Significance
The potential of nucleic acids as therapeutics is now well appreciated with the widespread use of mRNA and adenovirus-based vaccines for COVID-19. RNA and DNA have potent immunostimulatory activities which can be harnessed for vaccine efficacy. While AIM2 and cGAS represent important sensors of cytosolic dsDNA, additional sensors of dsDNA also exist in cells. By studying inflammasome responses in human keratinocytes in response to poly(dA:dT), a widely used dsDNA mimetic, we defined a pathway that did not involve either AIM2 or cGAS-STING. Instead, we identify the NLRP1 inflammasome as a sensor for poly(dA:dT)-induced inflammation in the skin. These studies further our understanding of sensors of poly(dA:dT), helping explain the potent immunogenicity of this vaccine adjuvant in humans.
Keywords: NLRP1, inflammasome, keratinocyte, AIM2, poly(dA:dT)
Abstract
The accrual of cytosolic DNA leads to transcription of type I IFNs, proteolytic maturation of the IL-1 family of cytokines, and pyroptotic cell death. Caspase-1 cleaves pro-IL1β to generate mature bioactive cytokine and gasdermin D which facilitates IL-1 release and pyroptotic cell death. Absent in melanoma-2 (AIM2) is a sensor of dsDNA leading to caspase-1 activation, although in human monocytes, cGAS-STING acting upstream of NLRP3 mediates the dsDNA-activated inflammasome response. In healthy human keratinocytes, AIM2 is not expressed yet caspase-1 is activated by the synthetic dsDNA mimetic poly(dA:dT). Here, we show that this response is not mediated by either AIM2 or the cGAS-STING-NLRP3 pathway and is instead dependent on NLRP1. Poly(dA:dT) is unique in its ability to activate NLRP1, as conventional linear dsDNAs fail to elicit NLRP1 activation. DsRNA was recently shown to activate NLRP1 and prior work has shown that poly(dA:dT) is transcribed into an RNA intermediate that stimulates the RNA sensor RIG-I. However, poly(dA:dT)-dependent RNA intermediates are insufficient to activate NLRP1. Instead, poly(dA:dT) results in oxidative nucleic acid damage and cellular stress, events which activate MAP3 kinases including ZAKα that converge on p38 to activate NLRP1. Collectively, this work defines a new activator of NLRP1, broadening our understanding of sensors that recognize poly(dA:dT) and advances the understanding of the immunostimulatory potential of this potent adjuvant.
Double-stranded DNA (dsDNA) is normally compartmentalized in the nucleus or within mitochondria, preventing access to cytosolic germline-encoded pattern recognition receptors of the innate immune system. Sensing of dsDNA by cytosolic DNA sensors (CDSs) such as cGAS promote a type I interferon response (1–3). Intracellular dsDNA can also be sensed by inflammasomes, which oligomerize in response to cognate pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) to promote inflammatory caspase cleavage, pyroptosis, and maturation and release of the IL-1 family of cytokines (4, 5).
AIM2 was the first sensor of dsDNA described and the first inflammasome shown to recognize dsDNA in murine bone marrow-derived macrophages (BMDMs) and human THP-1 cells (6–8). A synthetic dsDNA known as poly(dA:dT) (9) was instrumental in early studies that defined both cGAS-STING signaling and the AIM2 inflammasome pathway. Unlike genomic or plasmid DNA, poly(dA:dT) has a complex nonlinear branching structure (10, 11) and is also capable of activating RNA sensors including RIG-I through the generation of short dsRNA intermediates by RNA polymerase III (12). The detection of poly(dA:dT) by multiple innate immune sensors likely explains its potent immunogenicity and potential for use as an immunoadjuvant in human vaccine trials (13).
In humans, cytosolic dsDNA can also initiate AIM2-independent inflammasome activation. Transfected dsDNA in primary human monocytes and macrophages stimulates a cGAS-STING lysosomal cell death program with secondary NLRP3 activation with no evidence of AIM2 activation (14). Prompted by these observations, we sought to explore the contribution of AIM2 in other human cell types. Further, given the role of the cGAS-STING-NLRP3 pathway in human monocyte responses to dsDNA, we were also interested in testing if this STING-dependent pathway contributed to dsDNA-driven inflammasome responses in other nonmyeloid human cells.
Several studies have suggested that transfected poly(dA:dT) can activate the AIM2 inflammasome in human primary keratinocytes (HPKs) (15, 16). Psoriasis patients but not healthy controls express elevated levels of AIM2 in the skin (17). IFNγ and TNFα priming can enhance IL-1β release in poly(dA:dT)-transfected cells, but poly(dA:dT) transfection still elicits LDH cytotoxicity and IL-1β release even in unprimed keratinocytes (18).
In recent years, multiple groups have established that NLRP1 is basally expressed in keratinocytes at high levels (19, 20). Human NLRP1 senses a wide spectrum of different activating signals, including ultraviolet B (UVB) radiation (21, 22), viral 3C proteases (23–25), and long dsRNA (26). It is tonically suppressed by DPP9 (27–30) and TRX1 (31, 32), as well as by some viral proteins such as vaccinia virus (VACV) F1L (33). Inhibitors of DPP9 such as Val-boroPro (VbP) are potent activators of NLRP1, and the study of these molecules has provided key insights into NLRP1 activation in humans (27, 28, 34). NLRP1 activation is characterized by functional degradation through the proteasome, which releases a biologically active UPA-CARD (conserved in UNC5, PIDD, and ankyrins)-(caspase recruitment domain) C-terminal fragment that oligomerizes with ASC (apoptosis-associated speck-like protein containing a CARD) to initiate formation of a canonical inflammasome (35–38). Recently, ZAKα activation via ribosomal stalling (22) and p38/MAPK (39, 40) have been linked to NLRP1 inflammasome activation in response to multiple signals.
Due to the poor expression of AIM2 in healthy keratinocytes, we sought to investigate the mechanism of action by which a classical AIM2 ligand—poly(dA:dT)—activates the inflammasome in normal healthy human keratinocytes.
Results
Poly(dA:dT) Elicits an AIM2-Independent Inflammasome in Unprimed Keratinocytes.
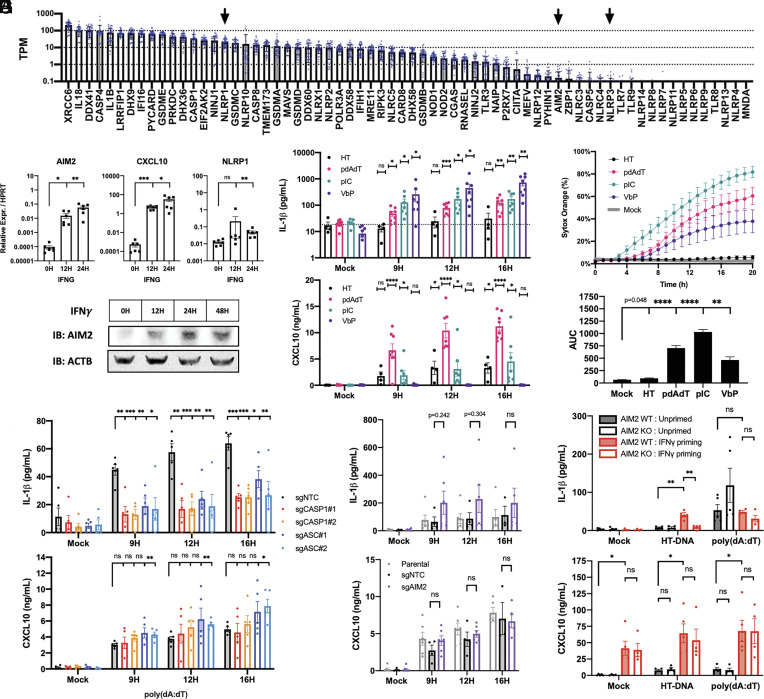
We first performed a meta-analysis of publicly available RNA-seq data of HPKs across 38 healthy donors and nine different studies (SI Appendix, Table S1) to survey inflammasome-related genes and nucleic acid sensors at greater sample size. NLRP1 was highly expressed in HPKs, whereas AIM2 and NLRP3 were barely detected (Fig. 1A) (Dataset S1). In cultured HPKs and an N/TERT2G immortalized keratinocyte cell line, AIM2 mRNA and protein were only induced after IFNγ stimulation (Fig. 1 B and C and SI Appendix, Fig. S1 A and B). Despite poor AIM2 expression in resting keratinocytes, transfected poly(dA:dT) elicited IL-1β release and cytotoxicity in both HPKs and N/TERT2G keratinocytes (Fig. 1 D and E and SI Appendix, Fig. S1 C and D). Notably, transfection of herring testes DNA (HT-DNA) and other linear dsDNA molecules including genomic DNA (gDNA) and a 44 bp dsDNA known as immunostimulatory DNA (ISD) were unable to induce IL-1β release and cytotoxicity in unprimed keratinocytes (Fig. 1 D and E and SI Appendix, Fig. S1 C–E), although all dsDNA molecules elicited CXCL10 production. In the absence of AIM2, most linear dsDNA are unable to elicit an inflammasome in human keratinocytes, but they are able to stimulate cGAS-STING to produce a type I interferon response. Poly(dA:dT) transfection elicited the strongest CXCL10 production, likely representative of the fact that poly(dA:dT) is recognized by both DNA and RNA sensing pathways (12).
Fig. 1.
Poly(dA:dT) but not HT-DNA transfection in unprimed keratinocytes elicits an AIM2 and STING-independent canonical inflammasome. (A) Publicly available RNA-seq datasets of healthy unstimulated HPKs (n = 38) were reanalyzed using Salmon. TPM for a list of relevant genes were plotted. (B and C) HPKs were treated with 100 ng/mL IFNγ for indicated time points and collected for qPCR (n = 6) or western blot. (D) HPKs were transfected with 1 µg/mL HT-DNA, poly(dA:dT), poly(I:C), or stimulated with 5 µM VbP and analyzed by ELISA (n = 4 to 8 donors) or (E) sytox orange time lapse imaging with AUC analysis (n = 6). (F and G) Monoclonal N/TERT2G knockout cell lines were transfected with 1 µg/mL poly(dA:dT) for indicated time points and supernatants were analyzed by ELISA (n = 5 and 6). (H) N/TERT2G wild type or AIM2 knockouts were primed with 100 ng/mL IFNγ for 24H and then transfected with 1 µg/mL HT-DNA or poly(dA:dT) for 12H followed by ELISA of supernatants (n = 4). Data represent mean ± SEM (A, B, and D–H) or a representative blot (C) from at least three independent experiments. If not otherwise indicated, statistical comparisons were made with respect to the mock-treated control. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student’s t test.
The capacity of poly(dA:dT) but not HT-DNA to elicit an inflammasome in unprimed keratinocytes implies that poly(dA:dT) has unique molecular features responsible for its immunostimulatory properties. Apart from its AT-rich sequence, poly(dA:dT) has a complex branching and nonlinear structure due to erratic hybridization of repetitive A-T sequences in higher-order aggregates (10). Treatment with S1 nuclease to digest branched DNA and ssDNA (SI Appendix, Fig. S1G) abrogated the immunogenicity of poly(dA:dT) but not HT-DNA, suggesting that the complex structure of poly(dA:dT) distinguishes it from linear dsDNA. As a control, DNase I digestion completely abrogated the capacity of poly(dA:dT) to elicit inflammasome activation (SI Appendix, Fig. S1F), verifying that this phenomenon is not attributable to contaminants.
Poly(dA:dT) transfection demonstrated a lytic phenotype that was morphologically similar to that seen with the dsRNA mimetic poly(I:C), albeit weaker with slightly delayed kinetics. Both synthetic molecules exhibited a bursting phenotype reminiscent of plasma membrane rupture (SI Appendix, Fig. S1H). In contrast, HPKs treated with the DPP9 inhibitor and small molecule NLRP1 activator VbP demonstrated a nonlytic morphology and mild membrane permeability to sytox orange dye (~40%) despite high levels of IL-1β release (Fig. 1E and SI Appendix, Fig. S1 D and H and Movie S1). These data suggest that NLRP1 activation in the context of VbP is a weak inducer of lytic membrane rupture in human keratinocytes, whereas cytotoxicity due to poly(dA:dT) and poly(I:C) transfection was more robust.
Utilizing a CRISPR/Cas9 approach in N/TERT2G keratinocytes, we found that poly(dA:dT)-induced IL-1β release required both caspase-1 (CASP1) and ASC (PYCARD) (Fig. 1F and SI Appendix, Fig. S1 I and J), which indicates that poly(dA:dT) activates a canonical inflammasome. These knockout keratinocytes had normal production of CXCL10, a cytokine that is downstream of cytosolic DNA sensors and type I interferons. Consistent with the fact that AIM2 is not expressed in resting keratinocytes, the poly(dA:dT)-induced IL-1β release was intact in cells lacking AIM2 (Fig. 1G and SI Appendix, Fig. S1K), indicating that another sensor is responsible for the poly(dA:dT) inflammasome in unprimed keratinocytes.
Priming N/TERT2G keratinocytes with IFNγ enabled IL-1β release in response to HT-DNA transfection in an AIM2-dependent manner (Fig. 1H), verifying that AIM2 is necessary to generate an inflammasome in response to linear dsDNA in immortalized keratinocytes. However, IFNγ priming had no significant effect on the poly(dA:dT) inflammasome response.
The poly(dA:dT) Inflammasome Is STING and NLRP3-Independent.
Given that STING and NLRP3 were implicated in the DNA-driven inflammasome response in primary human myeloid cells (14), we aimed to test whether the cGAS-STING-NLRP3 response occurs in keratinocytes. Consistent with published data, the NLRP3 inhibitor MCC950 completely abrogated the poly(dA:dT) inflammasome response in primary human monocytes (SI Appendix, Fig. S2A), as measured by processed IL-1β release in the supernatant. Furthermore, a high dose titration of a STING agonist diABZI was sufficient to activate an inflammasome in an NLRP3-dependent manner in monocytes (SI Appendix, Fig. S2B). However, equivalent doses of diABZI were unable to drive IL-1β release in keratinocytes, although diABZI could elicit a CXCL10 response (SI Appendix, Fig. S2C). This indicates that unlike in human monocytes, STING activation is insufficient to activate an inflammasome in keratinocytes.
We next generated CRISPR/Cas9 STING (STING1) knockout keratinocytes and observed that poly(dA:dT)-induced IL-1β release was intact in these cells (SI Appendix, Fig. S2 D, E and N), whereas CXCL10 release in response to conventional cGAS-STING ligands was abrogated. Pharmacologic inhibition of STING with H-151 yielded similar results (SI Appendix, Fig. S2F). Interestingly, poly(dA:dT)-induced CXCL10 release was also STING-independent, which could be a consequence of the fact that poly(dA:dT) is transcribed into short dsRNA intermediates that can be sensed by RIG-I (12). These data suggest that RNA sensing pathways may compensate for cGAS-STING signaling in poly(dA:dT)-treated keratinocytes, consistent with effects seen in other human cells. Finally, the NLRP3 inhibitor MCC950 has no effect on the poly(dA:dT) DNA response in keratinocytes (SI Appendix, Fig. S2G), and like other groups, we report that NLRP3 is not expressed in healthy keratinocytes (41).
Poly(dA:dT) Elicits a NLRP1-Dependent Inflammasome in Unprimed Keratinocytes.
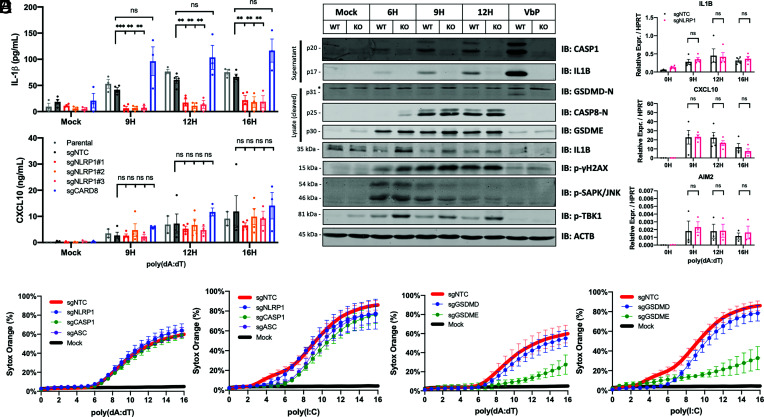
As described above, NLRP1 is highly expressed in keratinocytes at steady state, so we hypothesized that NLRP1 might be the poly(dA:dT) sensor in the context of healthy skin. To test this directly, we generated NLRP1 knockout keratinocytes and observed that poly(dA:dT)-induced IL-1β release was abrogated in these cells. Knocking out the NLRP1 homolog CARD8 had no impact on the IL-1β response (Fig. 2A and SI Appendix, Fig. S2N). Baseline expression of IL1B mRNA and pro-IL1β protein levels were not altered in the NLRP1 knockout keratinocytes (Fig. 2 B and C), indicating that NLRP1 acts at the level of IL1β maturation. In the setting of poly(dA:dT) transfection, NLRP1 knockout keratinocytes had impaired caspase-1 cleavage (p20) as well as processing of IL-1β (p17) and gasdermin D (p31). The effect of the NLRP1 knockout was restricted to caspase-1, IL-1β, and gasdermin D, as processing of caspase-8, gasdermin E and phosphorylation of TBK1, JNK, and γH2AX were all unaffected (Figs. 2B and 3A).
Fig. 2.
Poly(dA:dT) elicits NLRP1-dependent IL-1β release but NLRP1-independent cytotoxicity. (A and C) Monoclonal N/TERT2G knockout cell lines were transfected with 1 µg/mL poly(dA:dT) for indicated time points and samples were collected for supernatant ELISA or qPCR (n = 3 to 4). (B) N/TERT2G wild type or NLRP1 knockouts were transfected with 1 µg/mL poly(dA:dT) for indicated time points or stimulated with 5 µM VbP for 24H. Supernatants were precipitated for Western blot where indicated. (D and E) Monoclonal N/TERT2G knockout cell lines were transfected with 1 µg/mL poly(dA:dT) or poly(I:C) and subject to live cell imaging with sytox orange (n = 3). Data represent mean ± SEM (A and C–E) or a representative blot (B) from at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student’s t test.
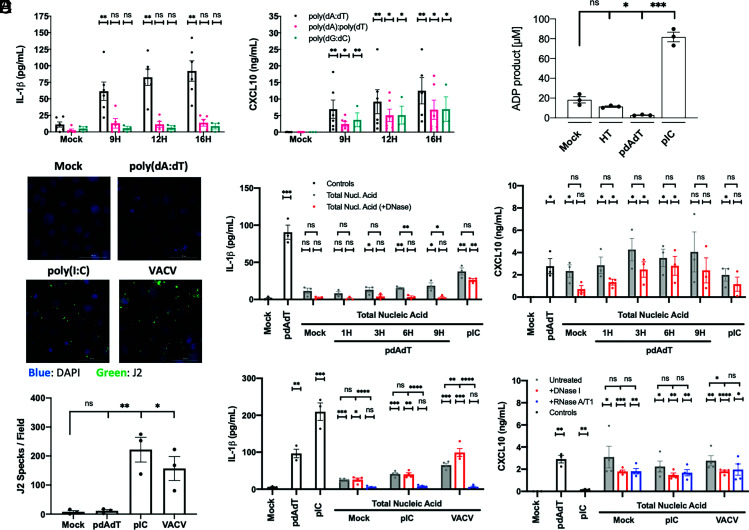
Fig. 3.
Poly(dA:dT) elicits NLRP1 through an indirect mechanism with limited evidence of a NLRP1-activating dsRNA intermediate. (A) N/TERT2G were transfected with 1 µg/mL of poly(dA:dT) heteropolymer, poly(dA):poly(dT) homopolymer, or poly(dG:dC) heteropolymer, and supernatants were analyzed by ELISA (n = 3 to 5). (B) HT-DNA, poly(dA:dT), or poly(I:C) were incubated with recombinant NLRP1ΔCARD, and ATP hydrolysis was measured (n = 3). (C and D) N/TERT2G were transfected with 1 µg/mL poly(dA:dT), poly(I:C), or stimulated with MOI 10 VACV for 4H. Cells were fixed, stained, and J2 positive specks were quantified (n = 3). (E and F) 293T cells were transfected with 2 µg/mL poly(dA:dT), poly(I:C), or stimulated with MOI 10 VACV for 4H, and total nucleic acid was isolated. Extracted nucleic acid was treated with DNase I or RNase A/T1 and transfected into N/TERT2G cells for 9H (n = 3 to 4). Data represent mean ± SEM (A, B, and D–F) or a representative image (C) from at least three independent experiments. If not otherwise indicated, statistical comparisons were made with respect to the mock-treated control. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student’s t test.
Gasdermin E and caspase-3 were a major drivers of cell death in poly(dA:dT)- and poly(I:C)-transfected N/TERT2G keratinocytes, as cytotoxicity was markedly delayed in gasdermin E and caspase-3 knockout keratinocytes (Fig. 2E and SI Appendix, Fig. S2J). Importantly, poly(dA:dT)- and poly(I:C)-induced cytotoxicity was largely independent of NLRP1, caspase-1, and ASC in these immortalized cells (Fig. 2D). The cytotoxicity in this context is likely attributable to dsRNA sensing pathways that induce cell death through apoptotic caspases (42). In comparison, VbP-induced cytotoxicity was dependent on NLRP1, ASC, caspase-1, and gasdermin D (SI Appendix, Fig. S2 I–K). Gasdermin E was not required for VbP-induced cytotoxicity or IL-1β release, which reflects the absence of active apoptotic caspases in VbP-stimulated keratinocytes (Fig. S2 K and L).
Poly(dA:dT)-induced IL-1β release was partially dependent on both gasdermin D and gasdermin E (SI Appendix, Fig. S2M) at early timepoints, suggesting that both pores are present in poly(dA:dT)-treated keratinocytes and that IL-1β can exit through either pore. The gasdermin D p31 fragment was not reproducibly detected in poly(dA:dT)-treated keratinocytes, but apoptotic caspases negatively regulate gasdermin D pore formation (43). Low levels of gasdermin D pores could still participate in cytokine release even if it is not the major driver of cell death.
NLRP1 activation occurs via functional degradation by the proteasome (44). Consistent with a role for NLRP1 in sensing poly(dA:dT), we also found that the proteasome inhibitors MG-132 and bortezomib blocked poly(dA:dT)-induced 1β release (SI Appendix, Fig. S2G). A pan-caspase inhibitor z-VAD-FMK also blocked the poly(dA:dT)-induced inflammasome, while a caspase-3 inhibitor z-DEVD-FMK and NLRP3 inhibitor MCC950 had no effect (SI Appendix, Fig. S2 G and H).
Poly(dA:dT) Activates the NLRP1 Inflammasome through an Indirect Mechanism.
We next wanted to understand how poly(dA:dT) activates NLRP1. Poly(I:C) and poly(dA:dT) are both high molecular weight nucleic acids with low sequence complexity, so we speculated that large nucleic acid aggregates could nucleate an inflammasome through electrostatic interactions. However, IL-1β release was not observed upon transfection of poly(dG:dC) and poly(dA):poly(dT) homopolymers (Fig. 3A), which suggests that nucleic acid structural complexity is not sufficient to explain the capacity of poly(dA:dT) to activate an inflammasome.
It was previously reported that both dsDNA and dsRNA bind the NLRP1 leucine-rich region (LRR), and but only dsRNA elicits NLRP1 ATPase activity. To test whether poly(dA:dT) directly activates NLRP1, we incubated poly(dA:dT) with purified recombinant NLRP1ΔCARD protein in vitro and monitored ATPase activity as previously described (45). Poly(dA:dT) and HT-DNA did not induce NLRP1 ATPase activity (Fig. 3B), whereas poly(I:C) catalyzed NLRP1 ATPase activity. These in vitro experiments suggest that poly(dA:dT) does not directly activate NLRP1.
RNA Isolated from poly(dA:dT)-Transfected Cells Is Not Sufficient to Activate an Inflammasome.
In the setting of poly(dA:dT) transfection, RNA polymerase III generates short dsRNA fragments that are capable of activating RNA sensors including RIG-I (12). We therefore hypothesized that a poly(dA:dT)-derived dsRNA intermediate could potentially activate the NLRP1 inflammasome. First, we assessed whether dsRNA levels were enhanced in poly(dA:dT) transfected cells by staining cells with the J2 antibody which recognizes dsRNA. Surprisingly, poly(dA:dT)-transfected cells did not stain positive with the J2 antibody (Fig. 3 C and D and SI Appendix, Fig. S3A), whereas VACV and poly(I:C) led to robust J2 staining. Importantly, the J2 antibody recognizes dsRNA over 40 bp in length, and it was previously reported that RNA polymerase III primarily generates short dsRNAs derived from poly(dA:dT) (12). In comparison, NLRP1 preferentially recognizes long dsRNA (26).
Despite the lack of long dsRNA, we tested whether any poly(dA:dT)-derived RNA was sufficient to activate NLRP1. An RNA polymerase III inhibitor ML-60218 (IC50 ~27 μM) was toxic in keratinocytes (SI Appendix, Fig. S3B), so we utilized an RNA transfer strategy to isolate RNA from poly(dA:dT)-treated cells and transfect it back into cells to monitor its immunostimulatory potential. Total RNA isolated from poly(dA:dT) transfected cells using trizol or silica-based spin columns was unable to elicit any IL-1β release in keratinocytes (SI Appendix, Fig. S3C). To address the possibility that DNA-RNA hybrids may be present in the system or that nucleic acids with a complex structure may not be efficiently isolated with acid-guanidinium-based techniques, we extracted total nucleic acid from poly(dA:dT) treated cells using phenol–chloroform precipitation and assessed if either the DNA or RNA component drives the IL-1β release phenotype by using nuclease depletion. DNase I treatment of poly(dA:dT) extracts abrogated IL-1β but not CXCL10 release, suggesting that poly(dA:dT)-derived RNA is sufficient to stimulate RIG-I but not NLRP1 (Fig. 3E).
Notably, our failure to demonstrate sufficiency through RNA transfer does not disprove the hypothesis that an RNA intermediate is required for poly(dA:dT)-induced NLRP1 inflammasome activation. Poly(dA:dT) could potentially generate unstable and transient dsRNA transcripts that are rapidly degraded or lost during nucleic acid isolation, which is incidentally also likely to disrupt the RNA tertiary structure. Alternatively, poly(dA:dT) could generate dsRNA below the detection limit of this assay, as we empirically determined that ~75% of input RNA is lost with each round of phenol–chloroform extraction.
We also tested if the dsRNA sensor PKR (EIF2AK2) could mediate the poly(dA:dT) inflammasome using an siRNA approach, but PKR knockdown using siRNA had no impact on poly(dA:dT)-induced IL-1β release (SI Appendix, Fig. S3 D and E).
As controls, J2 staining was observed with poly(I:C) transfection and VACV infection, and nucleic acid isolated from these cells elicited IL-1β release in an RNA-dependent manner (Fig. 3 C, D and F). It was previously reported that wild-type VACV encodes the virulence factor F1L which antagonizes the NLRP1 inflammasome (33). Indeed, wild-type VACV potently suppresses VbP-induced IL-1β release (SI Appendix, Fig. S3F), which suggests that the virus evolved to evade NLRP1 inflammasome activation. Vaccinia, which is derived from variola virus (smallpox), naturally infects human skin and has played an extensive role in human evolutionary history.
VACV ΔF1L mutants were able to elicit low-grade IL-1β release in an NLRP1-dependent manner (SI Appendix, Fig. S3G), but the ΔF1L virus still antagonized VbP-induced IL-1β release to some extent (SI Appendix, Fig. S3F). This suggests that VACV encodes additional evasion mechanisms in addition to F1L. Notably, VACVΔF1L was less potent than the wild-type virus at inhibiting NLRP1 at equivalent doses, and the highest doses of VACVΔF1L still demonstrated ~200 pg/mL IL-1β release during coinfection with VbP (SI Appendix, Fig. S3F).
Poly(dA:dT)-Induced Inflammasome Activation Is Correlated with Global Cellular Stress and Nucleic Acid Damage.
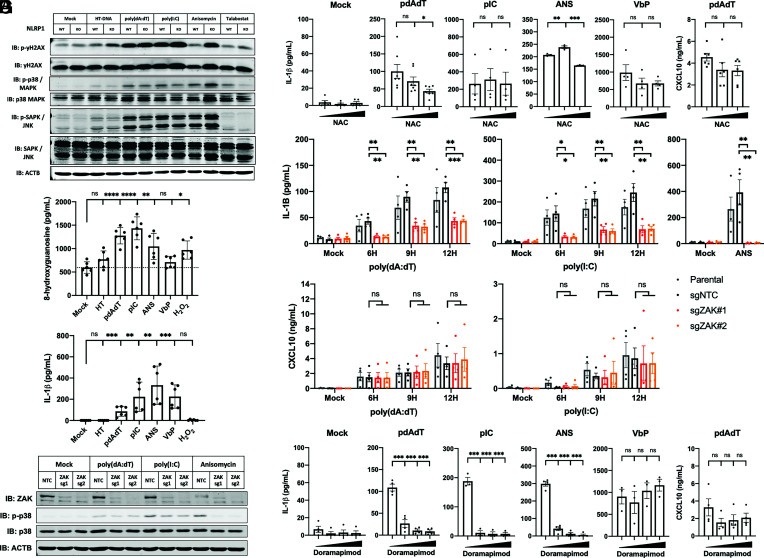
We next considered the possibility that a cellular stress signal induced by poly(dA:dT) leads to NLRP1 activation. Keratinocytes treated with poly(dA:dT), poly(I:C), and anisomycin (ANS) demonstrate activation of the p38-MAPK pathway not seen with VbP or HT-DNA treatment (Fig. 4A). VbP acts at a downstream step in the NLRP1 pathway, as DPP9 sequesters the biologically active NLRP1 C-terminus and directly regulates the final step of NLRP1 activation (38). In contrast, dsRNA and ribotoxic stress are proposed to act on the NLRP1 N terminus through a putative cellular kinase or E3 ubiquitin ligase. Poly(dA:dT), poly(I:C), and ANS all elicit cellular stress phenotypes as observed by keratinocyte morphology or measured by p38 and JNK phosphorylation (Fig. 4A). Phosphorylation of p38-MAPK is mediated by MAPK kinases (MAP2K) and MAP2K kinases (MAP3K) in response to a broad variety of potential stress signals ranging from osmotic shock, UV light, cytokines, growth factors, reactive oxygen species (ROS), endoplasmic reticulum (ER) stress, or calcium flux. ANS is a ribotoxin that induces ribosome stalling and triggers the MAP3K ZAKα, which was recently shown to mediate the NLRP1 inflammasome in the context of UVB irradiation (22).
Fig. 4.
Poly(dA:dT)-induced oxidative stress and p38/MAPK pathway activation is correlated with NLRP1 activation. (A) N/TERT2G cells were transfected with 9H 1 µg/mL HT-DNA, poly(dA:dT), poly(I:C), or stimulated with 6H 2 µM ANS, 24H 5 µM VbP, or 12H 10 µM H2O2. Lysates were analyzed by western blot. (B and C) N/TERT2G cells were stimulated as in (A) for 12H and supernatants were analyzed by 8-OHG or conventional ELISA (n = 6, 2 independent experiments). (D and G) N/TERT2G were pretreated with 1H 2.5/5 mM NAC or 0.1/1/10 µM doramapimod, and then transfected with 9H 1 µg/mL poly(dA:dT) or poly(I:C), 6H 2 µM ANS, or 12H 5 µM VbP. Supernatants were analyzed by ELISA (n = 3 to 4). (E and F) Polyclonal N/TERT2G ZAKα knockout cells were stimulated as in (A) for indicated time points, and supernatants were analyzed by ELISA (n = 4) or western blot. Data represent mean ± SEM (D, E, and G), a representative image (A) from at least three independent experiments, a representative image (F) from two independent experiments, or alternatively mean ± SD of pooled data from two experiments performed in triplicate (B and C). *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed Student’s t test.
We were especially intrigued by the presence of γH2AX phosphorylation, which is a sensitive marker of DNA damage in poly(dA:dT)-treated cells. In myeloid cells, a hyperinflammatory state is characterized by oxidative stress and production of ROS. ROS damages proteins, lipids, and DNAs but also cellular RNAs. Indeed, we measured elevated levels of 8-hydroxyguanosine oxidized DNA/RNA species in both poly(dA:dT) and poly(I:C) transfected keratinocytes, which correlated with phosphorylation of p38-MAPK and IL-1β release (Fig. 4 A–C).
Importantly, H2O2 treatment did not elicit IL-1β release, which suggests that ROS alone may not be sufficient to activate the NLRP1 inflammasome. It is possible that high-dose extracellular H2O2 elicits different types of oxidization in different targets, cellular compartments, and kinetics than poly(dA:dT) or poly(I:C) transfection. Notably, keratinocytes treated with high concentrations of the ROS scavenger N-Acetylcysteine (NAC) demonstrated a dose-dependent reduction in IL-1β release with poly(dA:dT) transfection but not VbP (Fig. 4D).
To determine whether kinases of the DNA or RNA damage response were involved in poly(dA:dT)-induced NLRP1 activation, we first tested an ATM inhibitor KU-55933 and DNA-PK inhibitor NU7441 and found that neither had an effect on poly(dA:dT)-induced IL-1β release (SI Appendix, Fig. S4 A and B). As reported by Robinson et al. (22), chemical DNA damaging agents such as etoposide do not elicit IL-1β release (22), suggesting that DNA damage is a consequence of poly(dA:dT) transfection but does not directly mediate activation of the NLRP1 inflammasome.
Given that p38-MAPK activation and nucleic acid damage are known features of the UVB NLRP1 inflammasome, we tested whether poly(dA:dT) and poly(I:C) activate ZAKα, the MAP3K that controls NLRP1 inflammasome responses due to ribosomal RNA damage. Intriguingly, the ZAK kinase inhibitors sorafenib (46) and PLX-4720 (47) could suppress poly(dA:dT), poly(I:C), and ANS-induced IL-1β release (SI Appendix, Fig. S4 C and D) but had no effect on VbP-induced IL-1β release. In ZAKα (MAP3K20) CRISPR/Cas9 knockout keratinocytes, poly(dA:dT), and poly(I:C)-induced IL-1β release was partially but significantly reduced (Fig. 4E and SI Appendix, Fig. S4F). ZAKα knockout keratinocytes also demonstrated residual p38 phosphorylation upon poly(dA:dT) and poly(I:C) transfection (Fig. 4F), whereas there was no residual IL-1β release or p38 phosphorylation in ANS-stimulated knockout keratinocytes. These data imply that multiple mechanisms contribute poly(dA:dT) and poly(I:C)-induced inflammasome activation, whereas the ANS-induced NLRP1 inflammasome is completely dependent on ZAKα.
Given that multiple kinase inhibitors potently inhibited the poly(dA:dT) and poly(I:C)-induced NLRP1 inflammasome while the effect in ZAKα knockout keratinocytes was incomplete, we speculated that additional stress-related MAP3K’s may regulate NLRP1 inflammasome activation in response to highly immunogenic nucleic acids such as poly(dA:dT) and poly(I:C). Small-molecule serine-threonine kinase inhibitors such as sorafenib and PLX-4720 are known to have many off-target effects and have activity on a wide spectrum of other cellular kinases.
Recent reports by Jenster et al. (40) have identified p38α and MKK3/6 as regulators of NLRP1 activation in response to multiple triggers, with the notable exception of VbP (40). We therefore tested whether inhibitors of the p38-MAPK pathway altered the poly(dA:dT)-induced response and found that second-generation p38-MAPK inhibitors doramapimod and SB-203580 suppressed poly(dA:dT)-, poly(I:C)-, and ANS-induced IL-1β release while having a marginal effect on cytotoxicity and the VbP-induced inflammasome (Fig. 4G and SI Appendix, Fig. S4 E–G). Taken together, these observations indicate that p38 is a central hub controlling activation of NLRP1 in human keratinocytes by multiple non-VbP ligands and offers evidence that immunogenic nucleic acids may stimulate the NLRP1 inflammasome through an indirect mechanism.
Discussion
Poly(dA:dT) is a potent immunostimulatory molecule with prospects as an adjuvant for vaccines and anti-tumor immunity. Multiple pattern recognition receptors mediate its immunostimulatory activity including cGAS/STING, AIM2, IFI16, DNA-PK, DAI (ZBP1), and KU70 which function in a cell type, context, and localization-dependent manner. Poly(dA:dT) can also be transcribed by RNA polymerase III into short cytosolic dsRNA which can be sensed by RIG-I. Activation of this collection of receptors results in pleiotropic responses including transcription of proinflammatory cytokines, interferons, apoptosis, pyroptosis, oxidative stress, ER stress, and arrest in mRNA translation. Here, we expand understanding of how poly(dA:dT) activates keratinocytes and define the NLRP1 inflammasome as a sensor of this immunostimulatory molecule in healthy skin.
Human NLRP1 is predominantly expressed in the epidermis and airway epithelium. Despite being the first inflammasome to be described, our understanding of NLRP1 biology and its activation is still not fully developed. This is in part due to poor NLRP1 conservation in mice and humans (48), its limited expression in myeloid cells, and until recently no clear activators of human NLRP1. The discovery of VbP—an inhibitor of DPP8/9—as an activator of NLRP1 laid the foundation for subsequent discoveries of additional NLRP1 activators. Recent studies have defined viral 3C proteases, long dsRNA, as well as UVB irradiation as bona fide activators. All known NLRP1 activators require a common intermediate step to achieve full inflammasome activation, namely the “functional degradation” of the auto-inhibitory N-terminal fragment. This liberates the C-terminal UPA-CARD fragment to initiate inflammasome assembly.
Like other activators, activation of NLRP1 by poly(dA:dT) also required functional degradation, as inhibitors of the proteosome abrogated downstream activation. Since dsRNA was recently defined as a ligand of NLRP1, we wanted to determine whether poly(dA:dT) activated NLRP1 through similar mechanisms. Activation of NLRP1 by dsRNA has been shown to occur through direct binding (26). The dsRNA mimetic poly(I:C) binds recombinant NLRP1 and stimulates NLRP1 ATPase activity in vitro. In contrast to poly(I:C), poly(dA:dT) failed to induce NLRP1 ATPase activity in vitro. Further, given prior observations that dsRNA could activate NLRP1 and earlier work demonstrating that poly(dA:dT) can be transcribed by RNA polymerase III into an dsRNA intermediate with RIG-I stimulatory activity, we tested the possibility that poly(dA:dT)-derived dsRNA intermediates were responsible for NLRP1 inflammasome activation. However, by staining cells with the J2 antibody, which recognizes dsRNA over 40 bp in length, we found no evidence of long dsRNA generated in poly(dA:dT) transfected keratinocytes. Furthermore, nucleic acids isolated from poly(dA:dT)-transfected cells failed to elicit NLRP1 inflammasome activation despite stimulating an interferon response.
Since the poly(dA:dT)-induced NLRP1 response was neither due to direct binding nor the generation of a transferrable long dsRNA intermediate, we speculated that poly(dA:dT) could elicit another response that is subsequently sensed by NLRP1. We found that poly(dA:dT) and poly(I:C) transfection resulted in nucleic acid damage and global cellular stress responses in keratinocytes. Activation of the p38-MAPK pathway has also been proposed to control NLRP1 inflammasome activation (39, 40). Poly(dA:dT) leads to p38-MAPK activation and pharmacologic inhibition of p38 suppressed NLRP1-mediated IL-1β release. Recent work from Zhong and colleagues have identified the stress-activated MAP3 kinase, ZAKα, as a key mediator of UVB-induced NLRP1 inflammasome activity in keratinocytes (22). UVB induces RNA damage and ribosome stalling which in turn activates ZAKα. ZAKα then phosphorylates NLRP1 leading to its activation. Interestingly, the poly(dA:dT)-induced NLRP1 response was only partially dependent on ZAKα, which indicates that multiple mechanisms contribute to NLRP1 inflammasome activation. We speculate that a currently unidentified kinase contributes to the poly(I:C)- and poly(dA:dT)-induced IL-1β release in addition to ZAKα.
Our studies also indicate that NLRP1 senses VACV, as reported previously. VACV is a DNA poxvirus that replicates in the cytoplasm and transcribes subgenomic RNAs with advanced secondary structure. Many DNA viruses generate dsRNA intermediates from DNA genomes, and AT-rich genomes are a feature of several pathogens. Evolutionarily, human keratinocytes are the natural host for VACV infection, and it is remarkable that VACV effectively evades multiple immune recognition pathways in keratinocytes. Wild-type VACV-infected keratinocytes generate neither interferon nor IL-1β release, but they express multiple virulence factors including F1L that antagonize host innate immune sensors. RNA isolated from VACV-infected cells was sufficient to elicit the inflammasome, which is otherwise potently suppressed. This speaks to VACV evolutionary adaption over human anthropological history, and the evolutionary pressure of smallpox may have contributed to the functional divergence of human NLRP1 from rodent alleles.
The capacity of a DNA virus to activate the NLRP1 inflammasome illustrates that human cells encode specialized mechanisms to distinguish pathogenic DNA from host DNA and sense foreign nucleic acids to trigger an inflammasome. Poly(dA:dT) reveals valuable insights about the sensing of nucleic acids from pathogens, and a combination of both secondary/tertiary structure and nucleic acid sequence are important for poly(dA:dT) immunogenicity across diverse pattern recognition receptors. Synthetic poly(dA:dT) provides a model for this pleiotropic response, and these properties can be leveraged in order to use this molecule as an immunoadjuvant in human patients.
Materials and Methods
Ethics.
Deidentified human blood products were obtained from Rhode Island Blood Center and foreskin tissue was obtained from UMass Memorial Hospital. Studies with human cells were conducted with approval from the Institutional Review Board of the University of Massachusetts Chan Medical School. All donors provided written informed consent. All studies were approved and monitored by the Institutional Biosafety Committee at University of Massachusetts Chan Medical School.
Human Samples.
HPKs were kindly provided by the laboratory of J.E. Harris. Primary keratinocytes were cultured from neonatal foreskin samples according to the previously described methods with some modifications (49–51). Briefly, after washing the samples with 70% ethanol and phosphate-buffered saline (PBS; Corning®), they were cut into small pieces. The samples were then incubated with 25 mg/mL Dispase® II (Roche) for 30 min at 37 °C followed by overnight at 4 °C. After separating the epidermis from each piece, they were incubated with 0.25% trypsin (Corning®) for 20 mins at 37 °C with vortexing every 5 min. The trypsin was neutralized with serum-containing media, and the cells were maintained in keratinocyte growth media (51) with 1% pen/strep and mitomycin C-treated J23T3 cells. After confirming the keratinocyte colonies, they were harvested and subcultured with EpiGROTM media (Sigma-Aldrich). Primary human peripheral blood mononuclear cells (PBMCs) were isolated from concentrated leukoreduction system (LRS) chambers purchased from Rhode Island Blood Center as previously described (52). In brief, CD14 positive monocytes were isolated from PBMCs by magnetic cell separation (MACS) using CD14 microbeads (Miltenyi #130-050-201). Primary monocytes were used in experiments within 16 h of isolation.
Cell Culture.
Lenti-X 293T (lab stock, Takarabio #632180), 3T3-J2 (Kerafast #EF3003), Hela (lab stock), and VeroE6 (lab stock) cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin. Immortalized N/TERT2G keratinocytes were provided by H. Rheinwald (Material Transfer Agreement) (53) and cultured in Keratinocyte SFM (Gibco #17005042). HPKs were isolated and maintained in an F-medium with 3T3 J2 fibroblasts as previously described (54). For experiments, HPKs were subcultured and plated in EpiGro media (EMD Millipore #SCMK001). Primary monocytes were cultured in RPMI-1640 media supplemented with 10% (v/v) fetal bovine serum, 1X MEM nonessential amino acids (Gibco #11140050), 1X GlutaMAX (Gibico #35050061), 1 mM sodium pyruvate (Gibco #11360070), 10 mM HEPES (Gibco #15630130), 100 U/mL penicillin, and 100 µg/mL streptomycin. Primary monocytes were primed with 2 µg/mL PAM3CSK4 for 3 h unless otherwise indicated. HPKs and N/TERT2G cells were unprimed and undifferentiated unless otherwise indicated. All cells were stimulated in experiments at 70 to 80% confluency. All transfections were performed using Lipofectamine 2000 according to the manufacturer’s recommendations.
Reagents.
The following chemicals and drugs were utilized in this study: Talabostat/VbP (MCE #HY-13233A), Lipofectamine 2000 (Invitrogen #11668019), double-stranded alternating copolymer poly(dA:dT) (pdAdT) (Sigma #P0883), poly(I:C) (pIC) (Invivogen #tlrl-picw), HT-DNA (Sigma #D6898), double-stranded homopolymer poly(dA):poly(dT) (Sigma #P9764), poly(dG:dC) (Invivogen #tlrl-pgcn), PAM3CSK4 (Invivogen #tlrl-pms), nigericin (Sigma #N7143), diABZI (MCE #HY-112921B), ANS (MCE #HY-18982), H2O2 (Sigma #H1009), MG-132 (MCE #HY-13259), Bortezomib (MCE #HY-10227), MCC950 (Invivogen #inh-mcc), z-VAD-FMK (Santa Cruz #sc-3067), z-DEVD-FMK (Santa Cruz #sc-311558), H-151 (MCE #HY-112693), NAC (Sigma #A9165), KU-44933 (Santa Cruz #sc-202963), NU-7441 (Tocris #3712), Sorafenib (Sigma #SML2633), PLX-4720 (MCE #HY-51424), Doramapimod (MCE #HY-10320), SB-202190 (MCE #HY-10295), and RNA Polymerase III inhibitor (Sigma #557403). gDNA was isolated from the genomic DNA of HEK293T cells. ISD was synthesized from custom oligos as previously described (55). Recombinant IFNγ was purchased from Peprotech (#300-02). DNase I (Bio-Rad #7326828), S1 Nuclease (Thermofisher #EN0321), and RNase A/T1 Cocktail (Thermofisher #AM2286) were purchased from the indicated vendors. VACV Copenhagen strain WT and ΔF1L were a kind gift of John Bell (56) and titered by plaque assay.
Plasmids.
LentiCas9-Blast (#52962), lentiGuide-Puro (#52963), psPAX2 (#12260), and pMD2.G (#12259) were purchased from Addgene.
CRISPR/Cas9 Knockout Keratinocytes.
N/TERT2G CRISPR/Cas9 AIM2, NLRP1, CARD8, STING, GSDMD, and GSDME monoclonal knockouts were generated using a liposomal system as previously described (57, 58). N/TERT2G CRISPR/Cas9 CASP1, CASP3, ASC, and ZAKα knockouts were created by first generating a N/TERT2G cell line stably expressing Cas9 using the LentiCas9-Blast system (59). Next, N/TERT2G-Cas9 were transduced with sgRNAs according to the LentiGuide-Puro system (59). Lentiviral supernatants were concentrated with a Lenti-X concentrator (Takarabio #631232) and applied to keratinocytes without polybrene. After 24 h, the media were changed and keratinocytes were selected with 15 µg/mL puromycin. Single-cell clones were isolated by limiting dilution for CASP1 and ASC knockouts. All knockout keratinocytes were validated by Sanger sequencing and/or immunoblot. CRISPR/Cas9 sgRNA sequences are available in SI Appendix, Table S3.
Cytokine Analysis.
Human IL-1 beta/IL-1F2 DuoSet Enzyme-Linked Immunosorbent Assay (ELISA) (R&D #DY201) and Human CXCL10/IP-10 DuoSet ELISA (R&D #DY266) were performed according to the manufacturer’s recommendations.
Immunoblotting.
Whole-cell lysates were prepared by lysing cells directly in Laemmli buffer, and samples were boiled at 95 °C for 15 min. For supernatant samples, methanol/chloroform precipitation was performed as previously described (60). Samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto 0.2 µm nitrocellulose membranes, blocked in 5% milk, and incubated with primary antibodies at a default dilution of 1:1,000 overnight. Corresponding Li-Cor IRDye conjugated secondary antibodies were utilized, and membranes were imaged using the Li-Cor Odyssey CLx instrument. Alternatively, chemiluminescent secondary antibodies were utilized and imaged on a Fuji LAS-3000 instrument. Human Caspase-1 (p20) mAb (Bally-1) was purchased from Adipogen. Human IL-1 beta/IL-1F2 Ab (#AF-201-NA) and Human NLRP1/NALP1 Ab (#AF6788) were purchased from R&D. AIM2 (D5X7K) Rabbit mAb (#12948), Cleaved Gasdermin D (Asp275) (E7H9G) Rabbit mAb (#36425), Cleaved Caspase-8 (Asp374) (18C8) Rabbit mAb (#9496), Caspase-3 Ab (#9662), Cleaved Caspase-3 (Asp175) Ab (#9661), Phospho-Histone H2A.X (Ser139) Ab (#2577), Histone H2A.X Ab (#2595), Phospho-p38 MAPK (Thr180/Tyr182) Ab (#9211), p38 MAPK Antibody (#9212), Phospho-SAPK/JNK (Thr183/Tyr185) (81E11) Rabbit mAb (#4668), SAPK/JNK Ab (#9252), Phospho-TBK1/NAK (Ser172) (D52C2) XP® Rabbit mAb (#5483), TBK1/NAK (E9H5S) Mouse mAb (#51872), and PKR Rabbit Ab (#3072) were purchased from Cell Signaling. Recombinant GSDMD mAb (#EPR19829/ab210070) and Recombinant DFNA5/GSDME mAb (#EPR19859/ab215191) were purchased from Abcam. ZAK Polyclonal Antibody (#A301-993A) was purchased from Thermofisher. Monoclonal Anti-β-Actin−Peroxidase Ab (#A3854) was purchased from Sigma.
RNA Isolation and qPCR.
Total RNA was extracted using the Aurum™ Total RNA Mini Kit (Bio-Rad #7326820), and cDNA was synthesized using the iScript™ cDNA Synthesis Kit (Bio-Rad #1708890). Quantitative PCR (qPCR) was performed using iTaq™ Universal SYBR® Green Supermix (Bio-Rad #1725125). Primer sequences utilized in this study are available in SI Appendix, Table S3.
Total Nucleic Acid Extraction.
Total RNA was isolated using QIAzol Lysis Reagent (Qiagen #79306) followed by one round of chloroform extraction. Total RNA was extracted using Direct-zol RNA Miniprep Kits (Zymo #R2050) according to the manufacturer’s recommendations. For total nucleic acid, cells were lysed using Zymo DNA/RNA lysis buffer (#D7001-1-50) and incubated for 1 h at room temperature with proteinase K (Lamda #DB0451). One round of phenol–chloroform precipitation was performed, followed by back-extraction of the organic layer, and then chloroform back-extraction to remove residual phenol. The aqueous layer was precipitated with 1.0 volume of ethanol with DNA/RNase-free GlycoBlue glycogen carrier (Invitrogen #AM9515) and spun at maximum speed at 4 °C at 15 min. The pellet washed three times with 70% ethanol, air dried, and then resuspended in appropriate buffers for enzymatic reactions.
Enzymatic Reactions.
All enzymatic reactions were performed in buffers according to the manufacturer’s recommendations. For poly(dA:dT) and HT-DNA digestion, nucleic acids were treated with DNase I and S1 nuclease at 37 °C for 1 h. Enzymes were inactivated with EDTA and heat, and the resulting solution was transfected into cells. Samples containing buffer/enzyme alone were used as mock transfection controls. For total nucleic acid depletion studies, nucleic acid was treated with DNase I in DNase I digestion buffer or RNase A/T1 in nuclease-free water (low salt conditions) at 37 °C for 30 min to deplete DNA or RNA, respectively.
ATPase Assay.
Assay was performed using the ADP-Glo kinase assay (Promega #V6930) as previously described (26). Expression and purification of recombinant NLRP1ΔCARD was previously described (26). In brief, recombinant NLRP1ΔCARD was incubated with indicated nucleic acids in ADP-Glo assay buffer. Luminescence was measured using a Spark 20M microplate reader (Tecan).
DNA/RNA Oxidative Damage.
DNA/RNA oxidative damage was measured using the DNA/RNA Oxidative Damage (High Sensitivity) ELISA Kit (Cayman # 589320) according to the manufacturer’s recommendations.
Time-Lapse Live Cell Imaging.
Keratinocytes were incubated in media containing 50 nM sytox orange (ThermoFischer #S11368) and 0.2 µg/mL Hoechst 34342 (Sigma #H3570). Cells were stimulated and imaged over time using the Biotek Cytation 5 Cell Imaging Multi-Mode Reader with fluorescent DAPI and RFP filter cubes at 37 °C with 5% CO2. Cellular analysis was performed with built-in Gen5 Cytation 5 software and DAPI/RFP double-positive cells were quantified with built-in software.
J2 Immunofluorescence.
Cells were plated in Lab-Tek Chamber Slides (Sigma #C7182) and stimulated. Cells were washed once with PBS and fixed with 4% paraformaldehyde for 15 min. Then, cells were washed twice with PBS, permeabilized with 0.2% Triton X-100 in PBS for 20 min, washed twice with PBS, and blocked with 1% bovine serum albumin (BSA) in PBS for 1 h. Cells were incubated with J2 primary antibody (gift of William McDougall) at 1:100 in 1% BSA for at least 1 h. Next, cells were washed twice with PBS and stained with goat anti-mouse Alexa Fluor 488 antibody (Thermofisher #A-11001) at 1:1,000 in 1% BSA for 1 h at room temperature. Then, cells were washed three times with PBS and stained with 2 µg/mL Hoechst 34342 (Sigma #H3570) for 10 min. Finally, the cells were washed two times with PBS and directly imaged using the Biotek Cytation 5 inverted fluorescent microscope using the Plan Fluorite WD 6.6 NA 0.45 20× objective lens. Nine images were taken per biological replicate and cellular analysis to quantify J2 specks was performed using built-in Gen5 Cytation 5 software.
siRNA Treatment.
Commercially available ON-TARGETplus EIF2AK2 (L-003527) and nontargeting control (D-001810) siRNA were purchased from Horizon Discovery Biosciences and transfected into keratinocytes using DharmaFECT 1 Transfection Reagent (T-2001) at 25 nM for 48 h according to the manufacturer’s recommendations.
RNA-seq Meta-Analysis.
Raw sequencing reads from publicly available RNA-seq data were downloaded from the NCBI GEO database (61) (SI Appendix, Table S1) and quantified using Salmon (62) (v1.7.0) with respect to the Ensembl GRCh38.p13 cDNA reference transcriptome (63) using default settings without decoy sequences. Estimated transcript per million (TPM) abundances were plotted.
Statistical Analysis.
Statistics were performed using GraphPad Prism software and represent a two-tailed Student’s t test unless otherwise indicated. In kinetic experiments, area-under-curve (AUC) was computed using GraphPad Prism. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (CSV)
N/TERT2G keratinocytes were transfected with 1 μg/mL poly(dA:dT) with 2 uL/mL lipofectamine 2000, or stimulated with 5 μM VbP. Cells were monitored by live-cell imaging using sytox orange dye with pictures taken at 1-hour intervals for 24 hours.
Acknowledgments
We would like to thank Dr. John Bell (Ottawa Hospital Research Institute) for providing vaccina virus Copenhagen strain ΔF1L, and Dr. William McDougall for providing the J2 antibody. Dr. Stefan Bauernfried and Dr. Veit Hornung generously performed the ATPase assay with recombinant NLRP1 protein. We would like to thank all members of the Fitzgerald laboratory, Dr. Megan Orzalli’s laboratory (UMass), and Dr. Mehdi Rashighi’s laboratory (UMass) for their continuous advice and guidance. This study was supported by NIH T32 grants (GM107000 and AI132152). J.E.G. and M.K.S. are supported by NIH P30 AR075043.
Author contributions
J.Y.Z., M.K.S., K.O., J.E.H., J.E.G., and K.A.F. designed research; J.Y.Z., M.K.S., and K.O. performed research; K.O., J.E.H., and J.E.G. contributed new reagents/analytic tools; J.Y.Z., M.K.S., J.E.H., J.E.G., and K.A.F. analyzed data; and J.Y.Z. and K.A.F. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
Reviewers: C.E.B., University of Cambridge; and J.T., The University of North Carolina at Chapel Hill.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. Previously published data were used for this work; publicly available RNA-seq datasets (SI Appendix, Table S1)were downloaded from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/) and used for meta-analysis.
Supporting Information
References
- 1.Sun L., Wu J., Du F., Chen X., Chen Z. J., Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu J., et al. , Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Q., Sun L., Chen Z. J., Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Sam Xiao T., Xiao Tsan Sam, address., The nucleic acid-sensing inflammasomes. Immunol. Rev. 265, 103–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rathinam V. A. K., Fitzgerald K. A., Inflammasome complexes: Emerging mechanisms and effector functions. Cell 165, 792–800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandes-Alnemri T., Yu J.-W., Datta P., Wu J., Alnemri E. S., AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rathinam V. A. K., et al. , The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hornung V., et al. , AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henner D., Further J. J., Primed and unprimed synthesis of poly (dA-dT) by calf thymus DNA polymerase alpha. J. Biol. Chem. 252, 1932–1937 (1977). [PubMed] [Google Scholar]
- 10.Sági J., et al. , Structures of poly(dA-dT, ip5dU) containing various small amounts of the antiherpetic 5-isopropyl-2’-deoxyuridine. Biochem. Biophys. Res. Commun. 185, 96–102 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Tanaka S., et al. , Synthesis of long Poly(dA). Poly(dT) DNA without structural defects using enzymatic reaction; tailored ligated Poly(dA).Poly(dT). Chem. Commun. 2, 2330–2331 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Ablasser A., et al. , RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 10, 1065–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbuto S., et al. , Induction of innate and adaptive immunity by delivery of poly dA:dT to dendritic cells. Nat. Chem. Biol. 9, 250–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaidt M. M., et al. , The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell 171, 1110–1124.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopfnagel V., Wittmann M., Werfel T., Human keratinocytes express AIM2 and respond to dsDNA with IL-1β secretion. Exp. Dermatol. 20, 1027–1029 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Reinholz M., et al. , HPV16 activates the AIM2 inflammasome in keratinocytes. Arch. Dermatol. Res. 305, 723–732 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Dombrowski Y., et al. , Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl. Med. 3, 82ra38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strittmatter G. E., et al. , IFN-γ primes keratinocytes for HSV-1-induced inflammasome activation. J. Invest. Dermatol. 136, 610–620 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Zhong F. L., et al. , Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167, 187–202.e17 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Burian M., Yazdi A. S., NLRP1 is the key inflammasome in primary human keratinocytes. J. Invest. Dermatol. 138, 2507–2510 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Fenini G., et al. , Genome editing of human primary keratinocytes by CRISPR/Cas9 reveals an essential role of the NLRP1 inflammasome in UVB sensing. J. Invest. Dermatol. 138, 2644–2652 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Robinson K. S., ZAKα-driven ribotoxic stress response activates the human NLRP1 inflammasome. Science 377, 328–335 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson K. S., et al. , Enteroviral 3C protease activates the human NLRP1 inflammasome in airway epithelia. Science 370 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Tsu B. V., et al. , Diverse viral proteases activate the NLRP1 inflammasome. Elife 10, e60609 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Planès R., et al. , Human NLRP1 is a sensor of pathogenic coronavirus 3CL proteases in lung epithelial cells. Mol. Cell 82, 2385–2400.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bauernfried S., Scherr M. J., Pichlmair A., Duderstadt K. E., Hornung V., Human NLRP1 is a sensor for double-stranded RNA. Science 371, eabd0811 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Gai K., et al. , DPP8/9 inhibitors are universal activators of functional NLRP1 alleles. Cell Death Dis. 10, 587 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okondo M. C., et al. , Inhibition of Dpp8/9 activates the Nlrp1b inflammasome. Cell Chem. Biol. 25, 262–267.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang M., et al. , Structural and biochemical mechanisms of NLRP1 inhibition by DPP9. Nature 592, 773–777 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong F. L., et al. , Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J. Biol. Chem. 293, 18864–18878 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ball D. P., et al. , Oxidized thioredoxin-1 restrains the NLRP1 inflammasome. Sci. Immunol. 7, eabm7200 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia J., Zhang X., Xu G., Zeng X., Li L., Thioredoxin-1 inhibits amyloid-β25-35-induced activation of NLRP1/caspase-1/GSDMD pyroptotic pathway in PC12 cells. Mol. Biol. Rep. 1, 1–8 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Gerlic M., et al. , Vaccinia virus F1L protein promotes virulence by inhibiting inflammasome activation. Proc. Natl. Acad. Sci. U.S.A. 110, 7808–7813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okondo M. C., et al. , DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat. Chem. Biol. 13, 46–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandstrom A., et al. , Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chui A. J., et al. , N-terminal degradation activates the NLRP1B inflammasome. Science 364, 82–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robert Hollingsworth L., et al. , Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes. Nat. Commun. 12, 189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollingsworth L. R., et al. , DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Nature 592, 778–783 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fenini G., et al. , The p38 mitogen-activated protein kinase critically regulates human keratinocyte inflammasome activation. J. Invest. Dermatol. 138, 1380–1390 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Jenster L.-M., et al. , P38 kinases mediate NLRP1 inflammasome activation after ribotoxic stress response and virus infection. J. Exp. Med. 220, e20220837 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong F. L., et al. , Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167, 187–202.e17 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Srivastava S. P., Kumar K. U., Kaufman R. J., Phosphorylation of eukaryotic translation initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 273, 2416–2423 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Taabazuing C. Y., Okondo M. C., Bachovchin D. A., Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem. Biol. 24, 507–514.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandstrom A., et al. , Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bauernfried S., Scherr M. J., Pichlmair A., Duderstadt K. E., Hornung V., Human NLRP1 is a sensor for double-stranded RNA. Science 371, eabd0811 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Vin H., et al. , Sorafenib suppresses JNK-dependent apoptosis through inhibition of ZAK. Mol. Cancer Ther. 13, 221–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maruyama T., et al. , ZAK inhibitor PLX4720 promotes extrusion of transformed cells via cell competition. iScience 23, 101327 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sand J., et al. , Expression of inflammasome proteins and inflammasome activation occurs in human, but not in murine keratinocytes. Cell Death Dis. 9, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitano Y., Okada N., Separation of the epidermal sheet by dispase. Br. J. Dermatol. 108, 555–560 (1983). [DOI] [PubMed] [Google Scholar]
- 50.Rheinwatd J. G., Green H., Seria cultivation of strains of human epidemal keratinocytes: The formation keratinizin colonies from single cells. Cell 6, 331–343 (1975), 10.1016/S0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 51.Ridky T. W., Chow J. M., Wong D. J., Khavari P. A., Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat. Med. 1612, 1450–1455 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agarwal S., et al. , The long non-coding RNA LUCAT1 is a negative feedback regulator of interferon responses in humans. Nat. Commun. 111, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickson M. A., et al. , Human keratinocytes that express hTERT and also bypass a p16 INK4a -enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20, 1436–1447 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orzalli M. H., et al. , Virus-mediated inactivation of anti-apoptotic Bcl-2 family members promotes gasdermin-E-dependent pyroptosis in barrier epithelial cells. Immunity 54, 1447–1462.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stetson D. B., Medzhitov R., Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24, 93–103 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Pelin A., et al. , Deletion of apoptosis inhibitor F1L in vaccinia virus increases safety and oncolysis for cancer therapy. Mol. Ther. Oncolytics 14, 246–252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swindell W. R., et al. , RNA-seq analysis of IL-1B and IL-36 responses in epidermal keratinocytes identifies a shared MyD88-dependent gene signature. Front. Immunol. 9, 80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarkar M. K., et al. , Photosensitivity and type I IFN responses in cutaneous lupus are driven by epidermal-derived interferon kappa. Ann. Rheum. Dis. 77, 1653–1664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jakobs C., Bartok E., Kubarenko A., Bauernfeind F., Hornung V., Immunoblotting for active caspase-1. Methods Mol. Biol. 1040, 103–115 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cunningham F., et al. , Ensembl 2022. Nucleic Acids Res. 50, D988–D995 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (CSV)
N/TERT2G keratinocytes were transfected with 1 μg/mL poly(dA:dT) with 2 uL/mL lipofectamine 2000, or stimulated with 5 μM VbP. Cells were monitored by live-cell imaging using sytox orange dye with pictures taken at 1-hour intervals for 24 hours.
Data Availability Statement
All study data are included in the article and/or SI Appendix. Previously published data were used for this work; publicly available RNA-seq datasets (SI Appendix, Table S1)were downloaded from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/) and used for meta-analysis.