The CRISPR/Cas12a system can be used in a wide range of organisms. However, in plants, efficient editing by Cas12a was hampered due to the lower temperatures required for plant cultivation (Malzahn et al., 2019). Recently, we engineered an improved, temperature‐tolerant variant of Cas12a from Lachnospiraceae bacterium, ttLbCas12a, which has shown dramatically increased editing efficiencies compared to LbCas12a in Arabidopsis at the standard cultivation temperature of 22 °C (Schindele and Puchta, 2020). Furthermore, we could show that, by using ttLbCas12a, gene targeting (GT) efficiencies can be elevated considerably, both in Arabidopsis and tobacco (Huang et al., 2021; Merker et al., 2020). Nevertheless, even when using ttLbCas12a, some target sites remain highly resistant to editing. Evidence is accumulating that especially heterochromatic sites are more recalcitrant to editing (Weiss et al., 2022). Recently, it was demonstrated that the incorporation of 13 introns into the Cas9 coding sequence dramatically improves the editing efficiency in plants (Grützner et al., 2021). Based on these findings, an intron‐containing ttLbCas12a construct (ttLbCas12a‐i) was established to investigate whether editing in plants could be further improved.
The ttLbCas12a‐i variant was generated similarly to the respective Cas9‐intron variant (Grützner et al., 2021). Short introns from Arabidopsis genes with canonical donor and acceptor sites (AGGT) were equally placed into the coding sequence with an average distance of 200–400 nt (exon size). Due to the smaller size of ttLbCas12a compared to SpCas9, only 10 introns were introduced into the coding sequence (Appendix S1). The resulting ttLbCas12a‐i ORF was optimized for improved splicing efficiency using the NetGene2 intron splice site prediction software (http://www.cbs.dtu.dk/services/NetGene2/). The intron‐containing ttLbCas12a coding sequence was cloned into a T‐DNA expression vector (Figure 1a, top) as previously described for ttLbCas12a (Schindele and Puchta, 2020). First, we selected seven different target sites to compare the editing efficiency of ttLbCas12a‐i with ttLbCas12a. Here, we mainly focused on target sites with low to medium editing efficiencies of ttLbCas12a. Two sites were located in the ECA3 gene and five in heterochromatic, pericentromeric DNA adjacent to centromere I, III and IV (Figure 1a, bottom). Using the floral dip method, Arabidopsis thaliana were transformed with T‐DNAs containing the respective crRNAs as well as the two variants. To determine the editing efficiency, 17–20 transgenic plants each were grown for 2 weeks at 22 °C. Subsequently, the gDNA of each individual whole plant was extracted and then assessed by TIDE analysis. As shown in Figure 1b,c, three target sites showed hardly any editing in case of ttLbCas12a (T1‐T3), another three showed low‐to‐medium editing efficiencies (T4‐T6) and one showed medium‐to‐high editing efficiencies (T7). The new construct ttLbCas12a‐i surpassed ttLbCas12a significantly in terms of editing efficiency at every single target. On average, ttLbCas12a‐i was between 1.3‐ and 6.8‐fold more efficient than ttLbCas12a. This improvement is also highlighted at the population level, where, for the majority of the respective targets and plants, higher and more consistent editing efficiencies could be observed in case of ttLbCas12a‐i. However, the most tremendous improvements were obtained in case of the three targets that had been barely edited by ttLbCas12a. By using ttLbCas12a‐i, all of these targets became accessible for editing. Beyond that, almost every single plant showed much higher editing efficiencies in case of ttLbCas12a‐i. Based on these results, we aimed to investigate whether also GT could be further improved by using ttLbCas12a‐i. Therefore, we performed GT experiments in Arabidopsis, using the ALS selection system and same setup as previously described for ttLbCas12a (Merker et al., 2020). Using the floral dip method, A. thaliana were transformed with T‐DNAs containing the respective GT constructs. For both variants, 100 transgenic plants each were grown at 22 °C, propagated and, in the following generation, analysed for GT events. We determined the percentage of lines revealing GT events and assessed the total GT efficiency in terms of percentage of GT events per line (Figure 1d). The percentage of lines with GT events was only slightly higher in case of ttLbCas12a‐i compared to ttLbCas12a. However, concerning the total GT efficiency, a dramatic increase could be observed in case of ttLbCas12a‐i compared with ttLbCas12a, which corresponded to a 2.7‐fold increase. If the GT efficiencies of the individual lines are taken into account, the difference between ttLbCas12a and ttLbCas12a‐i becomes even more apparent (Figure 1e): While in case of ttLbCas12a, less than a third of the positive lines showed GT efficiencies above 1%, in case of ttLbCas12a‐i, more than three‐quarter of the positive lines did. To analyse the nature of GT events, that is to assess whether GT events constitute perfect or ectopic GT events, molecular analysis of selected positive plants was performed (Figure 1d). It turned out that perfect GT events were obtained at similar frequencies by using either of the variants.
Figure 1.
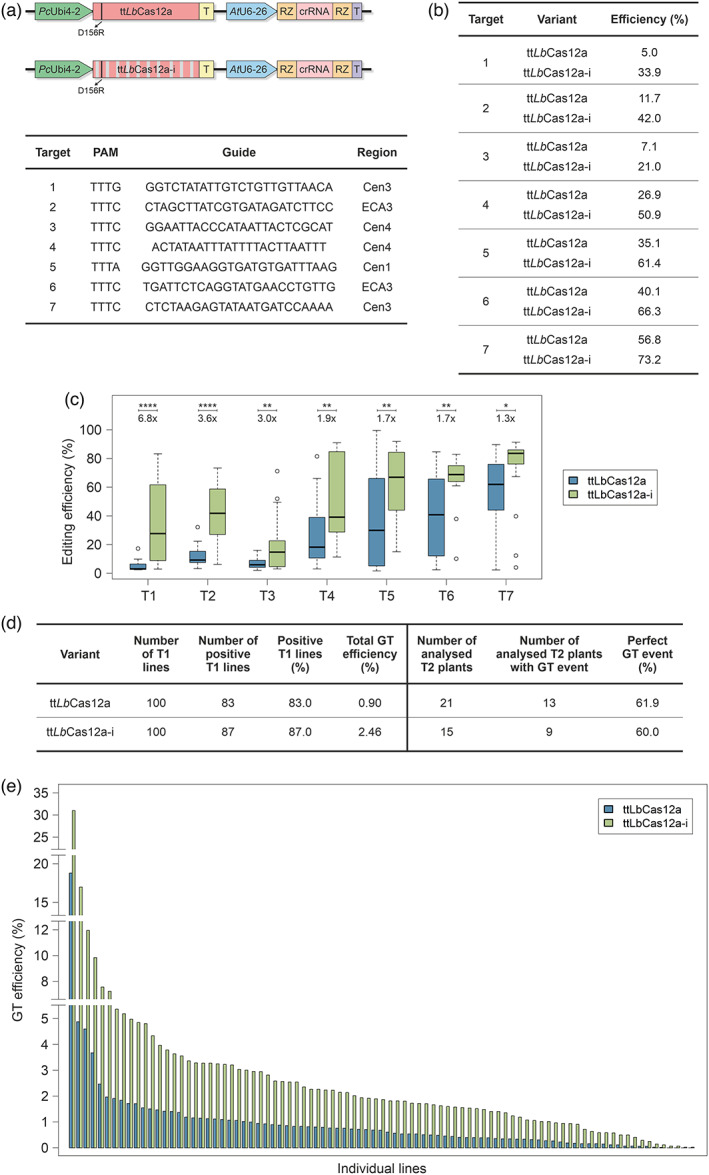
Improvement of gene editing and GT using ttLbCas12a‐i. (a) Cas12a expression cassettes for gene editing. The D156R mutation is highlighted, introns are represented by grey bars. The target sequences and regions are given. (b) Table showing the editing efficiencies determined by TIDE analysis as mean values for the respective target sites and variants. Editing efficiencies of the variants were determined for each of the tested target sites and individual transgenic plants grown at 22 °C. (c) Distribution of editing efficiencies of individual plants for both ttLbCas12a variants. Illustrated are the editing efficiencies of each of the analysed transgenic plants at the tested target sites (T1‐T7) for both variants at 22 °C. Data are presented as boxplots, each box represents the 25th and 75th percentile, the median is indicated by a black line, whiskers encompass the 1.5× interquartile range and data beyond, that is shown as outliers. Fold increases for ittLbCas12a‐i compared with ttLbCas12a at each target site are shown. T1: n = 20, T2: n = 20, T3: n = 17, T4: n = 17, T5: n = 18, T6: n = 20, T7: n = 20. P values were calculated using the one‐way ANOVA test: *P < 0.05, **P < 0.01, ****P < 0.0001. (d) Table showing the GT efficiencies and molecular analysis of GT events. The number of analysed T1 lines, the number and percentage of positive T1 lines and the total GT efficiency are shown on the left side of the table. The total GT efficiency is defined as the average of GT efficiencies of each individual line including the lines without GT events. The number of analysed positive T2 plants and the number and percentage of T2 plants with perfect GT events are shown on the right side of the table. (e) Distribution of GT efficiencies of the individual lines for both variants at 22 °C. The GT efficiencies of the individual positive lines are arranged in descending order for both variants. GT events were detected for 83 out of 100 and 87 out of 100 lines in case of ttLbCas12a and ttLbCas12a‐i, respectively.
Taken together, our results demonstrate that the introduction of introns into the coding sequence of ttLbCas12a dramatically improves both gene editing and gene targeting in Arabidopsis, so that even heterochromatic and other hardly accessible targets become editable. In an independent study, it has very recently been shown that the addition of 8 introns into the ttLbCas12a coding sequence can enhance editing efficiencies in barley (Lawrenson et al., 2022). Thus, in general, the inclusion of introns into ORFs of Cas nucleases seems to be a very promising way to enhance the editing efficiency in mono‐ as well as dicotyledonous plant species.
It has already been shown before that the inclusion of introns leads to higher mutation frequencies by Cas9 in Arabidopsis due to a related increase in protein levels (Grützner et al., 2021). We assume that the enhanced accessibility of heterochromatic sites to editing is also due to the presence of higher amounts of nuclease in the cells. This expression boost is promoter‐independent, making ttLbCas12a‐i a valuable novel tool for the plant community, enabling a wide variety of applications. It is especially promising in cases where the induction of several or multiple breaks with high efficiency is required, such as multiplexing. Moreover, the use of ttLbCas12a‐i might also improve gene editing efficiencies in other organisms that require cultivation temperatures similar to plants.
Conflict of interest
The authors declare no conflict of interest.
Author contribution
H.P. and A.T. conceived the research. P.S. and L.M. designed the analysis. T.S. designed the ttLbCas12a with introns. A.P. generated the original ttLbCas12a‐i construct. P.S. and L.M. executed the experiments. All authors wrote the article.
Supporting information
Appendix S1 Nucleotide sequence of the open reading frame of the intron‐containing ttLbCas12a‐i; all introns are highlighted in green.
Acknowledgements
We thank Aleyna Abinik and Carolin Brechtel for excellent technical assistance and Michelle Rönspies for critical reading of the manuscript. The ttLbCas12a‐i constructs (MoClo L0 Golden Gate module pAGT8163; AATG_ttLbCas12a‐i_TTCG) will be available via Addgene (ID 192453). Open Access funding enabled and organized by Projekt DEAL.
References
- Grützner, R. , Martin, P. , Horn, C. , Mortensen, S. , Cram, E.J. , Lee‐Parsons, C.W.T. , Stuttmann, J. et al. (2021) High‐efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun. 2, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T.‐K. , Armstrong, B. , Schindele, P. and Puchta, H. (2021) Efficient gene targeting in Nicotiana tabacum using CRISPR/SaCas9 and temperature tolerant LbCas12a. Plant Biotechnol. J. 19, 1314–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenson, T. , Hinchliffe, A. , Forner, M. and Harwood, W. (2022) Highly efficient genome editing in barley using novel Lb Cas12a variants and impact of sgRNA architecture. biorxiv, 10.1101/2022.04.28.489853v1 [DOI]
- Malzahn, A.A. , Tang, X. , Lee, K. , Ren, Q. , Sretenovic, S. , Zhang, Y. , Chen, H. et al. (2019) Application of CRISPR‐Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 17, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker, L. , Schindele, P. , Huang, T.‐K. , Wolter, F. and Puchta, H. (2020) Enhancing in planta gene targeting efficiencies in Arabidopsis using temperature‐tolerant CRISPR/LbCas12a. Plant Biotechnol. J. 18, 2382–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindele, P. and Puchta, H. (2020) Engineering CRISPR/LbCas12a for highly efficient, temperature‐tolerant plant gene editing. Plant Biotechnol. J. 18, 1118–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, T. , Crisp, P.A. , Rai, K.M. , Song, M. , Springer, N.M. and Zhang, F. (2022) Epigenetic features drastically impact CRISPR‐Cas9 efficacy in plants. Plant Physiol. 190, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Nucleotide sequence of the open reading frame of the intron‐containing ttLbCas12a‐i; all introns are highlighted in green.


