Significance
Antibiotic resistance is a global threat to public health and associated with the overuse of antibiotics. Although non-antibiotic drugs occupy 95% of the drug market, their impact on the emergence and spread of antibiotic resistance remains unclear. Here we demonstrate that antidepressants, one of the most frequently prescribed drugs, can induce antibiotic resistance and persistence. Such effects are associated with increased reactive oxygen species, enhanced stress signature responses, and stimulation of efflux pump expression. Mathematical modeling also supported a role for antidepressants in the occurrence of antibiotic-resistant mutants and persister cells. Considering the high consumption of antidepressants (16,850 kg annually in the United States alone), our findings highlight the need to re-evaluate the antibiotic-like side effects of antidepressants.
Keyword: antibiotic resistance, mutation, persistence, antidepressants
Abstract
Antibiotic resistance is an urgent threat to global health. Antidepressants are consumed in large quantities, with a similar pharmaceutical market share (4.8%) to antibiotics (5%). While antibiotics are acknowledged as the major driver of increasing antibiotic resistance, little attention is paid to the contribution of antidepressants in this process. Here, we demonstrate that antidepressants at clinically relevant concentrations induce resistance to multiple antibiotics, even following short periods of exposure. Antibiotic persistence was also enhanced. Phenotypic and genotypic analyses revealed the enhanced production of reactive oxygen species following exposure to antidepressants was directly associated with increased resistance. An enhanced stress signature response and stimulation of efflux pump expression were also associated with increased resistance and persistence. Mathematical modeling also predicted that antidepressants would accelerate the emergence of antibiotic-resistant bacteria, and persister cells would help to maintain the resistance. Overall, our findings highlight the antibiotic resistance risk caused by antidepressants.
Antibiotic resistance has emerged as a global crisis, causing increased mortality rates and healthcare costs (1, 2). It is well accepted that our overuse of antibiotics has generated strong selection pressure for the evolution of resistance and tolerance in bacteria (3–8). Non-antibiotic pharmaceuticals represent another category of compounds that occupy a huge share (~95%) of the drug market (9), yet their contribution to antibiotic resistance remains to be properly evaluated.
Among the non-antibiotic pharmaceuticals, antidepressants, the main drugs used for the treatment of depression, represented 4.8% of the market share in 2008 (10), placing them in the top five pharmaceutical categories and almost equal to antibiotics (5%) (9). The consumption of antidepressants has grown considerably among countries in the Organization for Economic Co-operation and Development, linked to an aging population and increased public awareness of psychiatric and mental health problems (11). More than 337 million prescriptions were written for antidepressant medications in 2016 in the United States alone, which corresponded to 16,850 kg (assuming each prescription refers to 50 mg) (10). Commonly prescribed antidepressants include selective serotonin reuptake inhibitors (SSRIs, e.g., sertraline, fluoxetine, escitalopram), serotonin norepinephrine reuptake inhibitors (SNRIs, e.g., duloxetine), norepinephrine dopamine reuptake inhibitors (e.g., bupropion), and atypical antidepressants (e.g., agomelatine) (12). Sertraline (first), escitalopram (third), bupropion (fifth), and duloxetine (tenth) ranked among the top ten most prescribed psychiatric medications in the United States. The daily dosage of antidepressants varies, ranging from 20 to 450 mg/day. Absorbed antidepressants can circulate in the human body with half-life from 1 to 2 h (agomelatine) to 144 h (fluoxetine) (13). Thus, antidepressants have been reported at a wide range of concentrations following consumption: for example, bupropion at a peak of 0.3 mg/L in plasma (14), escitalopram at 3 to 51 mg/L in the colon (15), and sertraline at 370 to 1,700 mg/L in the colon (15).
In addition to their specified treatment use, antidepressants have also been shown to modify the gut microbiota (16). Thus, we hypothesized that antidepressants, as one category of typical non-antibiotic pharmaceuticals, might contribute to the emergence of antibiotic resistance. Our previous work reported that fluoxetine could enhance bacterial resistance toward antibiotics (17), yet it is still unknown whether this response is drug-specific or it is a universal phenomenon. In addition, we only focused on mutants selected following exposure to the antidepressant fluoxetine, but the role of antidepressants in the emergence of persister cells was not investigated. More importantly, the underlying mechanisms by which antidepressants can mediate the spread of antibiotic resistance have not been revealed. To address these research gaps, we set up an evolutionary model to measure the development of resistance to multiple antibiotics and evaluate bacterial persistence following exposure to five commonly prescribed antidepressants at clinically relevant concentrations, including sertraline, escitalopram, bupropion, duloxetine, and agomelatine. We show that exposure to commonly prescribed antidepressants at clinically relevant concentrations led to enhanced bacterial persistence against the antibiotic ciprofloxacin. By applying plate culturing under both aerobic and anaerobic conditions, fluorescence-based flow cytometry detection, and genome-wide analyses, we found that exposure to antidepressants caused increased production of intracellular reactive oxygen species (ROS) and induced efflux pump expression, providing a direct correlation with antibiotic resistance and persistence phenotypes. In addition, antidepressants can promote conjugative gene transfer. Together, our findings describe a mechanism by which exposure to antidepressants can lead to enhanced resistance to antibiotics.
Results
Exposure to Antidepressants Can Induce Resistance to Multiple Antibiotics.
To test whether antidepressants induce bacterial resistance toward antibiotics, we exposed the E. coli K-12 strain MG1655 to five commonly prescribed antidepressants, including sertraline, duloxetine, bupropion, escitalopram, and agomelatine. The antidepressant concentrations covered low level (0.1 mg/L and 1 mg/L), medium level (10 mg/L), and high level (50 mg/L and 100 mg/L). During the 60-d exposure, bacterial resistance toward multiple antibiotics was tested by applying plate-counting periodically (Fig. 1A). The mutant colonies grown on antibiotic-selective plates were randomly selected to test their minimum inhibition concentration (MIC) against multiple antibiotics. The antibiotics employed in this study included amoxicillin, ampicillin, cephalexin, chloramphenicol, ciprofloxacin, colistin, erythromycin, kanamycin, levofloxacin, norfloxacin, roxithromycin, tetracycline, and trimethoprim. These antibiotics cover six categories, including β-lactams, phenicols, fluoroquinolones, macrolides, aminoglycosides, and tetracyclines, with the modes of action encompassing inhibition of DNA, protein, and cell wall synthesis (18, 19).
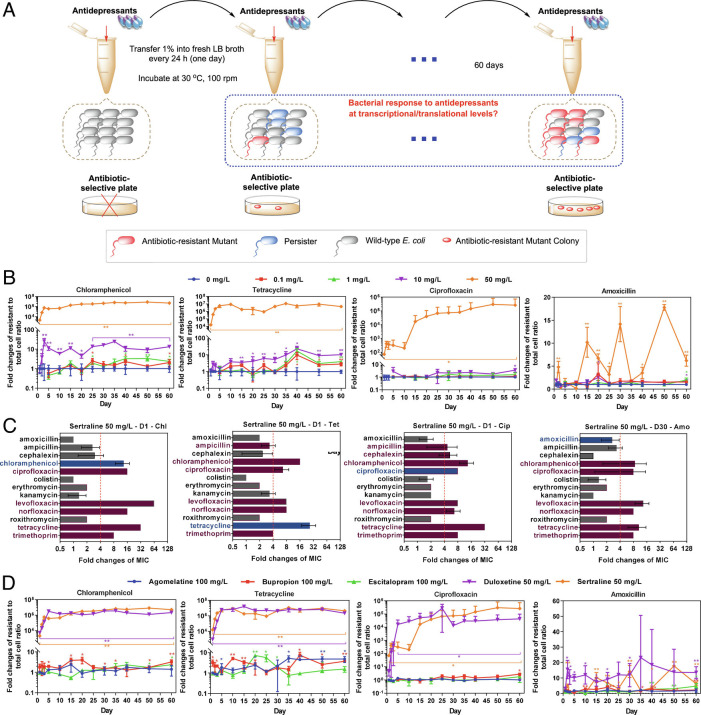
Fig. 1.
Antidepressants induce multidrug resistance. (A) Schematic design of the experiment. (B) Antidepressant sertraline increases ratio of resistant cell number to total cell number on chloramphenicol-, tetracycline-, ciprofloxacin-, and amoxicillin-selective plates. (C) Sertraline-induced mutants show enhancement on MICs toward multiple antibiotics. MIC fold changes higher than four folds were defined as significant enhancement (dotted red lines). Red bar: MIC fold changes higher than 4, gray bar: MIC fold changes less than 4, blue bar: the tested antibiotic same as the antibiotic-selective plate, no bar: no MIC fold change. (D) Fold change of ratio of resistant cell number to total cell number induced by the highest concentration of the other four antidepressants tested (100 mg/L agomelatine, 100 mg/L bupropion, 100 mg/L escitalopram, and 50 mg/L duloxetine). Data are shown as mean ± SD, n = 3 independent experiments. Significant differences between antidepressant-dosed samples and the non-antidepressant control are analyzed by independent sample t test with Benjamini–Hochberg multiple comparison testing, *P < 0.05, **P < 0.01. See also SI Appendix, Figs. S1 and S2.
We found that these antidepressants could induce multidrug resistance during the 60-d exposure. Among them, sertraline and duloxetine exhibited the most significant effects, resulting in high numbers of resistant cells (compared to total cells) over a short time period. Indeed, a high concentration of sertraline (50 mg/L) induced resistance to chloramphenicol, tetracycline, ciprofloxacin and the fold changes of the ratio of resistant cell number to total cell number were 2 × 103-, 2 × 104-, 71-fold, respectively, when compared with the non-antidepressant control group (Fig. 1B). For the ratio of resistant cell number to total cell number toward amoxicillin, a significant increase was observed after 30-d consecutive exposure to 50 mg/L sertraline. This may be due to the different drug types of these four antibiotics: chloramphenicol and tetracycline inhibit protein synthesis directly, ciprofloxacin inhibits DNA synthesis, while amoxicillin is a cell wall synthesis inhibitor (18), thus the time required for mutation appearing on amoxicillin-selective plate was longer. Moreover, based on the MIC measurement, we found that these mutants also showed significant enhancement toward other multiple antibiotics (Fig. 1C). For example, the mutant colonies exhibiting reduced susceptibility to chloramphenicol also exhibited reduced susceptibility to ciprofloxacin, levofloxacin, norfloxacin, tetracycline, and trimethoprim. Notably, the acquired resistance was heritable, as the enhanced MICs were stably maintained after the 5-d consecutive inoculation (i.e., 33 generations for E. coli) without any selective pressure from either antibiotic or antidepressant (SI Appendix, Table S1).
The resistance development was dose-dependent, in that lowering the concentration of sertraline prolonged the time for the occurrence of resistance. For example, at medium concentration (10 mg/L) sertraline induced significant enhancement of the ratio of resistant cell number to total cell number toward reduced susceptibility to chloramphenicol, tetracycline, amoxicillin, and ciprofloxacin in 3 d, 15 d, 35 d, and 60 d, respectively. In addition, based on the assessment by plate-counting, at low concentration (0.1 mg/L and 1 mg/L) sertraline also increased the ratio of resistant cell number to total cell number toward chloramphenicol and tetracycline within 60 d. However, the enhancement was not seen on ciprofloxacin- or amoxicillin-selective plates after 60-d consecutive exposure. The mutants on the selective plates showed phenotypic heritance of antibiotic resistance, which were seen from a significant enhancement of MICs compared with the wild-type E. coli, including the MICs toward the antibiotics ampicillin, levofloxacin, norfloxacin, and trimethoprim (SI Appendix, Table S1).
In addition to sertraline, duloxetine also exhibited similar effects on the induction of resistance to multiple antibiotics. Up to 5 × 103- and 896-fold increases in ratio of resistant cell number to total cell number toward chloramphenicol and tetracycline were detected following exposure of E. coli to high concentration level (50 mg/L) duloxetine after 1 d of exposure. Following exposure to duloxetine for 2 d, the ratio of resistant cell number to total cell number increased significantly toward amoxicillin (13-fold) and ciprofloxacin (123-fold). The ratios further increased when extending the exposure time to 60 d, resulting in increased ratio of resistant cell number to total cell number toward chloramphenicol (2 × 106-fold), tetracycline (2 × 106-fold), amoxicillin (15-fold), and ciprofloxacin (4 × 104-fold). The low concentration level applied in this study, i.e., 0.1 mg/L duloxetine, also enhanced the ratio of resistant cell number to total cell number toward both chloramphenicol and tetracycline significantly after 50 d (214-fold) (Fig. 1D and SI Appendix, Figs. S1 and S2).
In contrast, for the other three antidepressants examined (bupropion, escitalopram, and agomelatine), only high levels (50 mg/L and 100 mg/L) resulted in significantly increased resistance to chloramphenicol, tetracycline, and amoxicillin, with the highest fold change of eightfold comparing with the non-antidepressant control group (SI Appendix, Fig. S1).
Intra-ROS Generation Enhances the Resistant Cell Number.
We hypothesized the increased resistance to antibiotics observed following exposure to antidepressants may be associated with the production of hydroxyl radicals, including ROS (Fig. 2 A and B).
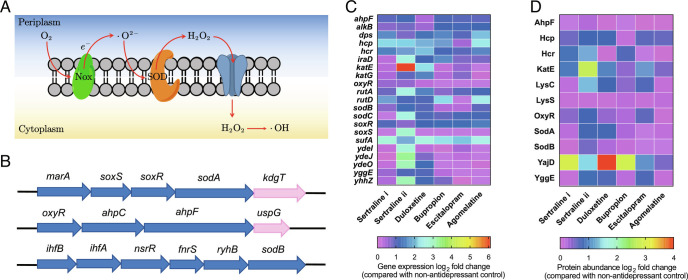
Fig. 2.
Antidepressants enhance ROS generation level. (A) Schematic ROS generation pathway of E. coli. (B) Genes responsible for oxidative stress and ROS generation. Genes are indicated as follows: blue, transcription factor binding site that activates transcription. Pink, transcription factor binding site that represses transcription. (C) Heatmap with log2 fold changes of key genes related to ROS under each antidepressant-dosage condition. (D) Heatmap with log2 fold changes of key proteins related to oxidative stress under each antidepressant-dosage condition. For C and D, sertraline i refers to 1 mg/L, and sertraline ii refers to 50 mg/L. The other four antidepressants are 50 mg/L. See also SI Appendix, Figs. S3 and S4.
We first employed flow cytometry to detect intracellular ROS generation using the 2′, 7′-dichlorofluorescein diacetate (DCFDA) dye. Compared with the non-antidepressant control group, both sertraline and duloxetine enhanced the ROS generation significantly with the lowest concentration, i.e., 0.1 mg/L (P = 0.0003–0.001). The fold change further increased when increasing the concentration to 50 mg/L: sertraline (1.9-fold) and duloxetine (4.4-fold) (SI Appendix, Fig. S3A). Bupropion only increased ROS generation 1.2-fold at the highest concentration applied (100 mg/L) (P = 0.028). In contrast, the other two antidepressants, escitalopram and agomelatine, did not lead to enhanced ROS production (SI Appendix, Fig. S3).
To further examine ROS induction, we conducted the experiments under anaerobic conditions, where ROS cannot be generated (20). As expected, no significant ROS generation enhancement was seen following exposure of E. coli to sertraline or duloxetine (SI Appendix, Fig. S3). Compared with the aerobic condition, ROS generation decreased significantly under the anaerobic condition (P = 0.000002–0.00003, SI Appendix, Fig. S3B). Moreover, when conducting the plate-counting experiments under anaerobic condition, the absolute mutant number on chloramphenicol-, tetracycline-, and ciprofloxacin- selective plates decreased significantly in the first 3 d (P = 0.00006–0.021). For example, following 1-d exposure to 50 mg/L sertraline anaerobically, the number of mutants grown on ciprofloxacin-selective agar decreased compared to the aerobic condition. Significant decrements were also detected with the exposure to 50 mg/L duloxetine.
Next, the transcriptomic and proteomic profile of E. coli following exposure to these antidepressants was tested by applying genome-wide mRNA and proteomic sequencing. Typical genes related to oxidative stress increased, for example, the regulation level of the gene katE increased drastically, and as high as 68.6-fold enhancement was detected after exposure to 50 mg/L sertraline (P < 0.05, q < 0.05, Fig. 2C and SI Appendix, Tables S2 and S3). The superoxide response regulator soxS (21) was up-regulated 7.3-fold after exposure to 50 mg/L sertraline (P < 0.05, q < 0.05). In addition, the redox-sensitive activator, soxR (21), was also significantly up-regulated following exposure to all five antidepressants. Correspondingly, the typical protein families related to oxidative stress, including Sod, Hcp, and KatE (21, 22), increased significantly after exposure to the antidepressants (q < 0.01, Fig. 2D). Compared to the non-antidepressant control group, a 2.0-fold and 7.5-fold increment was observed following exposure to 1 and 50 mg/L sertraline, respectively (q < 0.01). In contrast, agomelatine had limited effects on phenotypic mutation (1.3-fold).
As sertraline showed the highest ratio of resistant cell number to total cell number toward multiple antibiotics, bacterial genotypic responses to sertraline were tracked on a long-term scale. After 60-d exposure to 1 mg/L or 50 mg/L sertraline, the mutants grown on antibiotic-selective agar were cultured and exposed to sertraline. Compared to the non-sertraline control group, the abundance of oxidative stress-related proteins increased significantly; for example, expression of the oxidative stress defense protein YggE (23) was significantly increased, ranging from 1.2- to 2.9-fold. Consistent with this observation, significant upregulation of the superoxide response regulator soxS was also detected, with a 1.1- to 5.1-fold enhancement (P < 0.05, q < 0.05, SI Appendix, Tables S4 and S5).
Antidepressants Can Stimulate Efflux Pump Expression and Affect Membrane Integrity.
Efflux pump activation is a key resistance mechanism that enables bacteria to regulate their internal environment by removing toxic substances such as antibiotics (24, 25). We hypothesized that antidepressants might have triggered efflux pump expression, thus contributing to the induction of resistance to multiple antibiotics. We therefore examined the transcriptomic and proteomic responses based on known efflux pump transporters. Notably, the expression of key genes and proteins on resistance-nodulation-division (RND) type efflux pumps, ATP-binding cassette superfamily (ABC), and multi-antimicrobial extrusion protein family (MATE) showed significant variations when exposed to antidepressants (Fig. 3A and SI Appendix, Table S7). The enhancement caused by sertraline was generally dose-dependent, with the greatest increases induced by 50 mg/L sertraline (P < 0.05, q < 0.05, Fig. 3B and SI Appendix, Table S6). For example, MdtA expression increased 5.0-fold when exposed to 50 mg/L sertraline, while only a 1.2-fold increase was observed for 1 mg/L sertraline. In addition, after exposing to sertraline for a longer term (60 consecutive days), the mutants also showed increased transcription levels of the mdtABC genes, ranging from 1.2- to 63.3-fold (SI Appendix, Tables S8 and S9). These results indicate that sertraline stimulated multidrug efflux pump expression at both short-term and long-term levels. The abundances of major regulons for multidrug resistance, AcrAB, MdtA (21) increased under the effect of 1 mg/L or 50 mg/L sertraline, and 50 mg/L duloxetine. Compared to sertraline or duloxetine, the other three antidepressants (bupropion, escitalopram, and agomelatine) had a negligible effect on these genes, which corresponded to the observed mutant effects (Fig. 3C).
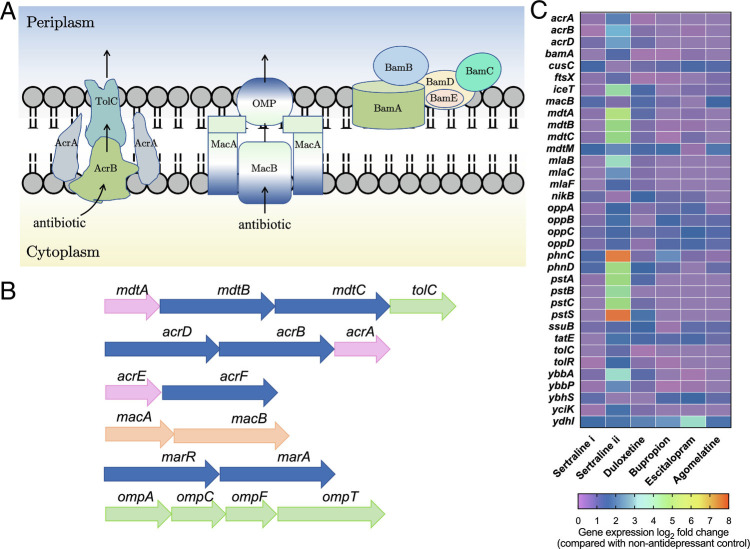
Fig. 3.
Antidepressants stimulate efflux pump and affect bacterial membrane. (A) Schematic major efflux pumps in E. coli. (B) Genes responsible for efflux pumps and outer membrane. Genes are indicated as follows: pink, membrane fusion protein gene. Blue, RND type efflux protein gene. Green, outer membrane protein gene. Orange, ABC superfamily transporter. (C) Heatmap with log2 fold changes of key genes related to efflux pump, including RND-type efflux pumps, ABC superfamily, and multi-antimicrobial extrusion protein family (MATE). Sertraline i refers to 1 mg/L, and sertraline ii refers to 50 mg/L. The other four antidepressants are 50 mg/L. See also SI Appendix, Fig. S4.
As each efflux transporter consists of the inner membrane transporter, the outer membrane channel, and the periplasmic lipoprotein, there is a close linkage between membrane proteins and the bacterial efflux pump (24, 26). In this study, we checked the abundance and regulation of membrane-related genes and proteins at the transcriptional level. We found that for membrane protein or porin-related genes, in total 430 genes were detected, and 50 mg/L sertraline or duloxetine resulted in the highest number of genes with significant variations (P < 0.05 and q < 0.05), which were 235 genes and 206 genes, respectively. For proteins, among the 141 membrane-related proteins detected, at least 127 differentially expressed proteins were identified following exposure to these antidepressants (q < 0.01). Notably, as one of the typical outer membrane-regulated genes, expression levels of ompF decreased significantly when challenged with antidepressants, and 50 mg/L sertraline or duloxetine showed the highest decrement, i.e., 10.0- and 9.2-fold decrement, respectively (P < 0.05, q < 0.05, Fig. 3C and SI Appendix, Tables S10 and S11). The down-regulated synthesis of OmpF results in the increased production of the main multidrug efflux pump AcrAB, thus inducing antibiotic resistance in E. coli (27). In addition, increased expression of OmpA was also observed, consistent with its role in decreased susceptibility to antibiotics (28). The mutants induced by sertraline following long-term exposure also showed significant variations of membrane proteins or porins (SI Appendix, Tables S12 and S13). Also, genes exbBD, fecA, and tonB down-regulated under the effect of antidepressants, TonB-related receptors in the outer membrane, and these receptors are responsible for transporting external chemicals like antibiotics (27). Interestingly, diazepam, another benzodiazepine drug, has also been shown to promote reduced susceptibility to antibiotics in E. coli and Klebsiella pneumoniae by depleting porin expression and inducing efflux systems previously (29).
Antidepressants Can Cause Chromosomal Mutations and Multidrug Resistance.
We hypothesized that the reduced susceptibility toward multiple antibiotics may also be associated with mutations on the chromosome. We therefore performed genome sequencing of the entire population following 15-d, 30-d, and 60-d sertraline or duloxetine exposure, as well as 30-d bupropion, escitalopram, and agomelatine exposure. In addition, individual antidepressant-exposed mutants selected from LB agar plates containing chloramphenicol, tetracycline, ciprofloxacin, or amoxicillin were sequenced. By comparing the sequencing results of the bacterial population or mutants with the wild-type E. coli MG1655, nonsynonymous single nucleotide polymorphisms (SNPs) were identified.
Serial passaging in the presence of all five antidepressants induced a range of mutations on E. coli MG1655 at the population level (Fig. 4A). Among them, nonsynonymous SNPs within the ompT, stfQ, stfR, and tfaD genes were identified in all nine populations. The ompT gene encodes as outer membrane porin, and it affects the antimicrobial susceptibility of E. coli (30). It is reported that mutations in ompT may reduce porin expression, thus leading to decreased susceptibility to multiple antibiotics, including chloramphenicol, ciprofloxacin, nalidixic acid, ampicillin, and cefotaxime (31). The other three most common mutated genes, stfQ, stfR, and tfaD, are linked to prophage or cryptic prophage, and we posit they may play a role in inducing oxidative stress as described previously (32). Nonsynonymous SNPs in marR were identified in all sequenced populations except those exposed to escitalopram and agomelatine. MarR is a transcriptional regulator of the MarAB efflux pump, reducing the accumulation of antibiotics, and conferring resistance to low-level antibiotics (33–35). This may explain the phenomenon that escitalopram and agomelatine had limited effects on the induction of bacterial multidrug resistance. Serial passaging in the presence of sertraline and duloxetine led to nonsynonymous SNPs in the acrR gene, which is responsible for multidrug transportation and efflux pump expression (36). This agrees with the phenotypic data that sertraline and duloxetine could cause significantly increased resistance.
Fig. 4.
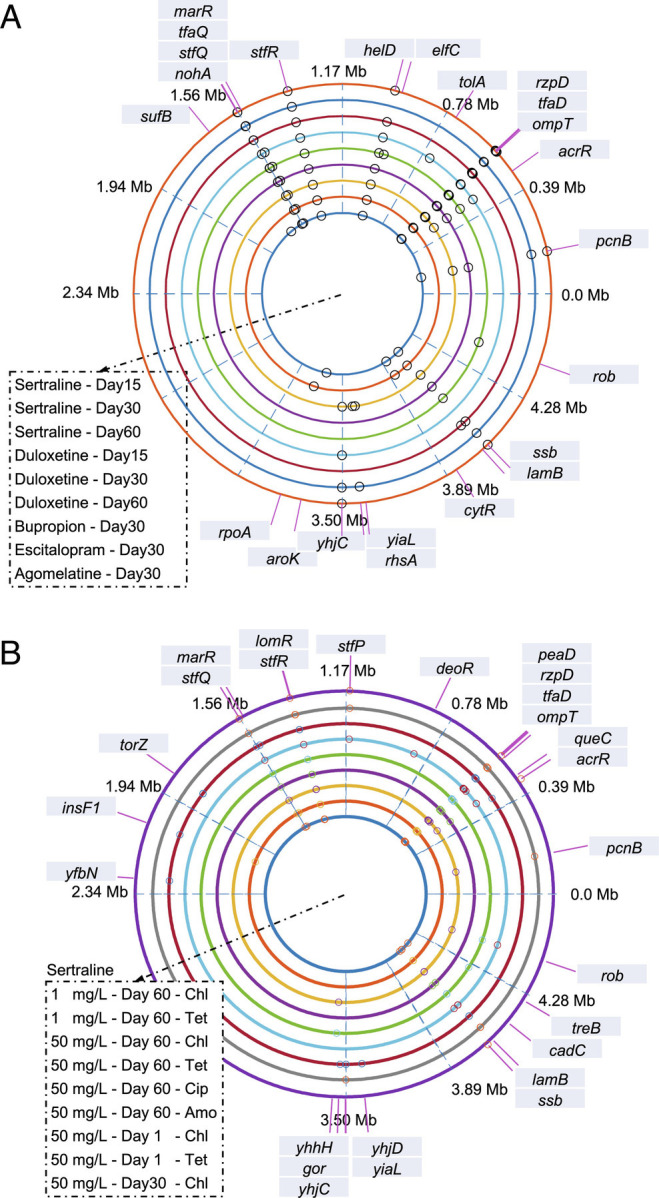
Antidepressants cause chromosomal mutations and multidrug resistance. Circos figure indicates the genes where SNPs happen chromosomally. (A) Bacterial population sequencing results. Circles from innermost to outermost are 50 mg/L sertraline 15 d-, 50 mg/L sertraline 30 d-, 50 mg/L sertraline 60 d-, 50 mg/L duloxetine 15 d-, 50 mg/L duloxetine 30 d-, 50 mg/L duloxetine 60 d-, 100 mg/L bupropion 30 d-, 100 mg/L escitalopram 30 d-, and 100 mg/L agomelatine 30 d-exposed bacterial population, respectively, with biological triplicate. (B) Mutant sequencing results. Circles from innermost to outermost are mutant strains of 1 mg/L sertraline-induced mutants on chloramphenicol-, tetracycline-selective plates (60-d exposure), 50 mg/L sertraline-induced mutants on chloramphenicol-, tetracycline-, ciprofloxacin-, amoxicillin-selective plates (60-d exposure), 50 mg/L sertraline-induced mutants on chloramphenicol-, tetracycline-selective plates (1-d exposure), and 50 mg/L sertraline-induced mutants on chloramphenicol-selective plate (30-d exposure), respectively, with biological triplicate. See also SI Appendix, Tables S14 and S15.
Regarding the sequenced mutant strains, all antidepressant-induced mutants possessed nonsynonymous SNPs in the ompT, rzpD, stfQ, and stfR genes (Fig. 4B). Among them, ompT, stfQ, and stfR were the same as those found in the population sequenced groups, suggesting these were dominant mutations induced in the population. Mutations were also found in the rzpD gene, which is located within a prophage; the same SNP was also found in heavy-metal stress-induced ciprofloxacin-resistant mutants (37), suggesting it may contribute to stress adaptation. Similar to the population sequencing results, nonsynonymous SNPs of marR and acrR were only shown in sertraline and duloxetine-induced mutants (SI Appendix, Tables S14 and S15).
Antidepressants Can Also Enhance the Persistence toward Antibiotics.
Persisters are dormant variants of wild-type bacteria, which are formed in microbial populations stochastically (38, 39). Persister cells are not resistant to antibiotics, but exhibit high tolerance toward antibiotics. In this study, we hypothesized that in addition to inducing resistance against multiple antibiotics, antidepressants may also promote bacterial persistence. To this end, after exposing to the antidepressants, the persister number toward antibiotic ciprofloxacin was measured (40). We found that 10 mg/L and 50 mg/L sertraline or duloxetine and 100 mg/L bupropion induced significant enhancement of the absolute number of persisters and the ratio of persister to total cell number (SI Appendix, Fig. S5, P = 0.000008–0.021). The fold change of absolute persister number reached as high as 52.3-fold comparing with the non-antidepressant control group, and the ratio of persister number to total cell number was up to 84.7-fold (Fig. 5 A and B). For antidepressants escitalopram and agomelatine, the highest fold change of persister number and ratio of persister number to total cell number was only up to 3.0-fold comparing with the non-antidepressant control. Extending the antidepressant exposure period to a consecutive 5-d period did not further increase the fold change of absolute persister number or ratio of persister number to total cell number significantly (P = 0.083–0.611, Fig. 5 C and D). Notably, the ciprofloxacin persister population generated rapidly (within 1 d) after exposure of E. coli to 10 mg/L sertraline or duloxetine; this occurred much faster than the ciprofloxacin-resistant bacteria formed (60 d). Also, the other three antidepressants could induce ciprofloxacin persister generation at medium and high concentrations within 5 d, while the ciprofloxacin-resistant mutant could not be induced in 60 d.
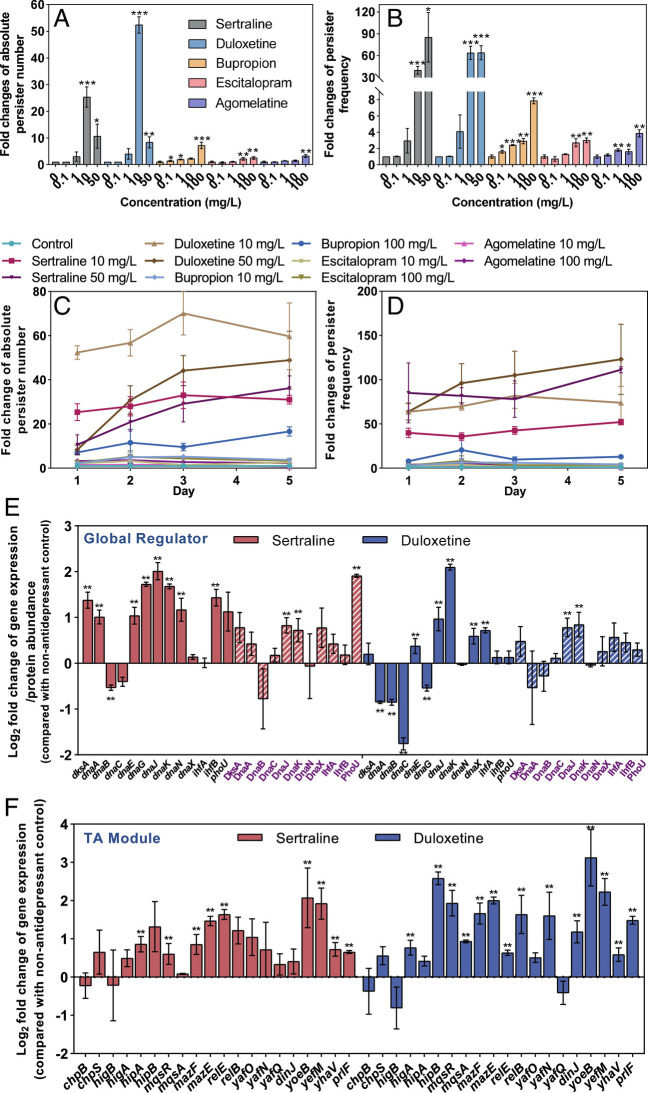
Fig. 5.
Antidepressants sertraline and duloxetine enhance bacterial persistence toward antibiotic ciprofloxacin. (A) Fold changes of absolute persister number when exposing wild-type E. coli to sertraline or duloxetine for 1 d. (B) Fold changes of ratio of persister number to total cell number when exposing wild-type E. coli to sertraline or duloxetine for 1 d. (C) Fold changes of absolute persister number when exposing wild-type E. coli to sertraline or duloxetine for a consecutive 5 d. (D) Fold changes of ratio of persister number to total cell number when exposing wild-type E. coli to sertraline or duloxetine for a consecutive 5 d. (E) Log2 fold changes of key genes/proteins related to bacterial persistence global regulator when exposing wild-type E. coli to 50 mg/L sertraline or duloxetine. Protein names are shown in purple font. (F) Log2 fold changes of detected genes relating to bacterial toxin–antitoxin (TA) modules when exposing wild-type E. coli to 50 mg/L sertraline or duloxetine. Data are shown as mean ± SD, n = 3 independent experiments. Significant differences between antidepressant-dosed samples and the non-antidepressant control are analyzed by independent sample t test with Benjamini–Hochberg multiple comparison testing, *P < 0.05, **P < 0.01, ***P < 0.001. See also SI Appendix, Fig. S5.
Genotypically, several global regulators have been reported to affect the expression of persister-related genes, and thus resulting in phenotypes of persistence. These global regulators include dnaKJ, ihfAB, dksA, spot, ssrS, phoU, ygfA (38). Genome-wide mRNA sequencing showed that both sertraline and duloxetine significantly enhanced expression levels of dnaKJ, phoU, and ihfA (P < 0.05, q < 0.05), with up to 4.3-fold increment compared to the control group without antidepressant treatment (Fig. 5E). The formation of persisters is also an evolutionary strategy to tolerate stress (41). Increases in persister number against antibiotics such as ampicillin, norfloxacin, and ciprofloxacin are linked to responses to stress (38, 42). In this study, we found antidepressants induced stress responses in E. coli, which was evidenced by the upregulation of the general stress response regulator rpoS and the umuCD SOS stress response genes (SI Appendix, Table S16).
Bacterial toxin–antitoxin (TA) modules are studied in the context of bacterial persister formation (38, 41). In this study, genes related to TA modules were also significantly up-regulated following exposure to sertraline or duloxetine. Modules including hipA-hipB, mazF-mazE, yoeB-yefM, and ypjF-yfjZ were significantly up-regulated (P < 0.05, q < 0.05), with a highest enhancement of 7.6-fold (Fig. 5F). In addition, the enhanced stress levels may cleave antitoxins and result in the excess of toxins (SI Appendix, Tables S16–S19) (43, 44).
Antidepressants Can Affect the Evolution of Persister and Resistant Bacteria.
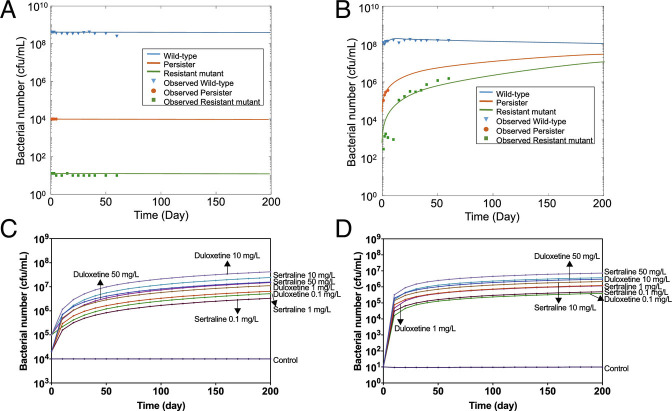
Generally, bacterial evolution through gene mutations is a slow and incremental process (45). In this study, we found that antidepressants can significantly promote the process of antibiotic resistance and persistence experimentally. To further predict their effects on long-term bacterial evolution, a mathematical model was proposed to describe the dynamic changes of the wild-type strain (i.e., antibiotic-sensitive bacterium), persister, and antibiotic-resistant mutant. In the ordinary differential equation model, W, P, and R represent the concentration of wild-type, persister, and antibiotic-resistant mutant, respectively. μ and d are the growth rate and death rate, respectively; η1 and λ1 refer to the evolving rate from wild-type to persister and wild-type to mutant, respectively; η2 and λ2 refer to the evolving rate from persister to wild-type and mutant to wild-type, respectively. Km is the environmental carrying capacity. Notably, the persister in the model was considered as phenotypically resistant, as no growth or death was shown, while antibiotic-resistant mutant was genetically resistant.
The experimentally measured numbers of wild-type (1 to 60 d), persister (1 to 5 d), and antibiotic-resistant mutant (1 to 60 d) toward ciprofloxacin were applied to calibrate the parameters, η1, η2, λ1, and λ2. The growth rate assays were applied to calibrate the parameters, μW, μR, dW, and dR. The trajectories of W, P, and R showed that when exposed to each antidepressant at their highest concentration continuously, wild-type bacteria would evolve to persisters and resistant bacteria (Fig. 6 A and B and SI Appendix, Fig. S6). Comparing the calibrated values of η1(10−9–10−4), λ1(10−10–10−6) under each condition (SI Appendix, Table S20), we found that it was easier for wild-type bacteria to become phenotypically persistence than directly evolve to antibiotic-resistant mutants. This corresponded with the previous findings that bacteria first have adaptive changes to antibiotic stress before a change in resistance happened (42), and persister is playing a key role in antibiotic-resistant bacteria formation (46). In addition, the calculated long-term stable values of wild-type, persister, and resistant bacteria varied under different conditions. Persister and resistant bacteria number increased with the increment of antidepressant concentrations (Fig. 6 C and D). Low concentration levels (0.1 mg/L and 1 mg/L) also imposed accumulative enhancement on persistence and resistance levels during the long-term period.
Fig. 6.
Antidepressants sertraline and duloxetine affect the evolution of persister and resistant bacteria. (A) Simulated trajectories of wild-type, persister, and antibiotic-resistant mutant numbers under the control condition (without any antidepressant dosage). (B) Simulated trajectories of wild-type, persister, and antibiotic-resistant mutant numbers after exposure of 50 mg/L sertraline. (C) Simulated trajectories of persister number after exposure of sertraline or duloxetine with different concentrations. (D) Simulated trajectories of antibiotic-resistant mutant number after exposure of sertraline or duloxetine with different concentrations. See also SI Appendix, Fig. S6 and Table S20.
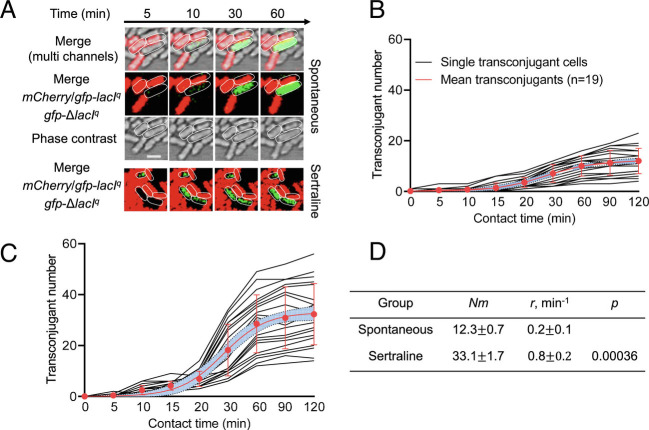
The Antidepressant Sertraline Promotes Conjugative Plasmid Transfer.
Multidrug resistance and persistence are the phenomenon of single clonal line, we further tracked the horizontal transfer of antibiotic resistance induced by the antidepressant sertraline. By applying microfluidic and confocal microscopy systems, we visualized plasmid conjugation in real time at the single-cell level. The time-lapse microscopy images showed that in the control group (without the addition of any antidepressants), the plasmid pKJK5 was successfully transferred to the recipient after 10 min of contact (Fig. 7 A and B). In contrast, under exposure to sertraline, the plasmid transfer process was significantly accelerated, and successful transfer was observed within 5 min (Fig. 7 A and C), with the transfer rate (r) increased ~4.6-fold (P < 0.001, Fig. 7D). Moreover, the number of transconjugants was dramatically increased by sertraline (~3.0-fold increase, P < 0.001), demonstrating that plasmid conjugation is significantly promoted in the presence of sertraline.
Fig. 7.
Real-time visualization of conjugative transfer by gfp-encoded pKJK5 plasmid. (A) Time-lapse microscopic images of conjugative transfer performed in a microfluidic chamber without antidepressant (spontaneous) or under the exposure of 10 mg/L sertraline. (B) Single-cell time-lapse quantification of transconjugant (green-fluorescent cell) number in the control group (without antidepressant). (C) Single-cell time-lapse quantification of transconjugant in the sertraline-exposed group. (D) Model fitting results of B and C. Nm is the predicted maximum transconjugant number, and r is the rate of conjugative transfer (transconjugants per minute). For B and C, each black line represents transconjugant number generated at different time points. The red line is the fitting curve of all black lines with standard deviations. The violet area is the 95% CI.
Discussion
In this work, we show that the commonly consumed antidepressants (e.g., SSRIs and SNRIs) can reduce susceptibility of E. coli to multiple antibiotics during a 60-d exposure period. Antimicrobial side effect of antidepressants and the required exposure time to trigger antibiotic resistance might vary, depending on antidepressant types and dosages. Strikingly, the antidepressants sertraline and duloxetine at clinically relevant concentrations in colon (e.g., 50 mg/L) (15) caused an effect after only 1 d of exposure. Compared to the non-antidepressant control, the ratio of resistant cell number to total cell number reached more than 10,000-fold based on colony enumeration. This was even higher than the mutant effects caused by several subinhibitory antibiotics, such as nalidixic acid, ampicillin, rifampicin, and carbenicillin (47, 48). This was consistent with our previous study, which showed that the antidepressant fluoxetine could induce resistance to multiple antibiotics at a concentration of 100 mg/L within 1 d of exposure (17). The susceptibility to multiple antibiotics was increased as high as 64-fold based on MIC measurements. The resistance mutations showed a stable heritance after 33 generations, indicating the antibiotic susceptibility did not revert to the non-susceptible state even if the exposure to antidepressants was removed, which is consistent with previous studies (3, 49). In addition to inducing resistance, exposure to sertraline and duloxetine for only 1 d also promoted bacterial persistence to the antibiotic ciprofloxacin. The developed persistence may help to facilitate the evolution of resistance (6). Unlike sertraline or duloxetine, the other three antidepressants (bupropion, escitalopram, and agomelatine) imposed limited effects on resistance, up to eightfold at high concentration level (i.e., 100 mg/L) according to colony counting, while at medium (10 mg/L) and low (0.1 mg/L, 1mg/L) concentrations they cannot enhance ratio of resistant cell number to total cell number. These resistance traits were also heritable.
Based on the phenotypic and genotypic analyses, we found that the enhanced ROS generation caused by sertraline and duloxetine was directly associated with induced resistance (Fig. 8). Under anaerobic conditions, ROS generation by the antidepressants sertraline and duloxetine was eliminated, leading to a significant reduction in the number of resistant mutants. Genotypically, proteins and genes related to ROS, including those for scavenging reactive species (AphF and SodBC) (21), activating redox sensors (soxR) (50), and regulating the superoxide response (soxS) (51), were significantly up-regulated by antidepressants sertraline and duloxetine (P < 0.05, q < 0.05). Notably, the other three antidepressants with limited effects on ratio of resistant cell number to total cell number (up to eightfold) did not alter the expression of these genes/proteins. Also, the radical-induced altered susceptibility to antibiotics was illustrated by sublethal antibiotics, as the formation of reactive species including ROS was promoted by antibiotics, and it may contribute directly to the evolution of multidrug resistance (52–54). To investigate whether selection might have biased antidepressant-increased mutation (e.g., by enriching for mutations conferring antibiotic resistance), we conducted fluctuation tests (55) by examining the spontaneous mutation rate against phage, a phenotype not under selection. The spontaneous phage resistance mutation rate of wild-type E. coli MG1655 was 7.25 ± 1.75 × 10−9, which was significantly (P = 0.003) lower than the one treated with 10 mg/L of sertraline (2.00 ± 0.29 × 10−8; SI Appendix, Figs. S7 and S8 and Table S21), suggesting that antidepressants indeed induce mutagenesis and increase mutation rate.
Fig. 8.
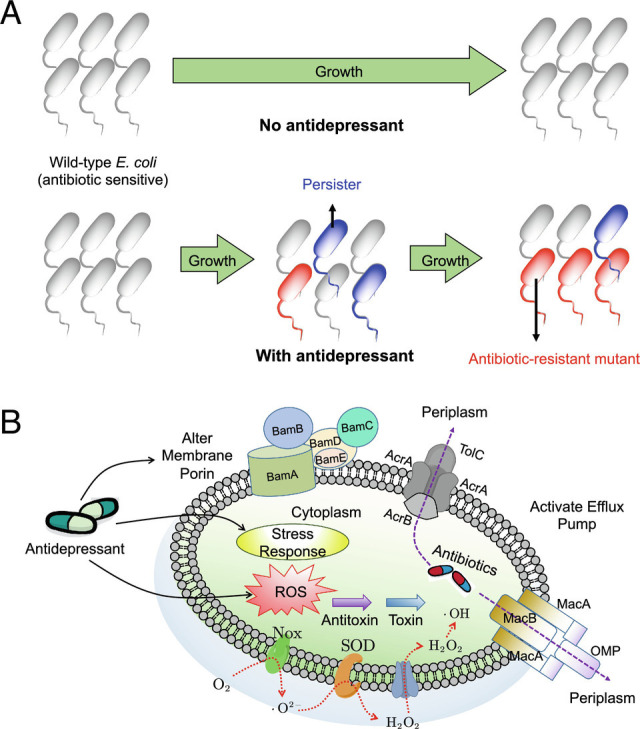
Antidepressants induce resistance to multiple antibiotics and enhance antibiotic persistence. (A) Overall bacterial evolution with and without antidepressant dosage. (B) Possible mechanisms illustrating the process: antidepressants increase bacterial intracellular ROS generation, enhance bacterial stress response, alter bacterial membrane, activate efflux pumps, and stimulate TA modules.
Efflux pump expression, as one of the main mechanisms for expelling antibiotics from the cell and reducing susceptibility to antibiotics (26, 30, 56), was stimulated by antidepressants sertraline and duloxetine. The activated RND, ABC, and MATE systems may link to the significant enhancement of antibiotic resistance levels. Maier et al. also showed that human-targeted drugs may boost antibiotic resistance by controlling efflux pump expression (57). Chromosomally, all tested antidepressants in this study can induce nonsynonymous SNPs on the outer membrane porin ompT, while sertraline and duloxetine can induce nonsynonymous SNPs in several other genes related to multidrug efflux pumps, including marR and acrR. This indicated a close relationship between antidepressants and antibiotic efflux pump expression. However, some environmental stresses, such as the up-regulated superoxide response regulator soxS, may also result in the increased production of the main multidrug efflux pump AcrAB (27, 58). More research is needed to demonstrate whether antidepressants activate antibiotic efflux pumps directly or via over-generated intracellular ROS. In addition, further studies are required to test whether the same effects would be seen on other microorganisms, for instance, those without an SOS response.
In addition to inducing antibiotic resistance, exposure to antidepressants sertraline and duloxetine at medium and high concentrations (10 mg/L and 50 mg/L) also promoted bacterial persistence toward ciprofloxacin within 1 d. As ciprofloxacin-resistant bacteria formed much later than the ciprofloxacin persisters, we hypothesized that persistence would evolve to resistance with an extended exposure time as previously reported (46, 59), and that persistence may also help maintain resistance when antidepressant exposure stopped (6). To this end, a mathematical model was proposed to simulate the population dynamic changes of wild-type, persister, and resistant bacteria when exposed to antidepressants. Simulation results indicated persister contributed to the evolution of resistant bacteria, with a dose-dependent response. Antidepressants sertraline and duloxetine with higher concentrations imposed a faster evolution rate for wild-type to persister or to resistant bacteria. However, all five antidepressants at low concentrations (0.1 mg/L and 1 mg/L) could also impose accumulated effects on evolution. Genetically, bacterial TA modules are related to persister formation, and we found that genes related to TA modules up-regulated when challenged with sertraline or duloxetine. This was consistent with previous studies that the transcriptomes of persister cells showed upregulations of TA modules in E. coli (60, 61). Also, the activated efflux pumps, bacterial stress response, and TA modules could result in the formation of biofilm, which shows a surprising ability to resist and tolerant killing by antibiotics (43, 62). Thus, although bacterial evolution is stochastic, antidepressant exposure could drive the bacterial evolution toward antibiotic resistance, contributing to the emergence of antibiotic resistance.
It is also regarded that both persistence and resistance are bacterial physiological states that can be triggered by enhanced bacterial stress levels and efflux pumps (63, 64). Thus, the enhanced persister and resistant bacteria induced by antidepressants may be due to the triggered responses to increased stress (including ROS). Antidepressants including SSRI and SNRI are shown to possess antimicrobial activity, which may associate with the stresses imposed on bacteria (65). Moreover, the stress may still exist after the exposure of antidepressants, resulting in continuous bacterial persistence and heritable resistance, like the effects imposed by antibiotics (66). The reductions in bacterial population size (bottlenecks) and varying selection levels imposed by drugs would also affect bacterial antibiotic resistance evolution during pharmaceutical treatment (67). Accordingly, the stress levels may be applied as an initial screen for the detection of non-antibiotic pharmaceuticals for their inducing antibiotic resistance ability. Notably, as the induction of multidrug resistance is antidepressant-dependent, this may be due to antidepressant type and chemical structure. For example, sertraline, duloxetine, and fluoxetine have significant effects on bacterial multidrug resistance, and they belong to SSRI or SNRI. Regarding chemical structure, all these three antidepressants have an amino group in the second from the last carbon chain, while bupropion, escitalopram, or agomelatine does not. Further studies should evaluate and reveal which of these function groups in antidepressants are associated with antibiotic-like characteristics, which might be useful to redesign or repurpose of antidepressants.
Although antibiotic resistance has become a major threat to human health worldwide, the phenomenon caused by antidepressants has been largely overlooked. In conclusion, our findings illustrate how antidepressants induce bacterial resistance and persistence to a range of antibiotics. The bacterial intracellular ROS caused by antidepressants plays a key role in antidepressant-induced bacterial evolution toward reduced antibiotic susceptibility. The antibiotic persistence helps to maintain bacterial resistance, and the persisters could evolve to resistant bacteria during the long-term evolutionary process. Also, the promoted plasmid-mediated conjugation by antidepressant sertraline at the single-cell level indicated antidepressants can facilitate the spread of antibiotic resistance through horizontal gene transfer. Collectively, our studies implicate antidepressants as a contributing driver of increasing antibiotic resistance. The findings might be also useful for pharmaceutical factories and public health organizations to comprehensively evaluate antimicrobial sides of antidepressants. Further studies will be required to validate the antibiotic-like roles of antidepressants through employing more bacterial strains or conducting in vivo studies of antibiotic resistance spread in the gut microbiota.
Methods
Bacterial Evolution under the Exposure of Antidepressants.
Wild-type E. coli MG1655 (ATCC 700926) from −80 °C was cultivated on Luria-Bertani (LB) agar plate to select a single isogenic strain. The selected single isogenic strain was then cultured overnight to obtain a concentration of 3 × 108 cfu/mL. The bacteria culture was then exposed to five antidepressants during a 60-d aerobic evolution. The antidepressants include sertraline, duloxetine, bupropion, escitalopram, and agomelatine. For sertraline and duloxetine, final concentrations were 0.1, 1, 10, 50 mg/L, and 0.1, 1, 10, 50, 100 mg/L were applied for bupropion, escitalopram, and agomelatine, which were clinically or environmentally relevant concentrations (15, 57, 68). The concentration of antidepressants was grouped as low level (0.1 mg/L and 1 mg/L), medium level (10 mg/L), and high level (50 mg/L and 100 mg/L). During the evolution period, 40 μL of the overnight grown wild-type E. coli was inoculated to 3.96 mL LB broth containing various concentrations of antidepressants. After incubation for 24 h (Day 1), 40 μL of the bacteria was transferred to 3.96 mL fresh LB broth containing the antidepressant with the same concentration as on the previous day and incubated for another 24 h (Day 2). The same transfer and culture process was repeated for 60 d (Day 1 to Day 60). Exceptionally, under the exposure of 50 mg/L sertraline and duloxetine, it took 3 d for wild-type E. coli to grow to the concentration of 108 cfu/mL. To make it consistent, the 3-d exposure to 50 mg/L sertraline and duloxetine was noted as Day 1, and the later transfer was completed every 24 h after Day 1. All of the incubation condition was 100 rpm at 30 °C.
Wild-type E. coli evolution was also conducted under a 5-d anaerobic condition. The single isogenic wild-type E. coli MG1655 was exposed to 50 mg/L sertraline and duloxetine. The evolution under the anaerobic condition was the same as the aerobic condition, except that oxygen in LB or phosphate buffered saline (PBS) was depleted and the experiments were conducted in an anaerobic chamber (Coy Laboratory Products Inc., United States).
Determinations of Antibiotic-Resistant Mutants and Ratio of Resistant Cell Number to Total Cell Number.
Antibiotics chloramphenicol, tetracycline, amoxicillin, and ciprofloxacin were applied to select the antibiotic-resistant mutants. These antibiotics belong to different categories, namely phenicol, tetracycline, β-lactam, and quinolone, respectively. In detail, LB agar plates containing either 16 mg/L chloramphenicol, 4 mg/L tetracycline, 8 mg/L amoxicillin, or 0.08 mg/L ciprofloxacin were employed to count the number of mutants toward corresponding antibiotics. The concentrations of these selective antibiotics were determined according to the colony number from the wild-type E. coli (control group, 108 cfu/mL). Generally, one–ten colonies could grow on these antibiotic-selective plates. During the exposure to antidepressants, the number of antibiotic-resistant mutants was determined by enumerating colonies on antibiotic-selective plates. In order to remove the antidepressants in LB broth, the bacteria was washed twice with PBS and resuspended in the same volume of PBS, before plating onto antibiotic-selective plates. For all the plating, 100 μL bacteria was applied (except when more than 500 colonies were grown on the plates, and serial dilution was applied). The incubation condition for antibiotic-selective plates was 30 °C for 48 h. Antibiotic-resistant mutants were counted on Day 1, 2, 3, 5, 10, 15, 20, 25, 30, 35, 40, 50, and 60. The corresponding optical density at 600 nm (OD600) values was measured, and the total bacterial number was calculated based on the relationship between OD600 and bacteria colony-forming unit.
MICs Testing toward Multiple Antibiotics.
The MIC was determined by microbroth dilution of OD600 values as described previously (69, 70). The MIC toward the corresponding antibiotic was determined as the lowest concentration that no growth of strains was detected. The antibiotic-resistant mutants were randomly picked from the antibiotic-selective plates, when the corresponding ratio of resistant cell number to total cell number increased significantly. The antibiotics for MIC testing included amoxicillin, ampicillin, cephalexin, chloramphenicol, ciprofloxacin, colistin, erythromycin, kanamycin, levofloxacin, norfloxacin, roxithromycin, tetracycline, and trimethoprim. These antibiotics covered all of the typical antibiotic classes (71). Fold changes of MIC for mutant strains were compared with the wild-type E. coli strain. MIC fold changes higher than four folds were defined as significant enhancement. All MIC assays were performed at least in biological triplicate.
Antibiotic Resistance Hereditary Stability Detection.
To test whether the antibiotic resistance exhibited is heritable, the randomly selected mutant colonies were cultivated in 5 mL LB broth without any antibiotic or antidepressant. After 1 d of culturing at 37 °C, 1% of the bacterial inoculum was transferred to fresh 5 mL LB broth without any antibiotic or antidepressant. Following 5-d transfer and culture, the MICs were tested based on the method above and labeled as MIC-D5. The MICs-D5 were compared with the initial MICs, to determine whether any significant variations were shown.
Persister Generation during the Evolution.
Persister cells were measured under the exposure of 0.1, 1, 10, 50 mg/L sertraline and duloxetine during a 5-d evolution. Bacteria was cultured and transferred every 24 h for a consecutive 5 d, except when exposed to 50 mg/L sertraline and duloxetine, as described above. After 1, 2, 3, and 5-d exposure to antidepressants, the bacteria was washed twice with PBS to remove the remaining antidepressants, and the total bacterial number was counted on LB agar plates. Then, the antidepressant-treated bacteria were cultured in LB broth without any antibiotic, or with 5 × MIC of ciprofloxacin (2.5 mg/L). After culturing at 30 °C 100 rpm for 24 h or 48 h, samples were washed twice with PBS to remove the remaining ciprofloxacin before plating on LB agar plates without any ciprofloxacin. Following incubation at 30 °C for 48 h, the colony number was counted, which included both the persister and mutant toward ciprofloxacin. The persister cell numbers were obtained by deducting the corresponding mutant number (40, 59).
Measurement of Intracellular ROS Generation under the Exposure of Antidepressants.
Bacterial intracellular ROS generation and cell membrane permeability were measured after the exposure to antidepressants. As bacteria generate ROS in a quick way, the ROS was measured after 2-h exposure to antidepressants by applying 2′- 7′-DCFDA dye as described previously (17). A CytoFLEX S flow cytometer (Beckman Coulter, United States) was applied for the measurement. In addition, the ROS generation caused by antidepressants under anaerobic conditions was measured, by depleting oxygen and employing an anaerobic chamber (Coy Laboratory Products Inc., United States).
Whole-Genome DNA Sequencing of Mutant Strains and SNPs Calling.
Genome-wide bacterial DNA sequencing was applied to locate where the SNP happened. Both the bacterial population and single strain were sequenced. For bacterial population sequencing, bacterial genomic DNA was extracted from the population exposed to 50 mg/L sertraline or duloxetine, or 100 mg/L bupropion or escitalopram or agomelatine, while the genomic DNA from wild-type E. coli MG1655 was the control. For single strain sequencing, bacterial genomic DNA was extracted from wild-type E. coli MG1655 (control) and antidepressant induced mutant strains. The mutant strains included the colonies grown on: a) chloramphenicol-selective plates after 1-d exposure to 50 mg/L sertraline, b) tetracycline-selective plates after 1-d exposure to 50 mg/L sertraline, c) chloramphenicol-selective plates after 30-d exposure to 50 mg/L sertraline, d) chloramphenicol-selective plates after 30-d exposure to 50 mg/L duloxetine, e) chloramphenicol-selective plates after 30-d exposure to 100 mg/L bupropion, f) chloramphenicol-selective plates after 30-d exposure to 100 mg/L escitalopram, g) chloramphenicol-selective plates after 60-d exposure to 1 mg/L sertraline, h) tetracycline-selective plates after 60-d exposure to 1 mg/L sertraline, i) chloramphenicol-selective plates after 60-d exposure to 50 mg/L sertraline, j) tetracycline-selective plates after 60-d exposure to 50 mg/L sertraline, k) ciprofloxacin-selective plates after 60-d exposure to 50 mg/L sertraline, and l) amoxicillin-selective plates after 60-d exposure to 50 mg/L sertraline. On each selective plate, three colonies were randomly picked and sequenced.
Bacterial genomic DNA was extracted and sequenced, followed by the SNP calling according to the standard best practice guide of Genome Analysis Toolkit (GATK, v4.1.4.1) (72). The obtained SNPs in each mutant strain were compared with those in the biological triplicate control group, and different nonsynonymous SNPs were regarded as where mutation happened. The details are shown in SI Appendix, Text S1.
Whole-Genome mRNA Sequencing under the Exposure of Antidepressants.
Genome-wide RNA sequencing was employed to track differences of gene expression of E. coli MG1655 when challenged with antidepressants. In detail, wild-type E. coli MG1655 was exposed to 50 mg/L antidepressants, sertraline, duloxetine, bupropion, escitalopram, and agomelatine, separately, for sertraline, in addition to 50 mg/L, 1 mg/L was also applied. The condition without any antidepressant dosage was the control. As bacteria respond quickly to external stress on the transcriptional level, an exposure time of 2 h was applied based on previous studies (17, 73).
In addition to the comparison group above, another comparison group was established to track whether the long-term exposure to antidepressants would affect bacterial transcriptional responses. Specifically, sertraline-induced mutants were selected, including the colonies grown on chloramphenicol, tetracycline, amoxicillin, or ciprofloxacin-selective plates after 60-d exposure to 1 mg/L or 50 mg/L sertraline, and the strains were the same as the ones for DNA sequencing. The mutant strains were exposed to 50 mg/L sertraline, in contrast, the condition without any sertraline exposure was the control. The exposure time was 2 h.
For both the comparison groups, after 2-h exposure to corresponding antidepressants, bacterial total RNA was extracted and sequenced as described previously (74). Gene expression levels between the antidepressant-dosed samples and the corresponding control sample were compared based on the bioinformatic pipeline reported in previous studies (75, 76). The details are shown in SI Appendix, Text S2.
Genome-Wide Proteomic Analyses under the Exposure of Antidepressants.
Proteomics analyses were conducted to reveal bacterial responses toward antidepressants on the translational level. Similar to the transcriptional analyses, two comparison groups were established. An exposure time of 8 h was applied based on previous studies (73, 77).
Bacterial total protein was digested, sequenced, and analyzed as described previously (70, 78). Abundance ratios of each protein between antidepressant-dosed samples and the corresponding control sample were calculated. The details are shown in SI Appendix, Text S3.
Quantifying Mathematical Modeling Parameters and Model Simulation.
Growth rate μ and death rate d of wild-type and antibiotic-resistant mutant bacteria were determined by measuring the growth curves under the exposure of various concentrations of antidepressants. Growth rates were calculated according to the modified Gompertz Model (79), and Curve Fitting Tool in MATLAB 2016b was applied (73) (results are shown in SI Appendix, Table S20). The details of measurement are shown in SI Appendix, Text S4.
During the model simulation, the global optimal values of parameters η, λ, and σ were calculated by ode45 function and genetic algorithm solver in the optimization toolbox 7.3 of MATLAB 2016b (80, 81).
Plasmid-Mediated Conjugation at the Single-Cell Level.
Microfluidic and confocal microscopy systems were applied to real-time visualize the conjugation process at the single-cell level, according to our previous work (82). The donor E. coli MG1655 carried pKJK5 plasmid encoding gfp fluorescence and its chromosome (at attTn7 site) was specifically integrated with the Tn7 lacIq-pLpp-mCherry-KmR region, which enables the donor to encode constitutive red fluorescence. The gfp gene was inserted into the plasmid pKJK5 and was under the control of a promoter operator repressed by lacIq operon. In this case, the donor strain only expresses red fluorescence. Once successful transfer of plasmid pKJK5 to the recipient cells, gfp gene expression is de-repressed and enables the new host to be green-fluorescent cells (become transconjugants).
Statistical Analysis.
All the experiments were conducted independently at least in biological triplicate. Mean ± SD was shown on all figures. SPSS for Mac version 25.0 was applied for data analysis. We applied independent sample t tests and “Benjamini–Hochberg” correction method for multiple comparisons (83). P values less than 0.05 were considered to be statistically significant.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We acknowledge the Australian Research Council for funding support through the Discovery Project (DP220101526). J.G. would like to thank the support by UQ Foundation Research Excellence Awards. Y.W. would like to thank the support from China Scholarship Council.
Author contributions
J.G., Y.W., and Z. Yu, designed research; Y.W., Z. Yu, P.D., J.L., and L.N. performed research; Y.W., Z. Yu, L.M., Z. Yuan., J.E., M.A.S., and J.G. analyzed data; Z. Yuan, J.E., M.A.S., and J.G. edited the paper; and Y.W. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All data generated during this study are available in publicly accessible databases. DNA sequencing data are accessible through Sequence Read Archive of NCBI (BioProject accession number PRJNA832085). RNA sequencing raw data are accessible through Gene Expression Omnibus of NCBI with accession number GSE201666. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (84) partner repository with the dataset identifier (PXD033554).
Supporting Information
References
- 1.Meredith H. R., Srimani J. K., Lee A. J., Lopatkin A. J., You L., Collective antibiotic tolerance: Mechanisms, dynamics and intervention. Nat. Chem. Biol. 11, 182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darboe S., et al. , Articles Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis Antimicrobial Resistance Collaborators. The Lancet 399, 629–655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson D. I., Hughes D., Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Baym M., Stone L. K., Kishony R., Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351, aad3292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair J. M., Webber M. A., Baylay A. J., Ogbolu D. O., Piddock L. J., Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Levin-Reisman I., et al. , Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Bengtsson-Palme J., Kristiansson E., Larsson D. J., Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42, 053 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long H., et al. , Antibiotic treatment enhances the genome-wide mutation rate of target cells. Proc. Natl. Acad. Sci. U.S.A. 113, 2498–2505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamad B., The antibiotics market. Nat. Rev. Drug. Discovery 9, 675–676 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Cipriani A., et al. , Comparative efficacy and acceptability of antidepressant drugs in the acute treatment of major depressive disorder: A network meta-analysis. Lancet 391, 1357–1366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.OECD, Health at a Glance: OECD Indicators (OECD, 2013). [Google Scholar]
- 12.Fournier J. C., et al. , Antidepressant drug effects and depression severity: A patient-level meta-analysis. Jama 303, 47–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marken P. A., Munro J. S., Selecting a selective serotonin reuptake inhibitor: Clinically important distinguishing features. Prim. Care Companion J. Clin. Psychiatry 2, 205–210 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coles R., Kharasch E. D., Stereoselective analysis of bupropion and hydroxybupropion in human plasma and urine by LC/MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life 857, 67–75 (2007). [DOI] [PubMed] [Google Scholar]
- 15.McGovern A. S., Hamlin A. S., Winter G., A review of the antimicrobial side of antidepressants and its putative implications on the gut microbiome. Aust. N. Z. J. Psychiatry 53, 1151–1166 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Lukić I., et al. , Antidepressants affect gut microbiota and Ruminococcus flavefaciens is able to abolish their effects on depressive-like behavior. Transl. Psychiatry 9, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin M., et al. , Antidepressant fluoxetine induces multiple antibiotics resistance in Escherichia coli via ROS-mediated mutagenesis. Environ. Int. 120, 421–430 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Kohanski M. A., Dwyer D. J., Collins J. J., How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 8, 423 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J., A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Keren I., Wu Y., Inocencio J., Mulcahy L. R., Lewis K., Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339, 1213–1216 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Pomposiello P. J., Bennik M. H., Demple B., Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183, 3890–3902 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seaver L. C., Imlay J. A., Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J. Bacteriol. 183, 7182–7189 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S., et al. , The gene yggE functions in restoring physiological defects of Escherichia coli cultivated under oxidative stress conditions. Appl. Environ. Microb. 71, 2762–2765 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikaido H., Prevention of drug access to bacterial targets–Permeability barrriers and active efflux. Science 264, 382–388 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Putman M., van Veen H. W., Konings W. N., Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. R 64, 672–693 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikaido H., Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol 178, 5853 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido H., Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. R. 67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi U., Lee C.-R., Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front Microbiol. 10, 953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavío M. M., et al. , Enhanced active efflux, repression of porin synthesis and development of Mar phenotype by diazepam in two enterobacteria strains. J. Med. Microbiol. 53, 1119–1122 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Fernández L., Hancock R. E., Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bore E., et al. , Adapted tolerance to benzalkonium chloride in Escherichia coli K-12 studied by transcriptome and proteome analyses. Microbiology 153, 935–946 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Wang X., et al. , Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 1, 1–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sulavik M. C., Gambino L. F., Miller P. F., The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: Prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1, 436–446 (1995). [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen S. P., Hächler H., Levy S., Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175, 1484–1492 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prajapat M. K., Jain K., Saini S., Control of marRAB Operon in Escherichia coli via autoactivation and autorepression. Biophys. J. 109, 1497–1508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma D., Alberti M., Lynch C., Nikaido H., Hearst J. E., The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19, 101–112 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Li X., et al. , Sub-lethal concentrations of heavy metals induce antibiotic resistance via mutagenesis. J. Hazard Mater. 369, 9–16 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Lewis K., Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Brauner A., Fridman O., Gefen O., Balaban N. Q., Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Conlon B. P., et al. , Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 1, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harms A., Maisonneuve E., Gerdes K., Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354, aaf4268 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Fridman O., Goldberg A., Ronin I., Shoresh N., Balaban N. Q., Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Page R., Peti W., Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Lewis K., Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64, 503–514 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lynch M., Evolution of the mutation rate. Trends Genet. 26, 345–352 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen N. R., Lobritz M. A., Collins J. J., Microbial persistence and the road to drug resistance. Cell Host Microbe 13, 632–642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charpentier X., Kay E., Schneider D., Shuman H. A., Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J. Bacteriol. 193, 1114–1121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutierrez A., et al. , β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat. Commun. 4, 1–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hershberg R., Antibiotic-independent adaptive effects of antibiotic resistance mutations. Trends Genet 33, 521–528 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Seo S. W., Kim D., Szubin R., Palsson B. O., Genome-wide reconstruction of OxyR and SoxRS transcriptional regulatory networks under oxidative stress in Escherichia coli K-12 MG1655. Cell Rep. 12, 1289–1299 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Martin R. G., Gillette W. K., Rhee S., Rosner J. L., Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: Sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34, 431–441 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Dwyer D. J., et al. , Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U.S.A. 111, 2100–2109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koutsolioutsou A., Martins E. A., White D., Levy S., Demple B., A soxRS-constitutive mutation contributing to antibiotic resistance in a clinical isolate of Salmonella enterica (serovar Typhimurium). Antimicrob. Agents Chemother. 45, 38–43 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kohanski M. A., DePristo M. A., Collins J. J., Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37, 311–320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lang G. I., "Measuring mutation rates using the Luria-Delbrück fluctuation assay" in Genome Instability (Springer, 2018), pp. 21–31. [DOI] [PubMed] [Google Scholar]
- 56.Poole K., Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemoth. 56, 20–51 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Maier L., et al. , Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holden E. R., Webber M. A., MarA, RamA, and SoxS as mediators of the stress response: Survival at a cost. Front Microbiol. 11, 828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Windels E. M., et al. , Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 13, 1239–1251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shah D., et al. , Persisters: A distinct physiological state of E. coli. BMC Microbiol. 6, 1–9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keren I., Shah D., Spoering A., Kaldalu N., Lewis K., Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol 186, 8172–8180 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adams K. N., et al. , Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 145, 39–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pu Y., et al. , Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol. Cell 62, 284–294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imlay J. A., The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munoz-Bellido J., Munoz-Criado S., Garcıa-Rodrıguez J., Antimicrobial activity of psychotropic drugs: Selective serotonin reuptake inhibitors. Int. J. Antimicrob. Ag 14, 177–180 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Rasouly A., Nudler E., Reactive oxygen species as the long arm of bactericidal antibiotics. Proc. Natl. Acad. Sci. U.S.A. 116, 9696–9698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niels Mahrt A. T., et al. , Bottleneck size and selection level reproducibly impact evolution of antibiotic resistance. Nat. Ecol. Evol. 5, 1233–1242 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ford A. T., Fong P. P., The effects of antidepressants appear to be rapid and at environmentally relevant concentrations. Environ. Toxicol Chem. 35, 794–798 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Imai Y., et al. , A new antibiotic selectively kills Gram-negative pathogens. Nature 576, 459–464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y., Lu J., Zhang S., Li J., Guo J., Non-antibiotic pharmaceuticals promote the transmission of multidrug resistance plasmids through intra- and intergenera conjugation. ISME J. 15, 2493–2508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walsh C., Antibiotics: Actions, Origins, Resistance (American Society for Microbiology, ASM, 2003). [Google Scholar]
- 72.McKenna A., et al. , The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y., et al. , Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial genera. ISME J. 13, 509–522 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu J., et al. , Triclosan at environmentally relevant concentrations promotes horizontal transfer of multidrug resistance genes within and across bacterial genera. Environ. Int. 121, 1217–1226 (2018). [DOI] [PubMed] [Google Scholar]
- 75.Lu J., Ding P., Wang Y., Guo J., Antidepressants promote the spread of extracellular antibiotic resistance genes via transformation. ISME Commun. 2, 1–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ding P., Lu J., Wang Y., Schembri M., Guo J., Antidepressants promote the spread of antibiotic resistance via horizontally conjugative gene transfer. Environ. Microbiol. 24, 5261–5276 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y., et al. , Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation. ISME J. 14, 2179–2196 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.HaileMariam M., et al. , S-Trap, an ultrafast sample-preparation approach for shotgun proteomics. J. Proteome Res. 17, 2917–2924 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Zwietering M., Jongenburger I., Rombouts F., Van’t Riet K., Modeling of the bacterial growth curve. Appl. Environ. Microb. 56, 1875–1881 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shampine L. F., Reichelt M. W., The matlab ode suite. Siam J. Sci. Comput. 18, 1–22 (1997). [Google Scholar]
- 81.Conn A. R., Gould N. I. M., Toint Ph. L., A globally convergent augmented Lagrangian barrier algorithm for optimization with general Inequality constraints and simple bounds. Math. Comput. 66, 261–288 (1997). [Google Scholar]
- 82.Yu Z., Wang Y., Lu J., Bond P. L., Guo J., Nonnutritive sweeteners can promote the dissemination of antibiotic resistance through conjugative gene transfer. ISME J. 15, 2117–2130 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R Stat. Soc. B Methodol. 57, 289–300 (1995). [Google Scholar]
- 84.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All data generated during this study are available in publicly accessible databases. DNA sequencing data are accessible through Sequence Read Archive of NCBI (BioProject accession number PRJNA832085). RNA sequencing raw data are accessible through Gene Expression Omnibus of NCBI with accession number GSE201666. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (84) partner repository with the dataset identifier (PXD033554).