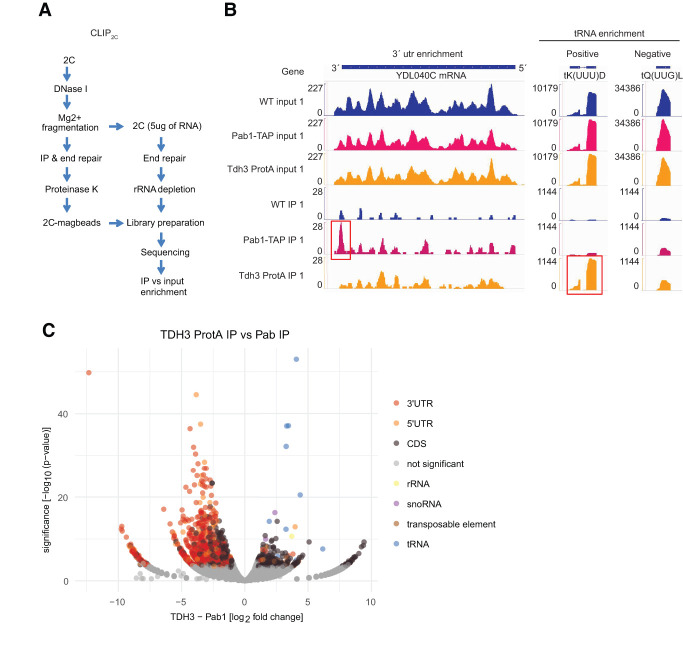
FIGURE 2.
CLIP2C identifies several tRNAs as specific GAPDH biding partners. (A) Schematic representation of CLIP2C workflow. (B) Read coverage along YDL040C, tK(UUU)D, and tQ(UUG)L genes after CLIP2C of Pab1-TAP and Tdh3-Protein A. An untagged WT strain was used as negative control. Scale of the y-axis for input and IPs samples were normalized to the highest value. Red boxes highlight statistically significant RNA target regions, the 3′UTR region of YDL040C for Pab1-TAP and tK(UUU)D for Tdh3. The binding of Tdh3 to tK(UUU)D was specific as alternative tRNA genes, like tQ(UUG)L, were not statistically enriched after CLIP2C. (C) Volcano plot displaying the log2 fold change in normalized read counts versus −log10 P-value after CLIP2C of Tdh3-Protein A and Pab1-TAP. Statistical significance was defined as log2FC ≥ 1 and P-value ≤ 0.05.