Highlights
-
•
Circular RNA circFBXO7 overexpression suppressed NSCLC cells proliferation.
-
•
Circular RNA circFBXO7 directly inhibited miR-296-3p as a ceRNA.
-
•
KLF15 bound to the promoter of CDKN1A gene directly and promoted its expression.
-
•
Mouse homologous circular RNA circFbxo7 overexpression also suppressed tumor growth.
-
•
The novel circFBXO7/miR-296-3p/KLF15/CDKN1A axis regulated NSCLC cell proliferation.
Keywords: Circular RNAs, miR-296-3p, Non-small cell lung cancer, KLF15, CDKN1A
Abstract
Background
Accumulating evidence indicates that circular RNAs (circRNAs) play important roles in various cancers. Hsa_circ_0008832 (circFBXO7) is a circRNA generated from the second exon of the human F-box only protein 7 (FBXO7). Mouse circFbxo7 is a circRNA generated from the second exon of mouse F-box only protein 7 (Fbxo7). The role of human circFBXO7 and mouse circFbxo7 in non-small cell lung cancer (NSCLC) has not been reported.
Methods
The expression of circFBXO7 was measured by quantitative real-time PCR. Survival analysis was performed to explore the association between the expression of circFBXO7 and the prognosis of patients with NSCLC. Lung cancer cell lines were transfected with plasmids. Cell proliferation, cell cycle, and tumorigenesis were evaluated to assess the effects of circFBXO7. Fluorescence in situ hybridization assay was used to identify the location of circFBXO7 and circFbxo7 in human and mouse lung cancer cells. Luciferase reporter assay was conducted to confirm the relationship between circFBXO7 and microRNA.
Results
In this study, we found that circFBXO7 was downregulated in NSCLC tissues and cell lines. NSCLC patients with high circFBXO7 expression had prolonged overall survival. Overexpression of circFBXO7 inhibited cell proliferation both in vitro and in vivo. Mechanistically, we demonstrated that circFBXO7 upregulated the expression of miR-296-3p target gene Krüppel-like factor 15 (KLF15) and KLF15 transactivated the expression of CDKN1A.
Conclusions
CircFBXO7 acts as a tumor suppressor by a novel circFBXO7/miR-296-3p/KLF15/CDKN1A axis, which may serve as a potential biomarker and therapeutic target for NSCLC.
Graphical abstract
Introduction
Lung cancer is a major cause of cancer death and is one of the most prevalent cancers worldwide [1]. In China also, lung cancer is the most common cancer [2]. Non-small cell lung cancer (NSCLC) accounts for approximately 80–85% of all cases of lung cancers, with lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and lung adenosquamous carcinoma (LASC) constituting the major histological subtypes [3]. Over the past decades, the development of targeted therapies has improved clinical outcomes in a subset of NSCLC patients [4]. Therefore, there is an urgent need to identify novel biomarkers and characterize of therapeutic targets for the early detection and advanced therapy of NSCLC.
It is now clear that only about 2% of human genes are protein-coding, with non-coding RNAs (ncRNAs) making up the majority of the eukaryotic transcriptome [5,6]. A growing number of studies have shown that ncRNAs play important roles in the development of different cancers [7], [8], [9], [10], [11]. Circular RNAs (circRNAs) are endogenous ncRNAs formed by covalently closed loops without 5′ end caps and 3′ Poly (A) tails [12]. Covalently closed loops are generated from a non-canonical splicing event called back-splicing, in which a downstream 5′ end is linked to an upstream 3′ end [13]. CircRNAs are mainly localized in the cytoplasm, originating from exons, introns, and exon-introns [12,14]. Although the expression levels of circRNAs are generally lower than that of their linear counterparts, circRNAs are more stable due to their covalent closed-loop structures [15]. The biological functions of circRNAs include competing endogenous RNA (ceRNA) by acting as miRNA sponges [16], interacting with RNA binding proteins [17], regulating the stability of mRNAs [18], modulating gene transcription [19], and translating proteins [20]. In lung cancer cell lines and tissues, a large number of circRNAs have been identified through high-throughput RNA sequencing and microarray analyses [21], [22], [23]. Accumulated evidence suggests that circRNAs play context-dependent roles in the pathological processes of NSCLC [24], [25], [26], [27], [28]. A previous study has reported that hsa_circ_0008832 is a significantly downregulated circRNA in gastric cancer tissues [29]. However, no studies have focused on the function of hsa_circ_0008832 in NSCLC.
In this study, we investigated the expression profiles of circRNAs in four NSCLC and paired normal lung tissues using circRNA sequencing (circRNA-seq). We identified a significantly downregulated circRNA, hsa_circ_0008832 (termed circFBXO7). CircFBXO7 inhibited lung cancer cell proliferation by acting as a ceRNA to upregulate Krüppel-like factor 15 (KLF15). KLF15 upregulated the expression of CDKN1A (p21) by acting as a transcription factor. Furthermore, we identified an orthologous mouse circRNA (termed circFbxo7) that inhibited tumor growth in C57BL/6 mice. Our results indicate that circFBXO7 plays an antitumor role in NSCLC and may be a potential therapeutic target for lung cancer.
Materials and methods
Patient tissues and cell lines
A total of 56 paired NSCLC tissues and adjacent normal lung tissues were obtained from surgical resections of patients without preoperative treatment from Union Hospital (Wuhan, China) and confirmed by pathological examination. The circRNA-seq of the four paired NSCLC samples was performed on the Illumina high-throughput circRNA sequencing platform (Novogene, China). The circRNA-seq data have been uploaded to National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/bioproject/PRJNA863919). Another 52 paired samples were used for quantitative real-time polymerase chain reaction (qRT-PCR) verification. All samples were stored at −80 °C. All specimens were collected between March 2018 and August 2020. Human materials were obtained with the consent of patients and approved by the Ethics Committees of Tongji Medical College, Huazhong University of Science and Technology. Human NSCLC cell lines (A549 and H226) and mouse Lewis lung carcinoma (LLC) cell lines were purchased from the American Type Culture Collection (ATCC). All cell lines tested negative for mycoplasma contamination. A549 and H226 cells were cultured in RPMI-1640 medium (Hyclone, USA) supplemented with 10% fetal bovine serum (FBS) (GIBCO, BRL). LLC cells were cultured in high-glucose DMEM (Hyclone, USA) supplemented with 10% FBS (GIBCO, BRL). All cells were cultured in a humidified incubator at 37 °C and 5% CO2.
Total RNAs and gDNA extraction
Total RNA from cells and tissues was extracted using RNAisoPlus reagent (Takara, Japan). Genomic DNA was extracted using a Genomic DNA Isolation Kit (Tsingke, China). Cytoplasmic and nuclear RNAs were extracted using a previously described method [30].
RT-PCR and qRT-PCR
PrimeSTAR® Max DNA Polymerase (Takara, Japan) was used for PCR. RNA was reverse-transcribed using HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, China). The PCR products of cDNA and gDNA were observed using 2% agarose gel electrophoresis. The expression levels of circRNAs, mRNAs, and microRNAs were normalized to U1 or GAPDH using the 2−ΔCt or 2−ΔΔCt method. The primers are listed in the supplementary materials (Table S1).
RNase R treatments
Total RNA (20 μg) was incubated for 15 min at 37 °C with or without RNase R (20 U/μl) (Epicenter Biotechnologies, USA) and subsequently purified using the RNeasy MinElute Cleaning Kit (QIAGEN, Hilden, Germany). The purified RNAs were reverse-transcribed using HiScript II Q RT SuperMix for qPCR (Vazyme, China) and then analyzed by qRT-PCR.
Actinomycin D assay
Total RNA was extracted after the H226 cells were exposed to 10 μg/ml actinomycin D (MCE, USA) at different time points. The stabilities of circFBXO7 and linear FBXO7 mRNA were analyzed using qRT-PCR.
RNA fluorescence in situ hybridization (FISH)
Fluorescence-labeled probes complementary to circFBXO7 back-spliced site sequences were synthesized by Tsingke (Wuhan, China). CircFBXO7 FISH was performed according to a previous study [31]. Briefly, the NSCLC cells were seeded on coverslips in 24 plates and cultured overnight, followed by fixation, permeabilization, and hybridization. After three washes with 2×SSC, the coverslips were sealed with antifade mounting medium containing 4′,6-Diamidino-2-Phenylindole (DAPI). Finally, the FISH images were acquired using a confocal laser scanning microscope (LSM 780, Carl Zeiss).
Construction and transfection of plasmids
To construct the human circFBXO7, human KLF15, and mouse circFbxo7 overexpression plasmids, human circFBXO7, human KLF15, and mouse circFbxo7 cDNAs were synthesized by Tsingke (Wuhan, China) and cloned into pcDNA3.1(+), circRNA mini vector (Addgene #60648), or pcDNA3.1(+) vector (Sigma-Aldrich). The plasmids were transfected with Neofect™ DNA transfection reagent (Wuhan, China) according to the manufacturer's instructions. The details of the plasmids are described in the supplementary materials (Table S1).
Cell counting Kit-8 (CCK-8) assay
Cell proliferation was tested using the CCK-8 cell counting kit (Vazyme, China) according to the manufacturer's instructions. An automatic microplate reader (BioTek, USA) was used to measure the absorbance at 450 nm.
Colony formation assay
Briefly, the NSCLC cells were plated in 6-well plates at a density of 300 cells per well; after 14 days of incubation at 37 °C in a humidified incubator, colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 30 min. Cell colony numbers were counted using ImageJ software (www.imagej.net/).
Flow cytometric analysis of cell cycle
Cells were fixed in 70% ethanol overnight at 4 °C and resuspended in PI/RNase staining buffer (BD Biosciences) following the manufacturer's instructions. Data were analyzed using FlowJo software (FlowJo, USA).
5-Ethynyl-2′-deoxyuridine (EdU) assay
Cell proliferation and DNA synthesis were investigated using the BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 594 (Beyotime, China). Cells transfected with the overexpression plasmid or negative control vector were incubated with EdU for 48 h. The cells were then fixed and stained. ImageJ software (www.imagej.net/) was used to count EdU-positive cells.
RNA sequencing
We isolated total RNA from NSCLC cells transfected with circFBXO7 overexpression vector or vector-NC and conducted mRNA sequencing using the Illumina HiSeq platform.
Dual-luciferase reporter system
The putative binding sites of miR-296-3p with wild-type circFBXO7 and 3 untranslated regions of KLF15 mRNA (KLF15-3`UTR) were amplified and inserted into the psiCHECK2™ plasmid (Promega, USA). Similarly, the mutated targeting sites of circFBXO7 (circFBXO7-mut) and KLF15-3`UTR (KLF15-3`UTR-mut) were cloned into the psiCHECK2™ plasmid (Promega, USA) to generate reporter vectors. Detailed sequences of mutated targeting sites of circFBXO7 and KLF15-3`UTR were provided in the supplementary materials (Table S1). The above vectors were co-transfected with miR-296-3p mimic and the negative control mimic (mimic NC) (Ribobio, China). To determine whether KLF15 directly binds to the promoter region of CDKN1A, dual-luciferase reporter vectors were constructed. The 2000-bp upstream region of the CDKN1A transcription start site (TSS) was cloned into a pGL3.0-basic luciferase plasmid (pGL3.0-CDKN1A). A dual-luciferase reporter assay kit (Vazyme, China) was used to measure the relative luciferase activity.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was conducted using the SimpleChIP® Enzymatic Chromatin IP Kit (CST, USA). Briefly, the NSCLC cells were subjected to crosslinking (10 min, room temperature) and then treated with the micrococcal nuclease to digest their DNA to a length of approximately 150–900 bp. KLF15 (ab2647, Abcam, UK) or a control IgG antibody was added to the chromatin samples. The PCR products for chromatin immunoprecipitation were observed using 2% agarose gel electrophoresis. Detailed primers for ChIP-PCR and quantitative ChIP-PCR (ChIP-qPCR) were provided in the supplementary materials (Table S1).
Immunofluorescence
NSCLC cells were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.1% TritonX-100 in PBS for 10 min, blocked with 2% bovine serum albumin (BSA) for 30 min, and incubated with KLF15 and CDKN1A antibodies overnight at 4 °C. After washing twice with PBS, the cells were incubated with the corresponding secondary antibody for 1 h at 37 °C, followed by sealing with an antifade mounting medium containing DAPI. Images were acquired using a confocal laser scanning microscope (LSM 780, Carl Zeiss).
Western blotting assays
Cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (RIPA, Service, China). The protein was prepared and quantified by bicinchoninic acid (BCA) analysis (Beyotime, China). The same amount of protein was separated by 15% SDS-PAGE and transferred onto a PVDF membrane (Millipore, Schwalbach, Germany). The blocked protein with 5% skim milk powder was incubated with primary antibodies, namely, anti-KLF15 (ab2647, Abcam, UK), anti-CDKN1A (10355-1-AP, Proteintech, USA), and anti-GAPDH (60004-1-Ig; Proteintech, USA) at 4 °C for 12 h. Then, the prepared membranes were incubated with secondary antibodies: HRP-conjugated secondary goat anti-mouse (SA00001-1, Proteintech, USA), goat anti-rabbit (SA00001-2, Proteintech, USA), and donkey anti-goat (SA00001-3, Proteintech, USA) for 2 h. Finally, the blots were detected using a high-sensitivity electrochemiluminescence (ECL) detection kit (Vazyme, China).
Subcutaneous tumor model
The subcutaneous tumor assay was approved by the Animal Health Committee of the Huazhong University of Science and Technology, Tongji Medical School. Male BALB/c nude mice (6–8 weeks old) and C57BL/6 wild-type mice (6–8 weeks old) were purchased from Beijing Charles River. NSCLC cells transfected with vector-NC, overexpression vectors, and silencing vectors were collected for subcutaneous injection into mice. Human NSCLC cells were injected subcutaneously into BALB/c nude mice (3 × 106 cells per mouse) and mouse LLC cells were injected subcutaneously into C57BL/6 wild-type mice (2 × 106 cells per mouse). After 4 weeks, the mice were sacrificed by cervical decapitation, and the tumors were resected for weight and volume assessment.
Immunohistochemistry (IHC)
The tumor tissues resected from the xenograft tumor model were fixed, dehydrated, and embedded in paraffin. Subsequently, 4-μm-thick sections were obtained and stained with specific antibodies against KLF15 (ab167192, Abcam, UK) and CDKN1A (10355-1-AP, Proteintech, USA).
Bioinformatic analysis
The circFBXO7 sequence was obtained from circBase (www.circbase.org/). TargetScan (www.targetscan.org/), miRanda database (www.miranda.org/), and RNAhybrid (www.bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) were used to predict the potential binding site of miR-296-3p with circFBXO7 and KLF15-3`UTR. The Cancer Genome Atlas (TCGA) LUAD database (https://portal.gdc.cancer.gov/) was used to explore the association between KLF15 and the prognosis of LUAD patients. The National Center of Biotechnology Information (NCBI) nucleotide BLAST was used to compare human circFBXO7 and mouse circFbxo7 sequences (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 8.0.2 (GraphPad Software, La Jolla, CA, USA) and R 4.0.1 (www.r-project.org/). Data from at least three independent experiments were expressed as the mean ± SD or median (interquartile range, IQR). Continuous data were compared using Student's t-test or Mann-Whitney U test if two groups were to be compared and one-way ANOVA or Kruskal-Wallis H-test for multiple groups. Differentially expressed circRNAs between four pairs of cancerous and normal tissues were identified using Mann-Whitney U test. Relationships were analyzed using Spearman's correlation analysis. Survival analysis was performed using the Kaplan-Meier method. All statistical tests were two-sided, and the statistical significance was set at P < 0.05.
Results
CircFBXO7 is downregulated in the NSCLC tissues
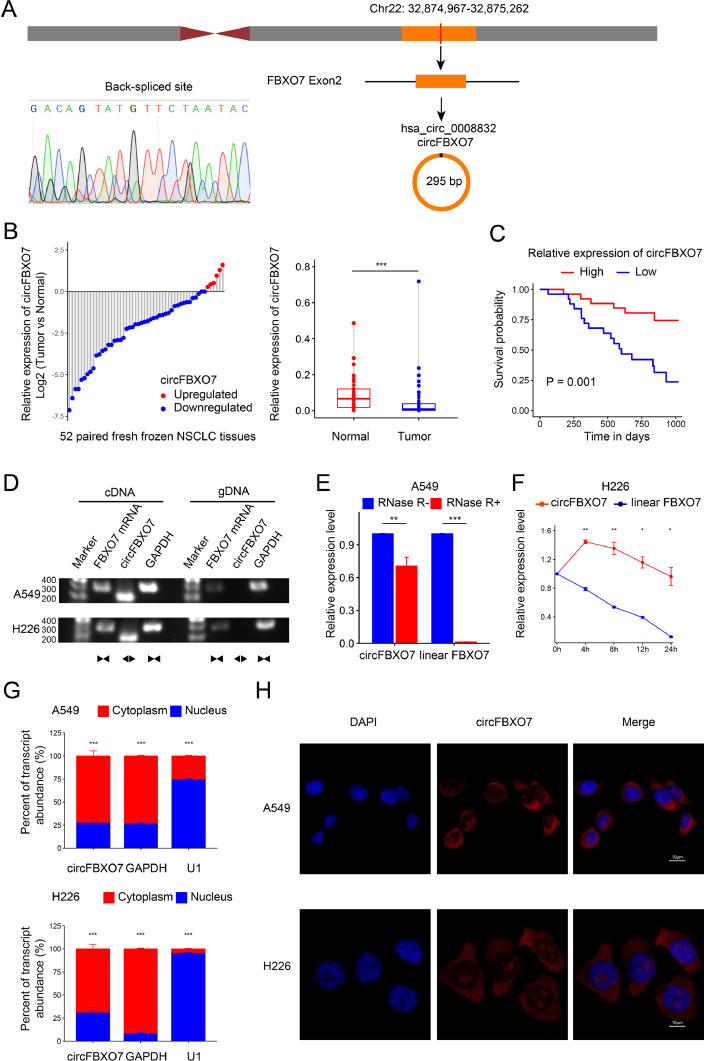
The expression profiles of circRNAs in four pairs of NSCLC and adjacent normal tissues were analyzed using the Illumina high-throughput circRNA sequencing platform. The cluster map for circRNA sequencing was shown in the supplementary materials (Fig. S1). The results demonstrated that among the circRNAs, circFBXO7 was significantly downregulated in NSCLC tissues than in their corresponding adjacent normal tissues. Other details for circRNA sequencing were shown in the supplementary materials (Fig. S2). CircFBXO7 is 295 bp in length and is derived from exon 2 of F-box only protein 7 (FBXO7) located on chromosome 22 (www.circbase.org/). Subsequently, the back-spliced site sequence of circFBXO7 was amplified using divergent primers and identified by Sanger sequencing (Fig. 1A). We then confirmed the downregulated expression of circFBXO7 in another 52 paired NSCLC tissues by qRT-PCR and found that the abundance of circFBXO7 was significantly lower in 88.5% (46/52) of the NSCLC tissues (Fig. 1B). The average expression of circFBXO7 was significantly higher in normal lung tissues (P < 0.001) (Fig. 1B). Kaplan-Meier analysis demonstrated that the overall survival of NSCLC patients with low circFBXO7 expression was significantly shorter than that of patients with high circFBXO7 expression (P = 0.001) (Fig. 1C). The relative expression of circFBXO7 in normal lung epithelial cell lines and different lung cancer cell lines was demonstrated in the supplementary materials. A549 and H226 cells were selected to conduct subsequent experiments because of their low expression of circFBXO7 (Fig. S3). RT-PCR analysis of cDNA and gDNA of A549 and H226 cells showed that divergent primers could only amplify circFBXO7 from cDNA, whereas linear FBXO7 mRNA could be amplified by convergent primers from both cDNA and gDNA (Fig. 1D). RNase R treatment and actinomycin D treatment revealed that circFBXO7 was more resistant to RNase R digestion (Fig. 1E) and more stable than linear FBXO7 mRNA (Fig. 1F). qRT-PCR analysis of cytoplasmic and nuclear fractions of A549 and H226 cells demonstrated that the circFBXO7 transcript was predominantly located in the cytoplasm (Fig. 1G). RNA-fluorescence in situ hybridization (RNA-FISH) also demonstrated that circFBXO7 was mostly expressed in the cytoplasm (Fig. 1H). These results indicate that circFBXO7 is a highly stable circular RNA that is downregulated in NSCLC tissues and is associated with the prognosis of NSCLC patients.
Fig. 1.
Identification and expression of circFBXO7 in NSCLC cells and tissues. (A) CircFBXO7 is constructed by the cyclization of exon 2 of FBXO7 from chromosome 22. Sanger sequencing confirmed the back-spliced site of circFBXO7. (B) qRT-PCR detected the differential relative expression of circFBXO7 in 52 paired NSCLC tissues and adjacent normal tissues. (C) The Kaplan-Meier method demonstrated the overall survival probability of 52 NSCLC patients with low and high expression of circFBXO7. (D) The existence of cricFBXO7 was confirmed in A549 and H226 cells by agarose gel electrophoresis. The divergent primers only amplified circFBXO7 in cDNA. (E) qRT-PCR for the abundance of circFBXO7 in A549 cells treated with RNase R. (F) qRT-PCR for the abundance of circFBXO7 and linear FBXO7 mRNA in H226 cells treated with Actinomycin D at different time points. (G) The abundance of circFBXO7 in the cytoplasmic and nuclear fractions of A549 and H226 cells. (H) Fluorescence in situ hybridization (FISH) showed the location of circFBXO7 A549 and H226 cancer cells. *P < 0.05, **P < 0.01, ***P < 0.001. NSCLC, non-small cell lung cancer.
CircFBXO7 suppresses lung cancer cell proliferation in vitro and in vivo
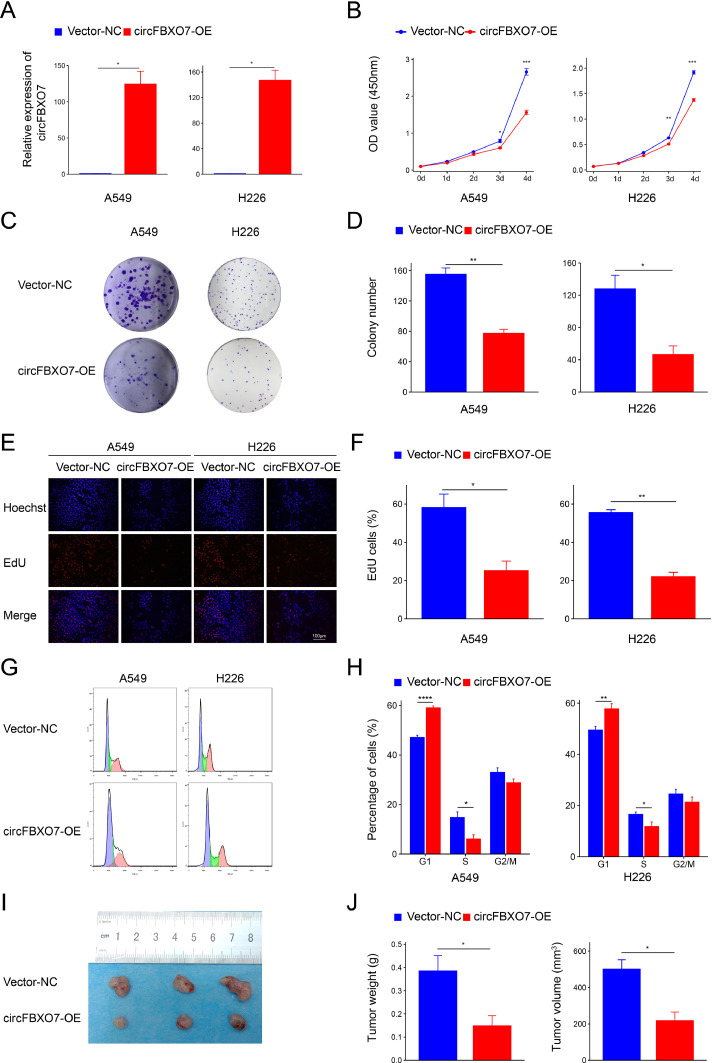
To explore the function of circFBXO7 in NSCLC, we constructed circFBXO7 overexpression plasmids (circFBXO7-OE). qRT-PCR analysis demonstrated that the expression of circFBXO7 was significantly increased in both A549 and H226 cells after transfection with circFBXO7-OE (Fig. 2A). CCK-8 assays (Fig. 2B), colony formation assays (Fig. 2C, D), and EdU assays (Fig. 2E, F) showed that overexpression of circFBXO7 suppressed cell proliferation in A549 and H226 cells. Flow cytometry analysis showed that the proportion of G1 phase cells was significantly increased, while the proportion of cells in the S-phase was significantly decreased after overexpression of circFBXO7 (Fig. 2G, H), indicating a cell cycle arrest at the G1/S phase. The BALB/c nude mice were injected with H226 cells transfected with circFBXO7-OE and a negative control vector (vector-NC). Four weeks after H226 cell injection, tumor masses were resected from nude mice and compared between circFBXO7-OE and vector-NC groups. The results showed that overexpression of circFBXO7 significantly reduced the weight and volume of tumors (Fig. 2I, J). These findings indicate that overexpression of circFBXO7 inhibits the growth of NSCLC cells in vitro and in vivo.
Fig. 2.
Overexpression of circFBXO7 inhibited NSCLC cell proliferation in vitro and in vivo. (A) The efficiency of transfected circFBXO7 overexpression vector (circFBXO7-OE) in A549 and H226 cells was detected by qRT-PCR. (B,C,D,E,F) The effect of circFBXO7-OE on A549 and H226 cell proliferation was detected by CCK-8 assays (B), colony formation assays (C, D), and EdU assays (E, F). Blue, Hoechst-stained nuclei; red, EdU-positive nuclei; scale bars =100 μm. (G, H) The percentages of cells in the G1, S, or G2 phases in A549 and H226 cells were detected by flow cytometry. CircFBXO7 arrested the A549 and H226 cell cycle at the G1/S phase. (I, J) Tumor weights and volumes of resected tumors were measured 4 weeks after inoculation of circFBXO7-transfected H226 cells (n = 3). ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. NSCLC, non-small cell lung cancer. Vector-NC, negative control vector.
CircFBXO7 upregulates the expression of KLF15 by sponging miR-296-3p
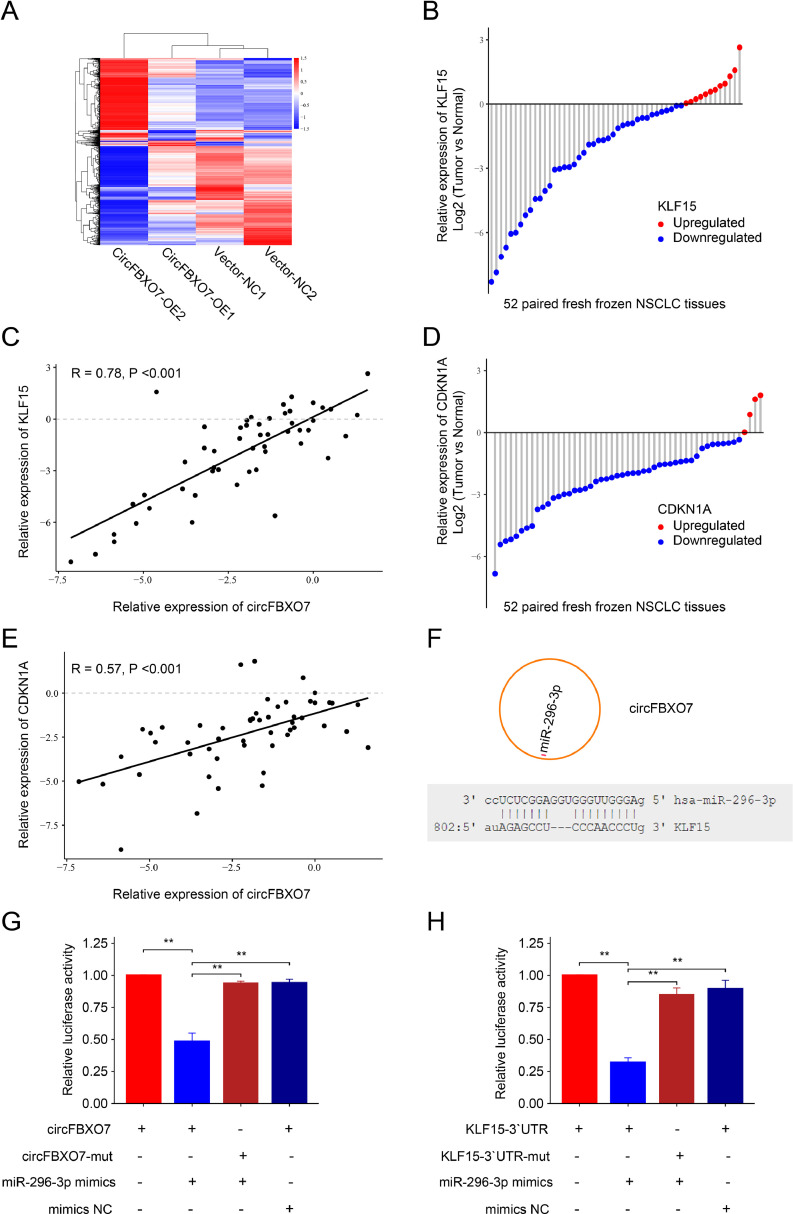
To explore the mechanism of circFBXO7 in NSCLC, we conducted mRNA sequencing in paired A549 and H226 cells transfected with circFBXO7-OE and vector-NC. Sequencing results (Fig. 3A) and bioinformatic analysis (Fig. S4) demonstrated significant changes in the expression of various genes after transfection. Among these genes, FBXO7 is the parent gene of circFBXO7. Additionally, a significant increase in the expression of KLF15 and CDKN1A was observed after circFBXO7-OE transfection (Fig. S4). The volcano diagram showed the upregulated and downregulated mRNAs (Fig. S5). Bioinformatic analysis of the TCGA LUAD dataset showed that the expression of KLF15 was significantly lower in tumor tissues and was associated with the overall survival of patients (Fig. S6). qRT-PCR analysis showed that KLF15 was relatively low expressed in 76.9% (40/52) of the NSCLC tissues included in this study (Fig. 3B). Pearson correlation analysis demonstrated that the expression of KLF15 was positively correlated with that of circFBXO7 (R = 0.78, P < 0.001) (Fig. 3C). Furthermore, qRT-PCR analysis showed that CDKN1A was relatively low expressed in 92.3% (48/52) of the NSCLC tissues (Fig. 3D). Also, the expression of CDKN1A was positively related with that of circFBXO7 (R = 0.57, P < 0.001) (Fig. 3E). According to the predicted results from the TargetScan (www.targetscan.org/), the miRanda database (www.miranda.org/), and the RNAhybrid (www.bibiserv.cebitec.uni-bielefeld.de/rnahybrid/), miR-296-3p had the potential to bind both circFBXO7 and KLF15-3`UTR (Fig. 3F and Fig. S7). However, no microRNA that could bind both circFBXO7 and CDKN1A-3`UTR was predicted, indicating that circFBXO7 might not upregulate CDKN1A expression directly through microRNAs (data was not shown). To validate the binding capability of the miR-296-3p to circFBXO7 and KLF15-3`UTR, luciferase reporter systems containing mutated predicted binding sites and wild-type of circFBXO7 and KLF15-3`UTR were constructed. The results demonstrated significant reductions in the luciferase activities of circFBXO7 and KLF15-3`UTR wild-type reporters when transfected with miR-296-3p mimics compared with the negative control mimics (mimics NC) or mutated luciferase reporter (Fig. 3G, H). In addition, the expression of CDKN1A and KLF15 was decreased after transfection of miR-296-3p mimics (Fig. S8). These findings indicate that circFBXO7 upregulates the expression of KLF15 by acting as a sponge of miR-296-3p.
Fig. 3.
CircFBXO7 upregulated the expression of KLF15 by sponging miR-296-3p. (A) Cluster heatmap of differentially expressed mRNAs in paired A549 and H226 cells transfected with circFBXO7 overexpression vector (circFBXO7-OE) and negative control vector (vector-NC). (B) The qRT-PCR analysis detected the differential abundance of KLF15 in 52 paired NSCLC and adjacent normal tissues. (C) The Pearson correlation analysis of KLF15 and circFBXO7 in 52 paired NSCLC and adjacent normal tissues. (D) The qRT-PCR analysis detected the differential abundance of CDKN1A in 52 paired NSCLC and adjacent normal tissues. (E) The Pearson correlation analysis of CDKN1A and circFBXO7 in 52 paired NSCLC and adjacent normal tissues. (F) A schematic model demonstrated the assumed binding sites of miR-296-3p on circFBXO7 and KLF15. (G, H) Dual-luciferase reporter activity of circFBXO7 and KLF15–3′UTR in H226 cells co-transfected with miR-296-3p mimics or mimics negative control (NC). *P < 0.05, **P < 0.01. NSCLC, non-small cell lung cancer.
KLF15 suppresses NSCLC cell proliferation by inducing a cell cycle arrest at the G1/S phase
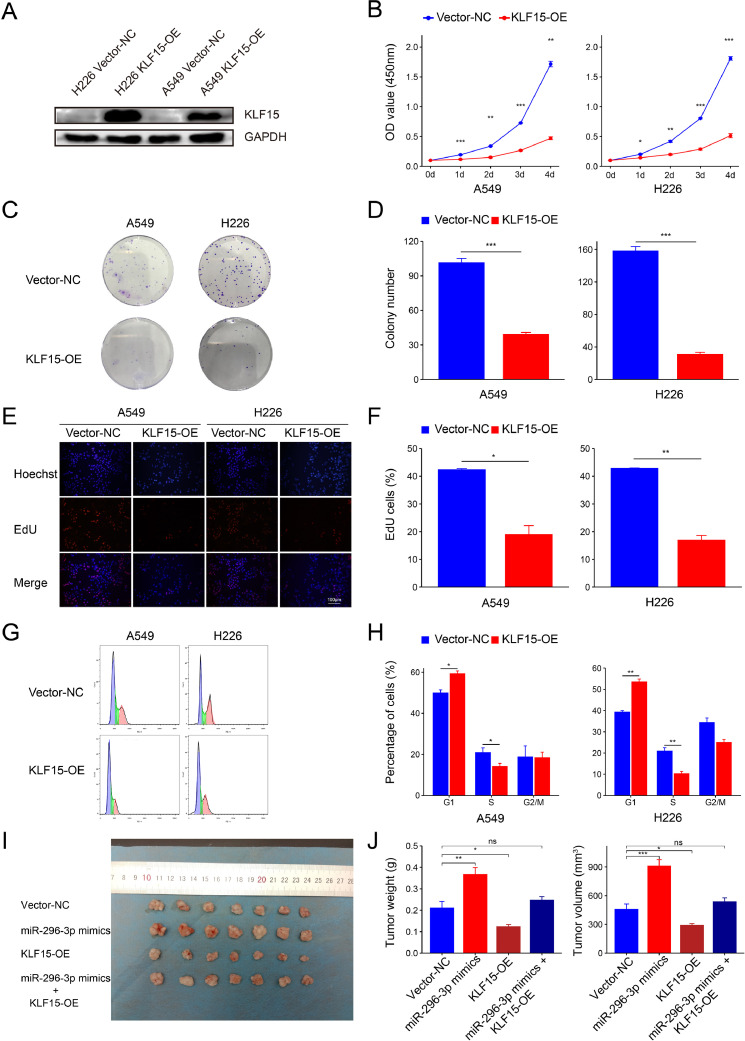
To confirm the function of KLF15 in NSCLC, we constructed KLF15 overexpression plasmids (KLF15-OE). Western blotting assay demonstrated significantly elevated levels of KLF15 protein in both A549 and H226 cells (Fig. 4A). CCK-8 assays (Fig. 4B), colony formation assays (Fig. 4C, D), and EdU assays (Fig. 4E, F) revealed that KLF15 overexpression suppressed cell proliferation in A549 and H226 cells. Flow cytometry analysis showed that the proportion of G1 phase cells significantly increased, while the proportion of cells in the S-phase significantly decreased (Fig. 4G, H), indicating that KLF15 induced cell cycle arrest at the G1/S phase. MiR-296-3p mimics were used to silence KLF15. BALB/c nude mice were injected with H226 cells transfected with the vector-NC, miR-296-3p mimics, KLF15-OE, and miR-296-3p mimics plus KLF15-OE. Four weeks after H226 cell injection, tumor masses from the different groups were resected and compared. Overexpression of KLF15 significantly inhibited tumor growth in vivo, whereas KLF15 silencing had the opposite effect. (Fig. 2I, J). These results indicate that KLF15 overexpression induces cell cycle arrest at the G1/S phase and inhibits NSCLC cell growth in vitro and in vivo which is consistent with previous studies [32]. Besides, these results suggest that miR-296-3p mimics inhibit the antitumor effect of KLF15 in vivo.
Fig. 4.
KLF15 inhibited NSCLC cell proliferation by inducing a cell cycle arrest at the G1/S phase. (A) The efficiency of transfected KLF15 overexpression vector (KLF15-OE) in A549 and H226 cells was detected by western blotting assay. (B,C,D,E,F) The effect of KLF15-OE on A549 and H226 cell proliferation was detected by CCK-8 assays (B), colony formation assays (C, D), and EdU assays (E, F). Blue, Hoechst-stained nuclei; red, EdU-positive nuclei; scale bars =100 μm. (G, H) The percentages of cells in the G1, S, or G2 phases in A549 and H226 cells were detected by flow cytometry. KLF15 arrested the A549 and H226 cell cycle at the G1/S phase. (I, J) Tumor weights and volumes of resected tumors were measured 4 weeks after inoculation of H226 cells transfected with negative control vector (vector-NC), miR-296-3p mimics, KLF15-OE, and miR-296-3p mimics plus KLF15-OE (n = 7). ns P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001. NSCLC, non-small cell lung cancer.
KLF15 transactivates CDKN1A by binding the promoter region of CDKN1A
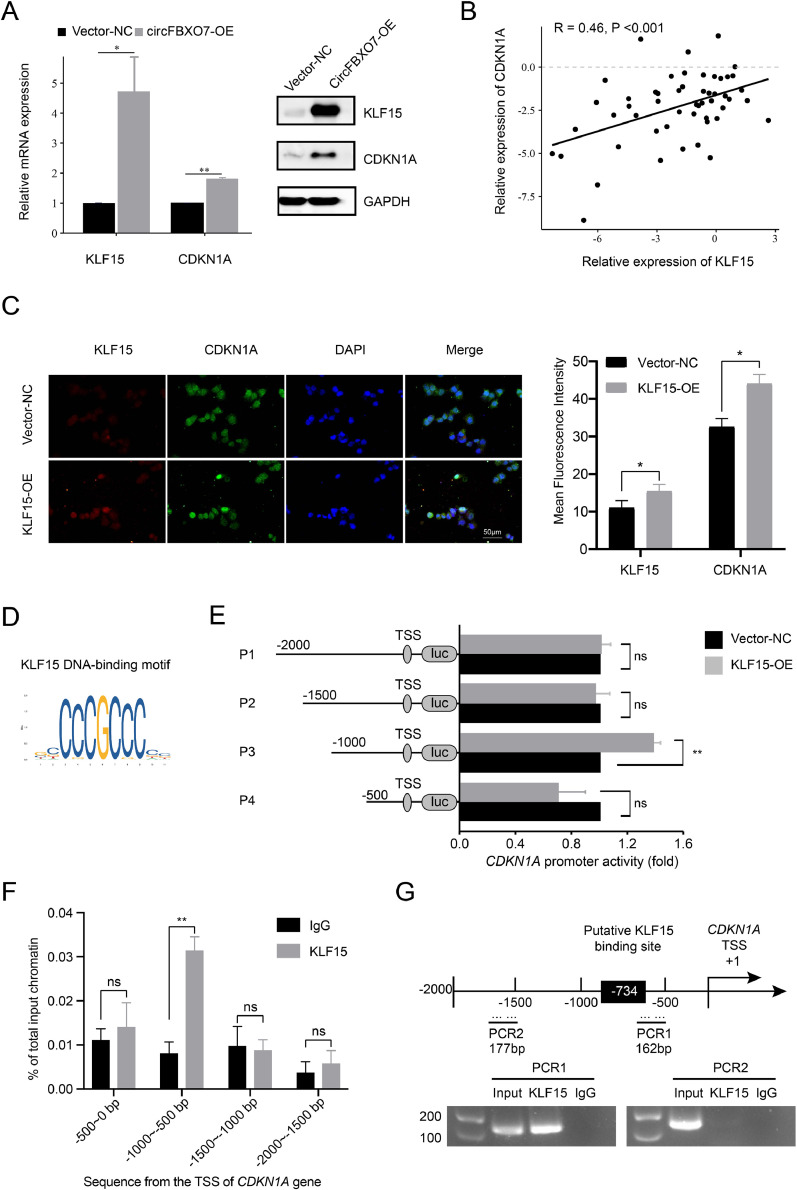
Western blotting assay and qRT-PCR analysis demonstrated that the mRNA expression and protein abundance of KLF15 and CDKN1A were significantly upregulated after circFBXO7-OE transfection (Fig. 5A). To validate the correlation between KLF15 and CDKN1A, the relative expression levels of KLF15 and CDKN1A in 52 paired NSCLC samples were detected by qRT-PCR (Fig. 5B). These results showed that the expression of CDKN1A was positively correlated with that of KLF15 (R = 0.46, P < 0.001) (Fig. 5B). Since KLF15 overexpression significantly upregulated CDKN1A (p21) (Fig. 5C), we speculated that KLF15 protein might transactivate CDKN1A gene by binding to the promoter region of it. The KLF15-binding site was presumed to be 734 bp in the 1000 bp upstream region of the CDKN1A TSS according to the JASPAR database (http://jaspar.genereg.net) (Fig. 5D). Dual-luciferase reporter vectors were constructed to determine whether KLF15 directly binds to the promoter region of CDKN1A. We cloned the 2000-bp upstream region of the CDKN1A TSS (−2000/+1 bp) into a pGL3.0-basic luciferase plasmid (pGL3.0-CDKN1A). To explore the core binding site within the CDKN1A promoter, the 2000-bp region was divided into four different lengths (P1: −2000/+1; P2: −1500/+1; P3: −1000/+1; P4: −500/+1). Transfection of pGL3.0-CDKN1A-P3 resulted in a significant increase in luciferase activity in H226 cells compared to the vector-NC (Fig. 5E), indicating that the sequence between nucleotides −1000 and +1 in the promoter of CDKN1A might contain binding sites for KLF15. ChIP-qPCR analysis revealed the presence of KLF15-binding sites in the promoter region (−1000 to −500) of the CDKN1A gene in H226 cells (Fig. 5F). Furthermore, ChIP assay was performed to determine the direct regulation of CDKN1A by KLF15. RT-PCR using specific primers for PCR1 (−833 to −672) and PCR2 (−1682 to −1505) was conducted to amplify the binding region (Fig. 5G). Results showed that KLF15 was strongly bound to PCR1 in the CDKN1A promoter region. Taken together, these findings suggest that KLF15 transactivates CDKN1A expression by binding to the promoter region of it.
Fig. 5.
KLF15 transactivated CDKN1A by binding the promoter region of it. (A) The western blotting assay and qRT-PCR analysis detected the increasing expression of KLF15 and CDKN1A in A549 cells after transfecting circFBXO7-OE. (B) The association between CDKN1A and KLF15 in 52 paired NSCLC tissues was detected by qRT-PCR. (C) The expression of CDKN1A protein after KLF15 overexpression was detected by immunofluorescence assay. (D) The putative transcription factor binding sites of KFL15 in the JASPAR database. (E) The CDKN1A promoter structure was constructed and relative luciferase activities were measured. (F) ChIP-qPCR analysis revealed the presence of KLF15-binding sites in the promoter region (−1000 to −500) of the CDKN1A gene in H226 cells. (G) Schematic model of the human CDKN1A promoter region in 2000 bp upstream or 500 bp downstream of transcription start site (TSS, designated as + 1). ChIP PCR products for putative KLF15 binding sites and an upstream region not expected to associate with KLF15 are depicted with bold lines. ns P > 0.05, *P < 0.05, ** P < 0.01.
Mouse homologous circular RNA circFbxo7 suppresses tumor growth
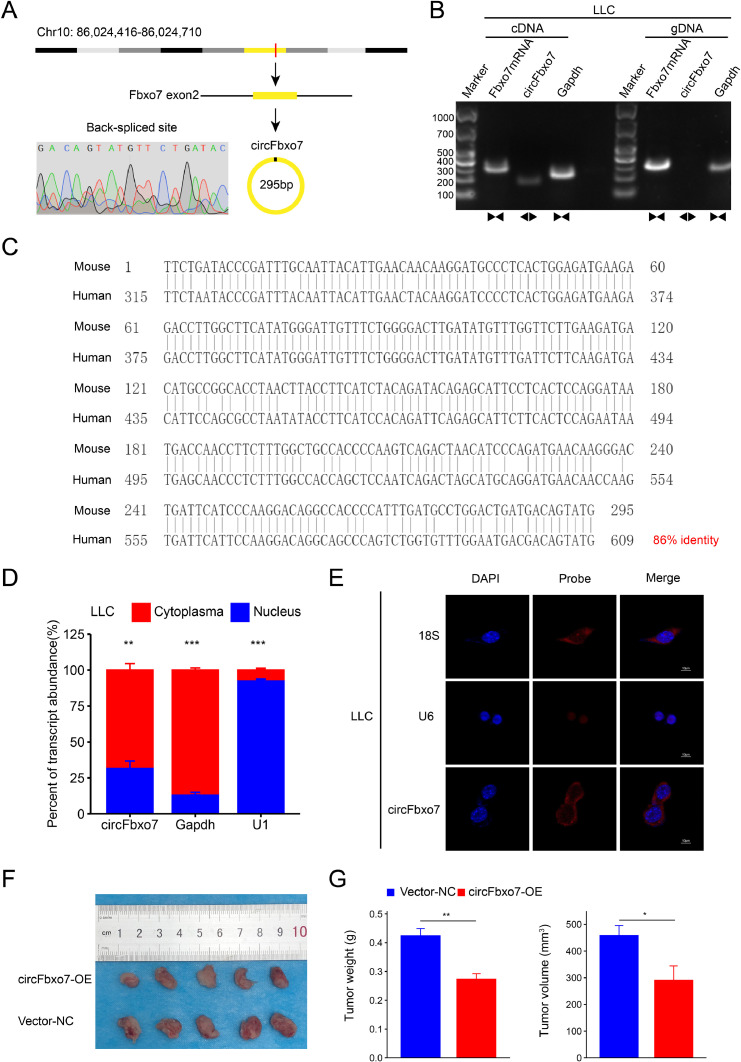
To explore whether there was a mouse ortholog of human circFBXO7, we amplified and identified the back-spliced site sequence of the putative mouse ortholog by Sanger sequencing. The results showed that there was an ortholog of circFBXO7 in mice, which was termed circFbxo7 (Fig. 6A, B). CircFbxo7 is 295 bp in length and is derived from exon 2 of mouse F-box only protein 7 (Fbxo7) located on chromosome 10 (Fig. 6A). Nucleotide BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that there was a close identity (86%) between human circFBXO7 and mouse circFbxo7 (Fig. 6C). qRT-PCR (Fig. 6D) and FISH (Fig. 6E) assays of LLC cells showed that the circFbxo7 transcript was predominantly located in the cytoplasm. To explore the function of circFbxo7 in mouse lung cancer, we constructed circFbxo7 overexpression plasmids (circFbxo7-OE) and established a wild-type mouse xenograft model by implanting LLC cells which were transfected with vector-NC or circFbxo7. Tumor weights and volumes were measured 4 weeks after the LLC cell injection. We found that overexpression of circFbxo7 significantly inhibited tumor growth in vivo (Fig. 6F, G). These findings indicate that mice express an ortholog of human circFBXO7 and mouse circFbxo7 also functions as a tumor growth suppressor.
Fig. 6.
Mouse homologous circular RNA circFbxo7 suppresses tumor growth. (A) CircFbxo7 is constructed by the cyclization of exon 2 of mouse Fbxo7 from chromosome 10. Sanger sequencing confirmed the back-spliced site of circFbxo7. (B) The existence of cricFbxo7 was confirmed in LLC cells by agarose gel electrophoresis. The divergent primers only amplified circFbxo7 in LLC cDNA. (C) Pairwise alignment of the human circFBXO7 and mouse circFbxo7 sequences. (D) The qRT-PCR analysis for the abundance of circFbxo7 in the cytoplasmic and nuclear fractions of LLC cells. (E) Fluorescence in situ hybridization showed the location of circFbxo7 in LLC cells. (F, G) Tumor weights and volumes of resected tumors from wild-type mice were measured after 4 weeks postinoculation of LLC cells transfected with circFbxo7-OE and vector-NC (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001. Vector-NC, negative control vector.
Discussion
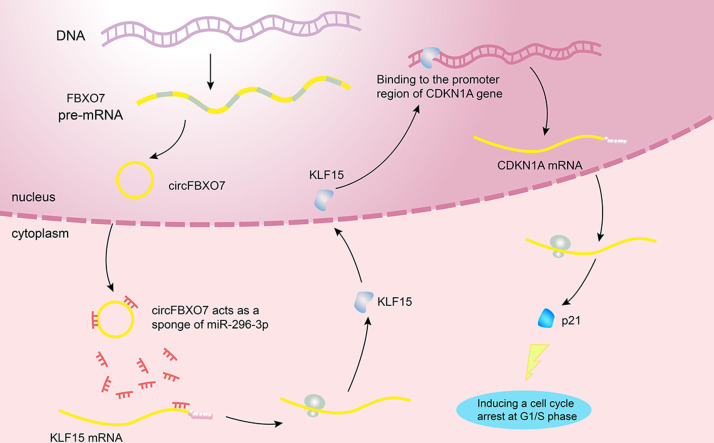
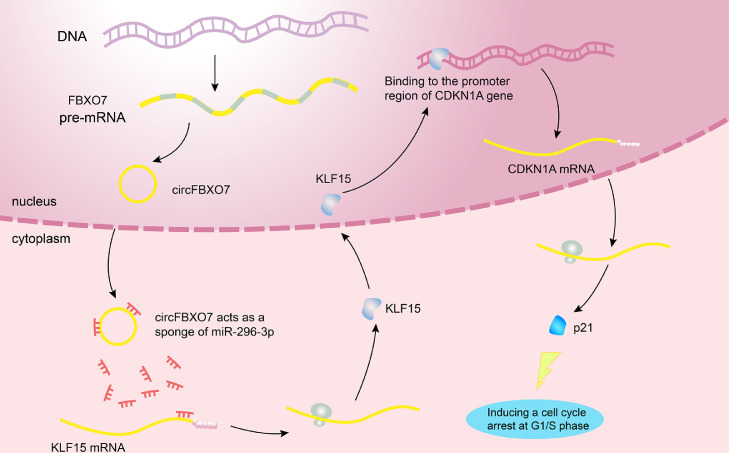
In this study, we demonstrated the role of circFBXO7 in human NSCLC for the first time (Fig. 7). Overexpression of circFBXO7 upregulated KLF15 mRNA levels by competitively binding to miR-296-3p. Elevated KLF15 protein transactivates CDKN1A by binding to the promoter region of it. The upregulation of CDKN1A protein inhibited NSCLC cell proliferation by inducing cell cycle arrest at the G1/S phase.
Fig. 7.
Hypothesis diagram illustrates the role of circFBXO7 in human NSCLC cells.
The ceRNA hypothesis has proposed that circRNAs mainly act as miRNA sponges to influence the function of miRNAs, and then regulate the stability of mRNAs that share similar miRNA response elements. Using nuclear-cytoplasmic fractionation assay and FISH, we found that circFBXO7 predominantly existed in the cytoplasm, suggesting its role in post-transcriptional regulation (Fig. 1). Based on the RNA-seq and bioinformatic analyses, we found that overexpression of circFBXO7 increased the expression of KLF15 (Fig. 3). Based on the bioinformatics analyses and the results of the dual-luciferase reporter system, we identified that miR-296-3p could bind both circFBXO7 and KLF15 (Fig. 3). Our results suggested that circFBXO7 functioned as a ceRNA to regulate the expression of KLF15 in lung cancer.
MiR-296-3p has been reported to be carcinogenic, and it has been reported to be either overexpressed or lost in different human cancer cell types, including LUAD, glioblastoma, clear cell renal cell carcinoma (ccRCC), and prostate cancer (PCa) [33], [34], [35], [36]. In ccRCC, miR-296-3p mimics promoted OSRC-2 cell migration and invasion [34]. In glioblastoma, miR-296-3p suppressed the growth and multi-drug resistance of tumors by targeting EAG1 [35]. In PCa, miR-296-3p increased the tolerance of cancer cells to NK cells and promoted cancer metastasis by targeting ICAM1 [36]. Moreover, miR-296-3p increased the epithelial-to-mesenchymal transition of peritoneal mesothelial cells via DOK2 in peritoneal fibrosis [37]. In NSCLC, miR-296-3p was found to be upregulated or downregulated in different studies [38]. Some studies suggested that miR-296-3p inhibited the proliferation of lung cancer, while other studies suggest that miR-296-3p was upregulated in lung cancer tissue and might play a role in promoting cancer [38]. However, studies about miR-296-3p are currently inconclusive. Our study revealed that miR-296-3p targeted KLF15 to undo the proliferation inhibition of KLF15 in NSCLC cells (Fig. 4). Therefore, miR-296-3p may function as a context-dependent onco-promoter or an onco-suppressor under different biological conditions, and further studies are needed to explore the multi-faceted functions of miR-296-3p [39].
The Krüppel-like factors (KLFs) belong to a specific family of proteins that are three C2H2-type zinc finger domains containing transcription factors. They can bind to GC-rich DNA motifs, indicating their essential roles in both enhancing and inhibiting transcription [40,41]. KLFs exert multiple functions in regulating cell cycle, proliferation, migration, invasion apoptosis, and differentiation, and they play crucial roles in the carcinogenesis of various cancers [42]. As a member of the KLF family proteins, KLF15 is expressed in multiple tissues, especially in the kidney, and is also known as kidney-enriched KLF [43]. More and more studies show that KLF15 plays an anti-tumor role in different cancers [44,45]. In LUAD, overexpression of KLF15 can arrest cell cycle at the G0/G1 phase and inhibit LUAD cell proliferation by upregulating CDKN1A/p21 and CDKN2A/p15 [32]. Survival analyses based on the TCGA LUAD database demonstrated that the low expression of KLF15 was associated with poor clinical outcomes. In our study, both the qRT-PCR results from clinical specimens and the profiles from the TCGA database indicated that the expression of KLF15 was downregulated in lung cancer tissues, suggesting that KLF15 was closely associated with the development of lung cancer. Our study also demonstrated that the expression of KLF15 was significantly positively correlated with the expression of CDKN1A (Fig. 5) and that KLF15 overexpression inhibited lung cancer cell proliferation by arresting the cell cycle at the G1/S phase (Fig. 4). These results are consistent with those of a previous study. Furthermore, based on the dual-luciferase assay and ChIP assay, we confirmed that KLF15 could bind to the DNA motif in the promoter of CDKN1A and acts as a transcription activator to upregulate the expression of CDKN1A (Fig. 5).
Our study identified a novel mouse circRNA, circFbxo7, which was generated from the second exon of mouse Fbxo7 (Fig. 6). Sequence analysis showed that there was a high identity between mouse circFbxo7 and human circFBXO7. Similar to human circFBXO7, mouse circFbxo7 mainly existed in the cytoplasm of LLC cells and significantly inhibited tumor proliferation in subcutaneously implanted tumor models in wild-type mice. However, further studies are required to explore the underlying mechanisms.
Conclusions
In conclusion, our study revealed that circFBXO7 was remarkably downregulated in NSCLC tissues and circFBXO7 significantly inhibited lung cancer proliferation via the circFBXO7/miR-296-3p/KLF15/CDKN1A axis. Our findings demonstrate that circFBXO7 plays a critical role in the growth of NSCLC and is a potential therapeutic target for NSCLC.
Funding
This work was supported in part by grants from the National Natural Science Foundation of China (No. 82170105, No. 82100111, and No. 81973990).
Availability of data and materials
All datasets enrolled in this study are available in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) databases, and the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/bioproject/PRJNA863919).
Ethics statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (2020–0120).
CRediT authorship contribution statement
Zi-Hao Wang: Conceptualization, Data curation, Investigation, Methodology, Validation, Visualization, Writing – original draft. Lin-Lin Ye: Conceptualization, Methodology, Writing – review & editing. Xuan Xiang: Conceptualization, Methodology, Validation, Visualization, Writing – review & editing. Xiao-Shan Wei: Conceptualization, Funding acquisition, Methodology, Writing – review & editing. Yi-Ran Niu: Conceptualization, Methodology, Writing – review & editing. Wen-Bei Peng: Conceptualization, Methodology, Writing – review & editing. Si-Yu Zhang: Project administration, Supervision, Writing – review & editing. Pei Zhang: Project administration, Supervision, Writing – review & editing. Qian-Qian Xue: Project administration, Supervision, Writing – review & editing. Hao-Lei Wang: Project administration, Supervision, Writing – review & editing. Yi-Heng Du: Resources, Writing – review & editing. Yao Liu: Resources, Writing – review & editing. Jia-Qi Ai: Methodology, Writing – review & editing, Writing – review & editing. Qiong Zhou: Conceptualization, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2023.101635.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Osmani L., et al. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin. Cancer Biol. 2018;52(Pt 1):103–109. doi: 10.1016/j.semcancer.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maemondo M. Tyrosine kinase inhibitors as first-line treatment in NSCLC. Lancet Oncol. 2016;17(5):541–543. doi: 10.1016/S1470-2045(16)30055-9. [DOI] [PubMed] [Google Scholar]
- 5.Mortoglou M., et al. Non-coding RNAs in pancreatic ductal adenocarcinoma: new approaches for better diagnosis and therapy. Transl. Oncol. 2021;14(7) doi: 10.1016/j.tranon.2021.101090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding L., et al. The emerging role of small non-coding RNA in renal cell carcinoma. Transl. Oncol. 2021;14(1) doi: 10.1016/j.tranon.2020.100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X., et al. Circular RNA 0010117 promotes aggressive glioblastoma behavior by regulating the miRNA-6779-5p/SPEN axis. Transl. Oncol. 2022;25 doi: 10.1016/j.tranon.2022.101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F., et al. Hsa_circ_0088212-mediated miR-520h/APOA1 axis inhibits osteosarcoma progression. Transl. Oncol. 2021;14(12) doi: 10.1016/j.tranon.2021.101219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu S.Y., et al. LncRNA GIHCG promoted the proliferation and migration of renal cell carcinoma through regulating miR-499a-5p/XIAP axis. Transl. Oncol. 2022;20 doi: 10.1016/j.tranon.2022.101356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng L., et al. Long non-coding FAM201A accelerates the tumorigenesis and progression of colorectal cancer through miR-3163/MACC1 axis. Transl. Oncol. 2022;25 doi: 10.1016/j.tranon.2022.101490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H., et al. Long noncoding RNA UCA1 promotes glutamine-driven anaplerosis of bladder cancer by interacting with hnRNP I/L to upregulate GPT2 expression. Transl. Oncol. 2022;17 doi: 10.1016/j.tranon.2022.101340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristensen L.S., et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y., et al. Circular RNAs: emerging cancer biomarkers and targets. J. Exp. Clin. Cancer Res. 2017;36(1):152. doi: 10.1186/s13046-017-0624-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng S., et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol. Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeck W.R., et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen T.B., et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Du W.W., et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng W.L., et al. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13(9):861–871. doi: 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 20.Legnini I., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panda A.C., et al. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2017;45(7):4021–4035. doi: 10.1093/nar/gkw1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu W., et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:18–27. doi: 10.1016/j.canlet.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Cheng Z., et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat. Commun. 2019;10(1):3200. doi: 10.1038/s41467-019-11162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao Y., Zhou Y., Hua Q. circRNA hsa_circ_0018414 inhibits the progression of LUAD by sponging miR-6807-3p and upregulating DKK1. Mol. Ther. Nucleic Acids. 2021;23:783–796. doi: 10.1016/j.omtn.2020.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu L., et al. Hypoxic tumor-derived exosomal circular RNA SETDB1 promotes invasive growth and EMT via the miR-7/Sp1 axis in lung adenocarcinoma. Mol. Ther. Nucleic Acids. 2021;23:1078–1092. doi: 10.1016/j.omtn.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Liang Y., et al. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol. Ther. Nucleic Acids. 2021;23:355–368. doi: 10.1016/j.omtn.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin M., et al. Upregulated circRNA ARHGAP10 Predicts an Unfavorable Prognosis in NSCLC through Regulation of the miR-150-5p/GLUT-1 Axis. Mol. Ther. Nucleic Acids. 2019;18:219–231. doi: 10.1016/j.omtn.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Song X., et al. circHMCU promotes proliferation and metastasis of breast cancer by sponging the let-7 family. Mol. Ther. Nucleic Acids. 2020;20:518–533. doi: 10.1016/j.omtn.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shao Y., et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173–1180. doi: 10.1002/cam4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rio D.C., et al. Preparation of cytoplasmic and nuclear RNA from tissue culture cells. Cold Spring Harb. Protoc. 2010;2010(6) doi: 10.1101/pdb.prot5441. pdb.prot5441. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H., et al. CircLIFR synergizes with MSH2 to attenuate chemoresistance via MutSα/ATM-p73 axis in bladder cancer. Mol. Cancer. 2021;20(1):70. doi: 10.1186/s12943-021-01360-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., et al. KLF15 suppresses cell growth and predicts prognosis in lung adenocarcinoma. Biomed. Pharmacother. 2018;106:672–677. doi: 10.1016/j.biopha.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Fu Q., et al. miRomics and proteomics reveal a miR-296-3p/PRKCA/FAK/Ras/c-Myc feedback loop modulated by HDGF/DDX5/β-catenin complex in lung adenocarcinoma. Clin. Cancer Res. 2017;23(20):6336–6350. doi: 10.1158/1078-0432.CCR-16-2813. [DOI] [PubMed] [Google Scholar]
- 34.Xue D., et al. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol. Cancer. 2019;18(1):151. doi: 10.1186/s12943-019-1072-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai Y., et al. MiR-296-3p regulates cell growth and multi-drug resistance of human glioblastoma by targeting ether-à-go-go (EAG1) Eur. J. Cancer. 2013;49(3):710–724. doi: 10.1016/j.ejca.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 36.Liu X., et al. MiRNA-296-3p-ICAM-1 axis promotes metastasis of prostate cancer by possible enhancing survival of natural killer cell-resistant circulating tumour cells. Cell Death Dis. 2013;4(11):e928. doi: 10.1038/cddis.2013.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X.W., Wang L., Ding H. Long noncoding RNA AK089579 inhibits epithelial-to-mesenchymal transition of peritoneal mesothelial cells by competitively binding to microRNA-296-3p via DOK2 in peritoneal fibrosis. FASEB J. 2019;33(4):5112–5125. doi: 10.1096/fj.201801111RR. [DOI] [PubMed] [Google Scholar]
- 38.Hu L., et al. Integrative microRNA and gene profiling data analysis reveals novel biomarkers and mechanisms for lung cancer. Oncotarget. 2016;7(8):8441–8454. doi: 10.18632/oncotarget.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The Roles of Hsa-Mir-296-3p in Cancers. Available from: http://atlasgeneticsoncology.org/Genes/GC_MIR296.html.
- 40.Dang D.T., Pevsner J., Yang V.W. The biology of the mammalian Krüppel-like family of transcription factors. Int. J. Biochem. Cell Biol. 2000;32(11–12):1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearson R., et al. Krüppel-like transcription factors: a functional family. Int. J. Biochem. Cell Biol. 2008;40(10):1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 42.Bureau C., et al. Expression and function of Kruppel Like-Factors (KLF) in carcinogenesis. Curr. Genom. 2009;10(5):353–360. doi: 10.2174/138920209788921010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida S., Sasaki S., Marumo F. Isolation of a novel zinc finger repressor that regulates the kidney-specific CLC-K1 promoter. Kidney Int. 2001;60(2):416–421. doi: 10.1046/j.1523-1755.2001.060002416.x. [DOI] [PubMed] [Google Scholar]
- 44.Sun C., et al. KLF15 inhibits cell proliferation in gastric cancer cells via up-regulating CDKN1A/p21 and CDKN1C/p57 expression. Dig. Dis. Sci. 2017;62(6):1518–1526. doi: 10.1007/s10620-017-4558-2. [DOI] [PubMed] [Google Scholar]
- 45.Sun C.X., et al. MiR-181a promotes cell proliferation and migration through targeting KLF15 in papillary thyroid cancer. Clin. Transl. Oncol. 2022;24(1):66–75. doi: 10.1007/s12094-021-02670-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets enrolled in this study are available in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/), The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) databases, and the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/bioproject/PRJNA863919).