Abstract
The design of efficient materials for intracellular protein delivery has attracted great interest in recent years; however, most current materials for this purpose are limited by poor serum stability due to the early release of cargoes triggered by abundant serum proteins. Here, we propose a light-activated crosslinking (LAC) strategy to prepare efficient polymers with excellent serum tolerance for intracellular protein delivery. A cationic dendrimer engineered with photoactivatable O-nitrobenzene moieties co-assembles with cargo proteins via ionic interactions, followed by light activation to yield aldehyde groups on the dendrimer and the formation of imine bonds with cargo proteins. The light-activated complexes show high stability in buffer and serum solutions, but dis-assemble under low pH conditions. As a result, the polymer successfully delivers cargo proteins green fluorescent protein and β-galactosidase into cells with maintained bioactivity even in the presence of 50% serum. The LAC strategy proposed in this study provides a new insight to improve the serum stability of polymers for intracellular protein delivery.
A light-activated crosslinking strategy was proposed to prepare a polymer with high robustness and serum stability for cytosol protein delivery. It can deliver proteins into cytosols with maintained bioactivities even in 50% FBS-containing medium.
Introduction
Protein therapeutics have attracted great interest in the pharmaceutical industry due to their advantages such as high potency and specificity, low adverse effects, limited immunogenicity, and faster approval compared to traditional small-molecule drugs.1 The sales of global therapeutic proteins have reached 98.1 billion dollars in 2021.2 However, protein therapeutics are hampered by two major limitations including susceptibility to enzymatic degradation and difficulty to cross biological barriers such as cell membranes.3–8 As a result, all current protein drugs have been developed based on extracellular targets.9,10
Several strategies have been proposed to achieve intracellular protein delivery.11–14 Physical methods such as electroporation and nanospearing that could transiently disrupt cell membranes are used to deliver proteins into cell cytosol,15 but these methods cannot be translated for in vivo protein delivery. Conjugation of cargo proteins with cell membrane permeable ligands is another widely used method for intracellular protein delivery.16–20 Proteins modified with cell penetrant peptides, cationic polymers, and amphiphilic molecules could be internalized by cells via endocytosis, but the method was associated with complicated synthesis and possibility of altered protein bioactivity after modification. Intracellular protein delivery using carriers such as nanoparticles and polymers is the most promising strategy in recent years.21–30 Cargo proteins were loaded by cell-permeable carriers via non-covalent interactions such as ionic and hydrophobic interactions for intracellular protein delivery.31–38 However, cargo proteins loaded by these carriers are easily released under physiological conditions due to competitive binding with abundant serum proteins. Strengthening the interactions with cargo proteins can improve the serum stability of carriers, but the delivery systems are usually associated with problems such as difficulty in intracellular protein release.39 It is highly desired to develop carriers that can deliver proteins inside cells in the presence of abundant serum proteins.40,41
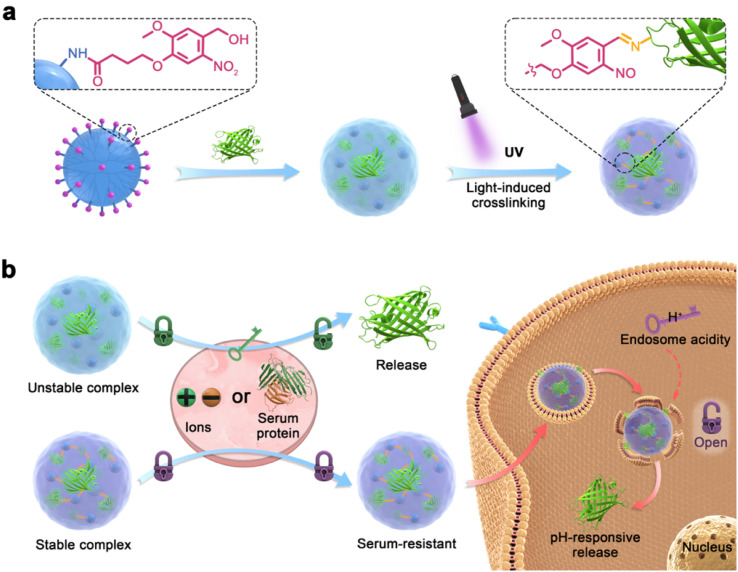
Here, we propose a light-activated crosslinking (LAC) strategy to prepare polymers with excellent serum tolerance for intracellular protein delivery (Fig. 1). O-Nitrobenzene (NB) and its derivatives were reported to be photo-activatable ligands that generate bioactive aldehyde groups under ultra-violet (UV) light irradiation,42–44 and the ligands were widely used for photo-activated ligation or bio-adhesion.45–50 The NB ligand (named NB-NHS) is modified on the surface of a cationic dendrimer to yield the protein-delivering material (abbreviated as ND, Fig. 1a). ND forms complexes with cargo proteins via non-covalent ionic and hydrophobic interactions, but the complexes are easily dis-assembled in the presence of salt ions and serum proteins due to competitive binding. After UV light (365 nm) irradiation, the ND complexes show strongly improved buffer and serum stabilities via the formation of dynamic covalent imine bonds between the polymer and cargo protein. After the complexes were endocytosed into cells, the imine bonds in the complexes will be cleaved by endolysosomal acidity, which triggers the release of bound proteins by ND (Fig. 1b). The study aims to provide a novel strategy to achieve efficient intracellular protein delivery in the presence of abundant serum proteins.
Fig. 1. The light-activated crosslinking (LAC) strategy to prepare polymers for intracellular protein delivery with serum stability. (a) Preparation of stable complexes by the LAC strategy. (b) Mechanism of the polymer during intracellular protein delivery.
Results and discussion
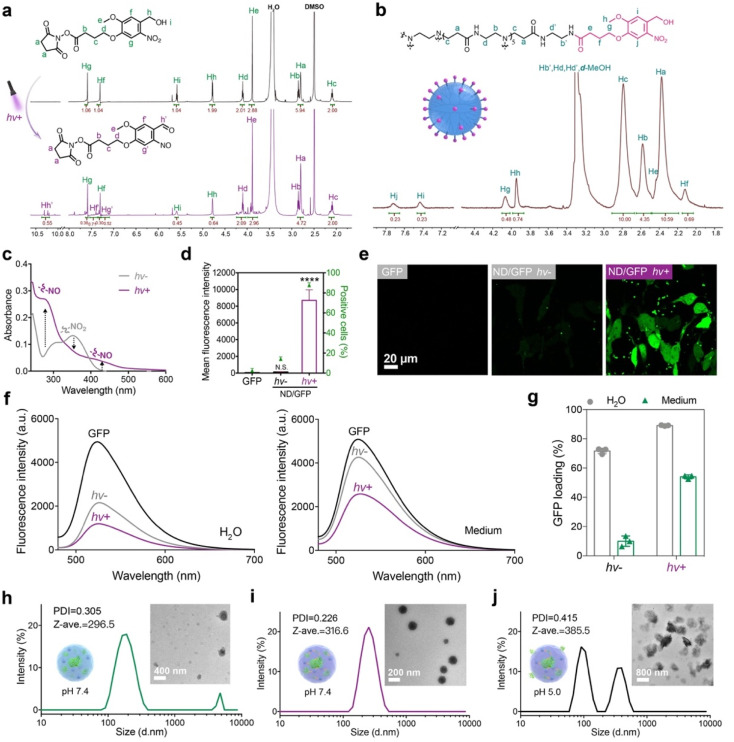
The photoreactive NB ligand was synthesized and characterized by 1H NMR (Fig. 2a), 13C NMR (Fig. S7†), and ESI-MS (Fig. S8†). After irradiation with 365 nm UV light at 10 mW cm−2 for 300 s, the proton signals Hg, Hf, Hh, and Hi on the NB ligand were greatly decreased, while new peaks Hg’, Hf’ and Hh’ appeared, suggesting that light irradiation activated the generation of aldehyde groups on NB-NHS. The NB ligand was then conjugated onto the surface of a generation 5 polyamidoamine dendrimer via amidation reactions, and the conjugate ND was characterized by 1H NMR (Fig. 2b). The average number of NB moieties modified on each dendrimer was measured to be about 11. The photoreactive properties of ND were further verified by UV-vis spectroscopy. After irradiation with UV light for 20 s, the absorbance peak for the NO2 group around 350 nm decreased, while peaks for the NO group at 280 nm and 430 nm greatly increased (Fig. 2c), which confirmed the photo-activatable properties of ND as proposed.
Fig. 2. Photo-activatable properties of the ND polymer in cytosolic protein delivery. (a) 1H NMR spectra of NB-NHS dissolved in DMSO-d6 before and after UV irradiation. (b) 1H NMR spectra of the ND polymer in methanol-d4. (c) UV-vis spectroscopy of the ND polymer before and after UV irradiation for 20 s. (d) Flow cytometry and (e) confocal images of 143B cells treated with ND/GFP complexes with or without UV irradiation. The concentrations of GFP and ND were 16 and 32 μg mL−1, respectively, and the cells were cultured for 24 h before measurement. Data are presented as mean ± standard deviation (s.d.) (n = 3). N.S.p > 0.05 and ****p < 0.0001, calculated by one-way ANOVA. (f) Fluorescence spectra and (g) GFP loading efficiency of the ND/GFP complexes before and after UV light activation in deionized (DI) water and culture media. The concentrations of GFP and ND were 4 and 8 μg mL−1, respectively. DLS and TEM images of ND/GFP complexes without light activation (h), ND/GFP complexes after light activation (i) and the sample of (i) at pH 5.0 (j) in culture media.
Green fluorescent protein (GFP) was first used as a model protein to evaluate the performance of the ND polymer in cytosolic protein delivery. ND failed to efficiently deliver GFP into 143B cells before LAC; however, the polymer showed significantly improved delivery efficacy when ND/GFP complexes were irradiated with 365 nm UV light at 10 mW cm−2 for 20 s (Fig. 2d and e). Further increase in irradiation time did not further increase the protein delivery efficacy (Fig. S9a and b†), indicating the fast transition of the benzyl alcohol moiety on NB into the benzyl aldehyde group after UV illumination. The irradiation time for LAC was fixed at 20 s for later studies. The yielding aldehyde groups on ND could react with lysine residues on GFP via the formation of dynamic covalent imine bonds, and improve the stability of ND/GFP complexes. We then measured the ND/GFP complexes in different solutions before and after LAC by fluorescence spectroscopy. In aqueous solution, ND interacts with GFP via ionic interactions and forms nanocomplexes in water, resulting in quenched GFP fluorescence. After light irradiation, the photoactivated aldehyde groups strengthened the interactions between ND and GFP via imine bonds, and thereby more efficient GFP fluorescence quenching was observed. In culture medium or PBS buffer, the ND/GFP complex is not stable due to the presence of highly abundant salt ions, but the complex still showed efficient fluorescence quenching after LAC (Fig. 2f and S9c†). The GFP loading efficiencies of the ND polymer were then quantitatively measured. As shown in Fig. 2g and S3d,† LAC treatment efficiently improved the GFP loading efficiency of ND, especially in culture medium and PBS buffer. The ND/GFP complexes before and after LAC were further measured by dynamic light scattering (DLS) and transmission electron microscopy (TEM). The results showed that LAC treatment facilitated the formation of uniform ND/GFP nanoparticles (Fig. 2h–j). The formed nanoparticles were unstable under pH 5.0, which is due to the acid-labile properties of imine bonds between ND and GFP. This behavior is beneficial for intracellular release of cargo proteins after localization in endo-lysosomes. These results together proved the beneficial role of LAC in improving the protein loading, complex stability, and intracellular delivery efficiency of ND/GFP complexes.
We further tested the possibility of the LAC-treated ND polymer in cytosolic protein delivery in the presence of serum proteins. As shown in Fig. 3a and b, the ND polymer after LAC efficiently delivered GFP into the cytosol of 143B cells in media containing 10–50% fetal bovine serum (FBS). The protein delivery efficacies of ND in media containing 10% and 20% FBS were scarcely changed compared to that in serum-free medium. Even in medium containing 50% FBS, the polymer still maintained relatively high delivery efficacy. In comparison, the commercial reagents for intracellular protein delivery such as PULSin and TransEx showed more than a 90% efficacy decrease in the presence of 10% FBS, suggesting the superior performance of the ND polymer under serum-containing conditions. We further tested the stability of the ND/GFP complex after LAC in serum-containing culture medium. As shown by confocal images in Fig. 3c, the LAC-treated complexes showed very similar structures and fluorescence signals in media containing 10–50% FBS compared to those in serum-free medium, suggesting that the bound GFP molecules in ND/GFP complexes were not replaced by the highly abundant serum proteins in media. TEM images also confirmed the excellent stability of LAC-treated ND/GFP nanoparticles in media containing FBS (Fig. S10a†). We further used fluorescence spectroscopy to reveal the stability of ND/GFP complexes in serum. As shown in Fig. 3d–e and S10b–c,† the fluorescence intensities of ND/GFP complexes without LAC were almost recovered when 50% FBS was added to DI water or culture medium, which is caused by GFP release from the complex due to competitive binding of serum proteins to ND. However, LAC treatment strongly improved the serum resistance of ND/GFP complexes as revealed by the slightly recovered GFP fluorescence intensity even in 50% FBS. In addition, the ND polymer after LAC treatment showed very similar GFP loading efficiencies in medium with or without FBS, while the commercial reagent TransEx showed greatly decreased protein loading in the presence of FBS (Fig. 3f). We next investigated the release responsiveness of cargo proteins from ND/GFP complexes via an agarose gel electrophoresis assay. As shown in Fig. 3g, LAC treated complexes showed minimal GFP release in the presence of 10% FBS at pH 7.4, while the released proteins were significantly increased when the pH was decreased to 6.0, suggesting pH-responsive release behavior of the delivery system. These results together confirmed the great potential of the LAC strategy in the improvement of protein delivery efficacy in serum-containing media and intracellular responsive release of cargo proteins.
Fig. 3. Serum stability and release responsiveness of the ND polymer in cytosolic protein delivery. Flow cytometry (a) and confocal images (b) of 143B cells treated with the ND/GFP complex after LAC in media containing different contents of FBS for 24 h (GFP, 16 μg mL−1; ND, 32 μg mL−1). Commercial reagents PULSin and TransEx were tested as controls. (c) Confocal images of ND/GFP complexes in culture medium containing different contents of FBS. Schematic illustration about fluorescence change (d) and fluorescence intensity (e) of ND/GFP complexes before and after LAC in culture medium containing 50% FBS. Free GFP was tested as a control. (f) GFP loading efficiency of ND after LAC in culture media containing different contents of FBS (GFP, 4 μg mL−1; ND, 8 μg mL−1). (g) Agarose gel electrophoresis of ND/GFP complexes under different conditions. All data are shown as mean ± s.d. (n = 3). N.S.p > 0.05, *p < 0.01, **p < 0.001, according to one-way ANOVA.
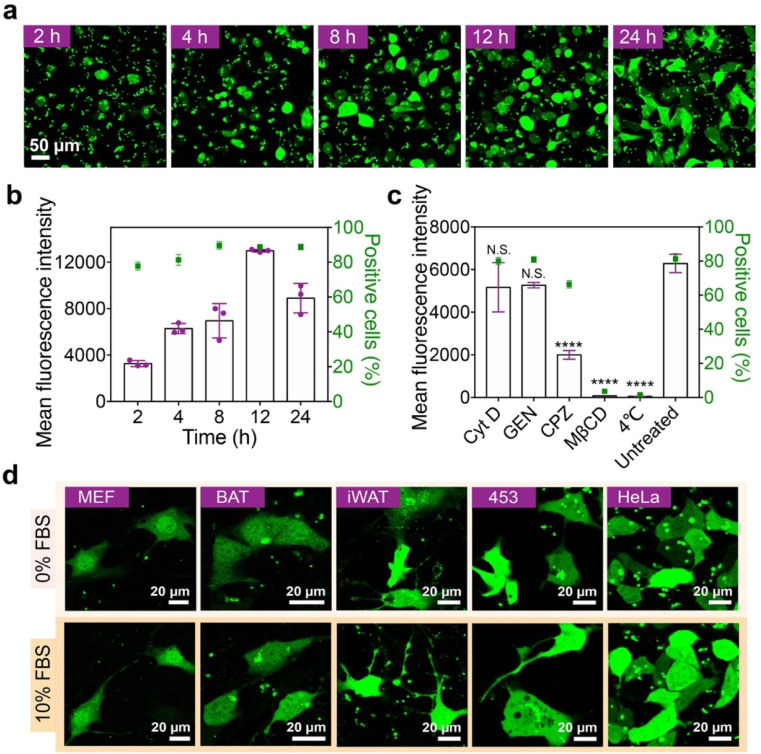
The effects of ND dose and incubation time on the GFP delivery efficacy were further investigated. The delivery efficacy of the ND polymer increases with increasing polymer dose, but a high polymer dose may lead to cytotoxicity in the treated cells (Fig. S11a–c†). According to the results, the ND polymer at 32 μg mL−1 with relatively high delivery efficacy and low cytotoxicity was chosen for later studies. The delivery efficacy of ND also increases along with the incubation time, and the highest efficacy is achieved at 12 h after incubation (Fig. 4a and b). Though the GFP fluorescence intensity of treated cells at 24 h was relatively lower, the delivered GFP molecules were more well distributed in the cells compared to those at 12 h. The internalization of ND/GFP complexes by 143B cells was efficiently inhibited at 4 °C or by pretreatment with methyl-β-cyclodextrin (MβCD) and chlorpromazine (CPZ) but scarcely affected by cytochalasin D (Cyt D) and genestein (GEN) (Fig. 4c), suggesting the involvement of lipid-raft and clathrin-dependent pathways during endocytosis of the nanoparticles. After treatment, the delivered ND/GFP nanoparticles were generally not co-localized with endolysosomes stained with Lysotracker red, suggesting efficient endosomal escape after 24 h incubation (Fig. S12†). Besides 143B cells, the LAC-treated ND polymer also efficiently delivered GFP into mouse embryonic fibroblast cells (MEF cells), mouse brown adipose tissue cells (BAT cells), mouse inguinal white adipose tissue cells (iWAT cells), human breast cancer cells (MDA-MB-453 cells), and HeLa cells in the presence of 10% serum (Fig. 4d), confirming the robustness of this material in cytosolic protein delivery.
Fig. 4. Behaviors of the ND polymer during intracellular GFP delivery. (a) Confocal images and (b) mean fluorescence intensity of 143B cells treated with ND/GFP complexes after LAC for 2–24 h. (c) Fluorescence intensity of 143B cells treated with ND/GFP complexes after LAC for 4 h at 37 °C. The cells were pre-treated with different endocytosis inhibitors before incubation, and the cells without inhibitor treatment (untreated) at 37 °C were tested as control. Data are shown as mean ± s.d. (n = 3). N.S.p > 0.05, ****p < 0.0001, one-way ANOVA. (d) Confocal images of different cells treated with ND/GFP complexes after LAC for 24 h in serum-free medium and 10% FBS-containing medium, respectively.
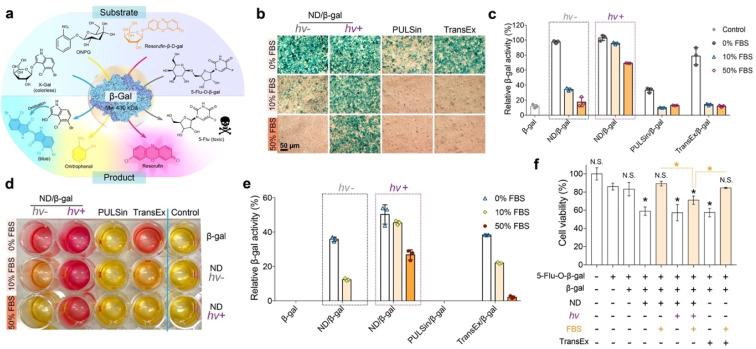
β-Galactosidase (β-gal) is capable of hydrolyzing the galactose residues from different substrates (Fig. 5a). As shown in Fig. S13,† the in vitro enzymatic activity of β-gal that complexed with the ND carrier after LAC would be partially inhibited in fresh medium (pH 7.4 or 6.0) or serum-containing medium (pH 7.4), while fully recovered in acidic serum-containing medium (pH 6.0). Hence, we presume that the cargo proteins can be released from the ND carrier and remain active after entering into cells. The efficacy of the ND polymer in the delivery of bioactive β-gal into 143B cells was next evaluated. The cells were first treated with ND/β-gal complexes with or without LAC for 24 h, and then with the substrates to analyze the delivery efficacy. First, a colorless substrate 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal) was used to stain the treated cells. As shown in Fig. 5b, the cells treated with ND/β-gal complexes (with or without LAC) and TransEx/β-gal complexes in serum-free media all showed obvious blue-colored products after X-gal staining; however, the number of blue-colored products was greatly decreased for ND/β-gal complexes without LAC and TransEx/β-gal complexes in serum-containing media. Similarly, the quantitative analysis via another substrate O-nitrophenyl-β-d-galactopyranoside (ONPG) showed superior β-gal delivery of the ND/β-gal complexes with LAC (Fig. 5c). Resorufin-β-d-galactopyranoside (resorufin-β-d-gal) can be hydrolyzed from non-fluorescent orange material into fluorescent red product by β-gal. After ordered treatment of the ND/β-gal complexes with LAC and resorufin-β-d-gal, the cells cultured with 0–50% serum were efficiently stained into red fluorescence (Fig. 5d). The relative quantitative result of resorufin-β-d-gal assay also demonstrated the maintained high β-gal activity for the ND/β-gal complexes with LAC in serum-containing media (Fig. 5e). 5-Fluorouridine-5′-O-β-d-galactopyranoside (5-flu-O-β-gal) is a low toxic compound, but it generates an anticancer drug 5-fluorouridine in the presence of β-gal. 5-Flu-O-β-gal was further used as the substrate to evaluate the amount of delivered β-gal into 143B cells. As shown in Fig. 5f, the cells treated with all the complexes in a serum-free medium showed obvious cytotoxicity after the addition of 5-flu-O-β-gal. However, the activities of ND/β-gal without LAC and TransEx/β-gal complexes were much decreased in serum-containing medium, while the activity for ND/β-gal complexes with LAC was maintained. These results together proved the important role of LAC treatment in improving the serum resistance of ND during intracellular protein delivery.
Fig. 5. Bioactive protein delivery with high serum-resistant performance. (a) Mechanisms of substrates catalyzed into products by β-gal. (b) Photographs of β-gal transfection efficacy through X-gal staining assay in 143B cells. (c) ONPG assay for determining β-gal activity after intracellular delivery by ND polymers. Cell lysate that added the same amount of β-gal was tested as 100% enzyme activity. (d) β-Gal enzyme activity of 143B cells, which were stained with resorufin-β-d-gal. (e) Quantitative analysis of resorufin-β-d-gal staining assay via a standard curve of different β-gal concentrations. (f) Viability of 143B cells treated with ND/β-gal complexes for 24 h, followed by 5-flu-O-β-gal treatment for another 24 h. Data are shown as mean ± s.d. (n = 3). N.S.p > 0.05 and *p < 0.05 were calculated by Student's t-test. Free β-gal was a negative control. PULSin and TransEx were used as positive controls.
Conclusions
In summary, we report a light-activated crosslinking strategy to achieve efficient intracellular protein delivery in serum-containing medium. The cationic polymer engineered with photoactivatable O-nitrobenzene moieties binds cargo proteins via ionic interactions to form nanoparticles. After light irradiation, the photoactivated aldehyde groups on the polymer further tag with amines on proteins via imine bonds to improve the complex stability. As a result, the polymer after light-activated crosslinking showed high efficacy in the delivery of cargo proteins into the cytosol of cells with maintained bioactivity. This study provides a versatile strategy to develop efficient polymers with high serum tolerance for intracellular protein delivery.
Data availability
The datasets supporting this article have been uploaded as part of the ESI.†
Author contributions
L. R. prepared and characterized the polymer, conducted the protein delivery experiments, and wrote the manuscript. L. J. synthesized and characterized the light-activatable NB ligand. Q. R. and J. L. performed part of the experiments on bioactive protein delivery and analyzed the mechanism and data. L. Z. and Y. C. designed and supervised the study, and wrote the manuscript.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (22135002) and the Basic Research Program of Science and Technology Commission of Shanghai Municipality (21JC1401800), and the National Key R&D Program of China, Synthetic Biology Research (No. 2019YFA0904500).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc05848k
References
- Leader B. Baca Q. J. Golan D. E. Nat. Rev. Drug Discovery. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- Sánchez-Trasviña C. Flores-Gatica M. Enriquez-Ochoa D. Rito-Palomares M. Mayolo-Deloisa K. Front. Bioeng. Biotechnol. 2021;9:717326. doi: 10.3389/fbioe.2021.717326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiper M. Niederreither K. Zuber G. Adv. Healthcare Mater. 2018;7:1701040. doi: 10.1002/adhm.201701040. [DOI] [PubMed] [Google Scholar]
- Luther D. C. Jeon T. Goswami R. Nagaraj H. Kim D. Lee Y.-W. Rotello V. M. Bioconjugate Chem. 2021;32:891–896. doi: 10.1021/acs.bioconjchem.1c00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X. Yu C. Wei J. Li L. Zhang C. Wu Q. Liu J. Yao S. Q. Huang W. Adv. Mater. 2019;31:1902791. doi: 10.1002/adma.201902791. [DOI] [PubMed] [Google Scholar]
- Lv J. Fan Q. Wang H. Cheng Y. Biomaterials. 2019;218:119358. doi: 10.1016/j.biomaterials.2019.119358. [DOI] [PubMed] [Google Scholar]
- Liu X. Wu F. Ji Y. Yin L. Bioconjugate Chem. 2019;30:305–324. doi: 10.1021/acs.bioconjchem.8b00750. [DOI] [PubMed] [Google Scholar]
- Xu J. Lv J. Zhuang Q. Yang Z. Cao Z. Xu L. Pei P. Wang C. Wu H. Dong Z. Chao Y. Wang C. Yang K. Peng R. Cheng Y. Liu Z. Nat. Nanotechnol. 2020;15:1043–1052. doi: 10.1038/s41565-020-00781-4. [DOI] [PubMed] [Google Scholar]
- Caravella J. Lugovskoy A. Curr. Opin. Chem. Biol. 2010;14:520–528. doi: 10.1016/j.cbpa.2010.06.175. [DOI] [PubMed] [Google Scholar]
- Mo R. Jiang T. Di J. Tai W. Gu Z. Chem. Soc. Rev. 2014;43:3595. doi: 10.1039/C3CS60436E. [DOI] [PubMed] [Google Scholar]
- Tai W. Zhao P. Gao X. Sci. Adv. 2020;6:eabb0310. doi: 10.1126/sciadv.abb0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. Li Z. Fan Q. Lv J. Li Y. Cheng Y. Adv. Mater. 2021:2104355. doi: 10.1002/adma.202104355. [DOI] [PubMed] [Google Scholar]
- Cheng Y. Chin. J. Chem. 2021;39:1443–1449. doi: 10.1002/cjoc.202000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J. Cheng Y. Chem. Soc. Rev. 2021;50:5435–5467. doi: 10.1039/D0CS00258E. [DOI] [PubMed] [Google Scholar]
- Cao Y. Ma E. Cestellos-Blanco S. Zhang B. Qiu R. Su Y. Doudna J. A. Yang P. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7899. doi: 10.1073/pnas.1818553116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J. Yu C. Li L. Yao S. Q. J. Am. Chem. Soc. 2015;137:12153–12160. doi: 10.1021/jacs.5b08130. [DOI] [PubMed] [Google Scholar]
- Qian L. Fu J. Yuan P. Du S. Huang W. Li L. Yao S. Q. Angew. Chem., Int. Ed. 2018;57:1532–1536. doi: 10.1002/anie.201711651. [DOI] [PubMed] [Google Scholar]
- Maier K. Wagner E. J. Am. Chem. Soc. 2012;134:10169–10173. doi: 10.1021/ja302705v. [DOI] [PubMed] [Google Scholar]
- Ng D. Y. W. Arzt M. Wu Y. Kuan S. L. Lamla M. Weil T. Angew. Chem., Int. Ed. 2014;53:324–328. doi: 10.1002/anie.201308533. [DOI] [PubMed] [Google Scholar]
- Mix K. A. Lomax J. E. Raines R. T. J. Am. Chem. Soc. 2017;139:14396–14398. doi: 10.1021/jacs.7b06597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzmann J. A. Luther D. C. Evans C. W. Jeon T. Jerome W. Gopalakrishnan S. Lee Y.-W. Norret M. Iyer K. S. Rotello V. M. J. Am. Chem. Soc. 2021;143:4758–4765. doi: 10.1021/jacs.1c00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-W. Luther D. C. Goswami R. Jeon T. Clark V. Elia J. Gopalakrishnan S. Rotello V. M. J. Am. Chem. Soc. 2020;142:4349–4355. doi: 10.1021/jacs.9b12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta K. Hu D. Zhao B. Ribbe A. E. Zhuang J. Thayumanavan S. J. Am. Chem. Soc. 2017;139:5676–5679. doi: 10.1021/jacs.7b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B. Ianosi-Irimie M. Thayumanavan S. ACS Nano. 2019;13:9408–9420. doi: 10.1021/acsnano.9b04198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. Zhang J. Meng F. Zhong Z. ACS Nano. 2018;12:11070–11079. doi: 10.1021/acsnano.8b05265. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Yang W. Zhang J. Meng F. Zhong Z. Adv. Mater. 2018;30:1800316. doi: 10.1002/adma.201800316. [DOI] [PubMed] [Google Scholar]
- Li G. Yuan S. Deng D. Ou T. Li Y. Sun R. Lei Q. Wang X. Shen W. Cheng Y. Liu Z. Wu S. Adv. Funct. Mater. 2019;29:1901932. doi: 10.1002/adfm.201901932. [DOI] [Google Scholar]
- Lv J. Wang H. Rong G. Cheng Y. Acc. Chem. Res. 2022;55:722–733. doi: 10.1021/acs.accounts.1c00766. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Gao X. Li Y. Lv J. Wang H. Cheng Y. CCS Chem. 2022 doi: 10.31635/ccschem.022.202202098. [DOI] [Google Scholar]
- Ren L. Gao Y. Cheng Y. Bioact. Mater. 2022;9:44–53. doi: 10.1016/j.bioactmat.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R. Wang M. Ray M. Jiang Y. Jiang Z. Xu Q. Rotello V. M. J. Am. Chem. Soc. 2017;139:8547–8551. doi: 10.1021/jacs.7b02801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mout R. Ray M. Yesilbag Tonga G. Lee Y.-W. Tay T. Sasaki K. Rotello V. M. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey N. D. Hango C. R. Minter L. M. Tew G. N. Bioconjugate Chem. 2018;29:2679–2690. doi: 10.1021/acs.bioconjchem.8b00363. [DOI] [PubMed] [Google Scholar]
- Sgolastra F. Backlund C. M. Ilker Ozay E. deRonde B. M. Minter L. M. Tew G. N. J. Controlled Release. 2017;254:131–136. doi: 10.1016/j.jconrel.2017.03.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Zhao Z. Wu F. Chen Y. Yin L. Adv. Mater. 2022;34:2108116. doi: 10.1002/adma.202108116. [DOI] [PubMed] [Google Scholar]
- Liu C. Wan T. Wang H. Zhang S. Ping Y. Cheng Y. Sci. Adv. 2019;5:eaaw8922. doi: 10.1126/sciadv.aaw8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L. Lv J. Wang H. Cheng Y. Angew. Chem., Int. Ed. 2020;59:4711–4719. doi: 10.1002/anie.201914970. [DOI] [PubMed] [Google Scholar]
- Zhang Z. Shen W. Ling J. Yan Y. Hu J. Cheng Y. Nat. Commun. 2018;9:1377. doi: 10.1038/s41467-018-03779-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Lv J. Gao P. Feng Q. Wang H. Cheng Y. Nano Lett. 2021;21:7855–7861. doi: 10.1021/acs.nanolett.1c03031. [DOI] [PubMed] [Google Scholar]
- Barrios A. Estrada M. Moon J. H. Angew. Chem., Int. Ed. 2022;61:e202116722. doi: 10.1002/anie.202116722. [DOI] [PubMed] [Google Scholar]
- Zhang S. Tan E. Wang R. Gao P. Wang H. Cheng Y. Nano Lett. 2022;22:8233–8240. doi: 10.1021/acs.nanolett.2c02948. [DOI] [PubMed] [Google Scholar]
- Zhang W. Bao B. Jiang F. Zhang Y. Zhou R. Lu Y. Lin S. Lin Q. Jiang X. Zhu L. Adv. Mater. 2021;33:2105667. doi: 10.1002/adma.202105667. [DOI] [PubMed] [Google Scholar]
- Ming Z. Fan J. Bao C. Xue Y. Lin Q. Zhu L. Adv. Funct. Mater. 2018;28:1706918. doi: 10.1002/adfm.201706918. [DOI] [Google Scholar]
- Hua Y. Xia H. Jia L. Zhao J. Zhao D. Yan X. Zhang Y. Tang S. Zhou G. Zhu L. Lin Q. Sci. Adv. 2021;7:eabg0628. doi: 10.1126/sciadv.abg0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. Yang H. Wang C. Xue Y. Wang X. Bao C. Zhu L. Chem. Sci. 2021;12:3627–3632. doi: 10.1039/D0SC06049F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. Birnbaum W. Anderski J. Picker M.-T. Mulac D. Langer K. Kuckling D. Biomacromolecules. 2018;19:4677–4690. doi: 10.1021/acs.biomac.8b01446. [DOI] [PubMed] [Google Scholar]
- Peng K. Tomatsu I. van den Broek B. Cui C. Korobko A. V. van Noort J. Meijer A. H. Spaink H. P. Kros A. Soft Matter. 2011;7:4881. doi: 10.1039/C1SM05291H. [DOI] [Google Scholar]
- Tan X. Li B. B. Lu X. Jia F. Santori C. Menon P. Li H. Zhang B. Zhao J. J. Zhang K. J. Am. Chem. Soc. 2015;137:6112–6115. doi: 10.1021/jacs.5b00795. [DOI] [PubMed] [Google Scholar]
- Agasti S. S. Chompoosor A. You C.-C. Ghosh P. Kim C. K. Rotello V. M. J. Am. Chem. Soc. 2009;131:5728–5729. doi: 10.1021/ja900591t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klán P. Šolomek T. Bochet C. G. Blanc A. Givens R. Rubina M. Popik V. Kostikov A. Wirz J. Chem. Rev. 2013;113:119–191. doi: 10.1021/cr300177k. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the ESI.†