Abstract
Objective.
Deficiency of adenosine deaminase 2 (DADA2) is a monogenic form of vasculitis that can resemble polyarteritis nodosa (PAN). This study was undertaken to identify potential disease-causing sequence variants in ADA2 in patients with idiopathic PAN, granulomatosis with polyangiitis (GPA), or microscopic polyangiitis (MPA).
Methods.
Patients with idiopathic PAN (n = 118) and patients with GPA or MPA (n = 1,107) were screened for rare nonsynonymous variants in ADA2 using DNA sequencing methods. ADA-2 enzyme activity was assessed in selected serum samples.
Results.
Nine of 118 patients with PAN (7.6%) were identified as having rare nonsynonymous variants in ADA2. Four patients (3.4%) were biallelic for pathogenic or likely pathogenic variants, and 5 patients (4.2%) were monoallelic carriers for 3 variants of uncertain significance and 2 likely pathogenic variants. Serum samples from 2 patients with PAN with biallelic variants were available and showed markedly reduced ADA-2 enzyme activity. ADA-2 enzyme testing of 86 additional patients revealed 1 individual with strongly reduced ADA-2 activity without detectable pathogenic variants. Patients with PAN and biallelic variants in ADA2 were younger at diagnosis than patients with 1 or no variant in ADA2, with no other clinical differences noted. None of the patients with GPA or MPA carried biallelic variants in ADA2.
Conclusion.
A subset of patients with idiopathic PAN meet genetic criteria for DADA2. Given that tumor necrosis factor inhibition is efficacious in DADA2 but is not conventional therapy for PAN, these findings suggest that ADA-2 testing should strongly be considered in patients with hepatitis B virus–negative idiopathic PAN.
INTRODUCTION
Polyarteritis nodosa (PAN), granulomatosis with polyangiitis (GPA), and microscopic polyangiitis (MPA) are vasculitides defined by inflammatory lesions and necrosis of the blood vessel wall leading to vascular stenosis, hemorrhage, and tissue ischemia (1). GPA and MPA are antineutrophil cytoplasmic antibody (ANCA)–associated small to medium-sized vessel vasculitides, while PAN is not associated with ANCA and mainly affects medium-sized arteries.
PAN has been causally linked to chronic hepatitis B virus (HBV) infection in a subset of patients, but the pathogenesis of idiopathic PAN, and of GPA and MPA, is still not clear (2). Discovery of molecular mechanisms of necrotizing vasculitides would improve the understanding of disease pathogenesis and may guide novel therapeutic strategies in specific subsets of patients.
Recently, pathogenic variants in the gene encoding adenosine deaminase 2 (ADA2, formerly known as CECR1) were discovered in a subset of patients diagnosed as having childhood-onset PAN. The disease, termed deficiency of adenosine deaminase 2 (DADA2), is a monogenic vasculopathy that often leads to vasculitis (3,4). DADA2 is caused by recessively inherited hypomorphic or loss-of-function variants in the ADA2 gene, which encodes a dimeric protein secreted by monocytes and macrophages. Although the ADA-2 protein is a low-affinity enzyme for the conversion of extracellular adenosine to inosine, its physiologic function is still incompletely understood.
Patients with DADA2 can have clinical and histologic disease features similar to those in patients with idiopathic PAN, including vasculitis of medium-sized arteries with predilection for the skin, peripheral nerves, gastrointestinal tract, and kidneys. However, hepatic disease, ischemic stroke in the deep brain nuclei, immunodeficiency, and bone marrow failure are more often seen in patients with DADA2 compared to idiopathic PAN. Differentiation between DADA2 and idiopathic PAN is clinically important, as treatment and monitoring strategies differ between the 2 conditions.
Recent studies have shown an increased prevalence of DADA2 in cohorts of patients with pediatric vasculitis, and a higher prevalence is also expected in patients with idiopathic PAN (5). However, the actual percentage of patients with DADA2 in cohorts of idiopathic small- and medium-vessel vasculitis is unknown.
The objectives of this study were to identify potential disease-causing sequence variants in ADA2 in large cohorts of patients with idiopathic PAN, GPA, and MPA, using DNA sequencing. Since DNA sequencing and enzymatic assays play complementary roles in establishing the diagnosis of DADA2, a protein functional test was also performed in all available PAN samples, and in patients with GPA or MPA who had any ADA2 variant.
PATIENTS AND METHODS
Patients and controls.
DNA, serum samples, and clinical data were collected from patients enrolled in observational, longitudinal cohort studies of PAN, GPA, and MPA conducted by the Vasculitis Clinical Research Consortium at 8 academic medical centers in the US and Canada. All patients were evaluated using standardized data collection forms. Patients with PAN met the American College of Rheumatology (ACR) 1990 classification criteria for PAN (6) and were excluded if there was a history of HBV infection. Patients with GPA met the modified ACR 1990 classification criteria for the disease, and patients with MPA fulfilled the 2012 revision of the International Chapel Hill Consensus Conference disease definitions for this condition (1,7,8). Because the initiation of these clinical protocols predated the discovery of DADA2 (3,4), some phenotypic data now understood to be associated with DADA2—most notably features related to immunodeficiency or bone marrow failure—were not specifically queried, although the investigator could write in any atypical features believed to be disease-associated. Patients with DADA2 who had biallelic ADA2 mutations, carriers with monoallelic mutations, and healthy controls were recruited at the National Institutes of Health (NIH) Clinical Center, as described in the Supplementary Methods (available on the Arthritis & Rheumatology website at http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract). All study participants provided consent for the use of their specimens and data. The local institutional review boards approved the study (see Supplementary Methods).
Genetic analysis.
Peripheral blood DNA samples (n = 118) from patients with idiopathic PAN were screened for variants in ADA2 by standard Sanger sequencing of the 9 coding exons. Copy number variations within the ADA2 gene were assessed for a subset of samples using a multiplex ligation-dependent probe amplification assay (MLPA) according to the manufacturer’s manual (MRC Holland).
For the sake of cost and efficiency, sequencing of the ADA2 gene in patients with GPA or MPA (n = 1,107) was performed using a next-generation target capture approach, as described in the Supplementary Methods (http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract). Identified variants were classified according to the American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) 2015 guidelines described in the Supplementary Methods (http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract).
Measurement of ADA-2 enzyme activity.
Serum ADA-2 activity relative to a control group was assessed with a spectrophotometric assay using a commercially available kit (Diazyme Laboratories) by adding ADA-1 inhibitor EHNA (100 μM) according to the protocol of the manufacturer (Sigma-Aldrich). An ADA calibrator (50.3 units/liter; Diazyme) was used to generate a linear slope of absorbance versus ADA-2 activity. To assay for reduced ADA-2 enzyme activity, serum samples were available for 88 patients with idiopathic PAN, 16 patients with GPA or MPA who had monoallelic ADA2 variants, and 35 randomly chosen patients with GPA/MPA who did not have ADA2 variants.
For a selected subset of ADA2-genotyped patients with idiopathic PAN (n = 11), along with various controls, ADA-2 enzymatic activity in stored serum was measured at a Clinical Laboratory Improvement Amendments (CLIA)–certified laboratory (described in Supplementary Methods, http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract). Reference ranges were determined for patients with DADA2, carriers, and healthy controls who had been identified based on clinical data and, in some cases, DNA sequencing.
Statistical analysis.
Differences between the groups were tested for statistical significance using the Kruskal-Wallis test, Mann-Whitney U test, or chi-square test, as appropriate (SPSS version 1.0.0.1213). P values less than 0.05 were considered significant.
RESULTS
PAN cohort.
One hundred eighteen patients with idiopathic PAN were included in this study. Demographic data on the cohort are summarized in Supplementary Table 1 (http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract). Samples for molecular genetic testing were available for all 118 patients, while serum samples were accessible for 88 patients (74.6%).
ADA2 gene sequencing.
Nine of 118 patients (7.6%) with PAN were identified as having rare ADA2 missense variants with a minor allele frequency of <0.005. Four patients (3.4%) were homozygous or compound heterozygous for pathogenic or likely pathogenic variants in ADA2, establishing a genetic diagnosis of DADA2 in these patients (Table 1). Of the 7 distinct variants present in these 4 patients, p.G47A (rs200930463), p.G47W (rs202134424), p.R169Q (rs77563738), p.E328K, p.F355L (rs116020027), and p.G383S (rs770689762) had previously been reported as causative for DADA2 (9–11). The remaining variant, p.P106S (rs747107966), was classified as likely pathogenic according to the ACMG/AMP 2015 guidelines.
Table 1.
Patients with PAN with rare variants in ADA2 and/or reduced ADA-2 enzyme activity*
| Patient | Sex | Age at onset, years | Age at diagnosis, years | Age at study entry, years | Nucleotide change | Amino acid substitution | MAF | ACMG/AMP classification | ADA-2 activity, mU/ml (HPLC) |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 2VAR-1 | M | 22.8 | 22.9 | 24.7 | c.140G>C | p.G47A | 6.01−05 | LP (PM1, PM2, PM5, PP1, PP5) | NA |
| 2VAR-1 | M | 22.8 | 22.9 | 24.7 | c.316C>T | p.P106S | 1.99−05 | LP (PM1, PM2, PP3, PP5) | NA |
| 2VAR-2 | F | 16.4 | 17.4 | 18.9 | c.982G>A | p.E328K | – | LP (PM1, PM2, PM5, PP3, PP5) | 2.2 |
| 2VAR-2 | F | 16.4 | 17.4 | 18.9 | c.1065C>A | p.F355L | 2.34−04 | LP (PM1, PM2, PM3, PP5) | 2.2 |
| 2VAR-3 | M | 10.1 | 24.4 | 34.4 | c.1147G>A | p.G383S | 3.58−05 | LP (PM1, PM2, PP3, PP5) | 0.1 |
| 2VAR-3 | M | 10.1 | 24.4 | 34.4 | c.1147G>A | p.G383S | 3.58−05 | LP (PM1, PM2, PP3, PP5) | 0.1 |
| 2VAR-4 | M | NA | 15.1 | NA | c.139G>T | p.G47W | 3.98−06 | LP (PM1, PM2, PM5, PP3, PP5) | NA |
| 2VAR-4 | M | NA | 15.1 | NA | c.506G>A | p.R169Q | 4.74−04 | P (PS3, PM2, PM3, PP1, PP3) | NA |
| 1VAR-1 | F | 34.4 | 34.4 | 34.4 | c.100C>T | p.R34W | 7.78−05 | LP (PM1, PM2, PM3) | NA |
| 1VAR-2 | F | 62.0 | 62.1 | 66.1 | c.194C>T | p.T65M | 6.29−04 | VUS (PM1, PM2) | 22.7 |
| 1VAR-3 | M | 10.6 | 10.6 | 17.8 | c.927G>A | p.M309I | 1.67−03 | VUS (PM2, BS1, BP6) | NA |
| 1VAR-4 | F | 49.5 | 49.6 | 53.4 | c.1045G>A | p.V349I | 2.14−03 | VUS (PM1, PM2, BP4, BP6) | 14.9 |
| 1VAR-5 | M | 48.4 | 48.4 | 48.5 | c.1358A>G | p.Y453C | 8.84−05 | LP (PM1, PM2, PP3, PP5) | 11 |
| 0VAR-1 | M | 59.3 | 60.3 | 60.9 | 3.3 | ||||
| 0VAR-2 | M | 50.3 | 51.3 | 54.2 | 6.4 | ||||
| 0VAR-3 | F | 65.3 | 65.6 | 71.2 | 6.8 | ||||
| 0VAR-4 | M | 63.3 | 64.2 | 71.6 | 7.5 | ||||
| 0VAR-5 | M | 49.0 | 49.0 | 50.8 | 8.3 | ||||
| 0VAR-6 | M | NA | 36.7 | 42.0 | 8.9 | ||||
For each of the patients with rare variants (VAR) in ADA2, sex, age at onset of symptoms, age at diagnosis of polyarteritis nodosa (PAN), and age at study entry are shown. For each of the identified variants, change in the coding sequence (nucleotide change), protein change (amino acid substitution), minor allele frequency (MAF) as reported in the Genome Aggregation Database, and the American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) classification analysis are shown. Adenosine deaminase 2 (ADA-2) activity in available serum samples was measured using a high-performance liquid chromatography (HPLC)–based method. For patients with reduced ADA-2 enzymatic activity without detectable variants, information on sex, age at onset of symptoms, age at diagnosis of PAN, age at study entry, and ADA-2 activity is shown. LP = likely pathogenic; PM = pathogenic moderate; PP = pathogenic supporting; NA = not available; P = pathogenic; PS = pathogenic strong; VUS = variant of uncertain significance; BS = benign strong; BP = benign supporting. For variant classification and for definition of PM1, PM2, PP3, PP5, BS1, BP4, and BP6, see Supplementary Methods, http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract. Reference values are as follows: deficiency of ADA-2 patients (n = 55), mean ± SD 0.4 ± 0.5 mU/ml (range 0–2.5); ADA-2 carriers (n = 46), mean ± SD 5.7 ± 1.9 mU/ml (range 2.9–11.4); healthy controls (n = 27 + pooled human plasma), mean ± SD 13.0 ± 5.1 mU/ml (range 4.7–27.2).
Five additional patients (4.2%) were carriers for the rare monoallelic variants p.R34W (rs750955849), p.T65M (rs61747288), p.M309I (rs146597836), p.V349I (rs74317375), and p.Y453C (rs376785840), of which p.R34W and p.Y453C were previously reported in patients with DADA2 (3,12) (Table 1). The remaining 3 variants, p.T65M, p.M309I, and p.V349I, are of unknown clinical significance according to the ACMG/AMP 2015 guidelines and were not previously associated with disease.
ADA-2 enzyme activity.
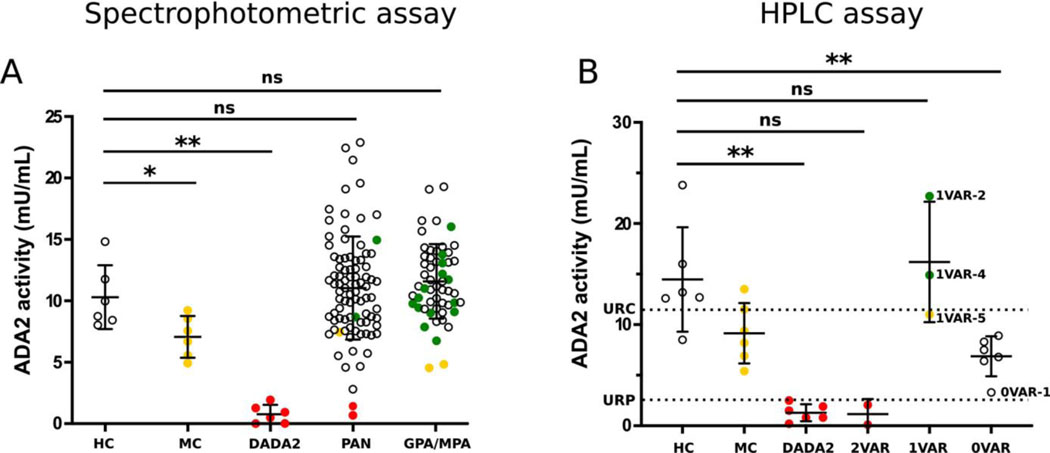
ADA-2 enzymatic activity was quantified by spectrophotometric assay on stored sera from 88 patients with idiopathic PAN (Figure 1A). To confirm the assessed ADA-2 activity and to validate the spectrophotometric assay, ADA-2 enzymatic activity was additionally quantified by a CLIA-certified high-performance liquid chromatography (HPLC)–based method in 11 patients (2 with biallelic variants, 3 with monoallelic variants, and 6 with no genetic variants but decreased enzymatic activity in the spectrophotometric assay) and 18 controls (6 healthy controls, 6 patients with DADA2, and 6 monoallelic carriers) (Figure 1B and Table 1).
Figure 1.
Adenosine deaminase 2 (ADA-2) levels in vasculitis. A, ADA-2 activity measured by spectrophotometric assay. ADA-2 activity was measured in serum from healthy controls (HCs; n = 6), monoallelic carriers (MCs; n = 6), and patients with deficiency of ADA-2 (DADA2; n = 6) from the National Institutes of Health (NIH) Clinical Center, and in serum from patients with idiopathic polyarteritis nodosa (PAN; n = 88) or granulomatosis with polyangiitis (GPA) or microscopic polyangiitis (MPA; n = 51). Among those with GPA/MPA, 16 patients had monoallelic ADA-2 variants, indicated by yellow and green circles, and 35 patients without a demonstrable variant were randomly chosen (open circles). B, ADA-2 activity measured by certified high-performance liquid chromatography (HPLC). ADA-2 activity was measured in serum from healthy controls (n = 6), monoallelic carriers (n = 6), and patients with DADA2 (n = 6) from the NIH Clinical Center, and in serum from patients with idiopathic PAN with biallelic variants (2VAR; n = 2), monoallelic variants (1VAR; n = 3), or no variant in ADA-2 (0VAR; n = 6, selected for low enzyme activity in A). Dashed lines show the upper limit of the reference range for patients with DADA2 (URP; 2.5 mU/ml) and the upper limit of the reference range for carriers (URC; 11.4 mU/ml), using HPLC. Bars show the mean ± SD. Subjects with 2 pathogenic or likely pathogenic variants are shown in red, those with 1 pathogenic or likely pathogenic variant are shown in yellow, those with 1 variant of unknown significance are shown in green, and those with no demonstrable variant are shown as open circles. * = P ≤ 0.05; ** = P ≤ 0.01 by Mann-Whitney U test. NS = not significant.
Serum samples were available for 2 of the patients with PAN who were homozygous or compound heterozygous for pathogenic or likely pathogenic variants in ADA2 (patients 2VAR-2 and 2VAR-3). These samples showed markedly reduced ADA-2 enzyme activity according to both assays, comparable to levels in DADA2 reference ranges for the HPLC testing laboratory (Figure 1B and Table 1). Sera from 3 monoallelic carriers with PAN (patients 1VAR-2, 1VAR-4, and 1VAR-5) showed ADA-2 activity in the normal range, also with both assays. The serum sample from the individual carrying the known pathogenic p.Y453C variant (1VAR-5) showed ADA-2 enzyme activity at the low end of the reference range for healthy controls and just below the upper end of the reference range for monoallelic carriers. This patient was 48 years old at the time of diagnosis and had classic features of PAN (e.g., fever, cutaneous ulcers, neuropathy) without additional features of DADA2 (e.g., stroke, livedoid rash).
With both assays, ADA-2 enzymatic testing revealed an additional patient with strongly reduced ADA-2 activity levels (patient 0VAR-1) (Figure 1B and Table 1), a man diagnosed as having PAN at age 60, with mesenteric and renal artery vasculitis, but none of the cutaneous, neurologic, or hepatic features of DADA2. There were 5 other patients with enzymatic activity in the range of carriers (0VAR-2 to 0VAR-6). Sequencing and copy number variation analyses of the ADA2 gene in these patients did not identify any pathogenic variants and suggest the possibility of deleterious deep intronic or regulatory variants undetectable by genomic sequencing or MLPA (Supplementary Figure 1A, http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract). Since circulating ADA inhibitors have been documented (13), the available serum sample from patient 0VAR-1 was tested in a mixing assay, but this failed to reveal evidence for a soluble inhibitor (Supplementary Figure 1B). Including the 3 patients with monoallelic variants, 80 of 88 available serum samples showed enzyme activity in the normal or high-normal range, using the spectrophotometric assay (Figure 1).
Clinical associations with ADA2 pathogenic variants.
Among patients with PAN, the median age at diagnosis for the 4 patients with biallelic ADA2 pathogenic or likely pathogenic variants was significantly lower (median 20.2 years [range 15.1–24.4]) compared to the 5 patients with monoallelic ADA2 variants (median 48.4 years [range 10.6–62.1]) or the 109 patients without ADA2 variants (median 46.9 years [range 11.4–75.3]; P = 0.01) (Table 2).
Table 2.
Comparison of clinical features among patients with polyarteritis nodosa with and without detectable mutations in ADA2*
| Biallelic ADA2 mutation (n = 4)† | Monoallelic ADA2 mutation (n = 5) | No ADA2 mutation (n = 109) | |
|---|---|---|---|
|
| |||
| Age at symptom onset, median (range) years | 16.4 (10.1–22.8)‡ | 48.4 (10.6–62.0) | 45.4 (9.1–74.7) |
| Age at diagnosis, median (range) years | 20.2 (15.1–24.4)§ | 48.4 (10.6–62.1) | 46.9 (11.4–75.3) |
| Age at study entry, median (range) years | 24.7 (18.9–34.4) | 48.5 (17.8–66.1) | 50.8 (17.5–78.7) |
| Female sex | 1 (25) | 3 (60) | 58 (53) |
| White | 3 (75) | 4 (80) | 98 (90) |
| Cardiac involvement | 0 (0) | 0 (0) | 13 (12) |
| Constitutional involvement | 3 (100) | 5 (100) | 75 (69) |
| Cutaneous involvement | 3 (100)¶ | 5 (100)# | 82 (75)** |
| Livedo reticularis | 1 (33) | 1 (20) | 24 (21) |
| Gastrointestinal involvement | 2 (67) | 2 (40) | 40 (37) |
| Hepatic involvement | 0 (0) | 0 (0) | 7 (6) |
| Kidney involvement | 1 (33) | 1 (20) | 28 (26) |
| Musculoskeletal involvement | 2 (67) | 3 (60) | 80 (73) |
| Nervous system involvement | 1 (33) | 3 (60) | 59 (54) |
| Stroke | 0 (0) | 0 (0) | 6 (5) |
| Ocular involvement | 1 (33) | 0 (0) | 10 (9) |
Except where indicated otherwise, values are the number (%) of patients.
Information on age at symptom onset, age at study entry, and pattern of organ involvement was not available for 1 patient with biallelic ADA2 mutations.
P = 0.04 versus the groups with monoallelic ADA2 mutation or no ADA2 mutation.
P = 0.01 versus the groups with monoallelic ADA2 mutation or no ADA2 mutation.
Included livedo reticularis (n = 1) and ulcers (n = 2).
Included livedo reticularis (n = 1), ulcers (n = 2), and nodules (n = 2).
Included livedo reticularis (n = 24), nodules (n = 40), purpura (n = 33), ulcers (n = 18), and digital infarction (n = 3).
Two patients (2VAR-1 and 2VAR-3) with biallelic ADA2 rare missense variants were diagnosed as having vasculitis in adulthood (Table 1). Clinical information was available for 3 of the 4 patients with biallelic ADA2 pathogenic or likely pathogenic variants, and there were no substantial differences in sex, ethnicity, or pattern of organ involvement between patients with and those without ADA2 variants (Table 2). Treatment information was available for 2 patients with biallelic ADA2 variants, neither of whom had previously received therapy with tumor necrosis factor (TNF) inhibitors.
GPA/MPA cohort.
There were 1,107 patients with GPA or MPA who were included in this study. Demographic data on these cohorts are summarized in Supplementary Table 1 (http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract ).
ADA2 gene sequencing.
Sequencing of ADA2 in patients with GPA or MPA did not identify any individuals with homozygous or compound heterozygous rare nonsynonymous genomic variants in ADA2, but identified 27 patients (2.4%) who carried monoallelic rare missense or canonical splice-site variants (Supplementary Table 2, http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract). Of the 27 identified variants, 3 (11.1%) were classified as likely pathogenic according to ACMG/AMP 2015 criteria.
ADA-2 enzyme activity.
Enzymatic testing was performed on serum samples from 51 patients with GPA or MPA (Figure 1A). Sixteen of these samples were from patients who carried monoallelic missense variants or canonical splice-site variants in ADA2 (Supplementary Table 2, http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract). Enzyme activity measurements identified 2 patients with ADA-2 enzyme activity slightly below the range in the 6 monoallelic carriers from the NIH Clinical Center, shown in Figure 1 (4.9–9.2 mU/ml), which is consistent with the likely pathogenic classification of p.P106S (GM6; 4.5 mU/ml) and c.1442+2T>G (GM24; 4.8 mU/ml) (Figure 1A; serum was not available for GM15 in Supplementary Table 2, http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract). The remaining serum samples from patients with GPA/MPA who had monoallelic variants showed ADA-2 activity in the normal to high-normal range of NIH controls (Figure 1A). With the spectrophotometric assay, enzymatic activity from 35 randomly chosen patients with GPA/MPA who did not have ADA2 variants was shown to be within or exceeding the range observed for the NIH healthy controls shown in Figure 1.
Clinical associations with ADA2 pathogenic variants.
There were no significant clinical differences, among patients with GPA/MPA, between those with and those without monoallelic ADA2 variants (Supplementary Table 3, http://onlinelibrary.wiley.com/doi/10.1002/art.41549/abstract).
DISCUSSION
Vasculitis is a group of multisystemic diseases of unknown etiology and significant morbidity and mortality. Discovery of additional causal mechanisms in necrotizing vasculitis would improve the understanding of disease pathogenesis and may inform development of novel therapeutic strategies in specific subsets of patients. This study used DNA sequencing to identify individuals with a genetic diagnosis of DADA2 in cohorts of patients with PAN, GPA, and MPA. The identification of biallelic ACMG/AMP-classified pathogenic or likely pathogenic variants in an individual establishes the genetic diagnosis of DADA2 (14,15).
Homozygous or compound heterozygous pathogenic or likely pathogenic variants in ADA2 were identified in 3.4% of patients diagnosed as having idiopathic PAN, including 2 patients who were diagnosed as having vasculitis in adulthood. Biallelic pathogenic or likely pathogenic variants in ADA2 were not found in any of 1,107 patients diagnosed as having GPA or MPA. Of the 27 identified monoallelic variants in ADA2, 3 were predicted to be likely pathogenic, a frequency comparable to that of such variants in ADA2 in the general population. There is a significant difference (P < 0.001) between the number of patients with 2 pathogenic or likely pathogenic variants in ADA2 in the PAN cohort (4 of 118) and the number of patients with 2 pathogenic or likely pathogenic variants in the GPA/MPA cohort (0 of 1,107). There remains the possibility that some patients with GPA/MPA have strongly reduced enzymatic activity without detectable germline pathogenic variants, as was recently described in a pediatric case (5).
This study also identified 1 patient (0VAR-1) in the PAN cohort who exhibited strongly reduced ADA-2 enzyme activity despite the absence of identifiable pathogenic variants. Patient 0VAR-1’s ADA-2 activity was higher than that in any of the 6 confirmed patients with DADA2 from the NIH cohort and was higher than activity assessed with the HPLC assay in any of the 55 patients with DADA2 tested in the CLIA reference laboratory; therefore, this patient cannot be definitively diagnosed as having DADA2 based on enzymatic activity. This finding is intriguing, since higher residual ADA-2 enzymatic activity in an in vitro transfection assay has recently been associated with an increased risk of developing vasculitis compared to other disease manifestations (10). It is possible that intronic or regulatory variants in this patient may account for the reduction in enzymatic activity. No evidence of an acquired inhibitor of ADA-2 was found in the available serum sample. As noted above, a child with GPA was found to have low ADA-2 enzymatic activity despite the absence of demonstrable coding or splice-site mutations (5).
The present study also identified 5 other patients with PAN with ADA-2 enzyme activity in the range observed in monoallelic carriers without detectable pathogenic variants, despite sequencing of the entire gene and MLPA analyses. The risk of disease in monoallelic carriers is still unclear, but a gene-dose effect in combination with other permissive genetic and environmental factors may lead to a decreased threshold for inflammatory manifestations. This finding is noteworthy since 2 carriers of the pathogenic ADA2 variant p.Y453C were reported to have late-onset lacunar strokes (3).
One limitation of this study is that even though this is one of the largest reported cohorts of patients with idiopathic PAN, the total sample size is still insufficient to permit identification of clinical features that might distinguish between PAN and DADA2. Because the PAN cohort was established long before the discovery of DADA2, clinical data regarding some of the key features of DADA2 were not specifically queried. Another limitation is that for some novel ADA2 variants identified in this cohort, even when applying the stringent ACMG/AMP criteria, it is still not possible to assess their pathogenicity in the absence of patient serum samples. As more new variants are identified in patients with clinical features of DADA2 and in available serum samples, it may be easier to evaluate their pathogenic significance.
Given the substantial degree of disease heterogeneity and potential overlap of clinical features between DADA2 and PAN, a diagnosis of DADA2 should be considered in patients clinically diagnosed as having or being evaluated for idiopathic PAN. A diagnosis can be made either genetically through the identification of biallelic ACMG/AMP-classified pathogenic or likely pathogenic variants or enzymatically through the detection of low ADA-2 enzymatic activity. These 2 methods are complementary and may need to be used concomitantly, at least in some cases. A limitation of genetic testing is that standard sequencing will fail to identify gene deletions/duplications in up to 3% of patients with DADA2 (16).
There are also limitations intrinsic to the interpretation of the enzymatic assay. In the CLIA-certified laboratory that was involved in the present investigation, a validated assay of plasma ADA-2 enzymatic activity has accurately demonstrated DADA2 in the 80% of cases in which results of ADA2 gene sequencing have been provided. The range of plasma ADA-2 activity has been established for authentic carriers, i.e., parents of patients with DADA2 and siblings shown to be carriers by gene sequence analysis. However, as there is overlap with the lowest plasma ADA-2 activity found in healthy controls, when enzyme activity is found to be within the carrier range, ADA2 gene sequence analysis is advised as necessary to establish heterozygote status. In true heterozygous carriers, certain factors, such as infection, may induce increased production of the ADA-2 protein from the wild-type allele (17). The situation may be further obscured if, as some postulate, there are biologic functions of the ADA-2 protein that are not measured by the enzymatic assay, which may affect the clinical phenotype. These considerations all underscore the notion that sequencing and functional assays are complementary and that both may be required for a complete clinical evaluation. There is a clear need for increased availability of CLIA-certified biochemical testing for DADA2 in the US.
The mainstay of treatment for idiopathic PAN consists of glucocorticoids and other immunosuppressive drugs and has improved the prognosis and survival rates of these patients (18). Unfortunately, treatment has considerable side effects, and disease relapse and mortality remain high. Importantly, several recent studies have demonstrated that conventional immunomodulatory therapy seems not to be beneficial in patients with DADA2, while anti-TNF treatment (etanercept, infliximab, adalimumab) appears to be highly effective in preventing strokes, normalizing inflammatory markers, and improving other PAN-like manifestations in these patients (19). None of the patients identified in the current study who had biallelic pathogenic variants in the ADA2 gene, and for whom treatment information was available, had received therapy with TNF inhibitors, as this class of medication is not commonly administered to patients diagnosed as having idiopathic PAN.
In summary, given the potential differences in approaches to treatment, clinical follow-up, prognosis, and consequences for other family members, the present findings suggest that ADA2 testing should strongly be considered in patients with HBV-negative idiopathic PAN. These data also suggest that ADA2 variants are unlikely to account for the molecular pathology of MPA and GPA. Going forward, it will be important to validate these findings prospectively in larger cohorts with paired analysis of genetic variants and enzyme activity.
Supplementary Material
Acknowledgments
The Vasculitis Clinical Research Consortium (VCRC) is part of the Rare Diseases Clinical Research Network, an initiative of the Office of Rare Diseases Research, National Center for Advancing Translational Science (NCATS). The VCRC is funded through a collaboration between the National Center for Advancing Translational Science and the National Institute of Arthritis and Musculoskeletal and Skin Diseases, NIH (grant U54-AR-057319) and has received funding from the National Center for Research Resources, NIH (grant U54RR-019497). This work was also supported by the National Human Genome Research Institute Intramural Research Program.
Dr. Khalidi has received consulting fees, speaking fees, and/or honoraria from Bristol Myers Squibb and Roche (less than $10,000 each). Dr. Pagnoux has received consulting fees, speaking fees, and/or honoraria from ChemoCentryx, Sanofi, Roche, and GlaxoSmithKline (less than $10,000 each) and research grants from Roche and GlaxoSmithKline. Dr. Springer has received research grants from InflaRx. Dr. Warrington has received consulting fees, speaking fees, and/or honoraria from Sanofi and Roche (less than $10,000 each) and research support from Eli Lilly, GlaxoSmithKline, and Roche. Dr. Merkel has received consulting fees, speaking fees, and/or honoraria from AbbVie, Biogen, CSL Behring, Janssen, Kiniksa, Sparrow, AstraZeneca, Boeringher-Ingelheim, Bristol Myers Squibb, Celgene, ChemoCentryx, Genentech/Roche, Genzyme/Sanofi, GlaxoSmithKline, and InflaRx (less than $10,000 each), research grants from AstraZeneca, Boeringher-Ingelheim, Bristol Myers Squibb, Celgene, ChemoCentryx, Genentech/Roche, Genzyme/Sanofi, GlaxoSmithKline, InflaRx, Kypha, and TerumoBCT, and royalties from UpToDate. No other disclosures relevant to this article were reported.
APPENDIX A: NISC COMPARATIVE SEQUENCING PROGRAM COLLABORATORS
Members of the NISC Comparative Sequencing Programn, in addition to the authors, are as follows: Beatrice B. Barnabas, MPH, MSc, Sean Black, MSc, Gerard G. Bouffard, PhD, Shelise Y. Brooks, BS, Holly Coleman, MSc, Lyudmila Dekhtyar, MSc, Joel Han, BS, Shi-ling Ho, BS, Juyun Kim, BS, Richelle Legaspi, MSc, Quino L. Maduro, BS, Catherine A. Masiello, MSc, Jennifer C. McDowell, PhD, Casandra Montemayor, MSc, James C. Mullikin, PhD, Morgan Park, PhD, Nancy L. Riebow, BS, Karen Schandler, MSc, Brian Schmidt, BS, Christina Sison, BS, Sirintorn Stantripop, BS, James W. Thomas, PhD, Pamela J. Thomas, PhD, Meghana Vemulapalli, MSc, Alice C. Young, BA.
REFERENCES
- 1.Jennette JC, Falk RJ, Bacon PA, Basu N, Cid MC, Ferrario F, et al. 2012 revised International Chapel Hill Consensus Conference nomenclature of vasculitides. Arthritis Rheum 2013;65:1–11. [DOI] [PubMed] [Google Scholar]
- 2.Pagnoux C, Seror R, Henegar C, Mahr A, Cohen P, Le Guern V, et al. Clinical features and outcomes in 348 patients with polyarteritis nodosa: a systematic retrospective study of patients diagnosed between 1963 and 2005 and entered into the French Vasculitis Study Group Database. Arthritis Rheum 2010;62:616–26. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Yang D, Ombrello AK, Zavialov AV, Toro C, Zavialov AV, et al. Early-onset stroke and vasculopathy associated with mutations in ADA2. N Engl J Med 2014;370:911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elkan PN, Pierce SB, Segel R, Walsh T, Barash J, Padeh S, et al. Mutant adenosine deaminase 2 in a polyarteritis nodosa vasculopathy. N Engl J Med 2014;370:921–31. [DOI] [PubMed] [Google Scholar]
- 5.Gibson KM, Morishita KA, Dancey P, Moorehead P, Drögemöller B, Han X, et al. Identification of novel adenosine deaminase 2 gene variants and varied clinical phenotype in pediatric vasculitis. Arthritis Rheumatol 2019;71:1747–55. [DOI] [PubMed] [Google Scholar]
- 6.Lightfoot RW Jr, Michel BA, Bloch DA, Hunder GG, Zvaifler NJ, McShane DJ, et al. The American College of Rheumatology 1990 criteria for the classification of polyarteritis nodosa. Arthritis Rheum 1990;33:1088–93. [DOI] [PubMed] [Google Scholar]
- 7.Leavitt RY, Fauci AS, Bloch DA, Michel BA, Hunder GG, Arend WP, et al. The American College of Rheumatology 1990 criteria for the classification of Wegener’s granulomatosis. Arthritis Rheum 1990;33:1101–7. [DOI] [PubMed] [Google Scholar]
- 8.WGET Research Group. Design of the Wegener’s Granulomatosis Etanercept Trial (WGET). Control Clin Trials 2002;23:450–68. [DOI] [PubMed] [Google Scholar]
- 9.Keer N, Hershfield M, Caskey T, Unizony S. Novel compound heterozygous variants in CECR1 gene associated with childhood onset polyarteritis nodosa and deficiency of ADA2. Rheumatology (Oxford) 2016;55:1145–7. [DOI] [PubMed] [Google Scholar]
- 10.Lee PY, Kellner ES, Huang Y, Furutani E, Huang Z, Bainter W, et al. Genotype and functional correlates of disease phenotype in deficiency of adenosine deaminase 2 (DADA2). J Allergy Clin Immunol 2020;145:1664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bras J, Guerreiro R, Santo GC. Mutant ADA2 in vasculopathies [letter]. N Engl J Med 2014;371:478–80. [DOI] [PubMed] [Google Scholar]
- 12.Kaljas Y, Liu C, Skaldin M, Wu C, Zhou Q, Lu Y, et al. Human adenosine deaminases ADA1 and ADA2 bind to different subsets of immune cells. Cell Mol Life Sci 2017;74:555–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaffee S, Mary A, Stiehm ER, Girault D, Fischer A, Hershfield MS. IgG antibody response to polyethylene glycol-modified adenosine deaminase in patients with adenosine deaminase deficiency. J Clin Invest 1992;89:1643–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rama M, Duflos C, Melki I, Bessis D, Bonhomme A, Martin H, et al. A decision tree for the genetic diagnosis of deficiency of adenosine deaminase 2 (DADA2): a French reference centres experience. Eur J Hum Genet 2018;26:960–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aksentijevich I, Moura NS, Barron K. Adenosine deaminase 2 deficiency. In: Ardinger HH, Pagon RA, Wallace SE, editors. GeneReviews®. Seattle (WA): University of Washington; 2019. URL: https://www.ncbi.nlm.nih.gov/books/NBK544951/. [Google Scholar]
- 17.Abdi M, Rahbari R, Khatooni Z, Naseri N, Najafi A, Khodadadi I. Serum adenosine deaminase (ADA) activity: a novel screening test to differentiate HIV monoinfection from HIV-HBV and HIV-HCV coinfections. J Clin Lab Anal 2016;30:200–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhtyar C, Guillevin L, Cid MC, Dasgupta B, de Groot K, Gross W, et al. EULAR recommendations for the management of primary small and medium vessel vasculitis. Ann Rheum Dis 2009; 68:310–7. [DOI] [PubMed] [Google Scholar]
- 19.Ombrello AK, Qin J, Hoffmann PM, Kumar P, Stone D, Jones A, et al. Treatment strategies for deficiency of adenosine deaminase 2 [letter]. N Engl J Med 2019;380:1582–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.