To the Editor: Hearing loss is the most common sensory disorder in humans. There is one case of congenital deafness among every 1000 newborns, and in 50% of cases, the deafness is hereditary. Deafness exhibits high genetic heterogeneity. To date, over 110 non-syndromic deafness genes have been identified (https://hereditaryhearingloss.org/). Lots of those genes can cause both autosomal-dominant hearing loss (ADNSHL) and autosomal-recessive non-syndromic hearing loss (ARNSHL) and TMC1 (encoding the transmembrane channel-like 1) is one of them. TMC1 (OMIM: 606706) is a member of the TMC family located at 9q21.13. The protein contains 760 amino acids and has six transmembrane regions. TMC1 is expressed in the inner and outer hair cells of the cochlea. A TMC1 mutation was first shown to cause deafness in 2002.[1] The prevalence of TMC1 variants ranged from 3.4% (19/557) among Pakistani ARNSHL families to 8.1% (7/86) in Turkish families. To date, around 20 hearing loss families associated with TMC1 variants have been reported in China.
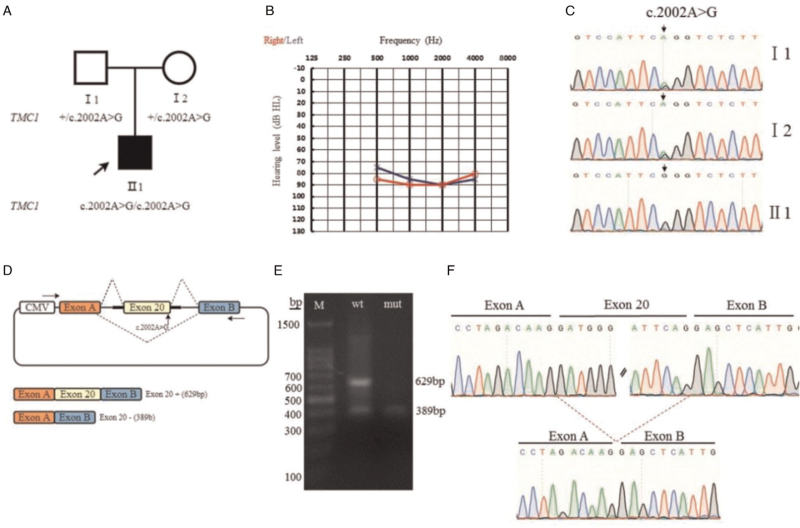
Here, we report a Chinese family with a boy affected by ARNSHL. The study complied with the Helsinki Declaration of the World Medical Association and was approved by the Medical Ethics Committee of the Second Affiliated Hospital of Zhengzhou University (No. 2018008). The subject's guardians volunteered to participate and signed an informed consent form. The proband was a 2-year-old boy with congenital deafness; his parents had normal phenotypes [Figure 1A]. We took a detailed medical history of the proband and performed audiological and radiological examinations. The parents were non-consanguineous and the proband had no history of noise exposure, ototoxic drug usage, external ear trauma, birth injury, hypoxia or asphyxia, or jaundice. No morphological changes in the inner ear or brain were detected by computed tomography (CT) of the temporal bone or magnetic resonance imaging. The auditory brainstem response (ABR) thresholds were 85 dBnHL (left ear) and 90 dBnHL (right ear), and the average auditory steady-state response (ASSR) binaural hearing thresholds were 75-85-90-85 dBnHL in the left ear and 85-90-90-80 dBnHL in the right ear [Figure 1B]. The distortion product otoacoustic emission test failed in both ears. The proband and his parents were subjected to genetic testing, i.e., targeted enrichment and high-throughput sequencing of 129 known deafness genes. We identified a novel variant c.2002A>G (NM_138691.3) in exon 20 of TMC1. The proband was homozygous, whereas the parents were heterozygous, for the variant. The variant was validated by Sanger sequencing of the proband and both parents [Figure 1C]. The primers [Supplementary Table 1] for the variant were designed by using the NCBIPrimer-BLAST software and synthesized by Shanghai Shangya Biotechnology Co., Ltd (Shanghai, China).
Figure 1.
The family pedigree, audiometric phenotypes, Sanger sequencing data, and minigene assay results. (A) The family pedigree. The arrow indicates the proband. Squares indicate males and circles females. White symbols indicate unaffected family members. (B) The audiogram of the proband. (C) Validation of candidate variants identified by Sanger sequencing. The proband is homozygous and the parents are heterozygous. The arrow indicates the variant site. (D) Schematic of minigene construction and the possible splicing modes. The arrow indicates the variant site. The minigene plasmid construct contained exon A, intron A, intron 19(197 bp), exon 20(240 bp), intron 20 (129 bp), intron B, and exon B (in that order). (E) The wild-type and mutant constructs were stably transfected into 293T cells and the exon 20 splicing pattern was analyzed via reverse transcription-polymerase chain reaction. The wild-type minigene (c.2002A) exhibited partial exon 20 inclusion, whereas the mutant minigene (c.2002A>G) showed almost complete skipping. M: size markers. (F) Sequencing of the splicing bands.
MaxEntScan predicted that the variant would affect splicing; the dbscSNV had ADA (based on adaptive boosting) and RF (based on random forest) scores >0.97. The minigene assay was used to evaluate the effect of the variant on splicing. The minigene plasmids used to test the variant in TMC1 exon 20 contained intron 19 (197 bp), exon 20 (240 bp), intron 20 (129 bp), and the universal sequence (exon A–intron A … intron B–exon B) [Figure 1D]. The wild-type and mutant recombinant plasmids were transiently transfected into the 293T cell line and total RNA was collected after 48 hours. The amplicon was sized by agarose electrophoresis and sequenced. The gel images had two bands of different sizes [Figure 1E]. The minigene splicing pattern of the normal construct revealed a major 629-bp fragment containing exon 20 and a minor 389-bp product lacking exon 20 [Figure 1E]. The gel image of the mutated construct displayed only a 389-bp transcript [Figure 1E]. The wild-type minigene exhibited partial exon 20-skipping, which could be caused by less-efficient splicing of minigene RNA. Importantly, however, the mutant minigene with the c.2002A>G variant showed complete (100%) exon-skipping, thus confirming the effect of the variant on exon skipping [Figure 1F]. The primer sequences are shown in Supplementary Table 2.
The variant is absent from the ExAC and gnomAD databases (ACMG PM2). The proband carries a TMC1 c.2002A>G homozygous variant, and both parents are carriers (ACMG PM3_supporting). In vitro functional tests showed that c.2002A>G affected splicing (ACMG PS3_moderate). TMC1 c.2002A>G (p.Ser668Gly) and c.2004T>G (p.Ser668Arg)[2] are two variants of the same amino acid, and the c.2004T>G variant is rated as “likely pathogenic” (ACMG PM5). Therefore, according to the ACMG genetic variation classification criteria and published guidelines,[3,4] the TMC1 c.2002A>G variant is likely pathogenic.
To date, sequence analysis of >15 ethnic groups worldwide has identified 120 TMC1 variants associated with NSHL (HGMD Professional 2020.4). Only 18 TMC1 variants associated with NSHL have been reported in China, of which 15 are missense/nonsense, 1 is a splicing variant, and 2 are small deletions.
DFNB7/11 deafness caused by TMC1 variants is often characterized by congenital or prelingual severe-to-profound deafness without abnormal vestibular function. All Chinese families reported on to date exhibited this clinical phenotype. However, De Heer et al.[5] reported a Dutch family with three affected individuals who had a homozygous splicing variant (c.1763+3A>G). The patient phenotypes were progressive and postlingual; the cause remains to be determined.
TMC1 variants can also cause ADNSHL (DFNA36). DFNA36 HL is often postlingual and initially affects high frequencies, gradually becoming severe-to-profound at all frequencies. However, DFNA36 families exhibit phenotypic variability in terms of the age of onset and rate of progression.[6]
To conclude, we report a novel variant of TMC1 in a Chinese family; we sequenced the 129 known deafness genes. Minigene assay confirmed that the variant caused exon skipping. Therefore, we conclude that the homozygous variant of the TMC1 gene was the cause of deafness in the proband; this further expands the mutational spectrum.
Acknowledgments
We sincerely thank our patient and his parents. We also thank the Supercomputing Center of Zhengzhou University for providing computational and storage resources.
Funding
The study was supported by a grant from the Collaborative Innovation Project of Zhengzhou (Zhengzhou University) (No. 18XTZX12004).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Zeng B, Xu H, Tian Y, Lin Q, Feng H, Zhang Z, Li S, Tang W. A novel splicing variant in the TMC1 gene causes non-syndromic hearing loss in a Chinese family. Chin Med J 2022;135:2631–2633. doi: 10.1097/CM9.0000000000001966
Beiping Zeng and Hongen Xu contributed equally to the work
Supplemental digital content is available for this article.
References
- 1.Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet 2002; 30:277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- 2.Santos RL, Wajid M, Khan MN, McArthur N, Pham TL, Bhatti A, et al. Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum Mutat 2005; 26:396.doi: 10.1002/humu.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oza AM, DiStefano MT, Hemphill SE, Cushman BJ, Grant AR, Siegert RK, et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat 2018; 39:1593–1613. doi: 10.1002/humu.23630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Heer AM, Collin RW, Huygen PL, Schraders M, Oostrik J, Rouwette M, et al. Progressive sensorineural hearing loss and normal vestibular function in a Dutch DFNB7/11 family with a novel mutation in TMC1. Audiol Neurootol 2011; 16:93–105. doi: 10.1159/000313282. [DOI] [PubMed] [Google Scholar]
- 6.Hilgert N, Monahan K, Kurima K, Li C, Friedman RA, Griffith AJ, et al. Amino acid 572 in TMC1: hot spot or critical functional residue for dominant mutations causing hearing impairment. J Hum Genet 2009; 54:188–190. doi: 10.1038/jhg.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.