Background:
Truncating variants in desmoplakin (DSPtv) are an important cause of arrhythmogenic cardiomyopathy; however the genetic architecture and genotype-specific risk factors are incompletely understood. We evaluated phenotype, risk factors for ventricular arrhythmias, and underlying genetics of DSPtv cardiomyopathy.
Methods:
Individuals with DSPtv and any cardiac phenotype, and their gene-positive family members were included from multiple international centers. Clinical data and family history information were collected. Event-free survival from ventricular arrhythmia was assessed. Variant location was compared between cases and controls, and literature review of reported DSPtv performed.
Results:
There were 98 probands and 72 family members (mean age at diagnosis 43±8 years, 59% women) with a DSPtv, of which 146 were considered clinically affected. Ventricular arrhythmia (sudden cardiac arrest, sustained ventricular tachycardia, appropriate implantable cardioverter defibrillator therapy) occurred in 56 (33%) individuals. DSPtv location and proband status were independent risk factors for ventricular arrhythmia. Further, gene region was important with variants in cases (cohort n=98; Clinvar n=167) more likely to occur in the regions resulting in nonsense mediated decay of both major DSP isoforms, compared with n=124 genome aggregation database control variants (148 [83.6%] versus 29 [16.4%]; P<0.0001).
Conclusions:
In the largest series of individuals with DSPtv, we demonstrate that variant location is a novel risk factor for ventricular arrhythmia, can inform variant interpretation, and provide critical insights to allow for precision-based clinical management.
Keywords: cardiomyopathies; primary; death, sudden cardiac; desmoplakins; genetic testing
Desmoplakin is a plakin family protein that anchors the desmosome to intermediate filaments and is abundant in tissues with greater mechanical stress such as the epidermis and myocardium.1,2 Genetic variants in the gene encoding desmoplakin (DSP) cause a range of cardio-cutaneous phenotypes, including arrhythmogenic cardiomyopathy (ACM), striate palmoplantar keratoderma, and lethal acantholytic epidermolysis bullosa in more severe cases.3 Truncating variants (DSPtv) that lead to putative loss of function via haploinsufficiency of the protein have been previously reported as causative of disease.2 DSP-null mice show extensive disruption of the cytoarchitecture and cell resilience in skin and heart tissue, with death in early development.4
Arrhythmogenic right ventricular cardiomyopathy (ARVC), the right dominant sub-form of ACM,2,5 is characterized by progressive loss and fibrofatty replacement of the ventricular myocardium.6 Diagnosis of ARVC can be challenging and 2010 Task Force Criteria consider electrical, structural (imaging and histological), and genetic characteristics.7 Historically, clinical descriptions of DSPtv were often based on ARVC cohorts, though growing recognition of left ventricular (LV) involvement has necessitated a shift to a broader phenotype description, ACM,8 encompassing left dominant arrhythmogenic cardiomyopathy and biventricular disease, with new Padua criteria proposed.9 Dilated cardiomyopathy and left dominant arrhythmogenic cardiomyopathy lie on a spectrum, with overlap in molecular causes. More recently, DSP has been definitely associated with both ARVC and Dilated cardiomyopathy by international gene curation expert panels10,11 In one of the largest studies to date, clinical characteristics of DSP variants in a population of 44 probands and 63 family members were reported as a distinct ACM characterized by LV fibrosis, myocardial inflammation and high incidence of ventricular arrhythmias.12 Biallelic DSP variants can give rise to Carvajal syndrome, characterized by woolly hair, palmoplantar keratoderma and development of ACM in childhood, and often due to homozygous or compound heterozygous DSPtv affecting the C-terminus.13
The N-terminal globular head of DSP is important in desmosome organization by binding plaque proteins such as plakophilin and plakoglobin, while the central rod domain contains a coiled-coil region.14 The C-terminal contains 3 plakin repeat domains, required for alignment and binding of intermediate filaments.15 Two predominant isoforms exist due to alternate splicing: DSPI (which is the longest isoform) and DSPII (which has a shortened central rod domain). DSPI and DSPII are expressed in equivalent levels in epidermis; however, DSPI is more prevalent in myocardium.16 Differences between the 2 isoforms relate to the rod domain size, considered important for self-association and formation of homo-dimers.17
Here we report an international cohort of individuals with a DSPtv. We describe the phenotype spectrum of DSPtv cardiomyopathy, family history characteristics, and provide insights into the genetic architecture of DSPtv cardiomyopathy and its relation to clinical phenotype.
Methods
Data are available by request to the corresponding author and adhering to site ethical approval. All aspects of the study were performed according to institutional human research ethics committee approval according to the local sites. Institutional ethics approval was granted by individual sites; including Sydney Local Health District, Royal Prince Alfred Hospital, Australia; Royal Brompton & Harefield Hospitals Cardiovascular Biobank (National Research Ethics Service), UK. Waiver of consent was granted at Stanford School of Medicine Internal Review Board, USA. All individual-level data were de-identified.
Detailed methods are available in the Supplemental Material.
Results
Study Population
Overall there were 98 probands (mean age at diagnosis 42±18 years; 59% women) and 72 family members identified (mean age at diagnosis 45±19 years; 61% women; Table 1). There were 95 probands with a cardiomyopathy and 3 with a primary cutaneous phenotype. Among family members, 48 of 72 were deemed affected, including 5 with a predominantly cutaneous phenotype. In total, 146 individuals were considered affected, including cardiomyopathy, ventricular arrhythmia and cutaneous phenotypes.
Table 1.
Clinical Characteristics by Proband Status and Sex
Sex Differences
Women were over-represented compared with men among affected individuals (86 [59%] versus 60 [41%]; Table 1). There was no difference in mean age at diagnosis between women and men (40±17 years versus 46±19 years; P=0.07). Myocarditis was more frequent in men (2/43 [5%] versus 6/25 [24%]; P=0.046) but this was not always reliably reported. Women had reduced LV ejection fraction on transthoracic echocardiography, but not cardiac magnetic resonance imaging (CMR) derived LV ejection fraction. Men had greater indexed right ventricular end diastolic volume (84±20 versus 100±27; P=0.01). No differences in clinical outcomes were reported between sexes. There was a comparable distribution of variants by gene region for men and women, as well as probands and affected relatives.
Electrophysiological Characteristics
There was a high rate of ventricular arrhythmia occurring in 56 (33%) individuals, including 46 (47%) probands and 10 (14%) family members. Ventricular arrhythmia included sudden cardiac death (SCD) in 13 (8%; 10 probands), resuscitated cardiac arrest in 15 (9%; 12 probands), appropriate implantable cardioverter defibrillator therapy in 16 (10%; 16 probands), and sustained ventricular tachycardia in 19 patients (11%; 15 probands); including 10 (14%) family members, and with some experiencing multiple events. Six probands experienced 2 ventricular arrhythmia episodes, initially having sustained ventricular tachycardia (n=4) or resuscitated cardiac arrest (n=2), followed by appropriate implantable cardioverter defibrillator therapy. SCD or resuscitated cardiac arrest was the presenting symptom in 24 (14%; 20 probands) patients. T wave inversion beyond V3 occurred in 33 (24%), low voltages in 47 (35%) and premature ventricular contractions in 56 (33%).
Imaging Characteristics
Echocardiographic and CMR characteristics are shown in Table 1. Signs of LV noncompaction were reported (n=22), with 6 having a ratio of noncompacted to compacted layer >2.3 on CMR. Four probands were reported to have hypertrophic cardiomyopathy (HCM) with ages at diagnosis ranging from 58 to 83 years, and LV hypertrophy measuring 26 mm, 16 mm, and an apical pattern in 2. DSPtv are not established as associated with HCM, and we consider it unlikely that these variants are causal for HCM for these 4 cases, but are reported as they met the pre-specified eligibility criteria. Late gadolinium enhancement (LGE) was reported in 59 (61%), and end-stage heart failure was reported in 10 (8%) patients. Two women developed disease while pregnant, 1 showed impaired LV function (LV ejection fraction <45%) at 32 weeks of gestation, while the other developed narrow complex tachycardia at 38 weeks of gestation with subsequent echocardiogram showing a dilated and impaired LV. A further 2 women developed disease during the postpartum period. Finally, another patient who died suddenly during pregnancy was identified to be positive for Parvovirus B19 on postmortem Parvo-polymerase chain reaction in myocardial tissue. Myocarditis was reported in 7 individuals on CMR (and another on postmortem investigation).
Genetic Analysis
A total of 69 distinct DSPtv were identified in the 98 probands (Table S1). Among the 69 DSPtv, there were 31 small insertions or deletions leading to a frameshift and downstream premature termination codon, 25 nonsense variants, 12 canonical splice-site altering variants and a large deletion of exons 5–24. Eleven (16%) variants were classified as pathogenic, 57 (83%) were classified as likely pathogenic and 1 (1%) was classified as a variant of uncertain significance. Two probands had a diagnosis of cardiomyopathy, with woolly hair and keratoderma (OMIM 605676) and were compound heterozygous, each carrying a DSPtv (p.Arg2229Serfs*32 or p.Tyr28Alafs*66) and a DSP splice site variant (the same c.273+5G>A in both). This splice site variant has an allele count of 79 in genome aggregation database (gnomAD), with an allele frequency of 0.028% and considered a variant of uncertain significance under a recessive inheritance model.
DSPtv Location
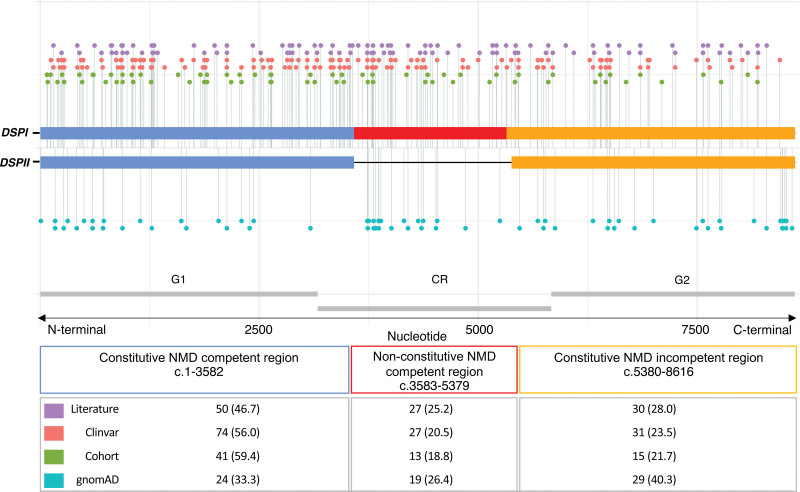
We investigated whether case and control variants localized to the specified gene regions, constitutive nonsense mediated decay (NMD) competent, nonconstitutive NMD-competent and constitutive NMD-incompetent (Figure 1). Pathogenic and likely pathogenic DSPtv submitted to ClinVar, as well as variants described above in the international cohort were included, giving a total of 265 cases. This included 69 unique DSPtv identified in 98 individuals in the international cohort and 134 unique DSPtv from 167 cases reported in ClinVar (Table S2). One variant reported in ClinVar was excluded from analysis given it resided in the small overlap region of exon 23 which is both nonconstitutive and predicted NMD-incompetent due to being <55 bp upstream of the last exon junction (DSP: c.5327_5330del; p.Glu1776Glyfs). Another was excluded after it was identified as a ClinVar entry for one of the cohort cases. Literature cases are shown in Figure 1 but not included in the analysis due to unquantified sample overlap. Variants observed in cases were compared with 72 unique DSPtv observed as 124 alleles in gnomAD controls (Table S3). Case variants were more frequently seen in the constitutive NMD-competent region compared with controls. Across the 3 gene regions (constitutive NMD-competent, nonconstitutive NMD-competent and constitutive NMD-incompetent, respectively), DSPtv were seen in 148 (56%), 59 (22%) and 58 (22%) cases compared with DSPtv observed in controls 29 (23%), 28 (23%) and 67 (54%), overall P<0.0001.
Figure 1.
Linear topology schematic showing distribution of DSPtv across key gene regions of major isoforms DSPI and DSPII. Case variants (Cohort, Clinvar, and Literature) are shown above and control variants below the line. The number of unique variants (per proband) from each source are shown in the table. CR indicates central fibrous rod domain; DSPtv, desmoplakin truncating variant; G1, globular 1; G2, globular 2; gnomAD, gnome aggregation database; and NMD, nonsense mediated decay.
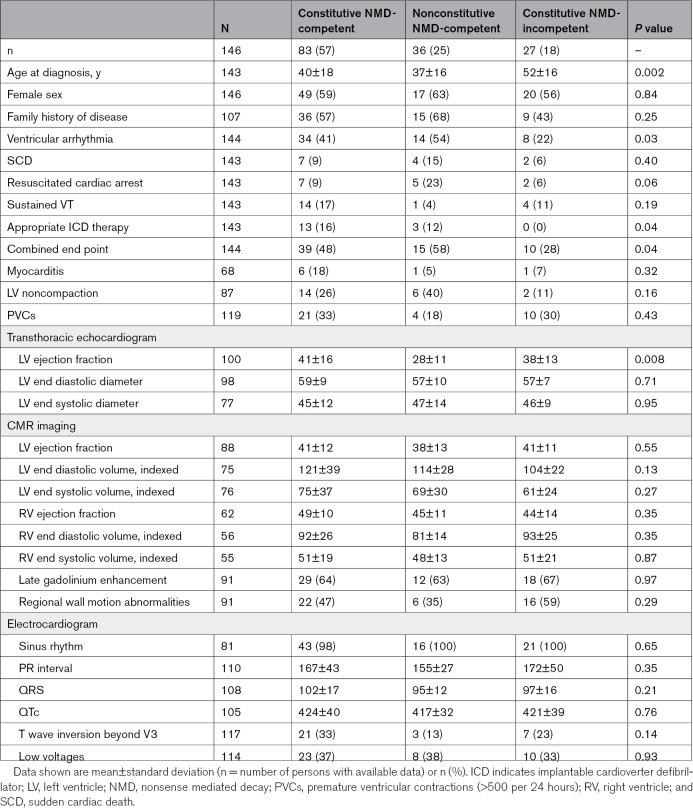
Clinical characteristics of patients with DSPtv in the 3 gene regions are shown in Table 2. Overall there were few significant differences between the patient groups based on gene region. Age at diagnosis was significantly younger in those with DSPtv in both NMD-competent regions (constitutive and nonconstitutive). Further, there was a greater risk of ventricular arrhythmia and risk of the combined end point in those with DSPtv in the constitutive and nonconstitutive NMD-competent regions.
Table 2.
Cardiac Investigation of Affected Individuals With DSPtv by Gene Region
Event-Free Survival From Ventricular Arrhythmia Based on Gene Region
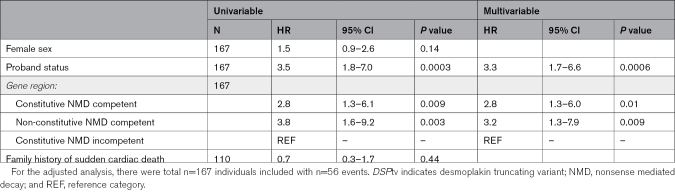
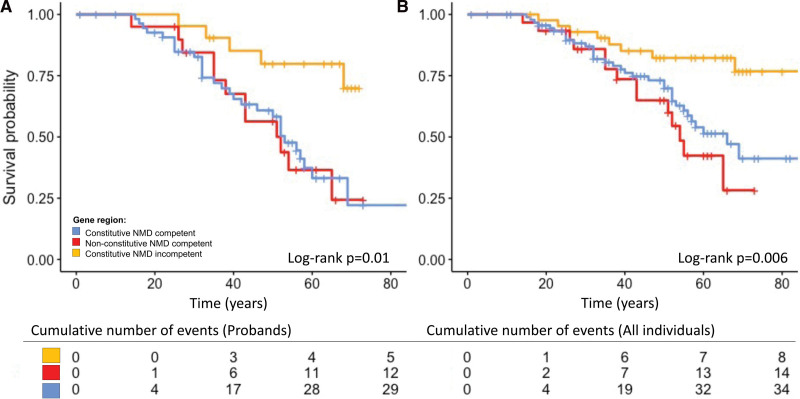
Information with regard to occurrence of ventricular arrhythmia or censoring was available for 167 individuals. There were 56 probands and family members who experienced a ventricular arrhythmia during their lifetime. Univariable Cox proportional hazards models showed gene region and proband status as significantly associated with worse survival from ventricular arrhythmias (Table 3; Figure 2). Adjusting for other variables, variants in the constitutive NMD competent region (HR, 2.8 [95% CI, 1.3–6.0]; P=0.01), nonconstitutive NMD-competent region (HR, 3.2 [95% CI 1.3–7.9]; P=0.009) and proband status (HR, 3.3 [95% CI, 1.7–6.6]; P=0.0006) remained significant independent life-time risk factors for ventricular arrhythmia (Table 3).
Table 3.
Lifetime Risk Factors for Ventricular Arrhythmia for Individuals With a DSPtv
Figure 2.
Independent life-time risk factor for ventricular arrhythmia. (A) Gene region including probands only. (B) Gene region including probands and affected family members. Time is given in years. NMD indicates nonsense mediated decay.
Cutaneous Phenotype
Cutaneous abnormalities were not systematically reported; however, notably 1 family with a DSPtv in the nonconstitutive NMD-competent region (cardiac isoform, DSPI) had an affected relative with hyperkeratosis and cardiomyopathy. An additional 13 individuals with DSPtv in the constitutive NMD-competent region and 5 in the constitutive NMD-incompetent region were reported with overt cardio-cutaneous features noted at clinical review. In 8 patients (5%; 3 probands) only cutaneous abnormalities were reported, and were the sole finding in 1 family following an autosomal dominant inheritance pattern.
Postmortem Findings and Cardiac Transplant Histology
Thirteen patients (8%; 10 probands) presented with SCD (Table S4). In all 13, a postmortem investigation was performed. The mean age at death was 26±11 years. Where recorded, the activity at time of death varied from exercise through to sleep. No decedent had a pre-morbid diagnosis of a cardiac condition. Nine decedents received a postmortem diagnosis of ARVC or probable ARVC. There was LV involvement in all cases and fibrosis and fatty infiltration commonly reported.
Two patients underwent a heart transplant due to end stage heart failure. Biventricular involvement was observed in both hearts, as were signs of LV noncompaction. One heart showed ARVC with septal involvement and replacement fibrosis in both ventricles and septum. The other heart showed LV noncompaction with notable right ventricular involvement consisting of fatty changes and atrophy.
Family History Characteristics
Among the probands, 49 (51%) had a documented family history of cardiomyopathy, while 16 (17%) had a family history of a suspicious SCD under the age of 40 years. Of the 72 family members with positive gene results included, 48 (67%) had overt disease, while 24 (33%) remained asymptomatic (mean age of 49±22 years and 15 [63%] were women). There were 8 family members aged 60 years or older (60–86 years; 5 women) with no clinical evidence of disease, suggesting incomplete penetrance. By gene region, there was no statistical difference in the proportion of probands with a positive family history (constitutive NMD-competent 27 [49%], nonconstitutive NMD-competent 13 [65%], constitutive NMD-incompetent 9 [43%], P=0.33).
Literature Review of Previously Reported DSPtv
Three hundred and fifteen studies were identified, 240 were screened, and 185 full texts were assessed for eligibility (85 were excluded from the final qualitative synthesis, including 66 that did not report any DSP variant, 2 where phenotype was not provided, 2 with no full-text article available, and 1 review; Figure S1). Of the 98 studies (describing both disease and genotype-first cohorts) included in the final selection, a total of 105 DSPtv in 143 probands from apparently unrelated families were reported, including 57 nonsense, 42 frameshift, and 6 splice site variants (Tables S5–S7). All reported variants were absent or very rare (allele count ≤2) in gnomAD and were classified as pathogenic or likely pathogenic. One variant (p.Thr2104fs*12) was present 13 times in gnomAD, however has strong evidence of pathogenicity and reported in a compound heterozygous state.
Both dominant and recessive patterns of inheritance of DSPtv were reported. Cascade genetic testing to confirm autosomal dominant inheritance was reported for only 19 DSPtv (dominant DSPtv) in 22 families. Families reported with autosomal dominant inheritance commonly demonstrated adult age of onset, incomplete penetrance and variable clinical expression. Of the 105 reported DSPtv, 26 were only identified in affected individuals with homozygous or compound heterozygous inheritance. Four DSPtv co-occurred in trans with one of 3 missense DSP variants (p.Ala2655Asp, p.Arg2366Cys, and p.Asn287Lys), each of which involved highly conserved residues within globular heads, are absent in gnomAD, and classified as likely pathogenic. There were 16 individuals with 23 DSPtv identified to have autosomal recessive disease, either homozygous (n=9) or compound heterozygous (n=7). In just those variants identified in a homozygous state there was only 1 (11%) in the constitutive NMD-competent region, 5 (56%) in the nonconstitutive NMD-competent region and 3 (33%) in the constitutive NMD-incompetent region.
Discussion
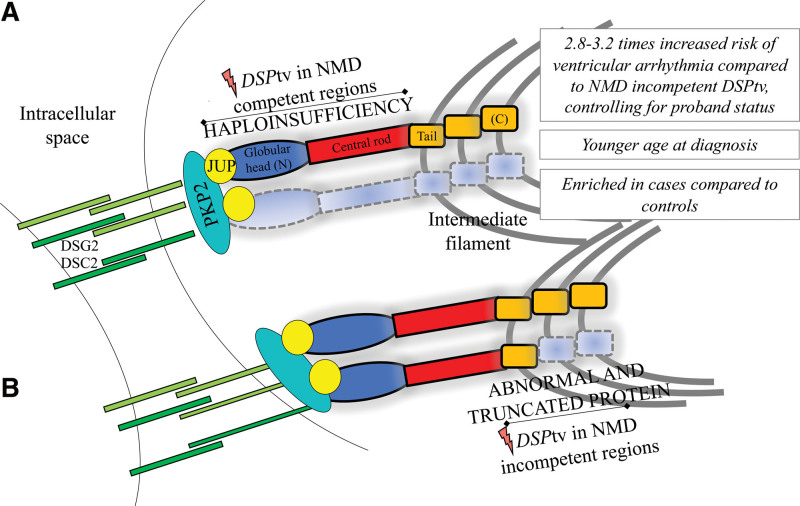
DSPtv lead to a distinct cardiomyopathy characterized by LV involvement and a high-risk of ventricular arrhythmia and SCD. We present a large international series of cases with DSPtv and demonstrate that the location of the DSPtv is a novel risk factor for ventricular arrhythmia (Figure 3). Truncating variants in the constitutive NMD-competent region were enriched in cases compared with controls, and predicted to result in NMD and haploinsufficiency of both DSPI and DSPII. Our findings highlight the importance of personalized medicine and the move towards gene-guided management of patients in the future.
Figure 3.
Summary of the key findings and illustration of the impact of DSPtv location on protein expression (for DSPI). DSC indicates desmocollin-2; DSG2, desmoglein-2; JUP, plakoglobin; PKP2, plakophilin-2; and NMD, nonsense mediated decay.
Ventricular arrhythmias occur frequently in patients with DSPtv cardiomyopathy, with one previous study reporting 23% presenting with SCD events.18 In our cohort, 47% of probands had ventricular arrhythmia either at presentation or during follow-up. In addition, 14% of relatives experienced ventricular arrhythmia, including 7% as their initial presenting symptom. This included 2 probands who presented with resuscitated cardiac arrest without any overt structural abnormalities of the heart, supporting the notion that life-threatening electrical phenotype can precede overt cardiac structural disease.19–21 While we were unable to robustly ascertain clinical risk factors due to the large proportion of cases who presented with ventricular arrhythmia (ie, without necessary pre-event clinical data), a recent series of 107 patients with any DSP variants (n=30 events) showed ventricular arrhythmias were associated with reduced LV ejection fraction, while premature ventricular contractions (>500 beats in 24 hours), LGE, and right ventricular dysfunction were not shown to be associated with ventricular arrhythmia.12 Family history of SCD has not previously been evaluated in this group, and we showed it is not associated with ventricular arrhythmia in our population.
Prior observation that DSPtv are predominantly associated with a left dominant form of ACM2,5,22 is in line with our findings. Recent examples of DSPtv presenting as recurrent myocarditis and acute myocardial infarction-like events have also been reported.23,24 Women were overrepresented in our population, but otherwise shared similar clinical characteristics compared with men, except reduced indexed right ventricular end diastolic diameter on CMR. This finding is in contrast to other reported inherited cardiomyopathy patient cohorts, where a higher prevalence of men is often reported.25–27 Of note, a recent report of ARVC presenting as clinical myocarditis showed disproportionately more women, with 10/11 having DSPtv.28 DSPtv cardiomyopathy patients frequently had low QRS voltage and negative T waves beyond V3. Low QRS voltage in limb leads have previously been shown to be associated with the presence and amount of LGE in a study of patients with ARVC.8 Regional wall motion abnormalities on CMR and epicardial to mid wall LGE patterns in the LV were frequently seen in our cohort. Septal LGE frequently occurs in patients with left dominant arrhythmogenic cardiomyopathy,15 and recent work has shown patients with DSP and FLNC ACM are more likely to have LGE, often with a ring-like pattern, compared with other Dilated cardiomyopathy genotypes.29 Four probands were reported to have HCM, however it should be noted that all 4 probands were men, presenting in older age and 3 had mild LV hypertrophy, all characteristics previously described in the nonfamilial sub-group of HCM.30 Previous assessment of the clinical validity of DSP variants causing HCM failed to identify sufficient evidence of gene-disease association.31 While our finding remains unclear, it seems reasonable to consider these clinical diagnoses as unrelated to the DSPtv.
A recent systematic evaluation of cutaneous abnormalities among DSPtv showed all patients expressed some degree of skin or hair abnormalities, except those with DSPtv in the nonconstitutive NMD-competent region (cardiac isoform, DSPI).32 Interestingly, we report 1 proband and their affected relative with palmoplantar keratoderma, with a DSPtv in the nonconstitutive NMD-competent region. Another study reported 10% of DSPtv had cutaneous disease only, while 12% were reported to have LV dominant ACM and cutaneous disease.5
We show DSPtv localized to the constitutive NMD-competent region, corresponding to the N-terminal globular head, were enriched in patients compared with controls, and this finding was replicated in the variants identified through literature review. This region plays a critical role in organization and assembly of the desmosomal complex by binding with plakophilin and plakoglobin. One previous report of DSP missense variants in patients with a clinical diagnosis of ARVC suggested a potential “hotspot” N-terminal region, with 8/17 (47%) missense variants localized to the N-terminal compared with 1/28 (4%) of controls (P<0.0008).33 Further, they concluded DSPtv were significantly more prevalent in ARVC cases than controls. Indeed, a recent study also showed clustering of missense variants in the N-terminal, but reported DSPtv to be more evenly distributed across the gene,12 potentially limited by sample size. Another showed enrichment of missense variants in the spectrin repeat domain, which is part of the constitutive NMD competent region.34 While it seems likely that truncating variants in the NMD-incompetent region escape NMD and have a later onset and less deleterious impact, functional work to date has shown highly variable pattern of protein expression representing both haploinsufficiency and dominant negative effects.35 Our literature review identified biallelic DSPtv localized more often to the constitutive NMD-incompetent region compared with dominant DSPtv, suggesting that single heterozygous DSPtv are more likely to cause disease when occurring in the NMD-competent regions. Further, very few cases with homozygous variants in the constitutive NMD-competent region have been reported, with 1 example of a sib pair with severe lethal acantholytic epidermolysis bullosa, who died at 1 and 3 days respectively.36 It seems unlikely these infants were DSP null, given DSP knockout mice show embryonic lethality,4 suggesting expression of low-level truncated protein may be able to rescue the phenotype to some degree. Taken together, identification of a DSPtv in the NMD-competent regions should be considered important and may prompt gene and disease-specific adaptation and use of the ACMG/AMP (American College of Medical Genetics and Genomics and Association for Molecular Pathology) criteria.37 We suggest DSPtv in this region be allocated very strong level of evidence, PVS1, when considering pathogenicity, when seen in an individual with a well characterized and concordant phenotype.
Study Limitations
This was a large retrospective cohort study, and while it was an international effort, differences in practices and data collection by site meant some variables were incomplete. Furthermore, the event rate and data missingness precluded more detailed risk factor analyses. Diagnosis was made by the referring clinician and most recruitment was from specialized tertiary referral centers and therefore likely represents more severe phenotypes. The literature review was limited by publication bias, and inconsistent reporting of clinical, family and genetic information.
Conclusions
We present a large international series of individuals with DSPtv and show gene region is a novel risk factor, specifically DSPtv leading to predicted NMD of truncated protein and haploinsufficiency of DSPI and/or DSPII is a risk factor for ventricular arrhythmias. By sub-typing disease by genotype there is increasing ability to offer precision medicine-based advice and therapies, and thereby improved outcomes for patients and their families.
Article Information
Sources of Funding
Dr Burns is the recipient of an Australia Postgraduate Award (APA). Dr Bagnall is supported by a grant from New South Wales Health. Dr Semsarian is the recipient of a National Health and Medical Research Council (NHMRC) Practitioner Fellowship (#1154992). Dr Ware is supported by the Wellcome Trust, the Medical Research Council (UK), the British Heart Foundation, the NIHR Royal Brompton Biomedical Research Unit, and the NIHR Imperial College Biomedical Research Centre. Drs van Tintelen, Wilde, Volders, van den Berg, and Hoorntje acknowledge the support from the Netherlands Cardiovascular Research Initiative, an initiative with support of the Dutch Heart Foundation (2014-40 DOSIS; 2012-10 PREDICT; 2018-30 PREDICT2; 2015-12 eDETECT). Dr Ingles is the recipient of an NHMRC Career Development Fellowship (#1162929). The other authors report no conflicts.
Disclosures
Dr Ingles receives research grant support from Bristol Myers Squibb, unrelated to this study. Dr Ware reports research grant support and consultancy fees from Bristol Myers Squibb, unrelated to this study. Dr Reuter is a consultant for My Gene Counsel. Dr Wheeler is a stockholder of Personalis Inc. The remaining authors have nothing to disclose.
Supplemental Material
Supplemental Material
Tables S1–S7
Figure S1
Supplementary Material
Nonstandard Abbreviations and Acronyms
- ACM
- arrhythmogenic cardiomyopathy
- ACMG/AMP
- American College of Medical Genetics and Genomics and Association for Molecular Pathology
- ARVC
- arrhythmogenic right ventricular cardiomyopathy
- CMR
- cardiac magnetic resonance imaging
- DSP
- desmoplakin
- DSPtv
- desmoplakin truncating variant
- HCM
- hypertrophic cardiomyopathy
- LGE
- late gadolinium enhancement
- LV
- left ventricular
- NMD
- nonsense mediated decay
- SCD
- sudden cardiac death
This article was sent to Ruth McPherson, MD, PhD, Guest Editor, for review by expert referees, editorial decision, and final disposition.
E.T. Hoorntje & C. Burns contributed equally.
J.S. Ware, J. Peter van Tintelen & J. Ingles contributed equally.
For Sources of Funding and Disclosures, see pages 77–78.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.121.003672.
Contributor Information
Edgar T. Hoorntje, Email: e.t.hoorntje@umcg.nl.
Charlotte Burns, Email: c.burns@centenary.org.au.
Luisa Marsili, Email: Luisa.MARSILI@chu-lille.fr.
Ben Corden, Email: bc291@doctors.org.uk.
Victoria N. Parikh, Email: vparikh@stanford.edu.
Gerard J. te Meerman, Email: g.j.te.meerman@umcg.nl.
Belinda Gray, Email: belinda.gray@sydney.edu.au.
Ahmet Adiyaman, Email: a.adiyaman@isala.nl.
Richard D. Bagnall, Email: r.bagnall@centenary.org.au.
Daniela Q.C.M. Barge-Schaapveld, Email: D.Q.C.M.Barge-Schaapveld@lumc.nl.
Maarten P. van den Berg, Email: m.p.van.den.berg@umcg.nl.
Marianne Bootsma, Email: m.bootsma@lumc.nl.
Laurens P. Bosman, Email: L.P.Bosman-3@umcutrecht.nl.
Gemma Correnti, Email: gemma.correnti@sa.gov.au.
Johan Duflou, Email: jduflou@forensicmedicine.com.au.
Ruben N. Eppinga, Email: r.n.eppinga@gmail.com.
Diane Fatkin, Email: d.fatkin@victorchang.edu.au.
Michael Fietz, Email: mfietz@illumina.com.
Eric Haan, Email: eric.haan@adelaide.edu.au.
Jan D.H. Jongbloed, Email: j.d.h.jongbloed@umcg.nl.
Arnaud D. Hauer, Email: arnaudhauer@hotmail.com.
Lien Lam, Email: hellolienlam@gmail.com.
Freyja H.M. van Lint, Email: F.H.M.vanLint@umcutrecht.nl.
Amrit Lota, Email: a.lota@rbht.nhs.uk.
Carlo Marcelis, Email: Carlo.marcelis@radboudumc.nl.
Hugh J. McCarthy, Email: hugh.mccarthy@health.nsw.gov.au.
Rogier A. Oldenburg, Email: r.oldenburg@erasmusmc.nl.
Nicholas Pachter, Email: Nicholas.Pachter@health.wa.gov.au.
R. Nils Planken, Email: r.n.planken@amc.uva.nl.
Chloe Reuter, Email: creuter@stanfordhealthcare.org.
Christopher Semsarian, Email: c.semsarian@centenary.org.au.
Jasper J. van der Smagt, Email: J.J.vanderSmagt@umcutrecht.nl.
Tina Thompson, Email: tina.thompson@mh.org.au.
Jitendra Vohra, Email: jitu@vohra1.com.
Paul G.A. Volders, Email: p.volders@maastrichtuniversity.nl.
Jaap I. van Waning, Email: j.vanwaning@erasmusmc.nl.
Nicola Whiffin, Email: nwhiffin@well.ox.ac.uk.
Arthur van den Wijngaard, Email: arthur.vdwijngaard@mumc.nl.
Ahmad S. Amin, Email: a.s.amin@amc.nl.
Arthur A.M. Wilde, Email: a.a.wilde@amc.uva.nl.
Gijs van Woerden, Email: g.van.woerden@umcg.nl.
Laura Yeates, Email: laura.yeates@populationgenomics.org.au.
Dominica Zentner, Email: dominica.zentner@mh.org.au.
Euan A. Ashley, Email: euan@stanford.edu.
Matthew T. Wheeler, Email: wheelerm@stanford.edu.
James S. Ware, Email: j.ware@imperial.ac.uk.
J. Peter van Tintelen, Email: j.p.vantintelen-3@umcutrecht.nl.
References
- 1.Corrado D, Basso C, Pilichou K, Thiene G. Molecular biology and clinical management of arrhythmogenic right ventricular cardiomyopathy/dysplasia. Heart. 2011;97:530–539. doi: 10.1136/hrt.2010.193276 [DOI] [PubMed] [Google Scholar]
- 2.Castelletti S, Vischer AS, Syrris P, Crotti L, Spazzolini C, Ghidoni A, Parati G, Jenkins S, Kotta MC, McKenna WJ, et al. Desmoplakin missense and non-missense mutations in arrhythmogenic right ventricular cardiomyopathy: genotype-phenotype correlation. Int J Cardiol. 2017;249:268–273. doi: 10.1016/j.ijcard.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 3.Bauce B, Basso C, Rampazzo A, Beffagna G, Daliento L, Frigo G, Malacrida S, Settimo L, Danieli G, Thiene G, et al. Clinical profile of four families with arrhythmogenic right ventricular cardiomyopathy caused by dominant desmoplakin mutations. Eur Heart J. 2005;26:1666–1675. doi: 10.1093/eurheartj/ehi341 [DOI] [PubMed] [Google Scholar]
- 4.Gallicano GI, Kouklis P, Bauer C, Yin M, Vasioukhin V, Degenstein L, Fuchs E. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J Cell Biol. 1998;143:2009–2022. doi: 10.1083/jcb.143.7.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Ayala JM, Gomez-Milanes I, Sanchez Munoz JJ, Ruiz-Espejo F, Ortiz M, Gonzalez-Carrillo J, Lopez-Cuenca D, Oliva-Sandoval MJ, Monserrat L, Valdes M, et al. Desmoplakin truncations and arrhythmogenic left ventricular cardiomyopathy: characterizing a phenotype. Europace.2014;16:1838–1846. doi: 10.1093/europace/euu128 [DOI] [PubMed] [Google Scholar]
- 6.Hoorntje ET, Te Rijdt WP, James CA, Pilichou K, Basso C, Judge DP, Bezzina CR, van Tintelen JP. Arrhythmogenic cardiomyopathy: pathology, genetics, and concepts in pathogenesis. Cardiovasc Res. 2017;113:1521–1531. doi: 10.1093/cvr/cvx150 [DOI] [PubMed] [Google Scholar]
- 7.Marcus FI, McKenna WJ, Sherrill D, Basso C, Bauce B, Bluemke DA, Calkins H, Corrado D, Cox MG, Daubert JP, et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation.2010;121:1533–1541. doi: 10.1161/CIRCULATIONAHA.108.840827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corrado D, van Tintelen PJ, McKenna WJ, Hauer RNW, Anastastakis A, Asimaki A, Basso C, Bauce B, Brunckhorst C, Bucciarelli-Ducci C, et al. ; International Experts. Arrhythmogenic right ventricular cardiomyopathy: evaluation of the current diagnostic criteria and differential diagnosis. Eur Heart J. 2020;41:1414–1429. doi: 10.1093/eurheartj/ehz669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corrado D, Perazzolo Marra M, Zorzi A, Beffagna G, Cipriani A, Lazzari M, Migliore F, Pilichou K, Rampazzo A, Rigato I, et al. Diagnosis of arrhythmogenic cardiomyopathy: the Padua criteria. Int J Cardiol. 2020;319:106–114. doi: 10.1016/j.ijcard.2020.06.005 [DOI] [PubMed] [Google Scholar]
- 10.James CA, Jongbloed JDH, Hershberger RE, Morales A, Judge DP, Syrris P, Pilichou K, Domingo AM, Murray B, Cadrin-Tourigny J, et al. International evidence based reappraisal of genes associated with arrhythmogenic right ventricular cardiomyopathy using the clinical genome resource framework. Circ Genom Precis Med.2021;14:e003273. doi: 10.1161/CIRCGEN.120.003273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan E, Peterson L, Ai T, Asatryan B, Bronicki L, Brown E, Celeghin R, Edwards M, Fan J, Ingles J, et al. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation.2021;144:7–19. doi: 10.1161/CIRCULATIONAHA.120.053033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith ED, Lakdawala NK, Papoutsidakis N, Aubert G, Mazzanti A, McCanta AC, Agarwal PP, Arscott P, Dellefave-Castillo LM, Vorovich EE, et al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation.2020;141:1872–1884. doi: 10.1161/CIRCULATIONAHA.119.044934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvajal-Huerta L. Epidermolytic palmoplantar keratoderma with woolly hair and dilated cardiomyopathy. J Am Acad Dermatol. 1998;39:418–421. doi: 10.1016/s0190-9622(98)70317-2 [DOI] [PubMed] [Google Scholar]
- 14.Green KJ, Stappenbeck TS, Parry DA, Virata ML. Structure of desmoplakin and its association with intermediate filaments. J Dermatol. 1992;19:765–769. doi: 10.1111/j.1346-8138.1992.tb03777.x [DOI] [PubMed] [Google Scholar]
- 15.Sen-Chowdhry S, Syrris P, Ward D, Asimaki A, Sevdalis E, McKenna WJ. Clinical and genetic characterization of families with arrhythmogenic right ventricular dysplasia/cardiomyopathy provides novel insights into patterns of disease expression. Circulation.2007;115:1710–1720. doi: 10.1161/CIRCULATIONAHA.106.660241 [DOI] [PubMed] [Google Scholar]
- 16.Uzumcu A, Norgett EE, Dindar A, Uyguner O, Nisli K, Kayserili H, Sahin SE, Dupont E, Severs NJ, Leigh IM, et al. Loss of desmoplakin isoform I causes early onset cardiomyopathy and heart failure in a Naxos-like syndrome. J Med Genet. 2006;43:e5. doi: 10.1136/jmg.2005.032904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Keefe EJ, Erickson HP, Bennett V. Desmoplakin I and desmoplakin II. Purification and characterization. J Biol Chem. 1989;264:8310–8318. [PubMed] [Google Scholar]
- 18.Bhonsale A, James CA, Tichnell C, Murray B, Madhavan S, Philips B, Russell SD, Abraham T, Tandri H, Judge DP, et al. Risk stratification in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated desmosomal mutation carriers. Circ Arrhythm Electrophysiol.2013;6:569–578. doi: 10.1161/CIRCEP.113.000233 [DOI] [PubMed] [Google Scholar]
- 19.Gomes J, Finlay M, Ahmed AK, Ciaccio EJ, Asimaki A, Saffitz JE, Quarta G, Nobles M, Syrris P, Chaubey S, et al. Electrophysiological abnormalities precede overt structural changes in arrhythmogenic right ventricular cardiomyopathy due to mutations in desmoplakin-a combined murine and human study. Eur Heart J. 2012;33:1942–1953. doi: 10.1093/eurheartj/ehr472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingles J, Bagnall RD, Yeates L, McGrady M, Berman Y, Whalley D, Duflou J, Semsarian C. Concealed arrhythmogenic right ventricular cardiomyopathy in sudden unexplained cardiac death events. Circ Genom Precis Med.2018;11:e002355. doi: 10.1161/CIRCGEN.118.002355 [DOI] [PubMed] [Google Scholar]
- 21.Cheung CC, Davies B, Krahn AD. Letter by Cheung et al Regarding Article, “Concealed arrhythmogenic right ventricular cardiomyopathy in sudden unexplained cardiac death events.”. Circ Genom Precis Med.2019;12:e002447. doi: 10.1161/CIRCGEN.118.002447 [DOI] [PubMed] [Google Scholar]
- 22.Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V, Zimbello R, Simionati B, Basso C, Thiene G, et al. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2002;71:1200–1206. doi: 10.1086/344208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh SM, Sharkey SW, Casey SA, Harris KM, Thaler CM, Chung M, Berg A, Bennett MK, Ducanson ER, Mackey-Bojack S, et al. Acute myocardial infarction-like events in related patients with a desmoplakin-associated arrhythmogenic cardiomyopathy. JACC Case Rep.2021;3:1667–1673. doi: 10.1016/j.jaccas.2021.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poller W, Haas J, Klingel K, Kuhnisch J, Gast M, Kaya Z, Escher F, Kayvanpour E, Degener F, Opgen-Rhein B, et al. Familial recurrent myocarditis triggered by exercise in patients with a truncating variant of the desmoplakin gene. J Am Heart Assoc.2020;9:e015289. doi: 10.1161/JAHA.119.015289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauce B, Frigo G, Marcus FI, Basso C, Rampazzo A, Maddalena F, Corrado D, Winnicki M, Daliento L, Rigato I, et al. Comparison of clinical features of arrhythmogenic right ventricular cardiomyopathy in men versus women. Am J Cardiol. 2008;102:1252–1257. doi: 10.1016/j.amjcard.2008.06.054 [DOI] [PubMed] [Google Scholar]
- 26.Fairweather D, Cooper LT, Jr, Blauwet LA. Sex and gender differences in myocarditis and dilated cardiomyopathy. Curr Probl Cardiol. 2013;38:7–46. doi: 10.1016/j.cpcardiol.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, Cirino AL, Fox JC, Lakdawala NK, Ware JS, et al. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation.2018;138:1387–1398. doi: 10.1161/CIRCULATIONAHA.117.033200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheel PJ, III, Murray B, Tichnell C, James CA, Tandri H, Calkins H, Chelko SP, Gilotra NA. Arrhythmogenic right ventricular cardiomyopathy presenting as clinical myocarditis in women. Am J Cardiol. 2021;145:128–134. doi: 10.1016/j.amjcard.2020.12.090 [DOI] [PubMed] [Google Scholar]
- 29.Augusto JB, Eiros R, Nakou E, Moura-Ferreira S, Treibel TA, Captur G, Akhtar MM, Protonotarios A, Gossios TD, Savvatis K, et al. Dilated cardiomyopathy and arrhythmogenic left ventricular cardiomyopathy: a comprehensive genotype-imaging phenotype study. Eur Heart J Cardiovasc Imaging.2020;21:326–336. doi: 10.1093/ehjci/jez188 [DOI] [PubMed] [Google Scholar]
- 30.Ingles J, Burns C, Bagnall RD, Lam L, Yeates L, Sarina T, Puranik R, Briffa T, Atherton JJ, Driscoll T, et al. Non-familial hypertrophic cardiomyopathy: prevalence, natural history, and clinical implications. Circ Cardiovasc Genet.2017;10:e001620. doi: 10.1161/CIRCGENETICS.116.001620 [DOI] [PubMed] [Google Scholar]
- 31.Ingles J, Goldstein J, Thaxton C, Caleshu C, Corty EW, Crowley SB, Dougherty K, Harrison SM, McGlaughon J, Milko LV, et al. Evaluating the clinical validity of hypertrophic cardiomyopathy genes. Circ Genom Precis Med.2019;12:e002460. doi: 10.1161/CIRCGEN.119.002460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maruthappu T, Posafalvi A, Castelletti S, Delaney PJ, Syrris P, O’Toole EA, Green KJ, Elliott PM, Lambiase PD, Tinker A, et al. Loss-of-function desmoplakin I and II mutations underlie dominant arrhythmogenic cardiomyopathy with a hair and skin phenotype. Br J Dermatol. 2019;180:1114–1122. doi: 10.1111/bjd.17388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapplinger JD, Landstrom AP, Salisbury BA, Callis TE, Pollevick GD, Tester DJ, Cox MG, Bhuiyan Z, Bikker H, Wiesfeld AC, et al. Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J Am Coll Cardiol. 2011;57:2317–2327. doi: 10.1016/j.jacc.2010.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grondin S, Wazirian AC, Jorda P, Terrone DG, Gagnon J, Robb L, Amyot J, Rivard L, Page S, Talajic M, et al. Missense variants in the spectrin repeat domain of DSP are associated with arrhythmogenic cardiomyopathy: a family report and systematic review. Am J Med Genet A. 2020;182:2359–2368. doi: 10.1002/ajmg.a.61799 [DOI] [PubMed] [Google Scholar]
- 35.Rasmussen TB, Hansen J, Nissen PH, Palmfeldt J, Dalager S, Jensen UB, Kim WY, Heickendorff L, Molgaard H, Jensen HK, et al. Protein expression studies of desmoplakin mutations in cardiomyopathy patients reveal different molecular disease mechanisms. Clin Genet. 2013;84:20–30 doi: 10.1111/cge.12056 [DOI] [PubMed] [Google Scholar]
- 36.Bolling MC, Veenstra MJ, Jonkman MF, Diercks GF, Curry CJ, Fisher J, Pas HH, Bruckner AL. Lethal acantholytic epidermolysis bullosa due to a novel homozygous deletion in DSP: expanding the phenotype and implications for desmoplakin function in skin and heart. Br J Dermatol. 2010;162:1388–1394. doi: 10.1111/j.1365-2133.2010.09668.x [DOI] [PubMed] [Google Scholar]
- 37.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins R, Laricchia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazzarotto F, Tayal U, Buchan RJ, Midwinter W, Wilk A, Whiffin N, Govind R, Mazaika E, de Marvao A, Dawes TJW, et al. Reevaluating the genetic contribution of monogenic dilated cardiomyopathy. Circulation. 2020;141:387–398. doi: 10.1161/CIRCULATIONAHA.119.037661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LG, Harrison SM; ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI)ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI). Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018;39:1517–1524. doi: 10.1002/humu.23626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lazzarini E, Jongbloed JD, Pilichou K, Thiene G, Basso C, Bikker H, Charbon B, Swertz M, van Tintelen JP, van der Zwaag PA. The ARVD/C genetic variants database: 2014 update. Hum Mutat. 2015;36:403–410. doi: 10.1002/humu.22765 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.