Abstract
Cyanobacteria are ideal candidates to use in developing carbon neutral and carbon negative technologies; they are efficient photosynthesizers and amenable to genetic manipulation. Over the past two decades, researchers have demonstrated that cyanobacteria can make sustainable, useful biomaterials, many of which are engineered living materials. However, we are only beginning to see such technologies applied at an industrial scale. In this review, we explore the ways in which synthetic biology tools enable the development of cyanobacteria-based biomaterials. First we give an overview of the ecological and biogeochemical importance of cyanobacteria and the work that has been done using cyanobacteria to create biomaterials so far. This is followed by a discussion of commonly used cyanobacteria strains and synthetic biology tools that exist to engineer cyanobacteria. Then, three case studies—bioconcrete, biocomposites, and biophotovoltaics—are explored as potential applications of synthetic biology in cyanobacteria-based materials. Finally, challenges and future directions of cyanobacterial biomaterials are discussed.
Keywords: Cyanobacteria, Synthetic biology, Engineered living materials, Biomaterials, Sustainability, Carbon sequestration
Graphical abstract
1. Introduction
Cyanobacteria have used photosynthesis to shape the biogeochemical cycles of Earth for billions of years. As the first organisms to evolve oxygenic photosynthesis, this phylum of photoautotrophic bacteria was responsible for the massive oxygenation of the planet two billion years ago which allowed for aerobic multicellular life to develop. Their photosynthetic metabolism significantly reduced the amount of CO2 in the atmosphere [1] in multiple, drastic planetary CO2 reduction events [2]. Today cyanobacteria are integral to many biogeochemical processes; they are estimated to be responsible for 25% of primary productivity in the ocean [3] and play an important role in nitrogen fixation and the burial of organic carbon in ocean sediments [4].
To combat the threat of climate change due to increasing levels of CO2 in the atmosphere, researchers are exploring alternative production methods, which use CO2 as an input, rather than an output [5]. Photosynthetic carbon fixation enables many proposed methods for Carbon Capture, Utilization, and Storage (CCUS) [6] by using atmospheric or emitted CO2 as a building block to create products. CO2 might be stored in products temporarily, as in biofuels, to replace fossil fuel emissions with a carbon neutral product, or could potentially be stored long-term, as in building materials, creating a net-negative drawdown of atmospheric CO2 [7].
Cyanobacteria are a prime candidate for biological CCUS technology development for several reasons. These include inexpensive feedstock requirements (they require only sunlight, CO2, water and a few nutrients), ease of genetic manipulation, native production of commercially interesting biomolecules [8], and fast growth rate. In addition to their carbon capture potential, cyanobacteria are more sustainable than traditional microbes used for bioproduction because they do not require sugar as a feedstock (feedstock sugar requires large amounts of arable land to grow). Agriculture is predicted to face more challenges as climate change intensifies, making this feature increasingly important [9,10]. Additionally, many cyanobacteria grow in seawater salinity, so they can be grown at-scale without using limited freshwater resources [11].
A number of material scientists have made progress developing cyanobacteria-based biomaterials with applications in carbon capture, construction, energy, and food production. Many of these materials could be considered engineered living materials (ELMs) as they utilise living cyanobacteria to perform “smart” functions: assembly, repair, and response to external stimuli [[12], [13], [14], [148]]. Researchers have capitalized on the natural biomineralization of cyanobacteria to produce regenerative building material made of a hydrogel-sand scaffold and Synechococcus elongatus PCC 7002 [15]. A network of Anabaena sp. cells and graphene nanoribbons has been 3D printed onto a fungal platform to produce a photocurrent [16]. A prototype of a textile-based cyanobacteria biocomposite to capture CO2 during wastewater treatment has been fabricated [17]. Some cyanobacteria-based products have already reached commercial-scale: Spira uses genetically engineered cyanobacteria to produce food dye and flavorings [18], Prometheus Materials makes concrete masonry units (cinder blocks) using biomineralizing cyanobacteria [19], Lumen Bioscience has developed biologic drugs that use cyanobacteria to deliver therapeutic molecules [20], and Photanol produces industrial biochemicals extracted from cyanobacteria [21]. At the same time, synthetic biology researchers have developed a suite of genetic techniques and computational tools to manipulate cyanobacteria, and there is a growing repository of standard biological parts, genome editing tools, and metabolic models for several strains [22].
So far, there has been little intersection of materials science and synthetic biology in the creation of cyanobacteria-based biomaterials [23,24]. In this review, we will explore the ways in which cyanobacteria-based biomaterials can be improved using synthetic biology, discussing existing genetic, genomic, and computational tools, three case studies of potential applications of synthetic biology in cyanobacteria-based biomaterials development (bioconcrete, biocomposites, and biophotovoltaics), and future directions of the growing field of cyanobacteria-based biomaterials (Fig. 1).
Fig. 1.
Schematic illustration of synthetic biology tools that enable development of cyanobacteria-based biomaterials from lab prototypes to industry scale materials. Biomaterials illustrated (from left to right): Biocomposite of cyanobacteria grown on a loofah scaffold for enhanced CO2 capture [25] could scale to become part of an industrial carbon capture system for scrubbing CO2 from flue gas; Bioconcrete block made from biomineralizing cyanobacteria, sand, and gelatin [15] could scale to become a sustainable structural material that can “regrow”; a 3D printed microarray that captures current from photosynthesis of cyanobacteria colonies [26] could scale to become an efficient biophotovoltaic energy source.
1.1. Cyanobacteria vs. other photosynthetic organisms
Several other photosynthetic organisms have been explored as potential photosynthetic chassis for biocarbon capture, notably plants and eukaryotic microalgae. Plants produce many useful biomolecules (terpenes, fatty acids, phenylpropanoids) [27], and their macroscopic structure can be utilized to easily form large biomaterials [28,29]. However, a plant-based carbon capture system at scale would require large amounts of arable land and their extremely slow growth makes them difficult to engineer [30]. The latter is also a limitation for macro-algae (e.g kelp), which have been investigated for their carbon sequestration abilities [31].
The term ‘microalgae’ often refers to unicellular eukaryotic algae, but may be used more broadly to describe both cyanobacteria and eukaryotic microalgae. This review will use the former definition. Microalgae and cyanobacteria share many of the same characteristics: photosynthesis, adaptability to diverse environmental conditions, and natural production of high-value molecules. They are often used for similar applications [32,33]. Sometimes they are even used together such as in a carbon-capture biocomposite made of a loofah scaffold and cyanobacteria and microalgae consortium [25]. However, each organism has distinct advantages. Cyanobacteria display the fastest photosynthetic growth rates measured [34] and their smaller genomes, lack of subcellular organization, and absence of epigenetic gene-silencing allows them to be genetically manipulated more easily [[35], [36], [37]]. Because they are easier to engineer, more genetic and computational tools have been developed for cyanobacteria than microalgae. Microalgae, on the other hand, may tolerate high light conditions and have more examples of growth at scale [38]. Other factors to consider include that cyanobacteria are reported to have high rates of UV-induced mutation [39] and some produce secondary metabolites that are harmful to humans under certain conditions [40].
2. The cyanobacteria synthetic biology toolkit
Engineering an organism beyond its natural capabilities is integral to developing innovative biomaterials. In the development of cyanobacteria-based biomaterials, desired engineered characteristics could range from the ability to produce non-native compounds, to changing phenotype based on environmental stimuli. The “design-build-test-learn” cycle of synthetic biology [41] is a model for an engineering workflow used to create new biological behavior and is made up of the following components: 1) a chassis/host cell with a characterized genome; 2) standardized biological parts compatible with the host; 3) genome editing to introduce biological parts and; 4) computational tools to aid interpretation of results and improve future designs [[42], [43], [44]]. Although not as characterized as tools for model organisms yeast and E. coli, the advances toward cyanobacteria synthetic biology toolkit are detailed below.
2.1. Choosing a cyanobacteria species
The phylum cyanobacteria is made up of over 6000 species [45] that have diverse phenotypes and natural environments. All cyanobacteria are gram-negative bacteria capable of performing oxygenic photosynthesis, but they vary widely in other traits: some are unicellular, some filamentous; some can form biofilms; some fix nitrogen; many prefer aquatic environments, but some are found in terrestrial environments as well. Only a few of these species have been developed as synthetic biology targets so far. Traditional model strains include Synechococcus elongatus PCC 7942, Synechococcus elongatus PCC 7002, and Synechocystis sp. 6803. Additionally, Anabaena sp. PCC 7120 has been commonly used as a model for filamentous nitrogen-fixing cyanobacteria. (See Table 1 for characteristics of commonly used strains).
Table 1.
Characteristics of common cyanobacteria strains.
| Strain | Doubling time (hours) | Morphology | Natural competency | Polyploidy (number of genome copies) | Salt tolerance | Metabolic model | Notes | Biomaterial references |
|---|---|---|---|---|---|---|---|---|
| Synechococcus elongatus PCC 7942 | 12 [56] | unicellular | conjugation, natural transformation, electroporation [22] | 3-6 [56] | freshwater [22] | yes [57] | carbon-capture biocomposite [25], bionic battery [58], 3D-printed biomaterial [59] |
|
| Synechococcus elongatus PCC 7002 | 3.5 [56] | unicellular | conjugation, natural transformation [22] | 6 [56] | marine [22] | yes [60] | bioconcrete [15] | |
| Synechocystis sp. PCC 6803 | 6 [56] | unicellular | conjugation, natural transformation, electroporation, ultra sonic transformation [22] | 60-225 [56] | freshwater [22] | yes [61] | biophotovoltaic [26], biophotovoltaic [62], bioconcrete [63] | |
| Anabaena (Nostoc) sp. PCC 7120 | 24 [56] | filamentous | conjugation, electroporation [22] | 8-10 [56] | freshwater [22] | yes [64] | nitrogen fixation ability [56] | 3D-printed biomaterial [16] |
| Synechococcus elongatus UTEX 2973 | 1.9 [34] | unicellular | conjugation [22], | 3-6 [22] | freshwater [22] | no | fastest growth rate under laboratory culture conditions [34] | |
| Synechococcus sp. PCC 11901 | 2-3 [51] | unicellular | natural transformation [51] | unknown | marine [51] | no | produces the most dry cell biomass under laboratory culture conditions [51] | |
| Synechococcus elongatus PCC 11801 | 2.3 [52] | unicellular | natural transformation, conjugation [52] | unknown | freshwater [52] | no | closely related to PCC 11802, recently reported to perform better under outdoor cultivation conditions [52] |
The genus Prochlorococcus exemplifies the need to continue development of genetic editing tools for cyanobacteria. Prochlorococcus are the most abundant cyanobacterium on earth [46], and they play a major role in the biological carbon pump [47] and as primary producers in ocean ecosystems. They also have the smallest genome of any photosynthetic organism, making them interesting models to study genome-reduction as a strategy in optimizing biotechnology [48]. However, after over a decade of work there has been little progress in developing tools that work to manipulate Prochlorococcus. This is due to their extremely slow doubling time, sensitivity to contamination and growth conditions, and resistance to transformation methods [49,50]. Given their ecological importance and desirable traits, continued efforts should be made to harness this recalcitrant genus.
Growth rate is a very important factor in choosing a chassis, as most model cyanobacteria have doubling times of 7–15 h, making it difficult to quickly prototype experiments [22]. A recent discovery of several species capable of doubling every ∼2 h (a rate comparable to the popular synthetic biology workhorse Saccharomyces cerevisiae), Synechococcus elongatus UTEX 2973 [34], Synechococcus elongatus PCC 11901 [51], and Synechococcus elongatus PCC 11801 and PCC 11802 [51,52], allows for a much faster cycle of “design-build-test-learn” in cyanobacterial synthetic biology. Polyploidy (having multiple genome copies in a cell) is a common trait in cyanobacteria, with most species having between 3 and 200 genome copies per cell [53]. The function of polyploidy has been associated with enhanced stress tolerance and increased metabolic output [54]. However, a cell with many genome copies is much more time consuming to genetically engineer, as it requires repeated selection steps in order to obtain a homozygous strain that has the desired edit in every genome copy. Advances have been made in alternative genetic editing methods that address the polyploid nature of cyanobacteria (see section 2.3.4. Genome Engineering). Continuing to make a strain that is tolerant of extreme environmental conditions—salinity, light, temperature, and moisture levels—is needed for eventual environmental use of cyanobacteria-based biomaterials. Salt tolerance is an especially important trait given that large-scale cyanobacteria growth will require large amounts of water. This could be solved by either using a marine strain as a chassis, or by optimizing a freshwater strain to tolerate high salinity (via overexpression of proteins involved in ion transport and molecular chaperones) [55].
2.2. A synthetic biology primer
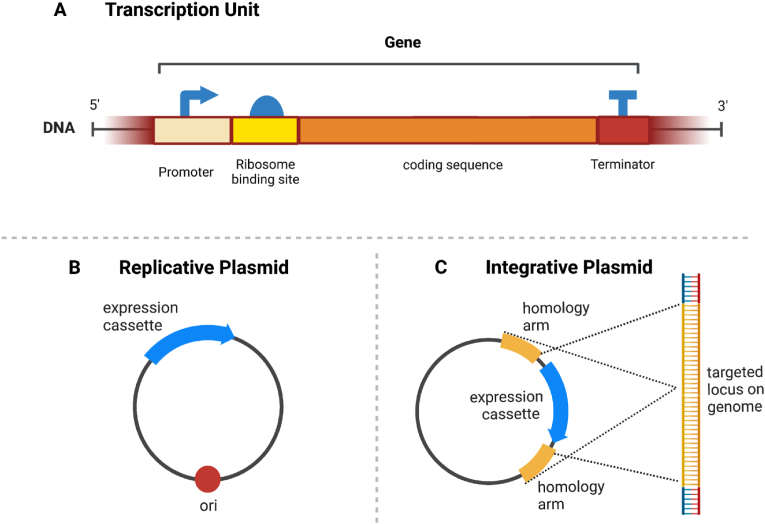
Synthetic biology, the rational engineering of cells to do useful things, is a bottom-up approach based on the foundational idea that standardized molecular “parts” can be developed from studying natural organisms [65]. These parts can be combined in circuits to control a cell's behavior, much like the programming of electronic circuits. The most basic parts—the promoter, RNA-coding sequence, and terminator—combine to form a “transcription unit,” a sequence of DNA that codes for the transcription of an RNA molecule (Fig. 2A). This RNA will then be translated into a protein which provides some functionality in the cell, or it will act to knock out the function of a native gene. Different combinations of these parts can produce complex synthetic devices: cellular logic gates, switches, amplifiers, memory elements and oscillators, as well as non-natural metabolism [66] .
Fig. 2.
Transcription unit and plasmid types. (A) Diagram of a transcription unit, or segment of DNA that encodes a RNA molecule. The unit contains a promoter, ribosome-binding site, RNA-coding sequence, and a terminator. (B) A replicative plasmid has an origin of replication that enables its independent replication in the host cell. (C) An integrative plasmid has homology arms that promote homologous recombination with a targeted region on the host genome.
The field is exemplified by two breakthrough genetic circuits developed in the early 2000s: the Collins group's construction of a genetic toggle switch that caused a cell to switch between two different states (e.g., producing or not producing a fluorescent protein) in response to external stimuli (e.g., heat) [67], and the Elowitz lab's “repressilator,” a three-part repressor system that induces periodic oscillations in production of a protein [68].
Today, synthetic biology has been used to create many successful commercial products, from medicines to food products to biomaterials [11]. However, compared to model organisms such as E. coli and S. cerevisiae, relatively few biological parts and assembly methods developed for cyanobacteria so synthetic biology in cyanobacteria has been limited. Additionally, the thousands of parts that have been characterized for model organisms (see the iGEM BioBrick database for a large collection [69]) often do not function in cyanobacteria [70].
The attraction in using cyanobacteria to convert CO2 into useful bioproducts has led to increased genetic tool developments in recent years, by adapting existing parts from other species or deriving novel parts from natural cyanobacteria sequences. Most tools have been characterized only in specific strains and are not guaranteed to function in diverse cyanobacteria. This host-dependency limits the speed at which novel chassis strains are developed, because each new species requires the testing and development of its own “toolkit” [[71], [72], [73]].
2.3. Synthetic biology in cyanobacteria
The below section is an overview of synthetic biology techniques developed in cyanobacteria. First, common biological parts and their use in cyanobacteria are discussed (promoters, terminators, ribosome-binding sites, selection markers, origins of replication, and homology arms). The next section highlights methods to assemble these parts into functional genetic vectors, deliver the vectors into the cell (transformation), and, once in the cell, to localize the vector to a specific target on the genome (homologous recombination and CRISPR). Finally, co-culture techniques and computational tools are discussed. For a thorough characterization of synthetic biology techniques characterized in cyanobacteria see Sengupta et al. and Sun et al. [33,74].
2.3.1. Standardized biological parts
Promoters are biological parts that control downstream gene expression and can either be constitutive or inducible under certain environmental conditions such as light, temperature, or the presence of a chemical. Promoters have been characterized in cyanobacteria that are induced by light intensity, CO2 level and time of day. Many of the promoters used in cyanobacteria have been adapted from the extensive promoter libraries for E. coli, for example a tetracycline and IPTG-regulated promoters have been adapted from E. coli and found to work in several cyanobacteria [72,74]. However, promoter function often varies between species, so cyanobacteria promoter libraries that have been tested on multiple strains need to be developed. A suite of inducible and constitutive promoters that function in multiple cyanobacteria was assembled as part of the CyanoGate kit [75].
Ribosome Binding Sites (RBS) are transcribed and recognized by ribosomes to initiate translation. RBS can be edited to affect translation efficiency, allowing for fine tuning the expression of a protein. The strength of 20 RBS sequences native to Synechocystis sp. 6803 has been determined [76]. Several online calculators exist for predicting RBS sequences. However, these tools were developed in E. coli and do not currently make predictions as reliably for cyanobacteria [77].
Terminators are sequences found downstream of a gene that end transcription of that gene by triggering the release of the gene transcript from the transcription complex. They have varying efficiencies, so it is important to test the strength of a terminator in your host organism. The strength of 34 terminators in Synechococcus elongatus UTEX 2973 and Synechocystis sp. PCC 6803 has recently been evaluated [78].
Selection Markers are genetic elements on a plasmid that allow for the selection of cells that have taken that plasmid. The most commonly used selection markers are antibiotic resistance genes; by including a gene that confers resistance to an antibiotic on a plasmid construct, transforming that plasmid into a population of cells, then treating the population with the antibiotic, only the cells that have taken up the plasmid will survive. Antibiotics commonly used in cyanobacteria include ampicillin (Ap), chloramphenicol (Cm), erythromycin (Em), gentamicin (Gm), kanamycin (Km), neomycin (Nm), spectinomycin (Sp), and streptomycin (Sm) [79]. Light sensitive antibiotics such as tetracycline (Tc) and rifampicin (Rf) are not effective in cyanobacteria given their high-light growth requirements [80].
Origins of Replication (ori) are sequence sites at which replication of a plasmid is initiated. Ori are included on plasmids to enable replication within the host without integrating into the host genome (Fig. 2B). Replicative plasmids are useful for rapid prototyping of a desired function and the chosen ori determines the copy number (number of plasmids per cell). However, the exact copy number of the introduced gene is variable in replicative plasmids so they cannot be used to achieve precise gene expression levels. Additionally, replicative plasmids need to be maintained with a selection marker to ensure they are passed down to daughter cells. Several plasmid origins of replication (the backbone of the plasmid that is recognized by host cell replication machinery) have been found that function well in cyanobacteria: RSF1010 across clades [81], pCA and pCB for Synechocystis sp. PCC 6803 [76], and pANS in Synechococcus elongatus PCC 7942 [82].
Homology Arms are sequences on a plasmid complementary to a target locus on a genome. They flank an expression cassette (a gene of interest and regulatory elements) to allow for its integration into the host genome at the locus (Fig. 2C). Integrative plasmids undergo homologous recombination to become a part of a cyanobacteria's genome. They are ideal to permanently introduce a function to a cyanobacteria cell line and to do gene knockout (as they replace native genes on the chromosome) for loss-of-function studies. For example, pSyn6 is a commonly used integrative vector in Synechococcus [83] and has built-in homology arms for integrating genetic payloads to a neutral site on the genome. However, it is more time-consuming to acquire homozygous, fully-segregated transformants using integrative plasmids as cyanobacteria are polyploid and several passages have to be taken to ensure the plasmid is integrated into all copies of the genome in a cell.
2.3.2. Assembly of functional constructs for genetic engineering
The parts described above can be assembled into genetic constructs (e.g plasmids) designed for specific functions within the host. Common DNA assembly methods are BioBrick Assembly [84], Gibson Assembly [85], and Golden Gate Assembly [86]. Two notable platforms for designing and assembling genetic vectors for cyanobacteria are CYANO-VECTOR [79] and CyanoGate [75]. In 2014, the cyanobacteria vector assembly platform CYANO-VECTOR [79] was developed using the BioBrick standard for compatible parts [84]. CYANO-VECTOR provides a platform for the construction of modular plasmids from a collection of biological parts including: origins of replication, antibiotic resistance markers, reporter genes, promoters, and RBSs. The parts have been tested in a range of cyanobacteria species (Anabaena sp. PCC 7120, Leptolyngbya sp. BL 0902, Nostoc punctiforme ATCC 29133, and Synechocystis sp. PCC 6803). CyanoGate is another modular platform that allows for standardized assembly of a library of genetic parts compatible with cyanobacteria [75]. Based on the plant Golden Gate MoClo kit, CyanoGate is a library of standardized promoters, terminators, antibiotic resistant markers, neutral sites (NS), and gene repression systems that can be assembled into vectors. Platforms like CYANO-VECTOR and CyanoGate provide the tools to do synthetic biology on characterized cyanobacteria strains. For example, Puzorjov et al. recently used CyanoGate to insert a phycobiliprotein gene (used to produce a high commodity dye derived from light harvesting complexes) from a thermophilic cyanobacteria into Synechocystis sp. PCC 6803, a mesophilic species more amenable to industrial production of the dye [87].
2.3.3. Construct delivery
There are several methods to deliver a DNA construct into a bacterial host. Some hosts can undergo natural transformation (they naturally uptake plasmids from the surrounding environment). Others require electroporation or chemical transformation to artificially increase the bacteria's membrane permeability. Conjugation, or the transfer of genetic material from one bacteria to another via direct cell to cell contact, is a technique used for hosts recalcitrant to other transformation methods. Some cyanobacteria (such as the popular Synechococcus elongatus PCC 7002, Synechococcus elongatus PCC 11901, Synechocystis sp. PCC 6803) are naturally competent, but others require facilitated transformation methods such as conjugation (Synechococcus elongatus UTEX 2973 and Anabaena sp. PCC 7120) or electroporation (Anabaena sp. PCC 7120). All of these methods rely on selective markers, of which there are limited characterized in cyanobacteria. Additionally, there is the worry of releasing antibiotic resistance genes into the environment for some applications. In response to this, there have been several markerless selection systems developed in cyanobacteria [88,89]. Free fatty acids have been used to select for markerless aas knockout mutants in both Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7002 [90]. And recently, a CRE recombinase in a two-plasmid system has been designed to generate markerless mutants in Synechocystis sp. PCC 6803 [91].
2.3.4. Genome engineering
Once a DNA construct is delivered into a cell, it can either continue to replicate independently of the host cell's genome (via a replicative plasmid) or it can be integrated into the host genome. Integration via homology arms on the plasmid is a simple way to integrate a gene of interest into a specific location on the genome, but homologous recombination has a low efficiency that requires several rounds of passaging to ensure every genome copy in cyanobacteria has been edited.
CRISPR-Cas9 is a promising genome-editing tool in cyanobacteria. It can simultaneously edit multiple genes and the double-stranded breaks it causes at a genome target site are toxic to cells, providing a counter selection strategy to allow for a faster segregation process [92]. CRISPR tools for cyanobacteria are in early stages of development; CRISPR-Cas9 has been used successfully to increase succinate production in Synechococcus elongatus PCC 7942 [93] and free fatty acid production in Synechococcus elongatus UTEX 2973 [94]; however, high levels of the Cas9 protein are toxic to most cyanobacteria [95]. Solutions to Cas9 toxicity have been explored. An alternative endonuclease, Cas12a, has been used successfully in Synechococcus elongatus UTEX 2973, Synechocystis sp. PCC 6803 and Anabaena sp. PCC 7120, though editing efficiencies are often lower than in comparable systems in other bacteria [96,97]. An inducible riboswitch-based CRISPR/Cas9 system was recently developed in Synechocystis sp. PCC 6803. The ability to tightly control of Cas9 expression allowed for reliable editing efficiency. CRISPR interference (CRISPRi) is another CRISPR-associated technology that utilizes a dead version of the Cas9 enzyme to inhibit expression of specific genes [92]. CRISPRi has been used in several cyanobacteria including Synechocystis sp. PCC 6803 [98], Synechococcus elongatus PCC 7942 [99], and Anabaena sp. PCC 7120 [100].
2.3.5. Community engineering and co-culture
When engineering cells for a specific function, a co-culture system, in which two or more microorganisms are grown together, can sometimes be advantageous over monoculture. It is difficult to optimize a single chassis for multiple objectives, since optimizing one trait (e.g. CO2 uptake in photosynthesis) may come at the expense of other desired metabolic traits (e.g., producing terpenes from the fixed carbon). Co-cultures allow for the achievement of complex production objectives because they allow for the division of labor and the optimization of different traits across the co-culture partners. They are also more resilient than monocultures to contamination and other environmental stress [101]. However, it is time-consuming to adjust the environmental co-culture conditions (temperature, pH, light) to allow for all species to grow successfully. Additionally, inoculation ratios of the partners need to be considered or the system will be unbalanced and one species will out-compete the others for resources,
Cyanobacteria co-cultures have been designed with yeast, microalgae, and other bacteria [[102], [103], [104]]. In a co-culture of Synechococcus elongatus PCC 7942 and various yeast strains, PCC 7942's ability to convert CO2 to sucrose was paired with the yeast's ability to ferment sucrose into biofuel precursor lipids. It was found that the co-culture produced more biomass and biofuel lipids than the PCC 7942 grown on its own and that the yeast limited the amount of toxic reactive oxygen species in the culture, thus facilitating long term PCC 7942 growth [105]. A bio-battery has been developed that uses a four species co-culture of Synechococcus elongatus PCC 7942, E. coli, Shewanella oneidensis, and Geobacter sulfurreducens to produce electricity by mimicking the photoelectric conversion in marine mats [58]. Recently, a “living” bandage made up of a hydrogel encapsulating a consortium of Synechococcus elongatus PCC 7942 and a heterotroph was used to promote wound healing [106].
2.3.6. Genome sequencing
Advances in the quality and affordability of genome sequencing has allowed for the ongoing discovery of novel cyanobacteria strains with useful traits. Recent developments in in situ portable NanoPore sequencing even allows for in the field sequencing [107]. The CyanoBase database contains genomes of over 376 cyanobacteria strains (86 complete and 290 draft) [108]. The Pasteur Culture collection of Cyanobacteria also contains over 100 sequenced genomes [109]. As sequencing has become increasingly routine, the public availability of more and more cyanobacteria species genomes is a necessary step to begin developing synthetic biology tools for these new species [34,51,52].
2.3.7. Computational simulation for metabolic engineering
Computational simulations can reduce the number of wet lab experimental iterations required for designing genetic engineering by predicting the metabolic manipulation required to achieve a specific phenotype. Genome-Scale Metabolic Models (GEMs) use the annotated genome of an organism to construct a simulated network of all reaction pathways within an organism [110]. A GEM can then be optimized (commonly using flux balance analysis [111]) to simulate the growth of the organism in a specific media and the organism's net metabolite secretion and consumption. Many cyanobacteria genomes have been sequenced. However, GEMs have been reconstructed for only a few model cyanobacteria species (see Hendry et al. for existing cyanobacteria GEMS [112]). Various design tools exist that determine which genetic modification (e.g., deleting genes, upregulating gene expressions) causes a desired phenotype in a GEM. These include OptGene, OptKnock, OptForce, and MOMA [112]. There are limitations to keep in mind when using GEMs as simulations of genetic modifications for an organism. GEMs simulate metabolic behavior and do not capture other factors such as gene regulation and environmental conditions. Additionally, as GEMs are derived from genome annotations, poor genomic annotations can lead to inaccurate predictions.
3. Cyanobacteria-based biomaterials: Three case studies
Now we will highlight three existing cyanobacteria-based biomaterial—bioconcrete, carbon capture biocomposites, and biophotovoltaics—and discuss how synthetic biology techniques can be used to optimize each biomaterial for industrial applications (Fig. 3).
Fig. 3.
Images and diagrams of three of existing cyanobacteria-based biomaterials: bioconcrete, a carbon capture biocomposite, and a biophotovoltaic. (A) Top: image of a bioconcrete block [15]. Bottom: diagram of internal structure of bioconcrete: cyanobacteria induced CaCO3 precipitation and the crosslinking of the gelatin scaffold. (B) Top: image of a loofah scaffold for cyanobacteria carbon capture [25]. Bottom: diagram of internal structure of the biocomposite: cyanobacteria cells trapped on the loofah surface by a latex coating (C) Top: image of a 3D printed microarray biophotovoltaic device [26]. Bottom: diagram of flow of electrons starting from water oxidation due to cyanobacterial photosynthesis, electrons, then collected at the anode and transferred to a cathode where oxygen is reduced.
3.1. Bioconcrete
Synechococcus induce biomineralization of CaCO3 because they naturally raise the pH of their extracellular environment during photosynthesis. This spontaneous CaCO3 precipitation has been used to bind a sand-hydrogel matrix resulting in a “bioconcrete” [15] (Fig. 3A). The bioconcrete could be cast in various molds, and biotic components could regenerate for three generations. The group showed that after splitting a brick in half, the second half grew back if additional abiotic material was added. The potential for bioconcrete as a sustainable building material is considerable given that traditional cement production contributes to 8% of anthropogenic CO2 emissions annually [113].
However, the existing material needs to be optimized in order to be competitive with traditional Portland cement. Specifically, the material's tensile strength could be improved by increasing CaCO3 precipitation further. One potential way to do this is to use carbonic anhydrase, a common enzyme that promotes CaCO3 precipitation. Engineering cyanobacteria to secrete carbonic anhydrase or to display the enzyme on the outer membrane could promote additional crystallization in the hydrogel matrix, strengthening the material. Another problem with the current material production method is that the Synechococcus requires a high moisture content to remain viable for regeneration (50–100% relative humidity). Genes associated with desiccation resistance in certain filamentous cyanobacteria have been characterized, and introducing them into Synechococcus could allow for the strain to remain viable in the cement for much longer [114]. Additionally, incorporating genetic circuits that are induced by external stimuli (such as red light [115] or pollutants [116]) into the cyanobacteria would allow for the development of a “smart” bioconcrete that can sense and respond to the environment.
3.2. Biocomposites for carbon capture
Given cyanobacteria's unparalleled photosynthetic efficiency, there have been many attempts at developing cyanobacteria cultivation systems for carbon sequestration. Where raceway ponds or photobioreactors cultivate cyanobacteria in a mixed, planktonic suspension, biocomposites are an alternative wherein a coating containing an organism is fixed to a non-living scaffold. Such alternative cultivation methods that intensify biological reactions (e.g., photosynthesis) [117,118]. A novel carbon-capture biocomposite has been designed that uses a bio-latex binder to encapsulate cyanobacteria onto a loofah sponge scaffold [25] (Fig. 3B). The porous nature of Loofah, or dried Luffa fruit, gives it a large high surface and excellent aeration for cyanobacterial photosynthesis. Simulating flue gas conditions, they found that this biocomposite allowed for Synechococcus to fix carbon 5–10 times more efficiently than suspension methods with less contamination and less water use. Their system used wildtype strains, but updating it with a strain engineered to sequester CO2 into a useful bioproduct (such as ethanol [119] or sucrose [120]) could make their biocomposite commercially viable. Alternatively, the system could be adapted for long term carbon sequestration and storage if the engineered cyanobacteria could use the fixed carbon to promote biomineralization or synthesizing recalcitrant biopolymers that are not easily biodegradable (see Section 3.1 for methods). One issue with the biocomposite is the weakening of adhesion and loss of some cells from the binder into the growth media after several weeks. This release from encapsulation decreases the carbon fixation efficiency of the bio-composite. A potential solution is to either use cyanobacteria that naturally form biofilms or to engineer the secretion of extracellular polymeric substances (EPS) in Synechococcus to promote adhesion.
3.3. Biophotovoltaics
In biofilms certain cyanobacteria transfer electrons across their membranes during photosynthesis, producing an electrical photocurrent. This photocurrent can be “harvested” when a biofilm is grown on an electrode [121]. 3D-printed electrode pillars have been fabricated to efficiently harness photocurrent from a biofilm of Synechocystis sp. PCC 6803 or Nostoc punctiforme. They used an aerosol jet printing method with indium tin oxide particle “ink” to print a novel branched pillar structure that allowed for light and electrolyte penetration throughout. Their photocurrent outputs are the highest that has been reported in semi-artificial photosynthesis systems, and rival the efficiency of existing biofuel systems [26] (Fig. 3C). In order to scale this technology to be competitive with other energy sources, improvements in electron transfer in these materials will need to be made. Work has already been done in genetically encoding outer membrane proteins that enhance extracellular electron transfer [122] and incorporating abiotic materials like carbon nanotubes to improve function [123].
4. Future directions
With continued work across the fields of materials science and synthetic biology, in the future cyanobacteria-based biomaterials can impact many sectors, from energy to agriculture, from medicine to construction (Fig. 4).
Fig. 4.
Schematic of future directions of cyanobactera-based biomaterials: expanded genetic toolbox, intracellular biomaterials, and carbon cycle engineering (A) Expansion of the cyanobacteria engineering toolbox. Cyanophages mediate horizontal gene transfer in the ecosystem and could be used for delivering genetic payloads.Transposon mutagenesis introduces random insertions on the genome, leading to disruption or activation of neighboring genes. Recombineering uses phage-derived proteins, including single-stranded DNA annealing protein (SSAP), to enable targetable, multiplex genome editing. (B) Intracellular biomaterial engineering. Cyanbacterial gas vesicles can be extracted (or expressed in living cells) and used as contrast agents in MRI scans. Genes that form the cyanobacterial carboxysome can be inserted into plants to increase the photosynthetic efficiency. (C) Altering the global carbon cycle with cyanobacteria biomass sinking in the ocean: engineering cyanobacteria to increase ocean alkalinity and capture CO2 as a CaCO3 shell or other recalcitrant biopolymers would cause cells to sink in the water column, eventually becoming trapped in deep sea ocean sediments and sequestering the carbon.
4.1. Expansion of the cyanobacteria engineering toolbox
Continued development of cyanobacteria engineering tools will allow us to manipulate these organisms more efficiently (Fig. 4A). Directed Evolution (DE) is a process of harnessing evolution to create optimized or novel biological functions. In classical DE, there are manual, iterative cycles of mutation and selection. Genes of interest can be mutagenized in vitro and the resulting variants inserted into host organisms where they are screened for desired function [124], or DE can be an untargeted way in which the entire organism's genome is mutagenized in vivo. DE has been used to increase high light tolerance, alcohol tolerance, and salt tolerance in cyanobacteria [[125], [126], [127]]. Traditional DE is a labor-intensive process, requiring iterative rounds of in vitro mutagenesis and selective cycles. To streamline this process, continuous DE techniques such as PACE (Phage Assisted Continuous Evolution) [128] have been developed that complete the mutation/selection cycle rapidly in vivo [129]. Perhaps high throughput culture platforms like the eVOLVER (which automates mating, temperature control, stirring, diluting and cleaning in yeast culture) could be adapted for use in cyanobacteria to automate the continuous DE process even further [130].
Additionally, current mutagenesis techniques for directed evolution (error prone PCR, and chemical mutagenesis) create point mutations within single genes or across genomes. Transposon mutagenesis allows for larger-scale mutations: entire gene knockouts or activation of naturally silenced genes [131,132] Transposon mutagenesis has already been shown to work in cyanobacteria, a transposon sequencing (TnSeq) technique was used to quickly screen Synechococcus elongatus PCC 7942 for essential and non essential genes [133], as well as enriching models of metabolism [57].
Expansion of rational genome engineering techniques is also needed. A recombination-mediated genetic engineering approach like multiplex automated genome engineering (MAGE) would allow for high throughput editing of multiple sites in the genome at once [134]. One relevant application of MAGE is it allows for the recoding of an organism's genome to rely on synthetic amino acids [135]. Recoding is a powerful biocontainment approach and could be applied as a biosafety measure to engineered living cyanobacteria-based materials. Exploiting the natural role of viruses in controlling cyanobacteria ecology and evolution through cyanophage mediated delivery of genes is a promising approach for genetic manipulation of cyanobacteria on an ecological scale. A preliminary cyanophage genetic engineering method has been developed [136].
4.2. Intracellular biomaterials
Cyanobacteria have microcompartments made up of a protein shell surrounding enzymes that serve a specific function, like an organelle in a eukaryotic cell. Two of these microcompartments, the carboxysome and gas vesicles, are strong candidates for biotechnological applications (Fig. 4B).
The carboxysome is an intracellular icosahedral protein compartment that concentrates CO2 near the carbon-fixing enzyme Rubisco, enabling efficient carbon fixation in cyanobacteria. There has been much interest in introducing carboxysomes into plant chloroplasts that could potentially improve carbon fixation in plants [137]. Recently, a minimal set of four carboxysome genes successfully produced simplified carboxysome structures when inserted into tobacco [138]. However, there is still much characterization of carboxysome associated proteins needed in order to construct functional carboxysomes in non-cyanobacteria. Carbonic anhydrase, the enzyme which interconverts carbon dioxide and bicarbonate within the carboxysome, has been explored as a possible component of biomimetic CO2 capture reactors [139].
Gas vesicles are small cylindrical protein structures in cells that provide buoyancy, allowing cyanobacteria to move vertically in water columns [140]. It has been found recently that purified vesicles from cyanobacteria can scatter sound, making them candidate contrast agents for non-invasive ultrasound imaging. There has been initial work done on using purified cyanobacteria gas vesicles in medical devices as a stable-high contrast agent for ultrasounds and MRIs [141]. Additionally, a recent in vivo study has shown gas vesicle's potential to track microbial cell location in live mammalian hosts [142]. Engineering cyanobacteria to regulate their production of gas vesicles in response to certain stimuli could allow for biomaterials with responsive buoyancy.
4.3. Global carbon cycle engineering
In the distant future, we could even use cyanobacteria to alter biogeochemical cycles in order to combat atmospheric CO2 imbalance on a global scale (Fig. 4C). The biological carbon pump is the sequestration of atmospheric carbon by photosynthetic organisms which then aggregate, and sink to the seafloor, where the carbon remains inert as recalcitrant biopolymers or precipitated CaCO3. This aggregation is facilitated by the secretion of transparent exopolymer particles (TEP) [143] which act as a glue binding cells and other particles. However, carbonate biomineralization could reduce the total alkalinity in the ocean and decrease dissolved carbon [146]. It is important to evaluate the global impact of engineered cyanobacteria in a contained environment before involving the open ocean [147].
Ideally, coupled with ocean alkalinity enhancement, engineered marine cyanobacteria could use enhanced recalcitrant biopolymer synthesis, tunable biomineralization, controllable buoyancy, and altered life cycle to promote fixed carbon being exported to the deep ocean. Potentially, engineered cyanophage could also increase the amount of marine cyanobacteria that are lysed, aggregate, then fall to the ocean floor as part of the “viral shunt” [47,144].
5. Conclusion
With more CO2 being emitted into the atmosphere each year, cyanobacteria-based biomaterials have the power to play an important role in mitigating the effects of climate change on our planet through carbon capture. But to fully unlock the potential of these powerful microbes, more work needs to be done in characterizing novel useful cyanobacteria and in developing standardized tools and parts that work for multiple species (especially ecologically relevant species like Prochlorococcus) [75]. Innovation in and integration of materials science and synthetic biology will allow for cyanobacteria-based biomaterials to develop from the lab bench to fully scaled industrial applications.
CRediT author statement
Isabella M. Goodchild-Michelman: Conceptualization, Writing - Original Draft, Visualization, George M. Church: Writing - Review & Editing, Max G. Schubert: Conceptualization, Writing - Review & Editing, Supervision, Tzu-Chieh Tang: Conceptualization, Writing - Review & Editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Funding for this work was provided by the US Department of Energy (DOE) under grant no. DE-FG02-02ER63445 and by the National Science Foundation (NSF) award no. MCB-2037995 (both to G.M.C.). The figures and graphical abstract were created using BioRender.com.
Contributor Information
Max G. Schubert, Email: mgschubert@gmail.com.
Tzu-Chieh Tang, Email: zijaytang@gmail.com.
Data availability
No data was used for the research described in the article.
References
- 1.Sánchez-Baracaldo P., Cardona T. On the origin of oxygenic photosynthesis and Cyanobacteria. New Phytol. 2020;225:1440–1446. doi: 10.1111/nph.16249. [DOI] [PubMed] [Google Scholar]
- 2.Li F.-W., Brouwer P., Carretero-Paulet L., Cheng S., de Vries J., Delaux P.-M., Eily A., Koppers N., Kuo L.-Y., Li Z., Simenc M., Small I., Wafula E., Angarita S., Barker M.S., Bräutigam A., dePamphilis C., Gould S., Hosmani P.S., Huang Y.-M., Huettel B., Kato Y., Liu X., Maere S., McDowell R., Mueller L.A., Nierop K.G.J., Rensing S.A., Robison T., Rothfels C.J., Sigel E.M., Song Y., Timilsena P.R., Van de Peer Y., Wang H., Wilhelmsson P.K.I., Wolf P.G., Xu X., Der J.P., Schluepmann H., Wong G.K.-S., Pryer K.M. Fern genomes elucidate land plant evolution and cyanobacterial symbioses. Nat. Plants. 2018;4:460–472. doi: 10.1038/s41477-018-0188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flombaum P., Gallegos J.L., Gordillo R.A., Rincón J., Zabala L.L., Jiao N., Karl D.M., Li W.K.W., Lomas M.W., Veneziano D., Vera C.S., Vrugt J.A., Martiny A.C. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sánchez-Baracaldo P., Bianchini G., Wilson J.D., Knoll A.H. Cyanobacteria and biogeochemical cycles through Earth history. Trends Microbiol. 2022;30:143–157. doi: 10.1016/j.tim.2021.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Campbell J.S., Foteinis S., Furey V., Hawrot O., Pike D., Aeschlimann S., Maesano C.N., Reginato P.L., Goodwin D.R., Looger L.L., Boyden E.S., Renforth P. Geochemical negative emissions technologies: Part I. Review. Front. Clim. 2022;4 https://www.frontiersin.org/articles/10.3389/fclim.2022.879133 accessed. [Google Scholar]
- 6.Gayathri R., Mahboob S., Govindarajan M., Al-Ghanim K.A., Ahmed Z., Al-Mulhm N., Vodovnik M., Vijayalakshmi S. A review on biological carbon sequestration: a sustainable solution for a cleaner air environment, less pollution and lower health risks. J. King Saud Univ. Sci. 2021;33 doi: 10.1016/j.jksus.2020.101282. [DOI] [Google Scholar]
- 7.Cuéllar-Franca R.M., Azapagic A. Carbon capture, storage and utilisation technologies: a critical analysis and comparison of their life cycle environmental impacts. J. CO2 Util. 2015;9:82–102. doi: 10.1016/j.jcou.2014.12.001. [DOI] [Google Scholar]
- 8.Martínez-Francés E., Escudero-Oñate C. IntechOpen; 2018. Cyanobacteria and Microalgae in the Production of Valuable Bioactive Compounds. [DOI] [Google Scholar]
- 9.Ortiz-Bobea A., Ault T.R., Carrillo C.M., Chambers R.G., Lobell D.B. Anthropogenic climate change has slowed global agricultural productivity growth. Nat. Clim. Change. 2021;11:306–312. doi: 10.1038/s41558-021-01000-1. [DOI] [Google Scholar]
- 10.Jägermeyr J., Müller C., Ruane A.C., Elliott J., Balkovic J., Castillo O., Faye B., Foster I., Folberth C., Franke J.A., Fuchs K., Guarin J.R., Heinke J., Hoogenboom G., Iizumi T., Jain A.K., Kelly D., Khabarov N., Lange S., Lin T.-S., Liu W., Mialyk O., Minoli S., Moyer E.J., Okada M., Phillips M., Porter C., Rabin S.S., Scheer C., Schneider J.M., Schyns J.F., Skalsky R., Smerald A., Stella T., Stephens H., Webber H., Zabel F., Rosenzweig C. Climate impacts on global agriculture emerge earlier in new generation of climate and crop models. Nat. Food. 2021;2:873–885. doi: 10.1038/s43016-021-00400-y. [DOI] [PubMed] [Google Scholar]
- 11.Voigt C.A. Synthetic biology 2020–2030: six commercially-available products that are changing our world. Nat. Commun. 2020;11:6379. doi: 10.1038/s41467-020-20122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert C., Ellis T. Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 2019;8:1–15. doi: 10.1021/acssynbio.8b00423. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigo-Navarro A., Sankaran S., Dalby M.J., del Campo A., Salmeron-Sanchez M. Engineered living biomaterials. Nat. Rev. Mater. 2021;6:1175–1190. doi: 10.1038/s41578-021-00350-8. [DOI] [Google Scholar]
- 14.Nguyen P.Q., Courchesne N.-M.D., Duraj-Thatte A., Praveschotinunt P., Joshi N.S. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 2018;30 doi: 10.1002/adma.201704847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heveran C.M., Williams S.L., Qiu J., Artier J., Hubler M.H., Cook S.M., Cameron J.C., Srubar W.V. Biomineralization and successive regeneration of engineered living building materials. Matter. 2020;2:481–494. doi: 10.1016/j.matt.2019.11.016. [DOI] [Google Scholar]
- 16.Balasubramanian S., Yu K., Meyer A.S., Karana E., Aubin-Tam M.-E. Bioprinting of regenerative photosynthetic living materials. Adv. Funct. Mater. 2021;31 doi: 10.1002/adfm.202011162. [DOI] [Google Scholar]
- 17.Hart R., In-na P., Kapralov M.V., Lee J.G.M., Caldwell G.S. Textile-based cyanobacteria biocomposites for potential environmental remediation applications. J. Appl. Phycol. 2021;33:1525–1540. doi: 10.1007/s10811-021-02410-6. [DOI] [Google Scholar]
- 18.Spira inc, Spira inc. https://www.spirainc.com/ (n.d.) accessed.
- 19.FAQ, Prometheus materials. (n.d.). https://prometheusmaterials.com/faq/(accessed September 27, 2022).
- 20.Homepage | lumen bioscience. https://www.lumen.bio/ (n.d.) accessed.
- 21.Photanol (n.d. https://photanol.com accessed.
- 22.Santos-Merino M., Singh A.K., Ducat D.C. New applications of synthetic biology tools for cyanobacterial metabolic engineering. Front. Bioeng. Biotechnol. 2019;7 doi: 10.3389/fbioe.2019.00033. https://www.frontiersin.org/articles/10.3389/fbioe.2019.00033 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu A.P., Appel E.A., Ashby P.D., Baker B.M., Franco E., Gu L., Haynes K., Joshi N.S., Kloxin A.M., Kouwer P.H.J., Mittal J., Morsut L., Noireaux V., Parekh S., Schulman R., Tang S.K.Y., Valentine M.T., Vega S.L., Weber W., Stephanopoulos N., Chaudhuri O. The living interface between synthetic biology and biomaterial design. Nat. Mater. 2022;21:390–397. doi: 10.1038/s41563-022-01231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang T.-C., An B., Huang Y., Vasikaran S., Wang Y., Jiang X., Lu T.K., Zhong C. Materials design by synthetic biology. Nat. Rev. Mater. 2021;6:332–350. doi: 10.1038/s41578-020-00265-w. [DOI] [Google Scholar]
- 25.In-na P., Umar A.A., Wallace A.D., Flickinger M.C., Caldwell G.S., Lee J.G.M. Loofah-based microalgae and cyanobacteria biocomposites for intensifying carbon dioxide capture. J. CO2 Util. 2020;42 doi: 10.1016/j.jcou.2020.101348. [DOI] [Google Scholar]
- 26.Chen X., Lawrence J.M., Wey L.T., Schertel L., Jing Q., Vignolini S., Howe C.J., Kar-Narayan S., Zhang J.Z. 3D-printed hierarchical pillar array electrodes for high-performance semi-artificial photosynthesis. Nat. Mater. 2022;21:811–818. doi: 10.1038/s41563-022-01205-5. [DOI] [PubMed] [Google Scholar]
- 27.Shih P.M. Towards a sustainable bio-based economy: redirecting primary metabolism to new products with plant synthetic biology. Plant Sci. 2018;273:84–91. doi: 10.1016/j.plantsci.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medford J.I., Prasad A. Plant synthetic biology takes root. Science. 2014;346:162–163. doi: 10.1126/science.1261140. [DOI] [PubMed] [Google Scholar]
- 29.Srubar W.V. Engineered living materials: taxonomies and emerging trends. Trends Biotechnol. 2021;39:574–583. doi: 10.1016/j.tibtech.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Rulli M.C., Bellomi D., Cazzoli A., De Carolis G., D'Odorico P. The water-land-food nexus of first-generation biofuels. Sci. Rep. 2016;6 doi: 10.1038/srep22521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurens L.M.L., Lane M., Nelson R.S. Sustainable seaweed biotechnology solutions for carbon capture, composition, and deconstruction. Trends Biotechnol. 2020;38:1232–1244. doi: 10.1016/j.tibtech.2020.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Mutale-Joan C., Sbabou L., Hicham E.A. Microalgae and cyanobacteria: how exploiting these microbial resources can address the underlying challenges related to food sources and sustainable agriculture: a review. J. Plant Growth Regul. 2022 doi: 10.1007/s00344-021-10534-9. [DOI] [Google Scholar]
- 33.Sengupta A., Pakrasi H.B., Wangikar P.P. Recent advances in synthetic biology of cyanobacteria. Appl. Microbiol. Biotechnol. 2018;102:5457–5471. doi: 10.1007/s00253-018-9046-x. [DOI] [PubMed] [Google Scholar]
- 34.Yu J., Liberton M., Cliften P.F., Head R.D., Jacobs J.M., Smith R.D., Koppenaal D.W., Brand J.J., Pakrasi H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci. Rep. 2015;5:8132. doi: 10.1038/srep08132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neupert J., Gallaher S.D., Lu Y., Strenkert D., Segal N., Barahimipour R., Fitz-Gibbon S.T., Schroda M., Merchant S.S., Bock R. An epigenetic gene silencing pathway selectively acting on transgenic DNA in the green alga Chlamydomonas. Nat. Commun. 2020;11:6269. doi: 10.1038/s41467-020-19983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.León R., Fernández E. In: Transgenic Microalgae as Green Cell Factories. León R., Galván A., Fernández E., editors. Springer; New York, NY: 2007. Nuclear transformation of eukaryotic microalgae; pp. 1–11. [DOI] [Google Scholar]
- 37.Allen D.T. National Academies of Sciences, Engineering, and Medicine; Washington, DC (United States): 2018. Gaseous Carbon Waste Streams Utilization Status and Research Needs. [DOI] [Google Scholar]
- 38.Lu Y., Zhang X., Gu X., Lin H., Melis A. Engineering microalgae: transition from empirical design to programmable cells. Crit. Rev. Biotechnol. 2021;41:1233–1256. doi: 10.1080/07388551.2021.1917507. [DOI] [PubMed] [Google Scholar]
- 39.Pathak J., Rajneesh, Singh P.R., Häder D.P., Sinha R.P. UV-induced DNA damage and repair: a cyanobacterial perspective. Plant Gene. 2019;19 doi: 10.1016/j.plgene.2019.100194. [DOI] [Google Scholar]
- 40.Du X., Liu H., Yuan L., Wang Y., Ma Y., Wang R., Chen X., Losiewicz M.D., Guo H., Zhang H. The diversity of cyanobacterial toxins on structural characterization, distribution and identification: a systematic review. Toxins. 2019;11:530. doi: 10.3390/toxins11090530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng F., Ellis T. The second decade of synthetic biology: 2010–2020. Nat. Commun. 2020;11:5174. doi: 10.1038/s41467-020-19092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., Zhang S., Young E.M., Jones T.S., Densmore D., Voigt C.A. Genetic circuit design automation for yeast. Nat. Microbiol. 2020;5:1349–1360. doi: 10.1038/s41564-020-0757-2. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen A.A.K., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., Ross D., Densmore D., Voigt C.A. Genetic circuit design automation. Science. 2016;352:aac7341. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- 44.Lippow S.M., Wittrup K.D., Tidor B. Computational design of antibody affinity improvement beyond in vivo maturation. Nat. Biotechnol. 2007;25:1171–1176. doi: 10.1038/nbt1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nabout J., Rocha B., Carneiro F., S. C.L How many species of Cyanobacteria are there? Using a discovery curve to predict the species number. Biodivers. Conserv. 2013;22 doi: 10.1007/s10531-013-0561-x. [DOI] [Google Scholar]
- 46.Biller S.J., Berube P.M., Lindell D., Chisholm S.W. Prochlorococcus: the structure and function of collective diversity. Nat. Rev. Microbiol. 2015;13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 47.De La Rocha C.L., Passow U. In: Treatise on Geochemistry. second ed. Holland H.D., Turekian K.K., editors. Elsevier; Oxford: 2014. 8.4 - the biological pump; pp. 93–122. [DOI] [Google Scholar]
- 48.Noack S., Baumgart M. Communities of niche-optimized strains: small-genome organism consortia in bioproduction. Trends Biotechnol. 2019;37:126–139. doi: 10.1016/j.tibtech.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 49.Laurenceau R., Bliem C., Osburne M.S., Becker J.W., Biller S.J., Cubillos-Ruiz A., Chisholm S.W. Toward a genetic system in the marine cyanobacterium Prochlorococcus. Access Microbiol. 2020;2 doi: 10.1099/acmi.0.000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tolonen A.C., Liszt G.B., Hess W.R. Genetic manipulation of Prochlorococcus strain MIT9313: green fluorescent protein expression from an RSF1010 plasmid and Tn5 transposition. Appl. Environ. Microbiol. 2006;72:7607–7613. doi: 10.1128/AEM.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Włodarczyk A., Selão T.T., Norling B., Nixon P.J. Newly discovered Synechococcus sp. PCC 11901 is a robust cyanobacterial strain for high biomass production. Commun. Biol. 2020;3:1–14. doi: 10.1038/s42003-020-0910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaiswal D., Sengupta A., Sohoni S., Sengupta S., Phadnavis A.G., Pakrasi H.B., Wangikar P.P. Genome features and biochemical characteristics of a robust, fast growing and naturally transformable cyanobacterium Synechococcus elongatus PCC 11801 isolated from India. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-34872-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Griese Marco, Lange Christian, Soppa Jörg. Ploidy in cyanobacteria. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Lett. 2011;323:124–131. doi: 10.1111/j.1574-6968.2011.02368.x. [DOI] [PubMed] [Google Scholar]
- 54.Schoenfelder K.P., Fox D.T. The expanding implications of polyploidy. J. Cell Biol. 2015;209:485–491. doi: 10.1083/jcb.201502016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui J., Sun T., Chen L., Zhang W. Engineering salt tolerance of photosynthetic cyanobacteria for seawater utilization. Biotechnol. Adv. 2020;43 doi: 10.1016/j.biotechadv.2020.107578. [DOI] [PubMed] [Google Scholar]
- 56.Ramey C.J., Barón-Sola Á., Aucoin H.R., Boyle N.R. Genome engineering in cyanobacteria: where we are and where we need to go. ACS Synth. Biol. 2015;4:1186–1196. doi: 10.1021/acssynbio.5b00043. [DOI] [PubMed] [Google Scholar]
- 57.Broddrick J.T., Rubin B.E., Welkie D.G., Du N., Mih N., Diamond S., Lee J.J., Golden S.S., Palsson B.O. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis. Proc. Natl. Acad. Sci. USA. 2016;113:E8344–E8353. doi: 10.1073/pnas.1613446113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu H., Xu L., Luan G., Zhan T., Kang Z., Li C., Lu X., Zhang X., Zhu Z., Zhang Y., Li Y. A miniaturized bionic ocean-battery mimicking the structure of marine microbial ecosystems. Nat. Commun. 2022;13:5608. doi: 10.1038/s41467-022-33358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Datta D., Weiss E.L., Wangpraseurt D., Hild E., Chen S., Golden J.W., Golden S.S., Pokorski J.K. 2023. Phenotypically complex living materials containing engineered cyanobacteria. 2023.01.26.525792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hendry J.I., Prasannan C.B., Joshi A., Dasgupta S., Wangikar P.P. Metabolic model of Synechococcus sp. PCC 7002: prediction of flux distribution and network modification for enhanced biofuel production. Bioresour. Technol. 2016;213:190–197. doi: 10.1016/j.biortech.2016.02.128. [DOI] [PubMed] [Google Scholar]
- 61.Sarkar D., Mueller T.J., Liu D., Pakrasi H.B., Maranas C.D. A diurnal flux balance model of Synechocystis sp. PCC 6803 metabolism. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1006692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sawa M., Fantuzzi A., Bombelli P., Howe C.J., Hellgardt K., Nixon P.J. Electricity generation from digitally printed cyanobacteria. Nat. Commun. 2017;8:1327. doi: 10.1038/s41467-017-01084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu T., Paulo C., Merroun M.L., Dittrich M. Potential application of biomineralization by Synechococcus PCC8806 for concrete restoration. Ecol. Eng. 2015;82:459–468. doi: 10.1016/j.ecoleng.2015.05.017. [DOI] [Google Scholar]
- 64.Comprehensively curated genome-scale two-cell model for the heterocystous cyanobacterium Anabaena sp. PCC 7120 | plant physiology | oxford academic. https://academic.oup.com/plphys/article/173/1/509/6116171 (n.d. accessed. [DOI] [PMC free article] [PubMed]
- 65.Cameron D.E., Bashor C.J., Collins J.J. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 66.Way J.C., Collins J.J., Keasling J.D., Silver P.A. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell. 2014;157:151–161. doi: 10.1016/j.cell.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 67.Gardner T.S., Cantor C.R., Collins J.J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 68.Elowitz M.B., Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 69.Smolke C.D. Building outside of the box: iGEM and the BioBricks foundation. Nat. Biotechnol. 2009;27:1099–1102. doi: 10.1038/nbt1209-1099. [DOI] [PubMed] [Google Scholar]
- 70.Huang H.-H., Camsund D., Lindblad P., Heidorn T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S., Sun T., Xu C., Chen L., Zhang W. Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab. Eng. 2018;48:163–174. doi: 10.1016/j.ymben.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 72.Kim W.J., Lee S.-M., Um Y., Sim S.J., Woo H.M. Development of SyneBrick vectors as a synthetic biology platform for gene expression in Synechococcus elongatus PCC 7942. Front. Plant Sci. 2017;8 doi: 10.3389/fpls.2017.00293. https://www.frontiersin.org/articles/10.3389/fpls.2017.00293 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mills L.A., Moreno-Cabezuelo J.Á., Włodarczyk A., Victoria A.J., Mejías R., Nenninger A., Moxon S., Bombelli P., Selão T.T., McCormick A.J., Lea-Smith D.J. Development of a biotechnology platform for the fast-growing cyanobacterium Synechococcus sp. PCC 11901. Biomolecules. 2022;12:872. doi: 10.3390/biom12070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun T., Li S., Song X., Diao J., Chen L., Zhang W. Toolboxes for cyanobacteria: recent advances and future direction. Biotechnol. Adv. 2018;36:1293–1307. doi: 10.1016/j.biotechadv.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 75.Vasudevan R., Gale G.A.R., Schiavon A.A., Puzorjov A., Malin J., Gillespie M.D., Vavitsas K., Zulkower V., Wang B., Howe C.J., Lea-Smith D.J., McCormick A.J. CyanoGate: a modular cloning suite for engineering cyanobacteria based on the plant MoClo syntax. Plant Physiol. 2019;180:39–55. doi: 10.1104/pp.18.01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu D., Pakrasi H.B. Exploring native genetic elements as plug-in tools for synthetic biology in the cyanobacterium Synechocystis sp. PCC 6803. Microb. Cell Factories. 2018;17:48. doi: 10.1186/s12934-018-0897-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thiel K., Mulaku E., Dandapani H., Nagy C., Aro E.-M., Kallio P. Translation efficiency of heterologous proteins is significantly affected by the genetic context of RBS sequences in engineered cyanobacterium Synechocystis sp. PCC 6803. Microb. Cell Factories. 2018;17:34. doi: 10.1186/s12934-018-0882-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gale G.A.R., Wang B., McCormick A.J. Evaluation and comparison of the efficiency of transcription terminators in different cyanobacterial species. Front. Microbiol. 2021:11. doi: 10.3389/fmicb.2020.624011. https://www.frontiersin.org/articles/10.3389/fmicb.2020.624011 accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taton A., Unglaub F., Wright N.E., Zeng W.Y., Paz-Yepes J., Brahamsha B., Palenik B., Peterson T.C., Haerizadeh F., Golden S.S., Golden J.W. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 2014;42:e136. doi: 10.1093/nar/gku673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Al-Haj L., Lui Y.T., Abed R.M.M., Gomaa M.A., Purton S. Cyanobacteria as chassis for industrial biotechnology: progress and prospects. Life. 2016;6:42. doi: 10.3390/life6040042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bishé B., Taton A., Golden J.W. Modification of RSF1010-based broad-host-range plasmids for improved conjugation and cyanobacterial bioprospecting. iScience. 2019;20:216–228. doi: 10.1016/j.isci.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Y., Taton A., Go M., London R.E., Pieper L.M., Golden S.S., J.W.Y. Golden, Self-replicating shuttle vectors based on pANS, a small endogenous plasmid of the unicellular cyanobacterium Synechococcus elongatus PCC 7942. Microbiology. 2016;162:2029–2041. doi: 10.1099/mic.0.000377. [DOI] [PubMed] [Google Scholar]
- 83.GeneArtTM Synechococcus protein expression vector. https://www.thermofisher.com/order/catalog/product/A24230 n.d. accessed.
- 84.Shetty R.P., Endy D., Knight T.F. Engineering BioBrick vectors from BioBrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 86.Engler C., Kandzia R., Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Puzorjov A., Dunn K.E., McCormick A.J. Production of thermostable phycocyanin in a mesophilic cyanobacterium. Metabol. Eng. Commun. 2021;13 doi: 10.1016/j.mec.2021.e00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsuoka M., Takahama K., T. Ogawa, Gene replacement in cyanobacteria mediated by a dominant streptomycin-sensitive rps12 gene that allows selection of mutants free from drug resistance markers. Microbiology. 2001;147:2077–2087. doi: 10.1099/00221287-147-8-2077. [DOI] [PubMed] [Google Scholar]
- 89.Begemann M.B., Zess E.K., Walters E.M., Schmitt E.F., Markley A.L., Pfleger B.F. An organic acid based counter selection system for cyanobacteria. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kojima K., Keta S., Uesaka K., Kato A., Takatani N., Ihara K., Omata T., Aichi M. A simple method for isolation and construction of markerless cyanobacterial mutants defective in acyl-acyl carrier protein synthetase. Appl. Microbiol. Biotechnol. 2016;100:10107–10113. doi: 10.1007/s00253-016-7850-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones C.M., Parrish S., Nielsen D.R. Exploiting polyploidy for markerless and plasmid-free genome engineering in cyanobacteria. ACS Synth. Biol. 2021;10:2371–2382. doi: 10.1021/acssynbio.1c00269. [DOI] [PubMed] [Google Scholar]
- 92.Behler J., Vijay D., Hess W.R., Akhtar M.K. CRISPR-based technologies for metabolic engineering in cyanobacteria. Trends Biotechnol. 2018;36:996–1010. doi: 10.1016/j.tibtech.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 93.Li H., Shen C.R., Huang C.-H., Sung L.-Y., Wu M.-Y., Hu Y.-C. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab. Eng. 2016;38:293–302. doi: 10.1016/j.ymben.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 94.Racharaks R., Arnold W., Peccia J. Development of CRISPR-Cas9 knock-in tools for free fatty acid production using the fast-growing cyanobacterial strain Synechococcus elongatus UTEX 2973. J. Microbiol. Methods. 2021;189 doi: 10.1016/j.mimet.2021.106315. [DOI] [PubMed] [Google Scholar]
- 95.Pattharaprachayakul N., Lee M., Incharoensakdi A., Woo H.M. Current understanding of the cyanobacterial CRISPR-Cas systems and development of the synthetic CRISPR-Cas systems for cyanobacteria. Enzym. Microb. Technol. 2020;140 doi: 10.1016/j.enzmictec.2020.109619. [DOI] [PubMed] [Google Scholar]
- 96.Ungerer J., Pakrasi H.B. Cpf1 is A versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci. Rep. 2016;6 doi: 10.1038/srep39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knoot C.J., Biswas S., Pakrasi H.B. Tunable repression of key photosynthetic processes using Cas12a CRISPR interference in the fast-growing cyanobacterium Synechococcus sp. UTEX 2973. ACS Synth. Biol. 2020;9:132–143. doi: 10.1021/acssynbio.9b00417. [DOI] [PubMed] [Google Scholar]
- 98.Liu D., Johnson V.M., Pakrasi H.B. A reversibly induced CRISPRi system targeting photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. ACS Synth. Biol. 2020;9:1441–1449. doi: 10.1021/acssynbio.0c00106. [DOI] [PubMed] [Google Scholar]
- 99.Choi S.Y., Woo H.M. CRISPRi-dCas12a: a dCas12a-mediated CRISPR interference for repression of multiple genes and metabolic engineering in cyanobacteria. ACS Synth. Biol. 2020;9:2351–2361. doi: 10.1021/acssynbio.0c00091. [DOI] [PubMed] [Google Scholar]
- 100.Higo A., Isu A., Fukaya Y., Ehira S., Hisabori T. Application of CRISPR interference for metabolic engineering of the heterocyst-forming multicellular cyanobacterium Anabaena sp. PCC 7120. Plant Cell Physiol. 2018;59:119–127. doi: 10.1093/pcp/pcx166. [DOI] [PubMed] [Google Scholar]
- 101.McCarty N.S., Ledesma-Amaro R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 2019;37:181–197. doi: 10.1016/j.tibtech.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith M.J., Francis M.B. A designed A. vinelandii–S. Elongatus coculture for chemical photoproduction from air, water, phosphate, and trace metals. ACS Synth. Biol. 2016;5:955–961. doi: 10.1021/acssynbio.6b00107. [DOI] [PubMed] [Google Scholar]
- 103.Perera I.A., Abinandan S., Subashchandrabose S.R., Venkateswarlu K., Naidu R., Megharaj M. Advances in the technologies for studying consortia of bacteria and cyanobacteria/microalgae in wastewaters. Crit. Rev. Biotechnol. 2019;39:709–731. doi: 10.1080/07388551.2019.1597828. [DOI] [PubMed] [Google Scholar]
- 104.Fedeson D.T., Saake P., Calero P., Nikel P.I., Ducat D.C. Biotransformation of 2,4-dinitrotoluene in a phototrophic co-culture of engineered Synechococcus elongatus and Pseudomonas putida. Microb. Biotechnol. 2020;13:997–1011. doi: 10.1111/1751-7915.13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li T., Li C.-T., Butler K., Hays S.G., Guarnieri M.T., Oyler G.A., Betenbaugh M.J. Mimicking lichens: incorporation of yeast strains together with sucrose-secreting cyanobacteria improves survival, growth, ROS removal, and lipid production in a stable mutualistic co-culture production platform. Biotechnol. Biofuels. 2017;10:55. doi: 10.1186/s13068-017-0736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li L., Yang C., Ma B., Lu S., Liu J., Pan Y., Wang X., Zhang Y., Wang H., Sun T., Liu D. Hydrogel-encapsulated engineered microbial consortium as a photoautotrophic “living material” for promoting skin wound healing. ACS Appl. Mater. Interfaces. 2023 doi: 10.1021/acsami.2c20399. [DOI] [PubMed] [Google Scholar]
- 107.Urban L., Holzer A., Baronas J.J., Hall M.B., Braeuninger-Weimer P., Scherm M.J., Kunz D.J., Perera S.N., Martin-Herranz D.E., Tipper E.T., Salter S.J., Stammnitz M.R. Freshwater monitoring by nanopore sequencing. Elife. 2021;10 doi: 10.7554/eLife.61504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fujisawa T., Narikawa R., Maeda S., Watanabe S., Kanesaki Y., Kobayashi K., Nomata J., Hanaoka M., Watanabe M., Ehira S., Suzuki E., Awai K., Nakamura Y. CyanoBase: a large-scale update on its 20th anniversary. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkw1131. D551–D554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cyanobacteria web site. https://webext.pasteur.fr/cyanobacteria/ n.d. accessed.
- 110.Thiele I., Palsson B.Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Orth J.D., Thiele I., Palsson B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hendry J.I., Bandyopadhyay A., Srinivasan S., Pakrasi H.B., Maranas C.D. Metabolic model guided strain design of cyanobacteria. Curr. Opin. Biotechnol. 2020;64:17–23. doi: 10.1016/j.copbio.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 113.Cement – analysis, IEA. https://www.iea.org/reports/cement n.d. accessed.
- 114.Murik O., Oren N., Shotland Y., Raanan H., Treves H., Kedem I., Keren N., Hagemann M., Pade N., Kaplan A. What distinguishes cyanobacteria able to revive after desiccation from those that cannot: the genome aspect. Environ. Microbiol. 2017;19:535–550. doi: 10.1111/1462-2920.13486. [DOI] [PubMed] [Google Scholar]
- 115.Kobayashi S., Nakajima M., Asano R., Ferreira E.A., Abe K., Tamagnini P., Atsumi S., Sode K. Application of an engineered chromatic acclimation sensor for red-light-regulated gene expression in cyanobacteria. Algal Res. 2019;44 doi: 10.1016/j.algal.2019.101691. [DOI] [Google Scholar]
- 116.Bilal M., Iqbal H.M.N. Microbial-derived biosensors for monitoring environmental contaminants: recent advances and future outlook. Process Saf. Environ. Protect. 2019;124:8–17. doi: 10.1016/j.psep.2019.01.032. [DOI] [Google Scholar]
- 117.Caldwell G.S., In-na P., Hart R., Sharp E., Stefanova A., Pickersgill M., Walker M., Unthank M., Perry J., Lee J.G.M. Immobilising microalgae and cyanobacteria as biocomposites: new opportunities to intensify algae biotechnology and bioprocessing. Energies. 2021;14:2566. doi: 10.3390/en14092566. [DOI] [Google Scholar]
- 118.Flickinger M.C., Bernal O.I., Schulte M.J., Broglie J.J., Duran C.J., Wallace A., Mooney C.B., Velev O.D. Biocoatings: challenges to expanding the functionality of waterborne latex coatings by incorporating concentrated living microorganisms. J. Coating Technol. Res. 2017;14:791–808. doi: 10.1007/s11998-017-9933-6. [DOI] [Google Scholar]
- 119.Chou H.-H., Su H.-Y., Chow T.-J., Lee T.-M., Cheng W.-H., Chang J.-S., Chen H.-J. Engineering cyanobacteria with enhanced growth in simulated flue gases for high-yield bioethanol production. Biochem. Eng. J. 2021;165 doi: 10.1016/j.bej.2020.107823. [DOI] [Google Scholar]
- 120.Lin P.-C., Zhang F., Pakrasi H.B. Enhanced production of sucrose in the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Sci. Rep. 2020;10:390. doi: 10.1038/s41598-019-57319-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wenzel T., Härtter D., Bombelli P., Howe C.J., Steiner U. Porous translucent electrodes enhance current generation from photosynthetic biofilms. Nat. Commun. 2018;9:1299. doi: 10.1038/s41467-018-03320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sekar N., Jain R., Yan Y., Ramasamy R.P. Enhanced photo-bioelectrochemical energy conversion by genetically engineered cyanobacteria. Biotechnol. Bioeng. 2016;113:675–679. doi: 10.1002/bit.25829. [DOI] [PubMed] [Google Scholar]
- 123.Antonucci A., Reggente M., Roullier C., Gillen A.J., Schuergers N., Zubkovs V., Lambert B.P., Mouhib M., Carata E., Dini L., Boghossian A.A. Carbon nanotube uptake in cyanobacteria for near-infrared imaging and enhanced bioelectricity generation in living photovoltaics. Nat. Nanotechnol. 2022:1–9. doi: 10.1038/s41565-022-01198-x. [DOI] [PubMed] [Google Scholar]
- 124.Arnold F.H. Design by directed evolution. Acc. Chem. Res. 1998;31:125–131. doi: 10.1021/ar960017f. [DOI] [Google Scholar]
- 125.Dann M., Ortiz E.M., Thomas M., Guljamow A., Lehmann M., Schaefer H., Leister D. Enhancing photosynthesis at high light levels by adaptive laboratory evolution. Nat. Plants. 2021;7:681–695. doi: 10.1038/s41477-021-00904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Srivastava V., Amanna R., Rowden S.J.L., Sengupta S., Madhu S., Howe C.J., Wangikar P.P. Adaptive laboratory evolution of the fast-growing cyanobacterium Synechococcus elongatus PCC 11801 for improved solvent tolerance. J. Biosci. Bioeng. 2021;131:491–500. doi: 10.1016/j.jbiosc.2020.11.012. [DOI] [PubMed] [Google Scholar]
- 127.Hu L., He J., Dong M., Tang X., Jiang P., Lei A., Wang J. Divergent metabolic and transcriptomic responses of Synechocystis sp. PCC 6803 to salt stress after adaptive laboratory evolution. Algal Res. 2020;47 doi: 10.1016/j.algal.2020.101856. [DOI] [Google Scholar]
- 128.A system for the continuous directed evolution of biomolecules | Nature. https://www.nature.com/articles/nature09929 accessed. [DOI] [PMC free article] [PubMed]
- 129.Molina R.S., Rix G., Mengiste A.A., Álvarez B., Seo D., Chen H., Hurtado J.E., Zhang Q., García-García J.D., Heins Z.J., Almhjell P.J., Arnold F.H., Khalil A.S., Hanson A.D., Dueber J.E., Schaffer D.V., Chen F., Kim S., Fernández L.Á., Shoulders M.D., Liu C.C. In vivo hypermutation and continuous evolution. Nat. Rev. Methods Primers. 2022;2:1–22. doi: 10.1038/s43586-022-00119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wong B.G., Mancuso C.P., Kiriakov S., Bashor C.J., Khalil A.S. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat. Biotechnol. 2018;36:614–623. doi: 10.1038/nbt.4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Emond S., Petek M., Kay E.J., Heames B., Devenish S.R.A., Tokuriki N., Hollfelder F. Accessing unexplored regions of sequence space in directed enzyme evolution via insertion/deletion mutagenesis. Nat. Commun. 2020;11:3469. doi: 10.1038/s41467-020-17061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cain A.K., Barquist L., Goodman A.L., Paulsen I.T., Parkhill J., van Opijnen T. A decade of advances in transposon-insertion sequencing. Nat. Rev. Genet. 2020;21:526–540. doi: 10.1038/s41576-020-0244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rubin B.E., Wetmore K.M., Price M.N., Diamond S., Shultzaberger R.K., Lowe L.C., Curtin G., Arkin A.P., Deutschbauer A., Golden S.S. The essential gene set of a photosynthetic organism. Proc. Natl. Acad. Sci. USA. 2015;112:E6634–E6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wannier T.M., Ciaccia P.N., Ellington A.D., Filsinger G.T., Isaacs F.J., Javanmardi K., Jones M.A., Kunjapur A.M., Nyerges A., Pal C., Schubert M.G., Church G.M., Recombineering, MAGE Nat. Rev. Methods Primers. 2021;1:1–24. doi: 10.1038/s43586-020-00006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rovner A.J., Haimovich A.D., Katz S.R., Li Z., Grome M.W., Gassaway B.M., Amiram M., Patel J.R., Gallagher R.R., Rinehart J., Isaacs F.J. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shitrit D., Hackl T., Laurenceau R., Raho N., Carlson M.C.G., Sabehi G., Schwartz D.A., Chisholm S.W., Lindell D. Genetic engineering of marine cyanophages reveals integration but not lysogeny in T7-like cyanophages. ISME J. 2022;16:488–499. doi: 10.1038/s41396-021-01085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Borden J.S., Savage D.F. New discoveries expand possibilities for carboxysome engineering. Curr. Opin. Microbiol. 2021;61:58–66. doi: 10.1016/j.mib.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Long B.M., Hee W.Y., Sharwood R.E., Rae B.D., Kaines S., Lim Y.-L., Nguyen N.D., Massey B., Bala S., von Caemmerer S., Badger M.R., Price G.D. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 2018;9:3570. doi: 10.1038/s41467-018-06044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Russo M.E., Capasso C., Marzocchella A., Salatino P. Immobilization of carbonic anhydrase for CO2 capture and utilization. Appl. Microbiol. Biotechnol. 2022;106:3419–3430. doi: 10.1007/s00253-022-11937-8. [DOI] [PubMed] [Google Scholar]
- 140.Hill A.M., Salmond G.P.C. Microbial gas vesicles as nanotechnology tools: exploiting intracellular organelles for translational utility in biotechnology, medicine and the environment. Microbiology (Read.) 2020;166:501–509. doi: 10.1099/mic.0.000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Farhadi A., Ho G., Kunth M., Ling B., Lakshmanan A., Lu G.J., Bourdeau R.W., Schröder L., Shapiro M.G. Recombinantly expressed gas vesicles as nanoscale contrast agents for ultrasound and hyperpolarized MRI. AIChE J. 2018;64:2927–2933. doi: 10.1002/aic.16138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bourdeau R.W., Lee-Gosselin A., Lakshmanan A., Farhadi A., Kumar S.R., Nety S.P., Shapiro M.G. Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature. 2018;553:86–90. doi: 10.1038/nature25021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gregson B.H., McKew B.A., Holland R.D., Nedwed T.J., Prince R.C., McGenity T.J. Marine oil snow, a microbial perspective. Front. Mar. Sci. 2021;8 https://www.frontiersin.org/articles/10.3389/fmars.2021.619484 accessed. [Google Scholar]
- 144.Zimmerman A.E., Howard-Varona C., Needham D.M., John S.G., Worden A.Z., Sullivan M.B., Waldbauer J.R., Coleman M.L. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat. Rev. Microbiol. 2020;18:21–34. doi: 10.1038/s41579-019-0270-x. [DOI] [PubMed] [Google Scholar]
- 146.Moras C.A., Bach L.T., Cyronak T., Joannes-Boyau R., Schulz K.G. Ocean alkalinity enhancement–avoiding runaway CaCO 3 precipitation during quick and hydrated lime dissolution. Biogeosciences. 2022;19(15):3537–3557. [Google Scholar]
- 147.Bach L.T., Gill S.J., Rickaby R.E., Gore S., Renforth P. CO2 removal with enhanced weathering and ocean alkalinity enhancement: potential risks and co-benefits for marine pelagic ecosystems. Frontiers in Climate. 2019;1:7. [Google Scholar]
- 148.An B., Wang Y., Huang Y., Wang X., Liu Y., Xun D.…Zhong C. Engineered Living Materials For Sustainability. Chemical Reviews. 2022 doi: 10.1021/acs.chemrev.2c00512. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.