Background:
The genetics of rheumatic heart disease (RHDGen) Network was developed to assist the discovery and validation of genetic variations and biomarkers of risk for rheumatic heart disease (RHD) in continental Africans, as a part of the global fight to control and eradicate rheumatic fever/RHD. Thus, we describe the rationale and design of the RHDGen study, comprising participants from 8 African countries.
Methods:
RHDGen screened potential participants using echocardiography, thereafter enrolling RHD cases and ethnically-matched controls for whom case characteristics were documented. Biological samples were collected for conducting genetic analyses, including a discovery case-control genome-wide association study (GWAS) and a replication trio family study. Additional biological samples were also collected, and processed, for the measurement of biomarker analytes and the biomarker analyses are underway.
Results:
Participants were enrolled into RHDGen between December 2012 and March 2018. For GWAS, 2548 RHD cases and 2261 controls (3301 women [69%]; mean age [SD], 37 [16.3] years) were available. RHD cases were predominantly Black (66%), Admixed (24%), and other ethnicities (10%). Among RHD cases, 34% were asymptomatic, 26% had prior valve surgery, and 23% had atrial fibrillation. The trio family replication arm included 116 RHD trio probands and 232 parents.
Conclusions:
RHDGen presents a rare opportunity to identify relevant patterns of genetic factors and biomarkers in Africans that may be associated with differential RHD risk. Furthermore, the RHDGen Network provides a platform for further work on fully elucidating the causes and mechanisms associated with RHD susceptibility and development.
Keywords: cardiovascular diseases, genetics, infections, rheumatic heart disease
Rheumatic heart disease (RHD) is a preventable sequela of rheumatic fever, characterized by permanent heart valve damage.1 RHD is the leading indication for cardiac surgery in the young (adolescents and young adults) in Sub-Saharan Africa, which carries a quarter of the global disease burden.1–3 Worldwide, RHD affects ≈40.5 million individuals, claiming up to 340 000 lives annually,4 the majority of whom live in low- and middle-income countries. The recognition of RHD as a common cause of heart failure, infective endocarditis, stroke, and maternal and perinatal mortality over the past 2 decades led to the reinstatement of RHD as a major public health priority.3 In 2018, the World Health Assembly passed a resolution on RHD mandating a coordinated global response.1,5,6
RHD’s persistence as a major public health issue in low- and middle-income countries is attributed to associated risk factors such as the lack of effective RHD prevention, control and elimination initiatives. Additionally, known risk factors for rheumatic fever (RHD’s prequel’s; eg, failure to drastically change socioeconomic factors [affecting those living in poverty and overcrowded dwellings], limited use/availability of antibiotics [especially, intramuscular Penicillin G Benzathine], and increased genetic susceptibility7) may also play a role. A recent systematic review indicated that rheumatic fever was heritable, reporting high odds of rheumatic fever among monozygotic twins with an estimated heritability of 60%.8 Furthermore, 60% of rheumatic fever patients develop RHD, and despite there being a proven association between group A Streptococcus infection and RHD, the triggered autoimmune process in RHD can occur autonomously after removing the stimulus. This suggests that after initiation of the autoimmune response via molecular mimicry, host factors (most likely genetic) play an important role in disease progression in susceptible individuals.9–11 Thus, using genetic studies, which could identify people at high risk, may help develop effective RHD prevention/control measures such as targeted approaches for RHD screening and treatments.8,12 Utilizing a hypothesis-free approach, such as a genome-wide association study (GWAS), may highlight key genetic risk factors associated with disease susceptibility.13
Here, we report on the rationale and design, and present baseline characteristics, of the Rheumatic Heart Disease Genetics (RHDGen) study with the primary objective to identify genetic variants affecting susceptibility and resistance to RHD in Africans. The secondary objective is to convey the polygenic nature of RHD susceptibility, its heritability in the study population, and to replicate any prior or novel GWAS findings. While seeking to identify genetic risk factors associated with RHD, and elucidate the pathogenesis of RHD susceptibility in Africans, a biorepository was formed, which will also serve as a resource for further studies, including biomarker analyses.
Methods
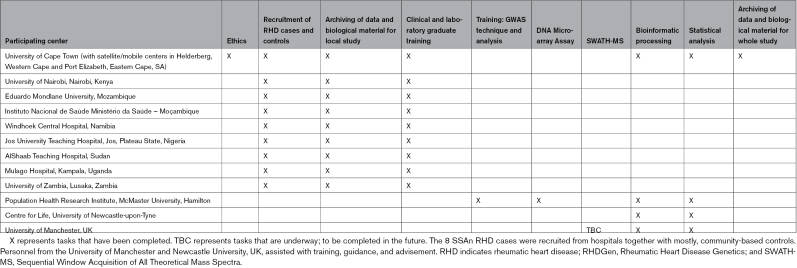
The development of the RHDGen Network (participating countries and sites: Figure 1 and Tables 1 and 2) and its related substudies was approved by appropriate institutional review committees, and all subjects provided written informed consent. Full details of data and methods used in this study are presented in the Supplemental Material and Methods. As per the American Heart Association’s TOP (Transparency and Openness Promotion) Guidelines, we declare that upon reasonable request data will be made available. Due to the sensitive nature of the data collected for this study, reasonable requests to access the dataset from qualified researchers trained in human subject confidentiality protocols may be sent to mark.engel@uct.ac.za and cc to taffymach@yahoo.com. The authors declare that all other supporting data are available within the article and the Supplemental Material.
Figure 1.
Location of the Rheumatic Heart Disease Genetics (RHDGen) recruitment site countries (yellow) and regional sites (dark blue).
Table 1.
Participating Centers for RHDGen and Scheduled Activities
Table 2.
RHDGen Countries, their Principal Investigator (PI), and Site Timelines

Results
Study Timeline and Baseline Clinical Characteristics
From December 31, 2012 to March 31, 2018, all GWAS participants and trio probands were screened by echocardiography. Preliminary data cleaning and analyses commenced May 15, 2017, until June 30, 2020. The GWAS arm included 2548 RHD cases and 2261 controls (3301 women [69%]; mean [SD] age, 37 [16.3] years), as per Table 3. RHD cases recruited were predominantly Black (66%), Admixed (23%), and other ethnicities classified as continental African citizens (11%); principal component analysis presented in Figure 2A. Baseline characteristics which included 34% of cases were asymptomatic, 26% of cases had valve surgery, and 23% of cases had atrial fibrillation. Most cases (41%) had a slight limitation in physical activity categorized by the New York Heart Association functional classification (Class II); presenting with mild symptoms (mild shortness of breath and/or angina) and slight limitation during ordinary activity.14 The trio family-based arm included 116 RHD trio probands and 232 matching parents; from the different ancestries across the 8 Sub-Saharan Africa countries in RHDGen (Figure 2B).
Table 3.
Clinical Characteristics of GWAS Participants

Figure 2.
An example of the principal component analysis (PCA) from GCTA for the participants of the RHDGen GWAS and the trio family study. NB: 1000G reference populations were from AFR (Black and African American), AMR (Ad Mixed American), EAS (East Asian), EUR (European) and SAS (South Asian) ancestries. From the 8 Sub-Saharan African countries in RHDGen, all populations self-reported as citizens/permanent residents of an African country. Hence, the “BLACK” group represents Black Africans, “ADMIXED” represents the “South African Coloureds” (SAC; ie, the same as the “Admixed Africans” description), “SUDANESE ARAB” are those who reside in Sudan and self-report being of Arab descent/ethnicity, “EUROPEAN” are those of European ancestry, “SOUTH ASIAN” are those of South Asian ancestry, “EAST ASIAN” are those of East Asian ancestry, and “NA” are those who did not self-report as identifying with any ethnicity/known ancestry.
Incidental Outcomes and Benefits
The RHDGen Network was part of the ASAP programme for ARF/RHD in Africa, which incorporates awareness-raising as 1 of 4 pillars to succesful eradication of RHD from Africa.15 Thus, fundamental to the RHDGen project is an overarching commitment to addressing the needs of this patient community in the different countries in which the research took place. Thus, a crucial component of our work is research aimed at making an impact as driven by the African Union communique.16
Thus far, the RHDGen Network has also developed additional indirect patient and community benefits, including increased access to RHD screening and patient care, developed and participated in patient/engagement events (eg, Listen to your heart).17 Hosted disease/genetics educational awareness activities, trained several experts in the field, as well as, advocated globally to change RHD from a neglected tropical disease status to a global health priority. In particular, our work on this and other RHD-related research projects has allowed us to work with WHO Africa to revise treatment guidelines for RHD in Africa. Furthermore, this research has allowed our research teams to engage with the World Heart Federation to lobby for greater focus on RHD and other heart conditions prevalent in the Global South.
Discussion
RHDGen is one of the largest case-control genetic association studies for RHD to date. It is also one of the first studies to evaluate the genetic susceptibility of RHD in continental Africans. While other genetic association studies have examined RHD in 4 other populations (Aboriginal Australians, Asians, Europeans, and Oceanians18–20), RHDGen is the first to do so in multiple African countries at a large scale.21
RHDGen had several unique and versatile features in its rationale, design, and execution. First, a resourceful aspect of this study was that the case report form was developed to include the majority of fields from other large RHD studies, like REMEDY (Global Rheumatic Heart Disease Registry). This allowed both current and future clinical/epidemiological and genetic questions to be addressed in the current study, as well as further research, such as meta-analyses with other epidemiological studies. Second, the presence of REMEDY IDs and participants may allow future prospective case analyses, as both participants are screened during REMEDY and RHDGen, as well as retrospective case-control analyses can be performed and genetic factors explored, too. Third, the transfer of the paper-based case report form to the electronic case report form in OpenClinica helped with real-time access to data and actionable visualizations, which made the dataset cleaner and improved workflows. Subsequently, another resourceful aspect of this study was the development of the RHD biobank, as it is a useful resource for future RHD work in Africans. Finally, another innovative aspect of this study is to use family-based genetic data for replication to minimize false positive rates.
RHDGen had several context-specific challenges due to using a family-based design for replication. Although family studies were previously the gold standard for genetic research due to their robustness, in practice they are now rarely used. This is due to the various challenges associated with recruiting complete families, often leading to smaller sample sizes than targeted.22 Similarly, RHDGen’s trio family recruitment numbers were significantly lower than anticipated; with only 440 families before QC (including parent–child duo families) versus the anticipated 2000. In our study across 8 different African countries, most issues arose from missing parental information.
Key factors that made complete trio recruitment difficult in RHDGen were: First, the African country with the largest GWAS recruitment numbers (ie, South Africa), had mostly female-headed households (57%) and elevated father absenteeism (≈14%) from a variety of reasons including possibly the high adult deaths from HIV/AIDs, since the 1980s. Second, trio probands were estranged from parents who worked as migrant workers, due to the impacts of the migrant labor system, for example, in South Africa under apartheid (1994), which prohibited male pass-controlled laborers from cohabiting with their spouse/child in work-related housing. This has been partially maintained in low income occupations, promoting offspring and working parent(s) estrangement/absenteeism.23 Third, distances between adult trio probands and their parents’ homes were far or inaccessible, significantly reducing dual parental recruitment. For instance, public health hospital systems and structures (eg, cardiac clinics) are often centralized and near richer neighborhoods (including even more private cardiac facilities), as CVD used to be thought to commonly affect only the rich. However, nowadays CVDs like RHD often occur in the children of the poor, who often live further away in impoverished, overcrowded townships/farm areas.
Eventually, toward the end of the study, a few community outreaches were attempted to increase trio family member recruitment. For example, Cape Town developed mobile cardiac clinic trips at Helderberg to increase trio recruitment. This outreach was a weekend mobile clinic site and a community-based outreach, enabling complete trio families to provide consent, be screened, provide blood, and be enrolled/included, all at once. Ultimately, the parents were mostly recruited in the countryside (ie, rural or farming areas); hence, these are poorly-resourced areas where improvization was needed and limited data were available. Hence, a polygenic transmission disequilibrium testing method was used for replication, to resolve the small sample size and limited parental information available.21,24 Thus, future family studies in Africa are recommended to include relevant contextual/tailored financial, transportation, and logistics considerations flexible enough to maximize the desired enrollment of the study participants.
Future Directions
We plan to carry out genetic and nongenetic follow-ups for further validation and replication with RHDGen; for instance, attempting collaborative meta-analyses, and mendelian randomization. Future research initiatives in RHD research are recommended to include functional validation through gene disruption studies, fine-mapping, and additional genotyping. Furthermore, a subset of RHDGen serum samples are currently undergoing SWATH-MS evaluation, which will represent the first RHD proteomic profiles in continental Africans. Thus, we invite investigators and funders with shared interests and resources to join and collectively enhance research efforts in RHDGen.
Conclusions
In summary, the RHDGen Network is a global collaboration among investigators who have recruited patients with RHD across Africa, seeking to gain a better understanding of RHD susceptibility, pathogenesis, and disease development. The RHDGen Network developed a unique network and biorepository to investigate RHD genetics and biomarkers in continental Africans. Ultimately, we hope that our work and collaborations can guide future RHD diagnostic, prevention, management and treatment options.
This research was funded in whole, or in part, by the Wellcome Trust [099313/B/12/A]. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Article version arising from this submission.
Article Information
Acknowledgments
In memory of deceased co-authors Bongani M Mayosi MD DPhil, Lungile Pepeta, MD and Veronica Francis, RN.
We would like to thank participants for being part of our study and the study planning, training, data entry, and cleaning staff of the Mayosi Research Group (MRG) Coordinating Office team, Department of Medicine, UCT, and collaborating sites. We acknowledge the UCT HICRA/CHI and its cardiovascular genetics (CVG) laboratory for assisting with all the preliminary wet laboratory work. We would also like to acknowledge PHRI and its CRLB-GMEL lab for assisting with the wet and dry laboratory work. Finally, we would also like to commend the Keavney Lab, Manchester, UK for their work on the proteomics; supported by the British Heart Foundation (BHF).
BMM (before his passing), BK, GP, HJC, RR, J.D-V, ML, TM, and MEE made substantial contributions to the conception and design of the RHDGen study. BMM, CC, MEE, SP C.H-H., SO, CM, JM, A.M., A.E-S., F.B-T., NL, MN, and LZ contributed to the acquisition of study participants and samples, as well as the development, management and completion of the RHDGen participant database. TM, BM, SP, MEE, and BMM reconciled the trios. BMM, GS, BM, and TM contributed to the laboratory studies by handling, acquiring, and processing RHDGen’s biological samples and developing, managing, and completing the RHDGen biological study samples database and the RHDGen biobank at HICRA & CHI, CVG laboratory, UCT. TM, MC, MEE, and GP performed and reviewed the statistics and bioinformatics analyses. TM, GP, MC, BK, HJC, MEE, and BMM managed the study results’ interpretation. TM wrote the first draft of the article under the supervision of MEE and GP. All authors contributed to revisions and approved the final version for publication, as per the international committee of medical journal editors (ICMJE) criteria.
Sources of Funding
RHDGen: The RHDGen Network was founded by funding awarded to Bongani Mayosi by the Wellcome Trust; described here: https://h3africa.org/index.php/consortium/the-rhdgen-network-genetics-of-rheumatic-heart-disease-and-molecular-epidemiology-of-streptococcus-pyogenes-pharyngitis/, https://app.dimensions.ai/details/grant/grant.3640606 and https://europepmc.org/grantfinder/grantdetails?query=pi%3A%22Mayosi%2BBM%22%2Bgid%3A%22099313%22%2Bga%3A%22Wellcome%20Trust%22. This research was funded in whole, or in part, by the Wellcome Trust [Grant number: 099313/B/12/A].
TM was supported by scholarships from the UCT (Mayosi Research Group Fellowships, the Crasnow Travel Fellowship, the Departmental Research Committee (DRC) of Medicine and the Pan African Society of Cardiology Award), Wellcome Trust, and Population Health Research Institute (PHRI) and McMaster University. LJZ Zühlke was supported by the South African Medical Research Council (SAMRC) through its Division of Research Capacity Development under the Mid-Career Scientist Programme. LJZ also received funding received from the National Research Foundation of South Africa (NRFSA), as well as the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement, via the African Research Leader Award (MR/S005242/1). MC is supported by a Canadian Institute of Health Research doctoral award (# 412329) and has received Bayer’s consulting fees. GP has received consulting fees from Sanofi, Bristol-Myers Squibb, Lexicomp, Amgen, and Bayer; has received support for research through his institution from Sanofi; and has received support from the Canada Research Chair in Genetic and Molecular Epidemiology, and CISCO Professorship in Integrated Health Systems. ME received support from the RHDGen Wellcome Trust grant and the South African National Research Foundation (NRF) # 116287, the American Heart Association, United States (Grant number: NW17SFRN33630027) and UCT. BK is supported by the BHF and is a BHF Professor of Cardiovascular Medicine and the Director of the Manchester BHF Accelerator, University of Manchester.
Disclosures
None.
Supplemental Material
Supplemental Methods
Supplemental Outcomes
Figures S1–S6
Supplementary Material
Nonstandard Abbreviations and Acronyms
- GWAS
- genome-wide association studies
- REMEDY
- Global Rheumatic Heart Disease Registry
- RHD
- rheumatic heart disease
- RHDGen
- the genetics of Rheumatic Heart Disease Network
G. Paré & M.E. Engel contributed equally.
A complete list of the investigators in The Genetics of Rheumatic Heart Disease (RHDGen) Network Consortium is provided in the Supplemental Appendix.
For Sources of Funding and Disclosures, see page 57.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.121.003641.
Contributor Information
Tafadzwa Machipisa, Email: Tafadzwa.Machipisa@phri.ca.
Chishala Chishala, Email: chishala.chishala@gmail.com.
Gasnat Shaboodien, Email: gasnat.shaboodien@uct.ac.za.
Liesl J. Zühlke, Email: liesl.zuhlke@uct.ac.za.
Babu Muhamed, Email: kambibabu@gmail.com.
Shahiemah Pandie, Email: shahiemah.pandie@uct.ac.za.
Jantina de Vries, Email: jantina.devries@uct.ac.za.
Nakita Laing, Email: n.verkijk@uct.ac.za.
Alexia Joachim, Email: alexia.joachim@uct.ac.za.
Rezeen Daniels, Email: rezeen.daniels@uct.ac.za.
Mpiko Ntsekhe, Email: mpiko.ntsekhe@uct.ac.za.
Christopher T. Hugo-Hamman, Email: christopher@hugo-hamman.com.
Bernard Gitura, Email: bgit27@gmail.com.
Stephen Ogendo, Email: swoogendo@gmail.com.
Peter Lwabi, Email: plwabi@yahoo.com.
Emmy Okello, Email: emmyoks@gmail.com.
Albertino Damasceno, Email: tino_7117@yahoo.com.br.
Celia Novela, Email: celia.novela@gmail.com.
Ana O. Mocumbi, Email: amocumbi@gmail.com.
Geoffrey Madeira, Email: geogirassol@gmail.com.
John Musuku, Email: jmusuku2001@yahoo.co.uk.
Agnes Mtaja, Email: msanida1@gmail.com.
Ahmed ElSayed, Email: asaelsayed@hotmail.com.
Huda H.M. Alhassan, Email: huda20@yahoo.com.
Fidelia Bode-Thomas, Email: bodefide@yahoo.com.
Christopher Yilgwan, Email: yilgwan@hotmail.com.
Ganiyu Amusa, Email: drganiamusa@gmail.com.
Esin Nkereuwem, Email: esinofils@yahoo.com.
Nicola Mulder, Email: nicola.mulder@uct.ac.za.
Raj Ramesar, Email: raj.ramesar@uct.ac.za.
Maia Lesosky, Email: maia.lesosky@uct.ac.za.
Heather J. Cordell, Email: heather.cordell@newcastle.ac.uk.
Michael Chong, Email: michael.chong@phri.ca.
Bernard Keavney, Email: bernard.keavney@manchester.ac.uk.
Guillaume Paré, Email: pareg@mcmaster.ca.
References
- 1.Beaton A, Kamalembo FB, Dale J, Kado JH, Karthikeyan G, Kazi DS, Longenecker CT, Mwangi J, Okello E, Ribeiro ALP. The American Heart Association’s call to action for reducing the global burden of rheumatic heart disease: a policy statement from the American Heart Association. Circulation. 2020;142:e358. doi: 10.1161/CIR.0000000000000922 [DOI] [PubMed] [Google Scholar]
- 2.Tissot C, da Cruz EM, Kalangos A, Buckvold S, Myers PO. Mitral Valve Anomalies and Related Disorders Critical Care of Children with Heart Disease. Springer; 2020. [Google Scholar]
- 3.Yuyun MF, Sliwa K, Kengne AP, Mocumbi AO, Bukhman G. Cardiovascular diseases in Sub-Saharan Africa compared to high-income countries: an epidemiological perspective. Glob Heart. 2020;15:15. doi: 10.5334/gh.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, Barengo NC, Beaton AZ, Benjamin EJ, Benziger CP, et al. ; GBD-NHLBI-JACC Global Burden of Cardiovascular Diseases Writing Group. Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 STUDY. J Am Coll Cardiol. 2020;76:2982–3021. doi: 10.1016/j.jacc.2020.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Rheumatic fever and rheumatic heart disease, 2018: Executive Board, 141st Session: Resolutions and decisions, annexes, summary records. Geneva, Switzerland. Available at: https://www.who.int/cardiovascular_diseases/publications/trs923/en/ [Google Scholar]
- 6.White A. WHO resolution on rheumatic heart disease. Eur Heart J. 2018;39:4233–4233. doi: 10.1093/eurheartj/ehy764 [DOI] [PubMed] [Google Scholar]
- 7.Abd El-Aal A. Mitral stenosis in Africa: Magnitude of the problem. E-Journal of Cardiology Practice. 2018;16:15–27 [Google Scholar]
- 8.Engel ME, Stander R, Vogel J, Adeyemo AA, Mayosi BM. Genetic susceptibility to acute rheumatic fever: a systematic review and meta-analysis of twin studies. PLoS One. 2011;6:e25326. doi: 1. Doi: 10.1371/journal.pone.0025326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferretti JJ, Stevens DL, Fischetti VA. Acute rheumatic fever and rheumatic heart disease. Sika-Paotonu D, Beaton A, Raghu A, Steer A, Carapetis J., eds. In: Streptococcus Pyogenes: Basic Biology to Clinical Manifestations [Internet]. Oklahoma City, OK: University of Oklahoma Health Sciences Center. 2017;1:1–32 [Google Scholar]
- 10.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X [DOI] [PubMed] [Google Scholar]
- 11.Abdallah AM, Abu-Madi M. The genetic control of the rheumatic heart: closing the genotype-phenotype gap. Front Med. 2021;8:611036. doi: 10.3389/fmed.2021.611036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chikowore T, Kamiza AB, Oduaran OH, Machipisa T, Fatumo S. Non-communicable diseases pandemic and precision medicine: is Africa ready? EBioMedicine. 2021;65:103260. doi: 10.1016/j.ebiom.2021.103260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1 [DOI] [PubMed] [Google Scholar]
- 14.Bredy C, Ministeri M, Kempny A, Alonso-Gonzalez R, Swan L, Uebing A, Diller G-P, Gatzoulis MA, Dimopoulos K. New York Heart Association (NYHA) classification in adults with congenital heart disease: relation to objective measures of exercise and outcome. Eur Heart J Qual Care Clin Outcomes. 2018;4:51–58. doi: 10.1093/ehjqcco/qcx031 [DOI] [PubMed] [Google Scholar]
- 15.Mayosi BM. A proposal for the eradication of rheumatic fever in our lifetime. S Afr Med J. 2006;96:229–230. [Google Scholar]
- 16.Watkins D, Zuhlke L, Engel M, Daniels R, Francis V, Shaboodien G, Mayosi BM, Kango M, Abul-Fadl A, Adeoye A. Seven key actions to eradicate rheumatic heart disease in Africa: the Addis Ababa communique. Cardiovasc J Afr. 2016;27:184. doi: 10.5830/CVJA-2015-090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zühlke L, Perkins S, Cembi S. Rheumatic heart disease patient event: Cape Town hosts 4th annual listen to my heart rheumatic heart disease for patients at the South African Heart Association meeting in 2017. Eur Heart J. 2018;39:1669–1671. doi: 10.1093/eurheartj/ehy199 [DOI] [PubMed] [Google Scholar]
- 18.Auckland K, Mittal B, Cairns BJ, Garg N, Kumar S, Mentzer AJ, Kado J, Perman ML, Steer AC, Hill AV, et al. The human leukocyte antigen locus and susceptibility to rheumatic heart disease in South Asians and Europeans. medRxiv. 2019;10:19003160. doi: 10.1038/s41598-020-65855-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parks T, Mirabel MM, Kado J, Auckland K, Nowak J, Rautanen A, Mentzer AJ, Marijon E, Jouven X, Perman ML, et al. ; Pacific Islands Rheumatic Heart Disease Genetics Network. Association between a common immunoglobulin heavy chain allele and rheumatic heart disease risk in Oceania. Nat Commun. 2017;8:14946. doi: 10.1038/ncomms14946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gray L-A, D’Antoine HA, Tong SY, McKinnon M, Bessarab D, Brown N, Reményi B, Steer A, Syn G, Blackwell JM. Genome-wide analysis of genetic risk factors for rheumatic heart disease in Aboriginal Australians provides support for pathogenic molecular mimicry. J Infect Dis. 2017;216:1460–1470. doi: 10.1093/infdis/jix497 [DOI] [PubMed] [Google Scholar]
- 21.Machipisa T, Chong M, Muhamed B, Chishala C, Shaboodien G, Pandie S, de Vries J, Laing N, Joachim A, Daniels R, et al. Association of novel locus with rheumatic heart disease in Black African individuals: findings from the RHDGen Study. JAMA Cardiol. 2021;6:1000. doi: 10.1001/jamacardio.2021.1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laird NM, Lange C. The role of family-based designs in genome-wide association studies. Stat Sci. 2009;24:388–397. doi: 10.1214/08-STS280 [Google Scholar]
- 23.Boone C. Land tenure regimes and state structure in rural Africa: implications for forms of resistance to large-scale land acquisitions by outsiders. J Contemp Afr Stud. 2015;33:171–190. doi: 10.1080/02589001.2015.1065576 [Google Scholar]
- 24.Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, Samocha KE, Goldstein JI, Okbay A, Bybjerg-Grauholm J, et al. ; iPSYCH-Broad Autism Group. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet. 2017;49:978–985. doi: 10.1038/ng.3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karthikeyan G, Zühlke L, Engel M, Rangarajan S, Yusuf S, Teo K, Mayosi BM. Rationale and design of a global rheumatic heart disease registry: the REMEDY study. Am Heart J. 2012;163:535–40.e1. doi: 10.1016/j.ahj.2012.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayosi B, Robertson K, Volmink J, Adebo W, Akinyore K, Amoah A, Bannerman C, Biesman-Simons S, Carapetis J, Cilliers A, et al. The Drakensberg declaration on the control of rheumatic fever and rheumatic heart disease in Africa. S Afr Med J. 2006;96:246. [PubMed] [Google Scholar]
- 27.Mweemba O, Musuku J, Mayosi BM, Parker M, Rutakumwa R, Seeley J, Tindana P, De Vries J. Use of broad consent and related procedures in genomics research: perspectives from research participants in the Genetics of Rheumatic Heart Disease (RHDGen) Study in a University Teaching Hospital in Zambia. Glob Bioeth. 2019;31:1–16. doi: 10.1080/11287462.2019.1592868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masiye F, Mayosi B, de Vries J. "I passed the test!" Evidence of diagnostic misconception in the recruitment of population controls for an H3Africa genomic study in Cape Town, South Africa. BMC Medical Ethics. 2017;18:12. doi: 10.1186/s12910-017-0175-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Vries J, Tindana P, Littler K, Ramsay M, Rotimi C, Abayomi A, Mulder N, Mayosi BM. The H3Africa policy framework: negotiating fairness in genomics. Trends Genet. 2015;31:117–119. doi: 10.1016/j.tig.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulder N, Abimiku A, Adebamowo SN, de Vries J, Matimba A, Olowoyo P, Ramsay M, Skelton M, Stein DJ. H3Africa: current perspectives. Pharmacogenomics Pers Med. 2018;11:59–66. doi: 10.2147/PGPM.S141546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tindana P, de Vries J. Broad consent for genomic research and biobanking: perspectives from low-and middle-income countries. Annu Rev Genomics Hum Genet. 2016;17:375–393. doi: 10.1146/annurev-genom-083115-022456 [DOI] [PubMed] [Google Scholar]
- 32.Grady C, Eckstein L, Berkman B, Brock D, Cook-Deegan R, Fullerton SM, Greely H, Hansson MG, Hull S, Kim S, et al. Broad consent for research with biological samples: workshop conclusions. Am J Bioeth. 2015;15:34–42. doi: 10.1080/15265161.2015.1062162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adebamowo SN, Francis V, Tambo E, Diallo SH, Landouré G, Nembaware V, Dareng E, Muhamed B, Odutola M, Akeredolu T, et al. Implementation of genomics research in Africa: challenges and recommendations. Glob Health Action. 2018;11:1419033. doi: 10.1080/16549716.2017.1419033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan JM. Ethnologue. Languages of Africa and Europe. Simons GF, Fennig CD, eds., In: Rocky Mountain Review of Language and Literature. 2018;72:223–225 [Google Scholar]
- 35.Di Carlo P, Good J, Ojong Diba R. Multilingualism in Rural Africa. Oxford University Press; 2019. [Google Scholar]
- 36.Reményi B, Wilson N, Steer A, Ferreira B, Kado J, Kumar K, Lawrenson J, Maguire G, Marijon E, Mirabel M, et al. World heart federation criteria for echocardiographic diagnosis of rheumatic heart disease—an evidence-based guideline. Nat Rev Cardiol. 2012;9:297–309. doi: 10.1038/nrcardio.2012.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavelaars M, Rousseau J, Parlayan C, de Ridder S, Verburg A, Ross R, Visser GR, Rotte A, Azevedo R, Boiten J-W. OpenClinica. Paper/Poster presented at: J Clin Bioinf.2015; [Google Scholar]
- 38.Machipisa T, Chong M, Muhamed B, Chishala C, Shaboodien G, Pandie S, de Vries J, Laing N, Joachim A, Daniels R, et al. Association of novel locus with rheumatic heart disease in black African individuals: findings from the RHDGen study. JAMA Cardiol. 2021;6:1000. doi: 10.1001/jamacardio.2021.1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becton, Dickinson and Company. BD Vacutainer Tube Guide. 2003. Available at: http://www.bd.com/vacutainer/pdfs/plus_plastic_tubes_wallchart_tubeguide_VS5229.pdf.
- 40.Bush V, Cohen R. The Evolution of Evacuated Blood Collection Tubes. Lab Med. 2003;34:304–310. doi: 10.1309/jcqe33nbyge0ffqr. [Google Scholar]
- 41.Provencher E, Remmel M, Hauch S, Otte M, Ullius A, Groelz D, Babayan A. Multimodal analysis of circulating tumor cell RNA, circulating cell-free DNA, and genomic DNA from a single blood sample collected into a PAXgene Blood ccfDNA Tube. Clin Cancer Res. 2020;26(11):Abstract nr PR11. [Google Scholar]
- 42.Ullius A, Provencher E, Voss T. Multimodal analysis of circulating cell-free RNA (ccfRNA), cell-free DNA (ccfDNA) and genomic DNA (gDNA) from blood samples collected in PAXgene Blood ccfDNA Tubes. In: AACR; 2020;15:1965–1965. [Google Scholar]
- 43.Warton K, Yuwono NL, Cowley MJ, McCabe MJ, So A, Ford CE. Evaluation of Streck BCT and PAXgene stabilised blood collection tubes for cell-free circulating DNA studies in plasma. Mol Diagn Ther. 2017;21:563–570. doi: 10.1007/s40291-017-0284-x [DOI] [PubMed] [Google Scholar]
- 44.Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, de Andrade M, Doheny KF, Haines JL, Hayes G, et al. Quality control procedures for genome wide association studies. Curr Protoc Hum Genet. 2011;Chapter 1(Supp1. 68):1.19. doi: 10.1002/0471142905.hg0119s68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilbert‐Diamond D, Moore JH. Analysis of gene–gene interactions. Curr Protoc Hum Genet. 2011;70:1.14. 11–11.14. 12. doi: 10.1002/0471142905.hg0114s70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717O111.016717. doi: 10.1074/mcp.o111.016717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGurk KA, Dagliati A, Chiasserini D, Lee D, Plant D, Baricevic-Jones I, Kelsall J, Eineman R, Reed R, Geary B, et al. The use of missing values in proteomic data-independent acquisition mass spectrometry to enable disease activity discrimination. Bioinformatics. 2019;36:2217–2223. doi: 10.1093/bioinformatics/btz898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salie MT, Yang J, Ramírez Medina CR, Zühlke LJ, Chishala C, Ntsekhe M, Gitura B, Ogendo S, Okello E, Lwabi P, et al. ; RHDGen Network Consortium. Data-independent acquisition mass spectrometry in severe rheumatic heart disease (RHD) identifies a proteomic signature showing ongoing inflammation and effectively classifying RHD cases. Clin Proteomics. 2022;19:7. doi: 10.1186/s12014-022-09345-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parker Z, Maslamoney S, Meintjes A, Botha G, Panji S, Hazelhurst S, Mulder N. Building infrastructure for African human genomic data management. Data Sci J. 2019;18:47. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.