Abstract
Background and Objectives
KCNH5 encodes the voltage-gated potassium channel EAG2/Kv10.2. We aimed to delineate the neurodevelopmental and epilepsy phenotypic spectrum associated with de novo KCNH5 variants.
Methods
We screened 893 individuals with developmental and epileptic encephalopathies for KCNH5 variants using targeted or exome sequencing. Additional individuals with KCNH5 variants were identified through an international collaboration. Clinical history, EEG, and imaging data were analyzed; seizure types and epilepsy syndromes were classified. We included 3 previously published individuals including additional phenotypic details.
Results
We report a cohort of 17 patients, including 9 with a recurrent de novo missense variant p.Arg327His, 4 with a recurrent missense variant p.Arg333His, and 4 additional novel missense variants. All variants were located in or near the functionally critical voltage-sensing or pore domains, absent in the general population, and classified as pathogenic or likely pathogenic using the American College of Medical Genetics and Genomics criteria. All individuals presented with epilepsy with a median seizure onset at 6 months. They had a wide range of seizure types, including focal and generalized seizures. Cognitive outcomes ranged from normal intellect to profound impairment. Individuals with the recurrent p.Arg333His variant had a self-limited drug-responsive focal or generalized epilepsy and normal intellect, whereas the recurrent p.Arg327His variant was associated with infantile-onset DEE. Two individuals with variants in the pore domain were more severely affected, with a neonatal-onset movement disorder, early-infantile DEE, profound disability, and childhood death.
Discussion
We describe a cohort of 17 individuals with pathogenic or likely pathogenic missense variants in the voltage-sensing and pore domains of Kv10.2, including 14 previously unreported individuals. We present evidence for a putative emerging genotype-phenotype correlation with a spectrum of epilepsy and cognitive outcomes. Overall, we expand the role of EAG proteins in human disease and establish KCNH5 as implicated in a spectrum of neurodevelopmental disorders and epilepsy.
The voltage-gated potassium channels are a large group of transmembrane proteins critical for controlling neuronal excitability and regulating electrophysiologic properties in the brain.1 The ether-a-go-go (EAG) subfamily consists of 2 members, EAG1/Kv10.1 encoded by KCNH1 and EAG2/Kv10.2 encoded by KCNH5. Pathogenic gain-of-function (GOF) variants in KCNH1 are implicated in Temple-Baraitser and Zimmermann-Laband syndromes2-4; epilepsy is a common feature of these overlapping syndromes.5 Individuals with variants in KCNH5 have been reported in large exome sequencing cohort studies of individuals with neurodevelopmental disorders (NDDs). Of the 3 previously reported individuals, 2 had a recurrent missense variant (p.Arg327His) associated with a developmental and epileptic encephalopathy (DEE) with onset prior to 1 year of age.6,7 The third had a neonatal-onset DEE and a different de novo missense variant (p.Ile463Thr).8 Despite these case reports, the KCNH5-associated phenotypes are not well delineated.
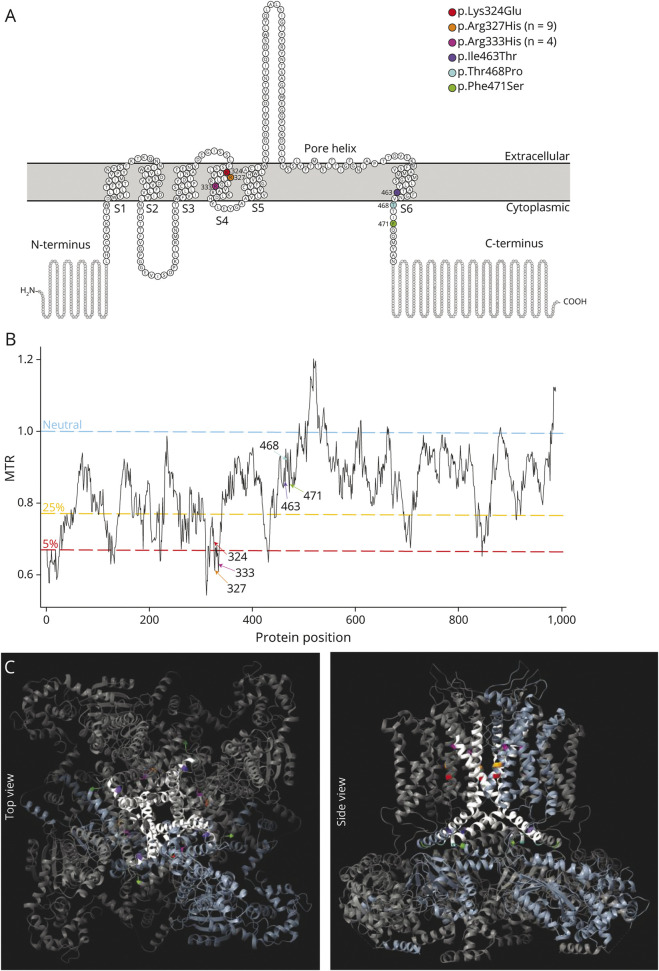
KCNH5 is expressed in excitatory neurons in upper layer IV of the cerebral cortex and the hippocampus.9,10 KCNH5 encodes for Kv10.2, which localizes to the somatodendritic region where it plays a role in controlling the electrical coupling between cell bodies and distal dendrites.10 Like most potassium channels, it consists of 4 transmembrane domains (S1–S4) that make up the voltage-sensing domain and 2 transmembrane domains and a reentrant loop (S5–S6) that comprise the pore module (Figure 1, A and C). The recurrent Kv10.2 p.Arg327His variant localizes to S4, where voltage-clamp analysis in a heterologous expression system demonstrated a hyperpolarized shift of voltage dependence of activation and acceleration of activation, consistent with a gain-of-function pathogenic mechanism.11
Figure 1. Location and Conservation of the 6 Unique De Novo KCNH5 Missense Variants.
(A) Schematic of Kv10.2 (EAG2), modified from Protter35 plot of Q8NCM2 (KCNH5_Human). Of the 6 transmembrane domains, S1-S4 make up the voltage-sensing domain, and S5-6 form the pore helix. The KCNH5 patient-specific variants identified in this study are indicated by their amino acid change. (B) Graph of missense tolerance ratio (MTR; y-axis) and protein position (x-axis), with variants indicated. MTR36 is a measure of protein-encoding cDNA sequence intolerance to missense variants. Variant positions with a value greater than 1 (blue line) is considered neutral; values below 1 are under constraint. Two variants (p.Arg327His and p.Arg333His) are in the 5th percentile of least tolerated missense alterations in the exome. The 5th and 25th percentiles are highlighted in red and yellow, respectively. (C) Locations of variants (color coded) are mapped onto the crystal structure of homotetrameric assembly of Kv10.1 (PDB5K7L); all variants are perfectly conserved between KCNH5 and KCNH1 and are mapped to the corresponding amino acid. The pore domain is highlighted in white, and a single tetrameric subunit is colored light blue. EAG = ether-a-go-go
Here we report a cohort of 17 individuals with de novo or inherited (n = 1) missense variants in the voltage-sensing and pore domains of KCNH5. We describe 14 new patients and provide additional phenotypic information for the 3 published individuals from large cohort studies.6-8 We analyze the spectrum of KCNH5-associated neurodevelopment disorders, particularly the epilepsy phenotypes, and discuss genotype-phenotype correlations.
Methods
Standard Protocol Approvals, Registrations, and Patient Consents
We performed KCNH5 targeted resequencing in 680 individuals with DEEs and exome sequencing in 213 individuals with DEEs, totaling 893 individuals. All individuals underwent clinical assessment, and medical records, EEG, and imaging data were obtained. Seizure types and epilepsy syndromes were classified according to the ILAE classification.12,13 Individuals with DEEs had been previously tested for pathogenic variants in the majority of known DEE genes (14 and unpublished data). The study was approved by the Austin Health Human Research Ethics Committee (H2007/02961), the New Zealand Health and Disability Ethics Committee (NTY/12/06/053/AM15), and the University Hospital of Lyon and Aix-Marseille University in France Ethics Committee (05/78, CPP Strasbourg Alsace 1). Written informed consent was provided by the patient or their parent or legal guardian in the case of minors or those with intellectual disability (ID).
We identified an additional 14 individuals with KCNH5 variants through GeneMatcher.15 These individuals were consented using research protocols approved by their local ethics committees. Their clinicians provided clinical information. For the 3 previously published individuals (probands 2, 10, and 15), we obtained additional clinical information. In total, the cohort consists of 19 individuals with KCNH5 missense variants, with 17 having variants localized in or near the voltage-sensing or pore domains.
KCNH5 Variant Identification and Analysis
We used molecular inversion probes (MIPs) to capture all exons and 5 base pairs of flanking intronic KCNH5 sequence; next-generation sequencing and data analysis were performed as described previously.14 We resequenced KCNH5, covering 95% of the gene at a depth of 50X (median across the cohort of 680 individuals). Exome sequencing (n = 213) was performed via Epi25.16 We considered only variants that were nonsynonymous, altered the acceptor/donor splice sites, or indels that disrupted the coding frame. Only variants that were not present in gnomAD17 and TOPMed18 were considered for segregation analysis. The variants in the 14 individuals recruited by the matchmaker exchange network15 were identified via clinical or research genome (n = 2), exome (n = 10), or gene panel (n = 2) sequencing.16 Library preparation and data analysis were performed using local protocols and computational pipelines. Variants were classified according to the American College of Medical Genetics and Genomics (ACMG) guidelines for variant classification.19
KCNH1 (EAG1/Kv10.1) and KCNH5 (EAG2/Kv10.2) have 72% identity at the amino acid level. The 3-dimensional structure of Kv10.2 has not yet been resolved; thus, to model the location of each of the identified missense variants, we used the Kv10.2 homology model generated from the Kv10.1 cryo-EM structure (5k7l.1.A) using UCSF ChimeraX.20
Data Availability
All data from this study are available in the manuscript or supplementary materials.
Results
Rare Missense Variants in KCNH5
We identified 2 individuals with the previously reported de novo missense variant p.Arg327His in 2 DEE cohorts (1/680 by MIPs and 1/213 by exome16). Through our international collaboration, we identified 5 additional individuals with this recurrent variant, which arose de novo in 4 individuals (Table 1). We also identified 5 additional missense variants in the voltage-sensing and pore domains of Kv10.2 (Figure 1). One variant (p.Arg333His) was present in 4 individuals; it arose de novo in 3 and was inherited from an affected mother in the fourth proband. The 4 other de novo missense variants were each observed once in single individuals.
Table 1.
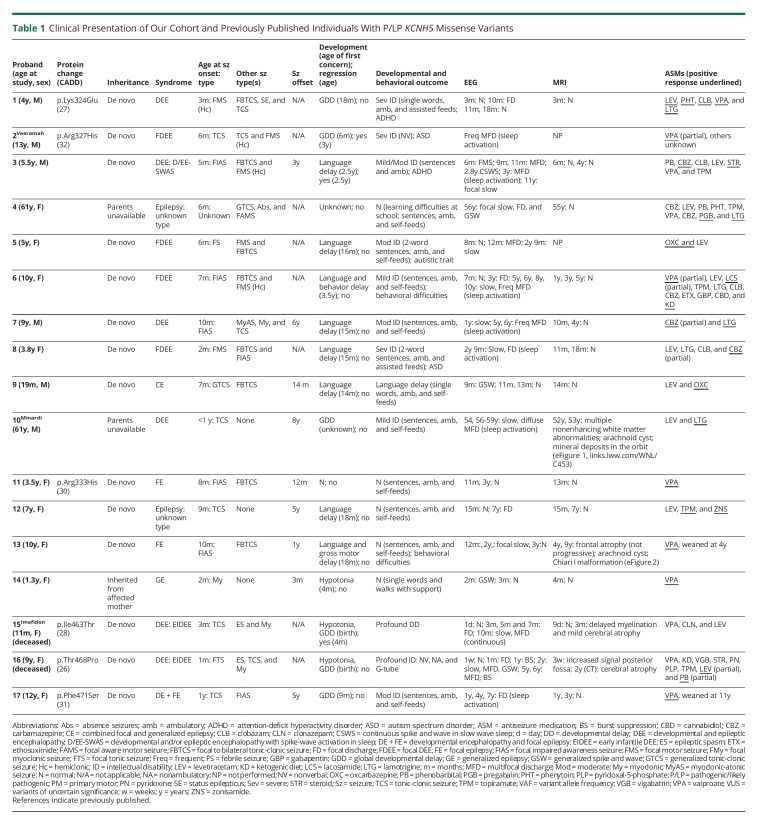
Clinical Presentation of Our Cohort and Previously Published Individuals With P/LP KCNH5 Missense Variants
The amino acids at all 6 variant positions (2 recurrent, 4 singletons) were conserved between Kv10 proteins. The missense variants had CADD21 scores of 26–32, conferring a high likelihood of deleteriousness (Figure 1B and Table 1). All variants were absent from the general population (gnomAD and TOPMed) and were classified as either pathogenic or likely pathogenic (P/LP) by the ACMG criteria (eTable 1, links.lww.com/WNL/C453).19
We also identified 2 de novo variants of uncertain significance (VUS) in the N-terminal (p.Leu181Pro) and C-terminal (p.Ile606Thr) of Kv10.2 in 2 individuals with NDDs but without seizures (eAppendix 1, links.lww.com/WNL/C453). These VUS were located outside of the functionally critical pore or voltage-sensing domains. One variant (p.Leu181Pro) did not segregate with the NDD in the family, and the individual with the p.Ile606Thr variant also carried a pathogenic 16p11.2 microdeletion that may explain some or all of their clinical features. Although these variants are included here for completeness, this study focuses on the 17 individuals with variants in the voltage-sensing or pore-forming domains.
Phenotypic Features in Individuals With KCNH5 Pathogenic or Likely Pathogenic Variants
All 17 individuals with pathogenic or likely pathogenic missense variants in the voltage-sensing or pore domains had epilepsy (Table 1). Moreover, the mother of proband 14 who carries the recurrent p.Arg333His variant also has epilepsy, although detailed clinical information was not available. The median age at seizure onset was 6 months (range: 1–12 months). A wide range of seizure types were seen: focal impaired awareness seizures (8), focal to bilateral tonic-clonic seizures (9), focal motor seizures (8), generalized myoclonic (4), generalized tonic-clonic seizure (2), myoclonic-atonic (1), absence seizure (1), unknown onset tonic-clonic seizures (6), and epileptic spasms (2). Nine (53%) individuals had a DEE. The remainder had focal epilepsy (3), generalized epilepsy (2), or there was inadequate information to make an epilepsy type diagnosis (3). Nine individuals (53%) achieved seizure freedom, with median seizure offset at 2.5 years (range: 3 months to 8 years).
Developmental concerns were noted at a median age of 15 months in 14 individuals (range birth to 3.5 years), predominantly affecting language (11). One child had normal development, and for 2 individuals, aged 61 years, early developmental information was limited. Cognitive outcome for the 10 individuals over 5 years of age ranged from normal (3/10) to mild (3/10), moderate (2/10), severe (1/10), and profound (1/10) ID. Two individuals died at ages 13 months and 9 years, respectively.
There were no consistent or significant dysmorphic features noted in these individuals. Proband 1 was reported to have long eyelashes and mild ptosis, whereas proband 2 had prominent ears, small hands, and a flexion contracture of the 4th and 5th digits of one hand. Significant sleep disorders were not noted in any individual.
For the 9 individuals with the recurrent voltage-sensing domain p.Arg327His variant, seizure onset occurred at a median age of 6 months (range 2–10 months). For the 8 individuals for whom an epilepsy syndrome diagnosis could be made, all but one, who was 19 months old, had a DEE (7/8), which was focal in 5 individuals. Proband 3 had the syndrome of epileptic encephalopathy with spike-and-wave activation in sleep, which was associated with language regression.12 All patients had EEGs, which captured sleep. Sleep activation of epileptiform discharges was noted in 6 other patients but was not associated with developmental regression. The cognitive outcome for those over 5 years of age with this recurrent variant ranged from normal with learning difficulties (1/6) to mild (3/6), moderate (1/6), or severe ID (1/6). For those in whom the complete antiseizure medication history was available, their epilepsy was drug resistant (8/9), with only 4/9 eventually becoming seizure-free. Brain MRI was normal in all apart from proband 10. He had no early imaging, but an MRI performed at 52 years demonstrated nonenhancing periventricular white matter abnormalities suggestive of demyelinating lesions. The lesions were stable on a repeat MRI 6 months later. Note was also made of an arachnoid cyst and mineral deposits in the orbit (eFigure 1, links.lww.com/WNL/C453).
The phenotype was much less severe for the 4 individuals with the second recurrent voltage-sensing domain p.Arg333His variant. This group had onset at a similar age (2–10 months) with either focal or generalized seizures. In contrast, their seizures were drug responsive, particularly to valproate (3/4), and all became seizure-free (aged 3 months to 5 years). One of the 4 individuals had mild motor and language delay at presentation, 1 had mild language delay, and 1 had only mild motor delay. All had normal cognitive development as assessed by their clinician when last seen at ages 1–10 years. MRI was normal apart from proband 13 who had mild frontal cortical atrophy, an arachnoid cyst, and a Chiari I malformation (eFigure 2, links.lww.com/WNL/C453).
Strikingly, probands 15 (p.Ile463Thr) and 16 (p.Thr468Pro), with missense variants in or at the junction of the S6 transmembrane pore-forming domain, had a much more severe phenotype. Both individuals presented with persistent nonepileptic myoclonus on day 1 of life and developed an early-infantile DEE with drug-resistant seizures by 3 months and eventual epileptic spasms. They had very limited developmental progression. Neuroimaging showed cortical atrophy. Proband 15 died at 13 months due to respiratory failure secondary to increased seizure activity. Proband 16 died at 9 years due to pneumonia.
There were 2 additional individuals with likely pathogenic variants and epilepsy. Proband 1, with p.Lys324Glu variant in the voltage-sensing domain, had a similar phenotype to individuals with the nearby recurrent p.Arg327His variant. This individual had a focal DEE with focal seizures beginning at 3 months and developmental delay evident by 18 months, evolving to severe ID. Proband 17 had the p.Phe471Ser variant in the C-terminal cytoplasmic domain, which was very close to the pore-forming domain. She had moderate ID, drug-responsive focal epilepsy, and a hyperkinetic movement disorder consisting of a tremor predominantly in the upper limbs and exacerbated when she was tired or unwell. She did not have an epileptic encephalopathy and presented a similar epilepsy phenotype to patients with the recurrent p.Arg333His variant. We provide clinical vignettes for 3 patients (probands 6, 13, and 16) to highlight the spectrum of phenotypes associated with KCNH5 pathogenic variants (eAppendix 2, Clinical Vignettes, links.lww.com/WNL/C453).
Discussion
Potassium channel subunits are one of the most important groups of genes associated with epilepsy, in particular with the most severe group of epilepsies, the DEEs.22 Here, we show that de novo pathogenic variants in KCNH5 in 17 patients are frequently associated with a DEE but may also cause self-limited epilepsies. We describe the phenotypic spectrum, with particular emphasis on the phenotype-genotype correlations and the putative effect of these missense variants on Kv10.2 channel function.
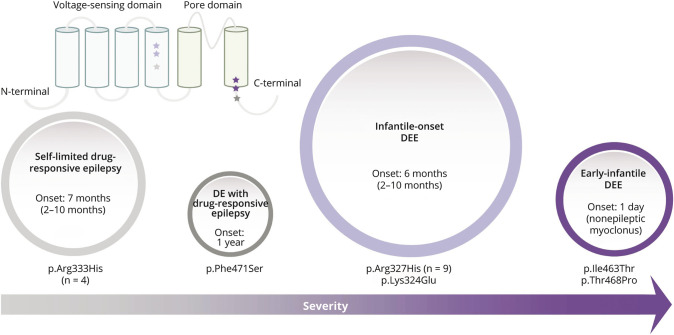
We highlight a genotype-phenotype correlation for variants across the protein (Figure 2). The p.Arg333His variant is associated with the mildest epilepsy phenotype, a drug-responsive self-limited focal or generalized epilepsy with normal cognitive outcome. Conversely, the nearby most common recurrent p.Arg327His variant has a far more severe epilepsy phenotype of DEE. Of interest, the nearby p.Lys324Glu voltage-sensing domain variant was also associated with a DEE. Individuals with these 2 variants predominantly presented with an infantile-onset DEE between 2 and 10 months of age with drug-resistant generalized and focal seizures. Early developmental concerns were noted with cognitive outcome ranging from learning difficulties to severe ID. Finally, 2 individuals with variants in the pore-forming S6 transmembrane domain (p.Ile463Thr and p.Thr468Pro) had an even more severe phenotype of neonatal-onset DEE with onset on day 1 of life resulting in profound disability and death.
Figure 2. Phenotypic Spectrum Associated With KCNH5 Missense Variants.
The number of individuals in the phenotype group is represented by circle size, and colors of each circle and star match the variant/phenotype class. The individuals with the p.Lys324Glu and p.Arg327His variants in the S4 transmembrane domain presented with an infantile-onset DEE with drug-resistant generalized and focal seizures, whereas the 4 individuals with the nearby recurrent p.Arg333His variant had drug-responsive seizures and became seizure-free. The 2 individuals with variants in or directly at the junction of the S6 transmembrane domain (p.Ile463Thr and p.Thr468Pro) are the most severely affected with a neonatal-onset movement disorder and early-infantile DEE. The single individual with the nearby p.Phe471Ser variant was less severely affected with moderate ID and drug-responsive seizures. Range is indicated in parentheses where applicable. DE = developmental encephalopathy; DEE = developmental and epileptic encephalopathy.
The p.Arg327His variant, located in the voltage-sensing S4 transmembrane domain, acts in a GOF manner.11 Multistate structural modeling demonstrated that the p.Arg327His variant favors channel opening, and voltage-clamp experiments showed a hyperpolarized shift of voltage dependence of activation and acceleration of activation.11 These changes in electrophysiologic properties are consistent with the GOF mechanism that is typically observed for several potassium channels implicated in DEEs, such as KCNH1,2 KCNT1,23 and KCNT2.24 A subset of epilepsy-associated variants in KCNA2, KCNB1, KCNQ2, and KCNQ3 have similarly been shown to act in a GOF manner.25-27 How these changes lead to epilepsy remains unresolved.28 We hypothesize that the new KCNH5 voltage-sensing and pore domain pathogenic variants described here also have a similar GOF mechanism to the p.Arg327His variant. This is supported by in silico data including the tolerance of KCNH5 to loss-of-function (truncating) variants in the general population. KCNH5 has a probability of loss-of-function intolerant (pLI) score of 0.0229. Epilepsy-associated genes that confer pathogenicity via haploinsufficiency typically have pLI scores ∼1, highlighting that loss of function is unlikely to be the pathogenic mechanism for KCNH5 variants. Conversely, missense variants in the gene are not as well tolerated, with a z-score of 2.51 (Figure 1B), suggesting selection against certain missense variants throughout human evolution.
The 2 most severely affected individuals carried missense variants (p.Ile463Thr and p.Thr468Pro) that cluster right at the junction of the S6 transmembrane domain and the early C-terminal cytoplasmic domain that form the channel pore. The 2 missense variants are located within 5 amino acids of each other right at the channel pore. Missense variants are similarly found in and just outside the S6 domain of the EAG1/Kv10 pore encoded by KCNH1 in individuals with Temple-Baraitser syndrome, which includes epilepsy as a key feature. Detailed electrophysiologic studies of these S6 missense variants in KCNH1 showed delayed deactivation and decreased threshold of activation in both human cells and Xenopus oocytes, consistent with a GOF mechanism.2,3 We hypothesize that the pore-forming KCNH5 missense variants act in a similar GOF manner. Like other voltage-gated potassium channels, Kv10.2 comprises 4 subunits that form a homotetramer. Any given tetramer could have between zero and 4 subunits containing the variant residue, and it is possible that the function of the resulting channels could be differentially disrupted. This principle is a challenge in ion channel physiology generally, and in particular, for epilepsy, new tools and models are currently being developed to address this challenge. Moreover, future studies will also be necessary to confirm the functional effect and determine whether missense pathogenic variants in the most central pore region have a more profound GOF effect on Kv10.2 function than those in the voltage-sensing domain or located further away from the pore.
De novo missense variants in KCNH1/Kv10.1 cause Temple-Baraitser syndrome (TBS) and Zimmermann-Laband syndrome (ZLS).2-4 Features of both include ID, epilepsy, facial dysmorphism, and nail hypoplasia.30 GOF KCNH1 missense variants have recently been reported in individuals with ID and epilepsy without the additional phenotypic features of TBS/ZLS.31,32 Individuals with both syndromic and nonsyndromic KCNH1-associated disorders with epilepsy have both focal and generalized seizures in infancy.5 Three of the 6 reported KCNH5 variants (p.Lys324Glu, p.Arg327His, and p.Ile463Thr) affect analogous KCNH1 amino acid residues that have been reported in individuals with KCNH1-associated NDDs,2,4,32 further highlighting the importance of these positions on channel function. KCNH1 and KCNH5 are predominantly expressed in the adult CNS (Human Protein Atlas33 and GTEx34); however, current expression data sets are largely limited to adult tissues. Particularly as all 6 amino acid residues affected by the nucleotide changes described here are conserved between KCNH1 and KCNH5 (eFigure 3, links.lww.com/WNL/C453), future data sets such as the Developmental Genotype-Tissue Expression initiative will be valuable for evaluating both the multisystem involvement of KCNH1-associated disorders and the CNS-limited KCNH5-related phenotypes.
Overall, we establish KCNH5 as a gene associated with epilepsy and neurodevelopmental phenotypes and identify an emerging genotype-phenotype correlation. We describe 17 individuals with pathogenic or likely pathogenic variants in KCNH5, including 4 novel variants. The p.Arg327His variant located in the voltage-sensing domain causes gain of channel function,11 and we hypothesize that the additional KCNH5 voltage-sensing and pore domain variants also lead to epilepsy via a similar GOF mechanism. We highlight an intriguing genotype-phenotype correlation with a spectrum of epilepsy and cognitive outcomes. These observations will be further supported by examining the functional effects associated with the location of different variants in Kv10.2, with potential therapeutic implications. Our study expands the role of EAG proteins in human disease, highlighting that KCNH5 variants are implicated in a spectrum of NDD and epilepsy phenotypes.
Accession Numbers
KCNH5 mRNA NM_139318.4 and protein NP_647479.2 sequence.
KCNH1 protein NP_758872.1.
Acknowledgment
The authors thank the patients and their families for participating in our research study. They also thank Jennifer Kearney for her valuable insight on ion channel genetics. They thank the Epi25 principal investigators, local staff from individual cohorts, and all patients with epilepsy who participated in the study for making possible this global collaboration and resource to advance epilepsy genetics research. This work is part of the Centers for Common Disease Genomics (CCDG) program, funded by the National Human Genome Research Institute (NHGRI) and the National Heart, Lung, and Blood Institute (NHLBI). CCDG-funded Epi25 research activities at the Broad Institute, including genomic data generation in the Broad Genomics Platform, are supported by NHGRI grant UM1 HG008895 (PIs: Eric Lander, Stacey Gabriel, Mark Daly, and Sekar Kathiresan). The Genome Sequencing Program efforts were also supported by NHGRI grant 5U01HG009088-02. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors thank the Stanley Center for Psychiatric Research at the Broad Institute for supporting the genomic data generation efforts.
Glossary
- ACMG
American College of Medical Genetics and Genomics
- DEE
developmental and epileptic encephalopathy
- EAG
ether-a-go-go
- ID
intellectual disability
- GOF
gain of function
- MIP
molecular inversion probe
- NDD
neurodevelopmental disorder
- VUS
variants of uncertain significance
Appendix. Authors
Study Funding
This work was supported by funding from the NIH (NINDS NS069605). H.C. Mefford is a recipient of the Burroughs Wellcome Fund Career Award for Medical Scientists. G.L. Carvill was funded by the Citizens United for Research in Epilepsy (CURE) Taking Flight Award and NIH (NINDS NS089858). I.E. Scheffer is supported by a National Health and Medical Research Council of Australia (NHMRC) Program grant, Investigator grant, as well as a Centre of Research Excellence and Synergy grant. L.G. Sadleir and G. Valles-Ibáñez were supported by Health Research Council of New Zealand and Cure Kids research grants. L. Sedlackova, K. Sterbova, M. Vlckova, P. Lassuthova, and A. Jahodova are supported by the Ministry of Health of the Czech Republic AZV NU20-04-00279.
Disclosure
L.G. Sadleir is a consultant for the Epilepsy Consortium and has received travel grants from Seqirus and Nutricia. She has received research grants and consultancy fees from Zynerba Pharmaceuticals. She has served on an Eisai Pharmaceuticals scientific advisory panel. R. Person, E. Torti, and K. McWalter are employees of GeneDx. B.K. Burton has received consulting fees and/or honoraria from Aeglea, Alexion, Applied Therapeutics, BioMarin, Capsida, Denali, Horizon, JCR Pharma, Moderna, SIO, Takeda, and Ultragenyx and has conducted clinical trials funded by BioMarin, Denali, JCR Pharma, Homology Medicines, Sangamo, and Ultragenyx. I.E. Scheffer has served on scientific advisory boards for BioMarin, Chiesi, Eisai, Encoded Therapeutics, GlaxoSmithKline, Knopp Biosciences, Nutricia, RogCon, Takeda Pharmaceuticals, UCB, and Xenon Pharmaceuticals; has received speaker honoraria from GlaxoSmithKline, UCB, BioMarin, Biocodex, Chiesi, Liva Nova, and Eisai; has received funding for travel from UCB, Biocodex, GlaxoSmithKline, BioMarin, and Eisai; has served as an investigator for Anavex Life Sciences, Cerecin Inc, Cereval Therapeutics, Eisai, Encoded Therapeutics, EpiMinder Inc, Epygenix, ES Therapeutics, GW Pharma, Marinus, Neurocrine BioSciences, Ovid Therapeutics, Takeda Pharmaceuticals, UCB, Ultragenyx, Xenon Pharmaceutical, Zogenix, and Zynerba; has consulted for Atheneum Partners, Care Beyond Diagnosis, Epilepsy Consortium, Ovid Therapeutics, UCB, and Zynerba Pharmaceuticals; and is a Non-Executive Director of Bellberry Ltd and a Director of the Australian Academy of Health and Medical Sciences and the Australian Council of Learned Academies Limited. She may accrue future revenue on pending patent WO61/010176 (filed: 2008): Therapeutic Compound; has a patent for SCN1A testing held by Bionomics Inc and licensed to various diagnostic companies; and has a patent molecular diagnostic/theranostic target for benign familial infantile epilepsy (BFIE) [PRRT2] 2011904493 & 2012900190 and PCT/AU2012/001321 (TECH ID:2012-009). The remaining authors have nothing relevant to disclose. Go to Neurology.org/N for full disclosures.
References
- 1.Bauer CK, Schwarz JR. Ether-a-go-go K(+) channels: effective modulators of neuronal excitability. J Physiol. 2018;596(5):769-783. doi: 10.1113/jp275477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons C, Rash LD, Crawford J, et al. Mutations in the voltage-gated potassium channel gene KCNH1 cause Temple-Baraitser syndrome and epilepsy. Nat Genet. 2015;47(1):73-77. doi: 10.1038/ng.3153 [DOI] [PubMed] [Google Scholar]
- 3.Kortum F, Caputo V, Bauer CK, et al. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat Genet. 2015;47(6):661-667. doi: 10.1038/ng.3282 [DOI] [PubMed] [Google Scholar]
- 4.Gripp KW, Smithson SF, Scurr IJ, et al. Syndromic disorders caused by gain-of-function variants in KCNH1, KCNK4, and KCNN3-a subgroup of K(+) channelopathies. Eur J Hum Genet. 2021;29(9):1384-1395. doi: 10.1038/s41431-021-00818-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mastrangelo M, Scheffer IE, Bramswig NC, et al. Epilepsy in KCNH1-related syndromes. Epileptic Disord. 2016;18(2):123-136. doi: 10.1684/epd.2016.0830 [DOI] [PubMed] [Google Scholar]
- 6.Veeramah KR, Johnstone L, Karafet TM, et al. Exome sequencing reveals new causal mutations in children with epileptic encephalopathies. Epilepsia. 2013;54(7):1270-1281. doi: 10.1111/epi.12201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minardi R, Licchetta L, Baroni MC, et al. Whole-exome sequencing in adult patients with developmental and epileptic encephalopathy: it is never too late. Clin Genet. 2020;98(5):477-485. doi: 10.1111/cge.13823 [DOI] [PubMed] [Google Scholar]
- 8.Imafidon ME, Sikkema-Raddatz B, Abbott KM, et al. Strategies in rapid genetic diagnostics of critically Ill children: experiences from a Dutch university hospital. Front Pediatr. 2021;9:600556. doi: 10.3389/fped.2021.600556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saganich MJ, de Miera EVS, Nadal MS, Baker H, Coetzee WA, Rudy B. Cloning of components of a novel subthreshold-activating K(+) channel with a unique pattern of expression in the cerebral cortex. J Neurosci. 1999;19(24):10789-10802. doi: 10.1523/jneurosci.19-24-10789.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeng CJ, Chang CC, Tang CY. Differential localization of rat Eag1 and Eag2 K+ channels in hippocampal neurons. Neuroreport 2005;16(3):229-233. doi: 10.1097/00001756-200502280-00005 [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Vasylyev DV, Dib-Hajj F, et al. Multistate structural modeling and voltage-clamp analysis of epilepsy/autism mutation Kv10.2-R327H demonstrate the role of this residue in stabilizing the channel closed state. J Neurosci. 33(42);2013:16586-16593. doi: 10.1523/jneurosci.2307-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheffer IE, Berkovic S, Capovilla G, et al. ILAE classification of the epilepsies: position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58(4):512-521. doi: 10.1111/epi.13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher RS, Cross JH, French JA, et al. Operational classification of seizure types by the international league against epilepsy: position paper of the ILAE commission for classification and terminology. Epilepsia. 2017;58(4):522-530. doi: 10.1111/epi.13670 [DOI] [PubMed] [Google Scholar]
- 14.Carvill GL, Heavin SB, Yendle SC, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45(7):825-830. doi: 10.1038/ng.2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928-930. doi: 10.1002/humu.22844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Epi25, Collaborative Electronic address sberkovic@unimelbeduau, Epi25 Collaborative. Ultra-rare genetic variation in the epilepsies: a whole-exome sequencing study of 17,606 individuals. Am J Hum Genet. 2019;105(2):267-282. doi: 10.1016/j.ajhg.2019.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.gnomAD [online]. Available at: gnomad.broadinstitute.org
- 18.TopMED [online]. Available at: bravo.sph.umich.edu/freeze3a/hg19/
- 19.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pettersen EF, Goddard TD, Huang CC, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70-82. doi: 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rentzsch P, Schubach M, Shendure J, Kircher M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13(1):31. doi: 10.1186/s13073-021-00835-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15(3):304-316. doi: 10.1016/s1474-4422(15)00250-1 [DOI] [PubMed] [Google Scholar]
- 23.Barcia G, Fleming MR, Deligniere A, et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat Genet. 2012;44(11):1255-1259. doi: 10.1038/ng.2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambrosino P, Soldovieri MV, Bast T, et al. De novo gain-of-function variants in KCNT2 as a novel cause of developmental and epileptic encephalopathy. Ann Neurol. 2018;83(6):1198-1204. doi: 10.1002/ana.25248 [DOI] [PubMed] [Google Scholar]
- 25.Miceli F, Soldovieri MV, Ambrosino P, et al. Early-onset epileptic encephalopathy caused by gain-of-function mutations in the voltage sensor of Kv7.2 and Kv7.3 potassium channel subunits. J Neurosci. 2015;35(9):3782-3793. doi: 10.1523/jneurosci.4423-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torkamani A, Bersell K, Jorge BS, et al. De novo KCNB1 mutations in epileptic encephalopathy. Ann Neurol. 2014;76(4):529-540. doi: 10.1002/ana.24263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Syrbe S, Hedrich UBS, Riesch E, et al. De novo loss- or gain-of-function mutations in KCNA2 cause epileptic encephalopathy. Nat Genet. 2015;47(4):393-399. doi: 10.1038/ng.3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niday Z, Tzingounis AV. Potassium Channel gain of function in epilepsy: an unresolved paradox. Neuroscientist. 2018;24(4):368-380. doi: 10.1177/1073858418763752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature. 2020;581(7809):434-443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton MJ, Suri M. “Electrifying dysmorphology”: potassium channelopathies causing dysmorphic syndromes. Adv Genet. 2020;105:137-174. doi: 10.1016/bs.adgen.2020.03.002 [DOI] [PubMed] [Google Scholar]
- 31.Aubert Mucca M, Patat O, Whalen S, et al. Patients with KCNH1-related intellectual disability without distinctive features of Zimmermann-Laband/Temple-Baraitser syndrome. J Med Genet. 2021;59(5):505-510. doi: 10.1136/jmedgenet-2020-107511 [DOI] [PubMed] [Google Scholar]
- 32.Fukai R, Saitsu H, Tsurusaki Y, et al. De novo KCNH1 mutations in four patients with syndromic developmental delay, hypotonia and seizures. J Hum Genet. 2016;61(5):381-387. doi: 10.1038/jhg.2016.1 [DOI] [PubMed] [Google Scholar]
- 33.The Human Protein Atlas. Updated December 7, 2022. proteinatlas.org/ [Google Scholar]
- 34.GTEx Portal. gtexportal.org/home/
- 35.Omasits U, Ahrens CH, Muller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30(6):884-886. doi: 10.1093/bioinformatics/btt607 [DOI] [PubMed] [Google Scholar]
- 36.Silk M, Petrovski S, Ascher DB. MTR-Viewer: identifying regions within genes under purifying selection. Nucleic Acids Res. 2019;47(W1):W121–W126. doi: 10.1093/nar/gkz457 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data from this study are available in the manuscript or supplementary materials.