Significance
The intestinal epithelium acts as the first barrier against the bacterial invasion of host tissues. Galectins are a major type of glycan-binding protein (GBP) that differ from other GBPs by being located mainly intracellularly. Galectin-4 is unique in that it is specifically expressed in the gastrointestinal tract. Here, we report that cytosolic galectin-4 enchains daughter bacteria and restricts their motility intracellularly and potentiates inflammasome activation in infected IECs. The results of our study demonstrate that cytosolic lectins can modulate the host response through interacting intracellularly with glycans on live microbes.
Keywords: galectin-4, lipopolysaccharide, Salmonella enterica, intestine
Abstract
Galectin-4, a member of the galectin family of animal glycan-binding proteins (GBPs), is specifically expressed in gastrointestinal epithelial cells and is known to be able to bind microbes. However, its function in host-gut microbe interactions remains unknown. Here, we show that intracellular galectin-4 in intestinal epithelial cells (IECs) coats cytosolic Salmonella enterica serovar Worthington and induces the formation of bacterial chains and aggregates. Galectin-4 enchains bacteria during their growth by binding to the O-antigen of lipopolysaccharides. Furthermore, the binding of galectin-4 to bacterial surfaces restricts intracellular bacterial motility. Galectin-4 enhances caspase-1 activation and mature IL-18 production in infected IECs especially when autophagy is inhibited. Finally, orally administered S. enterica serovar Worthington, which is recognized by human galectin-4 but not mouse galectin-4, translocated from the intestines to mesenteric lymph nodes less effectively in human galectin-4-transgenic mice than in littermate controls. Our results suggest that galectin-4 plays an important role in host-gut microbe interactions and prevents the dissemination of pathogens. The results of the study revealed a novel mechanism of host–microbe interactions that involves the direct binding of cytosolic lectins to glycans on intracellular microbes.
Intestinal homeostasis is controlled by interactions among the host immune system, diverse populations of microbes in the intestinal lumen, and intestinal epithelial cells (IECs). IECs are constantly exposed to many commensal microbes and are the primary sites of contact with invading and non-invading enteric pathogens. Therefore, IECs may exert critical innate immune functions to maintain the balance of gut microbiota composition and defend against pathogen attachment and invasion (1–3).
Various intracellular bacteria, such as Salmonella, can invade epithelial cells and initially reside and replicate in membrane-bound vacuoles (4). Although the majority of intracellular Salmonella stays in vacuoles, a small number can escape to the cytosol. IECs can mount the autophagy response by recognizing the disrupted bacteria-containing vacuoles or ubiquitinated cytosolic bacteria (5). While bacteria escaping to the cytosol can thereby be cleared, some populations remain and hyper-replicate, especially under conditions where the autophagy machinery is impaired (6, 7). It is known that IECs containing hyper-replicating bacteria may eventually extrude to the luminal space and undergo inflammatory cell death (pyroptosis) (8, 9). Consequently, cytosolic bacteria are released by these cells and may invade other IECs (9, 10).
Pyroptosis is mediated by inflammasome activation. Various inflammasome components are highly expressed in the intestinal epithelium, including nucleotide-binding oligomerization domain-like receptor 6 (NLR6), NLR1b, nucleotide-binding domain and leucine-rich repeat caspase recruitment domain 4 (NLRC4), ASC [apoptosis-associated speck-like protein containing a caspase-recruitment domain (CARD)], and caspase-1, caspase-4/5 (caspase-11 in mice) (11, 12). It is known that members of NAPI (NLR family of apoptosis inhibitory protein) sense bacterial flagellin or type III secretion system (T3SS) apparatus proteins and trigger NLRC4 to form an inflammasome together with ASC and pro-caspase1 (13, 14). Caspase-1 activation, associated with its autoproteolysis into enzymatically active p20 and p10 fragments, induces cleavage of the pro-form of IL-18 into an active form. In addition, noncanonical inflammasome activation is dependent on caspase-4 and triggered by cytosolic lipopolysaccharides (LPSs) (15, 16). Both caspase-1 and 4 can also induce cell death that is accompanied by IL-18 secretion (12, 17).
Glycans on microbial surfaces are important signatures that can be recognized by host glycan-binding proteins (GBPs) and are intimately involved in host–microbe interactions. LPSs are a major structure of the outer membrane of Gram-negative bacteria. It comprises a hydrophobic component named lipid A, a core oligosaccharide, and a glycan polymer with repetitive oligosaccharide units named O-antigen. Galectins are a major type of GBPs composed of conserved carbohydrate recognition domains (CRDs) that can recognize β-galactoside-based glycans. They are divided into three types based on their structure: the single-CRD type; chimeric type, which contains a non-lectin domain connected to a CRD; and tandem repeat type, which includes two distinct CRDs connected by a linker peptide (18). Galectins are mainly located in the cytosol, but can also be found in the nucleus. Although galectins are synthesized without a classical signal sequence, they can be released into the extracellular space via non-classical secretory pathways (19). Based on their localization, galectins exhibit both extracellular and intracellular functions, and the latter can be dependent or independent of glycan binding (20–23).
Galectin-4, a tandem-repeat-type galectin, is almost exclusively expressed in IECs within the gastrointestinal tract (24, 25). It was first discovered as a lectin of 36 kDa present in rat intestinal extracts and later determined to be a member of the galectin family (26, 27). Galectin-4 is an adherens junction protein and a significant component of lipid rafts in brush border membranes of the intestine (24, 28). It also binds to and recruits apical glycoproteins in human enterocyte HT-29 cells (29). The β-galactose-containing moiety is the basic component required for recognition by the galectin family of proteins. Galectin-4, however, possesses a unique carbohydrate-binding specificity, and can bind to O-linked sulfoglycans, sulfated glycosphingolipids, and biomolecules that contain no β-galactose residues, such as cholesterol 3-sulfate (30–32).
Several studies have demonstrated that galectins interact with LPS from various bacteria. For example, galectin-3 binds to Klebsiella pneumoniae and Salmonella minnesota R7 (33) and contributes to Helicobacter pylori’s colonization and adhesion to the gastric mucosa (34). Furthermore, galectin-4 and 8 are highly effective at killing Escherichia coli by binding to their cell surface O-antigen structures, which resemble the human blood group B antigen (35). However, with respect to intracellular galectin biology, the roles of host GBPs and microbial glycans in host-intracellular microbe interactions remain largely unexplored. We hypothesized that intracellular galectin-4 in IECs can readily recognize microbes that invade the cells, through binding to glycans on their surfaces.
In this study, we found that intracellular galectin-4 could bind to cytosolic Salmonella, resulting in their formation of chains and aggregates, and reduction in motility. In addition, galectin-4 amplified the inflammasome and IL-18 activation in IECs following infection with Salmonella especially when autophagy was inhibited. Furthermore, galectin-4 limited the bacterial spread from the intestinal lumen to the mesenteric lymph nodes (MLNs). We conclude that galectin-4 from IECs protects the host against pathogenic infections. Our study revealed a novel pathway in mucosal immunity that involves interaction between intracellular galectins and invading microbes.
Results
Intracellular Galectin-4 in IECs Coats Cytosolic Bacteria and Induces Their Chaining and Aggregation.
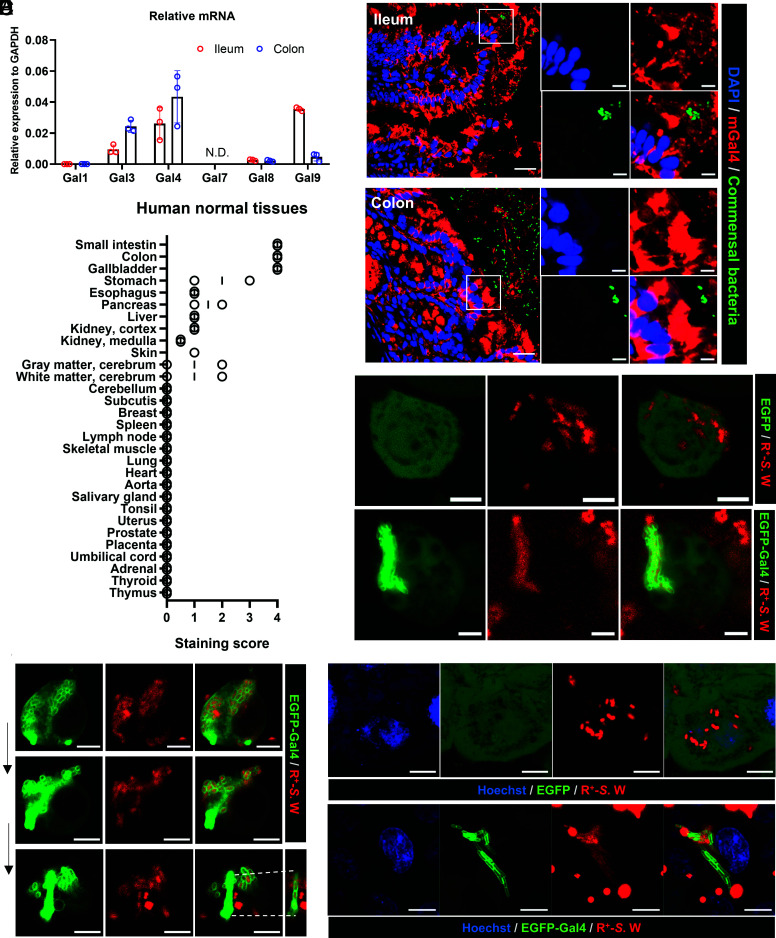
RNA was extracted from the ileum and colon of killed mice and the relative amounts of galectin mRNA were determined by real-time PCR. Various galectins were detected, including galectin-4, which was found to be highly expressed (Fig. 1A). Galectins were also detected in secretions from freshly isolated gastrointestinal tracts by enzyme-linked immunoassay; galectin-4 was particularly abundant (SI Appendix, Fig. S1). Immunostaining of intestinal tissues revealed that galectin-4 protein was present in the cytoplasmic compartment of epithelial cells of the ileum and colon, as well as associated with components in the luminal space that also contained commensal bacteria as indicated by fluorescence in situ hybridization (Fig. 1B). Next, we determined the galectin-4 expression levels in various normal human tissues by immunohistochemistry. We found that galectin-4 was highly expressed in epithelial cells of the small intestine, colon, and gallbladder (Fig. 1C). Since galectin-4 is abundantly present intracellularly, we investigated whether cytosolic galectin interacted with intracellular bacteria through lectin-glycan interactions.
Fig. 1.
Intracellular galectin-4 in IECs coats cytosolic bacteria and induces their chaining and aggregation. (A) The levels of galectin (Gal) mRNA relative to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in mouse ileum and colon tissues, as determined by real-time PCR. Data represent the mean ± SD, N = 3. N.D., not detected. (B) Immunofluorescence and fluorescence in situ hybridization staining of ileum and colon sections from a wild-type mouse. DAPI (blue), commensal bacteria detected by the universal bacterial probe (green), and anti-mouse galectin-4 (mGal4, red) staining. (Scale bars, 20 µm or 5 µm in zoomed images.) The images of isotype control staining conducted in the same experiment are shown in the SI Appendix, Fig. S2. (C) The scores of immunohistochemistry staining of galectin-4 in human normal tissues. (D) HT-29-EGFP and HT-29-EGFP-Gal4 cells both generated from HT-29 cells with endogenous galectin-4 deleted were treated with wortmannin and infected with RFP-expressing S. enterica serovar Worthington (R+-S.W). Representative Images obtained 10 h after infection are shown. Bacteria in HT-29-EGFP cells (Upper) appear as scattered red spots, while those in HT-29-EGFP-Gal4 cells (Lower) appear as compact green elongated aggregates (coated with EGFP-galectin-4). (Scale bars, 5 µm.) (E) Images of the same infected HT-29 cell were taken every 10 min by confocal microscopy after 12 h of infection. (Scale bars, 5 µm.) (F) HeLa-EGFP and HeLa-EGFP-Gal4 cells were infected with R+-S.W. Representative Images obtained 6 h after infection are shown. Bacteria in HeLa-EGFP cells (Upper) appear as scattered red spots, while those in HeLa-EGFP-Gal4 cells (Lower) appear as compact green elongated aggregates (coated with EGFP-galectin-4). Hoechst (blue), EGFP/EGFP-Gal4 (green), R+-S.W (red). (Scale bars, 5 µm.)
EGFP-galectin-4-expressing (HT-29-EGFP-Gal4) cells, as well as control cells expressing EGFP (HT-29-EGFP), were generated using the human colonic epithelial cell line HT-29, with the endogenous galectin-4 gene deleted via CRISPR (SI Appendix, Fig. S3A). Next, we infected HT-29-EGFP-Gal4 cells with S. enterica serovar Worthington, which contains polysaccharides classified as O13 antigens that are recognized by galectin-4, as determined by glycan array (36). We detected galectin-4-bound bacteria intracellularly (SI Appendix, Fig. S4A), but only in a small proportion of infected cells. This may be due to the low bacterial invasion rate in HT-29 cells. In addition, the intracellular pathogen can be eliminated via the autophagy machinery of the host cells (6, 37–39). Next, we increased the amount of S. enterica serovar Worthington with access to the cytosol by treating the cells with wortmannin, a phosphoinositide 3-kinase (PI3K) inhibitor known to inhibit autophagy (6, 7). We noted a large number of bacteria coated with galectin-4 starting at 6 h post-infection, and their amounts increased at 10 to 12 h post-infection. Specifically, chains and elongated aggregates of bacteria coated with galectin-4 (green colored in the Lower panel of Fig. 1D) were found in the cytosol of a subpopulation of HT-29-EGFP-Gal4 cells. This was in striking contrast to what was observed in control cells, where bacteria (RFP-expressing) were distributed in a scattered manner (red colored in the upper panel of Fig. 1D). The CFU number of intracellular bacteria was similar between HT-29-EGFP and HT-29-EGFP-Gal4 at 10 h post-infection (SI Appendix, Fig. S4B). Importantly, we found that Salmonella released from EGFP-galectin-4 expressing cells remained coated with galectin-4 and enchained (Fig. 1E), but not those from control cells. The basal expression of EGFP-galectin-4 and EGFP in uninfected cells is shown in SI Appendix, Fig. S3 A and B.
Due to the low invasion rate of S. enterica serovar Worthington in the HT-29 cell line, we also used the HeLa cell line, which is commonly used for studying Salmonella infection in epithelial cells; the proportion of cells containing hyper-replicating S. enterica serovar Typhimurium in the cytosol is around 10% of the infected cells (7, 40). Since HeLa cells do not express galectin-4, we transfected them with EGFP-galectin-4 (HeLa-EGFP-Gal4) (SI Appendix, Fig. S3C). Galectin-4-coated bacteria can be more easily detected without wortmannin treatment in HeLa-EGFP-Gal4 cells compared to HT-29-EGFP-Gal4 cells. Here, bacteria coated with galectin-4 were also arranged in chains and elongated aggregates (green colored in the Lower panel of Fig. 1F). In contrast, bacteria in HeLa-EGFP control cells showed a scattered-distribution pattern (red colored in the Upper panel of Fig. 1F). The basal expression of EGFP-galectin-4 and EGFP in uninfected cells is shown in SI Appendix, Fig. S3 C and D.
It is known that sifA mutant of Salmonella is more effective in escaping from the vacuoles following infection of epithelial cells and thus can replicate in the cytosol more readily (7, 41). Next, we infected HeLa-EGFP-Gal4 cells with sifA mutant of S. enterica serovar Worthington we generated and found that there was a higher percentage of cells containing cytosolic bacteria coated by galectin-4 compared to those infected with wild-type bacteria (SI Appendix, Fig. S5). In all the three systems described above, among galectin-4-expressing cells containing hyper-replicating bacteria in the cytosol, bacteria were coated with galectin-4 in nearly 100% of them. These results suggest that intracellular galectin-4 binds to cytosolic bacteria and induces the formation of bacterial chains and aggregates.
Galectin-4 Enchains Bacteria during Bacterial Growth by Binding to the O-antigen of LPS.
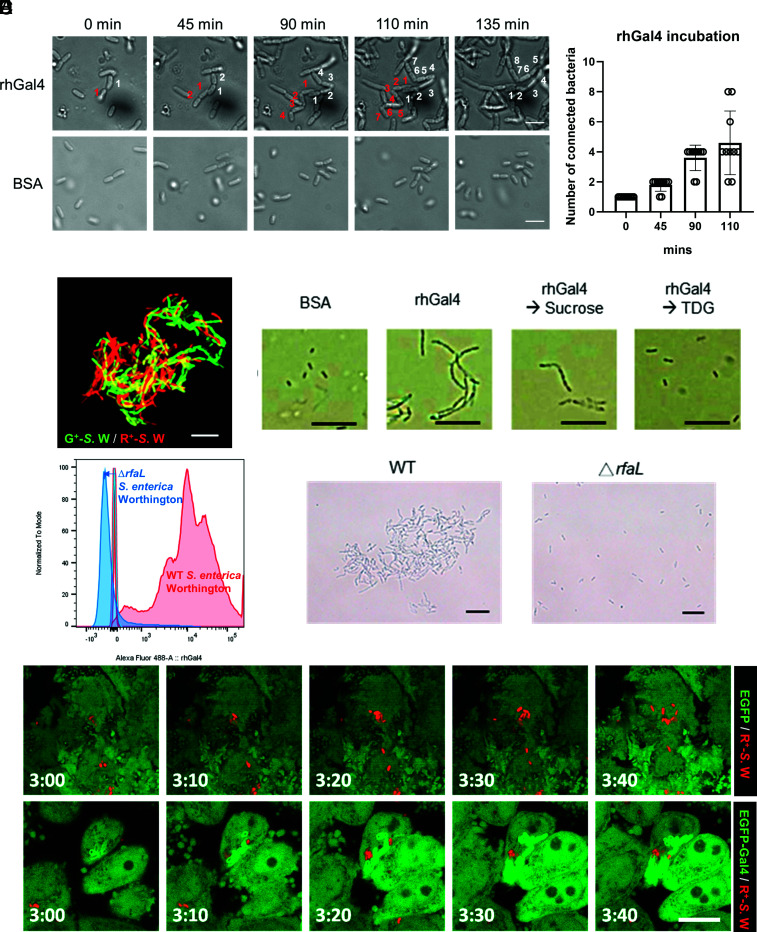
The above findings prompted us to confirm that galectin-4 could enchain growing bacteria. In the presence of human galectin-4, S. enterica serovar Worthington acquired a chaining phenotype during their growth in vitro, whereby the bacteria were connected end-to-end along the long axis (Fig. 2A and SI Appendix, Movie S1), but not in the presence of bovine serum albumin (BSA) (Fig. 2A and Movie S2). The number of connected bacteria derived from a single bacterium continued to increase throughout the period of incubation with rhGal4 (Fig. 2B). When GFP- and RFP-expressing S. enterica serovar Worthington were incubated with galectin-4, each of the individual chains contained mainly bacteria displaying the same color, suggesting that each chain was formed from the progeny of a single cell (Fig. 2C). Interestingly, although we found that a stable form of recombinant human galectin-9 (rhGal9null), lacking the linker region and resistant to proteolysis (42), also bound to S. enterica serovar Worthington (SI Appendix, Fig. S6A), it did not cause enchained growth of bacteria (SI Appendix, Fig. S6B). The bacterial chains induced by human galectin-4 could be disrupted by thiodigalactoside (TDG), which binds to galectins, but not by the negative control sucrose (Fig. 2D), suggesting that chain formation occurs through lectin-glycan interactions.
Fig. 2.
Galectin-4 enchains bacteria during bacterial growth by binding to the O-antigen of LPS. (A) Bacterial chains induced by galectin-4 are derived from the cell progeny. Images of S. enterica serovar Worthington cells incubated with 100 µg/mL recombinant human galectin-4 (rhGal4) or BSA from Movies S1 and S2, at different incubation time points. (Scale bars, 5 µm.) (B) The number of connected bacteria derived from a single bacterium was tracked throughout the period of incubation with rhGal4. Data represent the mean ± SD, N = 10. (C) GFP- and RFP-expressing bacteria (G+-S.W and R+-S.W) were incubated in LB media containing 50 µg/mL human galectin-4 at 37 °C for 2 h. Scale bars, 10 µm. (D) The chaining phenotype of S. enterica serovar Worthington induced by recombinant human galectin-4 (rhGal4) was disrupted by TDG, but not by sucrose. (Scale bars, 10 µm.) (E) Wild-type (WT) and rfaL deleted (ΔrfaL) S. enterica serovar Worthington were incubated with 2 µg/mL Alexa fluor 488-labeled human galectin-4 at 4 °C for 1 h. Galectin-4 binding was determined by flow cytometry. The images of bacteria were obtained after incubation in LB containing 50 µg/mL human galectin-4 at 37 °C for 2 h. (Scale bars, 10 µm.) (F) HeLa-EGFP and HeLa-EGFP-Gal4 cells were infected with R+-S.W. Live images were acquired at the indicated time points post-infection (h:min). EGFP/EGFP-Gal4 (green), R+-S.W (red). (Scale bars, 10 µm.)
Using biolayer interferometry (BLI), an optical biosensing technology for analysis of biomolecular interactions in real-time that can be used to quantify the binding strength, we demonstrated that galectin-4 strongly binds to LPS extracted from S. enterica serovar Worthington with KD < 1 × 10−10 M (SI Appendix, Fig. S7). O-antigens are a major component of LPS, present outside the external membrane of Gram-negative bacteria. Next, we generated S. enterica serovar Worthington lacking the O-antigen by deleting the rfaL (O-antigen ligase) gene (43). Galectin-4 was unable to bind to the rfaL mutant strain and neither bacterial chains nor clusters were observed (Fig. 2E). The results of our experiments suggest that bacterial chaining is due to the binding of galectin-4 to the O-antigen of LPS. Galectin-4 also enchained growing E. coli O19ab, O86, and S. enterica serovar Cubana (SI Appendix, Fig. S8A). The lowest concentration of galectin-4 that induced chaining was 0.78 µg/mL (~0.02 µM), as noted for E. coli O19ab; galectin-4 induced chaining of other bacteria listed above at higher concentrations (SI Appendix, Fig. S8A). Importantly, by time-lapse microscopy, we were also able to capture the enchained growth phenotype of cytosolic replicating bacteria caused by intracellular galectin-4 in HeLa-EGFP-Gal4 cells, but not in HeLa-EGFP control cells (Fig. 2F).
Galectin-4 in IECs Inhibits the Motility of Cytosolic Bacteria.
Galectin-4 was reported to bind specifically to E. coli expressing a human blood group B-like antigen through its C-terminal CRD and kill the bacteria (35). We found galectin-4 killed S.enterica serovar Worthington, which bears human blood group H antigens, under agitation, but not static culture conditions, after 1 h of incubation (SI Appendix, Fig. S9A). However, in the static culture for 2 h, along with the induction of the chaining phenotype, galectin-4 restricted the growth rate of wild-type bacteria compared to BSA and galectin-9, but did not affect that of rfaL mutant (SI Appendix, Fig. S9 B and C).
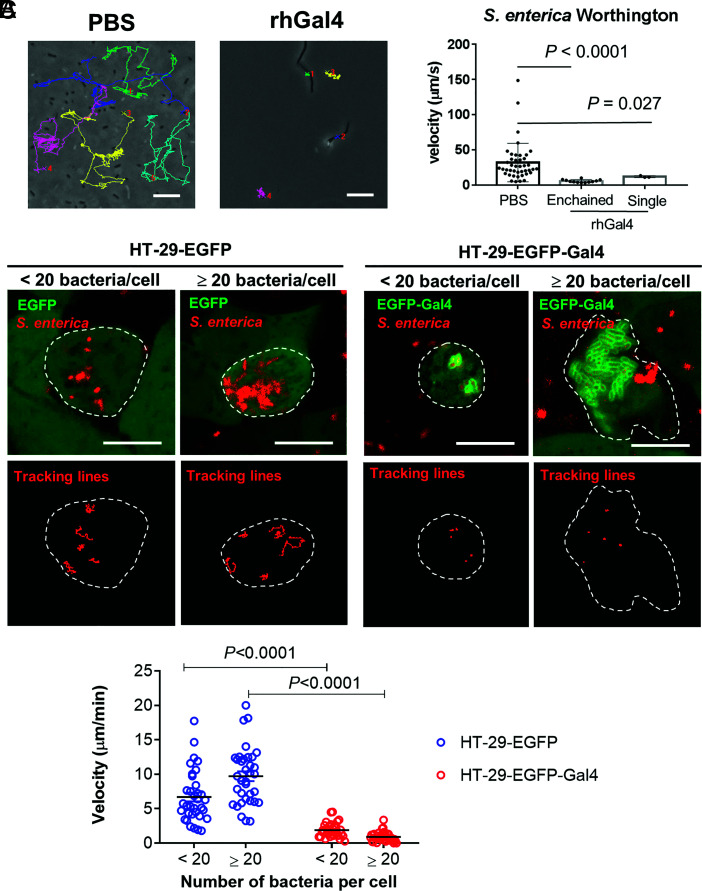
A time-lapse video revealed that the free-swimming speeds of individual and enchained S. enterica serovar Worthington significantly decreased when bacteria were cultured with human galectin-4 (Fig. 3A). The motility of a single bacterium was also reduced when bacteria were incubated with galectin-4 for only 10 min and not being enchained yet, compared to those incubated with BSA or galectin-9 (SI Appendix, Fig. S6C).
Fig. 3.
Galectin-4 in IECs inhibits the motility of cytosolic bacteria. (A) S. enterica serovar Worthington were incubated in LB containing 50 µg/mL recombinant human galectin-4 (rhGal4) or phosphate-buffered saline (PBS) at 37 °C for 2 h. (Scale bars, 10 µm.) Bacterial motility was tracked and calculated. rhGal4-enchained: enchained S. enterica serovar Worthington in LB containing rhGal4. rhGal4-single: single S. enterica serovar Worthington in LB containing rhGal4. Each dot represents an individual bacterium. Data represent the mean ± SD, and the P value was calculated using the Mann–Whitney test. (B) HT-29-EGFP and HT-29-EGFP-Gal4 cells both generated from HT-29 cells with endogenous galectin-4 deleted were treated with wortmannin and infected with RFP-expressing S. enterica serovar Worthington, MOI = 50. Time-lapse images were acquired 10 to 12 h post-infection by confocal microscopy. The images are representative of three biological replicates. The borders of the infected HT-29 cells are outlined with white dashed lines. (Scale bars, 10 µm.) The motility of an individual cytosolic bacterium was tracked (B) and calculated (C). (C) Each dot represents an individual bacterium. Three-to-five bacteria were randomly tracked in one cell. Data are representative of results from seven cells. Data represent the mean, and the P value was calculated by the unpaired t test with correction for multiple comparisons using the Holm–Sidak method.
We also tracked the fluorescently labeled intracellular S. enterica serovar Worthington in HT-29 cells with fewer than 20 bacteria or with more than 20 bacteria, respectively, using time-lapse microscopy. We found that in both groups, the motility of EGFP-galectin-4-coated bacteria was significantly diminished compared with that of their counterparts in control HT-29-EGFP cells (Fig. 3 B and C and Movies S3 and S4).
Galectin-4 Enhances Caspase-1 Activation and Mature IL-18 Production in Epithelial Cells after Bacterial Infection.
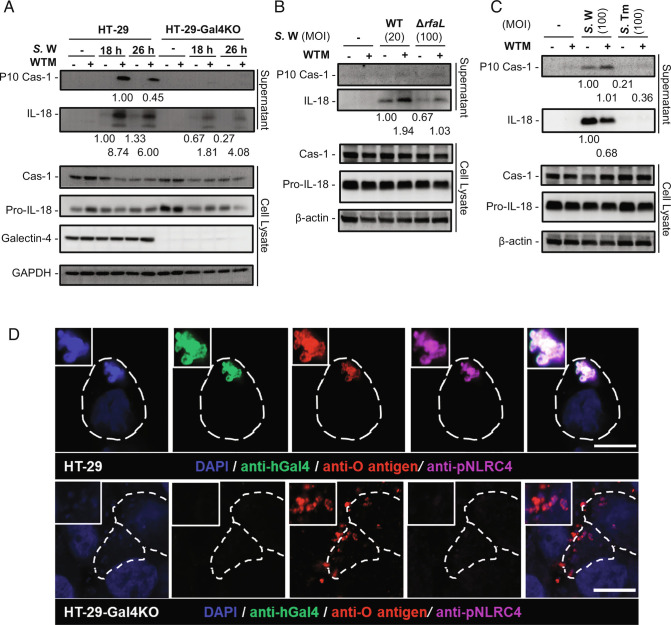
When Salmonella invades the IECs, the intracellular bacteria trigger NLRC4-NAIP inflammasome activation and activate caspase-1, leading to the maturation of pro-inflammatory cytokines (44). Next, we determined whether galectin-4 affected caspase-1 activation during S. enterica serovar Worthington infection. Caspase-1 activation, as indicated by the production of its cleavage fragment, p10, was observed in S. enterica serovar Worthington-infected HT-29 cells, but not in infected HT-29-galectin-4 knockout (Gal4-KO) cells following treatment with wortmannin (Fig. 4A). There was also a significantly higher amount of mature IL-18 release into the cell supernatant from the former than the latter (Fig. 4A), even in the group without wortmannin treatment (Fig. 4A). The enhanced inflammasome activation in HT-29 cells was not due to a higher initial number of bacteria escaping into the cytosol of host cells, as the number of cytosolic bacteria was similar between the two cell types (SI Appendix, Fig. S10A). Therefore, galectin-4 significantly enhances S. enterica serovar Worthington-induced caspase-1 activation and mature IL-18 production in cells, especially following the inhibition of autophagy.
Fig. 4.
Galectin-4 enhances mature IL-18 and caspase-1 (Cas-1) production from epithelial cells after bacterial infection especially following autophagy inhibition. (A) HT-29 and HT-29-Gal4KO cells treated with or without wortmannin (WTM) were infected with S. enterica serovar Worthington (S.W), MOI = 100. Supernatants and cell lysates were collected 18 and 26 h post-infection and analyzed by immunoblotting with the indicated antibodies. (B) HT-29 cells treated with or without WTM were infected with WT (MOI = 20) or ΔrfaL (MOI = 100) S.W. Supernatants and cell lysates were collected 24 h post-infection and analyzed by immunoblotting with the indicated antibodies. (C) HT-29 cells treated with or without WTM were infected with S.W or S. enterica serovar Typhimurium (S.Tm), MOI = 100. Supernatants and cell lysates were collected 24 h post-infection and analyzed by immunoblotting with the indicated antibodies. The membranes for supernatants in (A–C) were stripped and reprobed for IL-18 after initial probing for Cas-1. The relative amounts of P10 Cas-1 to Cas-1 and IL-18 to pro-IL-18, as determined by densitometric analysis, are indicated. The membrane for lysates in (A) was stripped and reprobed for GAPDH, Pro-IL-18, and then galectin-4 after initial probing for Cas-1. The membranes for lysates in (B and C) were stripped and reprobed for Pro-IL-18, and then β-actin after initial probing for Cas-1. In (A–C) the data are representative of the results from two independent experiments. (D) HT-29 and HT-29-Gal4KO cells treated with WTM were infected with S.W, MOI = 100. Confocal images of cells stained with S.W. DAPI (blue), anti-human galectin-4 (anti-hGal4, green), anti-O antigen of Salmonella (anti-O antigen, red), and anti-phospho-NLRC4 (anti-pNLRC4, purple). The borders of the infected HT-29 cells are outlined with white dashed lines. (Scale bars, 10 µm.)
We reasoned that galectin-4-enhanced inflammasome activation might be associated with the structural platform composed of galectin-4-coated bacteria. Because this platform involves lectin–glycan interactions, we investigated the contribution of the O-antigen, by using the rfaL mutant of S. enterica serovar Worthington. Compared to the wild-type strain, this mutant strain was much less potent at inducing caspase-1 activation and IL-18 maturation in HT-29 cells (SI Appendix, Fig. S10B). To exclude the possibility that diminished inflammasome activation was due to the lower number of rfaL mutants in the cytosol (SI Appendix, Fig. S10C), we standardized the number of cytosolic bacteria between the two strains by reducing the infection dose of the wild-type strain (SI Appendix, Fig. S10D). Under these conditions, HT-29 cells infected with rfaL mutant still exhibited lower levels of mature IL-18 in the supernatant compared to those infected with the wild-type strain (Fig. 4B). Importantly, infection with S. enterica serovar Typhimurium, the O-antigen of which is not recognized by galectin-4, did not induce IL-18 activation, despite their being present in the cytosol at numbers similar to those of S. enterica serovar Worthington (Fig. 4C and SI Appendix, Fig. S10E). These data suggest that inflammasome activation by galectin-4 is associated with the interaction between galectin-4 and the O-antigen.
Activated NLRC4, phosphorylation of NLRC4 on Ser-533 (45), was detected on the bacteria coated with endogenous galectin-4 in wild-type HT-29 cells but not in HT-29-Gal4-KO cells by immunofluorescence staining (Fig. 4D). In addition, activated NLRC4 on galectin-4-coated sifA mutant was observed in wild-type HT-29 without autophagy inhibition (SI Appendix, Fig. S11). The above results suggest that intracellular galectin-4-coated cytosolic S. enterica serovar Worthington act through NLRC4 to activate the inflammasome cascades. We were not able to detect bacteria coated with activated NLRC4 in infected HeLa cells, which may be due to the extremely low protein level of NLRC4 in this cell line (https://www.proteinatlas.org/ENSG00000091106-NLRC4/cell+line).
Galectin-4 Reduces Bacterial Dissemination In Vivo.
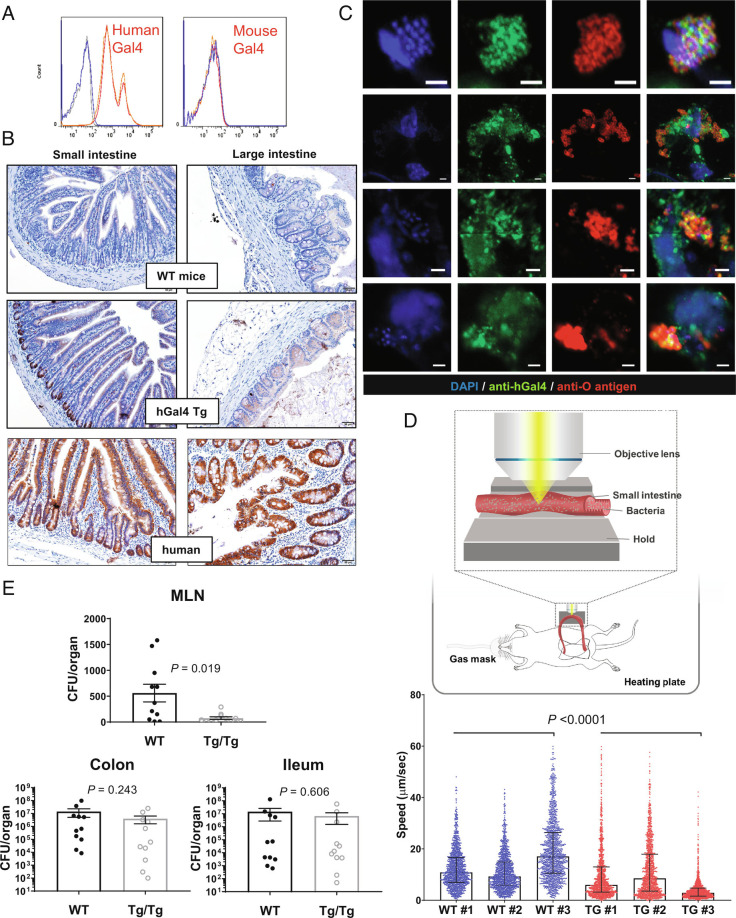
In contrast to human galectin-4, recombinant mouse galectin-4 did not bind to the S. enterica serovar Worthington (Fig. 5A), although it can bind to E. coli O19ab and induce bacterial chain formation like human galectin-4 (SI Appendix, Fig. S8B). We generated human galectin-4 transgenic (hGal4-Tg) mice to study the function of endogenous galectin-4 in intact mice during bacterial infection. Human galectin-4 was detected in the feces of hGal4-Tg mice but not in those of wild-type mice (SI Appendix, Fig. S12A). By immunohistochemistry, generally human galectin-4 levels in the epithelial cells of intestinal tissues of hGal4-Tg mice were not higher than those in normal human tissues, except in the crypts of the ileum (Fig. 5B). We infected hGal4-Tg mice by oral gavage with S. enterica serovar Worthington and found that endogenous human galectin-4 was detectable on the aggregated bacteria contained in dislodged cells (Fig. 5C) or in the luminal space (SI Appendix, Fig. S12D). The latter probably represent hyper-replicating S. released from dislodged pyroptotic cells. By contrast, only single Salmonella bacteria coated with galectin-4 were detectable 3 h after injection of GFP-expressing S. enterica serovar Worthington into the ileal loop of hGal4-Tg mice, when most bacteria remained in the luminal space and had not invaded IECs (SI Appendix, Fig. S13). Next, we tracked luminal GFP-expressing bacteria in the intact intestine using two-photon microscopy after injecting GFP-expressing S. enterica serovar Worthington directly into the ileum of the wild-type and hGal4-Tg mice. The free-swimming speeds of the bacteria were lower in the intestinal lumen of hGal4-Tg mice than in that of wild-type mice (Fig. 5D and Movie S5).
Fig. 5.
Galectin-4 reduces bacterial dissemination in vivo. (A) S. enteric serovar Worthington were incubated with 10 µg/mL Texas-Red-labeled recombinant human galectin-4 (Gal4) or 10 µg/mL of recombinant mouse galectin-4 at 4 °C for 1 h. The bacteria were washed with PBS (red line), 25 mM of TDG (blue line), or sucrose (orange line). Anti-mouse galectin-4 antibody was used to detect the binding of mouse galectin-4. Galectin-4 binding was determined by flow cytometry. (B) Images of immunohistochemistry staining of human galectin-4 in small and large intestines from wide-type (WT) mice, hGal4-Tg mice, and human normal tissues (from Fig. 1C). The images of isotype control staining conducted in the same experiment are shown in the SI Appendix, Fig. S12B. (C) Confocal images of ileum from hGal4-Tg mice 24 h after oral administration of S. enterica serovar Worthington stained with DAPI (blue), anti-human galectin-4 (anti-hGal4, green), anti-O antigen of Salmonella (anti-O antigen, red). (Scale bar, 2 µm.) The frequency of dislodged cells containing aggregated Salmonella coated with galectin-4 was approximately five cells per ileal transverse cross-section with a thickness of 5 µm (estimated from 15 sections of two mice). The image of isotype control staining conducted in the same experiment are shown in the SI Appendix, Fig. S12C. (D) GFP-expressing S. enterica serovar Worthington was injected directly into the ileum of anesthetized wild-type (WT) and hGal4-Tg (TG) mice. Luminal GFP-expressing bacteria were tracked individually in the intact intestine using two-photon microscopy. Data represent the mean ± SEM, and the P value was calculated using the Mann–Whitney test with the entire WT and TG datasets (three mice per group). (E) WT and homozygous hGal4-Tg mice (Tg/Tg) were treated with 2.5% DSS in their drinking water for 5 d. On day 7, mice were challenged orally with S. enterica serovar Worthington and then killed the next day. MLN, mesenteric lymph nodes. Data represent the mean ± SEM, and the P value was calculated using the Mann–Whitney test, N = 11 (pairs of littermates).
Since galectin-4 can hinder bacterial motility, promote inflammasome activation, and enhance IL-18 secretion, it may restrict bacterial dissemination or translocation, especially when intestinal barrier functions are compromised, such as under colitis conditions. We treated hGal4-Tg and wild-type mice with dextran sodium sulfate (DSS), a polymer known to affect IEC integrity and induce colitis (46). Afterward, mice were challenged orally with S. enterica serovar Worthington. Subsequently, fewer bacterial cells were noted to translocate from the intestinal lumen to the MLNs in hGal4-Tg mice compared to wild-type mice (Fig. 5E). In contrast, there was no statistically significant difference in bacterial load in the ileum and colon between these two groups of mice (Fig. 5E).
The fact fewer bacteria translocated to the MLNs in hGal4-Tg mice was not due to a difference in colitis severity between wild-type and hGal4-Tg mice after DSS treatment. For example, there were no statistically significant differences in weight loss or intestinal barrier permeability between the two groups of mice (SI Appendix, Fig. S14). These data suggest that endogenous galectin-4 impedes bacterial dissemination in vivo.
Discussion
Galectin-4, which is specifically expressed in the intestine, is abundant in the cytoplasmic compartment of IECs and binds to selective glycans on the bacterial surface. Thus, it is well-poised to serve as a cytosolic adaptor that recognizes glycans displayed on the surface of cytosolic bacteria. Here, we demonstrate that endogenous galectin-4 in intestinal cells binds to and enchains cytosolic Gram-negative bacteria, reduces their motility and modulates the innate immune response induced by these bacteria. Our data unraveled a novel host defense mechanism in mucosal immunity involving the interaction of cytosolic GBPs with glycans on live bacteria intracellularly.
A majority of studies investigating the roles of galectins in host–pathogen interactions have focused on their extracellular functions (47, 48); however, a significant number of intracellular functions of galectins have been demonstrated. For example, galectin-3 negatively regulates T cell activation and promotes HIV budding from T cells by stabilizing the association between Alix and HIV Gag protein (49, 50). Galectin-3 increases IL-1β production and NLRP3 inflammasome activation by promoting oligomerization of ASC in macrophages during influenza A virus H5N1 infection (51). Galectin-9 promotes the phagocytic function of dendritic cells by modulating the activity of Rac1, a Rho family GTPase (52). These functions involve interactions with intracellular regulators of the host cells and are glycan-independent, whereas the functions of galectin-4 reported herein involve interactions with intracellular pathogens and are glycan-dependent.
A critical function of intracellular galectins is the recognition of host glycans exposed to the cytosol when intracellular organelles are damaged. For example, galectin-8 binds to host N-glycans on damaged Salmonella-containing vacuoles and recruits NDP52 to activate autophagy in HeLa cells (37). Moreover, galectin-8 binds to host O-glycans on damaged lysosomes and in turn activates autophagy in gastric epithelial cells that are exposed to H. pylori (53). In contrast, galectin-3 inhibits autophagy by binding to host N-glycans on ruptured phagosomes in macrophages during Listeria monocytogenes infection (54). The morphology of the intracellular galectin-4-coated structures noted in the present study suggests that galectin-4 accumulates around the cytosolic bacteria and not around the damaged endosomes or lysosomes. It is to be noted also, Thurston et al. (37) screened a panel of human galectins, including galectin-1, 2, 3, 4, 7, 8, 9, 10, 12, 13, and 14, to examine their recruitment to bacteria-containing vacuoles in HeLa cells (37). Only galectin-3, 8, and 9, but not galectin-4 nor any of the others tested, were able to sense host glycans exposed on the ruptured membranes of vacuoles containing bacteria.
Time-lapse microscopy of infected cells revealed that the motility of galectin-4-coated S. enterica serovar Worthington in HT-29-EGFP-Gal4 cells was significantly lower than that of bacteria in control HT-29-EGFP cells. As Salmonella can replicate slowly in vacuoles (40), the possibility exists that in these experiments, vacuolar bacteria were also being monitored. Knodler et al. reported that two distinct types of intracellular bacterial populations reside in epithelial cells: the cytosolic population, which is motile and expresses T3SS-1, and the vacuolar population, which is immotile and expresses T3SS-2 (9). However, the detection of EGFP-galectin-4-coated bacteria indicated that these bacteria reached the cytosolic compartment. Therefore, we can state with confidence that cytosolic bacterial motility was reduced when coated with galectin-4. This may be due to the physical properties of cytosolic bacterial clumps connected by galectin-4 or direct effects on individual bacteria. In the cell-free system, the galectin-4 impaired motility of individual bacteria as well as enchained bacteria.
The proportion of cells containing galectin-4-coated bacteria depends on the ability of bacteria to invade the cells and escape from the vacuoles they are initially confined in. It is known that when bacteria escape into cytosol, they hyper-replicate. Hyper-replicating Salmonella induces pyroptosis and consequently extrusion or sloughing of infected epithelial cells, which causes the sustained release of Salmonella into the intestinal lumen in mice (9, 10). However, we found that HT-29 cells containing galectin-4-coated Salmonella were not stained positively by propidium iodide, indicating that galectin-4 recruitment to the cytosolic bacteria precedes pyroptosis. It has been shown that the released cytosolic Salmonella from extruded infected IECs cause persistent infection (10), because they express T3SS1 and flagella, crucial virulence factors for invading non-phagocytic cells (9). We noted that when bacteria that had replicated in the cytosol were released from the epithelial cells, they remained connected and coated with galectin-4. Typically, IECs containing intracellular Salmonella are rarely seen in wild-type mice subjected to infection with this species of bacteria through the oral route; the death and extrusion of infected IECs are essential mechanisms to maintain sterile intestinal epithelium after infection (10, 55, 56). We also observed galectin-4-coated Salmonella in the dislodged IECs and in the lumen of hGal4-Tg mice. The reduced motility and enchained growth of Salmonella caused by galectin-4 may play a role in preventing further infection of other cells.
We showed that galectin-4-induced inflammasome activation in bacteria-infected IECs was augmented by autophagy inhibition, which prevents the elimination of bacteria in host cells. This suggests that the number of bacteria that directly introduce flagellin or other pathogen-associated molecular patterns (PAMPs) into the cytoplasm is critical for triggering the inflammasome cascade in our infection model. Such a response was significantly lower in Gal4-KO HT-29 cells, indicating that galectin-4 is important for this response.
The compact bacterial aggregates and immotile status caused by the interaction between galectin-4 and the O-antigen may provide a stable and steady platform for high levels of PAMP components to initialize the inflammasome cascade. Our findings that IL-18 maturation was impaired in cells infected with rfaL mutant bacteria without O-antigens or S. Typhimurium containing O-antigens that are not recognized by galectin-4 suggest that such a platform may potentiate or stabilize inflammasome activation. We detected activated NLRC4 on the surface of galectin-4-coated bacteria in wild-type HT-29 cells, suggesting the activation of the inflammasome cascade involves NLRC4.
Cytosolic LPS can also trigger noncanonical inflammasome activation via caspase-4 (15, 16). We attempted to determine whether galectin-4 affects caspase-4 activation, especially since our group recently demonstrated that intracellular galectin-3 promotes caspase-4 activation in macrophages through binding to glycans on cytosolic LPS (57). However, caspase-4 activation in IECs was not consistently detected in our infection model. Wandel et al. showed that interferon-γ-inducing guanylate-binding proteins (GBPs) assembled on the surface of cytosol-invading Gram-negative bacteria, thus converting the bacterial surfaces into polyvalent signaling platforms, are essential for activating caspase-4 and IL-18 in epithelial cells (58). Those authors proposed that, in contrast to free cytosolic LPS, LPS present on the bacterial outer membrane does not activate caspase-4, because the membrane-embedded acyl chains of lipid A in LPS are not accessible to the ligand-binding CARD domain of caspase-4. GBP may alter the bacterial membrane and reveal hidden ligands, resulting in the activation of caspase-4. In future studies, the role of galectin-4 in caspase-4 activation during infection may be demonstrated using IECs treated with a combination of interferon-γ and autophagy inhibitors.
We found that galectin-4 decreased bacterial spreading from the intestinal lumen to MLNs in a mouse model of colitis following oral challenge with S. enterica serovar Worthington. We have examined the dissemination of S. enterica serovar Worthington in mice without DSS treatment, but did not detect bacteria that were translocated to MLNs, suggesting this strain of bacteria is not virulent enough to healthy mice. Mice with DSS-induced colitis serve as a model of inflammatory bowel disease (IBD) to study the pathogenicity of S. enterica serovar Worthington in hosts with underlying illnesses in the gastrointestinal tract. Studies have suggested that the ill effects of enteropathogenic infection are higher in IBD (59).
The decreased number of bacteria in MLNs of hGal4-Tg mice may be attributed to the reduced bacterial motility induced by galectin-4, or inflammasome activation and epithelium-derived IL-18, which contributes to host defense against pathogens (60), or both. However, expression of the human galectin-4 transgene in hGal4-Tg mice was controlled by the chicken β-actin promoter fused with the cytomegalovirus (CMV) enhancer (CAG) promoter, which efficiently and ubiquitously expresses the transgene (61). This is in contrast to wild-type mice and humans, in which galectin-4 mRNA is specifically expressed in the intestines (http://biogps.org). Thus, we cannot rule out the participation in our model system of galectin-4 in other dissemination pathways, which may reduce bacterial translocation to MLNs. For example, dendritic cells can capture and facilitate the transport of bacteria from the gut lumen to MLNs in vivo and transgenic galectin-4 in these cells may inhibit this process (62, 63).
The lack of functional autophagy machinery increases the number of bacteria escaping from bacteria-containing vacuoles and hyper-replicating in the cytosol of epithelial cells (38, 39). Autophagosome formation and maturation require phosphatidylinositol 3-phosphate, which is generated by PI3K. Inhibition of PI3K with wortmannin suppresses autophagy (6, 64). In our study, although we can observe hyper-replicating bacteria and galectin-4-coated cytosolic bacteria in cells without wortmannin, they could be much more easily detected in wortmannin-treated cells. Indicators of inflammasome activation were also more readily observed in wortmannin-treated HT-29 cells. Genome-wide association studies have revealed that polymorphisms in autophagy-related genes, such as ATG16L1 and IRGM, contribute to the pathogenesis of Crohn's disease, a type of IBD (65, 66). Studies have also shown that defects in these genes may impair the host response to bacterial infections (67, 68). Hence, we suspect that in IBD patients with impaired autophagic activation, the contribution of galectin-4 to host protection during bacterial infection is more significant and profound compared to that in healthy subjects.
Understanding the mechanisms through which IECs protect themselves against pathogens is essential for preventing and treating diseases in the intestinal tract. Our findings may apply to other Gram-negative bacteria such as Shigella boydii, Shigella dysenteriae, Providencia alcalifaciens, Klebsiella spp., and Proteus vulgaris, as galectin-4 can recognize specific serotypes of these bacteria (36, 69). Bacteria are rich in glycans on their surface and galectins are the only GBPs in the cytosol. Our results suggest that galectins may be a major component of intracellular immunity. The impact of galectins on host defense against pathogens in the gastrointestinal tract is an interesting and important subject worthy of further investigation.
Materials and Methods
See also SI Appendix, Extended Materials and Methods.
Bacteria.
S. enterica serovar Worthington (BCRC 15574) was purchased from the Bioresource Collection and Research Center in Taiwan. λ Red recombination was used to generate in-frame deletions of rfaL and sifA in S. enterica serovar Worthington, as described previously (70). The primers used for genetic manipulations and construct verification are listed in SI Appendix, Table S1. Clones with deletions were verified by PCR.
Live-Cell Imaging.
HT-29-EGFP, HT-29-EGFP-Gal4 cells both reconstituted from HT-29 cells with endogenous galectin-4 deleted were grown overnight on 35-mm glass culture dishes at a density of 1 × 106 cells. HeLa-EGFP and HeLa-EGFP-Gal4 cells were grown on a 35-mm dish at a density of 5 × 105 cells. RFP-expressing S. enterica serovar Worthington was grown overnight at 37 °C in LB and then the cultures were diluted 1:20 in fresh LB without antibiotics and incubated for 3 h at 37 °C with shaking at 200 RPM. RFP-expressing S. enterica serovar Worthington [multiplicity of infection (MOI) = 100] in OPTI-MEM was added to the cell culture, followed by centrifugation at 300 × g for 5 min. Cells were infected with bacteria for 1 h at 37 °C. Extracellular bacteria were killed by treatment with 100 µg/mL gentamicin for 30 min, followed by 10 µg/mL gentamicin throughout the rest of the experiments. Live-cell images were taken with an LSM 880 Laser Scanning Confocal Microscope (ZEISS) using a Plan Apochromat ×100 (1.4 NA) oil immersion objective in a 37 °C incubator chamber with 5% CO2 supplement. Images were processed using the ZEN Image Analysis module. In the wortmannin-treated HT-29 cells, 500 nM wortmannin was present throughout the indicated time periods, depending on the experiment. To track intracellular bacteria was conducted as previously described, with minor modifications (9). At least 10 cells infected with RFP-expressing S. enterica serovar Worthington were captured every 940 ms for 94 s (100 frames in total). The average instantaneous velocity of each motion of the bacterium was measured, and the resultant tracking data were analyzed using the Metamorph software (Molecular Devices).
Mice.
To generate human galectin-4 transgenic (hGal4-Tg) mice, coding sequences of human galectin-4 were cloned into the pCAGEN plasmid (Addgene 111160) and hGal4-Tg mice in a C57BL/6J background were produced by the Transgenic Core Facility at the Institute of Molecular Biology, Academia Sinica, Taiwan. Heterozygous mice were crossbred to generate wild-type, heterozygous, and homozygous transgenic mice.
Mouse Infection Model.
Control wild-type and hGal4-Tg/Tg littermates were obtained from hGal4-Tg heterozygous breeders. Male and female mice aged 6 to 12 wk were used in the experiments. Mice were administered 2.5% (weight/volume) DSS (molecular weight 36 to 50 kDa; MP Biomedicals) in drinking water for 5 d. The RFP-expressing S. enterica serovar Worthington was cultured overnight in LB containing chloramphenicol. Before infection, the overnight cultures were diluted 1:20 in fresh LB without antibiotics and incubated for 3 h at 37 °C with shaking at 200 RPM. The mice received 150 µL of bacterial suspension via orally gavage on day 7. At 24 h after infection, bacterial loads in tissues were measured in colony-forming units by plating serial dilutions of tissue homogenates on LB plates containing chloramphenicol. Ileum tissue was fixed in 4% paraformaldehyde overnight and embedded in optimum cutting temperature medium (Tissue-Tek) for immunofluorescent staining.
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism v.7, as described in the respective figure legends.
Supplementary Material
Appendix 01 (PDF)
Bacterial chains induced by galectin-4 are derived from cell progeny. Time-lapse imaging of S. enterica serovar Worthington cells incubated with 100 μg/mL of recombinant human galectin-4. Time indicated the real time (h:min:sec) on the day of the experiment.
Time-lapse imaging of S. enterica serovar Worthington cells incubated with 100 μg/mL BSA. Time indicated the real time (h:min:sec) on the day of the experiment.
Intracellular galectin-4 in IECs inhibits the motility of cytosolic bacteria (< 20 bacteria per cell). HT-29-EGFP and HT-29-EGFP-Gal4 cells were treated with wortmannin and infected with the RFP-expressing S. enterica serovar Worthington (MOI = 50). Time-lapse images were acquired 10 h post-infection using time-lapse microscopy.
Intracellular galectin-4 in IECs inhibits the motility of cytosolic bacteria (≥ 20 bacteria per cell). HT-29-EGFP and HT-29-EGFP-Gal4 cells were treated with wortmannin and then infected with the RFP-expressing S. enterica serovar Worthington (MOI = 50). Time-lapse images were acquired 10 h post-infection using time-lapse microscopy.
Galectin-4 restricts bacterial motility in the intestinal lumen. GFP-expressing S. enterica serovar Worthington was injected directly into the ileum of anesthetized wild-type or hGal4-Tg mice. Luminal GFP-expressing S. enterica serovar Worthington was tracked individually in intact intestines using two-photon microscopy.
Acknowledgments
We thank Dr. Ching-Liang Chu at National Taiwan University for providing the HT-29 cell line; Dr. Jr-Wen Shui at Academia Sinica for providing S. enterica serovar Typhimurium (SL1344); Dr. Yu-Ling Shih at the Institute of Biological Chemistry (IBC) in Academia Sinica for providing plasmids for λ Red recombination in bacteria; and Mr. Wei-Han Lin for the illustrations. We thank the IBC Glycobiology Core supported by Academia Sinica Core Facility and Innovative Instrument Project (AS-CFII-112-102) for providing the rhGal9null protein. We also thank the Transgenic Core, funded by AS-CFII-108-107, for the generation of hGal4-Tg mice, and the Light Microscopy Core Facility and the Flow Cytometric Cell Sorting Core Facility at the Institute of Biomedical Sciences, funded by AS-CFII-111-212, for cell sorting and technical support. This work was supported by Academia Sinica and the Ministry of Science and Technology in Taiwan (MOST 109-2320-B-001-024-MY3).
Author contributions
C.-S.L., T.-H.L., and F.-T.L. designed research; C.-S.L., T.-H.L., T.-J.T., D.-Y.C., and C.-I.Y. performed research; C.-H.L. and P.C. contributed new reagents/analytic tools; C.-S.L., T.-H.L., and T.-J.T. analyzed data; F.-T.L. contributed supervision and funding acquisition; and C.-S.L., T.-H.L., and F.-T.L. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Kayama H., Okumura R., Takeda K., Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu. Rev. Immunol. 38, 23–48 (2020). [DOI] [PubMed] [Google Scholar]
- 2.MacDonald T. T., Monteleone I., Fantini M. C., Monteleone G., Regulation of homeostasis and inflammation in the intestine. Gastroenterology 140, 1768–1775 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Maloy K. J., Powrie F., Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Steele-Mortimer O., The Salmonella-containing vacuole: Moving with the times. Curr. Opin. Microbiol. 11, 38–45 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu S., Shen Y., Zhang S., Xiao Y., Shi S., Salmonella interacts with autophagy to offense or defense. Front. Microbiol. 11, 721 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kageyama S., et al. , The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol. Biol. Cell 22, 2290–2300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knodler L. A., Nair V., Steele-Mortimer O., Quantitative assessment of cytosolic Salmonella in epithelial cells. PLoS One 9, e84681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rauch I., et al. , NAIP-NLRC4 Inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity 46, 649–659 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knodler L. A., et al. , Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U.S.A. 107, 17733–17738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong A., et al. , Cytosolic replication in epithelial cells fuels intestinal expansion and chronic fecal shedding of Salmonella Typhimurium. Cell Host Microbe 29, 1177–1185.e1176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sellin M. E., Maslowski K. M., Maloy K. J., Hardt W. D., Inflammasomes of the intestinal epithelium. Trends Immunol. 36, 442–450 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Winsor N., Krustev C., Bruce J., Philpott D. J., Girardin S. E., Canonical and noncanonical inflammasomes in intestinal epithelial cells. Cell Microbiol. 21, e13079 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Kofoed E. M., Vance R. E., Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y., et al. , The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Shi J., et al. , Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Kayagaki N., et al. , Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Lei-Leston A. C., Murphy A. G., Maloy K. J., Epithelial cell inflammasomes in intestinal immunity and inflammation. Front. Immunol. 8, 1168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirabayashi J., Kasai K., The family of metazoan metal-independent beta-galactoside-binding lectins: Structure, function and molecular evolution. Glycobiology 3, 297–304 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Liu F. T., Rabinovich G. A., Galectins as modulators of tumour progression. Nat. Rev. Cancer 5, 29–41 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Hong M. H., et al. , Intracellular galectins control cellular responses commensurate with cell surface carbohydrate composition. Glycobiology 30, 49–57 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Johannes L., Jacob R., Leffler H., Galectins at a glance. J. Cell Sci. 131, 208884 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Liu F. T., Patterson R. J., Wang J. L., Intracellular functions of galectins. Biochim. Biophys. Acta 1572, 263–273 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Rabinovich G. A., Toscano M. A., Turning "sweet" on immunity: Galectin-glycan interactions in immune tolerance and inflammation. Nat. Rev. Immunol. 9, 338–352 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Danielsen E. M., van Deurs B., Galectin-4 and small intestinal brush border enzymes form clusters. Mol. Biol. Cell 8, 2241–2251 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huflejt M. E., Leffler H., Galectin-4 in normal tissues and cancer. Glycoconj J. 20, 247–255 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Leffler H., Masiarz F. R., Barondes S. H., Soluble lactose-binding vertebrate lectins: A growing family. Biochemistry 28, 9222–9229 (1989). [DOI] [PubMed] [Google Scholar]
- 27.Oda Y., et al. , Soluble lactose-binding lectin from rat intestine with two different carbohydrate-binding domains in the same peptide chain. J. Biol. Chem. 268, 5929–5939 (1993). [PubMed] [Google Scholar]
- 28.Chiu M. L., Parry D. A., Feldman S. R., Klapper D. G., O’Keefe E. J., An adherens junction protein is a member of the family of lactose-binding lectins. J. Biol. Chem. 269, 31770–31776 (1994). [PubMed] [Google Scholar]
- 29.Stechly L., et al. , Galectin-4-regulated delivery of glycoproteins to the brush border membrane of enterocyte-like cells. Traffic 10, 438–450 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Ideo H., Seko A., Ohkura T., Matta K. L., Yamashita K., High-affinity binding of recombinant human galectin-4 to SO(3)(-)–>3Galbeta1–>3GalNAc pyranoside. Glycobiology 12, 199–208 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Ideo H., Seko A., Yamashita K., Galectin-4 binds to sulfated glycosphingolipids and carcinoembryonic antigen in patches on the cell surface of human colon adenocarcinoma cells. J. Biol. Chem. 280, 4730–4737 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Ideo H., Seko A., Yamashita K., Recognition mechanism of galectin-4 for cholesterol 3-sulfate. J. Biol. Chem. 282, 21081–21089 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Mey A., Leffler H., Hmama Z., Normier G., Revillard J. P., The animal lectin galectin-3 interacts with bacterial lipopolysaccharides via two independent sites. J. Immunol. 156, 1572–1577 (1996). [PubMed] [Google Scholar]
- 34.Fowler M., Thomas R. J., Atherton J., Roberts I. S., High N. J., Galectin-3 binds to Helicobacter pylori O-antigen: It is upregulated and rapidly secreted by gastric epithelial cells in response to Helicobacter pylori adhesion. Cell Microbiol. 8, 44–54 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Stowell S. R., et al. , Innate immune lectins kill bacteria expressing blood group antigen. Nat. Med. 16, 295–301 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knirel Y. A., et al. , Human tandem-repeat-type galectins bind bacterial non-betaGal polysaccharides. Glycoconj J. 31, 7–12 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Thurston T. L., Wandel M. P., von Muhlinen N., Foeglein A., Randow F., Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birmingham C. L., Smith A. C., Bakowski M. A., Yoshimori T., Brumell J. H., Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 281, 11374–11383 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Brumell J. H., Tang P., Zaharik M. L., Finlay B. B., Disruption of the Salmonella-containing vacuole leads to increased replication of Salmonella enterica serovar typhimurium in the cytosol of epithelial cells. Infect. Immun. 70, 3264–3270 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malik-Kale P., Winfree S., Steele-Mortimer O., The bimodal lifestyle of intracellular Salmonella in epithelial cells: Replication in the cytosol obscures defects in vacuolar replication. PLoS One 7, e38732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beuzón C. R., Salcedo S. P., Holden D. W., Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology 148, 2705–2715 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Nishi N., et al. , Development of highly stable galectins: Truncation of the linker peptide confers protease-resistance on tandem-repeat type galectins. FEBS Lett. 579, 2058–2064 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Zenk S. F., Jantsch J., Hensel M., Role of Salmonella enterica lipopolysaccharide in activation of dendritic cell functions and bacterial containment. J. Immunol. 183, 2697–2707 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Sellin M. E., et al. , Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe 16, 237–248 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Qu Y., et al. , Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490, 539–542 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Chassaing B., Aitken J. D., Malleshappa M., Vijay-Kumar M., Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 104, 11–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasta G. R., Roles of galectins in infection. Nat. Rev. Microbiol. 7, 424–438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baum L. G., Garner O. B., Schaefer K., Lee B., Microbe-host interactions are positively and negatively regulated by galectin-glycan interactions. Front. Immunol. 5, 284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S. F., et al. , Galectin-3 promotes HIV-1 budding via association with Alix and Gag p6. Glycobiology 24, 1022–1035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H. Y., et al. , Galectin-3 negatively regulates TCR-mediated CD4+ T-cell activation at the immunological synapse. Proc. Natl. Acad. Sci. U.S.A. 106, 14496–14501 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y. J., et al. , Galectin-3 enhances avian H5N1 influenza a virus-induced pulmonary inflammation by promoting NLRP3 inflammasome activation. Am. J. Pathol. 188, 1031–1042 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Querol Cano L., et al. , Intracellular galectin-9 controls dendritic cell function by maintaining plasma membrane rigidity. iScience 22, 240–255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li F. Y., et al. , Helicobacter pylori induces intracellular galectin-8 aggregation around damaged lysosomes within gastric epithelial cells in a host O-glycan-dependent manner. Glycobiology 29, 151–162 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Weng I. C., et al. , Cytosolic galectin-3 and -8 regulate antibacterial autophagy through differential recognition of host glycans on damaged phagosomes. Glycobiology 28, 392–405 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Knodler L. A., et al. , Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe 16, 249–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crowley S. M., et al. , Intestinal restriction of Salmonella typhimurium requires caspase-1 and caspase-11 epithelial intrinsic inflammasomes. PLoS Pathog. 16, e1008498 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lo T. H., et al. , Galectin-3 promotes noncanonical inflammasome activation through intracellular binding to lipopolysaccharide glycans. Proc. Natl. Acad. Sci. U.S.A. 118, e2026246118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wandel M. P., et al. , Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat. Immunol. 21, 880–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Axelrad J. E., et al. , Enteric infections are common in patients with flares of inflammatory bowel disease. Am. J. Gastroenterol. 113, 1530–1539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muñoz M., et al. , Interleukin-22 induces interleukin-18 expression from epithelial cells during intestinal infection. Immunity 42, 321–331 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Niwa H., Yamamura K., Miyazaki J., Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991). [DOI] [PubMed] [Google Scholar]
- 62.Bravo-Blas A., et al. , Salmonella enterica Serovar typhimurium travels to mesenteric lymph nodes both with host cells and autonomously. J. Immunol. 202, 260–267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tam M. A., Rydström A., Sundquist M., Wick M. J., Early cellular responses to Salmonella infection: Dendritic cells, monocytes, and more. Immunol. Rev. 225, 140–162 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Kabeya Y., et al. , LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo. J. 19, 5720–5728 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hampe J., et al. , A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007). [DOI] [PubMed] [Google Scholar]
- 66.McCarroll S. A., et al. , Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat. Genet. 40, 1107–1112 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuballa P., Huett A., Rioux J. D., Daly M. J., Xavier R. J., Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One 3, e3391 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lapaquette P., Glasser A. L., Huett A., Xavier R. J., Darfeuille-Michaud A., Crohn’s disease-associated adherent-invasive Escherichia coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 12, 99–113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stowell S. R., et al. , Microbial glycan microarrays define key features of host-microbial interactions. Nat. Chem. Biol. 10, 470–476 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Bacterial chains induced by galectin-4 are derived from cell progeny. Time-lapse imaging of S. enterica serovar Worthington cells incubated with 100 μg/mL of recombinant human galectin-4. Time indicated the real time (h:min:sec) on the day of the experiment.
Time-lapse imaging of S. enterica serovar Worthington cells incubated with 100 μg/mL BSA. Time indicated the real time (h:min:sec) on the day of the experiment.
Intracellular galectin-4 in IECs inhibits the motility of cytosolic bacteria (< 20 bacteria per cell). HT-29-EGFP and HT-29-EGFP-Gal4 cells were treated with wortmannin and infected with the RFP-expressing S. enterica serovar Worthington (MOI = 50). Time-lapse images were acquired 10 h post-infection using time-lapse microscopy.
Intracellular galectin-4 in IECs inhibits the motility of cytosolic bacteria (≥ 20 bacteria per cell). HT-29-EGFP and HT-29-EGFP-Gal4 cells were treated with wortmannin and then infected with the RFP-expressing S. enterica serovar Worthington (MOI = 50). Time-lapse images were acquired 10 h post-infection using time-lapse microscopy.
Galectin-4 restricts bacterial motility in the intestinal lumen. GFP-expressing S. enterica serovar Worthington was injected directly into the ileum of anesthetized wild-type or hGal4-Tg mice. Luminal GFP-expressing S. enterica serovar Worthington was tracked individually in intact intestines using two-photon microscopy.
Data Availability Statement
All study data are included in the article and/or SI Appendix.