Abstract
Background
Sepsis is a life-threatening condition that induce tens of million death each year, yet early diagnosis remains a formidable challenge. Many studies have focused on the diagnostic accuracy of microRNAs (miRNAs) for sepsis in recent years, particularly miR-155-5p, miR-21, miR-223-3p, miR-146a, and miR-125a. Thus, we conducted this meta-analysis to explore if miRNAs may be used as a biomarker for sepsis detection.
Methods
We searched PubMed, the Cochrane Central Register of Controlled Trials, EMBASE, and China National Knowledge Infrastructure through May 12, 2022. This meta-analysis was conducted using Meta-disc 1.4 and STATA 15.1 in a fixed/random-effect model.
Results
The analysis included a total of 50 relevant studies. The overall performance of total miRNAs detection was: pooled sensitivity, 0.76 (95% confidence interval [CI], 0.75 to 0.77); pooled specificity, 0.77 (95%CI, 0.75 to 0.78); and area under the summary receiver operating characteristic curves value (SROC), 0.86. The subgroup analysis suggested that detection in miR-155-5p group had the highest area under the curve (AUC) of SROC among all miRNAs: pooled sensitivity, 0.71 (95%CI, 0.67 to 0.75); pooled specificity, 0.82 (95%CI, 0.76 to 0.86); and SROC, 0.85. MiR-21, miR-223-3p, miR-146a, and miR-125a had SROC values of 0.67, 0.78, 0.69, and 0.74, respectively. The specimen type was found to be a source of heterogeneity in the meta-regression study. The SROC of serum was higher than that of plasma (0.87 and 0.83, respectively).
Conclusions
Our meta-analysis revealed that miRNAs, specifically miR-155-5p, could be useful biomarkers for detecting sepsis. A clinical serum specimen is also indicated for diagnostic purposes.
Introduction
Sepsis is a life-threatening disease that induce about 11 million death each year [1]. The main cause of sepsis is the maladjustment of the host’s response to infection [2]. Sepsis is treatable and timely implementation of targeted interventions can improve outcomes [3]. Meanwhile, delayed diagnosis is associated with increased mortality [4]. Therefore, early diagnosis of sepsis is the key to improve the survival rate. Nevertheless, sepsis is a heterogeneous syndrome and the diagnosis of sepsis detection is mainly according to the site of infection, etiology, onset time, and the patient’s profile [2]. However, traditional screening methods and biomarkers such as C-reactive protein (CRP) and procalcitonin (PCT) lack specificity, which leads to the early diagnosis of sepsis is still a formidable challenge. Therefore, it is important to find novel and reliable biomarkers for early diagnosis of sepsis.
MicroRNAs (miRNAs) are a type of small noncoding RNA with an average length of 18–25 nucleotides [5]. Previous research [6, 7] has discovered that circulating miRNAs can be used as biomarkers to detect various diseases. Vasilescu et al. [8] were the first to discover that plasma miR-150 was a potential biomarker for sepsis. Numerous studies [9–12] have since confirmed the significance of miRNAs in sepsis. Several miRNAs have been mentioned in several studies with varying diagnostic effectiveness. Shen et al. [13] conducted a meta-analysis in 2020 to validate the diagnostic accuracy of miRNAs for sepsis, and found that miR-223-3p might be used as a sepsis indicator. In recent years, however, a growing number of research have focused on the diagnostic accuracy of miRNAs for sepsis, particularly miR-155-5p [14–16], miR-21 [9, 10, 17], miR-146a [18–20], and miR-125a [21–23], but the results have been inconsistent. Thus, we collected all published case-control articles to gather evidence on the diagnostic accuracy of miRNAs for sepsis.
Materials and methods
Study protocol
This analysis was carried out according to a predetermined protocol, as recommended by Deeks [24]. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement (S1 Table) was used for data collection and analysis [25]. The ethics approval was not required because this was a comprehensive literature review. This study was registered with PROSPERO (CRD42022361151).
Search strategy
To find relevant studies, we searched PubMed, China National Knowledge Infrastructure (CNKI), EMBASE, and the Cochrane Central Register of Controlled Trials (CENTRAL) databases until May 12, 2022. Keyword search terms were (‘sepsis’ OR ‘pyemia’ OR ‘septicemia’) AND (‘MicroRNAs’ OR ‘miRNAs’ OR ‘MicroRNA’ OR ‘miRNA’). PubMed database was searched as follows: (Sepsis[MeSH Terms] OR pyemia OR septicemia) AND (MicroRNAs[MeSH Terms] OR miRNAs OR MicroRNA OR miRNA). Search terms for the CNKI, EMBASE and CENTRAL with corresponding publication numbers can be found in the S1 Appendix. Language was limited in English and Chinese.
Study selection
The titles and abstracts of studies were evaluated first. Full articles were then retrieved for potentially relevant research and reviewed for conformity with inclusion and exclusion criteria.
Criteria for inclusion: (1) all sepsis patients were confirmed by diagnosis criteria; (2) randomized or non-randomized controlled, cohort studies, clinical trials, evaluating the expression of miRNAs; (3) contained data of receiver operating characteristic (ROC) curve and the essential sample size, or the data of true positive (TP), false positive (FP), false negative (FN), and true negative (TN); (4) all studies had controls, including healthy people or infected patients; (5) full text published in English or Chinese.
Criteria for exclusion: (1) reviews, conferences articles, letters, or case reports without controls; (2) no adequate data to make a 2×2 table; (3) the total sample size of sepsis patients and controls included in the article was too small (n < 60); (4) duplicated studies.
Data collection and assessment of study quality
Two investigators (Yue Zhang and Xiaolan Zheng) independently reviewed study eligibility of studies at the title and abstract level, using the inclusion and exclusion criteria, with third reviewer (Yifei Li) determining the divergences and report quality. All papers that met all of the criteria for inclusion would be assessed further. According to the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) list [26], two investigators (Xiaolan Zheng and Sha Lin) independently assessed all enrolled studies, and any disagreements were resolved through discussion with a third reviewer (Yifei Li). Moreover, interrater reliability for the study selection was calculated by the kappa statistic. Besides, we extracted data from the figures using Photoshop CS6 (Adobe Systems Software Ireland Ltd) using the method given in our previous report [6]. Finally, two researchers (Xiaolan Zheng and Yue Zhang) retrieved data that may be used to determine TP, TN, FP, and FN, including sensitivity, specificity, and the number of patients and controls.
Evaluation indicators
Sensitivity, specificity, diagnostic odds ratio (DOR), and area under the summary ROC (SROC) curves values were all measured. The fraction of sepsis patients accurately identified by positive miRNA expression results was called sensitivity. Non-sepsis cases successfully recognized by negative miRNAs results were represented by specificity. DOR indicated that patients with positive results are much more likely to have sepsis than patients with negative results, and a higher DOR showed that the test had stronger discriminatory ability [27]. The area under the curve (AUC) value was analyzed as a global measurement of test performance, and the SROC curve was generated based on sensitivity and specificity. The greater the test performance, the closer the AUC value was to 1 [28].
Publication bias
Following funnel plots and the Deeks’ test, a quantitative analysis of all the publication bias was conducted using STATA 15.1 (Stata Corporation, College Station, Texas, USA). The possibility of publication bias was highlighted by an uneven distribution of data points with a quantified result of P < 0.05 [29].
Heterogeneity and meta-regression
Heterogeneity of pooling sensitivity and specificity was measured by x2 test, and the Cochran Q test was used to determine the heterogeneity of pooling DOR. When P < 0.05, statistically significant heterogeneity was present. In addition, inconsistency index (I2) test was used to determine the proportion of total variation between studies quantitatively. The I2 value ranged from 0 to 100%, with values of 25, 50, and 75% indicating low, moderate, and high heterogeneity, respectively [30]. When heterogeneity was detected, we used STATA 15.1 to do a meta-regression analysis to determine the source of heterogeneity. The correlation between possible factors and existent heterogeneities could be determined using meta-regression. When a significant difference is found, the factor should have a significant impact on the homogeneity of the included studies, with a P value of less than 0.05.
Sensitivity analysis and subgroup analysis
The sensitivity analysis for each study was carried using STATA 15.1 to quantify the impact of specific studies on the results. Meta-Disc 1.4 was used to perform subgroup analysis and detect threshold effects in studies.
Statistical analysis
For data processing and threshold analysis, Meta-Disc 1.4 was utilized [31]. Besides, publication bias was performed by STATA Version 15.1. Homogenous results were analyzed using the fixed effects model, while the heterogeneous (I2 > 50%) results utilized random effects model, and the data were presented using a forest map. Moreover, RevMan 5.4 (Cochrane, London, UK) was used to evaluate the quality of included studies based on QUADAS-2.
Trial sequential analysis
Trial sequential analysis (TSA) was performed by the TSA 0.9.5.10 beta (Copenhagen Trial Unit) to evaluate the stability of results and the required information size (RIS) [32]. In this meta-analysis, we set the type I error rate to 5%, the type II error rate to 20%, and the statistical power to be 80%, referring to the methods of previous studies [32–34]. If the results of TSA analysis show that the cumulative Z-curve crosses the trial sequential monitoring boundary or the required information size, this indicates that the statistical evidence of this meta-analysis is reliable. Otherwise, more studies must be conducted before conclusive results can be drawn [33].
Results
Search results
Initially, the search method retrieved 3560 potentially relevant studies, of which 233 studies were considered to read their whole studies after assessing titles and abstracts. However, due to article types, 21 studies were removed, and 113 studies lacked data on TP, TN, FP, and FN. Furthermore, 49 studies did not include a comparison of sepsis patients and controls. Fig 1 presents the study selection procedure. Finally, the meta-analysis comprised 50 studies [9–12, 14–23, 35–68], totaling 5225 sepsis patients and 4008 controls, and involving 48 miRNAs. Five miRNAs (miR-155-5p, miR-21, miR-223-3p, miR-146a, and miR-125a) were found to be implicated in more than two investigations. Furthermore, the age of the population was diverse. Nine studies focused on newborns less than 28 days old, four on children older than 1 month, and the remaining 37 on adults. Additionally, the sample types of 20 studies were plasma, 29 studies were serum, and one study was peripheral blood mononuclear cells (PBMC). Moreover, 46 studies from Asian (44 from China, one from India, and one from Vietnam), three from Africa (Egypt), and one from Europe (Germany). Furthermore, the 42 studies had a larger overall sample size (n ≥ 100) than the remaining eight (n < 100). Among the included articles, 44 studies followed the criteria for sepsis diagnosis were derived from sepsis 1.0 [69], sepsis 2.0 [70], sepsis 3.0 [2], while the remaining six studies did not give precise diagnostic criteria versions. In addition, 37 studies used healthy controls, whereas the remaining 13 studies used the infection controls, such as lung infection, pneumonia, and the systemic inflammatory response syndrome (SIRS). Besides, 40 studies used U6 as a qRT-PCR reference gene, eight literatures used non-U6 (miR-16-5p, SNORD61, cel-miR-39-3p, cel-miR-54), and two studies did not specify which reference gene was used. Kappa test showed the kappa value of agreement during the systematic searches was 0.87. Table 1 shows the essential characteristics of the studies that were included. The Data extracted from the included studies are shown in S2 Table.
Fig 1. Flow diagram of the study selection process.
Table 1. Characteristics of studies in meta-analysis.
| No. | First Author | Year | Country | Population | Type of control | Golden standard | Reference genes of qRT-PCR | Specimen | Case (male/female) | Control(male/female) | Selected miRNAs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Sankar S | 2022 | India | Neonates | HC | N/R | SNORD61 | Plasma | 42(24/18) | 42(23/19) | miR-21, miR-29a |
| 2 | Abdelaleem O | 2022 | Egypt | Neonates | HC | N/R | miR-16-5p | Serum | 90(33/57) | 90(29/61) | miR-34a-5p, miR-199a-3p |
| 3 | Wang H | 2021 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 146(87/59) | 60(37/23) | miR-223-3p |
| 4 | Deng Y | 2021 | China | Children | HC | Sepsis 3.0 | U6 | Plasma | 153(94/59) | 60(38/22) | miR-101-3p, miR-141-3p |
| 5 | Zhang S | 2021 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 90(53/37) | 92(47/45) | miR-940 |
| 6 | Zhang B | 2021 | China | Adults | HC | Sepsis 3.0 | U6 | Serum | 86(38/48) | 85(40/45) | miR-29c-3p |
| 7 | Yao J | 2021 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 151(113/38) | 15(10/5) | miR-3622b-3p, miR-519c-5p |
| 8 | Xu C | 2021 | China | Adults | Infection | Sepsis 3.0 | U6 | Serum | 69(35/34) | 59(30/29) | miR-21, miR-210 |
| 9 | Wang Q | 2021 | China | Adults | HC | Sepsis 3.0 | U6 | Serum | 80(39/41) | 72(28/44) | miR-378a-3p |
| 10 | Wang D | 2021 | China | Neonates | Infection | N/R | U6 | Serum | 72(39/33) | 56(32/24) | miR-1184 |
| 11 | Trung N | 2021 | Vietnam | Adults | HC | Sepsis 3.0 | miR-16-5p | Plasma | 130(114/16) | 82(58/24) | miR-146-3p, miR-147b, miR-155-5p, miR-223-3p |
| 12 | Sun B | 2021 | China | Adults | HC | Sepsis 3.0 | U6 | Serum | 108(55/53) | 101(54/47) | miR-486-5p |
| 13 | Mao Y | 2021 | China | Neonates | HC | N/R | U6 | Serum | 90(43/47) | 88(37/51) | miR-455-5p |
| 14 | Liu J | 2021 | China | Adults | HC | Sepsis 3.0 | U6 | Serum | 102(53/49) | 105(53/52) | miR-381-3p |
| 15 | Lin X | 2021 | China | Neonates | Infection | N/R | U6 | Serum | 98(46/52) | 50(22/28) | miR-141 |
| 16 | Li M | 2021 | China | Neonates | Infection | Sepsis 2.0 | U6 | Serum | 75(41/34) | 84(47/37) | miR-129-5p |
| 17 | Zhao J | 2020 | China | Children | HC | Sepsis 2.0 | U6 | PBMC | 90(55/35) | 60(35/25) | miR-466 |
| 18 | Yang Z | 2020 | China | Adults | HC | Sepsis 2.0 | U6 | Plasma | 120(81/39) | 120(68/52) | miR-103, miR-107 |
| 19 | Wang J | 2020 | China | Adults | HC | Sepsis 3.0 | N/R | Serum | 82(43/39) | 30(N/R) | miR-25 |
| 20 | Li H | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Serum | 30(19/11) | 30(17/13) | miR-150, miR-107 |
| 21 | Zhu X | 2020 | China | Adults | HC | Sepsis 2.0 | U6 | Plasma | 120(84/36) | 120(N/R) | miR-125a, miR-125b |
| 22 | Zhao D | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 150(98/52) | 150(92/58) | miR-125a, miR-125b |
| 23 | Yang Y | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 102(61/41) | 100(N/R) | miR-125a |
| 24 | Xu H | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Serum | 103(55/48) | 98(58/40) | miR-19b-3p |
| 25 | Wang H | 2020 | China | Adults | HC | Sepsis 2.0 | U6 | plasma | 132(86/46) | 131(76/55) | miR-146a |
| 26 | Wang H | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Serum | 98(56/42) | 65(38/27) | miR-451a |
| 27 | Sun B | 2020 | China | Adults | HC | Sepsis 1.0 | U6 | Serum | 110(69/41) | 89(58/31) | miR-328 |
| 28 | Salim R | 2020 | Egypt | Neonates | Infection | Sepsis 2.0 | U6 | Serum | 50(25/25) | 30(14/16) | miR-101, miR-187, miR-21 |
| 29 | Na L | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 219(143/76) | 219(N/R) | miR-21 |
| 30 | Liu W | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 196(130/66) | 196(119/77) | miR-125a |
| 31 | Liu G | 2020 | China | Neonates | Infection | Sepsis 2.0 | U6 | Serum | 102(53/49) | 50(28/22) | miR-181a |
| 32 | Lin R | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 208(137/71) | 210(134/76) | miR-126 |
| 33 | Dou H | 2020 | China | Adults | HC | Sepsis 2.0 | U6 | Serum | 203(117/86) | 100(64/36) | miR-155-5p, miR-143 |
| 34 | Chen W | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 104(62/42) | 100(N/R) | miR-146b |
| 35 | Chen L | 2020 | China | Adults | HC | Sepsis 3.0 | U6 | Plasma | 180(106/74) | 180(117/63) | miR-146a, miR-146b |
| 36 | Li W | 2019 | China | Adults | HC | N/R | U6 | Serum | 83(52/31) | 50(32/18) | miR-21 |
| 37 | Zhang W | 2019 | China | Adults | Infection | Sepsis 3.0 | U6 | Plasma | 44(34/10) | 52(31/21) | miR-7110-5p, miR-223-3p |
| 38 | Karam R | 2019 | Egypt | Children | HC | Sepsis 3.0 | U6 | Serum | 55(35/20) | 60(33/27) | miR-146a |
| 39 | Guo H | 2019 | China | Adults | HC | Sepsis 3.0 | U6 | Serum | 105(59/46) | 100(61/39) | miR-495 |
| 40 | Li J | 2018 | China | Adults | Infection | Sepsis 3.0 | U6 | Serum | 41(19/22) | 20(12/8) | miR-142-3p |
| 41 | Wu X | 2018 | China | Adults | HC | Sepsis 1.0 | U6 | Plasma | 187(125/62) | 186(112/72) | miR-223-3p |
| 42 | Rahmel T | 2018 | Germany | Adults | Infection | Sepsis 3.0 | cel-miR-54 | Serum | 108(64/44) | 20(9/11) | miR-122 |
| 43 | Chen C | 2018 | China | Children | Infection | Sepsis 3.0 | U6 | Serum | 60(36/24) | 25(14/11) | miR-126-3p |
| 44 | Chao L | 2018 | China | Adults | HC | Sepsis 3.0 | N/R | Serum | 105(59/46) | 35(N/R) | miR-155-5p, miR-133a-3p |
| 45 | Liu Z | 2017 | China | Adults | HC | Sepsis 3.0 | miR-16-5p | plasma | 103(47/56) | 30(N/R) | miR-122a, miR-146a, miR-155a |
| 46 | Lin H | 2017 | China | Adults | HC | Sepsis 3.0 | cel-miR-39-3p | plasma | 82(65/17) | 22(14/18) | miR-15b, miR-210, miR-486 |
| 47 | Han Y | 2016 | China | Adults | Infection | Sepsis 1.0 | U6 | Serum | 103(71/32) | 95(65/30) | miR-143 |
| 48 | Yao L | 2015 | China | Adults | Infection | Sepsis 2.0 | miR-16-5p | Serum | 70(36/34) | 30(12/18) | miR-25 |
| 49 | Wang X | 2015 | China | Neonates | Infection | Sepsis 2.0 | U6 | Serum | 46(27/19) | 41(25/16) | miR-15a, miR-15b, miR-16-5p, miR-223-3p |
| 50 | Deng J | 2013 | China | Adults | HC | Sepsis 2.0 | 5S rRNA | Serum | 52(42/10) | 23(N/R) | miR-122 |
HC = healthy control, miR = mircoRNA, PBMC = peripheral blood mononuclear cell, N/R = not report.
Study quality
The quality of the included studies was assessed using the QUADAS-2, and the results showed that the risk of bias for index test was high (S1 Fig).
Diagnostic accuracy of miRNAs
Total mixed miRNAs
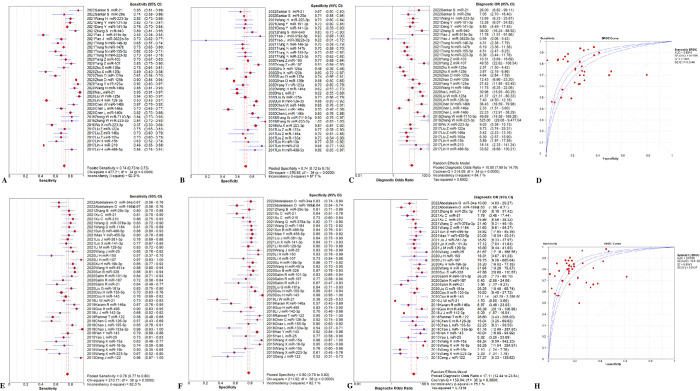
The overall diagnostic assessment of total mixed miRNAs (TmiRs) in identifying sepsis has been summarized in Fig 2. The summary sensitivity was 0.76 (95%CI, 0.75 to 0.77), and the pooled estimation revealed significant heterogeneity (P < 0.0001, x2 = 711.78, I2 = 89.6%, Fig 2A). Meanwhile, the summary specificity was 0.77 (95%CI, 0.75 to 0.78), and the pooled estimation also showed noticeable heterogeneity (P < 0.0001, x2 = 529.08, I2 = 86.0%, Fig 2B). In addition, the pooled DOR was 13.89 (95% CI, 11.05 to 17.47) with significant heterogeneity (P < 0.0001, Cochran-Q = 410.20, I2 = 82.0%, Fig 2C). The calculated AUC value was 0.86 ± 0.01 (Fig 2D). Besides, the result of threshold effect analysis suggested that the Spearman correlation coefficient was 0.155 and P = 0.185, which was indicating no threshold effect related to heterogeneity existed. Next, we ran a meta-regression analysis to see what factors might be causing the heterogeneities. Type of samples, region, total sample size, sepsis diagnostic criteria, qRT-PCR reference genes, population, miRNA expression level, and controls composition were all taken into account in the meta-regression to detect the origins of heterogeneities after reviewing the baseline data and the original data producing procedure. According to the findings (Fig 3), the type of samples may be a source of heterogeneity (P = 0.046, t = 2.03, 95%CI (1.01, 2.43), Fig 3H), While the remaining seven factors are not (P > 0.05, Fig 3A–3G).
Fig 2. Performance of total miRNAs detection for sepsis diagnosis.
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets. CI = confidence interval, DOR = diagnostic odds ratio, miR = mircoRNA, OR = odds ratio, SROC = summary receiver operating characteristic curves value.
Fig 3. The meta-regression of the enrolled studies.
(A) for the region, the meta-regression did not find it was a dramatic impact on the homogeneity of the enrolled studies, P = 0.296, t = -1.05, 95%CI (0.32, 1.42). (B) for the population, the meta-regression found no significant impact on the homogeneity of the enrolled studies, P = 0.519, t = 0.65, 95%CI (0.82, 1.47). (C) for the sepsis diagnostic criteria, the meta-regression did not detect it was a dramatic impact on the homogeneity of the enrolled studies, P = 0.331, t = -0.98, 95%CI (0.60, 1.19). (D) for the controls composition, the meta-regression did not detect it was a dramatic impact on the homogeneity of the enrolled studies, P = 0.76, t = 0.31, 95%CI (0.62, 1.91). (E) for the qRT-PCR reference genes, the meta-regression did not find it was a dramatic impact on the homogeneity of the enrolled studies, P = 0.553, t = -0.60, 95%CI (0.55, 1.38). (F) for the miRNA expression level, the meta-regression did not detect it was a dramatic impact on the homogeneity of the enrolled studies, P = 0.053, t = 1.97, 95%CI (0.99, 2.58). (G) for the total sample size, the meta-regression did not discover a significant impact on the homogeneity of the enrolled studies, P = 0.913, t = -0.11, 95%CI (0.51, 1.82). (H) for the type of samples, the meta-regression detected it was a dramatic impact on the homogeneity of the enrolled studies, P = 0.046, t = 2.03, 95%CI (1.01, 2.43).
Subgroup analysis
The plasma and serum groups were subdivided to see if they were the source of heterogeneity. In addition, the analysis was carried out for five miRNAs (miR-155-5p, miR-21, miR-223-3p, miR-146a, and miR-125a), which were studied in more than two separate investigations. Prior to subgroup analysis, Meta-Disc 1.4 was used to do threshold analysis on each group’s data, and the results indicated that there was no potential for a threshold effect. Then, we analyzed each subgroup. The results were shown in Table 2, Figs 4, 5 and S2–S5 Figs.
Table 2. Subgroup analysis results of included studies.
| Sensitivity (95% CI) | Specificity (95% CI) | DOR (95% CI) | SROC(AUC±SE) | |
|---|---|---|---|---|
| Type of samples | ||||
| Serum | 0.78(0.77–0.80) | 0.80(0.79–0.82) | 17.11(12.44–23.54) | 0.87±0.01 |
| P/I2 | < 0.0001/82% | < 0.0001/82.1% | < 0.0001/82% | - |
| Plasma | 0.74(0.73–0.75) | 0.74(0.72–0.75) | 10.80(7.89–14.79) | 0.83±0.02 |
| P/I2 | < 0.0001/92.9% | < 0.0001/87.7% | < 0.0001/84.1% | - |
| Type of miRNAs | ||||
| miR-155-5p | 0.71(0.67–0.75) | 0.82(0.76–0.86) | 11.90(4.07–34.76) | 0.85±0.08 |
| P/I2 | 0.0004/87.2% | 0.0213/74.0% | 0.0020/83.9% | - |
| miR-21 | 0.86(0.82–0.89) | 0.60(0.55–0.65) | 8.05(2.45–26.40) | 0.67±0.14 |
| P/I2 | < 0.0001/91.3% | 0.0169/66.8% | < 0.0001/89.6% | - |
| miR-223-3p | 0.70(0.66–0.74) | 0.81(0.77–0.85) | 8.82(4.41–17.66) | 0.78±0.06 |
| P/I2 | < 0.0001/86.6% | < 0.0001/90.2% | 0.0048/73.3% | - |
| miR-146a | 0.77(0.73–0.81) | 0.56(0.51–0.61) | 5.86(2.42–14.19) | 0.69±0.11 |
| P/I2 | 0.0001/85.9% | 0.0749/56.6% | 0.0002/85.0% | - |
| miR-125a | 0.81(0.77–0.84) | 0.67(0.63–0.71) | 10.54(2.88–38.56) | 0.74±0.12 |
| P/I2 | < 0.0001/95.8% | < 0.0001/87.9% | < 0.0001/94.4% | - |
CI = confidence interval, DOR = diagnostic odds ratio, SROC = summary receiver operating characteristic curves value, AUC = area under the curve, SE = standard error.
Fig 4. Performance of type of samples detection for sepsis diagnosis.
(A) Pooled sensitivity of plasma. (B) Pooled specificity of plasma. (C) Overall DOR of plasma. (D) The SROCs of plasma. (E) Pooled sensitivity of serum. (F) Pooled specificity of serum. (G) Overall DOR of serum. (H) The SROCs of serum. CI = confidence interval, DOR = diagnostic odds ratio, miR = mircoRNA, OR = odds ratio, SROC = summary receiver operating characteristic curves value.
Fig 5. Performance of miR-155-5p for sepsis diagnosis.
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets. CI = confidence interval, DOR = diagnostic odds ratio, miR = mircoRNA, OR = odds ratio, SROC = summary receiver operating characteristic curves value.
Sample type
The total diagnostic performance of the plasma group in detecting sepsis is shown in Fig 4A–4D. The pooled sensitivity was 0.74 (95%CI, 0.73 to 0.75), and the estimation showed significant heterogeneity (P < 0.0001, x2 = 477.71, I2 = 92.9%, Fig 4A). The summary specificity was 0.74 (95%CI, 0.72 to 0.75), and the pooled estimation showed high heterogeneity (P < 0.0001, x2 = 276.92, I2 = 87.7%, Fig 4B). The pooled DOR was 10.80 (95%CI, 7.89 to 14.79) with a noticeable heterogeneity (P < 0.0001, Cochran-Q = 214.09, I2 = 84.1%, Fig 4C). It revealed that the AUC value was 0.83 ± 0.02 (Fig 4D). Additionally, for the serum group, the overall sensitivity was 0.78 (95%CI, 0.77 to 0.80), and the pooled estimation showed significant heterogeneity (P < 0.0001, x2 = 210.71, I2 = 82.0%, Fig 4E). The summary specificity was 0.80 (95%CI, 0.79 to 0.82), and the pooled estimation showed high heterogeneity (P < 0.0001, x2 = 211.92, I2 = 82.1%, Fig 4F). Meanwhile, the pooled DOR was 17.11 (95%CI, 12.44 to 23.54) with a noticeable heterogeneity (P < 0.0001, Cochran-Q = 159.04, I2 = 76.1%, Fig 4G), and the AUC value was 0.87 ± 0.01 (Fig 4H).
miR-155-5p
Three studies [14–16] were included in examination to the overall diagnostic performance of miR-155-5p (Fig 5). The summary sensitivity was 0.71 (95%CI, 0.67 to 0.75), and the pooled estimation showed significant heterogeneity (P = 0.0004, x2 = 15.58, I2 = 87.2%, Fig 5A). Additionally, the summary specificity was 0.82 (95%CI, 0.76 to 0.86), and the pooled estimation showed moderate heterogeneity (P = 0.0213, x2 = 7.70, I2 = 74.0%, Fig 5B). The pooled DOR was 11.90 (95%CI, 4.07 to 34.76) with a noticeable heterogeneity (P = 0.0020, Cochran-Q = 12.43, I2 = 83.9%, Fig 5C). It revealed that the AUC was 0.85 ± 0.08 (Fig 5D).
miR-21
Five studies [9, 10, 17, 53, 59] were included to evaluate the overall diagnostic performance of miR-21 (S2 Fig). The summary sensitivity was 0.86 (95%CI, 0.82 to 0.89), and the specificity was 0.60 (95%CI, 0.55 to 0.65). Both pooled estimations showed significant heterogeneity (sensitivity: P < 0.0001, x2 = 45.85, I2 = 91.3%; specificity: P = 0.0169, x2 = 12.06, I2 = 66.8%, S2A and S2B Fig). The pooled DOR was 8.05 (95%CI, 2.45 to 26.40), with a noticeable heterogeneity (P < 0.0001, Cochran-Q = 38.52, I2 = 89.6%, S2C Fig). Besides, the AUC value was 0.67 ± 0.14 (S2D Fig).
miR-223-3p
Five studies [12, 14, 40, 58, 61] were included to analysis the overall diagnostic performance of miR-223-3p (S3 Fig). The summary sensitivity for the diagnostic performance of miR-223-3p was 0.70 (95%CI, 0.66 to 0.74), and the specificity was 0.81 (95%CI, 0.77 to 0.85). High heterogeneity showed in the results (sensitivity: P < 0.0001, x2 = 29.88, I2 = 86.6%; specificity: P < 0.0001, x2 = 40.71, I2 = 90.2%, S3A,and S3B Fig). The pooled DOR was 8.82 (95%CI, 4.41 to 17.66), with moderate heterogeneity (P = 0.0048, Cochran-Q = 14.97, I2 = 73.3%, S3C Fig), and the AUC value was 0.78 ± 0.06 (S3D Fig).
miR-146a
Four studies [18–20, 64] were included to assess the overall diagnostic performance of miR-146a (S4 Fig). The summary sensitivity was 0.77 (95%CI, 0.73 to 0.81), and the specificity was 0.56 (95%CI, 0.51 to 0.61). Both pooled estimations showed significant heterogeneity (sensitivity: P = 0.0001, x2 = 21.33, I2 = 85.9%; specificity: P = 0.0749, x2 = 6.91, I2 = 56.6%, S4A and S4B Fig). The pooled DOR was 5.86 (95%CI, 2.42 to 14.19), with high heterogeneity (P = 0.0002, Cochran-Q = 20.01, I2 = 85.0%, S4C Fig). Besides, the AUC value was 0.69 ± 0.11 (S4D Fig).
miR-125a
Four studies [21–23, 54] were included to analysis the overall diagnostic performance of miR-125a (S5 Fig). The summary sensitivity was 0.81 (95%CI, 0.77 to 0.84), and the specificity was 0.67 (95%CI, 0.63 to 0.71). High heterogeneity showed in the results (sensitivity: P < 0.0001, x2 = 70.70, I2 = 95.8%; specificity: P < 0.0001, x2 = 24.78, I2 = 87.9%, S5A and S5B Fig). The pooled DOR was 10.54 (95%CI, 2.88 to 38.56), with a noticeable heterogeneity (P < 0.0001, Cochran-Q = 53.83, I2 = 94.4%, S5C Fig), and the AUC value was 0.74 ± 0.12 (S5D Fig).
Sensitivity analysis and publication bias
We found no significant influence from any of the studies, and STATA 15.1 corroborated the TmiRs results (Fig 6A). Furthermore, funnel plots were utilized to assess publication bias in the included papers, and no significant publication biases were found (P = 0.859, 95% CI, -10.58 to 12.66) (Fig 6B).
Fig 6. Sensitivity analysis and publication bias of the individual trials on the results TmiRs.
(A) Sensitivity analysis for the result of TmiRs, (B) Funnel plot for the assessment of potential publication bias. DOR = diagnostic odds ratio, ESS = effective sample size, miR = mircoRNA, TmiRs = total mixed miRNAs.
Trial sequential analysis
TSA results demonstrated that the cumulative Z-score of TmiR, miR-155-5p, miR-223-3p, and miR-125a crossed its monitoring boundaries and reliable conclusions had been drawn. But the miR-21 and miR-146a did not reach the required sample size (S6 Fig).
Discussion
Many studies have been conducted to see if miRNAs may be utilized as biomarkers for sepsis since their emergence about a decade ago. In this meta-analysis, we enrolled 50 studies totaling 5225 sepsis patients and 4008 controls, involving 48 miRNAs. Finally, we discovered that TmiRs had a combined AUC of 0.86, with 0.76 pooled sensitivity and 0.77 specificity, indicating that miRNAs had a moderate diagnostic accuracy as a diagnostic biomarker in discriminating sepsis. In addition, we examined individual miRNAs in the overall miRNA library and discovered that miR-155-5p, miR-21, miR-223-3p, miR-146a, and miR-125a were the most often used in recent studies. We discovered that among all miRNAs, miR-155-5p had the highest AUC of SROC: pooled sensitivity, 0.71 (95%CI, 0.67 to 0.75); pooled specificity, 0.82 (95%CI, 0.76 to 0.86); and SROC, 0.85; indicating that miR-155-5p had reasonable diagnostic accuracy in identifying sepsis. To our knowledge, this is the first meta-analysis that focused on the accuracy of miR-155-5p, miR-21, miR-146a, and miR-125a in detecting sepsis. This meta-analysis was crucial in determining the potential for miRNAs to be utilized to diagnose sepsis.
It is worth noting that not every study provides cut-off value of miRNA. In addition, possibly due to factors such as qRT-PCR reference genes and the source of samples, the same miRNA was used in different papers with different cut-off values. Moreover, selecting the test cut-off to optimize sensitivity and/or specificity may lead to overestimation of test performance. These reasons may have contributed to the high risk of index test by QUADAS-2. Therefore, we did threshold analysis before each analysis and found no threshold effect, indicating that there was no threshold effect related to heterogeneity. Besides, as some pooled results showed large heterogeneity, type of samples, region, total sample size, sepsis diagnostic criteria, qRT-PCR reference genes, population, miRNA expression level, and controls composition were all analysis in the meta-regression to detect the origins of heterogeneity. Finally, the specimen type was discovered to be a source of heterogeneity, and the results of the following subgroup analysis revealed that serum miRNAs might be employed as sepsis diagnostic biomarkers compared to plasma (SROC: 0.87, 0.83, respectively).
Some studies [40, 44, 46] in our included literature focused on the ability of miRNA, PCT, CRP, and other markers to jointly diagnose sepsis. It showed that the diagnostic accuracy of combined markers was higher than that of single marker. Even so, we did not undertake a combined analysis of miRNAs and other indicators due to a lack of research and the various types of miRNA combinations discovered. Nonetheless, this approach to integrated diagnosis has a lot of promise. Previous studies [13, 71] have also indicated the importance of combined diagnosis in sepsis, however additional clinical investigations are needed in the future to clarify the signs of combined diagnosis.
In 2020, Shen et al. [13] conducted a meta-analysis in 2020 with a total of 22 studies, comprising 2210 sepsis and 1502 controls, to validate the diagnostic accuracy of miRNAs for sepsis, and discovered that miR-223-3p may be employed as a sepsis indicator. However, we discovered that miR-223-3p had worse diagnostic accuracy than miR-155-5p in our research with a wider literature and patient group (SROC: 0.78, 0.85, respectively). Nonetheless, the diagnostic accuracy of miR-223-3p was higher than that of miR-21, miR-146a, and miR-125a (SROC: 0.67, 0.69, and 0.74, respectively), which have received more attention in recent years. Therefore, the importance of miR-223-5p in sepsis detection cannot be overlooked. More crucially, the following are the benefits of our meta-analysis: First, this meta-analysis included more than twice as many studies as the previous one. Second, we tested the diagnostic accuracy of five miRNAs that may be used as sepsis biomarkers, and discovered that miR-155-5p was a crucial miRNA among the total miRNAs. Third, the subgroup analyses discovered that serum miRNAs had the best diagnostic accuracy. Moreover, we performed the TSA to evaluate the stability of results and the RIS, which showed the sample size included in the meta-analysis of TmiR, miR-155-5p, miR-223-3p, and miR-125a was sufficient and significant associations were observed.
There are also several limitations of this meta-analysis that must be addressed. First, our meta-analysis included 48 miRNA markers, with only 5 miRNA markers appearing in three to five of the publications included. Among them, the miR-155-5p showed the highest diagnostic ability. However, only three studies included to evaluate the diagnostic accuracy of miR-155-5p, which might lead to bias in meta-analysis results. Second, cross-comparisons between studies conducted by different laboratories are limited due to the lack of traditional methods for accurate and absolute quantification of miRNAs, as well as the inconsistent reference genes of qRT-PCR and sample types used by various laboratories, resulting in unconvincing results for the included studies.
Conclusions
In conclusion, our meta-analysis demonstrated that miRNAs, specifically miR-155-5p, could be valuable biomarkers for detecting sepsis. For diagnostic purposes, a clinical serum specimen is also required. To assess the usefulness of miRNA in the detection of sepsis in the future, more well-designed and harmonized clinical trials will be required.
Supporting information
(DOC)
(DOCX)
(DOCX)
(A) Methodological quality graph; (B) Methodological quality summary.
(PDF)
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets.
(PDF)
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets.
(PDF)
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets.
(PDF)
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets.
(PDF)
(A) TSA plot of TmiR for sepsis diagnosis. (B) TSA plot of miR-155-5p for sepsis diagnosis. (C) TSA plot of miR-21 for sepsis diagnosis. (D) TSA plot of miR-223-3p for sepsis diagnosis. (E) TSA plot of miR-146a for sepsis diagnosis. (F) TSA plot of miR-125a for sepsis diagnosis.
(PDF)
Abbreviations
- CRP
C-reactive protein
- PCT
procalcitonin
- miRNAs
microRNAs
- CI
confidence interval
- SROC
summary receiver operating characteristic curves value
- AUC
area under the curve
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- CNKI
China National Knowledge Infrastructure
- CENTRAL
Cochrane Central Register of Controlled Trials
- ROC
receiver operating characteristic
- TP
true positive false positive
- FP
false positive
- FN
false negative
- TN
true negative
- QUADAS
Quality Assessment of Diagnostic Accuracy Studies
- DOR
diagnostic odds ratio
- I2
inconsistency index
- TSA
trial sequential analysis
- RIS
required information size
- PBMC
peripheral blood mononuclear cells
- SIRS
systemic inflammatory response syndrome
- TmiRs
total mixed miRNAs
- OR
odds ratio
- ESS
effective sample size
- HC
healthy control
- N/R
not report
- SE
standard error
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet (London, England). 2020;395(10219):200–11. Epub 2020/01/20. doi: 10.1016/S0140-6736(19)32989-7 ; PubMed Central PMCID: PMC6970225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). Jama. 2016;315(8):801–10. Epub 2016/02/24. doi: 10.1001/jama.2016.0287 ; PubMed Central PMCID: PMC4968574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seymour CW, Gesten F, Prescott HC, Friedrich ME, Iwashyna TJ, Phillips GS, et al. Time to Treatment and Mortality during Mandated Emergency Care for Sepsis. The New England journal of medicine. 2017;376(23):2235–44. Epub 2017/05/23. doi: 10.1056/NEJMoa1703058 ; PubMed Central PMCID: PMC5538258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin JC, Spinella PC, Fitzgerald JC, Tucci M, Bush JL, Nadkarni VM, et al. New or Progressive Multiple Organ Dysfunction Syndrome in Pediatric Severe Sepsis: A Sepsis Phenotype With Higher Morbidity and Mortality. Pediatric critical care medicine: a journal of the Society of Critical Care Medicine and the World Federation of Pediatric Intensive and Critical Care Societies. 2017;18(1):8–16. Epub 2017/01/07. doi: 10.1097/PCC.0000000000000978 ; PubMed Central PMCID: PMC7261134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. International journal of genomics. 2014;2014:970607. Epub 2014/09/03. doi: 10.1155/2014/970607 ; PubMed Central PMCID: PMC4142390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng X, Zhang Y, Yue P, Liu L, Wang C, Zhou K, et al. Diagnostic significance of circulating miRNAs in systemic lupus erythematosus. PloS one. 2019;14(6):e0217523. Epub 2019/06/05. doi: 10.1371/journal.pone.0217523 ; PubMed Central PMCID: PMC6548426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng X, Li Y, Yue P, Ma F, Zhang Y, Wu G. Diagnostic significance of circulating miRNAs in Kawasaki disease in China: Current evidence based on a meta-analysis. Medicine. 2021;100(6):e24174. Epub 2021/02/14. doi: 10.1097/MD.0000000000024174 ; PubMed Central PMCID: PMC7886432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One. 2009;4(10):e7405. Epub 2009/10/14. doi: 10.1371/journal.pone.0007405 ; PubMed Central PMCID: PMC2756627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salim RF, Sobeih AA, Abd El Kareem HM. Evaluation of the clinical value of circulating miR-101, miR-187 and miR-21 in neonatal sepsis diagnosis and prognosis. Egyptian Journal of Medical Human Genetics. 2020;21(1). doi: 10.1186/s43042-020-00052-w [DOI] [Google Scholar]
- 10.Sankar S, Maruthai K, Bobby Z, Adhisivam B. MicroRNA Expression in Neonates with Late-onset Sepsis–A Cross-sectional Comparative Study. Immunological investigations. 2022. doi: 10.1080/08820139.2021.2020282 [DOI] [PubMed] [Google Scholar]
- 11.Wang D, Han L. Downregulation of miR-1184 serves as a diagnostic biomarker in neonatal sepsis and regulates LPS-induced inflammatory response by inhibiting IL-16 in monocytes. Experimental and therapeutic medicine. 2021;21(4):350. Epub 2021/03/19. doi: 10.3892/etm.2021.9781 ; PubMed Central PMCID: PMC7903473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Wang X, Liu X, Wang X, Xu J, Hou S, et al. miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. International journal of clinical and experimental medicine. 2015;8(4):5683–90. Epub 2015/07/02. ; PubMed Central PMCID: PMC4483976. [PMC free article] [PubMed] [Google Scholar]
- 13.Shen X, Zhang J, Huang Y, Tong J, Zhang L, Zhang Z, et al. Accuracy of circulating microRNAs in diagnosis of sepsis: a systematic review and meta-analysis. Journal of intensive care. 2020;8(1):84. Epub 2020/12/10. doi: 10.1186/s40560-020-00497-6 ; PubMed Central PMCID: PMC7607638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trung NT, Lien TT, Sang VV, Hoan NX, Manh ND, Thau NS, et al. Circulating miR-147b as a diagnostic marker for patients with bacterial sepsis and septic shock. PloS one. 2021;16(12):e0261228. Epub 2021/12/17. doi: 10.1371/journal.pone.0261228 ; PubMed Central PMCID: PMC8675720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dou H, Hu F, Wang W, Ling L, Wang D, Liu F. Serum MiR-155 and MiR-143 can be used as prognostic markers for severe sepsis/septic shock in the elderly. International journal of clinical and experimental medicine. 2020;13(6):3771–80. Epub 2020/06/30. [Google Scholar]
- 16.Chao L, Xiaopeng S, Nannan G, Hui P, Huali Z. Value of serum miR-155-5p and miR-133a-3p expression for the diagnosis and prognosis evaluation of sepsis. Zhonghua wei zhong bing ji jiu yi xue. 2018;28(8):694–8. doi: 10.3760/cma.j.issn.2095-4352.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 17.Xu C, Zhou G, Wang X, Zhang B, Zhao T, Wu L. Correlation analysis of serum miR-21 and miR-210 with hs-CRP, TNF-alpha, IL-6, and ICAM-1 in patients with sepsis after burns. Burns. 2021. Epub 2021/07/18. doi: 10.1016/j.burns.2021.05.026 . [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Liu L, Yang J, Wang Y. MicroRNA-146b correlates with decreased acute respiratory distress syndrome risk, reduced disease severity, and lower 28-day mortality in sepsis patients. Journal of clinical laboratory analysis. 2020;34(12):e23510. Epub 2020/08/28. doi: 10.1002/jcla.23510 ; PubMed Central PMCID: PMC7755760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Yu L, Zhang R, Zhu L, Shen W. Correlation of microRNA-146a/b with disease risk, biochemical indices, inflammatory cytokines, overall disease severity, and prognosis of sepsis. Medicine. 2020;99(22):e19754. Epub 2020/06/03. doi: 10.1097/MD.0000000000019754 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karam RA, Zidan HE, Karam NA, Abdel Rahman DM, El-Seifi OS. Diagnostic and prognostic significance of serum miRNA-146-a expression in Egyptian children with sepsis in a pediatric intensive care unit. The journal of gene medicine. 2019;21(11):e3128. Epub 2019/11/07. doi: 10.1002/jgm.3128 . [DOI] [PubMed] [Google Scholar]
- 21.Zhu X. MiR-125b but not miR-125a is upregulated and exhibits a trend to correlate with enhanced disease severity, inflammation, and increased mortality in sepsis patients. Journal of clinical laboratory analysis. 2020;34(3):e23094. Epub 2019/11/07. doi: 10.1002/jcla.23094 ; PubMed Central PMCID: PMC7083454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao D, Li S, Cui J, Wang L, Ma X, Li Y. Plasma miR-125a and miR-125b in sepsis: Correlation with disease risk, inflammation, severity, and prognosis. Journal of clinical laboratory analysis. 2020;34(2):e23036. Epub 2020/02/23. doi: 10.1002/jcla.23036 ; PubMed Central PMCID: PMC7031612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Yang L, Liu Z, Wang Y, Yang J. Long noncoding RNA NEAT 1 and its target microRNA-125a in sepsis: Correlation with acute respiratory distress syndrome risk, biochemical indexes, disease severity, and 28-day mortality. Journal of clinical laboratory analysis. 2020;34(12):e23509. Epub 2020/08/14. doi: 10.1002/jcla.23509 ; PubMed Central PMCID: PMC7755762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deeks JJ. Systematic reviews in health care: Systematic reviews of evaluations of diagnostic and screening tests. BMJ (Clinical research ed). 2001;323(7305):157–62. Epub 2001/07/21. doi: 10.1136/bmj.323.7305.157 ; PubMed Central PMCID: PMC1120791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS medicine. 2009;6(7):e1000097. Epub 2009/07/22. doi: 10.1371/journal.pmed.1000097 ; PubMed Central PMCID: PMC2707599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of internal medicine. 2011;155(8):529–36. Epub 2011/10/19. doi: 10.7326/0003-4819-155-8-201110180-00009 . [DOI] [PubMed] [Google Scholar]
- 27.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. Journal of clinical epidemiology. 2003;56(11):1129–35. Epub 2003/11/15. doi: 10.1016/s0895-4356(03)00177-x . [DOI] [PubMed] [Google Scholar]
- 28.Moses LE, Shapiro D, Littenberg B. Combining independent studies of a diagnostic test into a summary ROC curve: data-analytic approaches and some additional considerations. Statistics in medicine. 1993;12(14):1293–316. Epub 1993/07/30. doi: 10.1002/sim.4780121403 . [DOI] [PubMed] [Google Scholar]
- 29.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. Journal of clinical epidemiology. 2005;58(9):882–93. Epub 2005/08/09. doi: 10.1016/j.jclinepi.2005.01.016 . [DOI] [PubMed] [Google Scholar]
- 30.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in medicine. 2002;21(11):1539–58. Epub 2002/07/12. doi: 10.1002/sim.1186 . [DOI] [PubMed] [Google Scholar]
- 31.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC medical research methodology. 2006;6:31. Epub 2006/07/14. doi: 10.1186/1471-2288-6-31 ; PubMed Central PMCID: PMC1552081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brok J, Thorlund K, Gluud C, Wetterslev J. Trial sequential analysis reveals insufficient information size and potentially false positive results in many meta-analyses. Journal of clinical epidemiology. 2008;61(8):763–9. Epub 2008/04/16. doi: 10.1016/j.jclinepi.2007.10.007 . [DOI] [PubMed] [Google Scholar]
- 33.Huang J, Gao H, Tan HZ. SOX1 Promoter Hypermethylation as a Potential Biomarker for High-Grade Squamous Intraepithelial Neoplasia Lesion and Cervical Carcinoma: A Meta-Analysis With Trial Sequential Analysis. Frontiers in genetics. 2020;11:633. Epub 2020/08/28. doi: 10.3389/fgene.2020.00633 ; PubMed Central PMCID: PMC7411256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang KL, Wang SY, Lu WC, Chang YH, Su J, Lu YT. Effects of low-dose computed tomography on lung cancer screening: a systematic review, meta-analysis, and trial sequential analysis. BMC pulmonary medicine. 2019;19(1):126. Epub 2019/07/13. doi: 10.1186/s12890-019-0883-x ; PubMed Central PMCID: PMC6625016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdelaleem OO, Mohammed SR, El Sayed HS, Hussein SK, Ali DY, Abdelwahed MY, et al. Serum miR-34a-5p and miR-199a-3p as new biomarkers of neonatal sepsis. PloS one. 2022;17(1):e0262339. Epub 2022/01/07. doi: 10.1371/journal.pone.0262339 ; PubMed Central PMCID: PMC8735601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Wei Y, Liu J, Zhuang Y. Mir-940 serves as a diagnostic biomarker in patients with sepsis and regulates sepsis-induced inflammation and myocardial dysfunction. Journal of inflammation research. 2021;14:4567–74. doi: 10.2147/JIR.S316169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang B, Yu L, Sheng Y. Clinical value and role of microRNA-29c-3p in sepsis-induced inflammation and cardiac dysfunction. European journal of medical research. 2021;26(1):90. Epub 2021/08/12. doi: 10.1186/s40001-021-00566-y ; PubMed Central PMCID: PMC8353850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao J, Lui KY, Hu X, Liu E, Zhang T, Tong L, et al. Circulating microRNAs as novel diagnostic biomarkers and prognostic predictors for septic patients. Infection, Genetics and Evolution. 2021;95. doi: 10.1016/j.meegid.2021.105082 [DOI] [PubMed] [Google Scholar]
- 39.Wang Q, Liu K, Jin C. Clinical value of microRNA-378a-3p in sepsis and its role in sepsis-induced inflammation and cardiac dysfunction. Bioengineered. 2021;12(1):8496–504. Epub 2021/09/28. doi: 10.1080/21655979.2021.1985339 ; PubMed Central PMCID: PMC8806767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Xiao H, Chen X. Value of Combined Detection of Plasma miR-223-3p, PCT, IL-6 and CRP Levels in Experimental Diagnosis and Prognosis of Sepsis. Journal of Modern Laboratory Medicine. 2021;36(05):51–4+61. doi: 10.3969/j.issn.1671-7414.2021.05.011 [DOI] [Google Scholar]
- 41.Sun B, Guo S. miR-486-5p Serves as a Diagnostic Biomarker for Sepsis and Its Predictive Value for Clinical Outcomes. Journal of inflammation research. 2021;14:3687–95. Epub 2021/08/07. doi: 10.2147/JIR.S323433 ; PubMed Central PMCID: PMC8331108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao YY, Su C, Fang CC, Fan XP, Wang LP, Zhu SS, et al. Clinical significance of the serum miR-455-5p expression in patients with neonatal sepsis. Bioengineered. 2021;12(1):4174–82. Epub 2021/07/22. doi: 10.1080/21655979.2021.1955580 ; PubMed Central PMCID: PMC8806658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Yang Y, Lu R, Liu Q, Hong S, Zhang Z, et al. MicroRNA-381-3p signatures as a diagnostic marker in patients with sepsis and modulates sepsis-steered cardiac damage and inflammation by binding HMGB1. Bioengineered. 2021;12(2):11936–46. Epub 2021/11/18. doi: 10.1080/21655979.2021.2006967 ; PubMed Central PMCID: PMC8810158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin X, Wang Y. Mir-141 is negatively correlated with tlr4 in neonatal sepsis and regulates lps-induced inflammatory responses in monocytes. Brazilian Journal of Medical and Biological Research. 2021;54(7). doi: 10.1590/1414-431X2020e10603 PubMed Central PMCID: PMCThermo. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Li M, Huang X, Zhuo Q, Zhang J, Ju X. Clinical significance of miR-129-5p in patients with neonatal sepsis and its regulatory role in the LPS-induced inflammatory response. Bosnian journal of basic medical sciences. 2021;22(2):185–90. doi: 10.17305/bjbms.2020.5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng Y, Qiu C, Wu R, Kuang X, Lu L. Significances of the plasma expression of microRNA-101-3p and microRNA-141-3p in children with sepsis. Chinese Journal of Applied Clinical Pediatrics. 2021;36(18):1383–8. doi: 10.3760/cma.j.cn101070-20200429-00753 [DOI] [Google Scholar]
- 47.Zhao J, Dong G. Expression and diagnostic vaule of miRNA-466 and GPR 18 mRNA in peripheral blood mononuclear cells in children with sepsis. Int J Lab Med. 2020;41(20):2506–9. doi: 10.3969/j.issn.1673-4130.2020.20.018 [DOI] [Google Scholar]
- 48.Yang J, Zhang Y, Wang Q, Chen H, Feng Q, Zhou S. Correlations of microRNA-103 and microRNA-107 expressions with the clinical characteristics and prognosis of 120 cases of sepsis. Journal of Shandong University (Health Sciences). 2020;58(12):77–85+91. doi: 10.6040/j.issn.1671-7554.0.2020.0110 [DOI] [Google Scholar]
- 49.Xu H, Liu X, Ni H. Clinical significance of miR-19b-3p in patients with sepsis and its regulatory role in the LPS-induced inflammatory response. European journal of medical research. 2020;25(1):9. Epub 2020/03/20. doi: 10.1186/s40001-020-00408-3 ; PubMed Central PMCID: PMC7079357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Yan X, Wang H, Xu T. Evaluation vaule of serum microRNA-25 on severity and prognosis of sepsis patients. Journal of Clinical Emergency (China). 2020;21(06):493–8. doi: 10.13201/j.issn.1009-5918.2020.06.015 [DOI] [Google Scholar]
- 51.Wang H, Cui W, Qiao L, Hu G. Overexpression of miR-451a in sepsis and septic shock patients is involved in the regulation of sepsis-associated cardiac dysfunction and inflammation. Genetics and molecular biology. 2020;43(4):e20200009. Epub 2020/11/20. doi: 10.1590/1678-4685-GMB-2020-0009 ; PubMed Central PMCID: PMC7678258 that could be perceived as prejudicial to the impartiality of the reported research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun B, Luan C, Guo L, Zhang B, Liu Y. Low expression of microRNA-328 can predict sepsis and alleviate sepsis-induced cardiac dysfunction and inflammatory response. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas. 2020;53(8):e9501. Epub 2020/06/25. doi: 10.1590/1414-431X20209501 ; PubMed Central PMCID: PMC7307893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Na L, Ding H, Xing E, Zhang Y, Gao J, Liu B, et al. The predictive value of microRNA-21 for sepsis risk and its correlation with disease severity, systemic inflammation, and 28-day mortality in sepsis patients. Journal of clinical laboratory analysis. 2020;34(3):e23103. Epub 2019/11/30. doi: 10.1002/jcla.23103 ; PubMed Central PMCID: PMC7083453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu W, Geng F, Yu L. Long non-coding RNA MALAT1/microRNA 125a axis presents excellent value in discriminating sepsis patients and exhibits positive association with general disease severity, organ injury, inflammation level, and mortality in sepsis patients. Journal of clinical laboratory analysis. 2020;34(6). doi: 10.1002/jcla.23222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu G, Liu W, Guo J. Clinical significance of miR-181a in patients with neonatal sepsis and its regulatory role in the lipopolysaccharide-induced inflammatory response. Experimental and therapeutic medicine. 2020;19(3):1977–83. Epub 2020/02/28. doi: 10.3892/etm.2020.8408 ; PubMed Central PMCID: PMC7027122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin R, Hu H, Li L, Chen G, Luo L, Rao P. The potential of microRNA-126 in predicting disease risk, mortality of sepsis, and its correlation with inflammation and sepsis severity. Journal of clinical laboratory analysis. 2020;34(9). doi: 10.1002/jcla.23408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H. Preliminary study of miRNA in diagnosis and treatment of sepsis [硕士]: Guangzhou Medical University; 2020. [Google Scholar]
- 58.Zhang W, Jia J, Liu Z, Si D, Ma L, Zhang G. Circulating microRNAs as biomarkers for Sepsis secondary to pneumonia diagnosed via Sepsis 3.0. BMC pulmonary medicine. 2019;19(1):93. Epub 2019/05/16. doi: 10.1186/s12890-019-0836-4 ; PubMed Central PMCID: PMC6518454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, Zhang T, Pan W, Yang J. Clinical significance of serum microRNA-21 level change in sepsis Shandong Medical Journal. 2019;59(09):66–8. doi: 10.3969/j.issn.1002-266X.2019.09.018 [DOI] [Google Scholar]
- 60.Guo H, Tang L, Xu J, Lin C, Ling X, Lu C, et al. MicroRNA-495 serves as a diagnostic biomarker in patients with sepsis and regulates sepsis-induced inflammation and cardiac dysfunction. European journal of medical research. 2019;24(1):37. Epub 2019/11/28. doi: 10.1186/s40001-019-0396-3 ; PubMed Central PMCID: PMC6878688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu X, Yang J, Yu L, Long D. Plasma miRNA-223 correlates with risk, inflammatory markers as well as prognosis in sepsis patients. Medicine. 2018;97(27):e11352. Epub 2018/07/07. doi: 10.1097/MD.0000000000011352 ; PubMed Central PMCID: PMC6076081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahmel T, Schäfer ST, Frey UH, Adamzik M, Peters J. Increased circulating microrna-122 is a biomarker for discrimination and risk stratification in patients defined by sepsis-3 criteria. PloS one. 2018;13(5). doi: 10.1371/journal.pone.0197637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen C, Zhang L, Huang H, Liu S, Liang Y, Xu L, et al. Serum miR-126-3p level is down-regulated in sepsis patients. International journal of clinical and experimental pathology. 2018;11(5):2605–12. Epub 2018/05/01. ; PubMed Central PMCID: PMC6958305. [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Z. Value of plasma miRNA-122a, miRNA-146a and miRNA-155a expression for the diagnostic and prognostic evaluation of sepsis: Hebei University; 2017. [Google Scholar]
- 65.Lin H. Assessment on miR-15b, miR120 and miR-486 as elderly sepsis diagnosis and prognosis biomarkers: Guangzhou University of Chinese Medicine; 2017. [Google Scholar]
- 66.Han Y, Dai QC, Shen HL, Zhang XW. Diagnostic value of elevated serum miRNA-143 levels in sepsis. The Journal of international medical research. 2016;44(4):875–81. Epub 2016/05/27. doi: 10.1177/0300060516645003 ; PubMed Central PMCID: PMC5536632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yao L, Liu Z, Zhu J, Li B, Chai C, Tian Y. Clinical evaluation of circulating microRNA-25 level change in sepsis and its potential relationship with oxidative stress. International journal of clinical and experimental pathology. 2015;8(7):7675–84. Epub 2015/09/05. ; PubMed Central PMCID: PMC4555662. [PMC free article] [PubMed] [Google Scholar]
- 68.Deng J, Wang H, Su L, Zhang X, Xiao K, Jia Y, et al. Serum microRNA-122 as a specific marker of sepsis diagnosis. Med J Chin PAPF. 2013;24(05):383–6. doi: 10.14010/j.cnki.wjyx.2013.05.047 [DOI] [Google Scholar]
- 69.American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Critical care medicine. 1992;20(6):864–74. Epub 1992/06/01. . [PubMed] [Google Scholar]
- 70.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, et al. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Intensive Care Med. 2003;29(4):530–8. Epub 2003/03/29. doi: 10.1007/s00134-003-1662-x . [DOI] [PubMed] [Google Scholar]
- 71.Yang J, Liao Y, Dai Y, Hu L, Cai Y. Prediction of prognosis in sepsis patients by the SOFA score combined with miR-150. Advances in clinical and experimental medicine: official organ Wroclaw Medical University. 2022;31(1):9–15. Epub 2021/11/06. doi: 10.17219/acem/142536 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
(DOCX)
(A) Methodological quality graph; (B) Methodological quality summary.
(PDF)
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets.
(PDF)
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets.
(PDF)
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets.
(PDF)
(A) Pooled sensitivity. (B) Pooled specificity. (C) Overall DOR. (D) The SROCs for all datasets.
(PDF)
(A) TSA plot of TmiR for sepsis diagnosis. (B) TSA plot of miR-155-5p for sepsis diagnosis. (C) TSA plot of miR-21 for sepsis diagnosis. (D) TSA plot of miR-223-3p for sepsis diagnosis. (E) TSA plot of miR-146a for sepsis diagnosis. (F) TSA plot of miR-125a for sepsis diagnosis.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.