Summary
Molecular farming technology using transiently transformed Nicotiana plants offers an economical approach to the pharmaceutical industry to produce an array of protein targets including vaccine antigens and therapeutics. It can serve as a desirable alternative approach for those proteins that are challenging or too costly to produce in large quantities using other heterologous protein expression systems. However, since cost metrics are such a critical factor in selecting a production host, any system‐wide modifications that can increase recombinant protein yields are key to further improving the platform and making it applicable for a wider range of target molecules. Here, we report on the development of a new approach to improve target accumulation in an established plant‐based expression system that utilizes viral‐based vectors to mediate transient expression in Nicotiana benthamiana. We show that by engineering the host plant to support viral vectors to spread more effectively between host cells through plasmodesmata, protein target accumulation can be increased by up to approximately 60%.
Keywords: Molecular farming, plant‐based pharmaceutical, tobacco mosaic virus, viral expression vector, plasmodesmata, cell‐to‐cell movement
A transgenic N. benthamiana line (Sm5) engineered to facilitate cell‐to‐cell movement enhances the local and systemic spread of a fluorescently tagged TMV vector.
Introduction
Over the last three decades, plant systems have gradually gained attention as an alternative approach for heterologous protein production, especially in regard to proteins of pharmaceutical value. Early work focused primarily on the use of transgenic plants or plant cell cultures, with lead whole plant systems entering early phase clinical trials (Sohrab et al., 2017) and a plant cell system progressing all the way to product licensing and launch (Tekoah et al., 2015). Achieving economically competitive levels of expression has been a limitation with stable transgenic plant systems, although transplastomic systems have obtained much higher expression levels, for example, with Dengue virus antigens (van Eerde et al., 2019). Also, transgenic systems take extensive development times for each new target protein. Therefore, the main emphasis in the field has been on developing transient expression systems, primarily based on the use of Ti‐based or plant viral‐based vectors, predominantly Tobacco mosaic virus (TMV), introduced into the host Nicotiana species, most commonly through vacuum infiltration. Nicotiana benthamiana is the expression host of choice because this tobacco species is highly susceptible to several plant viruses, including TMV, and is susceptible to vacuum infiltration.
Key advances in the technology using the N. benthamiana system have centered around the development of higher performing vectors, such as hybrid vectors combining features of Agrobacterial and viral vectors (Marillonnet et al., 2005; Musiychuk et al., 2007), and more finely tuned plant expression hosts, such as those engineered to decorate heterologous proteins with glycans more representative of mammalian proteins (Montero‐Morales and Steinkellner, 2018). Using vacuum infiltration of N. benthamiana with Ti‐based or hybrid vectors, academic and commercial groups have advanced vaccine antigens into pivotal animal studies and early‐stage clinical trials (Chichester et al., 2012, 2018; Cummings et al., 2014; Landry et al., 2010; Pillet et al., 2016, 2018, 2019; Tuse et al., 2015; Ward et al., 2021a), with the most advanced examples progressing into Phase 3 clinical trials (Hager et al., 2022; Ward et al., 2020, 2021b), and the coronavirus example, Medicago's Covifenz, achieving regulatory approval in Canada.
To date, research on plant‐based transient expression technologies that include viral replication components have largely focused on the manipulation of the viral or hybrid vectors or on improvements to the transfection or transformation methodologies in an effort to expand the versatility of the vectors and increase the efficiency of viral infectivity and spread. Key advances have included optimizing vectors to increase their flexibility to accept larger open reading frames (ORFs) of interest, to co‐express multiple ORFs, as required for antibodies, to improve protein yields and to simplify the transfection process (Giritch et al., 2006; Hahn et al., 2015; Marillonnet et al., 2004; Shivprasad et al., 1999; Werner et al., 2011; Yusibov et al., 1999). By contrast, to the best of our knowledge, there has been no research to improve vector infectivity and dispersion in planta through engineering host modifications.
Plant viruses spread infectious materials between host cells by molecular movement through plasmodesmata. However, plants combat viral infection by recruiting an array of host defence proteins and inducing responses, which include altering plasmodesmal permeability (Lee and Lu, 2011). Among such host defence factors, Arabidopsis thaliana PLASMODESMATA‐LOCATED PROTEIN (PDLP) 5 is a highly potent plasmodesmal regulator that not only maintains basal plasmodesmal permeability but also acutely restricts overall molecular movement upon microbial pathogen infection (Cui and Lee, 2016; Lee et al., 2011; Lim et al., 2016; Wang et al., 2013). The mechanistic details behind PDLP5 function and its remarkable potency are not yet fully understood. However, our more recent findings have shown that PDLP5 forms homomeric multimers and heteromeric dimers/oligomers with other PDLP family members (Wang et al., 2020). This capability of forming multimers appears to be critical for PDLP5 activity, as alterations to the oligomeric state of PDLP5 result in a loss of its function regulating plasmodesmal permeability. In addition, PDLP5 deters systemic TMV spread by restricting the movement of the viral component p30 which is crucial for cell‐to‐cell transmission of the virus both in Arabidopsis and in N. benthamiana (Lee et al., 2011), which indicates that the mechanism by which PDLP5 regulates plasmodesmal permeability and restricts TMV movement is conserved between these plant species. Notably, while the presence of heterologously expressed wild‐type (WT) PDLP5 in N. benthamiana deters the movement of TMV, mutations of functional domains in PDLP5 result in the loss of this capability (Wang et al., 2020). These findings underscore that PDLP5 and its derivatives could serve as valuable molecular tools to manipulate TMV movement in Nicotiana expression systems.
In the current study, we describe an experimental approach, in which the N. benthamiana host plant system is genetically engineered to enhance the cell‐to‐cell movement of the TMV viral vector and hence to improve expression levels and target protein yields. We report that a semi‐dominant negative gain‐of‐function mutant form of PDLP5 enhances TMV infectivity and movement. Building upon this finding, we present proof‐of‐concept results to demonstrate: (i) how plasmodesmal connectivity can be manipulated in transgenic lines; (ii) how a viral vector can be utilized with these transgenic lines; and (iii) the efficacy of this system in expressing a reporter protein and two vaccine candidate antigens. Our results demonstrate that this novel approach is promising for improving yields for heterologous proteins and can likely be extended to other pharmaceutical protein targets.
Results
Ectopic expression of the PDLP5 mutant enhances viral spread in N. benthamiana
Mature PDLP5 protein consists of an N‐terminal extracellular domain of an unknown function and a transmembrane domain followed by a short cytosolic tail consisting of 14 amino acid residues (Figure 1a). Notably, PDLP5 contains three cysteine residues, C287, C288, and C298 in the cytosolic tail that are absent in seven other PDLP paralogs. We hypothesized that these cysteine residues have a functional significance that might pertain to the potency of PDLP5 as a plasmodesmal regulator. To test this hypothesis, we created three alanine substitution mutants targeting the cysteine residues of the cytosolic tail, namely 1C1A, 2C2A, and 3C3A, and fused them to Citrine yellow fluorescent protein (cYFP) (Figure 1a). These fusion constructs were transiently expressed in N. benthamiana leaves using agroinfiltration, and leaf epidermal cells were imaged using a confocal microscope. This experiment revealed that all three cysteine mutant fusion proteins were localized to plasmodesmata, similar to cYFP‐tagged intact PDLP5 (Figure 1b), which indicates that these cysteine residues are not critical for plasmodesmal localization.
Figure 1.
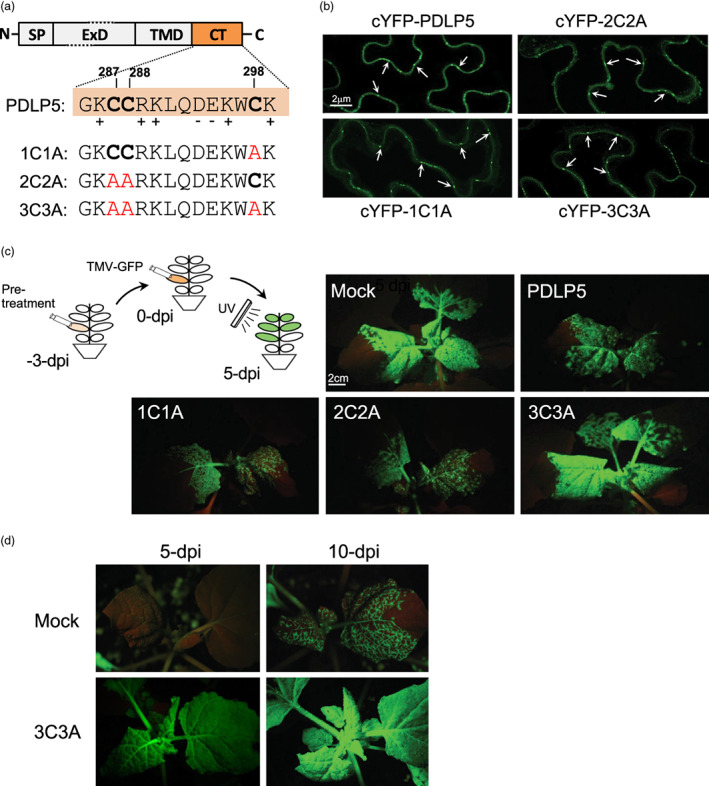
Transient expression of a PDLP5 mutant accelerates the systemic movement of TMV‐GFP in N. benthamiana. (a) Schematic representation of PDLP5 domain structure and the location of cysteine residues that are each mutated to alanine. (b) Representative confocal images showing that cysteine mutations do not alter plasmodesmal targeting of PDLP5. (c) A schematic diagram illustrating the experimental set‐up and representative photographs showing the effects of each PDLP5 mutant on the systemic movement of TMV‐GFP compared with those of mock (empty vector) and PDLP5 pre‐treatment. Two mature leaves (4th and 5th) of 4‐week‐old plants were agro inoculated by infiltrating them with the agrobacterial suspension diluted to OD600nm = 0.4. Three days after this pretreatment, the same leaves were infiltrated with Agrobacteria carrying TMV‐GFP vector, diluted to OD600nm = 0.2. The systemic spread of the vector was monitored 5 days following this step. (d) Photographs of systemic leaves showing that 3C3A enhances the spread of TMV‐GFP. Pre‐treatment and viral movement assays were performed as described for panel c except that the agrobacterial suspension carrying the TMV‐GFP vector was diluted to OD600nm = 0.004. The systemic spread of the vector was monitored over the next 5–10 days. Images in panels c and d were taken under a BlackRay UV lamp using a D3500 Nikon camera.
Next, to examine if the cysteine residues are crucial for PDLP5 activity, we performed Agrobacteria‐mediated viral movement assays using GFP‐tagged TMV (a recombinant TMV virus that moves systemically in N. benthamiana and expresses free GFP). We previously developed this viral movement assay to show that transient ectopic expression of PDLP5 in N. benthamiana leaves delays systemic TMV movement (Lee et al., 2011; Wang et al., 2020). Here, approximately 4‐week‐old N. benthamiana plants were infiltrated in two mature leaves with a mixture of two agrobacterial strains, one carrying a sequence encoding the silencing suppressor p19 and the other carrying a test sequence encoding either PDLP5 or mutant‐PDLP5 or carrying an empty vector (Figure 1c, schematic). Three days after this initial infiltration, the same leaves were infiltrated with another agrobacterial strain carrying TMV‐GFP at OD600nm = 0.2 to monitor viral spread. Five days later, systemic leaves were monitored under UV illumination to evaluate the effects of the specific pretreatments on the extent of virus spread. As expected, compared with empty vector (mock) control, ectopic expression of PDLP5 delayed systemic TMV movement (Figure 1c). Plants expressing 1C1A or 2C2A fusions also exhibited a similar delay in TMV movement, indicating that these mutations did not significantly affect PDLP5 activity (Figure 1c). In contrast, those expressing 3C3A showed no delay in viral movement, indicating that the cysteine residues may collectively be required for PDLP5 activity restricting TMV spread (Figure 1c). To further examine the effect of 3C3A on the systemic movement of TMV, we compared viral movement by introducing the Agrobacteria carrying the TMV‐GFP vector at a 50‐fold lower density (OD600nm = 0.004) and monitoring 5 and 10 days later. This experiment showed that compared with the mock treatment, 3C3A enhanced the systemic movement of TMV (Figure 1D).
Intrigued by the effect of 3C3A on facilitating TMV movement, we next examined how 3C3A may alter viral movement locally from cell to cell, closely monitoring viral movement in primary leaves that are infiltrated with Agrobacteria carrying TMV‐GFP at an earlier time point of 3 days (Figure 2a, schematic). Compared with the mock (empty vector) treatment, PDLP5‐expressing leaves formed a significantly lesser number of viral foci with low fluorescence intensities, which indicates that viral foci development was negatively impacted by PDLP5. This result is consistent with PDLP5's negative effect on systemic TMV spread. However, in stark contrast, 3C3A‐expressing leaves developed foci that were distinctively higher in number compared with the mock control (Figure 2a). This result suggests that the 3C3A mutation did not cause a loss of function of PDLP5 but rather conferred a gain of function. A later experiment showed that this gain‐of‐function by 3C3A is likely owing to its semi‐dominant negative capability; when expressed together with 3C3A, PDLP5's impact on TMV foci development was partially countered by 3C3A (Figure 2b).
Figure 2.
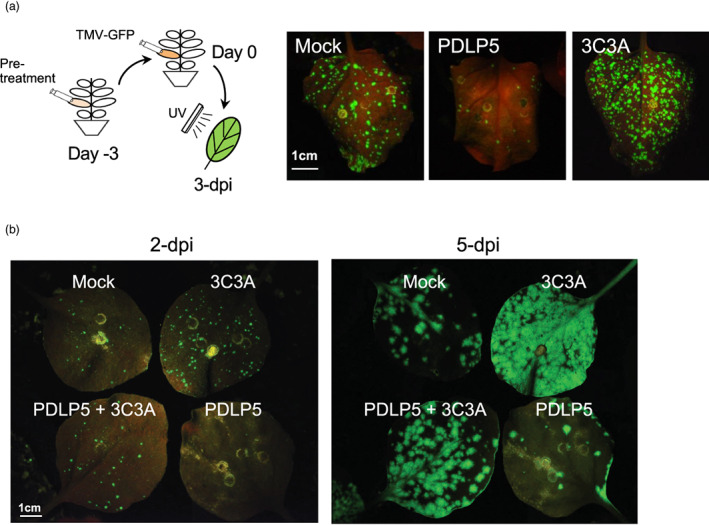
Transient expression of the PDLP5 mutant 3C3A accelerates foci development of TMV‐GFP in N. benthamiana. (a) A schematic diagram illustrating the experimental set‐up and representative photographs of leaves showing the effect of PDLP5 and 3C3A on TMV foci development. Agrobacterial strains used in pre‐treatment were diluted to OD600nm = 0.4 and that for TMV‐GFP to OD600nm = 0.02. (b) Representative fluorescent images showing the relative development of TMV‐GFP foci in leaves expressing PDLP5 or 3C3A or a 1:1 mixture of these. The final OD600nm of each agrobacterial strain is 0.5. Intact leaves were photographed without detaching from the plants. Mock, empty vector. Images were taken under a BlackRay UV lamp using a D3500 Nikon camera.
To assess whether the enhanced movement of TMV‐based vectors in the presence of 3C3A is limited to this virus or applies to other viral vectors, such as those derived from PVX that are also widely used for molecular farming applications, we introduced a PVX‐GFP vector following pre‐treatment of WT plants as described for the TMV‐GFP movement assay. This experiment revealed that 3C3A expression facilitated PVX‐GFP infection and movement both in local and systemic leaves (Figure S1a,b, respectively). This result indicates that the effect of ectopic expression of 3C3A on viral movement is likely generic.
Collectively, these results led to the identification of a PDLP5 mutant that promotes TMV invasiveness/foci development and viral movement, which therefore might serve as a useful tool to increase the yield of heterologous proteins expressed via a TMV‐based vector, and this PDLP5 mutant might also facilitate expression via other viral vectors.
Viral foci development and spread are enhanced in transgenic Nicotiana plants
To test the potential utility of the 3C3A mutant for TMV‐based heterologous expression for molecular farming purposes, we assessed the effect of an initial infiltration of N. benthamiana with Agrobacteria carrying 3C3A on the expression of GFP driven by a subsequent infiltration of Agrobacteria carrying TMV‐GFP. While an initial infiltration with PDLP5 appeared to slightly repress GFP expression, compared to a mock initial infiltration, an initial infiltration with 3C3A appeared to facilitate enhanced expression of GFP (Figure S2).
To overcome the requirement for multiple infiltrations and to attempt to establish host lines engineered for enhanced viral vector movement, we next generated transgenic N. benthamiana plants in which 3C3A was stably overexpressed (hereafter referred to as Sm5 plants). For this, a binary vector, pBI‐D‐PDLP5m5 carrying a 35 S promotor with dual enhancers and a 35 S terminator was constructed to drive the expression of 3C3A (Figure 3A). Following the co‐culture of leaf segments with an agrobacterial strain transformed with pBI‐D‐PDLP5m5, a total of 15 independent Sm5 lines were produced. These lines were tested for genomic integration of 3C3A (PCR) and expression of 3C3A transcripts (RT‐PCR) and T1 seedlings were tested for antibiotic resistance (Table S2). Among these, nine lines were chosen for TMV‐GFP foci growth analysis in T1 and T2 generations, which showed that they were all positive for the enhanced foci phenotype to varying degrees (Table S2).
Figure 3.
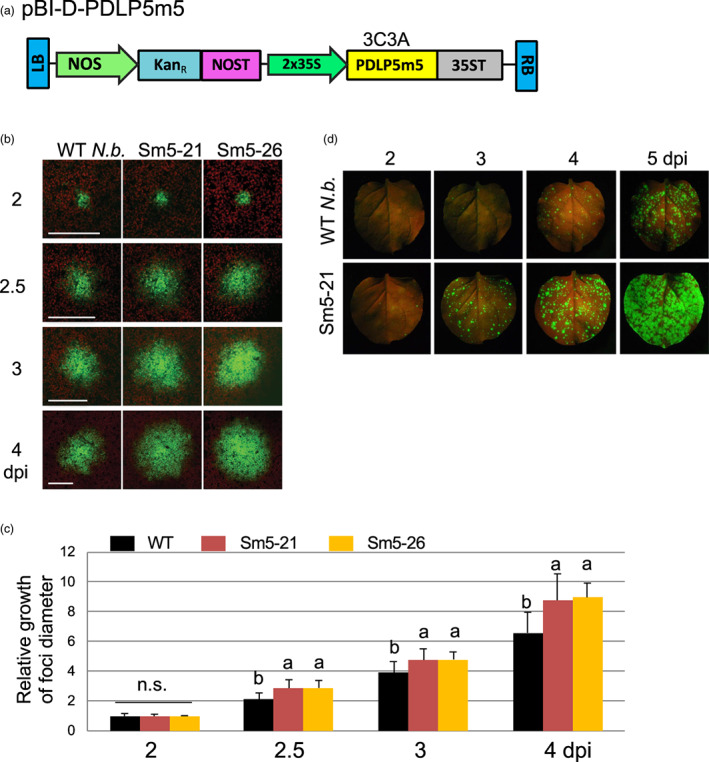
Transgenic Sm5 (3C3A) plants facilitate cell‐to‐cell viral movement. (a) Schematic representation of the T‐DNA portion of the plasmid pBI‐D‐PDLP5m5. LB and RB, left and right T‐DNA borders, respectively. NOS and NOST, Agrobacterium nopaline synthase promoter and terminator, respectively. KanR, neomycin phosphotransferase II, conferring resistance to kanamycin. 2x35S, two tandemly ordered Cauliflower mosaic virus 35 S promoters with the Tobacco etch virus leader sequence. 35ST, Cauliflower mosaic virus 35 S terminator. (b) Representative confocal images showing TMV‐GFP foci growth in select Sm5 lines compared to WT. The agrobacterial suspension was diluted to OD600nm = 0.001. Ten foci were selected for monitoring per line from multiple plant samples. Size bars, 100 μm. (c) Quantitative results of the foci growth assays. At least 10 foci selected from multiple plants were monitored per genetic background in each experiment and the experiments were repeated at least two times. Error bars, standard deviation. Levels not connected by the same letters are significantly different at α = 0.05 according to the LSD test following one‐way ANOVA. n.s., no significance. (d) Representative photographs showing TMV‐GFP foci development in Sm5‐21 compared to WT N.b. leaves. Mature leaves of 4‐week‐old plants were agro inoculated by infiltrating them with the agrobacterial suspension diluted to OD600nm = 0.001. Following agroinfiltration, the development of TMV‐GFP foci per leaf was monitored for 5 days following inoculation. Photos were taken under a BlackRay UV lamp using a D3500 Nikon camera.
For additional phenotyping, the two lines were chosen that showed the most enhanced foci phenotype (Sm5‐21 and ‐26), and these grew and developed similarly to WT plants, (Figure S3). These plants were infiltrated with Agrobacteria to monitor TMV‐GFP foci growth under a confocal microscope over a time course from 2‐ to 4‐days post infiltration (dpi). They all exhibited enhanced TMV foci growth from 2.5 dpi compared to WT plants, and by 4 dpi the differences were pronounced (Figure 3b,c). We performed a statistical analysis of the foci growth data with one‐way ANOVA. Reported P‐values for the dataset at 2‐, 2.5‐, 3‐ and 4‐dpi were 0.93, 4.7 e‐7, 5.5 e‐5 and 1.7 e‐6, respectively. To determine at which time point, Sm5 lines may significantly differ from WT, we then performed Fisher's LSD as a post hoc test. This analysis showed that while Sm5 lines were not significantly different from each other, they were significantly different from WT, starting from 2.5 dpi and onward. An additional analysis using Sm5‐21 plants showed that the TMV foci development was clearly enhanced at the macroscopic level (Figure 3d), further substantiating the microscopic data. Notably, this effect also extended to PVX. The enhanced movement of a PVX‐based vector that was seen in plants that had been transiently transformed with 3C3A (Figure S1a,b) was also seen in transgenic Sm5‐21 plants (Figure 1c), further supporting the generic effect of 3C3A on viral vector movement.
Collectively, these results corroborate that stable ectopic expression of 3C3A correlates with the rate of foci development and is thus an effective means to enhance viral infectivity and spread in N. benthamiana. Of note, 3C3A had no clearly discernable effect on the expression of GFP when the reporter was carried on a non‐viral binary vector (Figure S4). This is not surprising, since such non‐viral vectors do not typically replicate and spread cell‐to‐cell.
Expression of vaccine antigens is enhanced in Sm5 plants
To further test the efficacy of Sm5 plants in improving the accumulation of heterologously expressed proteins, we focused on two pharmaceutically relevant vaccine antigens, YFE‐1 and PA83. YFE‐1 comprises amino acids 286 to 682 of the polyprotein precursor of Yellow fever virus and PA83 comprises amino acids 30 to 764 of the protective antigen of Bacillus anthracis. These are major antigen targets for vaccine development for yellow fever (Tottey et al., 2018) and anthrax (Chichester et al., 2013), respectively, and the plant‐produced antigens have passed through extensive preclinical evaluation to demonstrate efficacy in animal models, with the PA83 antigen having completed a clinical trial (Paolino et al., 2022). Both proteins were expressed from hybrid vectors of the general type described in Musiychuk et al., 2007, combining TMV and agrobacterial plasmid features, and rely on TMV replication and movement protein functions for amplification and movement in infiltrated aerial plant tissues (Figure 4A). While processes for the expression of these two targets had already been extensively optimized to achieve the relatively high levels of expression necessary to advance them through preclinical development, further improvements in expression will further enhance the cost competitiveness of their production in plants, and we therefore tested their expression in transgenic plants expressing 3C3A.
Figure 4.
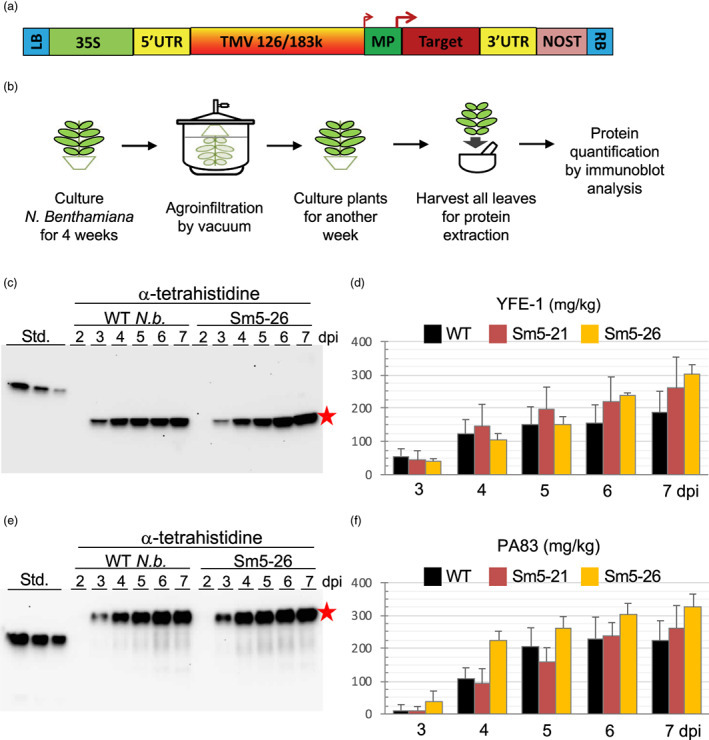
Expression of recombinant protein targets in transgenic 3C3A plants. (a) Schematic representation of the T‐DNA portion of the hybrid vector used to express the recombinant protein targets YFE‐1 and PA83. LB and RB, left and right T‐DNA borders, respectively. 35 S Cauliflower mosaic virus 35 S promoter. NOST, Agrobacterium nopaline synthase terminator. TMV, Tobacco mosaic virus genome including sequences encoding the viral replicase (126/183) and movement protein (MP) and subgenomic promoters (arrows) and 5′ and 3′ untranslated regions (5′UTR and 3′UTR). Target, sequence encoding YFE‐1 or PA83 inserted into the TMV genome in place of sequence encoding the TMV coat protein. (b) Schematic representation of the process used to uniformly introduce A. tumefaciens carrying genes encoding YFE‐1 and PA83 into N. benthamiana leaves and then assess expression levels for these targets. (c and d) Immunoblot analysis across a time course from 2–7 dpi for YFE‐1 expression following vacuum infiltration of WT and Sm5‐26 plants. (e and f) Immunoblot analysis across a time course from 2 to 7 dpi for PA83 expression following vacuum infiltration of WT and Sm5‐26 plants. As a positive control for immunoblotting, a recombinant protein with a poly‐histidine tag (Standard) was used. Red stars on panels c and e indicate the expressed recombinant proteins and error bars on panels d and f show standard deviation values. The expression data shown in panels d and f are from replicates of n = 2–6 and panels c and e show representative immunoblot images.
To express YFE‐1 and PA83, agrobacterial cultures carrying the vector constructs encoding these targets were introduced into 5–6‐week‐old WT, Sm5‐21 and ‐26 plants by vacuum infiltration, and all leaves were collected from 3‐ to 7‐days post‐infiltration, from which soluble protein was extracted and subjected to immunoblot analysis (Figure 4b). In WT plants, YFE‐1 protein was detected at approximately 56 mg/kg by 3‐dpi, increased to approximately 121 mg/kg by 4 dpi, but then only increased to approximately 184 mg/kg by 7‐dpi (Figures 4c,d and S5a). By contrast, although the two Sm5 lines showed similar or even slightly reduced levels of YFE‐1 to those for WT at 3‐dpi, target accumulation increased to a greater extent than for WT over the next 4 days, reaching approximately 260 and 302 mg/kg by 7‐dpi. In summary, Sm5‐21 and Sm5‐26 plants achieved an approximately 40% and 63% increase in accumulation of YFE‐1, respectively, compared to WT plants (Figure S5a).
PA83 accumulation in WT plants showed a broadly similar pattern to YFE‐1 accumulation, with an initial burst in expression through to 5‐dpi followed by only a minor further increase (Figure 4e,f). Interestingly, while target accumulation was similar in Sm5‐21 plants to WT plants, only showing about a 17% increase over WT plants at the peak of expression, Sm5‐26 plants showed a more striking improvement in the level of target accumulation, reaching over two‐fold higher expression by 4‐dpi and with peak target accumulation approximately 46% greater than for WT plants (approximately 326 compared to 224 mg/kg) (Figure S5a).
To assess if improved target accumulation is reflected in protein yield, vectors encoding YFE‐1 and PA83 were introduced into N. benthamiana plants by vacuum infiltration, 50 g of aerial tissue was harvested at 6‐dpi, soluble protein was extracted from plant tissue and the poly‐histidine tagged antigen targets were recovered by IMAC. Chromatography absorbance profiles showed a larger peak for the elution fraction for PA83 extracted from Sm5‐26 plants than from WT plants (Figure S5b). Quantification of YFE‐1 and PA83 recovered from infiltrated plants was achieved by comparing a dilution series of recovered protein to that of a protein standard on sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS–PAGE) gels stained with Coomassie blue (Figure S5c,d, respectively). Sm5‐26 plants could achieve an approximately 40% increase in recovery of YFE‐1 compared to WT plants, and an approximately 20% increase in recovery of PA83 compared to WT plants. These results indicate the potential for the Sm5 lines generated here for improved heterologous protein production in N. benthamiana.
Discussion
High‐performing hybrid expression vectors that utilize the viral replication machinery of TMV to achieve high levels of transcript typically carry a portion of the viral genome encoding a replicase and a movement protein. These vectors have a component of the genome encoding the coat protein that is non‐essential for replication or cell‐to‐cell movement and hence it is replaced with the ORF encoding the protein target to be expressed (Marillonnet et al., 2004, 2005; Musiychuk et al., 2007). While the coat protein is required for systemic movement, it is not required for the local cell‐to‐cell spread of the virus (Hilf and Dawson, 1993; Saito et al., 1990). Loss of systemic infection is tolerable because whole plant infection can be achieved using vacuum infiltration of intact plants with Agrobacteria carrying genes encoding viral components on a plasmid. This method of introducing vector sequences typically utilizes a high density of Agrobacterium cells to increase the coverage of infected host cells. However, higher concentrations of Agrobacteria can be detrimental to leaf tissue, leading to biotic stresses and even necrosis, resulting in undesirably low yields of heterologous proteins (Shamloul et al., 2014). Thus, further improvements to the expression system, allowing the vectors to more rapidly move cell‐to‐cell to cover all available leaf tissue, would potentially allow for reduced concentrations of Agrobacteria to be utilized.
To the best of our knowledge, the idea of exploiting a host mechanism through which molecular movement via plasmodesmata is regulated has not previously been applied to molecular farming. We acknowledge that finding functionally critical amino acid residues that confer a dominant‐negative gain‐of‐function modality to the master plasmodesmal regulator PDLP5 was a key to realizing this idea. Functional dissection of PDLP5 via domain mutagenesis indicates that the extracellular domain and the transmembrane domain are both required for its function of restricting molecular movement through plasmodesmata (Wang et al., 2020). However, introducing any of these mutations into PDLP5 led to the loss of its function. The exploration of targeting the C‐terminal cysteine residues for mutation was initially conceived as a part of efforts to identify amino acid residues that are vital for the plasmodesmal localization of PDLP5, as it was and is still unknown what molecular features of PDLP5 determine that localization. These cysteine residues were shown to be dispensable for localization but unexpectedly turned out to be quite potent in counteracting the endogenous mechanism that acts to keep viral movement at bay as much as possible. We speculate that these three cysteine residues likely participate in protein–protein interactions in the cytosol that might be required for downstream signalling pathways leading to changes in plasmodesmal permeability and/or immunity. The exact molecular mechanism behind the function of these residues warrants future investigation because this knowledge could lead to the identification of additional molecular players that might be even more potent in terms of altering plant intercellular communication, viral infectivity, and rate of viral spread in leaf tissue.
As demonstrated in the current study, manipulating plant intercellular trafficking systems seems to hold great potential to benefit molecular farming using viral vector systems for heterologous protein expression in N. benthamiana. Our protein production analysis provides a proof‐of‐concept for manipulating intracellular connectivity to enhance the production of pharmaceutically valuable proteins using TMV‐based vectors. Compared to non‐genetically engineered WT plants, our highest performing transgenic line produced from 46% to 63% more vaccine antigen over a 7‐day period. Such an improvement is significant for vaccine antigens where costs of production often dictate the feasibility of advancing lead candidates into clinical development and ultimately into large‐scale production and distribution. We speculate that the yield of other target proteins, such as antibodies, may also be improved using the combination of viral‐based vector and Sm5 lines. Examining how this system might benefit the production of high‐value proteins, especially those that are slow to accumulate or are toxic to the host plant, may also prove to be a fruitful endeavour. Furthermore, the utility of Sm5 lines might be extended to other (non‐TMV‐based) viral vector systems given that N. benthamiana is susceptible to many other plant viruses and these engineered plants may similarly facilitate the movement of other viral vectors and enhance infectivity. Indeed, we demonstrate here the enhanced movement of a PVX‐based vector. The general approach outlined in this study should also be applicable to alternative plant hosts (Green et al., 2009) when implemented as a convenient transient co‐infiltration scheme.
In conclusion, our current study provides experimental evidence that engineering host cell‐to‐cell movement to bolster viral vector infectivity and movement in host plants is a promising approach to improving viral vector‐mediated heterologous protein expression. Successful application of this approach to a wider range of protein targets and viral vector systems will further validate the concept of manipulating host plant cell‐to‐cell movement functions and such an approach may also be applicable to other plant viral‐based expression systems.
Materials and methods
Plant materials and growth conditions
For initial selection, transgenic N. benthamiana lines were germinated and selected on Murashige and Skoog plates containing 100 mg/L kanamycin, and transgenic seedlings were transferred into the soil after 10 days. For seed collection, these plants were transferred into larger pots and allowed to complete their life cycle in a greenhouse. For syringe infiltration and viral infection assays, plants were grown for 3–4 weeks in soil. For this purpose, WT and transgenic N. benthamiana seeds were first germinated in the dark for 5 days, and young seedlings were transferred into individual pots and grown in a Conviron walk‐in growth chamber (GR Series, Controlled Environments Inc.) at 24 °C, 70% humidity, and under 16 h light/8 h dark conditions. For vacuum infiltration, plants were grown for 5–6 weeks in a hydroponic system using rockwool as the support matrix (Shamloul et al., 2014).
Plasmid construction
Fluorescently tagged fusion constructs of PDLP5 (UniProt Q8GUJ2) or its cytoplasmic tail mutants were produced using overlapping PCR by Phusion high‐fidelity DNA polymerase (New England Biolabs), and purified DNA fragments were subsequently cloned into the Gateway entry vector pENTR/D‐TOPO (ThermoFisher Scientific) and destination vector pGWB. To create pBI‐D vector expression cassettes, sequences from the pBI121 binary vector were replaced by expression cassettes containing a Cauliflower mosaic virus (CaMV) 35 S promoter with dual enhancers, a 5′ nontranslated leader sequence from Tobacco etch virus (Carrington and Freed, 1990), and a CaMV 35 S terminator. The PDLP5 3C‐3A mutant was cloned into the binary vector pBI‐D, introduced into the Agrobacterium tumefaciens strain GV3101 by electroporation, and used to create transgenic N. benthamiana lines. Optimized Yellow fever virus envelope protein (YFE‐1) (UniProt P03314) and B. anthracis protective antigen (PA83) (UniProt P13423) genes were cloned into a pGreen‐based expression vector carrying TMV genome sequences from pBID4 (Musiychuk et al., 2007). The resulting constructs were introduced into the A. tumefaciens strain AGL1 by electroporation. All constructs were confirmed by Sanger sequencing before agro transformation. Additional information regarding plasmids and vectors used in the current study is provided in Table S1.
Agrobacterium‐mediated infiltration and plant transformation
Transformed Agrobacteria were cultured in Luria–Bertani (LB) liquid media at 28 °C and resuspended in infiltration buffer containing 10 mM MES, pH5.7, 10 mM MgCl2, and 200 μM acetosyringone. For subcellular localization, the concentration of resuspended Agrobacteria was adjusted to OD600nm = 0.5. For the viral infection study, two mature leaves of 4‐week‐old plants were infiltrated using a needleless syringe initially with Agrobacteria carrying genes encoding target proteins and the p19 silencing suppressor of Tomato bush stunt virus, each at OD600nm = 1.0 and mixed 1:1 for co‐infiltration. Three days later, the same leaves were subsequently infiltrated with Agrobacteria carrying TMV‐GFP at OD600nm = 0.2 for non‐microscopy visualization of expression and at OD600nm = 0.001 for visualization by microscopy, unless indicated otherwise in relevant figure legends. Infiltrated plants were covered and incubated in a walk‐in growth chamber overnight and uncovered 12–16 h later. Agrobacteria carrying plasmid constructs encoding Yellow fever virus and B. anthracis antigens were introduced into N. benthamiana by vacuum infiltration, as described previously using A. tumefaciens cultures at an OD600nm = 0.5 for the target and OD600nm = 0.1 for the P1/HC‐Pro silencing suppressor of Turnip mosaic virus (Shamloul et al., 2014). Transgenic N. benthamiana plants were generated by the transformation of leaf discs (Horsch et al., 1985).
Molecular analysis of transgenic plants
Genomic DNA and total RNA were isolated from WT and transgenic N. benthamiana plants regenerated on antibiotic selection media using DNeasy Plant and RNeasy Plant mini kits (Qiagen), respectively. To confirm transgene incorporation into the genome, PCR was performed on genomic DNA using A. thaliana PDLP5 gene‐specific primers, and to confirm transgene expression, RT‐PCR was performed on total RNA, also using A. thaliana PDLP5 gene‐specific primers. Segregation analysis was performed to identify transformed lines with a single copy of the transgene integrated into the plant genome. Transgenic plants were grown to maturity and self‐fertilized. Segregation analysis was performed on T1 seeds harvested from these plants. For each plant, 50 seeds were surface sterilized and allowed to germinate in the dark for 5 days on media supplemented with 100 mg/L kanamycin, before moving to the light. After 3–4 weeks, resistant seedlings were identified as having developed roots and green cotyledons and leaves. For seed sets showing a 3:1 ratio of resistant to sensitive seedlings, five resistant seedlings were grown to maturity and self‐fertilized. Segregation analysis was performed on T2 seeds harvested from these T1 plants, as above. Seed sets that were uniformly resistant to kanamycin were considered to be the progeny of homozygous T1 transgenic plants with a single site of transgene integration. This was then confirmed through a further generation of plant growth, self‐fertilization, and segregation analysis on T3 seed sets.
Confocal microscopy
Confocal images of subcellular localization and foci development were taken using a Zeiss LSM 510 META scanhead on a Zeiss LSM 5 DUO confocal microscope. Plant samples were placed into single‐well Lab‐Tek®II Chambers (#1.5 German Coverglass System, Cat# 155360), covered with coverslips, and flattened with a glass weight. YFP or GFP fluorescence was visualized using a C‐Apochromat X40/1.20‐W Korr UV–VIS‐IR objective with the 488 nm Argon laser and 505–550 nm band‐pass emission filter. For representative localization images, series of optical sections were acquired as Z‐stacks and rendered as 3‐D projections with a Zeiss LSM Image Examiner.
Viral infection assays
TMV‐GFP infection assays were performed using two mature leaves of each of 10 N. benthamiana plants. For WT plants, Agrobacteria carrying genes encoding PDLP5 or the 3C‐3A mutant construct were co‐infiltrated with Agrobacteria carrying the viral silencing suppressor protein p19. A subsequent infiltration using Agrobacteria carrying TMV‐GFP or PVX‐GFP (Table S1) was performed 3 days after the initial infiltration on the same leaves. For the initial infiltration, whole leaves were always fully infiltrated. The subsequent infiltration was done at a smaller but consistent volume across the plant samples to ensure it was done within the area of each leaf that received initial infiltration. To monitor systemic viral movement, lower leaves were either nearly fully infiltrated or spot infiltrated with Agrobacteria carrying the viral vector. For the spot infiltration, each leaf was inoculated with two injections as separate spots, each injection corresponding to 0.1 mL of agrobacterial suspension. To evaluate viral movement in transgenic plants, Sm5 plants were infiltrated only with Agrobacteria carrying a viral vector. The extent of local infectivity and systemic viral movement of the virus was recorded by photographing the plants under UV illumination through a deep yellow filter mounted on a Nikon D3100 DSLR camera. Alternatively, confocal microscopy (Zeiss LSM 5 DUO) was performed for GFP visualization using a 488 nm Argon laser and 505–550 nm band‐pass emission filter.
Protein extraction, purification, and quantification
The Yellow fever virus envelope protein antigen, YFE‐1, and the B. anthracis protective antigen, PA83, were targeted to the endoplasmic reticulum and carried poly‐histidine tags. For expression analyses, all leaves infiltrated with Agrobacteria were collected and ground for protein extraction. Soluble proteins were extracted in a Tris‐based extraction buffer for YFE‐1 or a phosphate‐based extraction buffer for PA83. Target expression levels were then assessed by SDS–PAGE followed by immunoblot analysis using a tetrahistidine‐specific monoclonal antibody (Qiagen), with a dilution series of a recombinant protein with a poly‐histidine tag providing a standard curve for quantification. For target protein purification, extracts were clarified by filtration through miracloth and centrifugation at 6800 g , and the target protein was recovered by immobilized metal affinity chromatography. YFE‐1 and PA83 yields were then assessed by SDS–PAGE followed by staining with Coomassie blue, with a dilution series of BSA providing a standard curve for quantification of an appropriate dilution of each target molecule that lay within the standard curve. The G:BOX Mini Gel and Blot Documentation System (Syngene) was used for target quantification by immunoblot analysis and SDS–PAGE.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Author contributions
X.W., A.I.P., S.J.S. and J.‐Y.L. contributed to the writing (X.W. and J.‐Y.L. drafted the manuscript; A.I.P. and S.J.S. wrote parts; J.‐Y.L. and S.J.S. revised and edited the manuscript). A.I.P. and J.‐Y.L. conceived the research idea; X.W., A.I.P., M.S. and M.A.R. conducted experiments; X.W., A.I.P., S.J.S. and J.‐Y.L. contributed to the experimental designs. X.W., A.I.P, S.J.S. and J.‐Y.L. analysed and interpreted experimental data. X.W. and A.I.P. are joint first authors.
Supporting information
Figure S1. Evaluation of the effect of 3C3A on PVX‐GFP movement in local and systemic leaves.
Figure S2. Evaluation of the effect of 3C3A by transient co‐expression on TMV vector‐driven protein accumulation.
Figure S3. Photos showing similar growth of N.b. Sm5 lines to WT plants.
Figure S4. Evaluation of non‐viral vector‐driven protein expression by Western blot analysis.
Figure S5. Evaluation of protein yields at a larger scale using vectors encoding YFE‐1 and PA83.
Table S1. Information related to vectors used in this study.
Table S2. A summary of the evaluation of transgenic Sm5 lines.
Acknowledgements
We thank J. Caplan (Univ. Delaware) for sharing the GV2260 agrobacterial cell carrying the TMV‐GFP virus. This research was supported primarily by Fraunhofer USA under a task agreement awarded to J.‐Y. Lee and A.I. Prokhnevsky made under a partnership agreement between Fraunhofer USA and the University of Delaware and partially by the National Science Foundation (awarded to J.‐Y. Lee: NSF MCB‐1820103 and IOS‐2054685).
References
- Carrington, J.C. and Freed, D.D. (1990) Cap‐independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J. Virol. 64, 1590–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichester, J.A. , Green, B.J. , Jones, R.M. , Shoji, Y. , Miura, K. , Long, C.A. , Lee, C.K. et al. (2018) Safety and immunogenicity of a plant‐produced Pfs25 virus‐like particle as a transmission blocking vaccine against malaria: A Phase 1 dose‐escalation study in healthy adults. Vaccine 36, 5865–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichester, J.A. , Jones, R.M. , Green, B.J. , Stow, M. , Miao, F. , Moonsammy, G. , Streatfield, S.J. et al. (2012) Safety and immunogenicity of a plant‐produced recombinant hemagglutinin‐based influenza vaccine (HAI‐05) derived from A/Indonesia/05/2005 (H5N1) influenza virus: a phase 1 randomized, double‐blind, placebo‐controlled, dose‐escalation study in healthy adults. Viruses 4, 3227–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichester, J.A. , Manceva, S.D. , Rhee, A. , Coffin, M.V. , Musiychuk, K. , Mett, V. , Shamloul, M. et al. (2013) A plant‐produced protective antigen vaccine confers protection in rabbits against a lethal aerosolized challenge with Bacillus anthracis Ames spores. Hum. Vaccin. Immunother. 9, 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, W. and Lee, J.Y. (2016) Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat Plants 2, 16034. [DOI] [PubMed] [Google Scholar]
- Cummings, J.F. , Guerrero, M.L. , Moon, J.E. , Waterman, P. , Nielsen, R.K. , Jefferson, S. , Gross, F.L. et al. (2014) Safety and immunogenicity of a plant‐produced recombinant monomer hemagglutinin‐based influenza vaccine derived from influenza A (H1N1)pdm09 virus: a Phase 1 dose‐escalation study in healthy adults. Vaccine 32, 2251–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giritch, A. , Marillonnet, S. , Engler, C. , van Eldik, G. , Botterman, J. , Klimyuk, V. and Gleba, Y. (2006) Rapid high‐yield expression of full‐size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc. Natl. Acad. Sci. U. S. A. 103, 14701–14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, B.J. , Fujiki, M. , Mett, V. , Kaczmarczyk, J. , Shamloul, M. , Musiychuk, K. , Underkoffler, S. et al. (2009) Transient protein expression in three Pisum sativum (green pea) varieties. Biotechnol. J. 4, 230–237. [DOI] [PubMed] [Google Scholar]
- Hager, K.J. , Perez Marc, G. , Gobeil, P. , Diaz, R.S. , Heizer, G. , Llapur, C. , Makarkov, A.I. et al. (2022) Efficacy and Safety of a Recombinant Plant‐Based Adjuvanted Covid‐19 Vaccine. N. Engl. J. Med. 386, 2084–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, S. , Giritch, A. , Bartels, D. , Bortesi, L. and Gleba, Y. (2015) A novel and fully scalable Agrobacterium spray‐based process for manufacturing cellulases and other cost‐sensitive proteins in plants. Plant Biotechnol. J. 13, 708–716. [DOI] [PubMed] [Google Scholar]
- Hilf, M.E. and Dawson, W.O. (1993) The tobamovirus capsid protein functions as a host‐specific determinant of long‐distance movement. Virology 193, 106–114. [DOI] [PubMed] [Google Scholar]
- Horsch, R.B. , Fry, J.E. , Hoffmann, N.L. , Eichholtz, D. , Rogers, S.G. and Fraley, R.T. (1985) A Simple and General‐Method for Transferring Genes into Plants. Science 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Landry, N. , Ward, B.J. , Trepanier, S. , Montomoli, E. , Dargis, M. , Lapini, G. and Vezina, L.P. (2010) Preclinical and clinical development of plant‐made virus‐like particle vaccine against avian H5N1 influenza. PLoS One 5, e15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y. and Lu, H. (2011) Plasmodesmata: the battleground against intruders. Trends Plant Sci. 16, 201–210. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y. , Wang, X. , Cui, W. , Sager, R. , Modla, S. , Czymmek, K. , Zybaliov, B. et al. (2011) A plasmodesmata‐localized protein mediates crosstalk between cell‐to‐cell communication and innate immunity in Arabidopsis. Plant Cell 23, 3353–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, G.H. , Shine, M.B. , de Lorenzo, L. , Yu, K. , Cui, W. , Navarre, D. , Hunt, A.G. et al. (2016) Plasmodesmata Localizing Proteins Regulate Transport and Signaling during Systemic Acquired Immunity in Plants. Cell Host Microbe 19, 541–549. [DOI] [PubMed] [Google Scholar]
- Marillonnet, S. , Giritch, A. , Gils, M. , Kandzia, R. , Klimyuk, V. and Gleba, Y. (2004) In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. U. S. A. 101, 6852–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillonnet, S. , Thoeringer, C. , Kandzia, R. , Klimyuk, V. and Gleba, Y. (2005) Systemic Agrobacterium tumefaciens‐mediated transfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 23, 718–723. [DOI] [PubMed] [Google Scholar]
- Montero‐Morales, L. and Steinkellner, H. (2018) Advanced Plant‐Based Glycan Engineering. Front. Bioeng. Biotechnol. 6, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiychuk, K. , Stephenson, N. , Bi, H. , Farrance, C.E. , Orozovic, G. , Brodelius, M. , Brodelius, P. et al. (2007) A launch vector for the production of vaccine antigens in plants. Influenza Other Respi. Viruses 1, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolino, K.M. , Regules, J.A. , Moon, J.E. , Ruck, R.C. , Bennett, J.W. , Remich, S.A. , Mills, K.T. et al. (2022) Safety and immunogenicity of a plant‐derived recombinant protective antigen (rPA)‐based vaccine against Bacillus anthracis: A Phase 1 dose‐escalation study in healthy adults. Vaccine 40, 1864–1871. [DOI] [PubMed] [Google Scholar]
- Pillet, S. , Aubin, E. , Trepanier, S. , Bussiere, D. , Dargis, M. , Poulin, J.F. , Yassine‐Diab, B. et al. (2016) A plant‐derived quadrivalent virus like particle influenza vaccine induces cross‐reactive antibody and T cell response in healthy adults. Clin. Immunol. 168, 72–87. [DOI] [PubMed] [Google Scholar]
- Pillet, S. , Aubin, E. , Trepanier, S. , Poulin, J.F. , Yassine‐Diab, B. , Ter Meulen, J. , Ward, B.J. et al. (2018) Humoral and cell‐mediated immune responses to H5N1 plant‐made virus‐like particle vaccine are differentially impacted by alum and GLA‐SE adjuvants in a Phase 2 clinical trial. NPJ Vaccines 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillet, S. , Couillard, J. , Trepanier, S. , Poulin, J.F. , Yassine‐Diab, B. , Guy, B. , Ward, B.J. et al. (2019) Immunogenicity and safety of a quadrivalent plant‐derived virus like particle influenza vaccine candidate‐Two randomized Phase II clinical trials in 18 to 49 and >/=50 years old adults. PLoS One 14, e0216533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, T. , Yamanaka, K. and Okada, Y. (1990) Long‐distance movement and viral assembly of tobacco mosaic virus mutants. Virology 176, 329–336. [DOI] [PubMed] [Google Scholar]
- Shamloul, M. , Trusa, J. , Mett, V. and Yusibov, V. (2014) Optimization and utilization of Agrobacterium‐mediated transient protein production in Nicotiana. J. Vis. Exp. 86, 51204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivprasad, S. , Pogue, G.P. , Lewandowski, D.J. , Hidalgo, J. , Donson, J. , Grill, L.K. and Dawson, W.O. (1999) Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus‐based vectors. Virology 255, 312–323. [DOI] [PubMed] [Google Scholar]
- Sohrab, S.S. , Suhail, M. , Kamal, M.A. , Husen, A. and Azhar, E.I. (2017) Recent Development and Future Prospects of Plant‐Based Vaccines. Curr. Drug Metab. 18, 831–841. [DOI] [PubMed] [Google Scholar]
- Tekoah, Y. , Shulman, A. , Kizhner, T. , Ruderfer, I. , Fux, L. , Nataf, Y. , Bartfeld, D. et al. (2015) Large‐scale production of pharmaceutical proteins in plant cell culture‐the Protalix experience. Plant Biotechnol. J. 13, 1199–1208. [DOI] [PubMed] [Google Scholar]
- Tottey, S. , Shoji, Y. , Jones, R.M. , Chichester, J.A. , Green, B.J. , Musiychuk, K. , Si, H. et al. (2018) Plant‐Produced Subunit Vaccine Candidates against Yellow Fever Induce Virus Neutralizing Antibodies and Confer Protection against Viral Challenge in Animal Models. Am. J. Trop. Med. Hyg. 98, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuse, D. , Ku, N. , Bendandi, M. , Becerra, C. , Collins, R., Jr. , Langford, N. , Sancho, S.I. et al. (2015) Clinical Safety and Immunogenicity of Tumor‐Targeted, Plant‐Made Id‐KLH Conjugate Vaccines for Follicular Lymphoma. Biomed. Res. Int. 2015, 648143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eerde, A. , Gottschamel, J. , Bock, R. , Hansen, K.E.A. , Munang'andu, H.M. , Daniell, H. and Liu Clarke, J. (2019) Production of tetravalent dengue virus envelope protein domain III based antigens in lettuce chloroplasts and immunologic analysis for future oral vaccine development. Plant Biotechnol. J. 17, 1408–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Robles Luna, G. , Arighi, C.N. and Lee, J.‐Y. (2020) An evolutionarily conserved motif is required for Plasmodesmata‐located protein 5 to regulate cell‐to‐cell movement. Communications Biology 3, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Sager, R. , Cui, W. , Zhang, C. , Lu, H. and Lee, J.Y. (2013) Salicylic acid regulates Plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell 25, 2315–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, B.J. , Gobeil, P. , Seguin, A. , Atkins, J. , Boulay, I. , Charbonneau, P.Y. , Couture, M. et al. (2021a) Phase 1 randomized trial of a plant‐derived virus‐like particle vaccine for COVID‐19. Nat. Med. 27, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, B.J. , Makarkov, A. , Seguin, A. , Pillet, S. , Trepanier, S. , Dhaliwall, J. , Libman, M.D. et al. (2020) Efficacy, immunogenicity, and safety of a plant‐derived, quadrivalent, virus‐like particle influenza vaccine in adults (18‐64 years) and older adults (>/=65 years): two multicentre, randomised phase 3 trials. Lancet 396, 1491–1503. [DOI] [PubMed] [Google Scholar]
- Ward, B.J. , Seguin, A. , Couillard, J. , Trepanier, S. and Landry, N. (2021b) Phase III: Randomized observer‐blind trial to evaluate lot‐to‐lot consistency of a new plant‐derived quadrivalent virus like particle influenza vaccine in adults 18‐49 years of age. Vaccine 39, 1528–1533. [DOI] [PubMed] [Google Scholar]
- Werner, S. , Breus, O. , Symonenko, Y. , Marillonnet, S. and Gleba, Y. (2011) High‐level recombinant protein expression in transgenic plants by using a double‐inducible viral vector. Proc. Natl. Acad. Sci. U. S. A. 108, 14061–14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusibov, V. , Shivprasad, S. , Turpen, T.H. , Dawson, W. and Koprowski, H. (1999) Plant viral vectors based on tobamoviruses. Curr. Top. Microbiol. Immunol. 240, 81–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Evaluation of the effect of 3C3A on PVX‐GFP movement in local and systemic leaves.
Figure S2. Evaluation of the effect of 3C3A by transient co‐expression on TMV vector‐driven protein accumulation.
Figure S3. Photos showing similar growth of N.b. Sm5 lines to WT plants.
Figure S4. Evaluation of non‐viral vector‐driven protein expression by Western blot analysis.
Figure S5. Evaluation of protein yields at a larger scale using vectors encoding YFE‐1 and PA83.
Table S1. Information related to vectors used in this study.
Table S2. A summary of the evaluation of transgenic Sm5 lines.


