Summary
Currently, feed enzymes are primarily obtained through fermentation of fungi, bacteria, and other microorganisms. Although the manufacturing technology for feed enzymes has evolved rapidly, the activities of these enzymes decline during the granulating process and the cost of application has increased over time. An alternative approach is the use of genetically modified plants containing complex feed enzymes for direct utilization in animal feedstuff. We co‐expressed three commonly used feed enzymes (phytase, β‐glucanase, and xylanase) in barley seeds using the Agrobacterium‐mediated transformation method and generated a new barley germplasm. The results showed that these enzymes were stable and had no effect on the development of the seeds. Supplementation of the basal diet of laying hens with only 8% of enzyme‐containing seeds decreased the quantities of indigestible carbohydrates, improved the availability of phosphorus, and reduced the impact of animal production on the environment to an extent similar to directly adding exogenous enzymes to the feed. Feeding enzyme‐containing seeds to layers significantly increased the strength of the eggshell and the weight of the eggs by 10.0%–11.3% and 5.6%–7.7% respectively. The intestinal microbiota obtained from layers fed with enzyme‐containing seeds was altered compared to controls and was dominated by Alispes and Rikenella. Therefore, the transgenic barley seeds produced in this study can be used as an ideal feedstuff for use in animal feed.
Keywords: recombinant enzymes, feed additive, feedstuff, nutrient utilization
Introduction
Cereals are the major contributors to energy and protein in livestock and poultry breeding. Barley is one of the major feed ingredients in animal diets and is primarily added as an energy source. It is worth noting that the energy value of barley depends on the starch content rather than on the indigestible non‐starch polysaccharide (NSP) content. However, barley contains mixed linked β(1‐3,1‐4)‐d‐glucan (β‐glucans) and arabinoxylans, the most important components of NSP in the cereal (Garcia‐Gimenez et al., 2019). A higher NSP content may decrease the utilization of energy and nutrients in single‐stomach animals because they do not produce sufficient endogenous enzymes that can efficiently degrade these components due to the structure of their gastrointestinal tract. It is difficult for domestic animals to digest and utilize these NSPs due to the high viscosity of the intestinal contents; in addition, NSPs act as anti‐nutritional factors that affect the digestion and absorption of nutrients, thereby resulting in mucus excretion and dysplasia in animals (Abdollahi et al., 2013).
As feed additives, enzymes are widely used in most animal diets. Adding an exogenous enzyme can help animals degrade the anti‐nutritional factors into small molecular fragments, release the encapsulated nutrients, reduce the viscosity of chyme, and ensure that the nutrients are fully digested and absorbed (Ward, 2021). For example, xylanase supplementation in swine diets improves the nutritional value (Moran et al., 2016), and xylanase and phytase supplementation in the de‐oiled rice bran (DORB)‐based diet improves the physiological status and growth performances of Labeo rohita (Ranjan et al., 2017, 2018a,b, 2021, 2022). However, although numerous enzymes are available from animals and plants, the cost of extraction is high, and the production is limited (Holme et al., 2012; Ramos and Malcata, 2017). At present, feed enzymes are usually obtained through the fermentation of fungi, bacteria, and other microorganisms. Microbial fermentation has many advantages, including the use of a wide source of raw materials, high yields, no seasonal restrictions, and is suitable for large‐scale production (Zhao et al., 2015). It should be noted that the manufacturing technology to isolate feed enzymes after fermentation not only increases the cost of application of feed enzymes but also results in a sharp decline in the activity of these enzymes. In addition, the raw materials are subjected to high temperatures, high humidity, and extrusion in the granulating process. Studies have shown that high temperatures can result in the breakage of some water‐dispersing bonds in the enzyme protein molecules, ultimately changing their molecular conformation (Kopriva and Chu, 2018; Borgi et al., 2015). An alternative approach is the use of genetically modified plants containing feed enzymes for direct utilization in animal feedstuff. Furthermore, the expression of recombinant enzymes in plants may provide a more cost‐effective choice than independent microbial fermentation by decreasing the mass transfer limitations of enzyme distribution over a complex polymeric substrate matrix and reducing the cost of granulation (Shen et al., 2012; Zhang et al., 2011).
The rapid development of plant genetic engineering technologies has resulted in well‐developed plant‐based bioproduction platforms for recombinant proteins. These platforms mainly include leaf‐based and seed‐based transgenic plants. Based on their functions and applications, plant‐made recombinant proteins are generally classified into two classes: therapeutic proteins and industrial enzymes (Xu et al., 2012). Tobacco and lettuce are the leading host plant for leaf‐based recombinant protein expression (Boyhan and Daniell, 2011). Indeed, enzymes introduced in tobacco and lettuce have been advanced to human clinical studies (Daniell et al., 2022a,b; Park et al., 2020; Tremblay et al., 2010). For example, secretory antibody variant of Guy's 13 was produced in field‐grown tobacco leaves (Xu et al., 2012). Several vaccine antigens were successfully bio‐produced via expression in lettuce (Kwon and Daniell, 2015). A recent study reported that a novel low‐cost, cold chain/poliovirus‐free, booster vaccine was bio‐produced in lettuce chloroplasts by expressing poliovirus capsid protein (VP1) fused with cholera non‐toxic B subunit (CTB) (Daniell et al., 2016). Besides therapeutic proteins, many industrial enzymes, including xylanase, glucanase, phytase, pectinases, lipase, mannanase, endoglucanase, and exoglucanase, have also been successfully expressed in leaf‐based plants, yielding a product with full function. For example, Pantaleoni et al. (2014) produced a thermostable xylanase in tobacco using chloroplast molecular farming; Ziegler et al. (2000) successfully expressed a thermostable endo‐1,4‐d‐glucanase from the Acidothermus cellulolyticus in the apoplast of tobacco resulting in the accumulation of large quantities of enzyme for commercial biomass conversion. Phytase was found to be biologically active and to accumulate up to 2.3% of the total soluble protein in tobacco leaves by overexpressing heat‐stable phytase from Aspergillus fumigatus (Wang et al., 2007). Daniell et al. (2019a,b) and Kumari et al. (2019) produced commercial leaf enzymes (pectinases, lipase, mannanase, endoglucanase, and exoglucanase) using a new leaf‐production platform. Although therapeutic proteins and industrial enzymes expressed in leaf‐based plants are stable at ambient temperature without losing their activity (Chan et al., 2016; Chatterjee et al., 2010; Daniell et al., 2016; Pantaleoni et al., 2014), there are many cases of recombinant proteins in leaves with accumulation‐associated issues. Product yields in field‐grown plants can be highly variable due to environmental impacts leading to increased consideration of growth in more controlled conditions (e.g., under plastic or in greenhouses). In addition, the harvested material has a limited shelf life and therefore, requires an immediate post‐harvest processing to assure the stability and quality of the protein (Xu et al., 2012).
Recombinant protein expression targeted to plant seeds can overcome limitations associated with leaf tissue (Boothe et al., 2010; Lau and Sun, 2009; Ma et al., 2003). Several cereal grains including rice, wheat, barley, soybean, and maize are commonly used as expression hosts (Ramessar et al., 2008a,b). The high protein content of seeds, ranging from 7% to 10%, has translated into high expression for several seed‐targeted proteins. Proteins expressed in seeds also are stable, and storage of upwards of 3 years at room temperature (longer with cold storage) resulted in minimal loss of protein activity. Advantages of a seed‐based production system include easy protein storage and the ability to apply the product directly to industrial processes, minimizing handling and enzyme manipulation (Boothe et al., 2010; Hood, 2002). Therefore, many feed enzymes have been expressed in seed‐based plants. For example, Zhang et al. (2013) reported that expression and localization of endo‐β‐1,4‐glucanase to various organelles in the embryonic tissue of transgenic maize seed was recovered at levels equal to commercial amounts. Corn seed‐specific overexpression of an Aspergillus niger phytase gene (PhyA2) was found to fully satisfy feed industry requirements (Chen et al., 2008). Expression of a xylanase transgene under the GluB‐1 promoter was only observed in the developing grains and not in the leaf, stem, or root tissues. Patel et al. (2000) found that xylanase was accumulated and stably preserved in the grain during grain maturation and post‐harvest storage.
Accumulating evidence suggests that a multi‐enzyme preparation, rather than a single enzyme, is suitable for the development of feed enzymes, as feed ingredients are structurally complex with various bonds to proteins, fatty acids, crude fibre, and other complex carbohydrates and polysaccharides (Choct, 2006; Ojha et al., 2019). For example, the supplementation of a mix of β‐glucanase and β‐xylanase in barley‐based diets increased nutrient utilization, thereby leading to improved growth performance in poultry (Munyaka et al., 2016). Therefore, simply adding xylanases in wheat‐based diets to degrade the arabinoxylan complexes may not provide complete benefits. However, a combination of xylanase and phytase in wheat‐based broiler rations was found to provide complete benefits regarding nutrients, utilization, and growth performance (Selle et al., 2009). This may be attributed to the fact that in multi‐enzyme preparations, the activity of one type of enzyme may be improved by the action of another enzyme by providing a larger substrate contact area or lowering the deleterious effects of the substrates on nutrient utilization. For example, Cowieson and Adeola (2005) reported that phytase had additive effects in nutritionally marginal broiler diets when mixed with protease and carbohydrase.
In recent years, many transgenic plants expressing single feed enzymes used as feed additives for direct utilization in animal feedstuff have achieved good results. For example, transgenic rice, corn, and canola overexpressing phytase from a bacterial origin supplemented in poultry feed resulted in significant improvements in nutritional value and a remarkable reduction in phosphorus excretion (Chen et al., 2013; Hamada et al., 2006; Hong et al., 2004; Nyannor et al., 2007; Zhang et al., 2000). Broiler chickens fed with β‐glucanase from transgenic potatoes expressing β‐glucanase from Fibrobacter succinogenes had significantly improved feed conversion and reduced ileal digesta viscosity (Baah et al., 2002). The expression of xylanase from Streptomyces olivaceoviridis in transgenic potatoes provides an alternative to present‐day xylanase additives to animal feed (Yang et al., 2007). More importantly, the use of barley grain as a tool for feed enzyme production has emerged as a favourable strategy as it is a plant storage organ rich in protein, ranging from 8% to 15% on a dry‐weight basis (Holme et al., 2017). In addition, certain feed enzymes in the barley grain can improve the quality of malt and feed. For example, Han et al. (2016) found that the overexpression of a barley (1,3;1,4)‐β‐glucanase isoenzyme EII gene (HvGlb2) resulted in significantly decreased amounts of (1,3;1,4)‐β‐d‐glucan in barley grains. Holme et al. (2017) developed a transgenic barley line in which the powdered straw was utilized directly as a supplement to poultry feed. Although previous work on the expression of single feed enzymes in barley grain has been successful (Han et al., 2016), only a small number of studies have explored production of multi‐enzymes in barley grains at commercial levels. At present, xylanase and β‐glucanase are the two main NSP‐digesting enzymes used in the animal feed industry. Xylanases break down arabinoxylans, which are ubiquitous in cereals, and also their by‐products, whereas β‐glucanase degrades β‐glucans and their by‐products, which are widespread in barley. Phytase is widely used as a feed additive to release the phosphate locked in phytate, the major storage form of phosphorus in cereal grains, ultimately making it available to monogastric animals. Therefore, the main objective of this study was to express a mixture of enzymes (phytase, β‐glucanase, and xylanase) in a barley seed‐based system with the overarching goals of enabling their high‐level accumulation in grains and reducing the cost of production to meet the cost target in the feed industry.
Results
Generation of transgenic plants
To the best of our knowledge, there are no reports concerning the expression of three or more enzymes in barley seed. Many factors, especially the gene modification and promoter choice for the generation of messenger RNA (mRNA), have significant impacts on achieving high concentrations of enzymes in seeds. Thus, all genes were modified with preferred plant codons chemically synthesized via the polymerase chain reaction (PCR)‐based accurate synthesis (PAS) method (Xiong et al., 2006). Moreover, the promoter of the glutelin GluB‐1 gene from rice and the NOS (nopaline synthase) terminator were used to construct the cassette of each enzyme gene. Subsequently, each gene was seamlessly connected with the promoter and terminator using the modified overlap‐extension technique (Peng et al., 2006; Figure 1a).
Figure 1.

(a) Schematic representation of recombinant vectors used for barley transformation, (b) Western blotting. WT, wild type; 565‐1, 564‐6, 565‐16: different lines of transgenic barley.
Transgenic barley plants were generated through co‐cultivation of mature zygotic immature feed barley with A. tumefaciens. More than 50 hygromycin‐resistant lines were recovered from somatic embryos. Finally, three transgenic lines were obtained through the identification of the trait segregation of the transgenic seeds.
Expression of enzymes in transgenic barley
The results of RT‐PCR analysis showed relatively high mRNA expression levels of YmAPPA, PceglA, and AkxynC in the mature endosperm of the three transgenic lines 565‐1, 564‐6, and 565‐16 (Figure S1). However, no mRNA of the three genes was detected in wild‐type (WT) barley.
Hetero‐expressed protein in transgenic lines was determined using a Western blot analysis. Surprisingly, phytase from seeds showed a large detectable band on the Western blot (approximately 60 kDa). After treatment with endoglycosidase H, the enzyme had approximately the same molecular weight as the wild‐type protein (46 kDa). Notably, the predicted phytase had a relatively high content of glycosylation sites, which promoted the stability of phytase when expressed in Pichia pastoris. The full‐length PceglA gene, as transformed into barley in this study, also produced a glycosylated protein that was larger than its calculated size. In addition, the AkxynC gene encoded a protein of approximately 24 kDa in barley seeds (Figure 1b).
Figure 2 shows the accumulated levels of the three enzymes in three transgenic lines. The results showed that seeds of transgenic line 565‐1 exhibited the highest phytase activity at 4000 U/kg (Figure 2b). All positive seeds in the three lines also showed higher activity of β‐glucanase and xylanase, with a value of ≥26 000 U/kg (Figure 2c,d). For the successful production of feed enzymes in barley grain, the stability of the enzyme in grains during post‐harvest storage is an important aspect. Figure 2 shows that the activities of all three feed enzymes were stable for at least 6 months, even at 37 °C. According to the quantity of feed enzymes in common complete feed, barley seeds added to feedstuff at a proportion of 8% met the above requirements (Slominski, 2011).
Figure 2.

Phytase (b), β‐glucanase (c), and xylanase (d) activities in barley seeds. (a) The colour of the three feed enzymes when the enzyme activity was measured. WT, wild type; 565‐1, 564‐6, 565‐16: different lines of transgenic barley; 565‐1 6 m, 564‐6 6 m, 565‐16 6 m: different lines of transgenic barley stored at 37 °C for at least 6 months.
Assessment of agronomic traits
Scanning electron microscopy (SEM) results showed that the transgenic barley was morphologically indistinguishable from wild‐type barley grown under identical conditions. Specifically, there were no differences in the starch structure of transgenic plants and wild‐type seeds (Figure 3a). Moreover, there was no significant difference in the 1000‐grain weight or seed germination rate between transgenic and wild‐type plants (Figure 3b,c). However, the contents of phytic acid and β‐glucan were significantly decreased in the transgenic barley (Figure 3d,e). Compared with wild‐type barley, the contents of phytic acid and β‐glucan in transgenic barley decreased by about 37% and 84% respectively. Protein is one of the main grain components that affects nutritional quality. The total protein was quantified to determine the effect of the increased feed enzymes in the transgenic seeds on the nutritional quality of the barley. As shown in Figure 3f, the contents of protein in the transgenic seeds were in the same ranges as in the WT seeds. The results indicated that the increased feed enzymes in the seeds at least did not decrease its nutritional quality.
Figure 3.
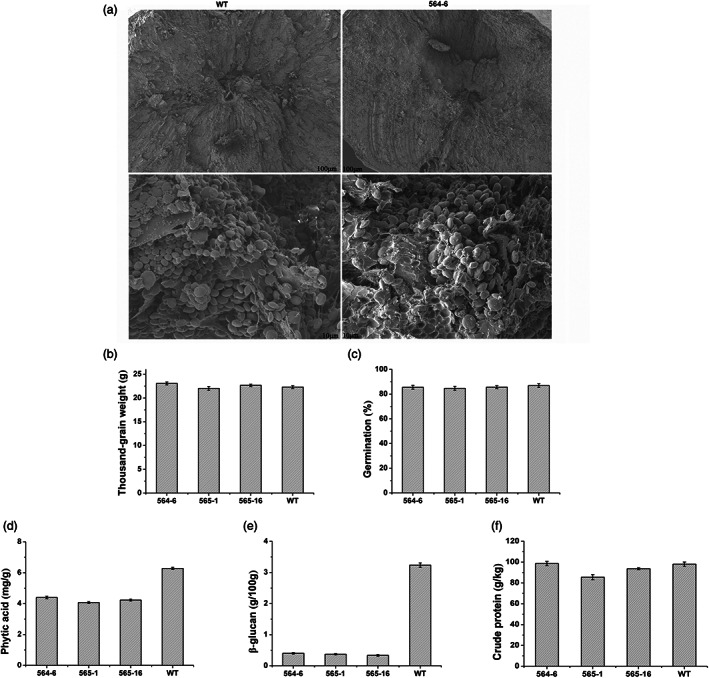
(a) Scanning electron microscope pictures of the barley grain sections. WT: wild type; 564‐6: transgenic barley; (b) 1000‐grain weight of seeds; (c) germination of seeds; (d) the content of phytic acid in seeds; (e) the content of β‐glucan in seeds; (f) the content of crude protein in seeds.
Effect of complex enzyme expression on animal growth performance
Over the years, microbially derived NSP enzymes and phytases have been widely used in the animal feed industry in the form of single or multi‐enzymes. Thus, an animal feeding trial was conducted to evaluate the effects on animal growth performance of the complex feed enzymes expressed in the 565‐16 transgenic barley inbred line. To this effect, flour obtained from transgenic seeds was added to basal animal diets. The results revealed that layers fed with a basal diet (control group) were inferior to those fed a basal diet in which the wild‐type barley was replaced with transgenic barley containing the enzymes (TBE group). The strength of the eggshells in the TBE group after 15–20 days of feeding increased by 10.0%–11.3% (Figure 4b), a level that was similar to that of the group fed a basic diet containing feed mixed with exogenous mix feed enzymes (EXE group; 250 U phytase, 2000 U β‐glucanase, and 2500 U xylanase). The weight of eggs from the TBE group also increased by 5.6%–7.7% compared to the control group (Figure 4a).
Figure 4.

(a) The weights of eggs; (b) the eggshell strength; (c) the relative digestibility of crude fibre (RDCF) and the content of crude fibre in faecal samples (CFF) among chickens fed with a basic diet, TBE, and EXE; (d) the relative digestibility of total phosphorus (RDTP) and the content of total phosphorus in faecal samples (PF) among layers fed with a basic diet, TBE, and EXE; (e) the relative digestibility of β‐glucan (RDG) and the content of β‐glucan in faecal samples (GF) among layers fed with a basic diet, TBE, and EXE; (f) the relative digestibility of crude protein (RDCP) and the content of crude protein in faecal samples (CPF) among layers fed with a basic diet, TBE, and EXE. TBE: fed with a basic diet that replaced the wild‐type barley with mixed enzyme transgenic barley. EXE: fed with basic diet containing feed mixed with exogenous enzymes (EXE group; 250 U phytase, 2000 U β‐glucanase, and 2500 U xylanase).
Next, we examined the relative digestibility and the contents of total phosphorus, β‐glucan, and crude fibre in the faeces obtained from the different groups (control group, EXE group, and TBE group) during the grower phases. The contents of crude fibre and phosphorus in the faeces of the control group were significantly higher than those in the other groups during the grower phases (P < 0.05; Figure 4c,d). After feeding with mixed enzyme barley (TBE group), the apparent relative digestibility of total phosphorus was improved compared to the control group (Figure 4d). The excretion of phosphorus in faeces obtained from the TBE group dropped by 50% or more compared to the control group. However, there was no significant difference between the TBE group and the EXE group after feeding for 28 days (Figure 4d). Collectively, these results suggest that the transgenic barley, when used as animal feed, was highly effective in reducing phosphorus excretion by layers. It should be noted that the β‐glucan content in the faeces of the TBE group was also significantly lower than that in the control group (Figure 4e). Although there was a remarkable difference in the β‐glucan content in the faeces between the EXE group and TBE group, the relative digestibility of β‐glucan between them was almost the same. In addition, the relative digestibility of crude protein between the EXE and TBE group was slightly higher than the control group because phytase acts on the protein‐phytate complex and makes the protein available for biological activities (Biradar et al., 2017; Figure 4f).
Effects of transgenic barley on intestinal microorganisms in laying hens
The experiment on the gut microbiome showed that the total quantities of Firmicutes and Bacteroidetes in the gut changed significantly, regardless of whether exogenous enzymes or transgenic barley seeds were added to the feed. A lower ratio of Firmicutes to Bacteroidetes species was found in the caecum of laying hens after feeding with the enzyme mix feed (Figure 5a). The ratio of Firmicutes/Bacteroidetes decreased from 63.7% to 41.1% or to 39% after the addition of enzymes to basal feed using transgenic barley seeds or exogenous enzymes respectively.
Figure 5.

(a) Relative abundance of bacterial taxa at the phylum taxonomic level among the control group (A1, A2, A3), EXE group (B1, B2, B3), and TBE group (C1, C2, C3). (b and c) Linear discriminant analysis effect size (LEfSe) for bacterial taxonomic differences between the control group and the TBE group; (d and e) LEfSe for KEGG pathways differences between the control group and the TBE group; (f) LEfSe for KEGG modules between the control group and the TBE group.
Furthermore, a logarithmic LDA (linear discriminant analysis) score cut‐off of 2.0 was employed to identify important taxonomic differences among the three groups. The LDA LEfSe (linear discriminant analysis effect size) analysis results showed a notable difference in the intestinal microorganisms between the TBE and control groups. The relative abundances of the Alispes and Rikenella genera were higher in the TBE group than in the control group, whereas the relative abundance of Treponema was higher in the controls than in the TBE group (LDA score > 2, Figure 5b,c). Notably, the specific bacterial species may be a forecaster of the response to a particular diet. No significant difference was found between the TBE and EXE groups.
Characterization of genes enriched in the gut microbiome of the TBE and control groups revealed a distinct nutrient acquisition of the gut microbiome that was largely influenced by the different dietary patterns of the host (Figure 5d–f). The gut microbiome of animals in the TBE group was enriched with genes for metabolism of aromatic amino acids and biosynthesis of vitamins B1 and B7. In addition, genes involved in ATP‐binding cassette (ABC) transporters and biosynthesis of antimicrobial peptides and antibiotics (e.g., ornithine and tetracenomycine biosynthesis), and the multidrug efflux pump were also enriched in the gut microbiome of the TBE group. Some enzymes involved in DNA repair were also enriched in the gut microbiome of the TBE group. Moreover, genes enriched in the gut microbiome of the control group were primarily genes targeting anaerobic energy production (methane metabolism), carbohydrate metabolism (glycolysis and Entner–Doudoroff pathways), and lipid metabolism, including enzymes for sphingosine and ceramide synthesis. The difference between genes enriched in the gut microbiome of the TBE and control groups may be caused by the decomposition of the phytic acid in the feed by the mixed enzyme barley, improving the absorption of nutrients in the intestinal tract of the animals, and thus resulting in a series of gene changes. For example, calcium phytate complexes react with fatty acids to form insoluble saponins in the intestine during the process of lipid digestion (Ellis et al., 1982). Phytates can also combine with starch, inhibiting the action of amylase, and reducing the digestibility of carbohydrates (Lenkova et al., 2020; Selle et al., 2000). Therefore, it is difficult for the control group to normally metabolize lipids and carbohydrates in the gut in control group due to the existence of phytic acid, while these nutrients can be normally metabolized in the TBE group due to phytase‐mediated decomposition of phytic acid. Also, beta‐glucan in damaged tissues, such as lymphoid tissue cells, accelerates the production of cytokines, making other drugs, including antibiotics, antifungal, and antiparasitic drugs, more effective (Angeli et al., 2006). Therefore, when β‐glucan was broken down by β‐glucanase in the TBE groups, these specific functions were affected. This resulted in the high expression of genes involved in the biosynthesis of antimicrobial peptides and antibiotics and some enzymes involved in DNA repair in the gut microbiome of TBE groups.
Discussion
Barley, one of the founder crops in agriculture, is the fourth most important cereal grain worldwide, with production exceeding 130 million metric tons annually (Mrízová et al., 2014). Notably, about 66% of the barley seed produced in the world is used as animal feed. However, the endosperm cell walls of barley grains are characterized by the presence of (1,3;1,4)‐β‐d‐glucans and arabinoxylans, two major anti‐nutritional materials in monogastric animals shown to increase chyme viscosity and reduce the rate of digestion and absorption of nutrients in the small intestine (Perera et al., 2019). In addition, about 70% of the total phosphate in seeds is stored as phytic acid. Most of this phytic acid is indigestible by monogastric animals, and thus is secreted with faeces and mixed with agricultural soils, where it eventually diffuses into the water environment as a contaminant, leading to algal growth and eutrophication. This may be attributed to the fact that monogastric animals lack an intestinal enzyme for liberation of phosphate groups from phytic acid (Gupta et al., 2015; Perera et al., 2018).
Given that feed ingredients are structurally complex, a multi‐enzyme preparation, rather than a single enzyme, provides a feasible approach to enhancing nutrient utilization in livestock and poultry diets (Juanpere et al., 2005). It is worth mentioning that if cereal seeds contain a sufficient number of different enzymes, there is no need for supplementation with microbial enzyme additives. Non‐supplemented feed significantly reduces feed costs and simplifies feed processing and diet formulation. However, there are only a few studies on the production of multiple feed enzymes in cereal seeds. We demonstrated in this study that transgenic barley seeds can express phytase, β‐glucanase, and xylanase together in endosperm without affecting seed germination. The phytase activity in transgenic seeds reached about 4000 U, whereas the β‐glucanase and xylanase activities reached about 26 000 U/kg seed. The feeding trials showed that addition of 8% transgenic seeds containing the three feed enzymes in the basal diet substituted for contents of multi‐enzyme additives for layers and achieved equivalent effects on egg quality and nutrient utilization.
Hood et al. (2012) reported that plant seeds have the advantages of protein stability, long storage time, easy transportation, and low cost. In this study, the activities of all three feed enzymes were stable for at least 6 months. Production of plant seeds with feed enzymes can reduce the cost of granulation and the loss of enzymes in the granulation process. In addition, the Western blot results showed that the transgenic barley expressed phytase and β‐glucanase at molecular weights greater than their calculated values, indicating different glycosylation patterns of both proteins in transgenic plants. Altogether, these advantages make the plant expression system very attractive for the production of industrial enzymes, particularly for those that do not require purification yet are required in large quantities.
There are three types of feed enzymes in livestock nutrition that are commonly used to degrade fibre, protein, starch, and phytate. Over the years, many enzymes have been used as feed additives, including cellulases, β‐glucanases, xylanases, phytases, proteases, lipases, and galactosidases (Adeola and Cowieson, 2011). This was largely due to the challenges in expressing a variety of enzymes in the same seed. In cereals, the endosperm is the major site of the grain reserve of nutrients and storage proteins (Shewry et al., 2002). Heterologous proteins are normally expressed at high levels and specifically in the endosperm. In this study, the promoter of rice GluB‐1, one of the most commonly used endosperm‐specific promoters, was used to control the expression of three enzymes, with the obtained transgenic barley seeds exhibiting high enzymatic activity (Qu and Takaiwa, 2004). Previous studies have demonstrated that other strong grain‐specific promoters of genes such as hordein B‐1 (HOR2‐4), hordein D (HOR3‐1), and γ‐hordothionin can achieve higher accumulation levels of proteins in seeds (Stahl et al., 2009a,b). This suggests that different promoters may achieve better results for the expression of a multi‐enzyme system in seeds. However, not all promoters can drive efficient expression of the enzyme due to the differences in promoter strength. Therefore, modification of promoters and signal peptide of seed‐specific genes by DNA shuffling will aid in the exploration of more suitable candidates for deposition of recombinant proteins in the protein bodies of developing endosperm cells (Lu and Jeffries, 2007). In addition, promoters from specific genes of other organs in seeds such as the α‐AMY promoter in the aleurone layer and barley germin gene Gerb promoter in the seed coat (Eskelin et al., 2009; Wu et al., 2000) were used to control the expression of feed enzymes and reduce the pressure of highly expressed proteins in seeds. Yang et al. (2012) proposed that reduction of endogenous seed storage proteins may be a new efficient strategy for enhancing the accumulation levels of desirable recombinant proteins.
Interestingly, we found that the richness and diversity of intestinal microbiota were altered in layers fed with transgenic barley compared to layers in the control group, a finding that was similar to layers fed with multi‐enzyme additives. Studies have demonstrated that diet performs a critical function in shaping and/or altering the microbiome, and diet‐induced alterations of microbial communities occurred in a rapid and reproducible manner (Kolodziejczyk et al., 2019; Yadav and Jha, 2019). Enzymes can modulate the gut microbiota by improving the digestibility of a number of feed components and by changing the nutritional content of the diet (Kogut, 2019). For example, phytase led to significant changes in the utilization of dietary limestone and phosphate. In addition, the carbohydrase enzymes eliminated the nutrient‐encapsulating effect of the cell wall by degrading specific chemical bonds in feedstuffs and partially breaking down polysaccharides, thereby increasing the availability of starches, amino acids, and minerals (Costa et al., 2008; Jha et al., 2015). Liu et al. (2012) reported that adding xylanase to a wheat‐based diet alleviated impairment of the intestinal mucosal barrier in birds challenged with Clostridium perfringens, a pathogen of gas gangrene. Another study revealed that phytase had a role in modulating the gut microbiota of chickens by increasing Lactobacillus sp. and Enterococcus sp. in the ileum (Ptak et al., 2015). Herein, we found that supplementation with multi‐enzymes generally enhanced Rikenellaceae (Alispes and Rikenella) bacteria in the caecum of chickens. Alistipes was enriched in a protein‐rich diet and regulated protein fermentation and synthesis of saturated fatty acids. This indicated that the basic microbiota of intestinal digestion was altered after supplementation with enzymes in the basal diet. Although there have been studies on the changes of intestinal microbiota in different diets (Ptak et al., 2015), the relationship between the diversity of species in the intestinal microbiota and the feeding efficiency of poultry is still unclear. Future studies should utilize molecular technologies to elucidate the mechanism through which enzymes exert changes on the intestinal microbiome. Specifically, the combined approach of metaproteomics, metatranscriptomics, and metabolomics is a useful tool for understanding the relationship between feed enzymes and the intestinal microbiota (Roberts et al., 2015).
In conclusion, the co‐expression of YmAPPA, PceglA, and AkxynC genes in barley seed resulted in remarkable increases in phytase, β‐glucanase, and xylanase activities. The supplementation with only 8% enzyme‐containing transgenic seeds in the basal diet of layers decreased indigestible carbohydrates, improved the availability of phosphorus, and reduced the impact of animal production on the environment. Collectively, our findings suggest that the transgenic barley seeds produced in this study can be used as an ideal feedstuff in animal feed.
Methods
Plant material, chemicals, and standards
Barley seed (cultivar Yangmai 3) was obtained from the plant genetic engineering laboratory of Shanghai Academy of Agricultural Sciences. The transgenic seedlings were transplanted to a paddy field in Baihe county, Qingpu district, Shanghai, China. Standard enzymes (phytase, β‐glucanase, and xylanase) and sodium phytate were purchased from Sigma Chemical Co., Ltd (St. Louis, MO, USA). All other chemicals were analytical grade and were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). All primers were purchased from Shanghai Sangon Biological Engineering Technology and Service Co., Ltd. (Shanghai, China). Agrobacterium tumefaciens strain EHA105 and Escherichia coli DH5α were obtained from the plant genetic engineering laboratory of Shanghai Academy of Agricultural Sciences. An RNA isolation kit was purchased from Fermentas International, Inc. (Burlington, ON, Canada), and a reverse Transcription System was purchased from Promega (Madison, WI). Twenty‐four 4‐week‐old black‐feathered laying hens (Xingyang) were obtained from the National Poultry Engineering Research Center of the Ministry of Agriculture (Shanghai, China).
Choice of enzymes and genes
The gene for a HAP phytases, YmAPPA, (Notes S1 and Figure S2) was chosen from the Enterobacteriaceae family member Yersinia mollaretii according to Shivange et al. (2012) due to its specific activity towards the natural substrate phytate, which is on average ten times higher than commonly used/reported fungal phytases. Furthermore, phytases can maintain stability in the gastrointestinal tract of animals largely due to their stability over broad pH ranges and pepsin resistance.
The gene of endo‐β‐1,3‐1,4‐glucanase (PceglA, Notes S1 and Figure S3) was isolated from Pectobacterium parmentieri according to Zhu et al. (2018). A comparison of its amino acid sequences with other β‐glucanase sequences from various sources showed that the enzyme exhibited 66% homology with lichenase obtained from Bacillus licheniformis (EC 3.2.1.73), an important glycosyl hydrolase in the hydrolysis of (1,3;1,4)‐β‐d‐glucans. The specific activity of the purified recombinant enzyme from P. parmentieri was higher than those produced by Bacillus spp. Moreover, the β‐glucanase was more thermotolerant than its barley counterparts, and the suitable range of pH for the enzyme was 3.5–4. Notably, the high thermostability and acidic catalyst make this enzyme appropriate for application in the feed industry.
The gene of xylanase (AkxynC, Notes S1 and Figure S4) from Aspergillus kawachii, which is used to make shochu (a Japanese traditional spirit), was selected for expression in barley according to Qiu et al. (2016). It should be noted that AkxynC belongs to the glycoside hydrolase family 11 and that the optimum pH of the extremely acidophilic xylanase was found to be 2.8, far lower than that of the bacterial enzyme. Furthermore, the enzyme showed high pepsin and trypsin resistance. To match the gastrointestinal pH, the enzyme was modified with two residue mutations (Thr64Ser and Asp48Asn), which resulted in its optimum pH shifting from 2.8 to 5.0 (Qiu et al., 2016).
Vector construction
To improve protein expression in the plants, all of the codons of each gene (YmAPPA, PceglA, and AkxynC) were first optimized for barley expression; the reverse repeat sequences, stem‐loop structures, and transcription termination signals in genes were eliminated to improve the stability of RNA; intron recognition sequences in the coding region were removed to avoid intron splicing, and the C and G oligonucleotides at the second and third sites of codons were modified as much as possible to avoid methylation in the plants. Then, all genes were chemically synthesized via the PCR‐based accurate synthesis (PAS) method according to Xiong et al. (2006). The first forward oligonucleotide at the most 5′‐end and the last reverse primer at the most 3'‐end were defined as outer oligonucleotides, whereas the remaining primers were defined as inner oligonucleotides. The PCR master mix contained 1–3 pmol of each of the inner oligonucleotides and 10–30 pmol of the outer oligonucleotides. PCR was conducted for 25 cycles using the following conditions: 94 °C for 30 s; 60 °C for 30 s; and 72 °C for 1–2 min, followed by extension at 72 °C for 10 min. Next, the chemically synthesized genes were placed under the control of the rice GluB‐1 promoter and the NOS terminator. Notably, each gene was seamlessly connected with the promoter and terminator to form gene expression cassettes. All expression cassettes were then assembled into the same expression vector, pYP694 (Tian et al., 2020) using isocaudomers. The final vector (pYP69‐YmAPPA‐PceglA‐AkxynC), named pAPPAXYNCEG, carried the three feed enzyme genes and a hygromycin resistance gene (hpt). The hpt gene was under the control of the CaMV35S promoter and was used as a selectable marker.
Agrobacterium‐mediated transformation of barley
Barley seeds (cultivar Yangmai 3) were grown in soil under natural conditions and harvested when the immature embryos (IEs) were approximately 1.5–2.5 mm in length. The immature embryos were isolated from the surface‐sterilized grain and then plated on callus induction medium. Next, the obtained calli were transformed via co‐cultivation with Agrobacterium tumefaciens strain EHA105 harbouring a binary expression plasmid (pAPPAXYNCEG) in accordance with the protocol described by Tingay et al. (1997). After 3 days of co‐cultivation at 24 °C in the dark, the calli were rinsed thoroughly with sterile distilled water, and then rinsed three to five times with 250 mg/L carbenicillin, blotted on filter paper, plated on the selection medium (callus induction medium supplemented with hygromycin), and incubated at 24 °C in low light for 8–10 weeks. The hygromycin‐resistant calli were then transferred to a regeneration medium. Finally, when the shoots reached 2–3 cm in length, they were transferred to a rooting medium. Next, the tissue culture‐derived plantlets were transplanted into soil in the glasshouse. Seeds from the T0 transgenic lines were sown on Murashige and Skoog (MS) medium supplemented with hygromycin to determine whether they were positive plants. Then, the polymerase chain reaction (PCR) was performed to determine the presence of the three genes to further screen the positive plants using DNA as a template. And, the T1 seeds of the transgenic lines were used to generate the T2 plants. This process was repeated until the T4 generation to obtain homozygous lines from the original plants. Each plant produced enough seeds for further analysis.
Gene expression analysis
The total RNA was extracted from 0.1 g of developing embryos (20 days after pollination) of transgenic barley lines (565‐1, 564‐6, 565‐16) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. First‐strand cDNA synthesis was performed using the PrimeScript RT‐PCR Kit (TaKaRa, Kyoto, Japan). For semi‐quantitative RT‐PCR detection of gene expression, 3 μg of total RNA was used for reverse transcription. PCR was performed using Actin2 as an internal control. The PCR was performed under the following conditions: 10 min at 94 °C, followed by 22 cycles of 30 s at 94 °C, 30 s at 56 °C, 30 s at 72 °C, and a final extension at 72 °C for 5 min. The sequences of primers used are listed in Table S1.
Western blot analysis
Randomly selected kernels were ground with a grinder (QianJiang instrument JSF‐13A; Hangzhou Qianjiang Instrument Equipment Co., Ltd., Hangzhou, China) followed by extraction of total proteins from the seed meal. Briefly, 20 mg of the seed powder was placed into 1.5‐mL microfuge tubes and extracted with 300 μL of extraction buffer (50 mm citric acid‐Na2HPO4, pH 3.5). The tubes were vibrated at room temperature for 1 h and centrifuged at 5000 g for 10 min. Next, supernatants were transferred into fresh tubes and incubated with a two‐fold volume of pre‐cooled acetone for 30 min. After centrifugation at 14 000 g for 10 min, the protein precipitate was dissolved in 30 mL of deionized water. The protein sample was then divided into two equal parts, one for deglycosylation with endo‐β‐N‐acetylglucosaminidase (Endo H) and the other remaining intact. Proteins extracted from untransformed Yangmai 3 were used as the negative control. Total protein concentrations in barley grains were determined using a Micro BCA Protein Assay Reagent Kit (Pierce, Rockford, IL, USA) according to the manufacturer's instructions.
The Western blot was probed with a polyclonal antibody raised in rabbits against the E. coli‐expressed enzyme. Three rabbits were injected with purified protein three times. After separation on SDS‐PAGE, proteins were transferred to a nitrocellulose (NC) membrane (Bio‐Rad, Hercules, CA, USA). The antibody specificity was verified and diluted by a factor of 2000 for Western blot analysis. The goat anti‐rabbit IgG labelled with alkaline phosphatase was used as the secondary antibody. The substrate electrochemiluminescence (ECL) method was used to develop and visualize the results under X‐ray.
Determination of the activities of phytase, β‐glucanase, and xylanase
To determine phytase activity, 100 mg of seed powder was placed in a 1.5‐mL Eppendorf tube, to which a 1 mL of extraction buffer (50 mM NaAc, 1 mM CaCl2, pH 5.5) was added. The tubes were mixed by vortexing at room temperature for 1 h, and then centrifuged at 5000 g for 10 min. Supernatants were transferred into fresh tubes and used for the determination of the phytase activity as described previously (Wyss et al., 1999b). Subsequently, 100 μL of the supernatant was mixed with 900 μL of 5 mM phytic acid and incubated at 37 °C for 30 min. The reaction was terminated with the addition of 1 mL of 15% (v/v) trichloroacetic acid (TCA). The released phosphate was quantified using a spectrophotometer with a solution containing 0.6 M H2SO4, 2% ascorbic acid, and 0.5% ammonium molybdate. Standard solutions of potassium phosphate were used as a reference. One unit (U) of phytase activity was defined as the amount of activity that liberated 1 μmol of phosphate per min at 37 °C.
The β‐Glucanase activity was determined by measuring the amount of reducing sugar released from lichenan using the 3,5‐dinitrosalicylic acid (DNS) method (Gusakov et al., 2011). The crude homogenate was mixed with DNS and boiled for 5 min. After the mixture was cooled, the amount of reducing sugar was determined by absorbance at 540 nm. One unit of enzymatic activity was defined as the amount of enzyme required to release 1 μmol of reducing sugar per minute from 1.0% lichenan in citric acid‐Na2HPO4 (50 mm, pH 3.5) at 60 °C for 10 min.
Xylanase activity was determined by measuring the amount of reducing sugar released from arabinoxylan as previously described (Fushinobu et al., 2011). Briefly, 10 μL of crude homogenate was mixed with 190 μL of xylan suspended in 50‐mm sodium citrate. The mixture was incubated at 40 °C for 30 min. Subsequently, the amount of released sugar was determined based on the absorbance at 490 nm. One unit of xylanase was defined as the amount that released 1 μmol of reducing sugar equivalents from xylan.
Assessment of enzyme stability in transgenic grain during storage
The stability of the enzyme was examined using the grains of the homozygous lines (565‐1, 564‐6, and 565‐16) stored at different temperatures, and the enzyme activity was analysed after various storage times.
Determination of phytic acid, β‐glucan, crude fibre, and total protein content
The amount of phytic acid was measured according to a previously published method (McKie and McCleary, 2016). Briefly, 30‐mg samples were extracted with 1 mL of 0.4 m HCl–15% TCA at room temperature for 3 h, and then centrifuged at 5000 g for 10 min. Next, 25 μL of the supernatant was mixed with 275 μL of 36.3 mm NaOH and 100 μL of 0.03% (w/v) FeCl3–0.3% sulfosalicylic acid. The sample was then centrifuged at 2000 g for 10 min, and its concentration was determined by measuring the absorbance at 500 nm. The same extract was used to determine the phosphate content.
The content of β‐d‐glucan was determined using a Mixed Linkage β‐Glucan Kit (Cat. K‐BGLU; Megazyme International, County Wicklow, Ireland). Before β‐Glucan detection, barley grains were ground in liquid nitrogen and dried at 60 °C for 3 days. Crude fibre in feed and faecal samples were quantified using the filtration method as detailed in a previous study (Lee and Hristov, 2013). Total protein was determined using the Kjeldahl method according to the AOAC (2007) method 979.09 with a conversion factor of 5.7.
Analysis of the morphology by scanning electron microscopy
The dry and mature seeds were selected and cleaned. Next, the seeds were cross‐cut and vacuum sprayed with gold. Then, the cross‐section of the seeds was turned upward and placed under a scanning electron microscope to observe and record images. Scanning electron micrographs were obtained using a scanning electron microscope (SEM; S4800; Hitachi, Tokyo, Japan) as previously reported (Tian et al., 2019).
Grain weight and germination rate of seeds
One thousand grains from each line were weighed to estimate the 1000‐grain weight.
One hundred grains from each line were placed at 25 °C in a dark chamber to allow germination. The germination rate was scored after 5 days. All analyses were performed in triplicate.
Feeding layers with barley seed rich in feed enzymes
Twenty‐four 4‐week‐old black‐feathered laying hens (Xingyang) were randomly and equally divided into three groups. The control group was fed with a basic diet containing 52% corn flour, 27% bean, 8% barley, 7.5% mineral elements, and 4% of a mixture of vitamins. The composition and nutritional level of the diet are shown in Table S2. An EXE diet was prepared by mixing 250 U phytase, 2000 U β‐glucanase, and 2500 U xylanase per kilogram. The diet for the test group was prepared by replacing wild‐type barley with transgenic barley 564‐16 (TBE).
Chickens were placed in three‐layer cages in a closed room. The laying hens were housed in a room controlled to a photoperiod of 16 h light : 8 h dark, a humidity setting of 45%–50%, and a temperature of 25 °C. The layers were given corresponding diets three times a day for 4 weeks while measuring the feed intake and body weight weekly. All excreta were collected and dried in a fan‐forced oven at 65 °C for 3 days, then ground to pass through a 1‐mm sieve for later analysis. Total excreta were collected from day 14 to day 28 for determination of apparent metabolizable nutrients (AMN) by measuring the quantity of diet and excreta gross nutrients as reported previously (Chen et al., 2013).
Sample collection and sequencing of the gut microbiome
To collect intestinal tissue samples from the layers, three chickens were randomly selected (9‐week‐old) from each group and slaughtered. The caecum was harvested and frozen in liquid nitrogen.
Bacterial cells were extracted from intestinal tissue samples, isolated through differential centrifugation, and then lysed. Microbial genome DNA was extracted using a QIAamp DNA stool mini kit (cat#51504; QIAGEN, Dusseldorf, Germany) following the manufacturer's instructions. Metagenomic DNA paired‐end libraries were prepared using an insert size of 150 base pairs (bp) and then sequenced on an Illumina HiSeq 2500. The quality of the sequenced raw data was evaluated using Fastqc. The average error rate of the clean reads was lower than 0.001. In addition, raw reads were cleaned to exclude adapter sequences and low‐quality sequences using Trimmomatic (v0.39). Short reads (length < 35 bp) and unpaired reads were also excluded to obtain data with clean reads.
For each sample, high‐quality reads were assembled into contigs using IDBA_UD (v1.1.2). The reads were comprehensively analysed to select the best k‐mers. Subsequently, gene prediction was performed on contigs larger than 100‐bp using Prodigal (v2.6). A non‐redundant gene catalogue was constructed using gene models predicted from each sample using CD‐HIT‐EST (v4.6.6), which used a greedy incremental clustering algorithm; the criterion of identity was >95%, and the overlap was >90% of the shorter genes (Hyatt et al., 2010).
Taxonomic classification and functional assignment of genes
DIAMOND (v0.8.20) alignment was performed against the NCBI‐NR database with an E‐value <1 × 10−5 and score > 60 for taxonomic assignments of protein sequences. The relative abundances of phylum, genus, species, Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologous group (KO), and eggNOG orthologous group (OG) relative abundance were calculated by summing the abundance of genes for each category per sample based on results of taxonomic assignments, KO, and OG annotations respectively. The relative gene abundance was also summarized into KEGG and eggNOG functional profiles for functional analysis (Jaime et al., 2016).
Differences were tested at the operational taxonomic units (OTUs), genus, and family levels. Features that passed the pairwise Wilcoxon test were considered successful biomarkers. The relative abundance of each taxon, KEGG, and module in TBE and controls were analysed using the non‐parametric Mann–Whitney U test (for two groups) or the Kruskal–Wallis test (for more than two groups). All experiments were run with the LEfSe α parameter for pairwise tests set to 0.05 for class normality and subclass tests, and the threshold of the logarithmic score of LDA analysis was set to 2.0 (Segata et al., 2011).
Statistical analysis
Statistical analyses were performed using SPSS 17.0 (SPSS Inc., Chicago, IL). The Origin 8.0 (OriginLab Co., Northampton, MA) software was used for mapping. Asterisks in the figures denote significant differences at *P < 0.05 or **P < 0.01.
Conflicts of interest
There are no conflicts of interest to declare.
Author contributions
Quan‐Hong Yao and Yong‐Sheng Tian designed the research. Ri‐He Peng, Wen‐Hui Zhang, Zhen‐Jun Li, Li‐Juan Wang, and Yong‐Dong Deng performed these experiments. Jing Xu, Bo‐Wang, Xiao‐Yan Fu, and Jian‐Jie Gao analysed these data. Hong‐Juan Han and Yu Wang performed the targeted metabolomics analyses. Ri‐He Peng wrote the manuscript.
Supporting information
Figure S1 Expression of the genes in transgenic barley by RT‐PCR using cDNA as template (1:565‐1; 2:564‐6; 3:565‐16; M: marker; W: wild type barley).
Figure S2 Alignment of amino acid sequences among Yersinia mollaretii (YmAPPA), Candidatus Hamiltonella (ChAPPA), Cronobacter malonaticus (CmAPPA), Citrobacter sp. KTE32 (CsAPPA), Escherichia albertii (EaAPPA), Hafnia alvei ATCC 51873 (HaAPPA), Yersinia aldovae ATCC 35236 (YaAPPA), Shigella boydii ATCC 9905 SGB (SbAPPA), Aspergillus niger (AnAPPA).
Figure S3 Alignment of amino acid sequences among Pectobacterium carotovorum (PceglAS), Stachybotrys chartarum (SceglAS), Gibberella zeae (GzeglAS), Streptomyces avermitilis (SaeglAS), Aspergillus oryzae (AoeglAS), Thermobispora bispora (TbeglAS).
Figure S4 Alignment of amino acid sequences among Aspergillus kawachii (AkxynCS), Bacillus sp (BsxynAS), Streptomyces sviceus(SsxynBS), Thermobacillus composti KWC4 (TcxynBS), Catenulispora acidiphila (CaxynBS), Thermobispora bispora (TbxynBS).
Notes S1 The sequence information after codon optimization as follow.
Table S1 Primers for molecular analysis in transgenic barley using cDNA as template.
Table S2 Ingredients and nutrients of diets for feeding trial.
Acknowledgements
This study was supported by the Key Project Fund of the Shanghai Municipal Committee of Agriculture (Tuizi 2022 1‐5); National Natural Science Foundation of China (32171977, 31901069); Shanghai Academic Technology Research Leader (20XD1432200; 19XD1432300); Talent Development Fund Project of Shanghai (2020101); The Innovation Team project of Shanghai Academy of Agricultural Sciences (2022) 005.
Contributor Information
Yong‐Sheng Tian, Email: tys810508@126.com.
Quan‐Hong Yao, Email: yaoquanhong_sh@aliyun.com.
References
- Abdollahi, M.R. , Ravindran, V. and Svihus, B. (2013) Pelleting of broiler diets: an overview with emphasis on pellet quality and nutritional value. Anim. Feed Sci. Technol. 179, 1–23. [Google Scholar]
- Adeola, O. and Cowieson, A.J. (2011) Board‐invited review: opportunities and challenges in using exogenous enzymes to improve nonruminant animal production. J. Anim. Sci. 89, 3189–3218. [DOI] [PubMed] [Google Scholar]
- Angeli, J.P.F. , Ribeiro, L.R. , Gonzaga, M.L.C. , Soares, S.A. , Ricardo, M.P.S.N. , Tsuboy, M.S. , Stid, R. et al. (2006) Protective effects of β‐glucan extracted from Agaricus brasiliensis against chemically induced DNA damage in human lymphocytes. Cell Biol. Toxicol. 22, 285–291. [DOI] [PubMed] [Google Scholar]
- AOAC (2007) Association of Official Analytical Chemist. Official Methods of Analysis, 18th edn. Washington, DC: AOAC. [Google Scholar]
- Baah, J. , Scott, T.A. , Kawchuk, L.M. , Armstrong, J.D. , Selinger, L.B. , Cheng, K.J. and Mcallister, L.B. (2002) Feeding value in broiler chicken diets of potato expressing a glucanase gene from Fibrobacter succinogenes. Can. J. Anim. Sci. 82, 111–113. [Google Scholar]
- Biradar, S. , Murthy, H.S. , Patil, P. , Jayaraj, E.G. and Tammegowda, N.K.B. (2017) Dietary supplementation of microbial phytase improves growth and protein efficiency ratio of freshwater prawn (Macrobrachium rosenbergii). Aquac. Int. 25, 567–575. [Google Scholar]
- Boothe, J. , Nykiforuk, C. , Shen, Y. , Zaplachinski, S. , Szarka, S. and Kuhlman, P. (2010) Seed‐based expression systems for plant molecular farming. Plant Biotechnol. J. 8, 588–606. [DOI] [PubMed] [Google Scholar]
- Borgi, M.A. , Boudebbouze, S. , Mkaouar, H. , Maguin, E. and Rhimi, M. (2015) Bacillus phytases: current status and future prospects. Bioengineered, 6, 233–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyhan, D. and Daniell, H. (2011) Low‐cost production of proinsulin in tobacco and lettuce chloroplasts for injectable or oral delivery of functional insulin and c‐peptide. Plant Biotechnol. J. 9, 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, H.T. , Xiao, Y. , Weldon, W.C. , Oberste, S.M. , Chumakov, K. and Daniell, H. (2016) Cold chain and virus‐free chloroplast‐made booster vaccine to confer immunity against different poliovirus serotypes. Plant Biotechnol. J. 14, 2190–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, A. , Das, N.C. , Raha, S. , Babbit, R. , Huang, Q. and Zaitlin, D. (2010) Production of xylanase in transgenic tobacco for industrial use in bioenergy and biofuel applications. In Vitro Cell. Dev. Biol. Plant 46, 198–209. [Google Scholar]
- Chen, R. , Xue, G. , Chen, P. , Yao, B. , Yang, W. , Ma, Q. , Fan, Y. et al. (2008) Transgenic maize plants expressing a fungal phytase gene. Transgenic Res. 17, 633–643. [DOI] [PubMed] [Google Scholar]
- Chen, R. , Zhang, C. , Yao, B. , Xue, G. , Yang, W. , Zhou, X. and Zhang, J. (2013) Corn seeds as bioreactors for the production of phytase in the feed industry. J. Biotechnol. 165, 120–126. [DOI] [PubMed] [Google Scholar]
- Choct, M. (2006) Enzymes for the feed industry: past, present and future. Worlds Poult. Sci. J. 62, 5–16. [Google Scholar]
- Costa, F.G. , Goulart, C.C. , Figueiredo, D.F. , Oliveira, C.F. and Silva, J.H. (2008) Economic and environmental impact of using exogenous enzymes on poultry feeding. Int J Poult Sci. 7, 311–314. [Google Scholar]
- Cowieson, A.J. and Adeola, O. (2005) Carbohydrase, protease and phytase have an additive beneficial effect in nutritionally marginal diets for broiler chicks. Poult. Sci. 84, 1860–1867. [DOI] [PubMed] [Google Scholar]
- Daniell, H. , Chan, H.T. and Pasoreck, E.K. (2016) Vaccination via chloroplast genetics: affordable protein drugs for the prevention and treatment of inherited or infectious human diseases. Annu. Rev. Genet. 50, 595–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Rai, V. and Xiao, Y. (2019a) Cold chain and virus‐free oral polio booster vaccine made in lettuce chloroplasts confers protection against all three poliovirus serotypes. Plant Biotechnol. J. 17, 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Ribeiro, T. , Lin, S. , Saha, P. and Agarwal, A. (2019b) Validation of leaf and microbial pectinases: commercial launching of a new platform technology. Plant Biotechnol. J. 17, 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Nair, S.K. , Esmaeili, N. , Wakade, G. , Shahid, N. and Ganesan, P.K. (2022a) Debulking SARS‐CoV‐2 in saliva using angiotensin converting enzyme 2 in chewing gum to decrease oral virus transmission and infection. Mol. Ther. 5, 1966–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell, H. , Nair, S.K. , Guan, H.C. , Guo, Y.W. and Kulchar, R.J. (2022b) Debulking different Corona (SARS‐CoV‐2 delta, omicron, OC43) and Influenza (H1N1, H3N2) virus strains by plant viral trap proteins in chewing gums to decrease infection and transmission. Biomaterials 288, 121671–121683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, R. , Morris, E.R. and Hill, A.D. (1982) Bioavailability to rats of iron and zinc in calcium‐iron‐phytate and calcium‐zinc‐phytate complexes. Nutr. Res. 2, 319–322. [Google Scholar]
- Eskelin, K. , Ritala, A. , Suntio, T. , Blumer, S. , Holkeri, H. and Wahlstrom, E.H. (2009) Production of a recombinant full‐length collagen type I alpha‐1 and of a 45‐kDa collagen type I alpha‐1 fragment in barley seeds. Plant Biotechnol. J. 7, 657–672. [DOI] [PubMed] [Google Scholar]
- Fushinobu, S. , Uno, T. , Kitaoka, M. , Hayashi, K. , Matsuzawa, H. and Wakagi, T. (2011) Mutational analysis of fungal family 11 xylanases on pH optimum determination. J. Appl. Glycosci. 58, 107–114. [Google Scholar]
- Garcia‐Gimenez, G. , Russell, J. , Aubert, M.K. , Fincher, G.B. , Burton, R.A. , Waugh, R. , Tucker, M.R. et al. (2019) Barley grain (1,3;1,4)‐β‐glucan content: effects of transcript and sequence variation in genes encoding the corresponding synthase and endohydrolase enzymes. Sci. Rep. 9, 17250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R.K. , Gangoliya, S.S. and Singh, N.K. (2015) Reduction of phytic acid and enhancement of bioavailable micronutrients in food grains. J. Food Sci. Technol. 52, 676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusakov, A.V. , Kondratyeva, E.G. and Sinitsyn, A.P. (2011) Comparison of two methods for assaying reducing sugars in the determination of carbohydrase activities. Int. J. Anal. Chem. 283, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, A. , Yamaguchi, K. , Harada, M. , Horiguchi, K. , Takahashi, T. and Honda, H. (2006) Recombinant, rice‐produced yeast phytase shows the ability to hydrolyze phytate derived from seed‐based feed, and extreme stability during ensilage treatment. Biosci. Biotechnol. Biochem. 70, 1524–1527. [DOI] [PubMed] [Google Scholar]
- Han, N. , Na, C.L. , Chai, Y.Q. , Chen, J.S. , Zhang, Z.B. , Bai, B. , Bian, H.W. et al. (2016) Over‐expression of (1,3;1,4)‐β‐D‐glucanase isoenzyme EII gene results in decreased (1,3;1,4)‐β‐D‐glucan content and increased starch level in barley grains. J. Sci. Food Agric. 97, 122–127. [DOI] [PubMed] [Google Scholar]
- Holme, I.B. , Dionisio, G. , Brinch‐Pedersen, H. , Wendt, T. , Madesen, C.K. , Vincze, E. and Holm, P.B. (2012) Cisgenic barley with improved phytase activity. Plant Biotechnol. J. 10, 237–247. [DOI] [PubMed] [Google Scholar]
- Holme, I.B. , Dionisio, G. , Madsen, C.K. and Brinch‐Pedersen, H. (2017) Barley HvPAPhy_a as transgene provides high and stable phytase activities in mature barley straw and in grains. Plant Biotechnol. J. 15, 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, C.Y. , Cheng, K.J. , Tseng, T.H. , Wang, C.S. , Liu, L.F. and Yu, S.M. (2004) Production of two highly active bacterial phytases with broad pH optima in germinated transgenic rice seeds. Transgenic Res. 13, 29–39. [DOI] [PubMed] [Google Scholar]
- Hood, E.E. (2002) From green plants to industrial enzymes. Enzyme Microb. Technol. 30, 279–283. [Google Scholar]
- Hood, E.E. , Devaiah, S.P. , Fake, G. , Egelkrout, E. , Teoh, K.T. , Requesens, D.V. , Hayden, C. et al. (2012) Manipulating corn germplasm to increase recombinant protein accumulation. Plant Biotechnol. J. 10, 20–30. [DOI] [PubMed] [Google Scholar]
- Hyatt, D. , Chen, G.L. , LoCascio, P.F. , Land, M.L. , Larimer, F.W. and Hauser, L.J. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11, 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaime, H.C. , Damian, S. , Kristoffer, F. , Helen, C. , Davide, H. and Walter, M.C. (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, R. , Woyengo, T.A. , Li, J. , Bedford, M.R. , Vasanthan, T. and Zijlstra, R.T. (2015) Enzymes enhance degradation of the fiber‐starch‐protein matrix of distillers dried grains with solubles as revealed by a porcine in vitro fermentation model and microscopy. J. Anim. Sci. 93, 1039–1051. [DOI] [PubMed] [Google Scholar]
- Juanpere, J. , Perez‐Vendrell, A.M. , Angula, E. and Brufau, J. (2005) Assessment of potential interaction between phytase and glycosidase enzyme supplementation on nutrient digestibility in broilers. Poult. Sci. 84, 571–580. [DOI] [PubMed] [Google Scholar]
- Kogut, M.H. (2019) The effect of microbiome modulation on the intestinal health of poultry. Anim. Feed Sci. Technol. 250, 32–40. [Google Scholar]
- Kolodziejczyk, A.A. , Zheng, D. and Elinav, E. (2019) Diet‐microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 17, 1–12. [DOI] [PubMed] [Google Scholar]
- Kopriva, S. and Chu, C. (2018) Are we ready to improve phosphorus homeostasis in rice? J. Exp. Bot. 69, 3515–3522. [DOI] [PubMed] [Google Scholar]
- Kumari, U. , Singh, R. , Ray, T. , Rana, S. , Saha, P. and Malhotra, K. (2019) Validation of leaf enzymes in the detergent and textile industries: launching of a new platform technology. Plant Biotechnol. J. 17, 1167–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, K.C. and Daniell, H. (2015) Low‐cost oral delivery of protein drugs bioencapsulated in plant cells. Plant Biotechnol. J. 13, 1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, O.S. and Sun, S.S.M. (2009) Plant seeds as bioreactors for recombinant protein production. Biotechnol. Adv. 27, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Lee, C. and Hristov, A.N. (2013) Short communication: evaluation of acid‐insoluble ash and indigestible neutral detergent fiber as total‐tract digestibility markers in dairy cows fed corn silage‐based diets. J. Dairy Sci. 96, 5295–5299. [DOI] [PubMed] [Google Scholar]
- Lenkova, T.N. , Egorov, I.A. , Egorova1, T.A. , Manukyan, V.A. and Vertiprakhov, V.G. (2020) Intestinal microbiota and broiler performance upon administration of phytase to increase phosphorus digestibility and nutrient utilization from feed. Agricult. Biol. 55, 406–416. [Google Scholar]
- Liu, D. , Guo, S. and Guo, Y. (2012) Xylanase supplementation to a wheat‐based diet alleviated the intestinal mucosal barrier impairment of broiler chickens challenged by Clostridium perfringens . Avian Pathol. 41, 291–298. [DOI] [PubMed] [Google Scholar]
- Lu, C. and Jeffries, T. (2007) Shuffling of promoters for multiple genes to optimize xylose fermentation in an engineered Saccharomyces cerevisiae strain. Appl. Environ. Microb. 73, 6072–6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.K. , Drake, P.M.W. and Christou, P. (2003) The production of recombinant pharmaceutical proteins in plants. Nat. Rev. Genet. 4, 794–805. [DOI] [PubMed] [Google Scholar]
- McKie, V.A. and McCleary, B.V. (2016) A novel and rapid colorimetric method for measuring total phosphorus and phytic acid in foods and animal feeds. J. AOAC Int. 99, 738–743. [DOI] [PubMed] [Google Scholar]
- Moran, K. , de Lange, C.F.M. , Ferket, P. , Fellner, V. , Wilcock, P. and van Heugten, E. (2016) Enzyme supplementation to improve the nutritional value of fibrous feed ingredients in swine diets fed in dry or liquid form. J. Anim. Sci. 94, 1031–1040. [DOI] [PubMed] [Google Scholar]
- Mrízová, K. , Holasková, E. , Oz, M.T. , Jiskrová, E. and Galuszka, P. (2014) Transgenic barley: a prospective tool for biotechnology and agriculture. Biotechnol. Adv. 32, 137–157. [DOI] [PubMed] [Google Scholar]
- Munyaka, P.M. , Nandha, N.K. , Kiarie, E. , Nyachoti, C.M. and Khafipour, E. (2016) Impact of combined β‐glucanase and xylanase enzymes on growth performance, nutrients utilization and gut microbiota in broiler chickens fed corn or wheat‐based diets. Poult. Sci. 95, 528–540. [DOI] [PubMed] [Google Scholar]
- Nyannor, E.K.D. , Williams, P. , Bedford, M.R. and Adeola, O. (2007) Corn expressing an Escherichia coli‐derived phytase gene: a proof‐of‐concept nutritional study in pigs. J. Anim. Sci. 85, 1946–1952. [DOI] [PubMed] [Google Scholar]
- Ojha, B.K. , Singh, P.K. and Shrivastava, N. (2019) Chapter 7 ‐ Enzymes in the animal feed industry. In Enzymes in Food Biotechnology ( Kuddus, M. , ed), pp. 93–109. Amsterdam: Academic Press. [Google Scholar]
- Pantaleoni, L. , Longoni, P. , Ferroni, L. , Baldisserotto, C. and Cella, R. (2014) Chloroplast molecular farming: efficient production of a thermostable xylanase by Nicotiana tabacum plants and long‐term conservation of the recombinant enzyme. Protoplasma 251, 639–648. [DOI] [PubMed] [Google Scholar]
- Park, J. , Yan, G. , Kwon, K.C. , Liu, M. , Gonnella, P.A. , Yang, S. and Daniell, H. (2020) Oral delivery of novel human IGF‐1 bioencapsulated in lettuce cells promotes musculoskeletal cell proliferation, differentiation and diabetic fracture healing. Biomaterials, 233, 119591–119602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, M. , Johnson, J. , Brettell, R. , Jacobsen, J. and Xue, G. (2000) Transgenic barley expressing a fungal xylanase gene in the endosperm of the developing grains. Mol. Breed. 6, 113–123. [Google Scholar]
- Peng, R.H. , Xiong, A.S. and Yao, Q.H. (2006) A direct and efficient PAGE‐mediated overlap extension PCR method for gene multiple‐site mutagenesis. Appl. Microbiol. Biotechnol. 73, 234–240. [DOI] [PubMed] [Google Scholar]
- Perera, I. , Seneweera, S. and Hirotsu, N. (2018) Manipulating the phytic acid content of rice grain toward improving micronutrient bioavailability. Rice, 11, 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera, W. , Abdollahi, M.R. , Ravindran, V. , Zaefarian, F. , Wester, T.J. and Ravindran, G. (2019) Nutritional evaluation of two barley cultivars, without and with carbohydrase supplementation, for broilers: metabolisable energy and standardised amino acid digestibility. Br. Poultry Sci. 60, 404–413. [DOI] [PubMed] [Google Scholar]
- Ptak, A. , Bedford, M.R. , Świątkiewicz, S. , Żyła, K. and Józefiak, D. (2015) Phytase modulates ileal microbiota and enhances growth performance of the broiler chickens. PLoS One, 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J. , Han, H.J. , Sun, B.H. , Chen, L. , Yu, C.Y. , Peng, R.H. and Yao, Q.H. (2016) Residue mutations of xylanase in Aspergillus kawachii alter its optimum pH. Microbiol. Res. 182, 1–7. [DOI] [PubMed] [Google Scholar]
- Qu, L. and Takaiwa, F. (2004) Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnol. J. 2, 113–125. [DOI] [PubMed] [Google Scholar]
- Ramessar, K. , Capell, T. and Christou, P. (2008a) Molecular pharming in cereal crops. Phytochem. Rev. 7, 579–592. [Google Scholar]
- Ramessar, K. , Sabalzaa, M. , Capell, T. and Christou, P. (2008b) Maize plants: an ideal production platform for effective and safe molecular pharming. Plant Sci. 174, 409–419. [Google Scholar]
- Ramos, O.S. and Malcata, F.X. (2017) Food‐grade enzymes. Comp. Biotechnol. 3, 587–603. [Google Scholar]
- Ranjan, A. , Sahu, N.P. and Deo, A.D. (2017) Xylanase and phytase supplementation in the de‐oiled rice bran (DORB) based diet improves the growth performance of Labeo rohita . Int. J. Curr. Microbiol. App. Sci. 6, 1493–1503. [Google Scholar]
- Ranjan, A. , Sahu, N.P. and Deo, A.D. (2018a) Comparative evaluation of fermented and non‐fermented de‐oiled rice bran with or without exogenous enzymes supplementation in the diet of Labeo rohita . Fish Physiol. Biochem. 44, 1037–1049. [DOI] [PubMed] [Google Scholar]
- Ranjan, A. , Kumar, S. , Sahu, N.P. and Kumar, S. (2018b) Comparative growth performance, in vivo digestibility and enzyme activities of Labeo rohita fed with DORB based formulated diet and commercial carp feed. Turkish J. Fish. Aquat. Sci. 18, 1025–1036. [Google Scholar]
- Ranjan, A. , Kumar, S. , Sahu, N.P. and Kumar, S. (2021) Exogenous phytase and xylanase supplementation of formulated diets for rohu (Labeo rohita): impact on haematology, histology and IGF I gene expression. Fish Physiol. Biochem. 47, 49–58. [DOI] [PubMed] [Google Scholar]
- Ranjan, A. , Kumar, S. and Sahu, N.P. (2022) Strategies for maximizing utilization of de‐oiled rice bran (DORB) in the fish feed. Aquac. Int. 30, 99–114. [Google Scholar]
- Roberts, T. , Wilson, J. , Guthrie, A. , Cookson, K. , Vancraeynest, D. and Schaeffer, J. (2015) New issues and science in broiler chicken intestinal health: emerging technology and alternative interventions. J. Appl. Poultry Res. 24, 257–266. [Google Scholar]
- Segata, N. , Izard, J. , Waldron, L. , Gevers, D. , Miropolsky, L. , Garrett, W.S. and Huttenhower, C. (2011) Metagenomic biomarker discovery and explanation. Genome Biol. 12, 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selle, P.H. , Ravindran, V. , Caldwell, A. and Bryden, W.L. (2000) Phytate and phytase: consequences for protein utilisation. Nutr. Res. Rev. 13, 255–278. [DOI] [PubMed] [Google Scholar]
- Selle, P.H. , Ravindran, V. and Partridge, G.G. (2009) Beneficial effects of xylanase and/or phytase inclusions on ileal amino acid digestibility, energy utilization, mineral retention and growth performance in wheat‐based broiler diets. Anim. Feed Sci. Technol. 53, 303–313. [Google Scholar]
- Shen, B. , Sun, X. , Zuo, X. , Shilling, T. , Apgar, J. and Ross, M. (2012) Engineering a thermoregulatedintein‐modified xylanase into maize for consolidated lignocellulosic biomass processing. Nat. Biotechnol. 30, 1131–1136. [DOI] [PubMed] [Google Scholar]
- Shewry, P.R. , Halford, N.G. , Belton, P.S. and Tatham, A.S. (2002) The structure and properties of gluten: an elastic protein from wheat grain. Philos. Trans. R Soc. Lond. 357, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivange, A.V. , Serwe, A. , Dennig, A. , Roccatano, D. , Haefner, S. and Schwaneberg, U. (2012) Directed evolution of a highly active Yersinia mollaretii phytase. Appl. Microbiol. Biotechnol. 95, 405–418. [DOI] [PubMed] [Google Scholar]
- Slominski, B.A. (2011) Recent advances in research on enzymes for poultry diets. Poult. Sci. 90, 2013–2023. [DOI] [PubMed] [Google Scholar]
- Stahl, R. , Dargatz, H. , Luhrs, R. and Berkemeyer, M. (2009a) Endosperm‐specific plant promoter for cultivated plants. US Patent 7576195 B2 .
- Stahl, R. , Luhrs, R. and Dargatz, H. (2009b) Thaumatin from transgenic barley. US Patent 20090031458 .
- Tian, Y.S. , Wang, B. , Peng, R.H. , Xu, J. , Li, T. , Fu, X.Y. , Xiong, A.S. et al. (2019) Enhancing carotenoid biosynthesis in rice endosperm by metabolic engineering. Plant Biotechnol. J. 17, 849–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y.S. , Wang, B. , Xu, J. , Fu, X.Y. , Gao, J.J. , Yao, Q.H. and Peng, R.H. (2020) Metabolic engineering of rice endosperm for betanin biosynthesis. New Phytol. 225, 1915–1922. [DOI] [PubMed] [Google Scholar]
- Tingay, S. , McElroy, D. , Kalla, R. , Fieg, S. , Wang, M.B. and Thornton, S. (1997) Agrobacterium tumefaciens mediated barley transformation. Plant J. 11, 1369–1376. [Google Scholar]
- Tremblay, R. , Wang, D. , Jevnikar, A.M. and Ma, S. (2010) Tobacco, a highly efficient green bioreactor for production of therapeutic proteins. Biotechnol. Adv. 28, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Gao, X. , Su, Q. , Wu, W. and An, L. (2007) Expression of heat stable phytase from Aspergillus fumigatus in tobacco (Nicotiana tabacum L. cv. NC89). Ind. J. Biochem. Biophys. 44, 26–30. [PubMed] [Google Scholar]
- Ward, N.E. (2021) Debranching enzymes in corn/soybean meal‐based poultry feeds: a review. Poult. Sci. 100, 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.P. , Druka, A. , Horvath, H. , Kleinhofs, A. , Kannangara, C.G. and Wettstein, D.V. (2000) Functional characterization of seed coat specific members of the barley germin gene family. Plant Physiol. Biochem. 38, 685–698. [Google Scholar]
- Wyss, M. , Brugger, R. , Kronenberger, A. , Remy, R. , Fimbel, R. , Oesterhelt, G. , Lehmann, M. et al. (1999b) Biophysical characterization of fungal phytases (myo‐inositol hexakisphosphate phosphohydrolases): molecular size, glycosylation pattern, and engineering of proteolytic resistance. Appl. Environ. Microbiol. 65, 359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, A.S. , Yao, Q.H. , Peng, R.H. , Duan, H. , Li, X. , Fan, H.Q. , Cheng, Z.M. et al. (2006) PCR‐based accurate synthesis of long DNA sequences. Nat. Protoc. 1, 791–797. [DOI] [PubMed] [Google Scholar]
- Xu, J.F. , Dolan, M.C. , Medrano, G. , Cramer, C.L. and Weathers, P.J. (2012) Green factory: plants as bioproduction platforms for recombinant proteins (special issue: production of recombinant proteins.). Biotechnol. Adv. 30, 1171–1184. [DOI] [PubMed] [Google Scholar]
- Yadav, S. and Jha, R. (2019) Strategies to modulate the intestinal microbiota and their effects on nutrient utilization, performance, and health of Poultry. J. Anim. Sci Biotech. 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P. , Wang, Y. , Bai, Y. , Meng, K. , Luo, H. and Yuan, T. (2007) Expression of xylanase with high specific activity from Streptomyces olivaceoviridis A1 in transgenic potato plants (Solanum tuberosum L.). Biotechnol. Lett. 29, 659–667. [DOI] [PubMed] [Google Scholar]
- Yang, L. , Hirose, S. , Takahashi, H. , Taiji Kawakatsu, T. and Takaiwa, F. (2012) Recombinant protein yield in rice seed is enhanced by specific suppression of endogenous seed proteins at the same deposit site. Plant Biotechnol. J. 10, 1035–1045. [DOI] [PubMed] [Google Scholar]
- Zhang, Z.B. , Kornegay, E.T. , Radcliffe, J.S. , Wilson, J.H. and Veit, H.P. (2000) Comparison of phytase from genetically engineered Aspergillus and canola in weaning pig diets. J. Anim. Sci. 78, 2868–2878. [DOI] [PubMed] [Google Scholar]
- Zhang, D. , Vanfossen, A.L. , Pagano, R.M. , Johnson, J.S. , Parker, M.H. and Pan, S. (2011) Consolidated pretreatment and hydrolysis of plant biomass expressing cell wall degrading enzymes. Bioenergy Res. 4, 276–286. [Google Scholar]
- Zhang, Y. , Xu, X. , Zhou, X. , Chen, R. and Yang, P. (2013) Overexpression of an acidic endo‐b‐1,3‐1,4‐glucanase in transgenic maize seed for direct utilization in animal feed. PLoS One, 8, e81993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H. , Tang, J. , Cao, L. , Jia, G. , Long, D. , Liu, G. , Chen, X. et al. (2015) Characterization of bioactive recombinant antimicrobial peptide parasin I fused with human lysozyme expressed in the yeast Pichia pastoris system. Enzyme Microb. Technol. 77, 61–67. [DOI] [PubMed] [Google Scholar]
- Zhu, Y.M. , He, Y.M. , Wang, B. , Peng, R.H. and Yao, Q.H. (2018) Characterization of a novel endo‐β‐1,4‐glucanase from the model soft rot phytopathogen Pectobacterium wasabiae SCC3193. J. Pure Appl. Microbiol. 12, 1–12. [Google Scholar]
- Ziegler, M.T. , Thomas, S.R. and Danna, K.J. (2000) Accumulation of a thermostable endo‐1,4‐β‐d‐glucanase in the apoplast of Arabidopsis thaliana leaves. Mol. Breed. 6, 37–46. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Expression of the genes in transgenic barley by RT‐PCR using cDNA as template (1:565‐1; 2:564‐6; 3:565‐16; M: marker; W: wild type barley).
Figure S2 Alignment of amino acid sequences among Yersinia mollaretii (YmAPPA), Candidatus Hamiltonella (ChAPPA), Cronobacter malonaticus (CmAPPA), Citrobacter sp. KTE32 (CsAPPA), Escherichia albertii (EaAPPA), Hafnia alvei ATCC 51873 (HaAPPA), Yersinia aldovae ATCC 35236 (YaAPPA), Shigella boydii ATCC 9905 SGB (SbAPPA), Aspergillus niger (AnAPPA).
Figure S3 Alignment of amino acid sequences among Pectobacterium carotovorum (PceglAS), Stachybotrys chartarum (SceglAS), Gibberella zeae (GzeglAS), Streptomyces avermitilis (SaeglAS), Aspergillus oryzae (AoeglAS), Thermobispora bispora (TbeglAS).
Figure S4 Alignment of amino acid sequences among Aspergillus kawachii (AkxynCS), Bacillus sp (BsxynAS), Streptomyces sviceus(SsxynBS), Thermobacillus composti KWC4 (TcxynBS), Catenulispora acidiphila (CaxynBS), Thermobispora bispora (TbxynBS).
Notes S1 The sequence information after codon optimization as follow.
Table S1 Primers for molecular analysis in transgenic barley using cDNA as template.
Table S2 Ingredients and nutrients of diets for feeding trial.


