Significance
Understanding the molecular mechanisms of SARS-CoV-2 viral pathogenesis and developing novel inhibitors are essential and urgent. Using genome-wide N6,2′-O-dimethyladenosine (m6Am)-exo-seq, RNA-seq analysis, and CRISPR/Cas9 genome editing, we discovered that phosphorylated CTD-interacting factor 1 (PCIF1), a cap-specific adenine N6-methyltransferase, plays a major role in facilitating infection of primary human lung epithelial cells and cell lines by SARS-CoV-2, variants of concern, and other coronaviruses. PCIF1 promotes infection by sustaining expression of the coronavirus receptors ACE2 and TMPRSS2 via m6Am-dependent mRNA stabilization. These results suggest a role for PCIF1 and cap m6Am in regulating SARS-CoV-2 susceptibility and identify a potential therapeutic target for prevention of infection.
Keywords: m6Am methylation, SARS-CoV-2, ACE2, NHBE cells, TMPRSS2
Abstract
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to be a major health problem worldwide. Due to the fast emergence of SARS-CoV-2 variants, understanding the molecular mechanisms of viral pathogenesis and developing novel inhibitors are essential and urgent. Here, we investigated the potential roles of N6,2′-O-dimethyladenosine (m6Am), one of the most abundant modifications of eukaryotic messenger ribonucleic acid (mRNAs), in SARS-CoV-2 infection of human cells. Using genome-wide m6Am-exo-seq, RNA sequencing analysis, and Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome editing, we demonstrate that phosphorylated C-terminal domain (CTD)-interacting factor 1 (PCIF1), a cap-specific adenine N6-methyltransferase, plays a major role in facilitating infection of primary human lung epithelial cells and cell lines by SARS-CoV-2, variants of concern, and other coronaviruses. We show that PCIF1 promotes infection by sustaining expression of the coronavirus receptors angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) via m6Am-dependent mRNA stabilization. In PCIF1-depleted cells, both ACE2/TMPRSS2 expression and viral infection are rescued by re-expression of wild-type, but not catalytically inactive, PCIF1. These findings suggest a role for PCIF1 and cap m6Am in regulating SARS-CoV-2 susceptibility and identify a potential therapeutic target for prevention of infection.
Despite the development and deployment of several effective vaccines, infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), continues to be a global public health threat. As of May 2022, SARS-CoV-2 has infected more than 534 million people and led to more than 6 million deaths worldwide (https://covid19.who.int). Emerging variants of concern (VOCs) such as Beta (B.1.351), Delta (B.1.617.2), and, more recently, Omicron (B.1.1.529) display partial resistance to neutralizing antibodies acquired naturally or via vaccination and continue to raise concern. Moreover, access to the current vaccines in many countries is limited by cost and/or distribution problems. Thus, there remains a need to take new approaches to the development of agents that prevent infection by SARS-CoV-2 and VOCs and/or treat the subsequent disease (1–5).
Entry of coronaviruses into human cells is initiated by viral spike proteins, which are composed of S1 and S2 subdomains. The S1 subunit contains the receptor-binding domain that determines the viral species specificity and tissue tropism. In the case of SARS-CoV-2, S1 binds to its specific cell-surface receptor, ACE2 (6), which exposes an S2′ site in the S2 subunit and enables its cleavage by the host enzyme TMPRSS2. In turn, exposure of a fusion peptide adjacent to the S2′ cleavage site initiates membrane fusion and pore formation, which is essential for viral entry. Most human coronaviruses, including SARS-CoV-2, SARS-CoV, Middle East respiratory syndrome (MERS)-CoV, HCoV-229E, HCoV-HKU1, and HCoV-OC43, rely on TMPRSS2 for proteolytic activation and viral entry into target cells (7–11). Therefore, ACE2 and TMPRSS2 represent promising targets for the development of pan-coronavirus antiviral therapeutic agents. Indeed, blocking antibodies, bioengineered mimics, and small-molecule inhibitors of these two molecules have shown excellent activity against anti-SARS-CoV-2 infection in vitro and in vivo (6, 9, 12–15).
N6-methyladenosine (m6A) is the most abundant and widely studied RNA modification and plays diverse roles in host–virus interactions (16, 17), including viral replication and assembly and the host antiviral immune response (18–20). We recently demonstrated that m6A methylation of SARS-CoV-2 genomic RNA decreases its recognition by the cytosolic pattern recognition receptor retinoic acid-inducible gene I (RIG-I) and subsequently attenuates activation of the type I interferon inflammatory response (21). N6,2′-O-dimethyladenosine at the 5′ mRNA cap (m6Am) is another abundant RNA modification that was recently reported to be catalyzed by phosphorylated CTD-interacting factor 1 (PCIF1) (22–25). PCIF1 has been shown to restrict HIV replication by enhancing expression of the host transcription factor ETS1, which binds to the HIV promoter and inhibits viral transcription (26). However, whether m6Am modification of mRNA encoding host proteins is involved in the life cycle of human coronaviruses, including SARS-CoV-2, is unclear.
In the present study, we investigated the potential influence of m6Am modification of host mRNAs on coronavirus infection by performing genome-wide RNA sequencing (RNA-seq) and m6Am-exo-seq of human lung cells. We found that PCIF1 plays an important role in facilitating infection by SARS-CoV-2 via stabilization of ACE2 and TMPRSS2 mRNA and maintenance of protein expression. We also demonstrate that PCIF1 depletion inhibits infection by SARS-CoV-2 and several other pathogenic human coronaviruses, suggesting that targeting of m6Am-modulating factors may represent an approach to combat infection by SARS-CoV-2 and other coronaviruses.
Results
PCIF1 Facilitates Entry of SARS-CoV-2 and VOCs into Human Lung Cells.
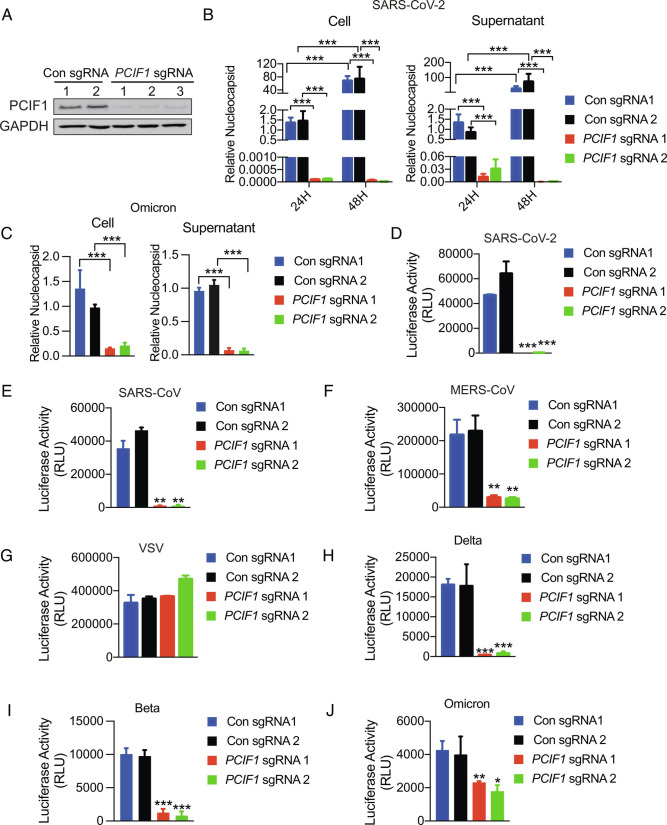
To investigate the potential role of m6Am-modified mRNA in SARS-CoV-2 infection, we first examined the effect of CRISPR/Cas9 sgRNA-mediated PCIF1 knockout (KO) on infection of the human lung adenocarcinoma cell line Calu-3 with authentic SARS-CoV-2 (strain USA-WA1/2020; hereafter referred to as parental SARS-CoV-2) or Omicron (strain BA.1). Efficient depletion of PCIF1 was confirmed by western blotting (Fig. 1A), and cellular proliferation was not significantly affected by PCIF1 depletion (SI Appendix, Fig. S1A). Control (0.5 × 106) or PCIF1-KO Calu-3 cells were seeded into the 24 wells and infected with SARS-CoV-2 and Omicron at a multiplicity of infection (MOI) of 0.1 for 1 h, and virus infection was then quantified by qRT-PCR analysis of nucleocapsid mRNA in the collected cells and culture supernatants at 24 or 48 h post-infection, respectively. Relative to the control cells, nucleocapsid mRNA levels were dramatically reduced in cells and supernatants following infection with SARS-CoV-2 (Fig. 1B) and Omicron (Fig. 1C), indicating that PCIF1 expression is essential for infection with these viruses. To determine which stage of the viral life cycle was disrupted by PCIF1 depletion, we infected control and PCIF1-KO Calu-3 cells with Vesicular stomatitis virus (VSV)-based pseudovirus-bearing SARS-CoV-2 spike protein and firefly luciferase, and viral entry was quantified after 24 h by measuring cell-associated luciferase activity. The results demonstrated that pseudovirus entry was virtually abolished by PCIF1 depletion (Fig. 1D), an observation confirmed by experiments with pseudoviruses with SARS-CoV-2 spike protein and Enhanced green fluorescent protein (EGFP) (SI Appendix, Fig. S1B). We next examined whether PCIF1 was essential for infection with other human coronaviruses by constructing pseudoviruses harboring luciferase and spike proteins from SARS-CoV, MERS-CoV, SARS-CoV-2 VOCs Delta, Beta, and Omicron, or VSV. This analysis demonstrated that PCIF1-KO significantly reduced infection of Calu-3 cells by all of the coronaviruses tested, although the effect on Omicron pseudovirus entry was relatively modest (Fig. 1 E–J and SI Appendix, Fig. S1 C–H). Interestingly, a possible explanation for the latter observation comes from two recent studies suggesting that Omicron is less dependent than other SARS-CoV-2 strains on TMPRSS2 for cell infection (27, 28). As expected, PCIF1-KO had no effect on entry of VSV pseudovirus, which employs low-density lipoprotein (LDL) receptors, and not ACE2 or TMPRSS2, as the cell receptors for entry (29) (Fig. 1G and SI Appendix, Fig. S1E). Moreover, the possibility of PCIF1 influencing the replication of VSV RNA genome from all the pseudoviruses was excluded.
Fig. 1.
Depletion of PCIF1 in human cells restricts coronavirus infection. A, Western blot analysis of Calu-3 cells subjected to CRISPR/Cas9 editing with PCIF1-targeting or non-targeting sgRNAs. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a loading control. B and C, qRT-PCR analysis of viral nucleocapsid mRNA in PCIF1-KO and control Calu-3 cells or culture supernatants after infection with authentic SARS-CoV-2 (B) or Omicron (C) at an MOI of 0.1 for 1 h. Data are expressed as the mean ± SD (n = 3). ***P < 0.001 by Student’s t test. D–J, Luciferase assays quantifying infection of PCIF1-KO and control Calu-3 cells with luciferase-expressing SARS-CoV-2 (D), SARS-CoV (E), MERS-CoV (F), VSV (G), Delta (H), Beta (I), and Omicron (J) pseudoviruses. Data are presented as the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test.
To confirm these observations with Calu-3 cells, we also performed shRNA-mediated knockdown (KD) of PCIF1 in Vero, a monkey kidney epithelial cell line, which very effectively diminished PCIF1 mRNA levels (SI Appendix, Fig. S2A), and cell proliferation was not obviously changed in PCIF1 KD cells compared to control (SI Appendix, Fig. S2B). Similar to the results with Calu-3 PCIF1-KO cells, PCIF1 KD in Vero cells resulted in significant suppression of infection by SARS-CoV-2, SARS-CoV, Delta, Beta, and Omicron pseudoviruses, but not VSV pseudovirus (SI Appendix, Fig. S2 C–H). Taken together, these data demonstrate that PCIF1 is essential for entry of a range of human coronaviruses into mammalian cells.
RNA-seq Identifies ACE2 and TMPRSS2 as Potential Targets of PCIF1.
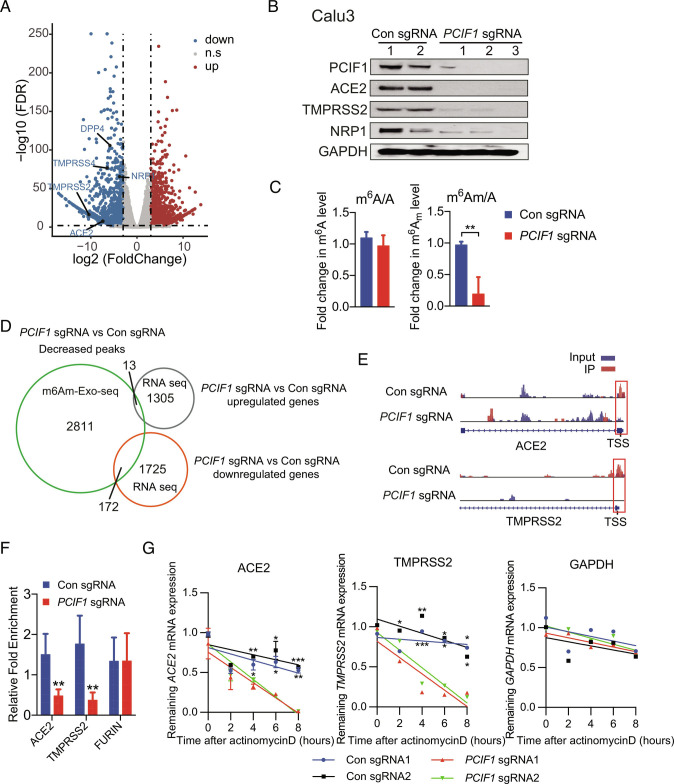
To better understand the molecular mechanisms by which PCIF1 facilitates cell entry by human coronaviruses, we performed RNA-seq of control and PCIF1-KO Calu-3 cells. Among the differentially expressed genes (DEGs), we identified several viral entry-related genes, namely ACE2, TMPRSS2, TMPRSS4, dipeptidyl peptidase 4 (DPP4), and neuropilin-1 (NRP1), that were significantly down-regulated in PCIF1-KO cells compared with control cells (Fig. 2A and Dataset S1). To validate these results, we performed qRT-PCR and western blot analysis to directly evaluate expression of these genes in control and PCIF1-KO Calu-3 cells and found significant down-regulation of all five genes at the transcript (SI Appendix, Fig. S3A) and protein (Fig. 2B) levels in PCIF1-KO cells compared with control cells. Similarly, qRT-PCR and western blot analyses demonstrated down-regulation of one or more of these genes in PCIF1-KD/KO Huh7 cells (a human hepatoma cell line; SI Appendix, Fig. S3 B and C) and PCIF1-KD Vero cells (SI Appendix, Fig. S3D).
Fig. 2.
Identification of PCIF1 target genes by RNA-seq and m6Am-exo-seq. A, Volcano plot of DEGs in PCIF1-KO Calu-3 cells compared with control cells. Significantly (q < 0.01, log2 fold change < −3 or > 3) down-regulated or up-regulated genes are shown in blue and red, respectively. B, Western blot analysis of the indicated proteins in PCIF1-KO and control Calu-3 cells. GAPDH served as a loading control. C, HPLC-MS/MS-based quantification of m6Am and m6A levels relative to unmodified adenosine (A) in mRNA from PCIF1-KO and control Calu-3 cells. Data are expressed as the mean ± SD (n = 3). **P < 0.01 by Student’s t test. D, Venn diagram showing overlap between genes identified by m6Am-exo-seq and RNA-seq as indicated. E, Genome browser views of m6Am sites in ACE2 and TMPRSS2 mRNAs from control and PCIF1-KO Calu-3 cells. Data are representative of duplicate samples. Scale for top tracks: 1 to 10/coverage. Scale for bottom tracks: 0 to 20/coverage. Read coverage of input sample and Immunoprecipitation (IP) sample are shown in blue and red, respectively. Red rectangles outline the m6Am peaks located near the TSS. F, m6Am-exo-qPCR analysis of m6Am enrichment in ACE2 and TMPRSS2 mRNAs from control and PCIF1-KO Calu-3 cells. FURIN served as a negative control. Data are presented as the mean ± SD (n = 3). **P < 0.01 by Student’s t test. G, qRT-PCR quantification of ACE2, TMPRSS2, and GAPDH mRNA in PCIF1-KO and control Calu-3 cells treated with actinomycin D for the indicated times. Data are presented as the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test.
We next investigated whether the altered expression of viral entry genes in PCIF1-depleted cells was a consequence of changes in m6Am methylation of the corresponding mRNAs. We first analyzed the ratios of m6A or m6Am to unmodified adenosine (m6A/A and m6Am/A, respectively) in total mRNA extracted from PCIF1-KO and control Calu-3 cells using high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS). Loss of PCIF1 significantly reduced m6Am/A, but not m6A/A (Fig. 2C), confirming that PCIF1 was at least partially responsible for m6Am deposition in Calu-3 cells, consistent with previous findings in human melanoma MEL624 cells (24). We then performed m6Am-exo-seq to identify m6Am-modified mRNAs and found a total of 2,811 m6Am peaks that were significantly decreased in PCIF1-KO cells relative to control cells (Fig. 2D and Dataset S2). A comparison of the m6Am-exo-seq peaks and RNA-seq genes revealed that 13 and 172 genes were up-regulated and down-regulated, respectively, by PCIF1-KO, identifying them as candidate PCIF1-mediated m6Am-modified genes (Fig. 2D and Dataset S3). Gene ontology analysis of the 172 down-regulated candidate genes demonstrated that they were enriched mainly in pathways associated with “inflammatory response”, “regulation of cell adhesion”, and “phosphoinositide 3-kinase (PI3K)-AKT-mammalian target of rapamycin (TOR) signaling” (SI Appendix, Fig. S3E). Of note, “SARS-CoV-2 and COVID-19 pathway”, which includes TMPRSS2 and ACE2, was the second most enriched pathway among the genes down-regulated by PCIF1-KO (SI Appendix, Fig. S3E). Further analysis of the m6Am-exo-seq data showed a clear reduction in m6Am peaks immediately 5′ to the transcription start sites (TSSs) of ACE2 and TMPRSS2 mRNA in PCIF1-KO Calu-3 cells compared with control cells (Fig. 2E). Finally, direct analysis of mRNAs immunoprecipitated by anti-m6A antibody using m6Am-exo-qPCR showed a significant reduction in m6Am content in ACE2 and TMPRSS2 mRNAs in PCIF1-KO cells compared with control Calu-3 cells (Fig. 2F), confirming the m6Am-exo-seq data.
To investigate the potential mechanism by which PCIF1-mediated m6Am deposition may affect ACE2 and TMPRSS2 mRNA expression, we measured the half-life of de novo synthesized ACE2 and TMPRSS2 mRNAs by qRT-PCR analysis of control and PCIF1-KO Calu-3 cells treated for various times with the transcriptional inhibitor actinomycin D. The results showed a significant time-dependent reduction in the levels of both ACE2 and TMPRSS2 mRNAs in PCIF1-KO cells compared with control cells (Fig. 2G), indicating that PCIF1 depletion destabilizes these mRNAs. Taken together, these results demonstrate that PCIF1-mediated m6Am deposition controls the stability of ACE2 and TMPRSS2 mRNAs.
PCIF1-Mediated Regulation of ACE2 and TMPRSS2 Expression Facilitates SARS-CoV-2 Entry.
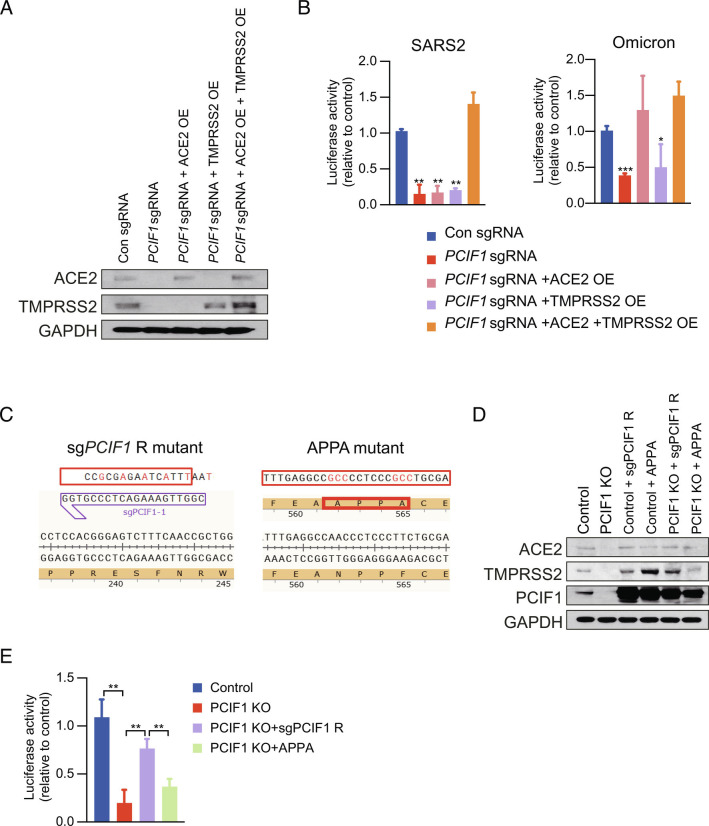
We next asked whether PCIF1-mediated control of ACE2 and TMPRSS2 mRNA stability was the mechanism by which PCIF1 modulates SARS-CoV-2 infection. To this end, we transfected control and PCIF1-KO Calu-3 cells with ACE2 and/or TMPRSS2 overexpression (OE) vectors and examined the effects on SARS-CoV-2 and Omicron pseudovirus entry. Western blot analysis confirmed that ACE2 and TMPRSS2 protein expression was restored in cells transfected with the corresponding OE vectors (Fig. 3A). Notably, co-expression of both ACE2 and TMPRSS2 in PCIF1-KO cells was required to restore SARS-CoV-2 pseudovirus infection to the level observed in control cells (Fig. 3 B, Left), whereas Omicron pseudovirus infection required ACE2 expression but was only marginally affected by restoration of TMPRSS2 expression (Fig. 3 B, Right). These results are consistent not only with our earlier observation that PCIF1-KO in Calu-3 cells had a more modest effect on entry of Omicron pseudovirus compared with other coronavirus pseudoviruses (Fig. 1J), but also with the recent report that Omicron may be less dependent than parental SARS-CoV-2 and other VOCs on TMPRSS2 for infection (30, 31). Thus, these data confirm that PCIF1-mediated regulation of ACE2 and TMPRSS2 expression levels directly modulates infection by SARS-CoV-2 and Omicron.
Fig. 3.
PCIF1 facilitates viral entry into Calu-3 cells by regulating ACE2 and TMPRSS2 expression. A, Western blot analysis of ACE2 and TMPRSS2 protein levels in control or PCIF1-KO Calu-3 cells transfected with ACE2 and/or TMPRSS2 OE vectors. GAPDH served as a loading control. B, Luciferase assays quantifying infection of control or PCIF1-KO Calu-3 cells with luciferase-encoding SARS-CoV-2 (Left) or Omicron (Right) pseudoviruses. Data are presented as the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test. C, Design of sgRNA-resistant (sgPCIF1-R) or inactive mutant (APPA) PCIF1 for rescue experiments. D, Western blot analysis of the indicated proteins in control or PCIF1-KO Calu-3 cells transfected with sgPCIF1-R or APPA OE vectors. E, Luciferase assay quantifying infection of the indicated Calu-3 cells with SARS-CoV-2 pseudovirus. Data are presented as the mean ± SD (n = 3). **P < 0.01 by Student’s t test.
To confirm that the methyltransferase activity of PCIF1 was necessary for regulation of ACE2 and TMPRSS2 expression and viral entry, we transfected control and PCIF1-KO Calu-3 cells with vectors encoding a sgRNA-resistant PCIF1 (sgPCIF1-R) or catalytically inactive mutant PCIF1 (APPA) (Fig. 3C). OE of sgPCIF1-R, but not of APPA, not only restored the expression of ACE2 and TMPRSS2 proteins in PCIF1-KO cells (Fig. 3D) but also rescued cell infection by SARS-CoV-2 pseudovirus (Fig. 3E). Therefore, SARS-CoV-2 infection depends critically on the 5′-cap methyltransferase activity of PCIF1 to sustain ACE2 and TMPRSS2 mRNA stability and protein expression.
PCIF1-Mediated Modulation of ACE2 and TMPRSS2 Expression Determines the Susceptibility of Primary Human Bronchial Epithelial Cells to SARS-CoV-2 Infection.
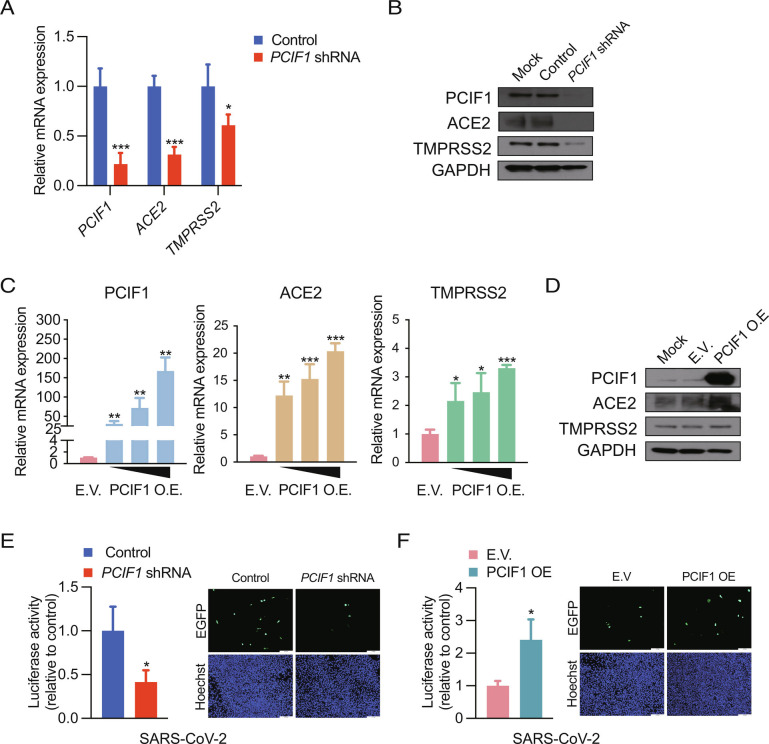
Having established the crucial role of PCIF1 in coronavirus infection of cell lines, we validated our findings in primary Normal human bronchial epithelial (NHBE) cells from healthy donors. Indeed, shRNA-mediated KD of PCIF1 in NHBE cells significantly decreased ACE2 and TMPRSS2 mRNA and protein levels (Fig. 4 A and B), and conversely, OE of PCIF1 significantly and dose-dependently increased ACE2 and TMPRSS2 mRNA and protein levels in NHBE cells (Fig. 4 C and D). Most importantly, infection of NHBE cells with either luciferase- or EGFP-expressing SARS-CoV-2 pseudoviruses was significantly decreased by PCIF1 KD (Fig. 4E) and significantly enhanced by PCIF1-OE (Fig. 4F) compared with control cells. Together, these data confirm the pathophysiological relevance of our findings by demonstrating the essential role of PCIF1 in facilitating SARS-CoV-2 infection of NHBE cells via maintenance of ACE2 and TMPRSS2 expression. Collectively, the results of this study identify the PCIF1–ACE2/TMPRSS2 axis as a critical regulator of human coronavirus entry into mammalian cells (Fig. 5).
Fig. 4.
PCIF1 regulates SARS-CoV-2 entry into primary NHBE cells by altering ACE2 and TMPRSS2 expression. A and B, qRT-PCR analysis (A) and western blot analysis (B) of PCIF1, ACE2, and TMPRSS2 mRNA and protein levels in control and PCIF1-KD NHBE cells. GAPDH served as a loading control. Data are presented as the mean ± SD (n = 3). *P < 0.05, ***P < 0.001 by Student’s t test. C and D, qRT-PCR analysis (C) and western blot analysis (D) of PCIF1, ACE2, and TMPRSS2 mRNA and protein levels in NHBE cells transfected with empty vector (EV) or a PCIF1-OE plasmid. Data are presented as the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t test. E and F, Luciferase activity (Left) and EGFP fluorescence microscopy imaging (Right) of PCIF1-KD (E) or PCIF1-OE (F) NHBE cells infected with SARS-CoV-2 pseudovirus. Nuclei were stained with Hoechst 33342. Data are presented as the mean ± SD (n = 3). *P < 0.05 by Student’s t test.
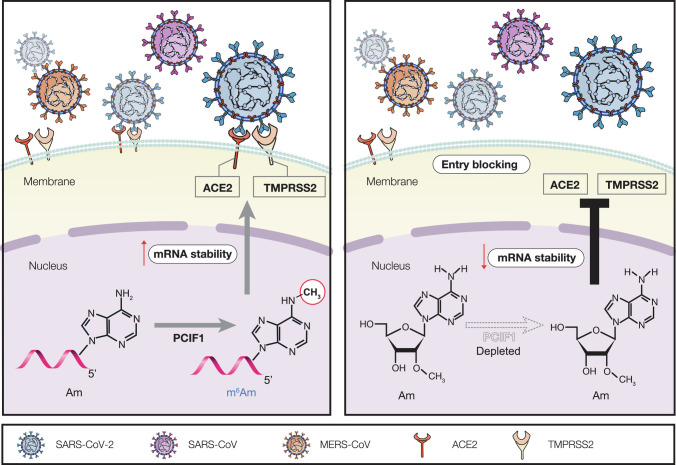
Fig. 5.
Schematic of the proposed functional and molecular mechanisms for PCIF1 involvement in virus infection.
Discussion
In this study, we demonstrated that PCIF1 regulates the entry of SARS-CoV-2 and other coronaviruses into human lung epithelial cells through its m6Am methyltransferase activity. Our analyses revealed that deposition of m6Am by PCIF1 promotes the stability of ACE2 and TMPRSS2 mRNAs, thereby sustaining the expression of these two key entry factors for many human coronaviruses, including SARS-CoV-2. Importantly, our findings were validated in primary NHBE cells and supported by the demonstration of positive correlations between PCIF1 and ACE2/TMPRSS2 expression levels in human lung tissues. Collectively, these results identify a novel pathway regulating SARS-CoV-2 entry into human lung epithelial cells and suggest a new approach to the development of agents to reduce or block SARS-CoV-2 infection.
Several studies have shown ACE2 transcription in human cells is regulated by multiple factors (31–33). Wei et al. performed a genome-wide CRISPR KO screen of Vero-E6 cells with authentic SARS-CoV-2 and identified the nucleoprotein high mobility group box protein 1 (HMGB1) as a regulator of chromatin accessibility at the ACE2 locus (31). In another genome-wide CRISPR KO of Calu-3 cells, Samelson et al. found that bromodomain-containing protein 2 (BRD2), a transcriptional regulator, promotes ACE2 transcription, and the authors further demonstrated that the BRD2 inhibitor ABBV-774 inhibited SARS-CoV-2 infection in vitro and in vivo (32). The transcriptional regulator GATA-binding protein 6 (GATA6) was shown to affect the susceptibility of Calu-3 cells to SARS-CoV-2 infection by directly binding to the ACE2 promoter (33). Thus, multiple host factors have been implicated in regulating cellular susceptibility to SARS-CoV-2 infection through modulation of ACE2 transcription. However, to the best of our knowledge, the present study is the first to identify posttranscriptional modulation of ACE2 and other SARS-CoV-2 entry factors and to demonstrate its critical role in permitting viral infection.
Approximately 30% of cellular mRNA is m6Am-modified at the cap structure (34), highlighting the functional significance of this modification. m6Am is known to increase the stability of a subset of host mRNAs and/or to decrease cap-dependent mRNA translation (23, 24); however, little is known about the role of m6Am on virus–host interactions. Our previous study revealed that PCIF1-mediated m6Am methylation of the transcriptional factor ETS1 enhanced its stability and restricted HIV infection of CD4+ T cells (35). Notably, ETS1 influenced viral genome transcription by direct binding to HIV promoters (26). Complementary to our previous study, we provide here a second example of the key role played by PCIF1-mediated m6Am deposition in modulating the stability of host mRNAs that are essential for viral entry into human cells. Strikingly, we showed that PCIF1-KO virtually abolished entry of both authentic and pseudotyped SARS-CoV-2 virus, as well as several SARS-CoV-2 VOCs and other human coronaviruses. We speculated that some essential entry factors for human coronaviruses including SARS-CoV-2 were regulated by PCIF1. Our hypothesis was confirmed that mRNA level and m6Am peak of 172 host genes were simultaneously down-regulated by PCIF1-KO and the key entry factors ACE2 and TMPRSS2 are in this list (Fig. 2D). ACE2 is the viral receptor and mediates the entry of SARS-related and some other human coronavirus (e.g., HCoV-NL63). Therefore, previous studies showing factors regulating ACE2 transcription can affect the entry of these ACE2-utilized coronaviruses (31). While TMPRSS2 is coronavirus-dependent entry factors (7–11), simultaneous regulation of both ACE2 and TMPRSS2 by PCIF1 can affect the susceptibility of a broader spectrum of human coronaviruses.
Two direct-acting antiviral drugs were recently granted emergency use authorization from the US Food and Drug Administration for the treatment of mild-to-moderate COVID-19 in certain subpopulations. Nirmatrelvir/ritonavir (Paxlovid; Pfizer) is a combination of a SARS-CoV-2 protease inhibitor (nirmatrelvir) and a pharmacokinetic boosting agent (ritonavir), and molnupiravir (Lagevirio; Merck) induces mutagenesis by viral RNA-dependent RNA polymerase (36, 37). However, a combination of the high intrinsic mutation rates of viruses and the emergence of new SARS-CoV-2 VOCs raises concerns about the development of drug-resistant strains (38). New approaches to antiviral drug development are needed that target host factors, thereby reducing the potential for drug resistance. Moreover, combination therapy with both viral- and host-targeted agents may have a synergistic effect and enhance the antiviral efficiency. The results of the present study suggest that inhibitors of the PCIF1–ACE2/TMPRSS2 axis may be effective for protection against currently circulating and emerging coronaviruses.
Methods
Cell Culture and Lentivirus Production and Transduction.
All studies were performed based on the approved Institutional Review Board (IRB) protocols by the University of California San Diego (UCSD). Calu-3 (HTB-55), Vero (CCL-81), and 293T (CRL-3216) cells were purchased from American Type Culture Collection. NHBE cells (CC-2540S) were obtained from Lonza. Huh7 was maintained in our lab. Calu-3 was grown in Minimum essential medium (MEM) (Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 1× nonessential amino acids (Gibco), and penicillin-streptomycin (100 IU/mL; Gibco). 293T, Vero, and Huh7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) supplemented with 10% FBS. NHBE cells were cultured in a human bronchial epithelial cell growth medium as previously described (39). All cells were maintained at 37 °C in a 5% CO2 atmosphere. 293T cells were seeded in 6-well plates and co-transfected with the packaging plasmids psPAX.2, pMD2.G, and CRISPRV2 (Addgene 52961) together with i) PCIF1 single-guide (sg) RNAs, ii) pLKO.1 (Addgene 8453) encoding PCIF1 shRNAs, or iii) pLVX vectors encoding PCIF1, sgPCIF1-R, or APPA using Lipofectamine 2000 Transfection Reagent (Life Technologies, 11668027) in Opti-MEM medium (Gibco) according to the manufacturers’ instructions. After 4 to 6 h at 37 °C, the supernatant was replaced with fresh medium, and the cells were incubated at 37 °C for an additional 48 h. The culture supernatant was harvested and used to infect cells by spin transduction. The gene-targeting sgRNAs used were PCIF1-sgRNA1: CGGTTGAAAGACTCCCGTGG, PCIF1-sgRNA2: TTCGGCGTGGGCCTCTACGA, and PCIF1-sgRNA3: ATTCACCAACCAGTCCCTGT. The PCIF1 shRNA target sequences were CATCCAGACCAATGCTGTCAT and CGTGGTCTGCATCCGGTATAA.
Western Blot Analysis.
Cells were lysed in Radioimmunoprecipitation (RIPA) buffer supplemented with protease inhibitors (Life Technologies, 87786) and centrifuged. Protein concentration in the supernatant was measured using a Bicinchoninic acid assay (BCA) protein assay kit (Bio-Rad). Proteins were resolved on 4 to 12% NuPAGE Bis-Tris gels (Fisher Scientific) and transferred to Polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Western blotting was performed using a Pierce Fast Western Blot Kit (Thermo Scientific) according to the manufacturer’s protocol with the following primary antibodies: GAPDH (Proteintech, HRP-60004), PCIF1 (Proteintech, 16082-1), ACE2 (Novus, NBP2-67692), TMPRSS2 (Abcam, ab92323), and NRP1 (Abcam, ab81321).
RNA Purification and qRT-PCR.
RNA was extracted using a Quick-RNA Miniprep Kit (Zymo Research, R1055) according to the manufacturer’s instructions. Purified RNA was reverse transcribed using an iScript c-DNA Synthesis Kit (Bio-Rad, 1708841) and then quantified with SsoAdvanced Universal SYBR Green PCR SuperMix (Bio-Rad, 1725270) on a LightCycler 480 PCR system. qPCR primers used are shown in SI Appendix, Table S1.
RNA-seq.
RNA was extracted from PCIF1-KO and control Calu-3 cells (three biological replicates for each) using a Quick-RNA Miniprep Kit. RNA-seq library preparation and sequencing were performed at the IGM Genomics Center, UCSD, using an Illumina NovaSeq 6000. For the analysis, paired-end reads were trimmed by cutadapt (v1.18) and then mapped to the human genome (hg38) using HISAT2 (v2.1.0). Transcripts were quantified using HTSeq (0.11.2), and DEGs were determined using DESeq2. DEGs with q < 0.01 and log2 fold change < −3 or > 3 were considered significant and selected for further analysis.
Packaging and Infection of SARS-CoV-2, VOCs, SARS-CoV, MERS-CoV, and VSV Pseudoviruses.
All the spike encoding plasmids below are codon optimized. The plasmids encoding spike of SARS-CoV-2 strains USA-WA1/2020 (WT), B. 1. 351 (Beta) were constructed in our lab. The spike encoding plasmids of B.1.617.2 (Delta) and B.1.1.529 (Omicron) were purchased from InvivoGen. The plasmids of SARS-CoV (CUHK-W1) spike and MERS-CoV (HCoV-EMC/2012) spike were obtained from Sino Biological. 293T cells were transfected with plasmids expressing the spike proteins of the indicated viruses (19 amino acids at the C termini of the spike proteins were deleted for SARS-CoV-2 WT and VOCs as reported previously (40, 41). After transfection for 24 h, 293T cells were infected with a VSV pseudovirus harboring firefly luciferase or EGFP at an MOI of 5 for 1 h. The cells were washed extensively to remove the added VSV pseudovirus. The supernatants containing pseudoviruses were then collected, centrifuged, and stored at −80 °C.
A total of 0.5 × 106 control and PCIF1-KO Calu-3 cells were infected with SARS-CoV-2, Delta, Beta, Omicron, SARS-CoV, MERS-CoV, or VSV pseudoviruses in serum-free medium for 2 h. The supernatants were then replaced with fresh media and the cells were incubated for an additional 24 h. Cells incubated with EGFP-expressing pseudoviruses were examined by fluorescence microscopy, and images were acquired from at least four random fields. Cells incubated with luciferase-expressing pseudoviruses were lysed, and luciferase activity was measured using a luciferase activity assay kit (Promega, E1500) according to the manufacturer’s instructions.
HPLC-MS/MS.
mRNA was purified from cells using a Magnetic mRNA Isolation Kit (New England Biolabs, S1550S) according to the manufacturer’s protocol. Aliquots (0.5 μg) of purified mRNA were de-capped by incubation with 1 U Cap-Clip (CellScript) at 37 °C for 60 min and then digested with 0.5 U nuclease P1 (Sigma) at 37 °C for 2 h. Ammonium bicarbonate and alkaline phosphatase (Sigma) were added, and the mixtures were incubated at 37 °C for an additional 2 h. All samples were then filtered (0.22-µm pore, Millipore) and analyzed by HPLC-MS/MS as previously described (21, 26).
m6Am-exo-seq and m6Am-exo-qPCR.
m6Am-exo-seq was performed according to protocols developed by the Shi group (24). Briefly, mRNA was extracted from PCIF1-KO and control Calu-3 cells using a Magnetic mRNA Isolation Kit. Aliquots (100 μg) of mRNA were fragmented using a Fragmentation Reagents Kit (Invitrogen, AM8740) according to the manufacturer’s protocol. Fragmented mRNAs were phosphorylated with T4 PNK (NEB, M0201S) and then dephosphorylated with Terminator 5′-Phosphate-Dependent Exonuclease (Lucigen). Finally, Cap-Clip (CellScript) was added to remove capped transcripts. Of the final uncapped fragmented mRNA preparation, 10% was reserved as input material, and the remaining 90% was subjected to immunoprecipitation with anti-m6A antibody (Abcam, ab151230). Immunoprecipitated RNA was eluted with RLT buffer (QIAGEN, 160051456), purified by ethanol precipitation, and prepared for library generation using a TruSeq mRNA library preparation kit (Illumina). Sequencing was performed at IGM Genomics Center, UCSD, using an Illumina NovaSeq 6000.
For m6Am-exo-qPCR, the same procedure as for m6Am-exo-seq was used through the anti-m6Am immunoprecipitation and RNA elution step. The eluted RNA and input samples were reverse transcribed and subjected to qPCR on a LightCycler 480 (Roche Diagnostics) using the primers listed in SI Appendix, Table S4.
m6Am-exo-seq Analysis.
Briefly, paired-end reads were trimmed using cutadapt and then mapped to the GRCh38 human genome using HISAT2 (v2.1.0). Sam files were converted to bam files and sorted using Samtools. TSS regions were extracted from TxDb.Hsapiens.UCSC.hg38.knownGene and corrected using the Cap Analysis of Gene Expression (CAGE) database. Reads within 100 nucleotides downstream of the TSS were then quantified by featureCount and normalized by the sequencing depth of each sample to obtain TSS reads of individual genes. The fold change in TSS reads was defined as
Differential P values were determined by hypergeometric tests. Genes with a fold change < 1 and differential P value < 0.05 in the PCIF1-KO sample were defined as cap m6Am-modified genes.
mRNA Stability.
Measurement of mRNA stability was performed according to a previously described protocol (42). Briefly, PCIF1-KO and control Calu-3 cells were treated with 5 µg/mL actinomycin D (Alfa Aesar, AAJ67160XF) for 0, 1, 2, 4, 6, or 8 h, and then ACE2, TMPRSS2, TMPRSS4, NRP1, and FURIN mRNAs were quantified by qRT-PCR using the primers listed in SI Appendix, Table S4.
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) Cellular Proliferation Assay.
A total of 5 × 103 cells were seeded into each well of 96-well plates and then performed the assay at the indicated time by using a CellTiter AQueous One Solution Cell Proliferation Assay Kit (Promega, G3580) following the manufacturer’s instructions. Absorbance at 490 nm was recorded.
Statistical Analysis.
Data are presented as the mean ± SD unless noted. Differences between group means were analyzed using Student’s t test with Prism 5.0 software (GraphPad). P values < 0.05 were considered significant.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (CSV)
Dataset S02 (CSV)
Dataset S03 (XLSX)
Acknowledgments
We thank Dr. Kristen Jepsen of the Institute of Genomic Medicine at UCSD for help with the HT-seq and members of the Rana lab for helpful discussions and advice. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281. This work was supported by a Career Award for Medical Scientists from the Burroughs Wellcome Fund USA and a grant from the National Institutes of Health USA (K08 AI130381) to A.F.C., and in part by grants from the National Institutes of Health (CA177322, DA039562, DA046171, and AI125103). G.M.G. was supported in part by NRSA institutional training grant (T32 ES018827).
Author contributions
T.M.R. designed research; L. Wang, S.W., L. Wu, W.L., W.B., A.E.C., and G.M.G. performed research; Y.W. and A.F.C. contributed new reagents/analytic tools; L. Wang, S.W., Y.W., A.F.C., and T.M.R. analyzed data; and L. Wang, S.W., and T.M.R. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
RNA-seq and m6Am-exo-seq data have been deposited at the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE208533 (43).
Supporting Information
References
- 1.Cao Y., et al. , Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663 (2022), 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dejnirattisai W., et al. , SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 185, 467–484.e415 (2022), 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M., et al. , The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell 185, 447–456.e411 (2022), 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L., et al. , Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 602, 676–681 (2022), 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 5.Viana R., et al. , Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 603, 679–686 (2022), 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du Y., et al. , A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants. Nat. Commun. 12, 5000 (2021), 10.1038/s41467-021-25331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertram S., et al. , TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 87, 6150–6160 (2013), 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glowacka I., et al. , Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85, 4122–4134 (2011), 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastard P., et al. , Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, eabd4585 (2020), 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J. E., et al. , Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U.S.A. 113, 12262–12267 (2016), 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirato K., Kawase M., Matsuyama S., Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 517, 9–15 (2018), 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glasgow A., et al. , Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 117, 28046–28055 (2020), 10.1073/pnas.2016093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahoney M., et al. , A novel class of TMPRSS2 inhibitors potently block SARS-CoV-2 and MERS-CoV viral entry and protect human epithelial lung cells. Proc. Natl. Acad. Sci. U.S.A. 118, e2108728118 (2021), 10.1073/pnas.2108728118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapira T., et al. , A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature 605, 340–348 (2022), 10.1038/s41586-022-04661-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z., et al. , Potent prophylactic and therapeutic efficacy of recombinant human ACE2-Fc against SARS-CoV-2 infection in vivo. Cell Discov. 7, 65 (2021), 10.1038/s41421-021-00302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominissini D., et al. , Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012), 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz S., et al. , Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 8, 284–296 (2014), 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gokhale N. S., et al. , N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665 (2016), 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lichinchi G., et al. , Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20, 666–673 (2016), 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu M., et al. , N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5, 584–598 (2020), 10.1038/s41564-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N., et al. , METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 35, 109091 (2021), 10.1016/j.celrep.2021.109091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akichika S., et al. , Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080 (2019), 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- 23.Boulias K., et al. , Identification of the m(6)Am methyltransferase PCIF1 reveals the location and functions of m(6)Am in the transcriptome. Mol. Cell 75, 631–643.e638 (2019), 10.1016/j.molcel.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sendinc E., et al. , PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell 75, 620–630.e629 (2019), 10.1016/j.molcel.2019.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun H., Zhang M., Li K., Bai D., Yi C., Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res. 29, 80–82 (2019), 10.1038/s41422-018-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q., et al. , HIV reprograms host m(6)Am RNA methylome by viral Vpr protein-mediated degradation of PCIF1. Nat. Commun. 12, 5543 (2021), 10.1038/s41467-021-25683-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng B., et al. , Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603, 706–714 (2022), 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao H., et al. , SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microbes Infect. 11, 277–283 (2022), 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M., LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 110, 7306–7311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inde Z., et al. , Age-dependent regulation of SARS-CoV-2 cell entry genes and cell death programs correlates with COVID-19 severity. Sci. Adv. 7, eabf8609 (2021), 10.1126/sciadv.abf8609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei J., et al. , Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 184, 76–91.e13 (2021), 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samelson A. J., et al. , BRD2 inhibition blocks SARS-CoV-2 infection by reducing transcription of the host cell receptor ACE2. Nat. Cell Biol. 24, 24–34 (2022), 10.1038/s41556-021-00821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Israeli M. A., et al. , CRISPR screens for host factors critical for infection by SARS-CoV-2 variants of concern identify GATA6 as a central modulator of ACE2. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2021.07.19.452809v2.abstract. (Accessed 18 January 2022). [Google Scholar]
- 34.Wei C., Gershowitz A., Moss B., N6, O2’-dimethyladenosine a novel methylated ribonucleoside next to the 5’ terminal of animal cell and virus mRNAs. Nature 257, 251–253 (1975), 10.1038/257251a0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q., et al. , HIV reprograms host m6Am RNA methylome by viral Vpr protein-mediated degradation of PCIF1. Nat. Commun. 12, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen D. R., et al. , An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 374, 1586–1593 (2021), 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 37.Wahl A., et al. , SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature 591, 451–457 (2021), 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chitalia V. C., Munawar A. H., A painful lesson from the COVID-19 pandemic: The need for broad-spectrum, host-directed antivirals. J. Transl. Med. 18, 390 (2020), 10.1186/s12967-020-02476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Millet J. K., Whittaker G. R., Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 111, 15214–15219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S., et al. , Cholesterol 25-hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 39, e106057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukushi S., et al. , Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Med. Virol. 86, 2269–2274 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Wang L., et al. , m6A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 39, e104514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L., PCIF1-mediated deposition of 5-cap N6,2-O-dimethyladenosine in ACE2 and TMPRSS2 mRNA regulates susceptibility to SARS-CoV-2 infection. GEO. (2022). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE208533. Deposited 19 July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (CSV)
Dataset S02 (CSV)
Dataset S03 (XLSX)
Data Availability Statement
RNA-seq and m6Am-exo-seq data have been deposited at the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE208533 (43).