Significance
PDV1 was thought to work corporately with PDV2 to recruit ARC5 to the chloroplast division site in the cytosol. However, we found that the major role of PDV1 is to interact with and regulate the stability of PARC6, a component of the Min system that function in the stroma to regulate the positioning of the division site. PDV1 inserts its C-terminal tail into the pocket-like structure of PARC6 to induce its dimerization. This interaction is regulated by redox and magnesium ions, which are affected by light. This work not only extends our understanding of the Min system from stroma to the intermembrane space but also uncovers the multilayer regulation of the PARC6–PDV1 pair and thus chloroplast division.
Keywords: chloroplast division, PARC6, PDV1, crystal structure, intermembrane space
Abstract
Chloroplast division involves the coordination of protein complexes from the stroma to the cytosol. The Min system of chloroplasts includes multiple stromal proteins that regulate the positioning of the division site. The outer envelope protein PLASTID DIVISION1 (PDV1) was previously reported to recruit the cytosolic chloroplast division protein ACCUMULATION AND REPLICATION OF CHLOROPLAST5 (ARC5). However, we show here that PDV1 is also important for the stability of the inner envelope chloroplast division protein PARALOG OF ARC6 (PARC6), a component of the Min system. We solved the structure of both the C-terminal domain of PARC6 and its complex with the C terminus of PDV1. The formation of an intramolecular disulfide bond within PARC6 under oxidized conditions prevents its interaction with PDV1. Interestingly, this disulfide bond can be reduced by light in planta, thus promoting PDV1–PARC6 interaction and chloroplast division. Interaction with PDV1 can induce the dimerization of PARC6, which is important for chloroplast division. Magnesium ions, whose concentration in chloroplasts increases upon light exposure, also promote the PARC6 dimerization. This study highlights the multilayer regulation of the PDV1–PARC6 interaction as well as chloroplast division.
Chloroplasts are plant-specific photosynthetic organelles that are encapsulated by double membranes. Chloroplasts have evolved from cyanobacteria and divide through binary fission (1). During the division of chloroplasts and cyanobacteria, protein complexes form at the division site (2–4) (Fig. 1). The GTPase FtsZ, which is related to tubulin, acts as a scaffold protein of the division complex (5, 6). FtsZ in plants is comprised of two subfamilies: FtsZ1 and FtsZ2. They coassemble into multimers to form a ring at the division site (7). The formation of this FtsZ ring is regulated by the Min system to ensure that only one FtsZ ring is formed in the center of the cyanobacterial cell or the chloroplast (3).
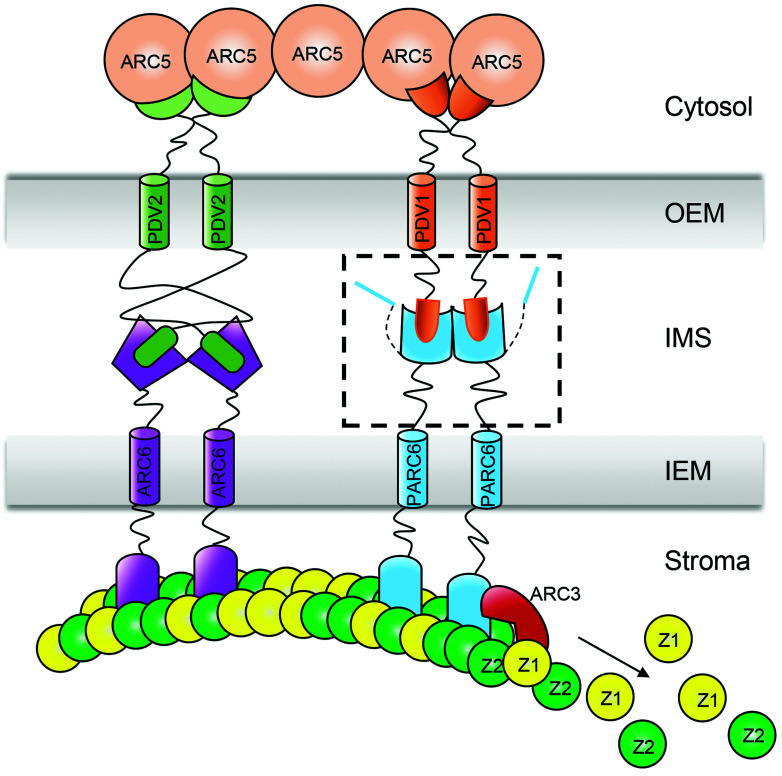
Fig. 1.
A schematic diagram of the topology and interaction of chloroplast division proteins at the division site. A box with dashed lines includes the C-terminal domain of PDV1 and the C-terminal domain of PARC6 in the IMS. The mechanism of the interaction between PDV1 and PARC6 is unclear before this study. Z1, FtsZ1; Z2, FtsZ2; OEM, outer envelope membrane; IMS, inter-membrane space; IEM, inner envelope membrane.
In chloroplasts, FtsZ is anchored to the inner membrane via an interaction between the C-terminal domain of FtsZ2 and the N-terminal domain of the transmembrane protein ACCUMULATION AND REPLICATION OF CHLOROPLAST6 (ARC6) (8) (Fig. 1). In arc6 mutants, FtsZ filaments are fragmented, suggesting that ARC6 is important for the assembly and stability of FtsZ rings (9). ARC6 is related to the cyanobacterial cell division protein Ftn2 (9, 10). The C-terminal domain of ARC6 interacts with the PLASTID DIVISION2 (PDV2) C terminus in the intermembrane space (IMS) (11). The chloroplast outer envelope protein PDV2 and its paralog PDV1 then recruit the cytosolic dynamin-related GTPase ARC5 to the chloroplast division site (12) (Fig. 1), which determines the rate of chloroplast division (13). Although it was widely believed that ARC6 may recruit PDV2 to the chloroplast division site, we have shown that PDV2 facilitates the dimerization of ARC6 and helps condense the chloroplast division ring (14). The C-terminus of FtsZ1 is distinct from that of FtsZ2. In angiosperms, it has a conserved Z1C motif with a weak membrane binding activity when exists as a monomer (15). However, FtsZ multimers with multiple Z1C motifs bind to membrane well. Z1C motif renders FtsZ filaments more liable to the regulation of the Min system at the nondivision site (15).
Over the course of evolution, the plant Min system has become more complex. The Min system of angiosperms includes MinD (16) and MinE (17), which are derived from cyanobacteria, as well as MULTIPLE CHLOROPLAST DIVISION SITE1 (MCD1) (18), ARC3 (19), and PARALOG OF ARC6 (PARC6) (20), which evolved later in land plants. PARC6, also named CHLOROPLAST DIVISION SITE POSITIONING1 (CDP1) (21), is a paralog of ARC6 with a similar topology (22). The role of PARC6 in chloroplast division is very different from that of ARC6. In the parc6 mutant, chloroplasts are heterogenous in size, and chloroplasts with multiple division sites and parallel FtsZ rings are frequently seen (20, 21), suggesting that PARC6 is a component of the Min system to prevent FtsZ ring formation outside of the division site. ARC3 is the central player of plant Min system (23). The N-terminal domain of PARC6 localizes to the stromal side of chloroplasts, where it interacts with both FtsZ2 and ARC3 (22, 24) (Fig. 1). By interacting with ARC3, PARC6 activates its function and promotes FtsZ ring remodeling at the division site (20–22, 24).
The C-terminal domain of PARC6 exists in the IMS and interacts with the C-terminal domain of PDV1 (22). The molecular mechanism by which PDV1 and PARC6 interact in the IMS has not been established. Light was shown to promote the division of chloroplasts in leaf cells (25). However, the underlying molecular mechanism has not been studied. In this study, we analyzed the working mechanism of PDV1 and PARC6 in the IMS in detail. We determined that PDV1 is not only important for the stability of PARC6 but also regulates its function by inducing its dimerization. The state of the PDV1–PARC6 complex is also affected by redox conditions and by magnesium ion (Mg2+) concentrations, both of which are regulated by light. These findings extend our understanding of the regulatory mechanism of the Min system for chloroplast division across the IMS.
Results
The pdv1 Mutant Has a Phenotype Similar to Min System Mutants.
ARC6 is the plant ortholog of the cyanobacterial cell division gene Ftn2 (9, 10), while PDV2 is a chloroplast division gene that evolved in land plants after the initial endosymbiosis event (12). Phylogenetic analysis of ARC6, PDV2, and related proteins in several species of land plants indicated that PDV1 and PDV2 define two separate clades, as did ARC6 and PARC6, with PDV1 and PARC6 being present across angiosperms (13, 20) (SI Appendix, Fig. S1 A and B and Table S1). As loss-of-function alleles in any one of these four genes have been reported to cause severe chloroplast division phenotypes in Arabidopsis (9, 12, 20, 21), ARC6, PDV2, and their paralogous proteins PARC6 and PDV1 are likely to perform nonredundant functions.
The pdv1 mutant was identified from a search for chloroplast division mutants in Arabidopsis with a phenotype similar to that of arc5, which is enlarged chloroplasts with a dumbbell shape (12). In our chloroplast division mutant screening, we isolated a null allele of PDV1, pdv1-3 (Fig. 2A and SI Appendix, Fig. S2). The mutation is the same as that of pdv1-1 (12). A careful observation of the chloroplast division phenotype of this mutant showed that besides their dumbbell shape, the chloroplasts of pdv1-3 are very heterogenous in size and often exhibit multiple division sites (Fig. 2 A and B). Immunofluorescence staining of FtsZ indicated that FtsZ filaments are often in parallel, which is similar to that of parc6 (Fig. 2C). These phenotypes of pdv1-3 were different from that of arc5 or pdv2 mutants but similar to that seen in mutants defective in the Min system, such as parc6 (Fig. 2 and SI Appendix, Fig. S3).
Fig. 2.
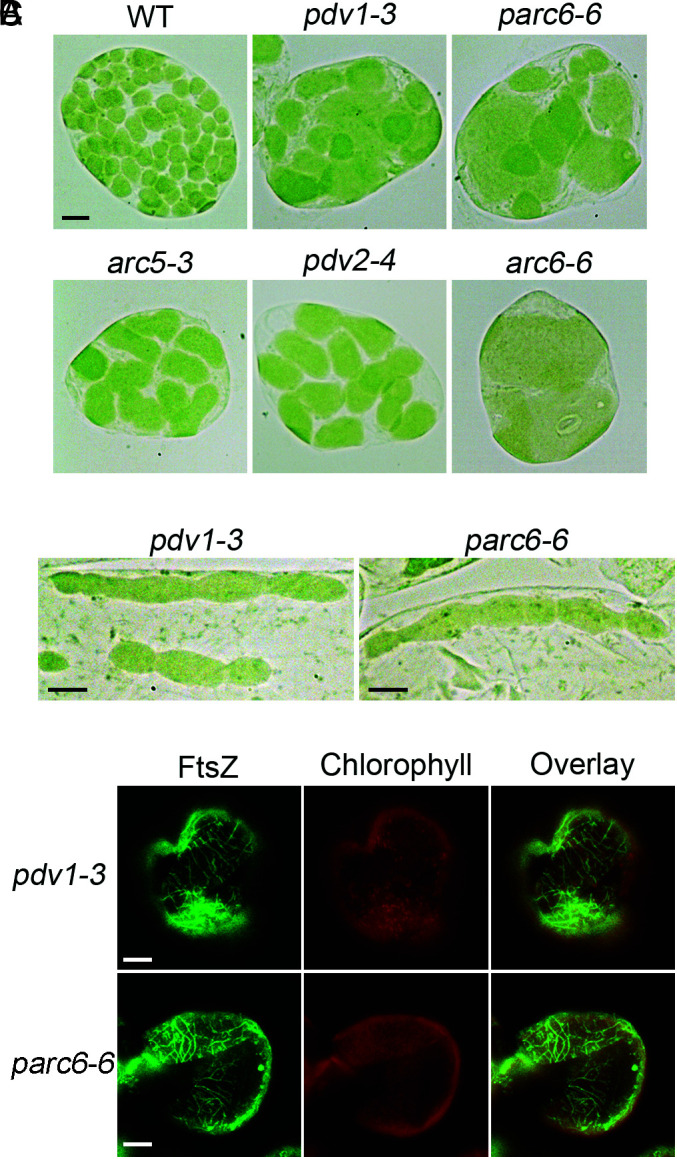
The pdv1 mutant exhibits phenotypes similar to that of parc6. (A) A comparison of the chloroplast division phenotypes. Cells are from mesophyll. WT, wild type. Bar, 10 μm. All images are at the same magnification. (B) pdv1 and parc6 have chloroplasts with multiple division sites. Cells are from bundle sheath. Bars, 10 μm. (C) Immunofluorescence staining of FtsZ filaments. Cells are from the mesophyll of mature leaves. Bars, 10 μm.
PDV1 Is Important for the Stability of PARC6.
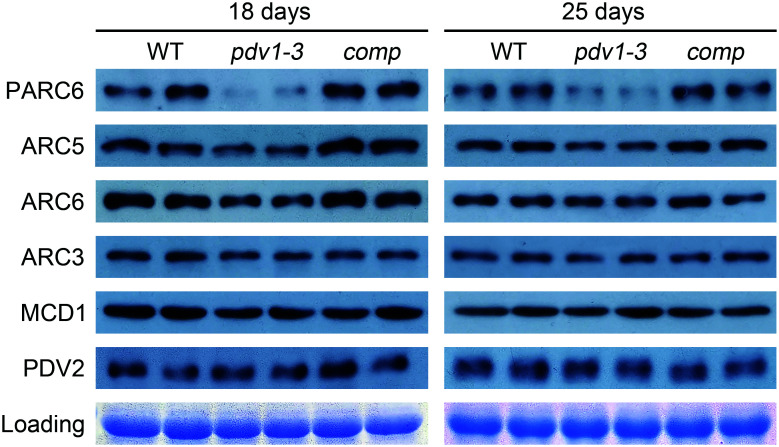
To explore the underlying cause of the pdv1-3 mutant phenotype, we performed immunoblot analyses of related chloroplast division proteins in wild-type (WT) Col-0, the pdv1-3 mutant, and a complemented line (Fig. 3). We first used 18-d-old plants, whose leaves are fast-growing (SI Appendix, Fig. S4). Notably, PARC6 abundance was much lower in the pdv1-3 mutant compared to the WT. ARC5 and ARC6 protein accumulation also decreased relative to the WT. By contrast, ARC3, MCD1, and PDV2 abundance was not affected in the pdv1-3 mutant. We also characterized protein abundance in 25-d-old plants, whose leaves are more mature (SI Appendix, Fig. S4), yielding results similar to those of 18-d-old plants (Fig. 3). End-point RT-PCR analyses of PARC6 and ARC5 transcript levels in the pdv1-3 mutant indicated no obvious differences between the mutant and the WT (SI Appendix, Fig. S5). These results suggest that PDV1 is important for the stability of PARC6.
Fig. 3.
PDV1 is important for the stability of PARC6. Immunoblot analysis of the abundance of various chloroplast division proteins in WT, pdv1, and complemented pdv1 plants (comp). Each genotype is represented by two replicates in the blots. Coomassie brilliant blue (CBB) staining of the sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels serves as a loading control.
Dimerization Is Important for PARC6 Function.
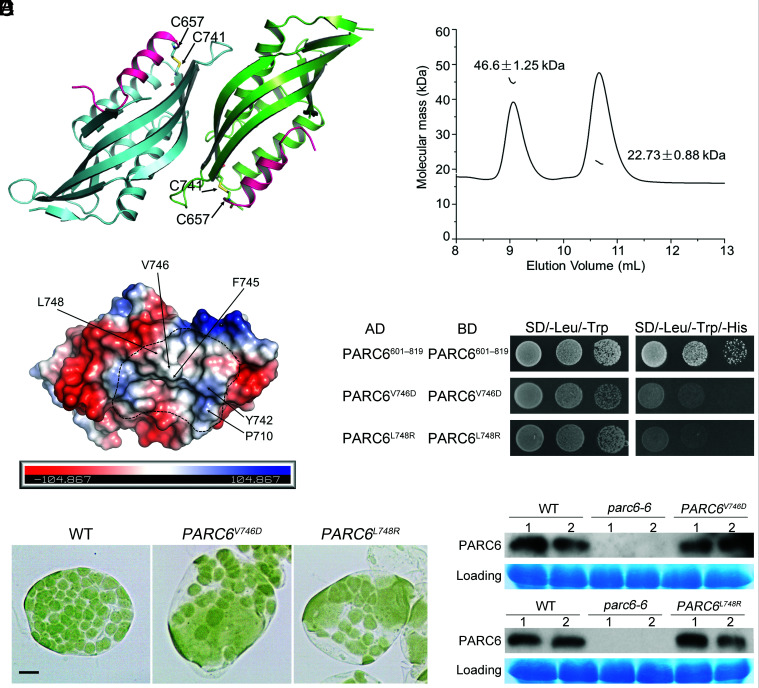
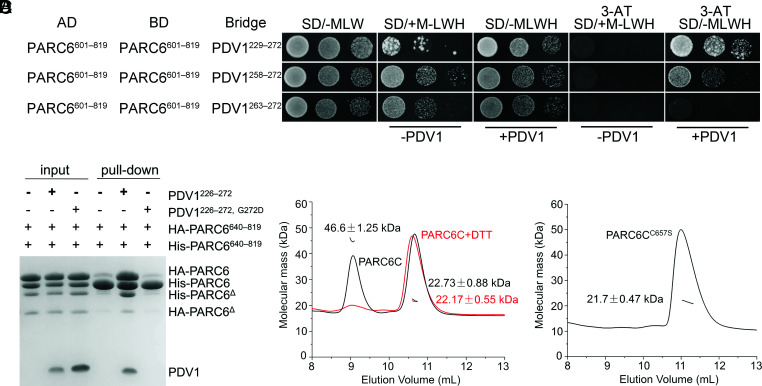
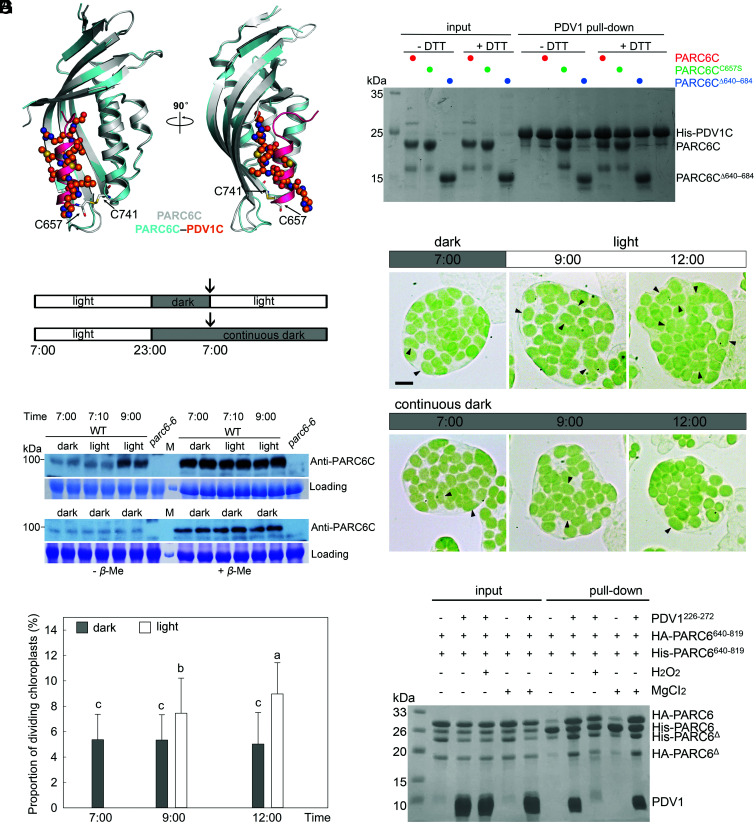
According to the sequence analysis, the C-terminal region of PARC6 in the IMS begins with the 596th amino acid residue and ends with the 819th amino acid residue. To explore how PARC6 and PDV1 function in the IMS, we tried different fragments of the C-terminal region of PARC6 in the IMS and finally solved the crystal structure of PARC6640–819 (PARC6C hereafter) at a resolution of 2.53 Å (Fig. 4A and SI Appendix, Table S2). The crystallographic data showed that one asymmetric unit contains two PARC6 molecules. Like the C-terminal domain of ARC6 (14), PARC6C was also characterized by a pocket on its surface. However, it is worth noting here that a short helix (I646 to H656) remained at the outlet of the pocket and served as a lid; in addition, the pocket was closed by the lid due to the formation of an intramolecular disulfide bond between cysteine residues C657 and C741 (Fig. 4A). To investigate the oligomeric state of PARC6C under oxidized conditions, we conducted a static light scattering (SLS) assay of the protein, which showed that PARC6C exists both as a dimer and a monomer in solution (Fig. 4B), suggesting that the binding interface of the two protomers in the crystal structure may reflect the dimer interface of PARC6C under oxidized conditions. The dimer interface was mainly composed of hydrophobic residues (Fig. 4C and SI Appendix, Fig. S6).
Fig. 4.
Dimerization is important for the function of PARC6. (A) Crystal structure of the PARC6C (residues 640 to 819) homodimer. The C-terminal region of PARC6 forms a pocket that is closed by a lid-like helix (shown in pink). The cysteine residues forming a disulfide bond (shown in yellow) in each protomer are shown as stick models. (B) SLS analysis of PARC6C in solution. PARC6C monomers and dimers coexist. (C) The self-interaction region of PARC6C is mainly hydrophobic. One PARC6C protomer is shown as an electrostatic model. The residues with hydrophobic side chains within the dimer interface are indicated. (D) Introducing hydrophilic residues in the self-interaction region of PARC6C abolishes its self-interaction in Y2H analysis. PARC6601–819 begins right after the transmembrane domain and contains PARC6C. PARC6601–639 serves as a linker. The negative controls are shown in SI Appendix, Fig. S9. (E) Disrupting the self-interaction of PARC6 affects chloroplast division. Two point mutants defective in self-interaction fail to complement the parc6-6 mutant phenotype. Bar, 10 μm. (F) Immunoblot analysis of the plants shown in E. CBB staining of the SDS-PAGE gels serves as a loading control.
To investigate the role that dimerization might play in the function of PARC6, we introduced several point mutations mapping to the dimer interface and tested dimerization of PARC6 in yeast two-hybrid (Y2H) assays. Mutating the residue at the interface to alanine had only a modest effect on the self-interaction of PARC6 (SI Appendix, Fig. S7). However, the introduction of charged residues in place of hydrophobic residues at the dimer interface abolished PARC6 self-interaction (Fig. 4D and SI Appendix, Figs. S8 and S9). These data thus suggest that the dimerization of PARC6 is driven by hydrophobic interaction at the interface.
To assess the phenotypic consequences of the loss of PARC6 dimerization in planta, we introduced constructs encoding two PARC6 variants impaired in self-interaction of PARC6C, V746D, and L748R, into the parc6-6 mutant. Importantly, neither variant rescued the mutant phenotype (Fig. 4E and SI Appendix, Fig. S10A). Immunoblot analysis indicated that the mutant proteins accumulate to levels close to that of WT (Fig. 4F). Taken together, the self-dimerization of PARC6 is important for its function.
Dissection of the Molecular Mechanism of the Interaction between PDV1 and PARC6.
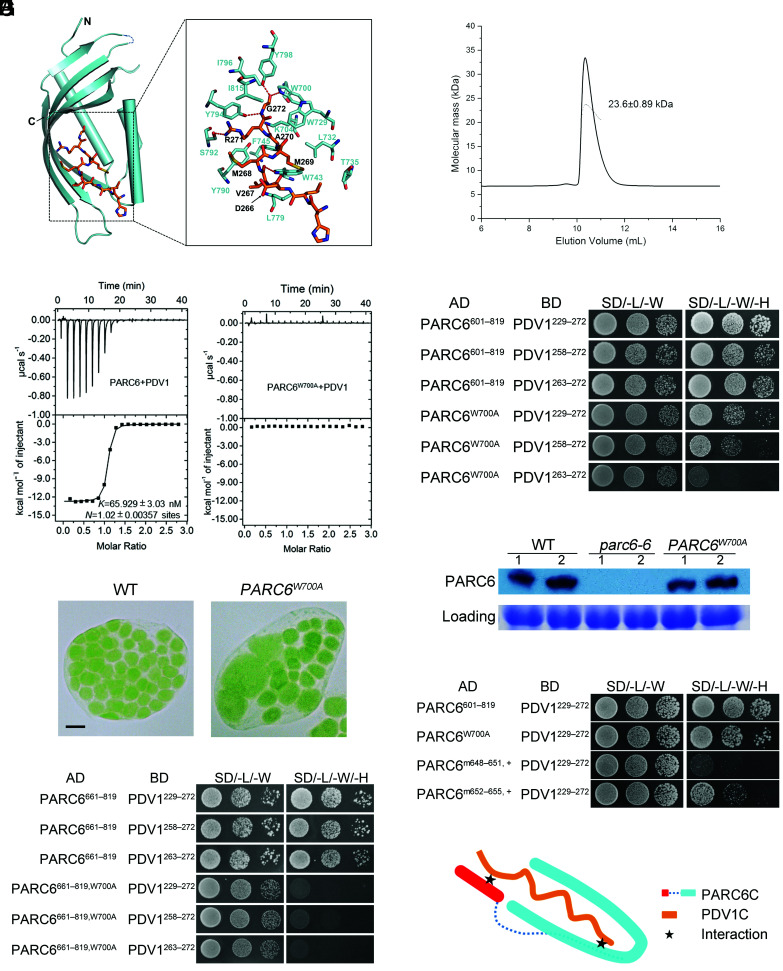
To investigate how PARC6 interacts with PDV1, we solved the crystal structure of the PARC6–PDV1 complex at a resolution of 2.894 Å (Fig. 5A and SI Appendix, Table S2). In light of the approach used in solving the structure of ARC6–PDV2, we fused the C-terminal peptide of PDV1 (PDV1C, PDV1263–272) to the C terminus of PARC6 (PARC6685–819) with the same 21-residue linker (STGNASDSSSDSSSSEGDGTV) used in the ARC6 study (14). We successfully solved the structure of the PARC6–PDV1 complex using the crystals obtained from this fusion protein (Fig. 5A and SI Appendix, Table S2). The crystallographic data showed that one asymmetric unit contains four PARC6–PDV1 complexes. To investigate its oligomeric state in solution, we also conducted an SLS analysis of the fusion protein, which revealed the exclusive monomeric nature of this fusion protein (Fig. 5B). During the course of our study, the structures of PARC6 and PARC6 complexed with PDV1 were also deposited to the Protein Data Bank (PDB codes 5U9L and 5U9O, respectively) by other groups. These two structures were very similar to those in our study with rmsd values of the Cα atoms of ~0.271 and 0.584 Å for PARC6C and PARC6C–PDV1C, respectively.
Fig. 5.
Two regions of PARC6 are involved in its interaction with PDV1. (A) Crystal structure of the PARC6685–819–PDV1263–272 fusion protein. The C-terminal end of PDV1 (PDV1C, residues 263 to 272, shown in orange) inserts into the pocket formed by the C-terminal region of PARC6 (shown in cyan). Residues involved in the interactions are shown as stick models. Hydrogen bonds are shown as red dashed lines. (B) SLS analysis of PARC6685–819–PDV1263–272 in solution. The fusion protein is only in a monomeric state. (C) ITC analysis of the interaction between PARC6 and PDV1. GST-tagged PARC6: residues 685 to 819. PDV1: residues 261 to 272. (D) Y2H analysis of the interaction between PARC6601–819, PARC6601–819, W700A, and PDV1. Various lengths of the C-terminal domain of PDV1 were tested. The negative controls are shown in SI Appendix, Fig. S11A. (E) The W700A mutation affects the function of PARC6. Left: WT cell; right: parc6-6 mutant transformed with PARC6W700A. Bar, 10 μm. (F) Immunoblot analysis of the seedlings shown in E. CBB staining of the SDS-PAGE gel serves as a loading control. (G) The W700A mutation abolishes the interaction between a truncated version of PARC6 (residues 661 to 819) and PDV1 in Y2H assays. This truncation lacks the “lid” (I646 to H656) of the PARC6 pocket and C657, which can form a disulfide bond with C741 and is essential for the closing of the “lid”. The negative controls are shown in SI Appendix, Fig. S11B. (H) Y2H analysis shows that the “lid” region of PARC6 is also important for the interaction between PARC6C and PDV1C. “m648–651, +”, VLID motif (residues 648 to 651) mutated to SDSA in the PARC6W700A mutant; “m652–655, +”, MLKM motif (residues 652 to 655) mutated to SDAS in the PARC6W700A mutant. The negative controls are shown in SI Appendix, Fig. S11C. (I) A working model of the interaction between PARC6C and PDV1C. There are two sites for the interaction (shown as stars): One is inside the pocket, and the other one is right outside of the pocket, which involves the lid of PARC6C and a region of PDV1C.
We noted the formation of several hydrogen bonds between PARC6 and PDV1. For example, the carboxyl group of the C-terminal residue G272 of PDV1 formed polar interactions with the sidechains of residues W700 and Y798 (Fig. 5A). Isothermal titration calorimetry (ITC) binding assays showed that mutating W700 to alanine (W700A) in PARC6 severely hinders its interaction with PDV1263–272 (Fig. 5C). In addition, the strength of the interaction between PARC6W700A and PDV1 gradually decreased as the length of PDV1 shortened, based on Y2H assays (Fig. 5D and SI Appendix, Fig. S11A). We introduced a construct encoding a mutant version of PARC6 harboring the W700A mutation (PARC6W700A) into the parc6-6 mutant to test the effects of the mutation in planta. None of the transformed plants were fully complemented, even when protein abundance was comparable to that of the WT (Fig. 5 E and F and SI Appendix, Fig. S10B). We conclude that the interaction between PARC6 and PDV1 is important for chloroplast division.
Interestingly, the interaction between PARC6W700A and PDV1 was completely abolished when the PARC6 fragment used in Y2H was shortened from residues 601-819 to 661-819 (Fig. 5G and SI Appendix, Fig. S11B), suggesting that the region of PARC6 between residues 601 and 660 is also involved in the interaction with PDV1. We determined that the lid of the PARC6C pocket (I646 to H656) has a hydrophobic surface (SI Appendix, Fig. S12 A and B), while the C terminus of PDV1 (residues 251 to 272) has a potential hydrophobic side (SI Appendix, Fig. S12C). We speculated that these two hydrophobic regions may interact with each other and thus mediate another interaction site between PARC6 and PDV1. To test this hypothesis, we mutated the hydrophobic region in the lid of PARC6C in the W700A mutant (Fig. 5H and SI Appendix, Fig. S11C). Indeed, these mutations reduced or even abrogated the interaction between PARC6C and PDV1, supporting our hypothesis.
The above data suggest that PDV1 and PARC6 interact via two sites: one inside the PARC6C pocket and the other right outside the pocket (Fig. 5I). The presence of two binding sites may also explain why the interaction between these two proteins is relatively resistant to single-site mutations in the PARC6C pocket (Fig. 5D).
Regulation of PARC6 Dimerization.
To investigate whether the PDV1–PARC6 interaction affects the self-interaction of PARC6, we performed yeast three-hybrid (Y3H) assays with PDV1 and PARC6 (Fig. 6A and SI Appendix, Fig. S13). The longest version of the PDV1 C-terminal region (residues 229 to 272) appeared to promote the self-interaction of PARC6. However, when the PDV1 fragment was shortened, the self-interaction of PARC6 decreased. This is more apparent when 3-amino-1,2,4-triazole (3-AT), an inhibitor of the reporter gene HIS3, was added for a stringent screening. Although PDV1263–272 interacted with PARC6, it failed to promote the self-interaction of PARC6, which was consistent with the fact that the fusion protein between PARC6685–819 and PDV1263–272 exists exclusively as a monomer (Fig. 5B). Furthermore, we conducted pull-down assays (Fig. 6B) and gel-filtration assays (SI Appendix, Fig. S14) to study the effect of PDV1 on the dimerization of PARC6 in solution. We added PDV1 peptide to PARC6 in a buffer containing the strong reducing agent dithiothreitol (DTT). Under these conditions, intact PDV1 peptide induced the dimerization of PARC6, but the G272D variant of PDV1 (pdv1-2) (12) did not. These data suggest that the interaction with PDV1 may promote the self-interaction of PARC6 only when the two interaction sites are present.
Fig. 6.
Dimerization of PARC6 is regulated by PDV1 and the redox status. (A) Y3H analysis shows that PDV1 promotes the self-interaction of PARC6. Various lengths of the C-terminal domain of PDV1 were tested. Methionine (M) suppresses the expression and accumulation of the bridge protein PDV1. 3-AT is an inhibitor of the enzyme encoded by the reporter gene HIS3. The negative controls are shown in SI Appendix, Fig. S13. (B) A pull-down assay shows that the interaction between PDV1 and PARC6 promotes the self-interaction of PARC6. The reaction buffer contains DTT (5 mM), which can reduce disulfide bonds and open the “lid”. Δ, a minor degradation product of the protein. (C) SLS analysis of PARC6C (residues 640 to 819) with or without 5 mM DTT. (D) SLS analysis of PARC6CC657S (residues 640 to 819) indicates that it is a monomer.
Interestingly, while PARC6C existed both as a dimer and a monomer (Fig. 6C), the C657S variant of PARC6C accumulated exclusively as a monomer in solution (Fig. 6D), suggesting that the intramolecular disulfide bond also plays an important role in the stabilization of the PARC6 dimer. In agreement, adding DTT to the buffer containing PARC6C also resulted in a monomer form of PARC6C (Fig. 6C).
Mutations of V746D and L748R were shown to abolish the self-interaction and probably the dimerization of PARC6 (Fig. 4D). We further tested whether these two mutations could also affect the interaction between PARC6 and PDV1 by Y2H analysis (SI Appendix, Fig. S15). Our result suggested that these two mutations had no effect on the interaction. Therefore, dimerization of PARC6 probably is not critical for its interaction with PDV1.
Taken together, these results indicate that the formation of the intramolecular disulfide bond under oxidized conditions or the interaction with PDV1 at two sites (the pocket and the lid) promotes the dimerization of PARC6, probably by stabilizing its protein structure and the dimerization interface.
Redox Regulation of PARC6.
As shown above, an intramolecular disulfide bond creates a lid over the PARC6C pocket (Fig. 4A). Structural superimposition between PARC6C and the PARC6C–PDV1C complex showed that the PDV1 peptide is located at the same position as the PARC6C lid (Fig. 7A), suggesting that the presence of the disulfide bond within PARC6C may block the insertion of PDV1 and prevent binding. In support of this idea, pull-down assays showed that PARC6C only interacts with PDV1 in the presence of DTT (Fig. 7B). Moreover, both PARC6CC657S and PARC6685–819 bound to PDV1 regardless of the presence of DTT (Fig. 7B), possibly because the PARC6C pocket cannot be closed in these contexts.
Fig. 7.
Redox state affects the disulfide bond in PARC6C and its interaction with PDV1C. (A) Crystal structure shows that the formation of a disulfide bond blocks the pocket required for PDV1–PARC6 interaction. Apo- and PDV1C-bound PARC6CΔ640–684 molecules are colored in gray and cyan, respectively. In apo-PARC6C, the “lid” helix is colored in pink, with the two cysteine residues forming the disulfide bond shown as stick models. PDV1C is shown as a ball-and-stick model. (B) Pull-down assay shows that the disulfide bond within PARC6C prevents PDV1–PARC6 interaction. His-SUMO tagged PDV1: residues 226 to 272. (C) WT plants were entrained with 16 h/8 h light/dark cycles (Top) and transferred to continuous dark (Bottom). The arrows indicate the start of the experiment. (D) Dark and light affect disulfide bond formation of PARC6C in Arabidopsis plants. Total proteins from the leaves of WT plants were extracted with buffer without β-mercaptoethanol and split into two aliquots, one of which received β-mercaptoethanol (5%) later. These two sets of samples were run in parallel on the same gel and were probed with anti-PARC6C antibodies. The disulfide bond blocks the antibody–antigen interaction. M, molecular weight markers. CBB staining of the SDS-PAGE gels serves as a loading control. (E) Chloroplast phenotype of mesophyll cells of 21-d-old plants at different time points. Dividing chloroplasts are indicated by arrows. Bar, 10 μm. (F) Proportion of dividing chloroplasts in the plants shown in E (n = 30 cells for each sample, P < 0.05). Chloroplasts with an aspect ratio larger than 1.5 are counted in F. For statistical analysis, different letters denote statistically significant (P < 0.05) differences between samples. Gray column, dark; white column, light. (G) A pull-down assay to show the effect of the oxidative condition on the PDV1–PARC6 interaction formed in the reduced state. HA-PARC6, His-PARC6, and PDV1 were first incubated in the presence of 5 mM DTT, and then the buffer was changed to one containing 0.5% H2O2 but no DTT. MgCl2 was added at a final concentration of 5 mM throughout the process where indicated. Δ, a minor degradation product of the protein.
Since the redox state of PARC6C affected the closure or opening of its lid and, thus, its interaction with PDV1, we investigated the redox state of PARC6C in planta. In the dark or shortly after transfer into light, PARC6C accumulated in a mostly oxidized state. Two hours after the onset of light exposure, PARC6C abundance in the reduced state increased (Fig. 7 C and D). In the meantime, more chloroplasts started to divide after light illumination (Fig. 7 E and F). Thus, light may promote chloroplast division by regulating the redox state of PARC6C.
To learn whether PDV1-induced dimerization of PARC6C is reversible during the dark, another pull-down assay was carried out (Fig. 7G). When the buffer was switched from reducing condition to oxidative condition, although PARC6C could still self-interact well, PDV1 was lost in the complex. This suggests that PARC6C could be oxidized again to close the lid and form a dimer in the dark.
Mg2+ Enhances the Dimerization of PARC6.
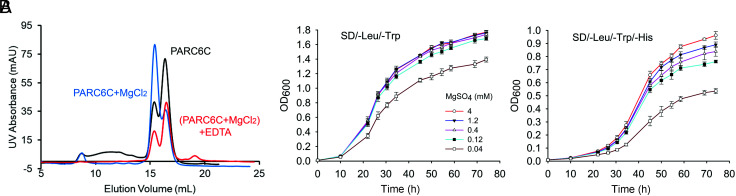
Because the PARC6C dimer interface is mostly surrounded by a negatively charged region (Fig. 4C), we reasoned that the dimerization of PARC6C may be enhanced by metal ions. Mg2+ is important for many physiological functions in chloroplasts, where it is maintained at a high concentration. We thus performed gel filtration experiments to test the effect of Mg2+ on PARC6C dimerization (Fig. 8A). Interestingly, adding Mg2+ to the buffer enhanced the dimerization of PARC6C, while the subsequent addition of ethylenediaminetetraacetic acid (EDTA) reversed the dimerization of PARC6C. Since PARC6C self-interacted in our Y2H assays, we conducted a Y2H experiment with various concentrations of Mg2+ added to the growth medium and measured optical density as an indication of interaction strength (Fig. 8B). The self-interaction of PARC6C was also decreased in the presence of lower Mg2+ concentrations. These data suggest that Mg2+ increases the dimerization potential of PARC6.
Fig. 8.
Magnesium ions affect the dimerization of PARC6C. (A) Gel infiltration analysis of PARC6C, PARC6C with 5 mM MgCl2, and PARC6C incubated with 5 mM MgCl2 before the addition of 5 mM EDTA. (B) Y2H analysis of the self-interaction of PARC6 in the presence of different Mg2+ concentrations. Yeast colonies harboring two plasmids, AD PARC6601–819 and BD PARC6601–819, were grown in liquid medium as indicated. OD600 values were measured at different time points to measure growth, which reflects the strength of the protein interaction.
In our pull-down assay, we also tested the effect of Mg2+ on the dimerization of fully reduced PARC6C, it turned out to have no effect (Fig. 7G). PDV1 can well induce the self-interaction of PARC6, and the addition of Mg2+ had no effect either (Fig. 7G). Thus, Mg2+ seems to only promote the dimerization of oxidized PARC6C.
Discussion
PDV1 was previously thought to only cooperate with PDV2 to recruit ARC5 to the chloroplast division site (12). Overexpression of PDV1 does not suppress but rather slightly promotes chloroplast division (13). PDV1 and ARC5 have been shown to interact (26), supporting the view that PDV1 helps recruit ARC5 to the division site. The lower ARC5 abundance seen in the pdv1 mutant is consistent with this notion (Fig. 3). However, GFP-ARC5 was shown to localize to the chloroplast division site in the pdv1 mutant, which exhibits a severe chloroplast division defect (12), suggesting that PDV1 may play other roles besides recruiting ARC5 to the chloroplast division site.
A careful characterization of our newly identified null pdv1 mutant allele revealed the striking similarity between its phenotypes and those of mutants defective in the Min system (16, 18, 20, 21, 27, 28), including parc6 mutants (Fig. 2). Furthermore, we showed that PARC6 abundance is very low in pdv1 (Fig. 3). Fluorescence signals from a GFP-PDV1 fusion protein have been reported to be difficult to visualize at the chloroplast division site in parc6 mutants (20), raising the possibility that PDV1 and PARC6 must both be present to accumulate. Their interaction may thus be important for the stability of the PDV1–PARC6 protein complex. Since PDV1 can also interact with PDV2 and ARC5, whose assembly into the division complex does not directly rely on PARC6, PDV1 may help bring PARC6 to the chloroplast division site after the assembly of the FtsZ–ARC6–PDV2–ARC5 complex to regulate chloroplast division.
Protein interaction assays indicated that PDV1 can induce the dimerization of PARC6 (Fig. 6 A and B), which is important for its function (Fig. 4). MinD, a key component of the bacterial and chloroplast Min system, also forms dimers. Interestingly, PDV2 can also induce the dimerization of ARC6, but in a different manner, as stated in the introduction (14). PDV2 and ARC6 are core components of the chloroplast division complex (3). Therefore, the formation of dimers may be a working state of PARC6 and other components of the Min system to efficiently regulate the dynamics of the chloroplast division complex.
The oligomeric state of PARC6C is redox regulated. Indeed, an intramolecular disulfide bond can form within PARC6C under oxidized conditions (Fig. 4A), bringing a helix-like lid to the outlet of the PARC6C pocket and thus blocking its interaction with PDV1 (Fig. 7 A, B, and G). When the disulfide bond is in its reduced form, the PARC6C outlet is open, allowing PDV1 to insert into the pocket of PARC6C for interaction (Fig. 7 A, B, and G). PARC6C tends to form a dimer, either when its intramolecular disulfide bond is formed (Fig. 4 A and B) or when the disulfide bond is disrupted and PDV1 is inserted to interact with PARC6 at two sites (Fig. 6 A and B). When the PARC6C lid is open without PDV1 (Fig. 6 C and D) or when PDV1 is inserted but the interaction at the outlet is missing (Figs. 5 and 6), PARC6C cannot form a dimer. Thus, the formation of the disulfide bond or the interaction at the outlet may tighten and stabilize the structure of PARC6C so it can dimerize. In the absence of PDV1, PARC6 abundance is low (Fig. 3), possibly because the opening and exposure of the hydrophobic lid and the loosening of the structure renders the protein unstable.
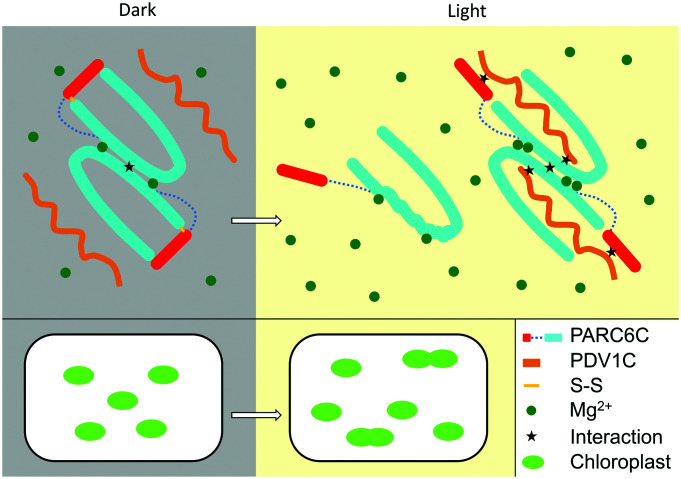
Light can increase the pool of reductants in chloroplasts (29–31), which diminishes or prevents the formation of the PARC6C disulfide bond (Figs. 7 and 9), promoting the interaction between PDV1 and PARC6, PARC6 dimerization, and hence chloroplast division (Figs. 7 and 9). Mg2+ is relatively abundant in chloroplasts, with estimated concentrations of 1 to 5 mM in the stroma and 30 to 50 mM in thylakoids (32). The self-interaction of PARC6C was enhanced by 5 mM Mg2+ (Figs. 8 and 9), close to the physiological range. Light was shown to increase the concentration of Mg2+ in chloroplasts (33). Thus, Mg2+ may be another factor utilized by light to promote chloroplast division. In summary, our study uncovers the multilayer regulation of the PARC6–PDV1 pair in the IMS and, thus, in chloroplast division.
Fig. 9.
A working model of the regulation of the PDV1–PARC6 interaction during light-regulated chloroplast division. In the dark, the intramolecular disulfide bond in PARC6C is oxidized. The PARC6C lid is thus closed and PARC6C forms a dimer. In the light, the intramolecular disulfide bond in PARC6C is reduced. The PARC6C lid is open, which destabilizes PARC6C and hampers its dimerization. However, interaction with PDV1C stabilizes the structure of PARC6C, promoting its dimerization. Mg2+ binding also promotes the dimerization of PARC6C. Light increases the concentration of Mg2+ in chloroplasts and further promotes the dimerization of PARC6C. Fluctuations in the redox state and Mg2+ concentrations modulate the light-mediated regulation of chloroplast division.
Materials and Methods
Bioinformatic Analysis of AtPARC6 and AtPDV1.
Protein sequences of AtPARC6, AtPDV1, and their homologs in other species, including Populus trichocarpa, Nicotiana tabacum, Oryza sativa, Zea mays, and Physcomitrium (Physcomitrella) patens et al., were downloaded from National Center for Biotechnology Information (NCBI) database through BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (SI Appendix, Table S1). Multiple protein sequence alignment was performed using BioEdit (https://bioedit.software.informer.com/), and phylogeny was analyzed using the neighbor-joining method of MEGA7 (https://www.megasoftware.net). Bootstrap values at the corresponding nodes were based on 1,000 bootstrap replications. Wenxiang diagram of protein sequences was performed as before (34, 35).
Plant Materials and Growth Conditions.
Arabidopsis thaliana plants used in this study were Columbia-0 (Col-0) ecotype. Complementation lines (pdv1-3 comp and parc6-6 comp) and transgenic plants containing mutant versions of PARC6 (PARC6V746D, PARC6L748R, and PARC6W700A) under control of the native promoter were obtained by Agrobacterium-mediated floral dipping method. Plants were grown in soil in a growth chamber at 21 °C ± 1 °C with 16 h light/8 h dark cycles.
Chloroplast Phenotype Analysis.
To characterize the chloroplast phenotype, fresh leaves of 4-week-old plants were fixed in 3.5% glutaraldehyde solution for 1 h in darkness. Then, the glutaraldehyde solution was replaced with 0.1 M Na2EDTA (pH = 9.0), and the leaves were placed in a water bath of 55 °C for 2 h. The chloroplast phenotype was observed with an Olympus CX21 (Olympus, Tokyo) microscope and captured with a USB2.0 digital camera (Changheng, Beijing). In order to determine the effect of light on chloroplast division, 18-d-old fast-growing WT plants were chosen for the analysis. Data were analyzed by one-way ANOVA using SPSS 22.0 (https://www.ibm.com/software/analytics/spss/). Significant differences among different treatments were given at P < 0.05. Plots were performed using SigmaPlot 12.5 software (https://sigmaplot.en.softonic.com/).
Identification of pdv1-3 and parc6-6 Mutants.
Mutants pdv1-3 and parc6-6 were obtained through ethyl methane sulfonate (EMS) mutagenesis. Plants with abnormal chloroplast division phenotype were screened by microscopy. The mutation sites of pdv1-3 and parc6-6 were identified by bulked segregant analysis and gene sequencing. pdv1-3 has a G to A mutation in PDV1 gene, which changes the 17th codon into a premature stop codon (SI Appendix, Fig. S2). parc6-6 has a G to A mutation in PARC6 gene, which changes the 338th codon into a premature stop codon (SI Appendix, Fig. S3). Complementation lines were obtained by transforming the WT PDV1 and PARC6 genes with their native promoters into pdv1-3 and parc6-6 plants, respectively.
Immunoblotting.
Proteins were extracted from fresh leaf tissues of 18- and 25-d-old plants (Fig. 3) and 3-wk-old plants (Figs. 4, 5 and 7), respectively. About 100 mg tissue sample was ground in liquid nitrogen and resuspended with 500 μL 2× SDS loading buffer (120 mM Tris-HCl pH 6.8, 10% glycerin, 10% β-mercaptoethanol, 2% SDS, bromophenol blue). The crude protein extracts were obtained through centrifugation. Total proteins were separated by a 10% SDS-PAGE gel and transferred onto a polyvinylidene difluoride membrane (Bio-Rad). After being blocked in a TBST buffer (10 mM Tris, 150 mM NaCl, 0.1% Tween-20) with 5% (w/v) fat-free milk for 2 h at room temperature, the membrane was probed with purified primary antibodies and HRP-conjugated goat anti-rabbit IgG secondary antibodies at a dilution of 1:10,000. Coomassie Brilliant blue (CBB) staining was used as the loading control.
Immunofluorescence Staining of FtsZ.
Immunofluorescence staining was performed as described previously (36). Four-week-old leaf tissues were obtained from WT, pdv1-3 and parc6-6 plants, respectively, and the protoplasts were isolated by enzyme digestion. After fixation, the protoplast suspensions were adsorbed on slides coated with poly L-lysine and incubated with anti-FtsZ2-1 polyclonal antibodies (1:100) followed by the addition of fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG secondary antibodies (1:100; Jiaxuan Biotech). Images were captured with a digital camera (E3ISPM) coupled with a fluorescent microscope (NE910, Nexcope, Ningbo, China).
RNA Extraction and Semiquantitative RT-PCR Analysis.
Total RNA of leaf tissues was extracted from 18- and 25-d-old plants using an RNApure total RNA isolation kit (Aidlab) and reversely transcribed with an M5 First Strand cDNA Synthesis Kit (Mei5bio). The cDNA templates, serially diluted three times with a dilution ratio of four, were used for PCR reactions. PP2AA3 was used as an internal control gene.
Y2H and Y3H Analyses.
The yeast strain AH109 was used for both Y2H and Y3H analyses, and the culture and transformation methods were in accordance with Clontech's instructions (http://www.clontech.com). For Y2H analysis, the C-terminal fragments of PARC6 and its mutants were amplified from Arabidopsis thaliana cDNA by PCR and cloned into pGADT7 vector or pGBKT7 vector. Similarly, the C-terminal fragments of PDV1 were cloned into pGBKT7. The pGADT7 and pGBKT7 constructs were cotransformed into AH109 by LiAc/SS-DNA/PEG method. Yeast cells were grown on a synthetic dropout medium either with (SD/-Trp-Leu) or without (SD/-Trp-Leu-His) Histidine. Positive interactions were detected by activation of the HIS3 reporter. The agar plates were incubated at 30 °C for 2 to 3 d.
For Y3H analysis, the C-terminal sequence of PARC6 was cloned into pGADT7 and the MCS I of pBridge vector, respectively. The C-terminal sequences of PDV1 were cloned into the MCS II of pBridge under the control of the Met25 promoter. pGADT7 and pBridge vectors were cotransformed into AH109 and cultured on SD/-MLW(-Met/-Leu/-Trp), SD/+M-LWH (+Met/-Leu/-Trp/-His), and SD/-MLWH (-Met/-Leu/-Trp/-His) medium at 30 °C for 3 to 5 d. Methionine was used to suppress the expression and accumulation of the bridge protein PDV1. For a stringent screening, 3 mM 3-AT was added.
Protein Expression and Purification.
Various segments of PARC6C and PDV1 were amplified by PCR and cloned into the pET28a vector. The protein was expressed and purified as previously described (37). Briefly, the proteins were expressed in Escherichia coli strain BL21 (DE3), and the bacteria were cultured at 18 °C overnight for the protein expression. The cell lysate was centrifuged to remove cell debris. Then, the supernatant was applied onto a self-packaged Ni-affinity column (2 mL Ni-NTA, Genscript). The fusion protein was eluted with elution buffer (50 mM Tris pH 8.0, 300 mM NaCl, 300 mM imidazole). The eluant of PARC6640–819 was concentrated and further purified using a Superdex-75 increase 10/300 GL (GE Healthcare) column equilibrated with a buffer containing 10 mM Tris-HCl pH 8.0 and 200 mM NaCl. The eluant of PARC6685–819–PDV1263–272 was concentrated and further purified using a Superdex-75 increase 10/300 GL (GE Healthcare) column equilibrated with a buffer containing 10 mM Tris-HCl pH 8.0, 200 mM NaCl, and 5 mM DTT. The purified protein was analyzed by SDS-PAGE. Fractions containing the target protein were pooled and concentrated.
Crystallization.
Crystals of PARC6640–819 were grown using the hanging-drop vapor diffusion method. The crystals were grown at 18 °C after mixing an equal volume of the PARC6640–819 protein (20 mg/mL in 10 mM Tris-HCl pH 8.0 and 200 mM NaCl) with a solution containing 0.2 M ammonium sulfate, 0.1 M MES pH 6.5, and 20% (w/v) PEG 8000. Crystals of PARC6685–819–PDV1263–272 fusion protein were obtained with a method similar to the above. Specifically, the reservoir solution contained 0.5 M potassium phosphate monobasic and 0.1 M sodium acetate pH 4.5.
X-ray Diffraction and Structural Determination.
The X-ray diffraction data were collected at the Shanghai Synchrotron Radiation Facility beamlines BL17U1 and BL19U1 and processed with the HKL2000 package (38). Both the structures were initially solved by molecular replacement with the structure of ARC6674–801 (PDB: 5HAD) as a template. The structures were refined manually with COOT (39) and PHENIX (40). The final structures were obtained through prudent refinement. The relevant details are summarized in SI Appendix, Table S2.
ITC Binding Assay.
The dissociation constants of binding reactions of PARC6C or PARC6C mutants with the PDV1 peptide were determined by ITC using a MicroCal ITC200 calorimeter (Malvern). Both proteins and peptides were desalted into the working buffer (20 mM HEPES pH 7.5 and 200 mM NaCl). The titration was carried out with 19 successive injections of 2 µL peptide at the 0.3 mM concentration, spaced 120 s apart, into the sample cell containing the PARC6C or PARC6C mutants with a concentration of 0.03 mM by 750 rpm at 25 °C. The Origin software (Origin 2018) was used for baseline correction, integration, and curve fitting to a single site-binding model.
In Vitro Pull-Down Assay.
The proteins used in the pull-down assay, such as His-tagged PARC6C, HA-tagged PARC6C, PDV1226–272, and its G272D mutant, were first exchanged into the buffer containing 10 mM Tris, pH 8.0, and 200 mM NaCl. The purified His-tagged PARC6C protein was used to pull down the HA-tagged PARC6C. The His-tagged PARC6C and the HA-tagged PARC6C protein were mixed at a molar ratio of 1:2 and complemented with the PDV1 peptide, and then, DTT was added to the system with a final concentration of 5 mM. After that, the mixture was loaded onto 30 µL Ni2+ resins and incubated at 4 °C for 1 h. The Ni2+ resins were first washed with buffer containing 50 mM Tris-HCl pH 8.0, 30 mM imidazole, and 300 mM NaCl five times and then, washed with buffer containing 50 mM Tris-HCl pH 8.0, 50 mM imidazole, and 300 mM NaCl once. The resins were added with the loading buffer and boiled at 100 °C for 5 min. Then, the samples were subjected to SDS-PAGE and visualized by CBB staining.
To test the effect of oxidative conditions on PDV1–PARC6 binding, pull-down assay under the reduction state (buffer containing 5 mM DTT) was performed first as described above. Subsequently, the buffer was replaced three times with a buffer containing 0.5% H2O2 and no DTT, and then the reaction system was incubated further for 3 h at 4°C. MgCl2 was added at a final concentration of 5 mM throughout the process when needed. Subsequent processing steps were performed as described above.
Multiangle Light Scattering.
Multiangle light scattering experiments of PARC6C or PARC6C mutant were performed in 10 mM Tris-HCl pH 8.0, 200 mM NaCl with a GE Healthcare Superdex-75 increase 10/300 GL size-exclusion column connected to the Wyatt DAWN HELEOS Laser photometer and Wyatt Optilab T-rEX differential refractometer. To compare the oxidized and reduced state of PARC6640–819, oxidized PARC6640–819 was incubated with 5 mM DTT on ice overnight and then multiangle light scattering experiments were performed with the same column. Wyatt ASTRA 7.3.2 software was used for the data analysis.
Gel Filtration-Binding Assay.
The PARC6640–819, HA-PARC6640–819, and PDV1226–272 purified as described above were subjected to gel filtration analysis (Superdex-75 increase 10/300 GL, GE Healthcare). They were mixed with a molar ratio of about 1:1:5 and incubated at 4 °C for 2 h before the gel filtration analysis in buffer containing 10 mM Tris-HCl pH 8.0, 200 mM NaCl, and 5 mM DTT. Samples of various fractions were also analyzed by SDS-PAGE.
The Effect of Mg2+ on the Dimerization of PARC6C.
Bacterial cells expressing PARC6640–819 were divided into two parts after being resuspended in lysis buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole and 1 mM phenylmethylsulfonyl fluoride). One part of the resuspended cells was supplemented with MgCl2 at a final concentration of 5 mM, which was designated as PARC6C + MgCl2 hereafter. PARC6C and PARC6 + MgCl2 were purified as described above, but 5 mM MgCl2 was included in the buffer throughout the purification of PARC6 + MgCl2. After purification and desalting to the working buffer containing 10 mM Tris-HCl pH 8.0 and 200 mM NaCl, half amount of PARC6+MgCl2 was incubated with 5 mM EDTA on ice overnight [designated as (PARC6 + MgCl2) + EDTA]. Finally, PARC6, PARC6 + MgCl2, and (PARC6 + MgCl2) + EDTA were analyzed by gel filtration (Superdex-75 increase 10/300 GL, GE Healthcare) in 10 mM Tris-HCl pH 8.0, 200 mM NaCl.
The effect of Mg2+ on the self-interaction PARC6C was also tested by a modified Y2H assay method. Four colonies with AD-PARC6601–819 and BD-PARC6601–819 were selected and grown in 2 mL liquid SD/-Trp-Leu medium to OD600 = 2. The cells were harvested by centrifugation, washed twice with double-distilled water to remove traces of Mg2+, and resuspended in double-distilled water. Yeast cells were grown in SD/-Trp-Leu and SD/-Trp-Leu-His liquid media containing various concentrations of MgSO4 (0.04, 0.12, 0.4, 1.2, and 4 mM) and with a starting OD600 of 0.01. Growth of the yeast cultures was monitored by following OD600 until the plateau phase. The growth curves of yeast were drawn using Sigmaplot 12.5 (https://sigmaplot.en.softonic.com/). Growth speeds of yeasts were taken as an indication of interaction strengths.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We would like to thank the staff at beamlines BL17U1 and BL19U1 of the Shanghai Synchrotron Radiation Facility for their assistance with data collection. We would like to thank the Tsinghua University Branch of China National Center for Protein Sciences Beijing and Shilong Fan for providing facility support for X-ray diffraction of the crystal samples. We acknowledge the Protein Preparation and Characterization Core Facility of Tsinghua University Branch of China National Center for Protein Sciences Beijing for providing the facility support. This work was supported by the Fundamental Research Funds for the Central Universities (2022BLRD14, 2019YC07) and the National Natural Science Foundation of China (31570182, 32070696).
Author contributions
H.G. and Y.F. designed research; Q.S., X.C., Z.L., C.A., J.H., Y.W., M.Q., T.G., W.C., and Y.Z. performed research; and Q.S., C.A., J.H., Y.F., and H.G. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Yue Feng, Email: fengyue@mail.buct.edu.cn.
Hongbo Gao, Email: gaohongbo@bjfu.edu.cn.
Data, Materials, and Software Availability
The accession number for the coordinates and structure factors of PARC6640–819 and PARC6685–819–PDV1263–272 are PDB (https://www.rcsb.org/): 6JZF and 6JZN, respectively. All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Osteryoung K. W., Nunnari J., The division of endosymbiotic organelles. Science 302, 1698–1704 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Osteryoung K. W., McAndrew R. S., The plastid division machine. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 315–333 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Osteryoung K. W., Pyke K. A., Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 65, 443–472 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Miyagishima S., Mechanism of plastid division: From a bacterium to an organelle. Plant Physiol. 155, 1533–1544 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida Y., Mogi Y., TerBush A. D., Osteryoung K. W., Chloroplast FtsZ assembles into a contractible ring via tubulin-like heteropolymerization. Nat. Plants 5, 119–119 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Chen C., MacCready J. S., Ducat D. C., Osteryoung K. W., The molecular machinery of chloroplast division. Plant Physiol. 176, 138–151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitha S., McAndrew R. S., Osteryoung K. W., FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153, 111–120 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maple J., Aldridge C., Moller S. G., Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J. Cell Mol. Biol. 43, 811–823 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Vitha S., et al. , ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15, 1918–1933 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koksharova O. A., Wolk C. P., A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J. Bacteriol. 184, 5524–5528 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glynn J. M., Froehlich J. E., Osteryoung K. W., Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20, 2460–2470 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyagishima S. Y., Froehlich J. E., Osteryoung K. W., PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18, 2517–2530 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okazaki K., et al. , The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21, 1769–1780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W. H., et al. , Structural insights into the coordination of plastid division by the ARC6-PDV2 complex. Nat. Plants 3, 17011 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Liu X., et al. , A novel amphiphilic motif at the C-terminus of FtsZ1 facilitates chloroplast division. Plant Cell 34, 419–432 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colletti K. S., et al. , A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10, 507–516 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Itoh R., Fujiwara M., Nagata N., Yoshida S., A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol. 127, 1644–1655 (2001). [PMC free article] [PubMed] [Google Scholar]
- 18.Nakanishi H., Suzuki K., Kabeya Y., Miyagishima S., Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr. Biol. 19, 151–156 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Shimada H., et al. , ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 45, 960–967 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Glynn J. M., et al. , PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 59, 700–711 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Zhang M., et al. , CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res. 19, 877–886 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Zhang M., Chen C., Froehlich J. E., TerBush A. D., Osteryoung K. W., Roles of arabidopsis PARC6 in coordination of the chloroplast division complex and negative regulation of FtsZ assembly. Plant Physiol. 170, 250–262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M., Schmitz A. J., Kadirjan-Kalbach D. K., Terbush A. D., Osteryoung K. W., Chloroplast division protein ARC3 regulates chloroplast FtsZ-ring assembly and positioning in arabidopsis through interaction with FtsZ2. Plant Cell 25, 1787–1802 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C., Cao L., Yang Y., Porter K. J., Osteryoung K. W., ARC3 activation by PARC6 promotes FtsZ-ring remodeling at the chloroplast division site. Plant Cell 31, 862–885 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashimoto H., Possingham J. V., Effect of light on the chloroplast division cycle and DNA synthesis in cultured leaf discs of spinach. Plant Physiol. 89, 1178–1183 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holtsmark I., et al. , Plastid division control: The PDV proteins regulate DRP5B dynamin activity. Plant Mol. Biol. 82, 255–266 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Pan D., et al. , Genetic mapping and isolation of two arc3 alleles in Arabidopsis. Plant Cell Rep. 32, 173–182 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Pyke K. A., Leech R. M., A genetic analysis of chloroplast division and expansion in arabidopsis thaliana. Plant Physiol. 104, 201–207 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cejudo F. J., Ojeda V., Delgado-Requerey V., Gonzalez M., Perez-Ruiz J. M., Chloroplast redox regulatory mechanisms in plant adaptation to light and darkness. Front. Plant Sci. 10, 380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cejudo F. J., Gonzalez M. C., Perez-Ruiz J. M., Redox regulation of chloroplast metabolism. Plant Physiol. 186, 9–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmer D., et al. , Topology of the redox network during induction of photosynthesis as revealed by time-resolved proteomics in tobacco. Sci. Adv. 7, eabi8307 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermans C., Conn S. J., Chen J. G., Xiao Q. Y., Verbruggen N., An update on magnesium homeostasis mechanisms in plants. Metallomics 5, 1170–1183 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Li J., et al. , Diel magnesium fluctuations in chloroplasts contribute to photosynthesis in rice. Nat. Plants 6, 848–859 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Zhou G. P., The disposition of the LZCC protein residues in wenxiang diagram provides new insights into the protein-protein interaction mechanism. J. Theor. Biol. 284, 142–148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou G. P., Chen D., Liao S., Huang R. B., Recent progresses in studying helix-helix interactions in proteins by incorporating the wenxiang diagram into the NMR spectroscopy. Curr. Top. Med. Chem. 16, 581–590 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Li Y. Q., Sun Q. Q., Feng Y., Liu X. M., Gao H. B., An improved immunofluorescence staining method for chloroplast proteins. Plant Cell Rep. 35, 2285–2293 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Yang L., et al. , Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24. Nat. Commun. 13, 1931 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams P. D., et al. , PHENIX: Building new software for automated crystallographic structure determination. Acta crystallogr. D. Biol. Crystallogr. 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The accession number for the coordinates and structure factors of PARC6640–819 and PARC6685–819–PDV1263–272 are PDB (https://www.rcsb.org/): 6JZF and 6JZN, respectively. All study data are included in the article and/or SI Appendix.