Abstract
Apoptosis has been widely studied from mammals to insects. Inhibitor of apoptosis (IAP) protein is a negative regulator of apoptosis. Recent studies suggest that iap genes could be excellent targets for RNA interference (RNAi)-mediated control of insect pests. However, not much is known about iap genes in one of the well-known insect model species, Tribolium castaneum. The orthologues of five iap genes were identified in T. castaneum by searching its genome at NCBI (https://www.ncbi.nlm.nih.gov/) and UniProt (https://www.uniprot.org/) databases using Drosophila melanogaster and Aedes aegypti IAP protein sequences as queries. RNAi assays were performed in T. castaneum cell line (TcA) and larvae. The knockdown of iap1 gene induced a distinct apoptotic phenotype in TcA cells and induced 91% mortality in T. castaneum larvae. Whereas, knockdown of iap5 resulted in a decrease in cell proliferation in TcA cells and developmental defects in T. castaneum larvae which led to 100% mortality. Knockdown of the other three iap genes identified did not cause a significant effect on cells or insects. These data increase our understanding of iap genes in insects and provide opportunities for developing iap1 and iap5 as targets for RNAi-based insect pest control.
Keywords: iap, red flour beetle, RNA interference, survivin, Tribolium castaneum
1 ∣. INTRODUCTION
Apoptosis, the programmed cell death, is an evolutionarily conserved pathway of cell suicide and determines the rate of turnover of the cells (Vaux, Haecker, & Strasser, 1994). Living multicellular organisms look quite constant at the organismal level, but when it comes to its cellular level, many cells die and are replaced by new cells constantly to maintain an equilibrium (Elmore, 2007). This dynamic and highly regulated process involves numerous genes and signaling pathways, creating a balance between activation and inhibition of cell death. In insects, apoptosis is an essential process in cellular activity and plays an important role in many processes in development, tissue homeostasis, and innate antiviral response (Miura, 2011; Parthasarathy, Tan, Bai, & Palli, 2008).
Not all inhibitor of apoptosis (IAP) proteins inhibit the apoptosis pathway, but some cooperate with caspases through the baculovirus IAP repeat (BIR) domain. iap genes were first discovered in baculoviruses known to inhibit the host insect cell's suicide response (Crook, Clem, & Miller, 1993). Later, iap genes were identified in many eukaryotic organisms, including mammals and insects (Hay, Wassarman, & Rubin, 1995; Liston et al., 1996; Srinivasula & Ashwell, 2008). The IAP family proteins contain highly conserved protein interaction motifs, including the BIR (Birnbaum, Clem, & Miller, 1994; Crook et al., 1993). A typical IAP protein contains one to three BIR domains. The BIRs function as protein recognition and protein–protein interaction sites (Srinivasula & Ashwell, 2008). Therefore, IAP family proteins are called BIR-containing proteins (BIRPs or BIRCs), which include BIRC1 (NAIP), BIRC2 (human IAP2, cellular IAP1, cIAP1), BIRC3 (human IAP1, cellular IAP2, cIAP2), BIRC4 (X-linked IAP, xIAP), BIRC5 (survivin), BIRC6 (BIR-containing ubiquitin-conjugating enzyme, apollon), BIRC7 (livin/melanoma-IAP, ML-IAP, KIAP), and BIRC8 (testis-specific IAP, Ts-IAP, hILP-2; Saleem et al., 2013; Silke & Vaux, 2001; Srinivasula & Ashwell, 2008; Verhagen, Coulson, & Vaux, 2001). In Drosophila, a known model insect, only four BIRs were found: Deterin, DIAP1, DIAP2, and DBruce (Apollon; Srinivasula & Ashwell, 2008), suggesting that different organisms would have different numbers of BIRCs.
BIR proteins play major roles in apoptosis and cell-cycle function. BIR proteins (BIRC1, 2, 3, 4, 7, and 8) act on inhibiting caspases activation (Saleem et al., 2013). BIRC5 and 6 act on regulating cytokinin and mitotic spindle formation to inhibit apoptosis (Saleem et al., 2013; Silke & Vaux, 2001). In cells, overexpression of iap gene blocks apoptosis; knockdown of iap gene expression promotes apoptosis (Hawkins, Ekert, Uren, Holmgreen, & Vaux, 1998; Hawkins, Uren, Hacker, Medcalf, & Vaux, 1996; Lu, Wang, He, & Xi, 2008). However, not much is known about the function of BIRC5 in insects. Even in the well-known insect model, Drosophila melanogaster, BIRC5 was not annotated and the function of BIRC5 is not known in Tribolium castaneum.
RNA interference (RNAi) is a posttranscriptional gene silencing mechanism in which exogenous double-stranded RNA (dsRNA) triggers the suppression of the gene expression or messenger RNA (mRNA) translation (Fire et al., 1998) Applications of RNAi include cancer treatment in the medical field to insect pest control in agriculture (Andersen, Howard, & Kjems, 2009; Barnard, Nijhof, Fick, Stutzer, & Maritz-Olivier, 2012; Gaur & Rossi, 2006; Huvenne & Smagghe, 2010; Kim & Rossi, 2008; Morris & Rossi, 2006; Palli, 2014; Reidhaar-Olson & Rondinone, 2009; K. Y. Zhu & Palli, 2020). Due to its relatively high sensitivity to RNAi and well-annotated genome sequences, T. castaneum became an insect model to study the function of genes (Arakane, Muthukrishnan, Beeman, Kanost, & Kramer, 2005; Knorr et al., 2018; Richards et al., 2008; Tomoyasu et al., 2008; F. Zhu et al., 2010). Even though T. castaneum is one of the important insect models and many genes have been annotated and functionally verified, no studies on the iap genes in this insect have been conducted. Therefore, the goal of these studies was to perform a thorough annotation of T. castaneum iap genes and determine the function of these genes. Five iap genes were identified in the Tribolium genome by searching National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) and UniProt (https://www.uniprot.org/) databases using D. melanogaster and Aedes aegypti IAP protein sequences as queries (Puglise, Estep, & Becnel, 2016). Then, in vitro and in vivo RNAi assays were performed to determine if any of these genes are involved in inhibition of apoptosis. The results showed that two of the five iap genes tested caused detectable phenotypes in cells and larvae.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Insect rearing and cell culture
The GA-1 strain of the red flour beetle, T. castaneum was reared on organic wheat flour (Heartland mill) mixed with 10% baker's yeast. The insects were maintained in the dark at 30°C and 65 ± 5% relative humidity. The BCIRL-TcA-CLG1 (TcA) cells were routinely maintained at 27°C in EX-CELL 420 (Sigma-Aldrich) medium with 10% heat-inactivated fetal bovine serum (Seradigm) and antibiotics.
2.2 ∣. Gene identification
Tribolium iap genes were obtained from homology searches by running BLASTp and tBLASTn using the NCBI BLAST service (http://www.ncbi.nlm.nih.gov/) and Uniport BLAST service (http://www.uniprot.org/). The Tribolium caspase genes were selected from the Flybase database (http://flybase.org/) and homology searches into the Tribolium genome database in NCBI. The dsRNA (400–500 bp) and quantitative reverse-transcription polymerase chain reaction (qRT-PCR; 80–120 bp) primers were designed from these sequences. The target sites for qRT-PCR did not overlap with dsRNA amplicon.
2.3 ∣. In vivo and in vitro RNAi assay
Methods for in vitro dsRNA synthesis have been described in our recent paper (Yoon, Shukla, Gong, Mogilicherla, & Palli, 2016). For in vitro RNAi assay, 25,000 TCA cells/well were seeded in 96 well plates. The equal amount (1 μg/μl) of dsRNA was added to cell and pictures were taken on 6th day after dsRNA treatment. The same amount of luciferase dsRNA was used as a negative control. For in vivo RNAi assay, 200 ng of dsRNA was injected into the dorsal side of the abdomen using a Drummond Nanoject III fitted with 3.5″ glass capillary tube, pulled by a needle puller (Model P-1000, Sutter Instruments). Newly molted last instar larvae were injected. After injection, the insects were reared under standard conditions. Mortality was recorded after 14 days. Those insects showing distinct phenotypes after dsIAP1 treatment were treated with phosphate-buffered saline (PBS) + 0.1% trypan blue solution to identify the excreted substance. The phenotype of the quiescent stage larvae and adults injected with dsTcIAP5 were photographed using a digital microscope system (UNITRON Z850 Stereo Microscope, CB-ZM deep focus extension module, QuickPHOTO industrial 3.1 program, Canon EOS Rebel T5i camera).
2.4 ∣. Knockdown efficiency studies
Methods for total RNA extraction, PCR, and quantitative real-time PCR (qRT- PCR) were described in our previous paper (Yoon, Shukla et al., 2016). Abour 200 ng of dsIAP was injected into early last instar larva. The larvae were collected on 2nd day after dsRNA injection, and RNA was extracted. Knockdown efficiency was determined by quantifying TcIAP5 mRNA levels in dsTcIAP5 treatment and dsLUC (control) treated insects using qRT-PCR and gene-specific primers (Table 1). Ribosomal Protein 49 (RP49) was used as an internal control.
TABLE 1.
List of the primers used in this study
| Primers | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| TcIAP1_dsR | CAAAGCGGGCTTCTACTTCT | TTCGTCCTTCCAGTCCCTAA |
| TcIAP2_dsR | ACTACGGCGACCAAGTAATG | GACTCCCAGACAGTGCAAAT |
| TcVIAP_dsR | CCAATGAGGACACTGAATGGA | CAGGAACTTAACAGCTGGGTAT |
| TcIAP5_dsR | GTTGAACCGTTGGTTGTGTTT | GGATTTCTCGGGCTTGTTCT |
| TcIAP6_dsR | GGCCTTCAAACAGACCTACA | CAAACGACTAAATGGACACACG |
| TcIAP1_qRT | TGTCGGCGAGTAATGTAACC | AAATCTGTCGCAAATTCTGCAT |
| TcIAP2_qRT | CATCGGCTTGAATGTCGAAAG | TCGCTGTGCTGGAGATTG |
| TcVIAP_qRT | GTGGGTCCAGTCGAACTTAG | TCGGCTTTGGGTCTTCTTT |
| TcIAP5_qRT | AAGAGTTGTGGATAGAGCATT | ACATGCACACCTTATTCTAACT |
| TcIAP6_qRT | CTACGATGCCACACCTTTGA | CCAAACAAGCTAAGAGCAAGTG |
Note: T7 promoter sequences were added at the 5′ end of dsRNA (dsR) primers.
Abbreviation: dsRNA, double-stranded RNA.
2.5 ∣. Expression profile of iap1 and iap5 genes
Samples were collected at 24 hr intervals during penultimate larva, final instar larva, and pupal stages. Total RNA was extracted from pools of two larvae/pupae for each time interval. RP49 was used as an internal control for measuring the relative mRNA levels.
2.6 ∣. Statistics
For testing the knockdown efficiency and mRNA level of iap genes, Student's t test was used to compare the gene expression difference between the control and treatment groups. A p-value ≤ .05 between groups was considered as significantly different. The analysis of variance method was used to analyze the significance of differences between the samples among the developmental stages (JMP 11.0 software, SAS, Cary, NC).
3 ∣. RESULTS
3.1 ∣. Annotation of iap genes and their domain structures
We identified orthologues of five iap genes from NCBI with the following annotations: inhibitor of apoptosis 1 (LOC663941), inhibitor of apoptosis 2 (LOC663905), viral IAP-associated factor homolog (LOC656000), baculovirus IAP repeat-containing protein 5.2 (LOC100141706), and baculovirus IAP repeat-containing protein 6 (LOC662712; Table 2). Uniport database contained duplicated or limited annotations (Table 2). For example, apoptosis 2 inhibitor-like protein (6WAP2) contained the information of IAP1 (LOC663941), but Inhibitor of apoptosis 1 protein (G8G3Y5) has limited or almost no information.
TABLE 2.
List of Tribolium castaneum iap genes and their annotated information
| Name | TC number |
NCBI | UniProt | |
|---|---|---|---|---|
| TcIAP1 | TC001192 | Inhibitor of apoptosis 1 | Apoptosis 2 inhibitor-like protein | Inhibitor of apoptosis 1 protein |
| LOC663941 | D6WAP2 | G8G3Y5 | ||
| TcIAP2 | TC001189 | Inhibitor of apoptosis 2 | Apoptosis 2 inhibitor-like protein | |
| LOC663905 | D6WAN9 | |||
| TcVIAP | TC003566 | Viral IAP-associated factor homolog | Viral IAP-associated factor homolog-like protein | |
| LOC656000 | D6WHT2 | |||
| TcIAP5 | TC002709 | Baculovirus IAP repeat-containing protein 5.2 | Apoptosis 2 inhibitor-like protein | |
| LOC100141706 | D6WDU5 | |||
| TcIAP6 | TC009848 | Baculovirus IAP repeat-containing protein 6 | Uncharacterized protein | |
| LOC662712 | D6WQ00 |
Abbreviations: IAP, inhibitor of apoptosis; NCBI, National Center for Biotechnology Information.
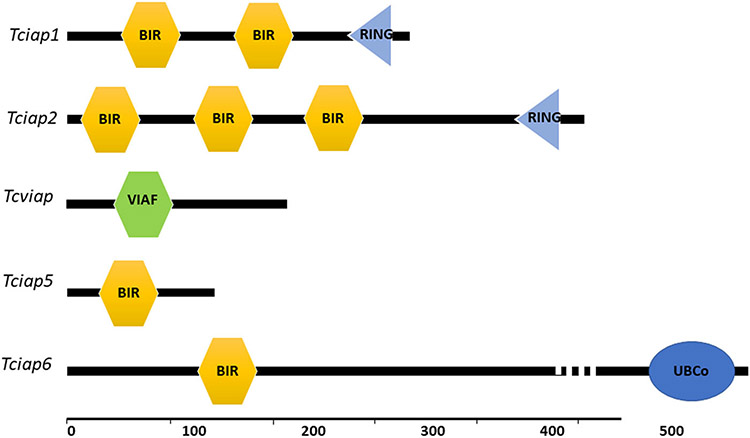
The IAP proteins identified contained one to three BIR domains, except the vIAP, which contained a viral inhibitor of apoptosis-associated factor (VIAF) domain (Figure 1). Other than BIR domains, IAP1, and IAP2 contain RING domains different from other IAPs. IAP5 and IAP6 contained only one BIR domain. The IAP6 contains ubiquitin-conjugated enzyme sites at the 3′ end of the sequence that includes the E3 interaction residue (Figure 1).
FIGURE 1.
Domain structure of Tribolium IAPs. The baculovirus IAP repeat (BIR) and the ring (RING) domains and viral inhibitor of apoptosis-associated factors (VIAF) are marked. IAP, inhibitors of apoptosis protein
3.2 ∣. Knockdown of iap genes in TcA cells
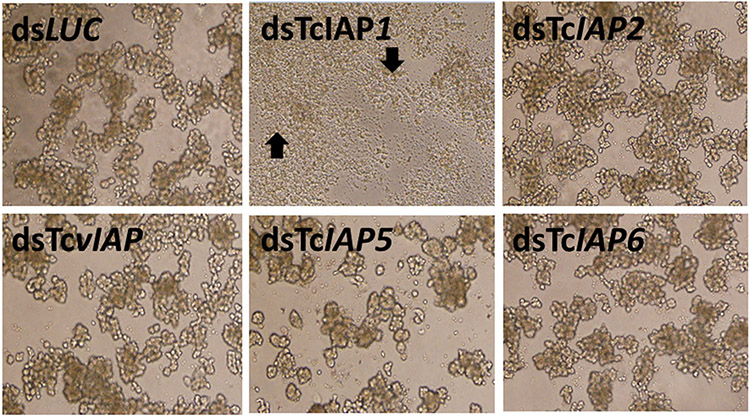
The TcA cells were exposed to 1 μl of 1 μg/μl concentration in vitro synthesized dsRNA targeting each iap gene. Within 2 days after the addition of dsTcIAP1, the cells started showing apoptosis phenotype. Pictures taken on the sixth day after the addition of dsRNA are shown in Figure 2 under the inverted microscope (Olympus IX71). The dsRNAs targeting the other four iap genes did not show apoptosis phenotype. However, the number of cells in wells containing dsTcIAP5 was lower when compared with the cells exposed to dsRNA targeting the luciferase gene (dsLUC) as a control. These data suggest that only TcIAP1 protein inhibits apoptosis and TcIAP5 may affect cell proliferation. The other three IAPs tested did not show any visible effect on TcA cells. This experiment was repeated at least three times with three technical replications.
FIGURE 2.
RNA interference in TcA cells. About 25,000 TcA cells/well were seeded in each well of 96-well plates. An equal amount (1 μg) of dsRNA targeting each iap gene was added to each well, and pictures were taken on the 6th day after treatment with dsRNA. dsRNA targeting the luciferase gene was used as a control. The arrows point to apoptotic bodies. dsRNA, double-stranded RNA
3.3 ∣. In vivo RNAi experiments
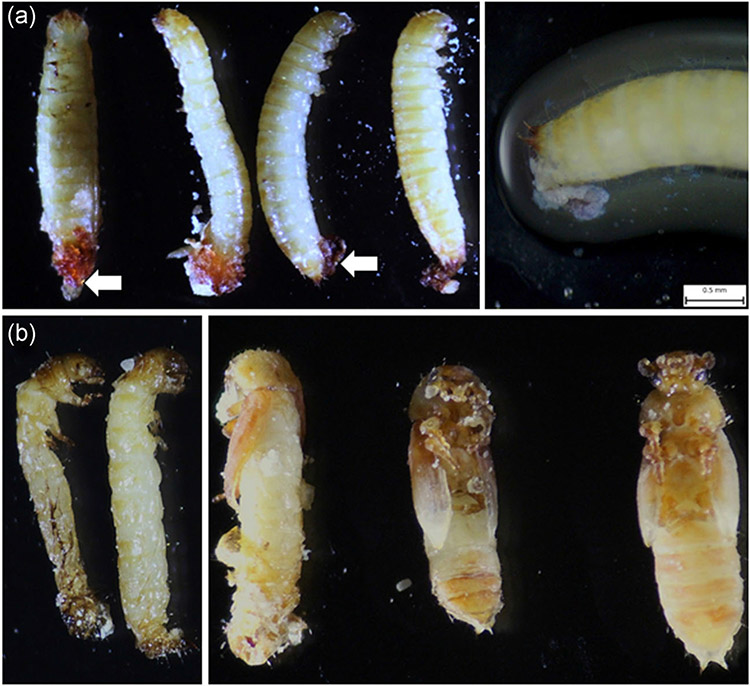
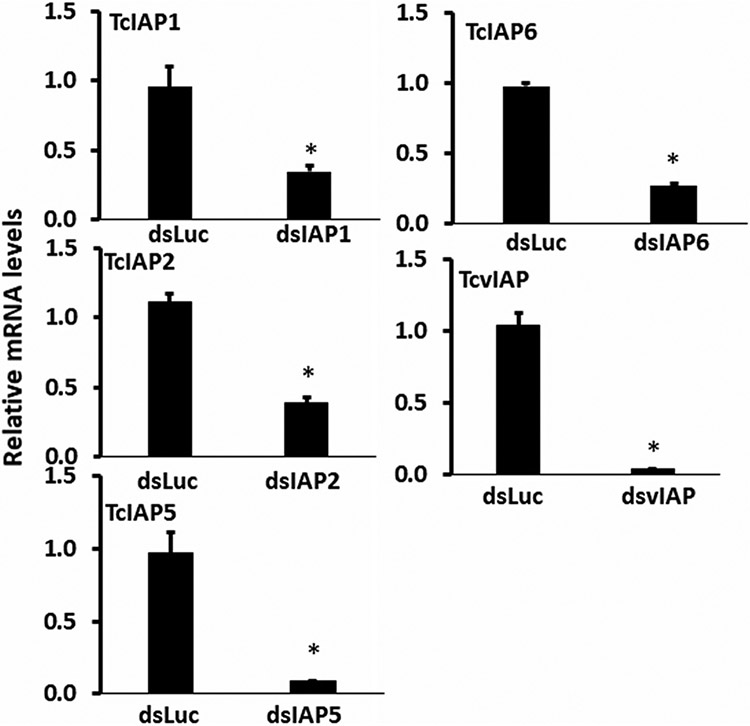
We injected 200 ng of dsRNA targeting each of the five iap genes into early last instar larvae and recorded mortality on 13th day after injection. The dsTcIAP1 injected larvae died during the quiescent stage (Figure 3). Knockdown of iap1 made the larvae excrete brown substance from their anus. To determine if the brown substance contains discarded midgut tissue, we stained the substance with 0.1% trypan blue dissolved in PBS solution. The substance slowly liquefied in the solution and contained cellular material stained blue suggesting that the cells sluffed off from the alimentary canal are in the brown substance excreted (Figure 3). In the case of iap5 knockdown, 75% of larvae died during the prepupal stage exhibiting the hunchback shape with the dorsal split phenotype; most of these insects stuck in the exuvia and did not complete pupal ecdysis (Figure 3). Some of them survived through the larval stage, but got stuck in the pupal stage and/or became defective adults. Knockdown of the rest of the iap genes did not induce any mortality (Table 3). To determine knockdown efficiency of dsTcIAPs, the iap mRNA levels were determined in the larvae injected with dsRNA targeting iap genes or the luciferase dsRNA used as a control. dsRNA targeting all five iap genes showed a significant knockdown of its target gene (Figure 4).
FIGURE 3.
Phenotypes induced by dsRNA targeting iap1 and iap5 genes. (a) About 200 ng dsTcIAP1 injected larvae on the 7th day after treatment were photographed under a microscope. The white arrows point to the brown substance excreted by the larvae. The picture on the right shows the same larva soaked in PBS + 0.1% trypan blue solution to visualize the extruded alimentary canal. (b) Phenotypes of larvae injected with dsRNA targeting iap5 gene. The picture on the left shows the larvae died during the prepupal stage. The picture on the right shows dead pupae and adults developed from larvae injected with dsRNA targeting iap5 gene. dsRNA, double-stranded RNA; PBS, phosphate-buffered saline
TABLE 3.
RNA interference in vivo
| dsRNA | Mortality (%)* | Comments |
|---|---|---|
| LUC | 5 | A negative control |
| TcIAP1 | 91 | All of them died during the quiescent stage |
| TcIAP2 | 0 | |
| TcVIAP | 26 | |
| TcIAP5 | 100 | 24% Died as pupa and adults |
| 76% Died during the larval stage | ||
| TcIAP6 | 15 |
Abbreviation: dsRNA, double-stranded RNA.
The early last instar Tribolium castaneum larvae were injected with 200 ng dsRNA targeting each iap gene and the Luciferase gene as a negative control. The mortality was recorded on the 13th day after injection (N = 20).
FIGURE 4.
Confirmation of Knockdown of each gene by RT-qPCR. About 200 ng dsRNA targeting each iap gene was injected into larvae and the larvae were collected on 2nd day after injection. Total RNA was isolated and used to determine relative iap mRNA levels by RT-qPCR. The dsLUC injected larvae were used as control. The gene coding for Ribosomal Protein 49 (RP49) was used as a reference. Statistical significance was analyzed with a Student's t test (*p < .05). dsRNA, double-stranded RNA; mRNA, messenger RNA; qRT-PCR, quantitative reverse-transcription polymerase chain reaction
3.4 ∣. Expression profiles of iap1 and iap5 genes
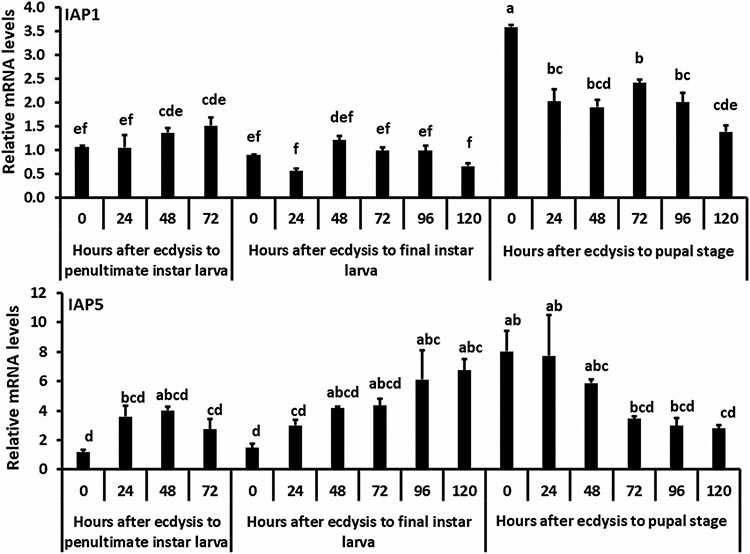
Since we observed the notable phenotypes in larvae after knockdown iap1 and iap5 genes, we thought that examining their gene expression patterns during the larval stage may help to understand the function of these proteins. Staged insects were collected at 24 hr intervals from the penultimate larval stage to the pupal stage. As shown in Figure 5, lower levels of iap1 mRNA were detected until the beginning of the pupal stage suggesting that apoptosis is actively occurring during the larval stage. The iap5 mRNA levels gradually increased from the last instar larval stage to 24 hr after ecdysis to the pupal stage (Figure 5).
FIGURE 5.
The iap1 and iap5 mRNA levels during penultimate and last instar larval and pupal stages. Samples were collected at 24 hr intervals during penultimate larval, final instar larval, and pupal stages. The RP49 mRNA levels were used as an internal control for measuring the relative mRNA levels. Mean + SE (n = 3) are shown. The letters indicate significant differences between the samples among the developmental stages by ANOVA (JMP 11.0 software, SAS, Cary, NC). ANOVA, analysis of variance; mRNA, messenger RNA; RP49, Ribosomal Protein 49; SE, standard error
3.5 ∣. The target specificity of dsRNAs in knocking down iap genes
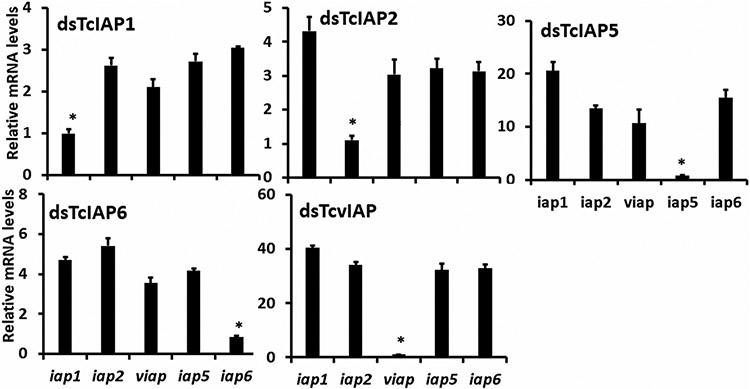
We tested to determine whether knockdown of one of the iap genes affects the expression of other iap genes. The larvae were treated with dsRNA targeting each iap gene, 2 days after the injection of dsRNA, total RNA was isolated and used to quantify mRNA levels of all iap genes. As expected, dsRNA targeting each iap gene induced knockdown of its target gene (Figure 6). No significant change in mRNA levels of the other four iap genes was detected in larvae injected with dsRNA targeting each one of the five iap genes (Figure 6).
FIGURE 6.
The target specificity of dsRNAs in knocking down iap genes. About 200 ng of each dsRNA was injected into larvae and the larvae were collected on 2nd day after injection. The total RNA was isolated and used to measure relative mRNA levels. The ANOVA method was used to analyze the significance of differences between the samples (JMP 11.0 software, SAS, Cary, NC). Mean + SE (n = 3) are shown. ANOVA, analysis of variance; dsRNAs, dsRNA, double-stranded RNAs; mRNA, messenger RNA. *p < .001
4 ∣. DISCUSSION
T. castaneum has been a valuable model for RNAi research due to its ease of injection, strong and systemic RNAi response, ease of rearing in large numbers, and availability of a wealth of genetic and genome resources. Moreover, with the increasing use of iap genes in various experiments such as the RNAi of RNAi assay, as a mortality marker, an RNAi-based pest control target, and in resistance studies (Cao, Gatehouse, & Fitches, 2018; Dhandapani, Gurusamy, Howell, & Palli, 2019; Igaki, Yamamoto-Goto, Tokushige, Kanda, & Miura, 2002; Mogilicherla, Howell, & Palli, 2018; Powell et al., 2017; Rodrigues, Dhandapani, Duan, & Palli, 2017; Walker & Allen, 2011; Yoon, Mogilicherla et al., 2018; Yoon, Shukla, et al., 2016), it is important to identify iap genes and their functions in insects.
During insect development, cells divide, die, and differentiate to accommodate changes needed to progress from larva to pupa to adult. The mRNA levels of iap1 are low during the larval stage and increased during the early pupal stage (Figure 5a). This expression pattern agrees with a recent report on iap1 expression in T. castaneum (Cao et al., 2018). In general, lack of IAP1 protein leads to apoptosis, therefore, there may be cells in the larvae undergoing apoptosis. Larval midgut remodeling involves the death of larval cells and the differentiation of stem cells into pupal/adult midgut (Wu, Parthasarathy, Bai, & Palli, 2006). The iap1 knockdown in T. castaneum larvae resulted in their death and excretion of the alimentary canal cells (Figure 3a). The excretion may be due to the increase in apoptosis of midgut cells induced by a reduction in levels of IAP1 in the midgut cells.
The function of IAP5 (baculovirus inhibitor of apoptosis repeat-containing 5, BIRC5), also known as survivin, has been extensively studied in mammals by both biochemical and genetic approaches in the fields of cell-death pathway, mechanisms of cell-cycle progression and microtubule stability (Altieri, 2003; Saleem et al., 2013). Interestingly, we observed the same effects in our in vitro and in vivo assays using an insect model. However, the specific functions of TcIAP5 in insects remain obscure. IAP5 in A. aegypti mosquitoes is transcriptionally active during programmed autophagy and upregulated after the blood meal (Eng, van Zuylen, & Severson, 2016). Its function in the postembryonic development of the midgut in a lepidopteran insect, Helicoverpa armigera was reported previously (He, Hou, Wang, & Zhao, 2012). The IAP5 in Cydia pomonella granulosis virus stimulates the antiapoptotic activity of CpIAP3 (Vilaplana & O'Reilly, 2003). Taken together these data point to a critical role for TcIAP5 in cell-death machinery and cell proliferation.
In this study, we identified five iap genes in Tribolium genome. RNAi studies showed that TcIAP1 and TcIAP5 play vital roles in apoptosis and cell proliferation, respectively. The in vivo RNAi studies showed that knockdown of iap1 resulted in mortality during the quiescent stage, and knockdown of iap5 showed developmental abnormalities, which led to death. This paper not only broadens our knowledge of IAPs but also identified iap1 and iap5 as potential insecticidal targets for RNAi-based insect control.
ACKNOWLEDGMENTS
Supported by grants from the National Institutes of Health (GM070559-14 and 1R21AI131427-01), the National Science Foundation (Industry/University Cooperative Research Centers, the Center for Arthropod Management Technologies under Grant IIP-1821936), and the National Institute of Food and Agriculture, US Department of Agriculture (under HATCH Project 2353057000).
Footnotes
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
REFERENCES
- Altieri DC (2003). Validating survivin as a cancer therapeutic target. Nature Reviews Cancer, 3(1), 46–54. 10.1038/nrc968 [DOI] [PubMed] [Google Scholar]
- Andersen MØ, Howard KA, & Kjems J (2009). RNAi using a chitosan/siRNA nanoparticle system: In vitro and in vivo applications. In Rondinone CM & Reidhaar-Olson JF (Eds.), Therapeutic applications of RNAi: Methods and protocols (pp. 77–86). Totowa, NJ: Humana Press. [Google Scholar]
- Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, & Kramer KJ (2005). Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proceedings of the National Academy of Sciences of the United States of America, 102(32), 11337–11342. 10.1073/pnas.0504982102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard A-C, Nijhof AM, Fick W, Stutzer C, & Maritz-Olivier C (2012). RNAi in arthropods: Insight into the machinery and applications for understanding the pathogen-vector interface. Genes, 3(4), 702–741. 10.3390/genes3040702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum MJ, Clem RJ, & Miller LK (1994). An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. Journal of Virology, 68(4), 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Gatehouse JA, & Fitches EC (2018). A systematic study of RNAi effects and dsRNA stability in Tribolium castaneum and Acyrthosiphon pisum, following injection and ingestion of analogous dsRNAs. International Journal of Molecular Science, 19(4), 1079. 10.3390/ijms19041079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, & Miller LK (1993). An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. Journal of Virology, 67(4), 2168–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhandapani RK, Gurusamy D, Howell JL, & Palli SR (2019). Development of CS-TPP-dsRNA nanoparticles to enhance RNAi efficiency in the yellow fever mosquito, Aedes aegypti. Scientific Reports, 9(1), 8775. 10.1038/s41598-019-45019-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S (2007). Apoptosis in cancer. Apoptosis: a review of programmed cell death, 35(4), 495–516. 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng MW, vanZuylen MN, & Severson DW (2016). Apoptosis-related genes control autophagy and influence DENV-2 infection in the mosquito vector, Aedes aegypti. Insect Biochemistry and Molecular Biology, 76, 70–83. 10.1016/j.ibmb.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, & Mello CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391(6669), 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Gaur RK, & Rossi JJ (2006). The diversity of RNAi and its applications. Biotechniques, 40(4S), S4–S5. 10.2144/000112168 [DOI] [PubMed] [Google Scholar]
- Hawkins CJ, Ekert PG, Uren AG, Holmgreen SP, & Vaux DL (1998). Anti-apoptotic potential of insect cellular and viral IAPs in mammalian cells. Cell Death Differentiation, 5(7), 569–576. 10.1038/sj.cdd.4400389 [DOI] [PubMed] [Google Scholar]
- Hawkins CJ, Uren AG, Hacker G, Medcalf RL, & Vaux DL (1996). Inhibition of interleukin 1 beta-converting enzyme-mediated apoptosis of mammalian cells by baculovirus IAP. Proceedings of the National Academy of Sciences of the United States of America, 93(24), 13786–13790. 10.1073/pnas.93.24.13786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, & Rubin GM (1995). Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell, 83(7), 1253–1262. 10.1016/0092-8674(95)90150-7 [DOI] [PubMed] [Google Scholar]
- He HJ, Hou L, Wang JX, & Zhao XF (2012). The apoptosis inhibitor survivin prevents insect midgut from cell death during postembryonic development. Molecular Biology Reports, 39(2), 1691–1699. 10.1007/s11033-011-0909-9 [DOI] [PubMed] [Google Scholar]
- Huvenne H, & Smagghe G (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. Journal of Insect Physiology, 56(3), 227–235. 10.1016/j.jinsphys.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Igaki T, Yamamoto-Goto Y, Tokushige N, Kanda H, & Miura M (2002). Down-regulation of DIAP1 triggers a novel Drosophila cell death pathway mediated by Dark and DRONC. Journal of Biological Chemistry, 277(26), 23103–23106. 10.1074/jbc.C200222200 [DOI] [PubMed] [Google Scholar]
- Kim D, & Rossi J (2008). RNAi mechanisms and applications. Biotechniques, 44(5), 613–616. 10.2144/000112792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr E, Fishilevich E, Tenbusch L, Frey MLF, Rangasamy M, Billion A, … Narva KE (2018). Gene silencing in Tribolium castaneum as a tool for the targeted identification of candidate RNAi targets in crop pests. Scientific Reports, 8(1), 2061. 10.1038/s41598-018-20416-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, … Korneluk RG (1996). Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature, 379(6563), 349–353. 10.1038/379349a0 [DOI] [PubMed] [Google Scholar]
- Lu YH, Wang K, He R, & Xi T (2008). Knockdown of survivin and upregulation of p53 gene expression by small interfering RNA induces apoptosis in human gastric carcinoma cell line SGC-823. Cancer Biotherapy Radiopharmocology, 23(6), 727–734. 10.1089/cbr.2008.0503 [DOI] [PubMed] [Google Scholar]
- Miura M (2011). Active participation of cell death in development and organismal homeostasis. Development Growth and Differentiation, 53(2), 125–136. 10.1111/j.1440-169X.2010.01228.x [DOI] [PubMed] [Google Scholar]
- Mogilicherla K, Howell JL, & Palli SR (2018). Improving RNAi in the brown marmorated stink bug: Identification of target genes and reference genes for RT-qPCR. Scientific Reports, 8, 3720. 10.1038/s41598-018-22035-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV, & Rossi JJ (2006). Antiviral applications of RNAi. Current Opinion in Molecular Therapeutics, 8(2), 115–121. [PubMed] [Google Scholar]
- Palli RS (2014). RNA interference in Colorado potato beetle: Steps toward development of dsRNA as a commercial insecticide. Current Opinion in Insect Science, 6, 1–8. 10.1016/j.cois.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R, Tan A, Bai H, & Palli SR (2008). Transcription factor broad suppresses precocious development of adult structures during larval-pupal metamorphosis in the red flour beetle, Tribolium castaneum. Mechanisms of Development, 125(3-4), 299–313. 10.1016/j.mod.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell M, Pyati P, Cao M, Bell H, Gatehouse JA, & Fitches E (2017). Insecticidal effects of dsRNA targeting the Diap1 gene in dipteran pests. Scientific Reports, 7(1):15147. 10.1038/s41598-017-15534-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglise JM, Estep AS, & Becnel JJ (2016). Expression profiles and RNAi silencing of inhibitor of apoptosis transcripts in Aedes, Anopheles, and Culex mosquitoes (Diptera: Culicidae). Journal of Medical Entomology, 53(2), 304–314. doi.org/ 10.1093/jme/tjv191 [DOI] [PubMed] [Google Scholar]
- Reidhaar-Olson JF, & Rondinone CM (2009). Therapeutic applications of RNAi. Methods and protocols. Preface. Methods in Molecular Biology, 555, v–vi. [PubMed] [Google Scholar]
- Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, … Bucher G (2008). The genome of the model beetle and pest Tribolium castaneum. Nature, 452(7190), 949–955. 10.1038/nature06784 [DOI] [PubMed] [Google Scholar]
- Rodrigues TB, Dhandapani RK, Duan JJ, & Palli SR (2017). RNA interference in the Asian longhorned beetle: Identification of key RNAi genes and reference genes for RT-qPCR. Scientific Reports, 7, 8913. 10.1038/s41598-017-08813-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M, Qadir MI, Perveen N, Ahmad B, Saleem U, Irshad T, & Ahmad B (2013). Inhibitors of apoptotic proteins: New targets for anticancer therapy. Chemistry Biology of Drug Design, 82(3), 243–251. 10.1111/cbdd.12176 [DOI] [PubMed] [Google Scholar]
- Silke J, & Vaux DL (2001). Two kinds of BIR-containing protein - inhibitors of apoptosis, or required for mitosis. Journal of Cell Science, 114(Pt 10), 1821–1827. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, & Ashwell JD (2008). IAPs: What's in a name? Molecular Cell, 30(2), 123–135. 10.1016/j.molcel.2008.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D, & Bucher G (2008). Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biology, 9(1), R10. doi:ARTN R10 10.1186/gb-2008-9-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Haecker G, & Strasser A (1994). An evolutionary perspective on apoptosis. Cell, 76(5), 777–779. 10.1016/0092-8674(94)90350-6 [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Coulson EJ, & Vaux DL (2001). Inhibitor of apoptosis proteins and their relatives: IAPs and other BIRPs. Genome Biology, 2(7):reviews3009.1. 10.1186/gb-2001-2-7-reviews3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaplana L, & O'Reilly DR (2003). Functional interaction between Cydia pomonella granulovirus IAP proteins. Virus Research, 92(1), 107–111. 10.1016/s0168-1702(02)00324-6 [DOI] [PubMed] [Google Scholar]
- Walker WB III, & Allen ML (2011). RNA interference-mediated knockdown of IAP in Lygus lineolaris induces mortality in adult and pre-adult life stages. Entomologia Experimentalis et Applicata, 138(2), 83–92. 10.1111/j.1570-7458.2010.01078.x [DOI] [Google Scholar]
- Wu Y, Parthasarathy R, Bai H, & Palli SR (2006). Mechanisms of midgut remodeling: Juvenile hormone analog methoprene blocks midgut metamorphosis by modulating ecdysone action. Mechanisms of Development, 123(7), 530–547. 10.1016/j.mod.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Yoon JS, Mogilicherla K, Gurusamy D, Chen X, Chereddy S, & Palli SR (2018). Double-stranded RNA binding protein, Staufen, is required for the initiation of RNAi in coleopteran insects. Proceedings of the National Academy of Sciences of the United States of America, 115(33), 8334–8339. 10.1073/pnas.1809381115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JS, Shukla JN, Gong ZJ, Mogilicherla K, & Palli SR (2016). RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: Identification of key contributors. Insect Biochemistry and Molecular Biology, 78, 78–88. 10.1016/j.ibmb.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Zhu F, Parthasarathy R, Bai H, Woithe K, Kaussmann M, Nauen R, … Palli SR (2010). A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proceedings of the National Academy of Sciences of the United States of America, 107(19), 8557–8562. 10.1073/pnas1000059107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu KY, & Palli SR (2020). Mechanisms, applications, and challenges of insect RNA interference. Annual Review of Entomology, 65, 293–311. 10.1146/annurev-ento-011019-025224 [DOI] [PMC free article] [PubMed] [Google Scholar]