukhari et al. 2016). The m7G-cap binding in this mechanism is executed by the deadenylase PARN (Fig. 3A). In the RNAi pathway, however, miRNAs target AGO2 to inhibit mRNA translation and then PARN to induce mRNA decay via deadenylation and subsequent decapping in P-bodies. If a similar complex can stimulate translation, the decapping activity should likely be switched off. Whether this is the case and which protein determines the fate of the AGO2-PARN-mRNA complex is not yet established. A poly(A)-tail is also reversibly shortened during the circadian cycle concomitantly with the mTOR inactivation, and eIF4G2 depletion causes a lengthening of the circadian rhythm in Drosophila (Bradley et al. 2012), suggesting that eIF4G2 operates there in a similar fashion.
FIGURE 3.
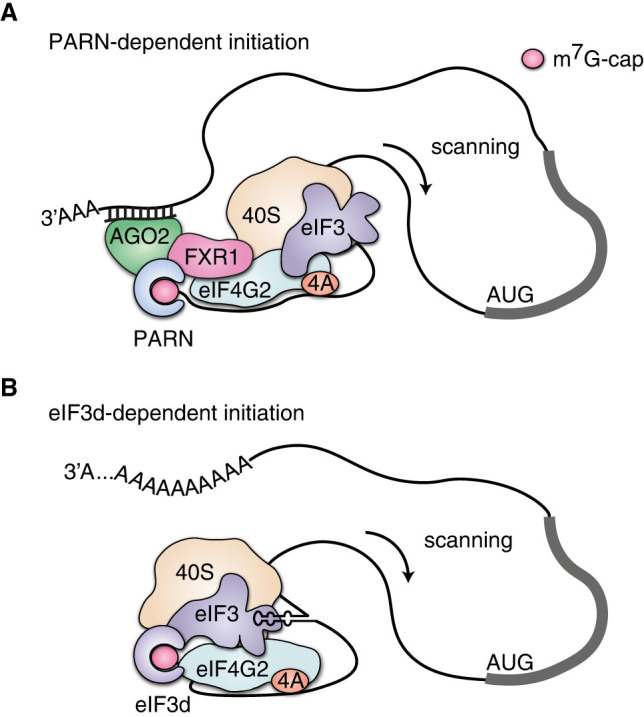
Unconventional cap-dependent initiation modes that involve eIF4G2. (A) PARN-dependent initiation. The miRNA induces mRNA deadenylation by PARN, which also binds to the m7G-cap and is found in the complex with AGO2, FXR1, and eIF4G2. Presumably, eIF4G2 facilitates further scanning. (B) eIF3d-dependent initiation. eIF3 interacts with the special structure in the 5′ UTR of mRNA; the eIF3d subunit specifically binds the m7G-cap and eIF4G2, which participates in subsequent scanning.
Another eIF4G2 partner in the discussed mechanism is FXR1. Notably, FXR1 was found in a complex with eIF4G2 in many reports aimed to identify eIF4G2 protein partners, but the same applies to eIF4G1 (Sugiyama et al. 2017; de la Parra et al. 2018). Moreover, FXR1 is one of the major constituents of stress granules (Ivanov et al. 2018) and, like eIF4G1, eIF4G2 is also found in stress granules (Jain et al. 2016; Anders et al. 2018).
The other proposed mechanism is an eIF3d-dependent recruitment of the 40S ribosomal subunit to the m7G-cap. eIF3 specifically binds some 5′ UTRs (e.g., from JUN mRNA), and its eIF3d subunit possibly binds to the m7G-cap at the 5′ end of these transcripts (Lee et al. 2016). This case is fundamentally different from canonical cap-dependent translation, in which the primary act is the interaction of eIF4E with the m7G-cap, followed by the subsequent assembly of the preinitiation complex at the 5′ end of an mRNA. Here, the primary recognition is most likely not the interaction of eIF3d with the m7G-cap but rather the interaction of whole eIF3 with the 5′ UTR of JUN mRNA. As a result of this interaction, the m7G-cap finds itself on the eIF3d subunit. Therefore, translation initiation of JUN mRNA turns out to be cap-dependent (Lee et al. 2015), and hence the corresponding mechanism does not fit the CITE definition.
When the group led by Robert Schneider discovered that eIF3d cross-links to eIF4G2, a model of eIF3d-mediated cap-dependent translation initiation emerged (Fig. 3B). According to this model, eIF3d recognizes the m7G-cap and brings eIF4G2 rather than eIF4G1 (de la Parra et al. 2018). Unlike the canonical mechanism, where eIF3 first binds to the 40S subunit, eIF4G2 and eIF3 may form an RNP that specifically attracts the small subunit to this particular mRNA. The model implies that eIF3d/eIF4G2-dependent translation occurs when eIF4E-dependent translation is inhibited. For instance, eIF4G2/eIF3d double knockdown and simultaneous overexpression of 4E-BP1 specifically reduce the synthesis of some cellular proteins (JUN, CDK12, and MMP1) in MDA-MB-231 cells.
More recently, the same group studied eIF4G2's role in differentiation of regulatory T cells under conditions of suppressed eIF4E-dependent translation, which is the natural state of these cells prior to their activation (Volta et al. 2021). The researchers again suggested participation of the eIF3d/eIF4G2 complex in translation initiation under such conditions. While the eIF4G2 input in translation indeed increases under conditions of limiting eIF4G1 or mTOR inactivation (Ramírez-Valle et al. 2008; Smirnova et al. 2022), the exact role of eIF3d in this process remains ambiguous. Nonetheless, this work proved the necessity of eIF4G2 for differentiation of regulatory T cells and translation of respective specific mRNAs under conditions of suppressed protein synthesis.
eIF4G2 in conventional cap-dependent translation
The majority of validated eIF4G2-dependent reporter mRNAs show rather high cap-dependencies (Smirnova et al. 2019, 2022) and are sensitive to eIF4E inactivation caused by PP242-mediated inhibition of mTOR kinase or overexpression of 4E-BP1 (Weber et al. 2022). Similarly, eIF4G2 knockdown does not affect the response of eIF4G2 targets to mTOR inactivation. Therefore, under normal conditions, eIF4G2 is involved in canonical cap-dependent translation, presumably downstream from m7G-cap recognition. Moreover, the degree of eIF4G2-dependence of target mRNAs does not correlate in any way with their sensitivity to the mTOR inhibitor PP242. The majority of eIF4G2 targets were found to be as sensitive to PP242 as control β-globin reporter mRNA, while others (PPFIA4 and Maf1) were weakly inhibited by this drug. In the latter case, the small ribosomal subunit can be recruited to mRNAs through a CITE mechanism. Presumably, the putative CITEs of PPFIA4 and Maf1 interact with eIF4G1, rather than with eIF4G2. Otherwise, these two targets would show a particularly strong drop in translation upon eIF4G2 depletion in the presence of PP242, but this was not observed (Smirnova et al. 2022).
Strikingly, deletion of uAUG from eIF4G2 mRNA targets largely relieves the need for this initiation factor, while the insertion of an uORF into the 5′ UTR of an eIF4G2-independent messenger results in the requirement for this protein (Smirnova et al. 2022). Moreover, the uORFs found in the eIF4G2 target mRNAs remain translated when eIF4G2 is lacking (Weber et al. 2022; Smirnova et al. 2022), and a bioinformatic analysis indeed shows an increase in the ratio of uORF/mORF translation in the eIF4G2-depleted cells (David et al. 2022). Since there are only two mechanisms that provide translation of the downstream ORFs, that is, leaky scanning and reinitiation, eIF4G2 should promote at least one of these processes. In our hands, eIF4G2 affected leaky scanning (Smirnova et al. 2022). To definitively distinguish between leaky scanning and reinitiation, we mutated stop codons of uORFs in several eIF4G2-dependent 5′ UTRs so that these extended uORFs substantially overlapped out-of-frame with the luciferase ORF (Smirnova et al. 2022), and translation of these mRNAs remained dependent on eIF4G2. Thus, in the case of these mRNAs, eIF4G2 promotes ribosomal scanning inside and/or downstream from the uORFs rather than reinitiation. Moreover, an uAUG context enhancement inhibited translation initiation at the main AUG and did not alter the need for eIF4G2. Taken together, all of our data suggest that eIF4G2 bolsters translation by stimulating leaky scanning of a subset of mRNAs.
A recent preprint showed that the eIF4G2-interacting proteins PRRC2A, PRRC2B, and PRRC2C also promote leaky scanning on certain, but not all, mRNAs (Bohlen et al. 2022). Strikingly, subsets of mRNAs that require eIF4G2 or PRRC2A-C for their translation at least partially overlap (Bohlen et al. 2022; Smirnova et al. 2022). Future research will show how these proteins cooperate in facilitating leaky scanning.
In another study, eIF4G2 was suggested to promote reinitiation (Weber et al. 2022). This apparent discrepancy could have a variety of mutually nonexclusive causes. First, it is quite possible that eIF4G2 indeed promotes reinitiation on certain mRNAs and leaky scanning on others. Second, Weber and colleagues studied mRNAs with multiple uORFs, and found that extending upstream uORFs may interfere with translation of downstream uORFs and thereby with leaky scanning through them.
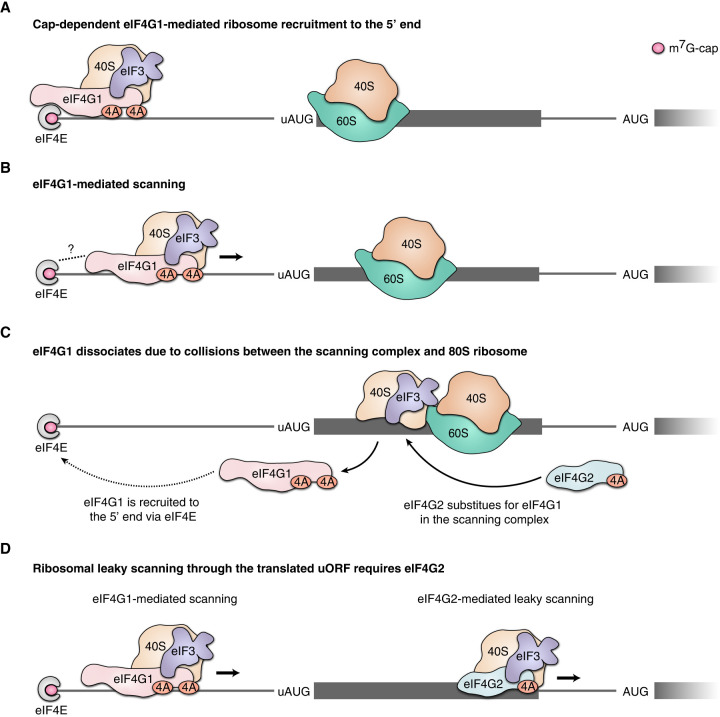
To explain how scanning through a translated uORF results in the need for eIF4G2, we proposed a model, which we tentatively named “a swap of horses.” It postulates that eIF4G2 substitutes for eIF4G1 once the latter dissociates from the scanning complex as a result of collisions of initiating, and/or elongating, and/or terminating 80S ribosomes with scanning complexes that have leaked through the uAUG (Fig. 4). Indeed, in certain cases, uORFs encoding as little as three amino acids are sufficient to promote eIF4G2-dependence, but in other cases, eIF4G2-dependence positively correlates with the uORF length (Smirnova et al. 2022). These data show that elongating ribosomes can definitely cause interference, but initiating or even terminating ribosomes can cause it as well.
FIGURE 4.
Proposed mechanism of eIF4G2's role in conventional cap-dependent translation. The interference of scanning complexes with 80S ribosomes on an uORF leads to the need for eIF4G2. (A) The small ribosome subunit is attached to the 5′ end of mRNA via interaction between m7G-cap and eIF4F. The 80S ribosome is depicted translating the uORF. (B) eIF4G1 facilitates scanning from the very 5′ end. (C) A collision between the scanning complex and 80S ribosome within the uORF forces a partial loss of eIF4G1 that is brought back to the m7G-cap via interaction with eIF4E. The role of helicase is then assumed by eIF4G2–eIF4A. (D) eIF4G1 is involved in scanning from the very 5′ end, and eIF4G2 participates in scanning after the eIF4G1 loss and thus mediates leaky scanning.
Apparently, this interference can occur when a faster ribosome complex bumps into a slower downstream ribosome or another obstacle. The most likely impediments for the scanning complex are the slower post-scanning initiating (Wang et al. 2022) or terminating ribosomes. Collisions between scanning and elongating ribosomes can also occur. Scanning complexes have been shown to move faster than elongating ribosomes (Vassilenko et al. 2011). All translation elongation rate estimates fall in the ballpark of around 5 codons per second (Gerashchenko et al. 2021), while attempts to estimate a scanning rate are less coherent. A recent observation of extremely fast scanning (over 100 nt/sec) (Wang et al. 2022) suggests that a scanning complex can potentially catch up ribosomes that translate the uORF. From a structural point of view, eIF4G1 resides on the periphery of the 48S complex, equidistant from either the entry and the exit of the mRNA channel (Querido et al. 2020), and any frontal or rearal bumping can arguably make eIF4G1 dissociate.
The exchange mechanism should respond to changes in eIF4G1 concentration, and the data on double eIF4G1/eIF4G2 knockdown also support the idea that eIF4G2 can replace eIF4G1 during scanning. The limited availability of eIF4G1 makes translation of eIF4G2 nontarget mRNAs at least partially dependent on eIF4G2 and vice versa (Smirnova et al. 2022), and a synergy in eIF4G1/eIF4G2 depletion was previously reported (Ramírez-Valle et al. 2008). This mechanism rescues, at least partially, leaky scanning through uORFs, thereby increasing the number of scanning complexes that reach the main start codon. This can be crucial for higher eukaryotes, whose mRNAs are rich in uAUGs. So, it is eIF4G1 that loads the preinitiation complex to the 5′ end of an mRNA, and then eIF4G2 participates in scanning downstream from m7G-cap recognition. However, uORFs are certainly not the only obstacle to the scanning ribosomes, causing eIF4G1 to be replaced by eIF4G2. For example, the PCBP2 mRNA does not contain any uORF, but its translation nevertheless strongly depends on eIF4G2 (Smirnova et al. 2019). Secondary structures and mRNA-binding proteins could be proposed as such impediments to scanning complexes. These obstacles may also be the reason for an augmentation in eIF4G2 dependence as the 5′ UTR length increases.
eIF4G2 SIGNIFICANCE IN CELL FATE
eIF4G2 depletion often reduces the viability, protein synthesis level, and growth rate of cells in culture but does not have a critical effect under normal conditions (Lee and McCormick 2006; Lewis et al. 2008; Marash et al. 2008; Ramírez-Valle et al. 2008; Thoreen et al. 2012). In this regard, many research groups have suggested that eIF4G2's role in translation may be more pronounced under specific conditions, such as apoptosis, differentiation, arrest at certain phases of the cell cycle, etc.
Apoptosis
The research group of Adi Kimchi was one of the groups that discovered eIF4G2 as a novel translation factor (Levy-Strumpf et al. 1997). They also showed that eIF4G2 is cleaved at the DETD792 site to form an 86 kDa fragment (p86; eIF4G21–792) in SKW 6.4 cells (human B cells) during apoptosis, and caspase inhibitors prevent this (Henis-Korenblit et al. 2000). Upon induction of apoptosis, p86 is also formed in NB4 cells, and the eIF4G2 knockdown reduces the level of apoptosis (Ozpolat et al. 2008). According to some reports, p86 specifically stimulates the expression of proapoptotic proteins (c-Myc, XIAP, Apaf-1, and eIF4G2) (Henis-Korenblit et al. 2000, 2002; Nevins et al. 2003), but the authors attribute these effects to a specific p86 involvement in IRES-mediated translation. The problems with the methodology for identifying cellular IRES have been discussed above.
The cleavage of eIF4G2 occurs not only during apoptosis. Treatment of HEK293T cells with thapsigargin (a calcium pump inhibitor) or tunicamycin (an inhibitor of protein glycosylation), which both cause endoplasmic reticulum stress, also leads to a caspase-dependent limited proteolysis of eIF4G2 (Warnakulasuriyarachchi et al. 2004), while the overall eIF4G2 level is increased (Lewis et al. 2008). Translation of the proapoptotic HIAP2 protein is induced by ER stress in the eIF4G2-depleted cells to a lesser degree than in control cells (Lewis et al. 2008); therefore, eIF4G2 may be involved in the translational response to ER stress.
If the above facts speak in favor of a proapoptotic function of eIF4G2, some studies came to the opposite conclusion. It was shown that in the eIF4G2-depleted HeLa cells, the expression of anti-apoptotic BCL2 and CDK1 proteins decreased, while the level of proapoptotic Bax (interacts with BCL2), on the contrary, increased significantly. In the absence of eIF4G2, an arrest in the M-phase (but not the S-phase) increases the activity of caspase (Marash et al. 2008; Liberman et al. 2009), demonstrating eIF4G2's participation in counteracting apoptosis. Overexpression of either p97 or p86 isoforms attenuates cisplatin-induced apoptosis (Gao et al. 2012). It is known that eIF4G2 is less susceptible to proteasomal degradation than eIF4G1, and eIF4G2 depletion reduces cell survival under oxidative stress, also indicating the antiapoptotic function of eIF4G2 (Alard et al. 2019). But there is no contradiction in these data. First, it is known that at the initial stages of apoptosis, the cell has not yet made a final decision about its fate, so expression of anti-apoptotic proteins can reverse the response to stress, and second, there is nothing strange in the fact that one protein can promote translation of different mRNAs encoding anti- and proapoptotic proteins.
Cellular differentiation
Yamanaka's group obtained homozygous mouse embryonic stem cells (mESC) with eIF4G2 knockout but failed to obtain homozygous (eIF4G2–/–) mice, since such embryos failed to form mesoderm (Yamanaka et al. 2000). At the same time, homozygous (eIF4G2–/–) mESCs did not differentiate after retinoic acid (RA) treatment and did not form mesodermal tissues when injected into mice. Later, the same research group demonstrated that eIF4G2–/– mESCs have expression profiles similar to those of wild-type cells incapable of differentiation (after treatment with Erk and Gsk3b kinase inhibitors) (Sugiyama et al. 2017). They also studied the fruit fly eIF4G2 orthologue (dNAT1) and showed that the W2 domain deletion (i.e., similar to the apoptotic p86 fragment described above) was lethal as the elongation of the germ layer was impaired (Yoshikane et al. 2007). In addition, approximately one-quarter of Danio rerio embryos with eIF4G2 knockdown also failed to develop mesoderm (Nousch et al. 2007).
Retinoic acid-induced differentiation in NB4 cells (acute promyelocytic leukemia) increased the level of eIF4G2 protein (Harris et al. 2004; Ozpolat et al. 2007), and its knockdown inhibited the differentiation (Ozpolat et al. 2008). The human embryonic stem cells (hESC) depleted of eIF4G2 also did not differentiate properly upon retinoic acid treatment: an increased expression of pluripotency genes and a decreased expression of differentiation markers were observed. The same pattern was demonstrated for the embryonic bodies that normally undergo spontaneous differentiation, but they continued to express pluripotency genes at day 21 under eIF4G2 depletion. In addition, they did not show any histological signs of differentiation, such as pseudoepithelial and cavity formation (Yoffe et al. 2016). eIF4G2 is also required for the artificial differentiation of naïve T lymphocytes into regulatory T cells (Volta et al. 2021).
Expression of eIF4G2 is enhanced during myogenic differentiation, and its knockdown inhibits the formation of myotubes from cultured myoblasts (Sanson et al. 2020). This was attributed to reduced translation of Usmg5 mRNA. In neurons, BDNF stimulates eIF4G2 mRNA transcription (Seo et al. 2019), whereas eIF4G2 knockdown in axons leads to a decrease in β-actin, HIF1A and COX IV protein levels (Kar et al. 2013).
It was also shown that during oocyte fertilization in sea urchins, some mRNAs, including one coding for eIF4G2, are directed to polysomes, while mRNAs that encode the canonical translation factors eIF4E1, eIF4A, eIF4G1, PABP, and 4E-BP are absent from there (Chassé et al. 2019). This indirectly confirms the need for eIF4G2 for embryonic development processes, including differentiation.
Thus, it can be confidently stated that eIF4G2 is required for cell differentiation and mesoderm formation during the development of many organisms, but it remains unclear whether this is the primary function of eIF4G2 (if such a thing as a “primary function of eIF4G2” exists) and what the corresponding molecular mechanism is.
Translation of specific mRNAs
In search of eIF4G2-dependent processes and the conditions under which this factor is possibly activated, some researchers have tried different approaches. In this regard, immunoprecipitation, mRNA sequencing from polysomes (sequencing of a heavy fraction of lysate in a sucrose gradient containing mRNAs covered with ribosomes, i.e., actively translated mRNAs; polysome profiling) (Mašek et al. 2011), and ribosome profiling (sequencing of mRNA fragments protected by the ribosome) (Ingolia et al. 2009) were used to identify mRNAs whose translation is specifically dependent on eIF4G2, and what they have in common.
The researchers in Adi Kimchi's group immunoprecipitated overexpressed FLAG-p86 from HEK293T cells. The coprecipitated mRNAs were turned into labeled cDNAs and hybridized on a chip with 200 cDNAs of genes related to the cell cycle and apoptosis. The 13 mRNAs for which the signal was at least two times higher than the control belonged to the TNF, NF-κB pathway, or IGF cascade family groups. One of the identified mRNAs was CDK1, whose binding to eIF4G2 was also confirmed by other methods (Marash et al. 2008).
It is clear that the most informative way to identify genes whose translation requires a particular protein is the quantitative sequencing of translated mRNAs in control cells and those depleted of the target protein under normal and/or stress conditions. This allows the identification of genes whose translation efficiency significantly changes upon the depletion. The mRNA sequencing method from the polysomal fraction was then applied to the wild-type and eIF4G2 permanently knocked-down hESC cells by Adi Kimchi's group (Yoffe et al. 2016). One hundred and twenty two genes were found whose translation efficiency decreased by 1.5 times or more. The ontological analysis showed the enrichment of targets in mitochondrial proteins, especially components of the oxidative respiratory chain, proteins involved in translation, including many ribosomal proteins and mRNA splicing factors; and proteins that participate in embryogenesis and differentiation (HMGN3, ANAPC5, ZDHHC4).
The polysome mRNA sequencing was used by the group led by Robert Schneider. They demonstrated that about 10% of genes were translated significantly worse with the induced eIF4G2 knockdown in MDA-MB-231 cells (de la Parra et al. 2018). These genes are mostly related to cell death and survival, cell organization, cell motility, and DNA repair.
Ribosome profiling is a more accurate and informative modification of the mRNA sequencing method from the polysomal fraction. In this approach, polysomes are treated by a ribonuclease, and only mRNA fragments protected by the ribosomes remain intact and are then sequenced. Thus, it is possible not only to determine the efficiency of mRNA translation more accurately but also to detect the distribution of ribosomes along the transcript. This is crucial for research on uORF-mediated translational control because translation of uORF and main ORF may follow distinct patterns of regulation. For example, if an uORF translation does not depend on eIF4G2 and is efficient, this mRNA will be detected at the polysome fraction despite a decrease in the main ORF translation. Yamanaka's research group applied ribosomal profiling to wild-type and eIF4G2 knockout mESCs (Sugiyama et al. 2017). Upon the knockout, 14 eIF4G2 target mRNAs with significantly decreased translation efficiency (including Map3k3 and Sos1) and 4 mRNAs with significantly increased translation efficiency (including Sorbs2, Xaf1) were identified. Overexpression of one of eIF4G2's translational targets, Map3k3, in knockout cells caused cell differentiation, which is consistent with the above data on eIF4G2's role in this process.
Further prospects for research of eIF4G2's role in translation initiation
Despite recent advances in understanding the mechanics of eIF4G2, many question marks over the protein remain. First, we do not know why certain mRNAs containing uORFs demand eIF4G2 while the others do not. This may well be related to the question of how sequences near the uORFs affect the requirement for eIF4G2. Ultimately, analysis of those eIF4G2-dependent 5′ UTRs that do not contain uORFs, for example, PCBP2 (Smirnova et al. 2019, 2022), KMT2D (David et al. 2022), and Sos1 (Sugiyama et al. 2017) will be important. It is quite possible that these 5′ UTRs contain yet unidentified non-AUG initiation codons. If they do not, then we need to look for other elements of the 5′ UTR structure that can cause the substitution of eIF4G1 with eIF4G2. Second, the eIF4G2 mRNA targets seem to have rather long 5′ UTRs. Although this may be a coincidence (there is a higher probability of meeting an uAUG in a longer 5′ UTR), this also may reflect a higher probability of eIF4G1 dissociation from the scanning complexes that are moving away from the m7G-cap. Other determinants of eIF4G2-dependence should be identified, and their structures should be established. Some nucleotide sequences may act as barriers to scanning in the case of a few eIF4G2 targets, as some of them are impressive in their abundance of G and C residues, putative G quadruplexes, and G, C, and polypyrimidine tracts (see, for instance, the 5′ UTRs of PCBP2, PHD2, FOXP3, HMGN3, and PRICKLE1 mRNAs). In other cases, secondary and ternary structures may initiate the assembly of scanning complexes in a CITE-mode with eIF4G2 participation or increase a local eIF4G2 concentration so that dissociated eIF4G1 could be readily replaced. Indeed, high affinity sites for eIF4G1 and eIF4G2 have recently been demonstrated for the 5′ UTRs of some animal mRNAs (Haizel et al. 2020). Understanding the dynamics of the interaction between m7G-cap and eIF4F during scanning could also be instrumental in revealing the mechanics of eIF4G2-mediated initiation.
The role of eIF2–eIF4G2 interaction remains unclear. Interestingly, eIF4G1 binds eIF1 in yeast (He et al. 2003; Singh et al. 2012) and in humans (Sinvani et al. 2015; Sehrawat et al. 2022), and in both cases this interaction is required for stringency in initiation site selection (He et al. 2003; Sehrawat et al. 2022). eIF4G2 apparently lacks the eIF1 binding site; thus, the eIF4G2-driven scanning complexes may have altered codon stringency recognition properties. On the other hand, human eIF5, unlike its yeast counterpart, cannot bind to both eIF1 and eIF2β simultaneously (Luna et al. 2012); thus, the eIF2β–eIF4G2 interaction likely interferes with eIF2β–eIF5 binding and can contribute to start-codon selection by eIF4G2-driven complexes. Moreover, the apoptotic carboxy-terminally truncated eIF4G2 fragment (p86) lacks the eIF2-binding site, but we still do not know what the difference between the activities of p86 and full-length forms of eIF4G2 is.
The eIF3–eIF4G2 interaction is also tricky. It is still not established whether the binding of eIF4G1 and eIF4G2 to eIF3 is mutually exclusive, and which part of eIF4G2 binds eIF3 is unknown. When eIF4G2 is brought to an mRNA via cap-bound eIF3d, there is no explanation why eIF3d brings eIF4G2 and not eIF4G1. Finally, given the suggested role of eIF4G2 in scanning, it is not entirely clear why eIF4G2 depletion enhances certain mRNAs’ translation.
It would be very helpful to assemble 48S initiation complexes on eIF4G2-dependent 5′ UTRs using complete cell extracts or proteins from the ribosomal salt wash fraction and study their protein composition. In this way, it might be possible to identify specific eIF4G2 partners. Cell lines expressing eIF4G2 and eIF4G1 with affinity tags would be helpful for immunoprecipitation of the corresponding scanning complexes. Modern NGS and omics technologies are valuable for identifying eIF4G2 translational targets and potential partners, but the detailed molecular mechanism of the protein function can best be established in subsequent in vitro experiments involving the assembly of functioning systems from completely purified components.
Clearly, canonical cap-dependent ribosomal scanning is the major mechanism of translation initiation in higher eukaryotes. However, minor pathways or deviations from the basic mechanism that only marginally manifest themselves under normal conditions exist and are not easy to address. Different poorly characterized eIF4G homologs and/or yet-to-be discovered proteins may play a role in alternative translation initiation, particularly under temporary or chronic stress.
ACKNOWLEDGMENTS
This work was supported by a grant from the Russian Science Foundation (19-14-00152) to Ivan N. Shatsky.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079462.122.
Freely available online through the RNA Open Access option.
REFERENCES
- Abastado JP, Miller PF, Jackson BM, Hinnebusch AG. 1991. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol 11: 486–496. 10.1128/mcb.11.1.486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alard A, Marboeuf C, Fabre B, Jean C, Martineau Y, Lopez F, Vende P, Poncet D, Schneider RJ, Bousquet C, et al. 2019. Differential regulation of the three eukaryotic mRNA translation initiation factor (eIF) 4Gs by the proteasome. Front Genet 10: 254. 10.3389/fgene.2019.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, Verzini S, Qian S-B, Ignatova Z. 2018. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci Alliance 1: e201800113. 10.26508/lsa.201800113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, Dmitriev SE, Terenin IM, Prassolov VS, Merrick WC, Shatsky IN. 2009. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res 37: 6135–6147. 10.1093/nar/gkp665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O'Connor PBF, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. 2015. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife 4: e03971. 10.7554/elife.03971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce AJD, Noderer WL, Wang CL. 2017. Complete motif analysis of sequence requirements for translation initiation at non-AUG start codons. Nucleic Acids Res 46: 985–994. 10.1093/nar/gkx1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanzino BC, Jue H, Miller CM, Cheung E, Fuchs G, Fraser CS. 2018. Molecular mechanism of poliovirus Sabin vaccine strain attenuation. J Biol Chem 293: 15471–15482. 10.1074/jbc.ra118.004913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badura M, Braunstein S, Zavadil J, Schneider RJ. 2012. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci 109: 18767–18772. 10.1073/pnas.1203853109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biziaev NS, Egorova TV, Alkalaeva EZ. 2022. Dynamics of eukaryotic mRNA structure during translation. Mol Biol 56: 382–394. 10.1134/s0026893322030037 [DOI] [PubMed] [Google Scholar]
- Bohlen J, Fenzl K, Kramer G, Bukau B, Teleman AA. 2020. Selective 40S footprinting reveals cap-tethered ribosome scanning in human cells. Mol Cell 79: 561–574.e5. 10.1016/j.molcel.2020.06.005 [DOI] [PubMed] [Google Scholar]
- Bohlen J, Roiuk M, Teleman AA. 2022. PRRC2 proteins regulate translation initiation by promoting leaky scanning. Biorxiv 10.1101/2022.11.11.516176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley S, Narayanan S, Rosbash M. 2012. NAT1/DAP5/p97 and atypical translational control in the Drosophila circadian oscillator. Genetics 192: 943–957. 10.1534/genetics.112.143248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MC, Bryant JD, Dobrikova EY, Shveygert M, Bradrick SS, Chandramohan V, Bigner DD, Gromeier M. 2014. Induction of viral, 7-methyl-guanosine cap-independent translation and oncolysis by mitogen-activated protein kinase-interacting kinase-mediated effects on the serine/arginine-rich protein kinase. J Virol 88: 13135–13148. 10.1128/jvi.01883-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant JD, Brown MC, Dobrikov MI, Dobrikova EY, Gemberling SL, Zhang Q, Gromeier M. 2018. Regulation of hypoxia-inducible factor 1α during hypoxia by DAP5-induced translation of PHD2. Mol Cell Biol 38: e00647-17. 10.1128/mcb.00647-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari SIA, Truesdell SS, Lee S, Kollu S, Classon A, Boukhali M, Jain E, Mortensen RD, Yanagiya A, Sadreyev RI, et al. 2016. A specialized mechanism of translation mediated by FXR1a-associated microRNP in cellular quiescence. Mol Cell 61: 760–773. 10.1016/j.molcel.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassé H, Boulben S, Cormier P, Morales J. 2019. Translational control of canonical and non-canonical translation initiation factors at the sea urchin egg to embryo transition. Int J Mol Sci 20: 626. 10.3390/ijms20030626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau DHW, Yuan J, Zhang H, Cheung P, Lim T, Liu Z, Sall A, Yang D. 2007. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis 12: 513–524. 10.1007/s10495-006-0013-0 [DOI] [PubMed] [Google Scholar]
- Chen R, Wang SK, Belk JA, Amaya L, Li Z, Cardenas A, Abe BT, Chen C-K, Wender PA, Chang HY. 2022. Engineering circular RNA for enhanced protein production. Nat Biotechnol 1–11. 10.1038/s41587-022-01393-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras V, Richardson MA, Hao E, Keiper BD. 2008. Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death Differ 15: 1232–1242. 10.1038/cdd.2008.46 [DOI] [PubMed] [Google Scholar]
- Dave P, George B, Raheja H, Rani P, Behera P, Das S. 2019. The mammalian host protein DAP5 facilitates the initial round of translation of Coxsackievirus B3 RNA. J Biol Chem 294: 15386–15394. 10.1074/jbc.ra119.009000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M, Olender T, Mizrahi O, Weingarten-Gabbay S, Friedlander G, Meril S, Goldberg N, Savidor A, Levin Y, Salomon V, et al. 2022. DAP5 drives translation of specific mRNA targets with upstream ORFs in human embryonic stem cells. RNA 28: 1325–1336. 10.1261/rna.079194.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Parra C, Ernlund A, Alard A, Ruggles K, Ueberheide B, Schneider RJ. 2018. A widespread alternate form of cap-dependent mRNA translation initiation. Nat Commun 9: 3068. 10.1038/s41467-018-05539-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Jia M-Z, Gong W. 2010. Crystal structure of the C-terminal region of human p97/DAP5. Proteins 78: 2385–2390. 10.1002/prot.22735 [DOI] [PubMed] [Google Scholar]
- Friedrich D, Marintchev A, Arthanari H. 2022. The metaphorical Swiss army knife: the multitude and diverse roles of HEAT domains in eukaryotic translation initiation. Nucleic Acids Res 50: 5424–5442. 10.1093/nar/gkac342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Cai G, Ning Y, Liu L, Yang J, Dong D, Fu B, Lu Y, Cui S, Chen X. 2012. DAP5 ameliorates cisplatin-induced apoptosis of renal tubular cells. Am J Nephrol 35: 456–465. 10.1159/000338302 [DOI] [PubMed] [Google Scholar]
- Gerashchenko MV, Peterfi Z, Yim SH, Gladyshev VN. 2021. Translation elongation rate varies among organs and decreases with age. Nucleic Acids Res 49: e9. 10.1093/nar/gkaa1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A-C, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Gene Dev 15: 2852–2864. 10.1101/gad.912401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer C, Altmann M, Lee HS, Blanc A, Deshmukh M, Woolford JL, Trachsel H, Sonenberg N. 1993. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol Cell Biol 13: 4860–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio ED, Preiss T, Hentze MW. 1999. Translation driven by an eIF4G core domain in vivo. EMBO J 18: 4865–4874. 10.1093/emboj/18.17.4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. 1983. New initiation factor activity required for globin mRNA translation. J Biol Chem 258: 5804–5810. [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N. 1995. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J 14: 5701–5709. 10.1002/j.1460-2075.1995.tb00257.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haizel SA, Bhardwaj U, Gonzalez RL, Mitra S, Goss DJ. 2020. 5′-UTR recruitment of the translation initiation factor eIF4GI or DAP5 drives cap-independent translation of a subset of human mRNAs. J Biol Chem 295: 11693–11706. 10.1074/jbc.ra120.013678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PJ, Ye X, Qiu Y, Zhang HM, Hemida MG, Wang F, Lim T, Gu A, Cho B, Kim H, et al. 2016. Cleavage of DAP5 by coxsackievirus B3 2A protease facilitates viral replication and enhances apoptosis by altering translation of IRES-containing genes. Cell Death Differ 23: 828–840. 10.1038/cdd.2015.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MN, Ozpolat B, Abdi F, Gu S, Legler A, Mawuenyega, KG, Tirado-Gomez M, Lopez-Berestein G, Chen X. 2004. Comparative proteomic analysis of all-trans-retinoic acid treatment reveals systematic posttranscriptional control mechanisms in acute promyelocytic leukemia. Blood 104: 1314–1323. 10.1182/blood-2004-01-0046 [DOI] [PubMed] [Google Scholar]
- Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC. 2006. mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J 25: 1659–1668. 10.1038/sj.emboj.7601047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, von der Haar T, Singh CR, Ii M, Li B, Hinnebusch AG, McCarthy JEG, Asano K. 2003. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol Cell Biol 23: 5431–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit S, Strumpf NL, Goldstaub D, Kimchi A. 2000. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol 20: 496–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A. 2002. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc Natl Acad Sci 99: 5400–5405. 10.1073/pnas.082102499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83: 779–812. 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. 2016. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352: 1413–1416. 10.1126/science.aad9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JJD, Wang M, Audas TE, Kwon D, Carlsson SK, Timpano S, Evagelou SL, Brothers S, Gonzalgo ML, Krieger JR, et al. 2016. Systemic reprogramming of translation efficiencies on oxygen stimulus. Cell Rep 14: 1293–1300. 10.1016/j.celrep.2016.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. 2018. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20: 285–295. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundsdoerfer P, Thoma C, Hentze MW. 2005. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc Natl Acad Sci 102: 13421–13426. 10.1073/pnas.0506536102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Sonenberg N. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol 17: 6940–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H, Olsen HS, Sonenberg N. 1997. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J 16: 817–825. 10.1093/emboj/16.4.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhak DN, Tyanova S, Cox J, Borner GH. 2016. Global, quantitative and dynamic mapping of protein subcellular localization. Elife 5: e16950. 10.7554/elife.16950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF. 2010. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci 107: 18056–18060. 10.1073/pnas.1009269107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, Anderson P. 2018. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol 11: a032813. 10.1101/cshperspect.a032813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ. 2013. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol 5: a011569. 10.1101/cshperspect.a011569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. 2016. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar AN, MacGibeny MA, Gervasi NM, Gioio AE, Kaplan BB. 2013. Intra-axonal synthesis of eukaryotic translation initiation factors regulates local protein synthesis and axon growth in rat sympathetic neurons. J Neurosci 33: 7165–7174. 10.1523/jneurosci.2040-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Goldman DH, Choi J, Nwaezeapu C, Liang D, Green KM, Goldstrohm AC, Todd PK, Green R, Wilusz JE. 2019. Ribosome queuing enables non-AUG translation to be resistant to multiple protein synthesis inhibitors. Gene Dev 33: 871–885. 10.1101/gad.324715.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Fabian JR, Pavitt GD, Hinnebusch AG, Jefferson LS. 1998. Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2α. J Biol Chem 273: 12841–12845. 10.1074/jbc.273.21.12841 [DOI] [PubMed] [Google Scholar]
- Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 2001. New ways of initiating translation in eukaryotes? Mol Cell Biol 21: 1899–1907. 10.1128/mcb.21.6.1899-1907.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratzat H, Mackens-Kiani T, Ameismeier M, Potocnjak M, Cheng J, Dacheux E, Namane A, Berninghausen O, Herzog F, Fromont-Racine M, et al. 2021. A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes. EMBO J 40: e105179. 10.15252/embj.2020105179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases implications for cap-dependent and cap-independent translational initiation (∗). J Biol Chem 270: 21975–21983. 10.1074/jbc.270.37.21975 [DOI] [PubMed] [Google Scholar]
- Lee SH, McCormick F. 2006. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J 25: 4008–4019. 10.1038/sj.emboj.7601268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-Y, Cevallos RC, Jan E. 2009. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2α phosphorylation. J Biol Chem 284: 6661–6673. 10.1074/jbc.m806735200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Kranzusch PJ, Cate JHD. 2015. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522: 111–114. 10.1038/nature14267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Doudna JA, Cate JHD. 2016. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536: 96–99. 10.1038/nature18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JWB, Rhoads RE. 2006. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 281: 22917–22932. 10.1074/jbc.m605418200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Strumpf N, Deiss LP, Berissi H, Kimchi A. 1997. DAP-5, a novel homolog of eukaryotic translation initiation factor 4G isolated as a putative modulator of gamma interferon-induced programmed cell death. Mol Cell Biol 17: 1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM, Cerquozzi S, Graber TE, Ungureanu NH, Andrews M, Holcík M. 2008. The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Res 36: 168–178. 10.1093/nar/gkm1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman N, Dym O, Unger T, Albeck S, Peleg Y, Jacobovitch Y, Branzburg A, Eisenstein M, Marash L, Kimchi A. 2008. The crystal structure of the C-terminal DAP5/p97 domain sheds light on the molecular basis for its processing by caspase cleavage. J Mol Biol 383: 539–548. 10.1016/j.jmb.2008.08.013 [DOI] [PubMed] [Google Scholar]
- Liberman N, Marash L, Kimchi A. 2009. The translation initiation factor DAP5 is a regulator of cell survival during mitosis. Cell Cycle 8: 204–209. [DOI] [PubMed] [Google Scholar]
- Liberman N, Gandin V, Svitkin YV, David M, Virgili G, Jaramillo M, Holcík M, Nagar B, Kimchi A, Sonenberg N. 2015. DAP5 associates with eIF2β and eIF4AI to promote internal ribosome entry site driven translation. Nucleic Acids Res 43: 3764–3775. 10.1093/nar/gkv205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP. 2012. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40: 2898–2906. 10.1093/nar/gkr1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167: 27–33. 10.1083/jcb.200408003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus MA, Akabayov B, Milbradt AG, Luna LE, Seo H-C, et al. 2012. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Rep 1: 689–702. 10.1016/j.celrep.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailliot J, Martin F. 2018. Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdiscip Rev RNA 9. 10.1002/wrna.1458 [DOI] [PubMed] [Google Scholar]
- Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, Kimchi A. 2008. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell 30: 447–459. 10.1016/j.molcel.2008.03.018 [DOI] [PubMed] [Google Scholar]
- Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu A-B, Imataka H, Gehring K, Sonenberg N. 2008. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol 28: 6658–6667. 10.1128/mcb.00738-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mašek T, Valášek L, Pospíšek M. 2011. Polysome analysis and RNA purification from sucrose gradients. Methods Mol Biology Clifton N J 703: 293–309. 10.1007/978-1-59745-248-9_20 [DOI] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S-B, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras M, Miller WA, Truniger V, Aranda MA. 2017. Non-canonical translation in plant RNA viruses. Front Plant Sci 8: 494. 10.3389/fpls.2017.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen RD, Serra M, Steitz JA, Vasudevan S. 2011. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA–protein complexes (microRNPs). Proc Natl Acad Sci 108: 8281–8286. 10.1073/pnas.1105401108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S, Mann M. 2011. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol 7: 548. 10.1038/msb.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins TA, Harder ZM, Korneluk RG, Holcík M. 2003. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J Biol Chem 278: 3572–3579. 10.1074/jbc.m206781200 [DOI] [PubMed] [Google Scholar]
- Noderer WL, Flockhart RJ, Bhaduri A, de Arce AJD, Zhang J, Khavari PA, Wang CL. 2014. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol 10: 748. 10.15252/msb.20145136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M, Reed V, Bryson-Richardson RJ, Currie PD, Preiss T. 2007. The eIF4G-homolog p97 can activate translation independent of caspase cleavage. RNA 13: 374–384. 10.1261/rna.372307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozpolat B, Akar U, Steiner M, Zorrilla-Calancha I, Tirado-Gomez M, Colburn N, Danilenko M, Kornblau S, Berestein GL. 2007. Programmed cell death-4 tumor suppressor protein contributes to retinoic acid-induced terminal granulocytic differentiation of human myeloid leukemia cells. Mol Cancer Res 5: 95–108. 10.1158/1541-7786.mcr-06-0125 [DOI] [PubMed] [Google Scholar]
- Ozpolat B, Akar U, Zorrilla-Calancha I, Vivas-Mejia P, Acevedo-Alvarez M, Lopez-Berestein G. 2008. Death-associated protein 5 (DAP5/p97/NAT1) contributes to retinoic acid-induced granulocytic differentiation and arsenic trioxide-induced apoptosis in acute promyelocytic leukemia. Apoptosis 13: 915–928. 10.1007/s10495-008-0222-9 [DOI] [PubMed] [Google Scholar]
- Park E-H, Zhang F, Warringer J, Sunnerhagen P, Hinnebusch AG. 2011. Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics 12: 68. 10.1186/1471-2164-12-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. 10.1038/334320a0 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci 98: 7029–7036. 10.1073/pnas.111145798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S, Imataka H, Gingras A, Fukunaga R, Hunter T, Sonenberg N. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J 18: 270–279. 10.1093/emboj/18.1.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido JB, Sokabe M, Kraatz S, Gordiyenko Y, Skehel JM, Fraser CS, Ramakrishnan V. 2020. Structure of a human 48S translational initiation complex. Science 369: 1220–1227. 10.1126/science.aba4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. 2008. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol 181: 293–307. 10.1083/jcb.200710215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson M, Hong AV, Massourides E, Bourg N, Suel L, Amor F, Corre G, Bénit P, Barthélémy I, Blot S, et al. 2020. miR-379 links glucocorticoid treatment with mitochondrial response in Duchenne muscular dystrophy. Sci Rep 10: 9139. 10.1038/s41598-020-66016-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Sehrawat U, Haimov O, Weiss B, Harush AT-B, Ashkenazi S, Plotnikov A, Noiman T, Leshkowitz D, Stelzer G, Dikstein R. 2022. Inhibitors of eIF4G1–eIF1 uncover its regulatory role of ER/UPR stress-response genes independent of eIF2α-phosphorylation. Proc National Acad Sci 119: e2120339119. 10.1073/pnas.2120339119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J-Y, Jung Y, Kim D-Y, Ryu HG, Lee J, Kim SW, Kim K-T. 2019. DAP5 increases axonal outgrowth of hippocampal neurons by enhancing the cap-independent translation of DSCR1.4 mRNA. Cell Death Dis 10: 49. 10.1038/s41419-018-1299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaughnessy JD, Jenkins NA, Copeland NG. 1997. cDNA cloning, expression analysis, and chromosomal localization of a gene with high homology to wheat eIF-(iso)4F and mammalian eIF-4G. Genomics 39: 192–197. 10.1006/geno.1996.4502 [DOI] [PubMed] [Google Scholar]
- Shen L, Pelletier J. 2020. General and target-specific DExD/H RNA helicases in eukaryotic translation initiation. Int J Mol Sci 21: 4402. 10.3390/ijms21124402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokikh NE, Preiss T. 2018. Translation initiation by cap-dependent ribosome recruitment: recent insights and open questions. Wiley Interdiscip Rev RNA 9: e1473. 10.1002/wrna.1473 [DOI] [PubMed] [Google Scholar]
- Shirokikh NE, Dutikova YS, Staroverova MA, Hannan RD, Preiss T. 2019. Migration of small ribosomal subunits on the 5′ untranslated regions of capped messenger RNA. Int J Mol Sci 20: 4464. 10.3390/ijms20184464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Brito Querido J, Myasnikov AG, Mancera-Martinez E, Renaud A, Kuhn L, Hashem Y. 2016. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol Cell 63: 206–217. 10.1016/j.molcel.2016.05.033 [DOI] [PubMed] [Google Scholar]
- Singh CR, Watanabe R, Chowdhury W, Hiraishi H, Murai MJ, Yamamoto Y, Miles D, Ikeda Y, Asano M, Asano K. 2012. Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S preinitiation complex and promotes its shift to the initiation mode. Mol Cell Biol 32: 3978–3989. 10.1128/mcb.00376-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. 2015. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab 21: 479–492. 10.1016/j.cmet.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Smirnova VV, Shestakova ED, Bikmetov DV, Chugunova AA, Osterman IA, Serebryakova MV, Sergeeva OV, Zatsepin TS, Shatsky IN, Terenin IM. 2019. eIF4G2 balances its own mRNA translation via a PCBP2-based feedback loop. RNA 25: 757–767. 10.1261/rna.065623.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova VV, Shestakova ED, Nogina DS, Mishchenko PA, Prikazchikova TA, Zatsepin TS, Kulakovskiy IV, Shatsky IN, Terenin IM. 2022. Ribosomal leaky scanning through a translated uORF requires eIF4G2. Nucleic Acids Res 50: 1111–1127. 10.1093/nar/gkab1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin II, Vassilenko KS, Terenin IM, Kalinina NO, Agol VI, Dmitriev SE. 2021. Non-canonical translation initiation mechanisms employed by eukaryotic viral mRNAs. Biochem Biokhimiia 86: 1060–1094. 10.1134/s0006297921090042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spijker HM, Stackpole EE, Almeida S, Katsara O, Liu B, Shen K, Schneider RJ, Gao F-B, Richter JD. 2021. Ribosome profiling reveals novel regulation of C9ORF72 GGGGCC repeat-containing RNA translation. RNA 28: 123–138. 10.1261/rna.078963.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Budak G, Dash S, Lachke SA, Janga SC. 2017. Transcriptome analysis of developing lens reveals abundance of novel transcripts and extensive splicing alterations. Sci Rep 7: 11572. 10.1038/s41598-017-10615-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama H, Takahashi K, Yamamoto T, Iwasaki M, Narita M, Nakamura M, Rand TA, Nakagawa M, Watanabe A, Yamanaka S. 2017. Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc Natl Acad Sci 114: 340–345. 10.1073/pnas.1617234114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Maruyama M, Tokuzawa Y, Murakami M, Oda Y, Yoshikane N, Makabe KW, Ichisaka T, Yamanaka S. 2005. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2). Genomics 85: 360–371. 10.1016/j.ygeno.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Tang L, Morris J, Wan J, Moore C, Fujita Y, Gillaspie S, Aube E, Nanda J, Marques M, Jangal M, et al. 2017. Competition between translation initiation factor eIF5 and its mimic protein 5MP determines non-AUG initiation rate genome-wide. Nucleic Acids Res 45: 11941–11953. 10.1093/nar/gkx808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. 2014. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev 28: 357–371. 10.1101/gad.231407.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Andreev DE, Dmitriev SE, Shatsky IN. 2013. A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res 41: 1807–1816. 10.1093/nar/gks1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Smirnova VV, Andreev DE, Dmitriev SE, Shatsky IN. 2017. A researcher's guide to the galaxy of IRESs. Cell Mol Life Sci 74: 1431–1455. 10.1007/s00018-016-2409-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoteo GD, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, Incarnato D, Oliviero S, Fatica A, Morlando M, et al. 2020. Modulation of circRNA metabolism by m6A modification. Cell Rep 31: 107641. 10.1016/j.celrep.2020.107641 [DOI] [PubMed] [Google Scholar]
- Tong J, Lu J, Mao X, Zhu Z, Wang Y, Lou M, Zhang K. 2022. Circular RNA-UBE2D2 accelerates the proliferation and metastasis of non-small cell lung cancer cells via modulating microRNA-376a-3p/Eukaryotic Translation Initiation Factor 4γ2 axis. Bioengineered 13: 5942–5953. 10.1080/21655979.2022.2027068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilenko KS, Alekhina OM, Dmitriev SE, Shatsky IN, Spirin AS. 2011. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res 39: 5555–5567. 10.1093/nar/gkr147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science 318: 1931–1934. 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci 101: 11269–11274. 10.1073/pnas.0400541101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, Kieft JS, Rissland OS. 2018. Revisiting the closed-loop model and the nature of mRNA 5′–3′ communication. Mol Cell 72: 805–812. 10.1016/j.molcel.2018.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa N, Do A, Hershey JWB, Fraser CS. 2013. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem 288: 32932–32940. 10.1074/jbc.m113.517011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgili G, Frank F, Feoktistova K, Sawicki M, Sonenberg N, Fraser CS, Nagar B. 2013. Structural analysis of the DAP5 MIF4G domain and its interaction with eIF4A. Structure 21: 517–527. 10.1016/j.str.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volta V, Pérez-Baos S, de la Parra C, Katsara O, Ernlund A, Dornbaum S, Schneider RJ. 2021. A DAP5/eIF3d alternate mRNA translation mechanism promotes differentiation and immune suppression by human regulatory T cells. Nat Commun 12: 6979. 10.1038/s41467-021-27087-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss MRH, van Diest PJ, Raman V. 2017. Targeting RNA helicases in cancer: the translation trap. Biochim Biophys Acta 1868: 510–520. 10.1016/j.bbcan.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shin B-S, Alvarado C, Kim J-R, Bohlen J, Dever TE, Puglisi JD. 2022. Rapid 40S scanning and its regulation by mRNA structure during eukaryotic translation initiation. Cell 185: 4474–4487.e17. 10.1016/j.cell.2022.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnakulasuriyarachchi D, Cerquozzi S, Cheung HH, Holcík M. 2004. Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J Biol Chem 279: 17148–17157. 10.1074/jbc.m308737200 [DOI] [PubMed] [Google Scholar]
- Weber R, Kleemann L, Hirschberg I, Chung M-Y, Valkov E, Igreja C. 2022. DAP5 enables main ORF translation on mRNAs with structured and uORF-containing 5′ leaders. Nat Commun 13: 7510. 10.1038/s41467-022-35019-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Khan D, Liberman N, Yoffe Y, Bialik S, Das S, Oren M, Kimchi A. 2014. The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 33: 611–618. 10.1038/onc.2012.626 [DOI] [PubMed] [Google Scholar]
- Wu R, Li A, Sun B, Sun J-G, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q, et al. 2019. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res 29: 23–41. 10.1038/s41422-018-0113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Huangyang P, Wang Y, Xue L, Devericks E, Nguyen HG, Yu X, Oses-Prieto JA, Burlingame AL, Miglani S, et al. 2021. ERα is an RNA-binding protein sustaining tumor cell survival and drug resistance. Cell 184: 5215–5229.e17. 10.1016/j.cell.2021.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Unbehaun A, Spahn CMT. 2017. Ribosomal chamber music: toward an understanding of IRES mechanisms. Trends Biochem Sci 42: 655–668. 10.1016/j.tibs.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Poksay KS, Arnold KS, Innerarity TL. 1997. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev 11: 321–333. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Zhang XY, Maeda M, Miura K, Wang S, Farese RV, Iwao H, Innerarity TL. 2000. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J 19: 5533–5541. 10.1093/emboj/19.20.5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen L-L, Wang Y, et al. 2017. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27: 626–641. 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoffe Y, David M, Kalaora R, Povodovski L, Friedlander G, Feldmesser E, Ainbinder E, Saada A, Bialik S, Kimchi A. 2016. Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev 30: 1991–2004. 10.1101/gad.285239.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikane N, Nakamura N, Ueda R, Ueno N, Yamanaka S, Nakamura M. 2007. Drosophila NAT1, a homolog of the vertebrate translational regulator NAT1/DAP5/p97, is required for embryonic germband extension and metamorphosis. Dev Growth Differ 49: 623–634. 10.1111/j.1440-169x.2007.00956.x [DOI] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. 2008. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem 283: 7064–7073. 10.1074/jbc.m708530200 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian S-B. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]