Abstract
Xanthoria elegans is a lichen symbiosis, that inhabits extreme environments and can absorb UV-B. We reported the de novo sequencing and assembly of X. elegans genome. The whole genome was approximately 44.63 Mb, with a GC content of 40.69%. Genome assembly generated 207 scaffolds with an N50 length of 563,100 bp, N90 length of 122,672 bp. The genome comprised 9,581 genes, some encoded enzymes involved in the secondary metabolism such as terpene, polyketides. To further understand the UV-B absorbing and adaptability to extreme environments mechanisms of X. elegans, we searched the secondary metabolites genes and gene-cluster from the genome using genome-mining and bioinformatics analysis. The results revealed that 7 NR-PKSs, 12 HR-PKSs and 2 hybrid PKS-PKSs from X. elegans were isolated, they belong to Type I PKS (T1PKS) according to the domain architecture; phylogenetic analysis and BGCs comparison linked the putative products to two NR-PKSs and three HR-PKSs, the putative products of two NR-PKSs were emodin xanthrone (most likely parietin) and mycophelonic acid, the putative products of three HR-PKSs were soppilines, (+)-asperlin and macrolactone brefeldin A, respectively. 5 PKSs from X. elegans build a correlation between the SMs carbon skeleton and PKS genes based on the domain architecture, phylogenetic and BGC comparison. Although the function of 16 PKSs remains unclear, the findings emphasize that the genes from X. elegans represent an unexploited source of novel polyketide and utilization of lichen gene resources.
Keywords: Xanthoria elegans, DNA sequencing, genome-mining, polyketide synthase
1. Introduction
Lichens are stable, self-sustaining and mutualistic complex symbiotic associations between lichen-forming fungi (LFF) and photoautotrophic algae or cyanobacteria partners [1]. Lichens can live all over the world, including in extremely harsh habitats [2]. Lichen symbionts produce more than 1000 known secondary metabolites (SMs), these SMs accumulated in the cortical or medullary layers of lichen thalli, and provide self-protection including anti herbivory, antilarval actions or nematicidal and photoprotection against intense radiation [3]. Many of these SMs are polyketides such as anthraquinones, depsides, depsidones and dibenzofurans (for example usnic acid), and lichen-derived polyketides are produced by LFF [4]. Polyketides are low molecular weight compounds and typically aromatic polyphenols with varying levels of oxidative modifications, they have been shown an impressive range of biological activities such as antibiotics, antifungal, anti-inflammatory, antioxidant, antiviral and pharmaceutical activities [5].
Various polyketides were isolated from lichens, for example, the polyketides of Ramalina terebrata including parietin, usnic acid, and atraric acid were extracted by methanol solvent [6]; the identification of usnic acid biosynthesis in Cladonia uncialis was characterized [7]; the screening capacity of melanin in the upper cortices of Cetraria islandica was isolated [8]. The anthraquinone like parietin, fallacinal, teloschistin, parietinic acid and emodin were isolated and characterized from the upper cortex of genus Xanthoria [9], and usnic acid in X. elegans was determined by HPLC [10]. Parietin is the most common anthraquinone in X. parietina [11] and X. elegans (syn. Rusavskia elegans) [8], and parietin plays a role of photoprotection through absorbing UV-B and blue light [12] and has biological activities such as antibacterial, antifungal and antiproliferative [11]. Polyketides in lichens are mainly synthesized by polyketide synthase (PKSs) via malonyl CoA and acetate CoA pathways and are primarily governed by large multifunctional type I PKSs through serial reactions [13]. PKSs can assemble the carbon backbones of many secondary metabolites, and often cluster with other secondary pathway genes [14]. Iterative type I PKSs (T1PKSs) contain multi-functional domains, including keto synthase domain (KS), acyltransferase (AT), an acyl carrier protein (ACP), ketoreductase (KR), dehydratase (DH), enoyl reductase (ER), thioesterase (TE) domains, C-methyltransferase (MeT) [15]. The domains of KS, AT and ACP in the T1PKSs can make up the minimal configuration PKS, and the other domains are optional [16]. According to the absence or presence of beta-keto processing domains such as ER, KR and DH, iterative type I PKSs can be subdivided into non-reducing PKS (NR-PKS), partially reducing PKS (PR-PKS) and highly reducing PKS (HR-PKS) [17], in addition, there are some polyketide synthase and non-ribosomal peptide synthetase (PKS-NRPS) hybrid enzymes [18].
The polyketides of lichens are difficult to obtain for the limit of slow-growth and difficult cultivation [5], with the development of sequencing technology, specific polyketide biosynthesis has become a possible way to solve this problem [4]. And the isolation and characterization of specific PKS genes or related gene clusters are required by genome-mining from the increasing draft genomes of LFF [7,19]. For example, 32 PKS genes in the genome of C. uncialis [7], 9 polyketide synthase (PKS) genes in the genome of X. parietina [1] were isolated. The grayanic acid biosynthetic gene cluster (BGC) was identified in the genome of C. grayi in combination with phylogenetic analysis, the domain architecture, and the correlation of mRNA levels with metabolites production [14]. The genome of LFF X. elegans was sequenced and analyzed to ascertain the biological features and the traits of lichenization, furthermore, we would identify the orthologous genes of PKSs for biosynthesis genes of the polyketide in the genome of X. elegans.
2. Materials and methods
2.1. Sample collection and lichen-forming fungi isolation
Xanthoria elegans lichen species were collected from the Laojunshan National Park in Lijiang in the northwestern Yunnan Province in China in September 2015. Lichen-forming fungi (LFF) strains of X. elegans were performed by the isolation method of algae and fungi [20], and the isolations were cultured in the MYA media (including maltose extract 20 g/L, yeast powder 2 g/L, agarose 8 g/L) at 28 °C for 2 weeks.
2.2. Genome sequencing, assembly and evaluation
The NGS library was loaded and sequenced on the Illumina HiSeq platform. The Illumina HiSeq paired-end 150 bp libraries were constructed with an insertion size of 400 bp. Raw data are available in GenBank. The raw reads with a high proportion of Ns (ambiguous bases) and low-quality bases were filtered out using SOAPec (v2.0) and obtained clean data [21]. The genome size of X. elegans was estimated using K-mer frequency and calculated using a short insertion sequence before genome assembly, and its formula is G=(N×(L-K + 1)-B)/D (G, genome size; L, the total number of Reads; L, the average length of Reads; K, K-mer; B, low-frequency K-mer; D, the peak of K-mer distribution); according to the ratio of hetero-peak height/homo-peak height could reflect individual heterozygosity [19]. The pair-end Illumima Hiseq library was used to draw 17-mer frequency plots. JELLYFISH (2.0.0) was used to calculate the frequencies. Illumima Hiseq genome sequencing pair-end reads were assembled by A5-MiSeq v20150522 de novo and checked the results using software pilon (v1.18), which adopts the BUSCO (Benchmarking Universal Single-Copy Orthologs, http://busco.ezlab.org, v3.0.2) program to estimate the integrity and continuance of the genome and construct contigs. Then, we checked the full genome alignment using Mauve and MUMmer full genome blast.
2.3. Identification of repetitive elements and non-coding RNA genes
Repetitive sequences were identified using multiple tools. Transposable elements (TEs) were identified by aligning against the Repbase database using RepeatMasker (v4.0.5) [22] with parameters “-species is fungi, search engine is rmblast2.2.27+”, and the Repbase version is 20150807 [23]. Meanwhile, the de novo repeat library was detected using the de novo search program RECON (V1.0.8, http://selab.janelia.org/recon.html) and RepeatScout (version1.0.5, http://repeatscout.bioprojects.org/) of the software RepeatModler (v1.0.4, http://repeatmasker.org/RepeatModeler.html) with default parameters.
For non-coding RNA (ncRNA), the tRNA genes were predicted using tRNAscan-SE (V1.3.1) with default parameters [24]. The rRNA genes were identified using RNAmmer (v1.2) [25]. The prediction of other ncRNAs was obtained after aligning against the Rfam database with a blast [26].
2.4. Gene prediction, genome annotation and evaluation
The genome of X. elegans was annotated using RNA data of X. elegans (PRJNA916377, SRR22904707) as auxiliary reference. The genome prediction of X. elegans mapped the transcript to the genome using PASA (Program to Assemble Spliced Alignments) to execute structural annotation of genes; first, the clean data of next-generation transcriptome sequences were de novo assembled and obtained “Trinity.fasta” using software Trinity(v2.5.1, the parameter is set to “-jaccard_clip”), and the alignment of genome assembly and next-generation transcriptome using TopHat (v2.1.1, parameter is set to “tophat -p 16”), genome-guided “Trinity.GG.fasta” was obtained using Trinity (v2.5.1, the parameter is set to “-genome_guided_bam, -genome_guided_max_intron 10,000”), and then de novo assembly result “Trinity.fasta” created list “tdn.accs”, the transcript assembled by Trinity mapped reference genome again using PASA and obtained comprehensive transcriptome database and integrated the gene prediction result. And lastly, to improve the accuracy of gene prediction, the present study adopted three steps to predict the gene models, (1) de novo prediction of the gene model was predicted using Augustus (v3.03), glimmerHMM (V3.0.1) and SNAP (v2006-07-28) [27–29]; (2) homologous prediction of relative species was used by software exonerate (v2.2.0) after aligning against the protein sequences of relative species; (3) the gene prediction was integrated with de novo gene prediction and the homologous prediction of relative species using EVidenceModeler (v r2012-06-25) [30].
The predicted genes were aligned to the KEGG, Swiss-Prot, KOG, NCBI nr, CAZy and GO databases using BLASTALL with the parameters “-p blastp -e 1e-5 -F F -a 4 -m 8” [31]. The X. elegans assembly was uploaded to the antiSMASH (v6.0) website to identify the secondary metabolite gene cluster [32].
2.5. Prediction of PKS genes in X. elegans
To characterize the potential PKS genes or gene clusters, the draft contig file from the genome of X. elegans was submitted to antiSMASH (v6.0) using the default parameters with relaxed detection strictness using a rule-based detection method [32]. Additionally, we implemented bioinformatics analysis to verify the PKS genes of antiSMASH-defined gene clusters and the domain in these clusters; the candidate PKS genes were verified by BLASTx analysis, NCBI conserved domain search program and Pfam database. The standard using identifying the polyketide synthase modules and domains both should meet an e-value ≥0.01, the alignment length of amino acids was greater than 100 and the identity was ≥35%.
2.6. Determination of the secondary metabolites catalyzed by candidate PKS modules or gene cluster
To identify the genes flanking the PKS clusters, Blast searches and antiSMASH were performed to link the known secondary metabolites and related PKS gene clusters [32]. The possible compounds of the candidate PKS genes could be identified from the contigs of X. elegans using antiSMASH program and compared with the reported related gene cluster to identify the corresponding gene cluster of X. elegans.
2.7. Phylogenetic analysis of the complete candidate PKS from the genome of X. elegans
To identify the evolutionary and the specific PKS related to the corresponding lichen compound, two phylogenetic trees of the concatenated protein sequences containing conserved KS domains of 40 NR-PKSs and 54 HR-PKSs were reconstructed, respectively. In the construction process of NR-PKS phylogenetic tree, a total of 40 amino acid NR-PKSs including 7 amino acid sequences of NR-PKSs from X. elegans, 24 NR-PKSs that have been linked to known compounds in nonlichenized fungi, and 9 NR-PKSs from other LFF were retrieved from NCBI. In the HR-PKSs of phylogenetic analysis, 12 HR-PKSs from X. elegans, 28 HR-PKSs linking with known compounds and 14 other HR-PKSs from LFF were retrieved from NCBI. All the PKSs were aligned by ClustalW (using MEGA X), adopting default parameters, and then the phylogenetic tree was constructed using Maximum-Likelihood method, 1000 bootstraps and other default parameters.
3. Results
3.1. General features of the X. elegans genome
The genome of X. elegans was sequenced using Illumima HiSeq platform. The total reads were 9,812 Mb in length, representing an approximate 252-fold sequence coverage, and deposited in NCBI (PRJNA511131, SAUG00000000.1). The results of K-mer genome survey analysis estimate the genome size of X. elegans to be 51.62 Mb (Table 1). K-mer analysis with a length of 17-mers indicated that the genome of X. elegans had a heterozygous rate of 0.01% and a K-mer repetitive sequence content of about 46.72%, and its accuracy rate is 99.93%. BUSCO integrity assessment was conducted using the genome database (fungi_odb9, http://busco.ezlab.org), and 98.60% complete BUSCOs (747 genes) were found, and 98.30% complete and single-copy BUSCOs (745 genes) were found, 100% total BUSCOS groups (758 genes) were found in the genome of X. elegans, and this reflected the high integrity of assembly results (Table 2). In total, 9812 Mb of clean data were obtained, from which a 44.63 Mb assembly was obtained. The genome consisted of 207 scaffolds with N50 of 563, 100 bp, N90 of 122,672 bp, and 40.69% G + C content (Table 1). A total of 9,976 protein-coding genes were predicted in the X. elegans genome, with 1594.5 bp, an average CDS length 1463.9 bp, an average of 2.7 exons per gene, an average exon length of 529.7 bp, an intron average length of 74 bp, and CDSs accounted for 29.67% of the X. elegans genome (Table 1).
Table 1.
Main features of the Xanthoria elegans genome.
| Characteristics | Value |
|---|---|
| Raw data | |
| High-quality (HQ) data (bp) | 9,812,213,868 |
| Coverage | 252× |
| Genome size (Mb) | 51.62 |
| Revised genome size (Mb) | 51.58 |
| Assembly total sequences length (bp) | 44,625,953 |
| HQ data (%) | 96.16 |
| HQ reads number | 63,728,134 |
| HQ reads (%) | 98.07 |
| Heterozygous rate (%) | 0.01 |
| Repeat (%) | 46.72 |
| Accuracy rate (%) | 99.93 |
| Scaffold | |
| Total number | 207 |
| Total length (bp) | 44,625,953 |
| N50 (bp) | 563,100 |
| N90 (bp) | 122,672 |
| Maximum sequence length (bp) | 1,247,775, |
| Min sequence length (bp) | 1,011 |
| GC content (%) | 40.69 |
| Genome | |
| Gene total length (bp) | 14,604,751 |
| Gene number | 9,976 |
| Gene length/genome (%) | 35.65 |
| Average gene length (bp) | 1,594.5 |
| Average CDS length (bp) | 1463.9 |
| Average exons length (bp) | 529.7 |
| Average exons per gene | 2.7 |
| Average introns length (bp) | 74 |
| CDSs percentage of genome (%) | 32.72 |
Table 2.
The integrity assement of genome assembly.
| Property | Number | Percent (%) |
|---|---|---|
| Complete BUSCOs | 747 | 98.60% |
| Complete and single-copy BUSCOs | 745 | 98.30% |
| Complete and duplicated BUSCOs | 2 | 0.30% |
| Fragmented BUSCOss | 1 | 0.10% |
| Missing BUSCOs | 10 | 1.30% |
| Total BUSCO groups searched | 758 | 100% |
A blast search of repeat sequences produced 750,617 bp, covering 1.68% of the X. elegans genome; meanwhile, interspersed nuclear elements and tandem repeats accounted for 1.63% and 0.05%, respectively. Approximately 1.43% of the genome was long terminal repeats (LTR), 0.13% was DNA transposons and 0.0.2% was unclassified. The proportion of tandem repeats in the assembled genome was 0.05%, while satellite DNA and simple repeats accounted for 0.01% and 0.04%, respectively (Table 3).
Table 3.
The statistics of different repeats in Xanthoria elegans genome.
| Number of elements | Length occupied (bp) | Percentage of sequence (%) | |
|---|---|---|---|
| LTR elements | 1538 | 636,338 | 1.43 |
| non-LTR elements | 314 | 22,294 | 0.05 |
| DNA transposons | 762 | 59,170 | 0.13 |
| Unclassified | 83 | 8,965 | 0.02 |
| Satellites | 43 | 4,507 | 0.01 |
| Simple repeats | 156 | 19,629 | 0.04 |
| Total | 2896 | 750,903 | 1.68 |
LTR: long terminal repeat.
3.2. Functional annotation
There were 9976 gene models predicted in the different databases with a total sequence length of 14,604,751 bp, accounting for 35.65% of the whole genome with an average sequence length of 1,594.5 bp (Table 1). We predicted 67 tRNAs (6,637 bp), 9 rRNAs (9,632 bp) and 62 ncRNAs (8,579 bp) (Table 3). And the 56 tRNAs were anticodon tRNAs corresponding to the 20 common amino acids codons.
The genome annotation of X. elegans was performed with the NCBI nr, EggNOg, KEGG, Swiss-Prot, GO and CAZy databases. In the nr databases, with a total of 8604 genes annotated (accounting for 86.25% of the total predicted genes); in the KEGG database, with a total of 9,791 genes annotated (accounting for 98.15% of the total predicted genes); in the GO database, with a total of 5608 genes (accounting for 56.21% of the total predicted genes); in the EggNOg databases, with a total of 7612 genes (accounting for 76.30% of the total predicted genes); in the Swiss-Prot database, with a total of 6046 genes (accounting for 60.61% of the total predicted genes) (Table 4).
Table 4.
Summary of Xanthoria elegans gene annotations.
| Database used for gene/ protein annotation | Number | Percentage (%) |
|---|---|---|
| NCBI nr | 7785 | 81.25 |
| GO | 6757 | 70.53 |
| KEGG | 8716 | 90.97 |
| KOG | 7266 | 75.84 |
| Swiss-Prot | 5645 | 58.92 |
| CAZy | 331 | 3.45 |
NCBI nr: National Center for Biotechnology Information non-reduced protein database; EggNOg: Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups; GO: Gene Ontology database; KEGG: Kyoto Encyclopedia of Genes and Genomes pathway database; CAZy: Carbohydrate-Active enZYmes Database.
KEGG enrichment analysis revealed that 9791 genes that corresponded to KEGG pathways were enriched in 380 metabolic pathways, and related to metabolism including amino acid metabolism (449 genes), metabolism of cofactors and vitamins (156 genes), metabolism of terpenoids and polyketides (45 genes), nucleotide metabolism (83 genes), energy metabolism (129 genes), carbohydrate metabolism (349 genes), glycan biosynthesis and metabolism (97 genes), lipid metabolism (237 genes), other secondary metabolism (66 genes), xenobiotics biodegradation and metabolism (100 genes) and unclassified (125 genes); genetic information processing and signaling and cellular process (Table 5, Figure 1).
Table 5.
The metabolism genes count based on KEGG database.
| Metabolism category in KEGG | Count |
|---|---|
| Amino acid metabolism | 369 |
| Metabolism of other amino acids | 80 |
| Biosynthesis of other secondary metabolites | 66 |
| Carbohydrate metabolism | 349 |
| Energy metabolism | 129 |
| Glycan biosynthesis and metabolism | 97 |
| Lipid metabolism | 237 |
| Metabolism of cofactors and vitamins | 156 |
| Metabolism of terpenoids and polyketides | 45 |
| Nucleotide metabolism | 83 |
| Xenobiotics biodegradation and metabolism | 100 |
| Unclassified | 125 |
| Total | 1836 |
Figure 1.
Kyoto Encyclopedia of Genes and Genomes pathway annotation of Xanthoria elegans genes.
A total of 353 CAZyme-encoding gene models were assigned in the X. elegans genome, including 87 glycosyltransferases, 67 carbohydrate esterases, 65 auxiliary activities, 13 carbohydrate-binding modules and 121 glycoside hydrolases (Table 6).
Table 6.
The genes class definition based on CAZy database.
| Class definition | Gene count |
|---|---|
| Glycosyl transferases | 87 |
| Polysaccharide lyases | 0 |
| Carbohydrate esterases | 67 |
| Auxiliary activities | 65 |
| Carbohydrate-binding modules | 13 |
| Glycoside hydrolases | 121 |
| Total | 353 |
3.3. Identification of PKS genes and domains organization from the genome of X. elegans
A total of 21 PKS genes from the genome of X. elegans was predicted by comparing the results of antiSMASH, Blast and Conserved domains analysis, they all have the KS, AT and ACP domains. Of them, 7 NR-PKSs genes named XePKS1 to XePKS7, 12 HR-PKSs genes named XePKS8 to XePKS19, and 2 hybrid PKS-NRPSs named XePKS20 to XePKS21 were isolated from the genome (Figure 2), and there were no partially PKS. There were 2 NR-PKSs, 7 HR-PKSs and 2 hybrid PKS-NRPSs with MeT domain.
Figure 2.
Domain organization of PKS protein from the genome of the lichen-forming fungi Xanthoria elegans. For distinguishing the organisational differences and similarities between the different PKS domains, each type of domain is revealed in a different colour box.
3.4. Phylogenetic analysis of PKS corresponding for lichen substances
In the maximum-likelihood phylogeny of NR-PKSs with a 40 NR-PKSs, the phylogenetic tree is divided into 6 subclades based on the protein sequence similarity and the PKS domain organization (Figure 3): the result revealed that XePKS6 was in group IV, the domain architecture of NR-IV is SAT-KS-AT-PT-ACP-TE, and the product of Fusarium fujikuroi FSR1 (CCE67070.1) was fusarubin, which is a highly pigmented naphthoquinone, and the compound of XePKS6 could be naphthoquinone like fusarubin; XePKS2 clustered in group V, the domain architecture of NR-V is SAT-KS-AT-PT-ACP without TE domain, and the products of A. nidulans PkgA (XP_664675.1) and A. fumigatus EncA (XP_746435.1) were alternariol and endocrocin, respectively, these two compounds both belong to anthraquinone, the putative compound of XePKS2 is a kind of anthraquinone; and PKS genes in this group were suggested to be involved in the biosynthesis mycophenolic acid such as Penicillium brevicompactum MpaC (ADY00130.1), the product of XePKS1 might be mycophenolic acid, and XePKS5 might be mycophenolic acid with methyl group; XePKS7 was in group VII, its domain organization was SAT-KS-AT-ACP-ACP-R, and the compounds in this group could be proposed to produce azaphilones [33], the compound might be a azaphilones; XePKS3 was in group IX, its domain organization was SAT-KS-AT-ACP-ACP-MeT-TE, and the compound of Acremonium egyptiacum AscC (A0A455R5P9.1) was characterized to be asocholrin by heterologous expression, which is a prenylated aryl-aldehyde and a key pathway intermediate for many fungal meroterpenoids [34], and the putative compound of XePKS3 is a prenylated aryl-aldehyde.
Figure 3.
Phylogenetic analysis of nonreducing PKSs proteins. A maximum likelihood tree of 40 NR-PKSs including known compounds of 24 non-lichenized fungal NR-PKSs with known SMs [4], 7 NR-PKSs from Xanthoria elegans and 9 lichen-forming fungi PKSs were reconstructed using concatenated sequences of KS domains.
For our phylogenetic analysis, the fungal HR-PKSs were divided into six groups (Figure 4), XePKS17, XePKS9, XePKS14, XePKS16 and XePKS10 were in group I, this group was the largest group and these 5 genes in this group were nested in different clades including Ia and Ib, but the compound in this group is still unclear; the domain organization was KS-AT-DH-MeT-ER-KR-ACP, and XePKS14 and XePKS16 did not contain MeT domain. XePKS18 and XePKS12 were in clade VII, and there were characterized sequences of A. terreus (LovF, Q9Y7D5.1) and P. citrinum (mlcB, Q8J0F5.1), which indicate participating in the biosynthesis of the diketide portion of lovastatin and citrinin. Clade IV and V were sister groups, clade IV was divided into two subclades and contained a conserved MeT domain, including XePKS15 in clade IVa and XePKS13 and XePKS11 in clade IVb; the gene sol1 from A. oryzae produced solanapyrone was identified in clade IVa; a novel alkylresorcinols soppilines biosynthesis gene pspA (P. soppi, P0DUK1.1) was characterized; the putative product of XePKS15 was solanapyrone analogs, and the putative products of XePKS13 and XePKS11 were the toxin soppilines analogs according to the phylogenetic analysis; XePKS19 and XePKS18 were in clade V, BreA gene from P. brefeldianum in this group was experimentally identified to brefeldin A, which was a macrolactone, and their putative products were macrolactones.
Figure 4.
Phylogenetic tree of fungal highly reducing PKSs (HR-PKSs) proteins. A maximum likelihood tree of 54 HR-PKSs including 12 HR-PKSs from X. elegans and 42 HR-PKSs from lichen-forming fungus and other fungus PKSs was reconstructed using concatenated sequences of KS domains.
3.5. Prediction of PKS gene clusters in the genome of X. elegans
The results of PKS gene clusters by antiSMASH and Blast searches revealed that there were five gene clusters. The gene cluster comparison of A. fumigatus (CM000172.1), A. nidulans (BN001304.1) and X. elegans (Scaffold 24) revealed that (Figure 5) the three gene clusters had the same core genes including an NR-PKS and a metallo-β-hydrolase gene, and the product of BGC A. fumigatus (CM000172.1) identified to be endocrocin by gene knock out; and A. nidulans (BN001304.1) was alternariol by verifying promoter replacement experiments, and these two compounds belong to anthraquinones. And the flanking genes of the three core genes were different, the gene cluster of scaffold 24 containing XePKS2 might be an anthraquinone.
Figure 5.
The comparison of anthraquinone gene cluster organization in different fungi. The colorful arrows denote genes that are required for anthraquinones production. Black arrows denote genes that are part of the anthraquinone biosynthetic gene cluster. MFS: major facilitator superfamily.
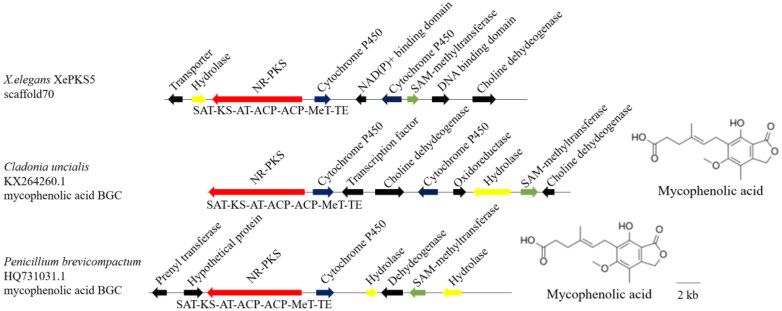
In the biosynthesis of mycophenolic acid, the gene clusters contained two core genes encoding an NR-PKS and a cytochrome P450 were required in C. uncialis and P. brevicompactum, and the same core genes were in X. elegans scaffold 70 (Figure 6); however, the flanking genes adjacent to the core genes from the three fungi were different, which suggested that they could produce the same carbon skeleton but different attached groups; the X. elegans scaffold 70 could produce mycophenolic acid analogs.
Figure 6.
The comparison of usnic acid gene cluster organization in different fungi. The colorful arrows denote genes that are required for usnic acid production. Black arrows denote genes that are part of the usinic acid biosynthetic gene cluster.
There were three core genes containing an HR-PKS, a T3PKS and a cytochrome P450 gene in P. soppi, Letharia columbiana (JACCJC010000048) and X. elegans (scaffold 51) (Figure 7), among them the cryptic psp BGC in P. soppi was identified by heterologous expression in A. oryzae, and the product of this cluster was characterized as Z, E, Z-triene (novel alkylresorcinols named soppilines) [35]; and the putative BGC contained gene XePKS13 belongs to alkylresorcinols.
Figure 7.
The comparison of alkylresorcinols gene clusters organization in Penicillium soppi, Letharia columbiana and Xanthoria elegans. The colorful arrows denote genes that are required for anthraquinones production. Black arrows denote genes that are part of the anthraquinone biosynthetic gene cluster. T3PKS: type III PKS.
There are core genes including HR-PKS, a hydrolase, a cytochrome b5 and two cytochrome P450 genes in A. nidulans (BN001306.1), Xylaria acuta (MU383237.1) and X.elegans (scaffold 84) (Figure 8). The product of BGC containing gene XePKS16 might be (+)-asperlin analogs.
Figure 8.
The comparison of the gene cluster organization between Aspergillus nidulans, Xylaria acuta and Xanthoria elegans. The colorful arrows denote genes that are required for anthraquinones production. Black arrows denote genes that are part of the biosynthetic gene cluster. FACL: fatty acid CoA ligase.
The brefeldin A BGC composed of HR-PKS, 4 cytochrome P450s and a hydrolase gene is necessary for the biosynthesis of relatively longer (>C10) acyclic polyketide brefeldin A identifying by reconstruction in P. brefeldianum [36]; there is HR-PKS, two hydrolases and two cytochrome P450 genes in Monosporascus sp. (QJOB01000139.1), and there is a hydrolase, a two hydrolase and cytochrome P450 genes in X. elegans (Figure 9). The core enzymes from the three fungi are similar, and the putative product of BGC scaffold 134 containing XePKS19 might be acylcyclic polyketide.
Figure 9.
The comparison of macrolactone gene cluster organization between Penicillium brefeldianum, Monosporascus sp. and Xanthoria elegans. The colorful arrows denote genes that are required for anthraquinones production. Black arrows denote genes that are part of the macrolactone biosynthetic gene cluster.
4. Discussion
In the genomic era, the number of available genomes is increasing, genome mining joined to synthetic biology applies to a significant help in specific compound discovery, and genome comparison is widely applied. Meanwhile, we compared the genome of closely related species of X. elegans and their protein-coding genes, their genomes and relative information from the NCBI genome database found that the genome total length of X. mediterranea was 32.48 Mb and its protein-coding genes were 8998 (JALAII000000000); the genome total length of X. aureola was 38.52 Mb and its protein-coding genes were 9875 (JALAIA000000000); the genome total length of X. steineri was 30.64 Mb and its protein-coding genes were 9445 (JALAIK000000000). Although the genomes total length of Xanthoria had a large difference, and their number of protein-coding genes was similar to X. elegans, so we could deduce the protein-coding genes number of genus Xanthoria has similar protein-coding genes. In the present study, genome survey estimation showed that the heterozygous rate of X. elegans genome was 0.01%; the genome integrity assessment of X. elegans showed that its genome total length has 44.63 Mb and its genome integrity and continuity is very well, and no contamination/multiple origins, can be used for subsequent analysis.
Lichens, especially in LFF, could produce a lot of SMs with interesting biological activity, and some of them such as anthraquinones, aflatoxin, depsidones have been used as commercial drugs [1]. These SMs are mainly synthesized by PKSs via malonyl CoA and acetate CoA pathways [37]. Genetic tools for studying LFF could avail to identify the PKSs, the BGCs and industrial applications of lichen substances. Genome analysis of LFF indicated that there are 13-34 complete PKS genes in their genome, including C. metacorallifera with 31 putative PKSs, Endocarpon pusillum with 15 PKS genes, C. macilenta with 34 PKSs, Umbilicaria muehlenbergii with 20 PKS genes [38]. In the present study, the repertoires of polyketide biosynthetic genes from LFF X. elegans were investigated, and 7 NR-PKSs, 12 HR-PKSs and 2 hybrid PKS-NRPS were found; yet Erken et al. [1] found 3 NR-PKSs, 6 PR-PKSs and 4 HR-PKSs in C. metacorallifera, 3 NR-PKSs, 6 PR-PKSs and 4 HR-PKSs in E. pusillum from their genomes. Different lichens and its habitat resulted in the various number of PKSs in different LFF fungus.
The iterative type I PKSs can be divided into different major groups based on the protein sequences similarity and PKS domain architecture, and they have similar domain structure and similar chemical characteristics in the same group [4,39]. For example, Kroken et al. [39] found different groups were predicted to synthesize various polyketides; Kim et al. [4] identified the putative atranorin PKS gene (atr1) by phylogenetic analysis and heterologous expression, linked atr1 to a depside atranorin. The phylogenetic tree showed that XePKS2 was in group V and had the same domain organization as other PKSs in this group, two characterized nonlichenized fungal PKSs, A. nidulans PkgA (XP_664675.1) [33] and A. fumigatus EncA (XP_746435.1) [40], their products were alternariol and endocrodin, respectively; together with the compound isolation in X. elegans, this lichen produces many anthraquinones like parietin, fallacinal, erythroglaycin, teloschistin, parietinic acid and emodin [9], the major lichen substances in the cortical layer is parietin [8,12]; the results of gene cluster analysis between X. elegans (scaffold 24), A. fumigatus (CM000172.1) and A. nidulans (BN001304.1) revealed that the 3 gene clusters have the same core enzymes containing an NR-PKS and a metallo-β-hydrolase; we could predict that the product of XePKS2 is an emodin anthrone. XePKS1 and XePKS5 were in group VI, sequences located in group VI are largely characterized by having a methylation domain (MeT) between the ACP domain and the N-terminal domain [41], XePKS5 had the domain organization SAT-KS-AT-ACP-ACP-MeT-TE, XePKS1 didn’t have the MeT domain, and this phenomenon of MeT domain loss in group VI have been observed [4], genes from this group were suggested to be involved in the biosynthesis of usnic acid [41]; and usnic acid as a UV-B absorbing polyketide is an important pigment and has been isolated by HPLC (high-performance liquid chromatography) [10]; NR-PKS and cytochrome P450 were required during the mycophenolic acid biosynthesis, and the core genes existed in all three gene clusters, we could deduce that the product of XePKS5 is methyl-mycophenolic acid, and the product of XePKS1 is mycophenolic acid without methyl group.
Besides KS, AT and ACP domains in HR-PKS, ketoreductase (KR), dehydratase (DH) and enoyl reductase (ER) domains are added to process the β-keto [33]. In the present study, 12 HR-PKSs were isolated, eight of them had all three domains, and four of them lacked the DH domain (Figure 1). The polyketide with branched methyl groups was introduced by the catalysis of methyltransferase (MeT), and most of HR-PKSs in fungus lack a chain-release domain, therefore the chain-release mechanism is not available as limited information [42]. HR-PKS genes are involved in synthesizing various reduced linear polyketides [43]. Combined with the domain architecture, phylogenetic analysis and gene cluster comparison of different fungi, the PKS genes can be distinguished from different groups and produced compounds [43,44]. In the HR-PKS phylogenetic tree, the products of HR-PKS genes in group I were not unclear, but the gene cluster comparison found that the XePKS16 located in group I have the same core genes with A. nidulans (BN001306.1) and X. acuta (MU383237.1), among them, the product of BGC A. nidulans (BN001306.1) (+)-asperlin was identified by activating the hybrid transcription factor and gene deletion [18], the domain organization of all three HR-PKSs in the gene cluster have the same domain architecture KS-AT-DH-ER-KR-ACP, and the putative product of XePKS16 might be a toxin like (+)-asperlin. XePKS13 is located in group IV, and a previous study revealed genes in this group may contain a conserved domain MeT, and a novel alkylresorcinols toxin soppilines biosynthesis gene pspA (P. soppi, P0DUK1.1) was characterized by the heterologous expression [35], XePKS13 and pspA have a close relationship and they have the same core genes according to the results of BGC comparison, and the putative product of XePKS3 may be a toxin like soppilines. XePKS19 was in group V, an HR-PKS gene BreA produced brefeldin A was identified [36], the BGC comparison results showed them have the core genes containing HR-PKS, cytochrome P450s and hydrolase gene and the domains architecture of XePKS19 and BreA was KS-AT-MeT-ER-KR-ACP, and the putative product of XePKS19 might be a macrolactone. Compared to the NR-PKSs, less is known about the possible compounds of HR-PKSs [43], and unlike what has been shown in previous studies, all studied HR-PKSs and the candidate HR-PKSs from X. elegans lacked a second ACP domain or other domains like DH or MeT [4,43]. In the present study, XePKS11, XePKS12, XePKS13 and XePKS15 have revealed they lacked the DH domain, and XePKS14 and XePKS16 in group I lacked the MeT domain. This phenomenon of domain loss in fungi was not unique, some studies showed TE domain loss in PR-PKS [45], MeT and ER domain was absent in the HR-PKSs [39]. It is known that fungal reduced polyketides show vast differences as a result of PKS structure. Additionally, the two hybrid PKS-NRPS of phylogenetic analysis was not performed, and their compounds were unknown. Due to non-reducing and reduced polyketides in X. elegans, it can absorb UV light and defend herbivore, fungus and bacteria, and survive in the extreme habitat.
The PKSs from X. elegans were isolated by genome mining and analyzed with bioinformatics including phylogenetic, domain organization and gene cluster comparison, some correlation between the SMs carbon skeleton and specific PKS genes primarily built, and provided some basis on heterologous expression about the specific PKS genes. In general, the PKS mining from the genome of X. elegans will be helpful to understand the reason that this lichen could live in the extreme habitat, and then, the induction factors of polyketides produced in this lichen could be clarified.
Funding Statement
This work was supported by a grant from the National Natural Science Foundation of China (31860177), General Project of Basic Research Program in Yunnan Province (202101AT070218), the Reserve Talents for Young and Middle-aged Academic and Technical Leaders of the Yunnan Province (202205AC160044).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Erken MT, Cansaran-Duman D, Tanman U.. In silico prediction of type I PKS gene modules in nine lichenized fungi. Biotechnol Biotec Eq. 2021;35(1):376–383. [Google Scholar]
- 2.Elkhateeb WA, Ghwas DEE, Daba GM.. Lichens uses surprising uses of lichens that improve human life. J Biomed Res Environ Sci. 2022;3(2):189–194. [Google Scholar]
- 3.Calcott MJ, Ackerley DF, Knight A, et al. Secondary metabolism in the lichen symbiosis. Chem Soc Rev. 2018;47(5):1730–1760. [DOI] [PubMed] [Google Scholar]
- 4.Kim W, Liu R, Woo S, et al. Linking a gene cluster to atranorin, a major cortical substance of lichens, through genetic dereplication and heterologous expression. mBio. 2021;12(3):e01111-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devashree PA, Dikshit A.. Lichens: fungal symbionts and their secondary metabolites. In: Joginder S and Praveen G, editors.. New and future developments in microbial biotechnology and bioengineering. Amsterdam: Elsevier Science B.V; 2021. p. 107–115. [Google Scholar]
- 6.Cornejo A, Salgado F, Caballero J, et al. Secondary metabolites in Ramalina terebrata detected by UHPLC/ESI/MS/MS and identification of parietin as tau protein inhibitor. Int J Mol Sci. 2016;17(8):1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdel-Hameed M, Bertrand RL, Piercey-Normore MD, et al. Putative identification of the usnic acid biosynthetic gene cluster by de novo whole-genome sequencing of a lichen-forming fungus. Fungal Biol. 2016;120(3):306–316. [DOI] [PubMed] [Google Scholar]
- 8.Nybakken L, Solhaug KA, Bilger W, et al. The lichens Xanthoria elegans and Cetraria islandica maintain a high protection against UV-B radiation in arctic habitats. Oecologia. 2004;140(2):211–216. [DOI] [PubMed] [Google Scholar]
- 9.Wang HY, Li HM, Shi N, et al. The HPLC analysis of the lichen substances in five species of Xanthoria (Ascomycota). Mycosystema. 2003;22(4):536–541. [Google Scholar]
- 10.Mamut R, Abbas A.. The determination of usnic acid by HPLC method in Xanthoria elegans and their antibacterial activity. Food Sci Technol. 2012;37(8):216–219. [Google Scholar]
- 11.Basile A, Rigano D, Loppi S, et al. Antiproliferative, antibacterial and antifungal activity of the lichen Xanthoria parietina and its secondary metabolite parietin. Int J Mol Sci. 2015;16(4):7861–7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gausla Y, Ustvedt EM.. Is parietin a UV-B or a blue-light screening pigment in the lichen Xanthoria parietina? Photochem Photobiol Sci. 2003;2(4):424–432. [DOI] [PubMed] [Google Scholar]
- 13.Gagunashvili AN, Davidsson SP, Jonsson ZO, et al. Cloning and heterologous transcription of a polyketide synthase gene from the lichen Solorina crocea. Mycol Res. 2009;113(3):354–363. [DOI] [PubMed] [Google Scholar]
- 14.Armaleo D, Sun X, Culberson C.. Insights from the first putative biosynthetic gene cluster for a lichen depside and depsidone. Mycologia. 2011;103(4):741–754. [DOI] [PubMed] [Google Scholar]
- 15.Sabatini M, Comba S, Altabe S, et al. Biochemical characterization of the minimal domains of an iterative eukaryotic polyketide synthase. Febs J. 2018;285(23):4494–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5(13):2010–2026. [DOI] [PubMed] [Google Scholar]
- 17.Kage H, Riva E, Parascandolo JS, et al. Chemical chain termination resolves the timing of ketoreduction in a partially reducing iterative type I polyketide synthase. Org Biomol Chem. 2015;13(47):11414–11417. [DOI] [PubMed] [Google Scholar]
- 18.Grau MF, Entwistle R, Chiang YM, et al. Hybrid transcription factor engineering activates the silent secondary metabolite gene cluster for (+)-asperlin in Aspergillus nidulans. ACS Chem Biol. 2018;13(11):3193–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SY, Choi J, Lee GW, et al. Draft genome sequence of Umbilicaria muehlenbergii KoLRILF000956, a lichen-forming fungus amenable to genetic manipulation. Genome Announc. 2014;2(2):e00357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto Y, Mizuguchi R, Yamada Y.. Tissue cultures of Usnea rubescens and Ramalina yasudae and production of usnic acid in their cultures. Agric Biol Chem. 1985;49(11):3347–3348. [Google Scholar]
- 21.Luo R, Liu B, Xie Y, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2015;4(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bao W, Kojima K, Kohany O.. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapitonov V, Jurka J.. A universal classification of eukaryotic transposable elements implemented in repbase. Nat Rev Genet. 2008;9(5):411–412. [DOI] [PubMed] [Google Scholar]
- 24.Lowe T, Eddy S.. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagesen K, Hallin P, Rodland E, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalvari I, Nawrocki E, Argasinska J, et al. Non-coding RNA analysis using the rfam database. Curr Protoc Bioinformatics. 2018;62(1):e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanke M, Morgenstern B.. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33(2):465–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majoros WH, Pertea M, Salzberg SL.. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20(16):2878–2879. [DOI] [PubMed] [Google Scholar]
- 29.Korf I. Gene finding in novel genomes. BMC Bioinf. 2004;5(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas BJ, Salzberg SL, Zhu W, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9(1):R7–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D, Yu J, Ma C, et al. Genomic analysis of the mycoparasite Pestalotiopsis sp. PG52. Pol J Microbiol. 2021;70(2):189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blin K, Shaw S, Kloosterman AM, et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49(W1):W29–W35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahuja M, Chiang YM, Chang SL, et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134(19):8212–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araki Y, Awakawa T, Matsuzaki M, et al. Complete biosynthetic pathways of ascofuranone and ascochlorin in Acremonium egyptiacum. Proc Natl Acad Sci U S A. 2019;116(17):8269–8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko A, Morishita Y, Tsukada K, et al. Post-genomic approach based discovery of alkylresorcinols from a cricket-associated fungus, Penicillium soppi. Org Biomol Chem. 2019;17(21):5239–5243. [DOI] [PubMed] [Google Scholar]
- 36.Zabala AO, Chooi YH, Choi MS, et al. Fungal polyketide synthase product chain-length control by partnering thiohydrolase. ACS Chem Biol. 2014;9(7):1576–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen HT, Ketha A, Kukavica B, et al. Anti-inflammatory potential of lichens and its substances. In: Vinay Bharadwaj T, editor. Inflammatory bowel disease. Reno (NV): MedDocs Publishers; 2021. p. 1–9. [Google Scholar]
- 38.Wang Y, Geng C, Yuan X, et al. Identification of a putative polyketide synthase gene involved in usnic acid biosynthesis in the lichen Nephromopsis pallescens. PLoS One. 2018;13(7):e0199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroken S, Glass NL, Taylor JW, et al. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A. 2003;100(26):15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim FY, Hou Y, Chen Y, et al. Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus. Appl Environ Microbiol. 2012;78(12):4117–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pizarro D, Divakar PK, Grewe F, et al. Genome-wide analysis of biosynthetic gene cluster reveals correlated gene loss with absence of usnic acid in lichen-forming fungi. Genome Biol Evol. 2020;12(10):1858–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ugai T, Minami A, Fujii R, et al. Heterologous expression of highly reducing polyketide synthase involved in betaenone biosynthesis. Chem Commun. 2015;51(10):1878–1881. [DOI] [PubMed] [Google Scholar]
- 43.Gerasimova JV, Beck A, Werth S, et al. High diversity of type I polyketide genes in Bacidia rubella as revealed by the comparative analysis of 23 lichen genomes. J Fungi. 2022;8(5):449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Punya J, Swangmaneecharern P, Pinsupa S, et al. Phylogeny of type I polyketide synthases (PKSs) in fungal entomopathogens and expression analysis of PKS genes in Beauveria bassiana BCC 2660. Fungal Biol. 2015;119(6):538–550. [DOI] [PubMed] [Google Scholar]
- 45.Gallo A, Ferrara M, Perrone G.. Phylogenetic study of polyketide synthases and nonribosomal peptide synthetases involved in the biosynthesis of mycotoxins. Toxins. 2013;5(4):717–742. [DOI] [PMC free article] [PubMed] [Google Scholar]