Abstract
Antrodia cinnamomea, an edible and medicinal fungus with significant economic value and application prospects, is rich in terpenoids, benzenoids, lignans, polysaccharides, and benzoquinone, succinic and maleic derivatives. In this study, the transcriptome of A. cinnamomea cultured on the wood substrates of Cinnamomum glanduliferum (YZM), C. camphora (XZM), and C. kanehirae (NZM) was sequenced using the high-throughput sequencing technology Illumina HiSeq 2000, and the data were assembled by de novo strategy to obtain 78,729 Unigenes with an N50 of 4,463 bp. Compared with public databases, about 11,435, 6,947, and 5,994 Unigenes were annotated to the Non-Redundant (NR), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genome (KEGG), respectively. The comprehensive analysis of the mycelium terpene biosynthesis-related genes in A. cinnamomea revealed that the expression of acetyl-CoA acetyltransferase (AACT), acyl-CoA dehydrogenase (MCAD), 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA), mevalonate pyrophosphate decarboxylase (MVD), and isopentenyl diphosphate isomerase (IDI) was significantly higher on NZM compared to the other two wood substrates. Similarly, the expression of geranylgeranyltransferase (GGT) was significantly higher on YZM compared to NZM and XZM, and the expression of farnesyl transferase (FTase) was significantly higher on XZM. Furthermore, the expressions of 2,3-oxidized squalene cyclase (OCS), squalene synthase (SQS), and squalene epoxidase (SE) were significantly higher on NZM. Overall, this study provides a potential approach to explore the molecular regulation mechanism of terpenoid biosynthesis in A. cinnamomea.
Keywords: Antrodia cinnamomea, de novo transcriptome, biosynthesis, terpenoid, qRT-PCR
1. Introduction
Antrodia cinnamomea (Syn. Antrodia camphorata and Taiwanofungus camphorata) belonging to the Polyporaceae family, is an extremely rare medicinal fungus restricted to Taiwan’s endemic host tree C. kanehirai [1], and has been widely used in Taiwan and China [2]. The crude extracts and active ingredients of A. cinnamomea pose various biological activities, such as antitumor [3], antioxidant [4], and liver protection effects [5]. So far, seventy-eight compounds have been identified in the fruiting bodies and mycelium of A. cinnamomea [6], including sesquiterpene [7], diterpenes [8], triterpenes, and sterols [9,10], with triterpenoids being the primary active compound, contributing to the bitter taste of A. cinnamomea [11,12].
Terpenoids comprised of isoprene (C5) units, and their derivatives and are one of the major secondary metabolites of fungi and are widely distributed in nature [13]. Terpenoid is primarily synthesized by the C5 precursors, such as isopentenyl diphosphate (IPP) and dimethyl allyl diphosphate (DMAPP) through the mevalonate (MVA) and methyl tetrahydroxyl 4-phosphate (MEP) pathways [14]. Lu et al. [15] elucidated the position and role of AACT, hydroxymethylglutaryl CoA reductase (HMGR), MVD, IDI, and other key enzymes in the chitosan and CaCl2 to liquid fermented A. cinnamomea increases the mycelial biomass and triterpenoids content [16]. Ma et al. [17] found that the addition of citrus peel extracts rich in essential oils increased the production of triterpenoids by ten folds. Previous studies have analyzed the volatile compounds in A. cinnamomea, such as hemipterpenes α-Cadinol, T-Muurolol, T-Cadinol, and α-Terpineol using organic solvents and water vapor distillation and found that the fermentation broth of A. cinnamomea substrates and mycelium contains many active substances [18]. Related studies have found that the bioactive triterpenoids in A. cinnamomea are abundant and structurally unique, mainly the ergostane and lanostane types [11].
Several studies have reported that the cultivation of substrates affects fungal metabolite synthesis. A. cinnamomea grown on different wood substrates poses different metabolite compositions and produces different metabolites when cultured in non-primary host wood or organs [19]. Yang et al. [20] used oats, wheat, buckwheat, and barley as the substrates for solid-state fermentation of A. cinnamomea, which significantly enhanced the production of intracellular polyphenols and triterpenoids, and secondary metabolites. Lin [21] found that many active secondary metabolites could be isolated from the mycelium or substrates of A. cinnamomea. Some researchers have demonstrated that the metabolic profile of A. cinnamomea fruit bodies cultured on A. cinnamomea wood substrates significantly differ from A. cinnamomea fruit bodies cultured on other wood substrates, expressing abundant metabolites, especially ergostane and lanostane triterpenes, benzene compounds, and polyacetylene [22].
However, so far, no studies have reported the mycelium of A. cinnamomea cultured on different wood substrates. Therefore, in this study, the transcriptome of the A. cinnamomea mycelium cultured in different wood substrates was sequenced using Illumina 2 000 to analyze the genes involved in terpenoid biosynthesis. The study results will provide a foundation for the functional identification of terpenoid biosynthetic genes, synthetic pathways, and molecular breeding.
2. Materials and methods
2.1. Antrodia cinnamomea mycelia
The strain was provided by the Green Development Research Institute of Southwest Forestry University and numbered BSNZZ. The wood substrates were mainly based on three species of camphor plants, including C. glanduliferum, C. camphora, and C. kanehirae. All woods used in this study were identified by the Institute of Green Development of Southwest Forestry University. The strains were inoculated on the outer walls of different camphor plant substrates, and incubated for 270 d in a constant temperature incubator at 26 °C with 95% humidity. After the mycelium of A. cinnamomea covered the wood substrates’ surface, 0.5 cm2 of the mycelium was taken under sterile conditions and transferred to a 2 mL centrifuge tube. 3 replicates per sample, labeled, and stored at −80 °C for later use.
2.2. Determination of biomass
A certain area of the mycelium cultured on NZM, XZM and NZM was taken to measure the biomass using the colony diameter, and the influence of different conditions on the biomass of the strains was analyzed.
2.3. Determination of triterpene content
Standard curve production: 10.08 mg of oleanolic acid standard dried to constant weight was weighed in a 25 mL volumetric flask, dissolved in 80% ethanol and fixed at a concentration of 0.4032 mg/mL. The oleanolic acid standard solution (0.00, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80 mL) was absorbed into the graduated test tube and placed in a water bath at 50 ∼ 60 °C. After the solvent was dried, 0.3 mL of 5% vanillin glacial acetic acid solution and 1 mL perchloric acid solution were added, respectively. After cooling, 10 mL of glacial acetic acid was added and shaken vigorously. The blank was used as the control, and the absorbance was measured at 550 nm. The concentration of oleanolic acid (mg/g) was used as the abscissa and the absorbance value was used as the ordinate to plot the standard curve and obtain the regression equation.
Sample extraction: The samples were weighed and mixed with an appropriate amount of 50% ethanol. Then, the samples were sonicated for 30 min, left for 48 h, and filtered. Later, the filter and filter residues were washed with water fractional times, concentrated, and dried until no ethanol taste was left. The filtrate was transferred to a separating funnel, extracted with trichloromethane, and the extract was collected, and merged into a 50 mL volumetric flask. Afterward, the volume was fixed and shaken to obtain the test solution. An appropriate amount of test solution and reference series solution was taken in a 10 mL colorimetric tube, dried, and cooled, followed by the addition of 0.2 mL of vanillin glacial acetic acid solution and 0.8 mL of perchloric acid and vigorous shaking. It was kept warm in a water bath at 70 °C for 15 min, cooled in an ice bath immediately, and removed. Finally, 5 mL of acetic acid was added, shaken well at a wavelength of 550 nm, and the absorbance was measured to, plot the standard curve.
2.4. RNA extraction and sequencing library construction
RNA was extracted from three species of A. cinnamomea mycelium samples, namely YZM, XZM, and NZM, using the TransZol Up Plus RNA Kit (Transgen Biotech, Beijing, China). About 3 μg of each sample was added to the NA 6000 Pico chip (Agilent Bioanalyzer) for RNA identification. The sequencing library was constructed using the NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA). The mRNA was enriched with polyA structure in the total RNA by Oligo (dT) magnetic beads. The RNA fragments with a length of about 300 bp were interrupted by ion interruption. The first strand of cDNA with 6-base random primers and reverse transcriptase was synthesized using RNA as a template. The second strand of cDNA was synthesized using the first strand of cDNA as a template. After the library was constructed, the library fragment enrichment was carried out by PCR amplification, the library size was selected according to the fragment size at 450 bp, and the library was quality checked by an Agilent 2 100 Bioanalyzer. The library was constructed by Pythenos Biotechnology for Paired-end (PE) sequencing using the Illumina HiSeq 2 000 sequencing platform with independent sample sequencing depths of 6 G.
2.5. Transcriptome assembly and functional annotation
The raw reads in the FASTQ format were processed by an internal Perl script that obtained clean reads by filtering the readings containing joints, poly-N, and low-quality reads and calculated the Q20, Q30, and GC contents in the clean reads. Subsequent analyses were performed using high-quality clean reads. The assembly was performed using Trinity software (Http//trinityrnaseq.github.io/), which concatenates the reads with a certain length of overlap into fragments without N into Contigs, and then connects different Contigs from the same transcript to obtain a non-redundant sequence that cannot be extended at both ends. Sequence similarity was performed on Unigene using the sequence comparison method, and NR, GO, KEGG, and evolutionary genealogy of genes (eggNOG) (E-value< 10-5) were used to annotate the function of Unigene of A. cinnamomea.
2.6. Functional analysis of differential genes
The readings mapped to each gene were counted using HTSeq v0.6.1, and the fragments per kilobase of transcript per million mapped fragments (FPKM) were used to calculate the expression level of each transcript in different samples. The gene length and sequencing depth were normalized using FPKM. According to the FPKM value of gene expression, the differential expression of multiple genes between different samples was calculated using DEseq. The threshold was set to qvalue < 0.001 and log2Ratio ≥ 1 to screen the differentially expressed genes (DEGs). GOseq was used for GO enrichment analysis, and the significantly enriched GO pathways were screened. The hypergeometric tests were applied in the KEGG Pathways to detect the pathways with the most significant enrichment of DEGs compared with the assembled transcriptomes’ background.
2.7. Real-time quantitative fluorescence PCR
Specific primers (Table S1) were designed to detect acetyl CoA synthase (ACS13), AACT2, Acetyl-CoA carboxylase (ACC1), acetyl-CoA terpenoid cyclases (AOC), HMG-CoA, MVD, OCS, SQS, SE, and Hydroxymethylglutaryl CoA synthase (HMGS2) genes of A. cinnamomea by fluorescence quantitative PCR using a PCR thermal cycler (ABI 7 300; Applied Biosystems, Foster City, CA, USA) to validate the results of transcriptome sequencing and the expression pattern of target genes. The PCR reaction system conditions were as follows: denaturation procedure (94 °C, 2 min), amplification quantification procedure was repeated 40 times (94 °C, 15 s; 58 °C, 15 s; 72 °C, 45 s), and finally, 72 °C, 10 min. The qPCR analysis was performed for 3 independent biological replicates of each sample and 3 technical replicates of each biological replicate.
3. Results
3.1. Growth rate and biomass
A. cinnamomea mycelia showed different growth rates on different wood substrates. After 30 days of growth on NZM and XZM, the wood was almost completely coated with white hyphae, while YZM was partially coated with white hyphae. A. cinnamomea showed that after 90 days of growth on NZM and XZM, the mycelia began to be white and gradually changed to orange with more bright color. Afterward, the mycelia of YZM became heavy and changed to orange-red color. The mycelia of YZM became thick but partially covered the wood. Compared with YZM, NZM and XZM grew significantly faster (Figure 1). The biomass of A. cinnamomea mycelium growing on YZM was 0.323 g/cm2. The biomass of A. cinnamomea mycelium growing on XZM was the least, accounting for 0.305 g/cm2. A. cinnamomea mycelium grown on NZM was between two i.e., 0.310 g/cm2. The growth rate and biomass of A. cinnamomea mycelium cultured on different wood substrates showed significant differences, indicating that different cultivation conditions affect the A. cinnamomea mycelium growth and secondary metabolites production.
Figure 1.
Growth of mycelia on different wood substrates of Antrodia cinnamomea. (A) NZM 90 d; (B) XZM 90 d; (C) YZM 90 d.
3.2. Triterpene content of A. cinnamomea
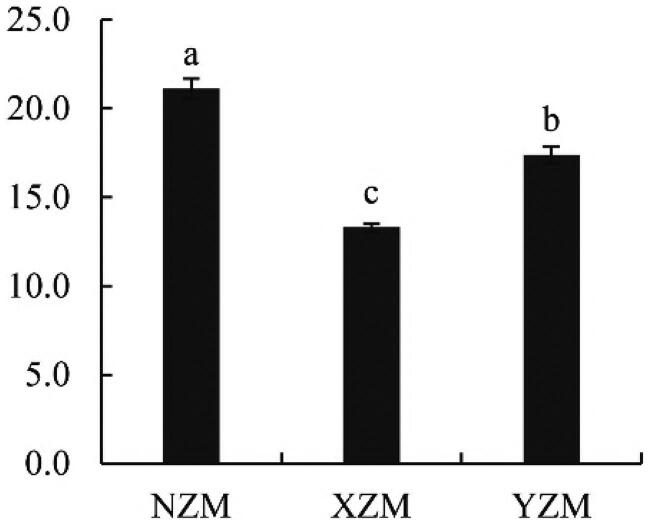
The triterpenes content in A. cinnamomea was determined by vanillin - glacial acetic acid chromogenic method using oleanolic acid as the standard, and the standard curve equation was y = 0.0082x-0.0087 (r = 0.9995). The triterpenes content was calculated, as shown in Figure 2. Triterpenes are the primary active substances in A. cinnamomea. The mycelia of A. cinnamomea grown on three kinds of wood substrates contained different triterpene contents. The contents of NZM, XZM, and YZM triterpenes were 21.1 mg/g, 17.4 mg/g, and 13.3 mg/g. From the perspective of triterpenoid content, NZM triterpenoid content was the highest, followed by XZM and YZM. Different culture methods had significant effects on the triterpenoid contents of A. cinnamomea mycelium and may affect the synthesis of terpenoids to different degrees.
Figure 2.
Triterpene content of Antrodia cinnamomea. Data are expressed as the average of three biological replicates.Different letters indicate significant differences at p < 0.05 (Duncan’s multiple range test).
3.3. Sequencing and assembly of the transcriptome data from Antrodia cinnamomea
Based on the Illumina HiSeq 2 000 high-throughput sequencing platform of Shanghai Personal Biotechnology Co., Ltd., the transcriptome sequencing of A. cinnamomea mycelium cultured on different camphor wood substrates, namely YZM, XZM, and NZM was performed. Each substrate was treated with 3 biological replicates and 3 biological replicates were mixed sequencing (Table S2). A total of 153,448,716 raw reads were obtained. After removing the sequence joint and low-quality reads, 144,241,396 clean reads were obtained, accounting for 93.9% of the raw reads, and the average proportions of Q20 and Q30 were 97.6% and 93.9%, respectively. The sequenced clean data was assembled by splicing using Trinity software (Table 1). A total of 78,729 transcripts, and a total of 18,391 Unites, accounting for 23.4% of the total transcripts, were obtained. The total sequence length was 223,972,512 bp, the average length was 2844.9 bp, and the N50 was 4,463 bp.
Table 1.
De novo assembly of the transcriptome of Antrodia cinnamomea.
| Transcript | Unigene | |
|---|---|---|
| Total Length (bp) | 223972512.00 | 28026308.00 |
| Sequence Number | 78729.00 | 18391.00 |
| Max. Length (bp) | 23996.00 | 23996.00 |
| Mean Length (bp) | 2844.85 | 1523.91 |
| N50 (bp) | 4463.00 | 3502.00 |
| N50 Sequence No. | 15982.00 | 2329.00 |
| N90 (bp) | 1489.00 | 469.00 |
| N90 Sequence No. | 48643.00 | 10761.00 |
| GC% | 51.84 | 51.79 |
N50 (bp): Arrange all the sequences from the longest to the shortest, add the sequence lengths in that order, and when the sum reaches 50% of the total length of the sequence, the length of the last sequence; N90 (bp): Arrange all the sequences from the longest to the shortest, add the sequence lengths in that order, and when the sum reaches 90% of the total length of the sequence, the length of the last sequence; N50 Sequence No.: The total number of sequences with length greater than N50; N90 Sequence No.: The total number of sequences whose length is greater than N90; GC%: GC content of the sequence.
3.4. Functional annotation of the unigenes from A. cinnamomea
The Unigenes obtained by assembling the transcriptome of A. cinnamomea were functionally annotated with the NR, GO, KEGG, and eggNOG databases, the gene information was comprehensively obtained, and the annotations of each database were counted (Table S3). The NR and eggNOG databases showed the most successful Unigenes of about 11,435 and 10,539, respectively, accounting for 62.18% and 57.31% of the assembled transcripts. Of which, 6,947 (37.77%) transcripts were annotated with the GO database, and 5,994 (32.59%) transcripts were annotated with the KEGG database.
The Unigene sequences were compared with the NR database using BLAST, and 11,435 Unigenes were recorded in the NR database, accounting for 64.45% of the total Unigenes. The results of matching annotation by species are depicted in Figure S1A. The Unigenes sequenced and assembled by A. cinnamomea showed similarity with other sequences at 6,094 or 53.3%, presumably containing its own gene sequences different from most species. It was followed by Fibroporia radiculosa with 1,662 Unigenes at 14.54%, and Laetiporus sulphureus with 1,506 Unigenes at 13.17%, with the remaining nearly 1/5 of Unigene distributed in more than 2,171 other species. The distribution of matching sequence similarities (Figure S1B) showed that the sequences with 60% to 80% similarities accounted for 37.88%, the sequences with 40% to 60% similarity accounted for 27.63%, 20.72% similarity> 80% to 95%, and 9.95% similarity <40%. It can be seen from the E-value distribution (Figure S1C) that in the annotated Unigenes, 1/3 (29.69%) of the E-values were between 0 and 1e-100, 16.01% were between 1e-100 and 1e-60, and 16.54% were at e = 0. The similarity of matches and E-value distribution results showed that A. cinnamomea had a high degree of matching with the NR library of NCBI.
The GO classification is an international standardized classification system applicable to various species and can define and describe the genes. The GO functional analysis of the Unigene of A. cinnamomea mycelium was performed. About 6,947 Unigenes were annotated and divided into 67 branches in three broad categories: biological processes, cell composition, and molecular function (Figure S2). The number of genes in each class showed that in the biological process, the metabolic process, cellular process, and single-organism process were higher, and the cell, cell part, organelle, and membrane accounted for a relatively higher proportion of the cell components. The molecular function classification was abundant in catalyst activity and binding.
The KEGG database can be used to classify the genes according to the pathways involved or their functions and analyze the metabolic pathways of genes in the cells and their functions. A total of 5,994 Unigenes were annotated with the KEGG database for metabolic classification analysis. They were classified into 5 categories and 35 subclasses (Figure 3). The results showed that among the five categories, the metabolism-related pathways accounted for the largest proportion (41.83%), which could be subdivided into 13 subcategories, mainly carbohydrate metabolism, overview, energy metabolism, amino acid metabolism, and lipid metabolism. It was followed by the Organic Systems-related pathways, accounting for 17.09%, which could be subdivided into 10 subcategories, dominated by the immune system and the endocrine system. In contrast, the Environmental Information Processing, Cellular Processes, and Genetic Information Processing-related pathways accounted for a relatively small proportion, including only 3, 4, and 5 subcategories, respectively. The combined results of GO and KEGG database annotation revealed that A. cinnamomea had active metabolic activity and synthesized various metabolites.
Figure 3.
The KEGG metabolic pathway map of Antrodia cinnamomea Unigene transcriptome.
The genes were annotated with eggNOG comparisons, and the eggNOG number of the best comparison results was assigned to the corresponding genes. A total of 10,539 Unigenes (Figure S3) were assembled from the transcriptome sequencing results of A. cinnamomea: of which, the gene with the unknown function was the largest (2,622), accounting for 19.48%, which was speculated to be the specific gene of A. cinnamomea. It was followed by General function prediction only, accounting for 15.37%, with 2,068 Unigenes, Posttranslational modification, protein turnover, chaperones, and Energy production, and conversion classes with 1006 and 795, accounting for 7.9% and 5.9%, respectively. Additionally, 590 (4.38%) Unigenes participated in the synthesis, transport, and catabolism of secondary metabolites. Further analysis of the function of these genes helped to clarify the molecular mechanisms of the biosynthesis of these potent components of A. cinnamomea. However, only 9 Unigenes were associated with Cell motility and only 10 with Extracellular structures. These results demonstrated that the Unigenes covered various transcripts of the A. cinnamomea genome.
3.5. Differential expression analysis of terpene metabolism
Based on the KEGG terpene metabolic pathway depicted in Venn (Figure 4), the differential expression analysis was performed using the mycelium database of A. cinnamomea, and the transcripts of adjusted P-value < 0.05 and multiple differences (log2FC) >1 were designated as the transcripts with significant differential expression. In the three comparison groups, about 1,387 differential genes were identified, and the mycelium of A. cinnamomea cultured on XZM and NZM showed a total of 497 differentially expressed transcripts. Of which, 346 were upregulated, and 151 were down-regulated. Similarly, the number of mycelia cultured on YNM and NZM was 572, with 389 upregulated transcripts and 183 down-regulated transcripts. The number of mycelia cultured on YNM and XZM showed a total of 318 differentially expressed transcripts of A. cinnamomea; of which, 164 transcripts were upregulated and 154 were down-regulated, showing 3 upregulated differential genes and 3 downregulated differential genes in these 3 groups. This result indicated that the cultivation of A. cinnamomea on different wood substrates could regulate the mycelial gene expression and increase the number of upregulated DEGs under different culture conditions. These DEGs might be the key factors affecting the terpenoid genes of A. cinnamomea grown on different wood substrates.
Figure 4.
Venn diagram of differentially expressed genes in the transcriptome of Antrodia cinnamomea. The sum of the numbers in each circle represents the total number of differential genes in the comparison combination, and the overlap of the circles represents the common differential genes between the two comparison groups. (A)Venn diagrams showing the up-regulated DEGs; (B)Venn diagrams showing the down-regulated DEGs.
3.6. Biosynthesis of terpenoids and related genes in the A. cinnamomea mycelia
According to the molecular pathway of terpenoid biosynthesis, 37 Unigenes (Table S4) of the synthetic MVA pathway were identified in the transcriptome data of A. cinnamomea; of which, the number of DEGs on NZM was significantly higher than XZM and YZM. This result indicated that NZM had a greater influence on the terpene metabolic pathway of A. cinnamomea mycelium. The expression profiles of terpene-related transcripts of A. cinnamomea were obtained according to the differential expression patterns of the terpenoid metabolic pathways of A. cinnamomea grown on three species of wood substrates, and the expression interaction heat map was plotted (Figure 5).
Figure 5.
Interactive heat map of transcriptome expression of terpenoid biosynthetases encoded by Antrodia cinnamomea transcriptome. Each column represents an experimental sample (NZM, XZM, and YZM) and each row represents a gene. High expression levels are shown in red. Low expression levels are shown in blue.
According to the annotation results, the synthesis pathway of terpenoids could be divided into three stages: synthesis of terpenoid precursors, sesquiterpene synthesis pathway, and synthesis of the triterpenoid skeleton. The enzymes associated with the biosynthesis of precursor substances in the upstream stage include AACT of the MVA pathway, HMGS, HMGR, mevalonate kinase (MVK), and IDI. The differential expression analysis showed that the expression of ACS4 gene in the precursor pathway on YZM was higher than that of NZM and XZM, indicating that both were involved in the transcriptional regulation of YZM. The expression of AACT2, MCAD, HMG-CoA, MVD, and IDI in NZM was significantly higher than that of the other two wood substrates, and the MVA pathway gene was highly expressed on NZM, which might promote mycelial terpenoids biosynthesis.
The midstream stage was passed by geranyl pyrophosphate synthase (GPPS), farnesyl diphosphate synthase (FPPS), geranylgeranyl pyrophosphate synthase (GGPPS), squalene synthase (SS), and squalene epoxidase (SE), and the formation of α - amyrin was catalyzed by amyrin synthase (AS). The results showed that the expression of GGT1 on YZM was significantly higher than that of NZM and XZM, and the expression of FTase2 on XZM was higher, while the expressions of OCS, SQS, and SE were higher on NZM than that of XZM and YZM. The expression of TPS was higher on these three wood substrates. The expression level of YZM was 3 times higher than that of NZM. Under the culture conditions of NZM, the expression of FTase2 was down-regulated, and the expression of TPS, SQS, OCS, and SE was upregulated. Therefore, it was speculated that the sesquiterpene and triterpenes biosynthesis were positively regulated under NZM. Meanwhile, the expression of MVD was upregulated under the culture conditions of NZM, which is a key enzyme in the MVA pathway, indicating that NZM promoted the positive regulation of terpenoids in A. cinnamomea mycelium.
3.7. Validation of the RNA-Seq gene expression data by QRT-PCR
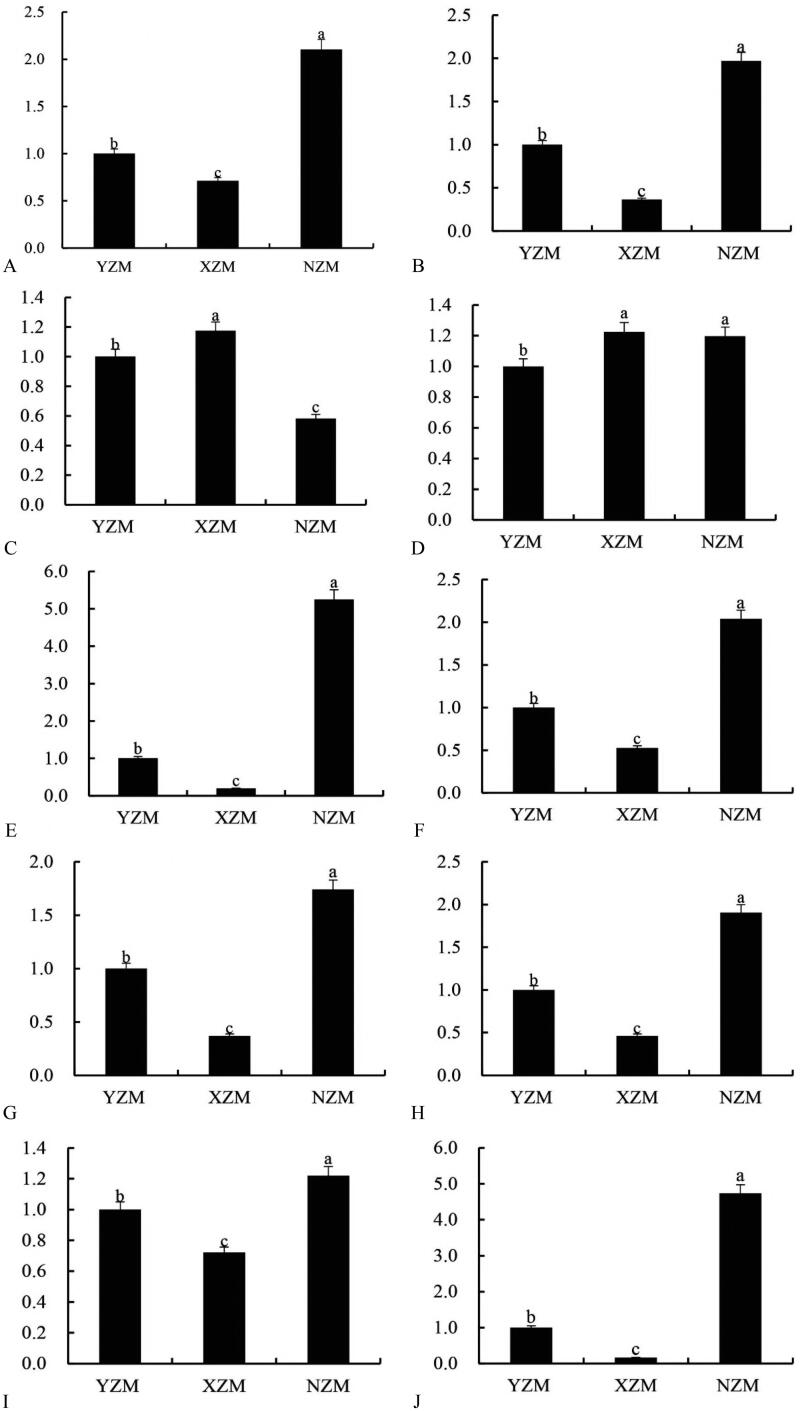
The specific expression of terpene metabolism-related genes on NZM, XZM, and YZM was analyzed using QRT-PCR (Figure 6). The results showed that the relative expression of 10 genes was consistent with the transcriptome expression profiling trend. Compared with YZM, the expression of ACS13, AACT2, AOC, HMG-CoA, MVD, OCS, SQS, SE, and HMGS2 in the mycelium cultured on NZM was up-regulated, indicating that NZM was involved in the positive regulation of terpenoid synthesis. However, under XZM culture, the expression of ACS13, AACT2, HMG-CoA, MVD, OCS, SQS, SE, and HMGS2 was down-regulated, which might inhibit the transcriptional regulation of XZM. Overall, the terpenoids were found in the mycelium of A. cinnamomea cultured on NZM, and multiple key response genes in the terpenoid pathway were involved in the regulation at a transcriptional level.
Figure 6.
Relative expression of differentially expressed genes by qRT-PCR. (A) ACS13; (B) AACT2; (C) ACC1; (D) AOC; (E) HMG-CoA; (F) MVD; (G) OCS; (H) SQS; (I) SE; (J) HMGS2; qRT-PCR: real-time quantitative reverse transcription PCR. Error bars indicate the standard deviation of three independent biological replicates. Different letters indicate significant differences at p < 0.05.
4. Discussion
The RNA-Seq high-throughput sequencing is a rapid technology to interpret the non-model organism gene information, discover new genes, and study the gene functions. Besides, it is effective in analyzing the differently expressed genes between species and new gene function tests due to its uniform read order, coverage, and comparable data quality [23]. So far, many studies have successively applied transcriptome sequencing techniques to analyze the gene function annotation and metabolic pathways of Coprinus cinereus [24], Pleurotus ostreatus [25,26], Ganoderma lucidum [27], Lentinula edodes [28], Flammulina velutipes [29], A.cinnamomea [15] and other fungi. Yu et al. [30] obtained many functional genes in the mycelium of Ganoderma lucidum through transcriptome sequencing. Huang et al. [31] identified the genes associated with sterol biosynthesis in the mycelium of Phellinus linteus through transcriptome sequencing. Plaza et al. [32] sequenced the transcriptomes of the vegetative hyphae and protoplasmic stages of C. cinereus and found a conservative transcriptional circuit in the transcription loop during the fruiting body formation, containing a protein upregulation with the Velvet domain.
The fungi have potential drug-active constituents with diverse and unique secondary metabolites. As such, analyzing the changes in terpenoid metabolic genes could provide insights into the key synthetic genes and associated metabolites of A.cinnamomea [33]. Accumulating studies have reported that different culture media affect the metabolism of A. cinnamomea, including its compounds and genes. Meng et al. added various vegetable oils to the fermentation broth of A. cinnamomea to improve the growth of fungi and the production of terpenoids and found that the biomass and triterpenoid content increased with the addition of corn oil, while the triterpenoids content increased by four folds compared with the blank control. Furthermore, the transcriptome difference analysis showed that the corn oil was significantly enriched in several metabolic pathways, including glycolysis/gluconeogenesis, propionic acid metabolism, and transmembrane hydrophobic protein [34]. In addition, Antro Quinonol was synthesized by adding coenzyme Q and p-hydroxybenzoic acid to A. cinnamomea mycelium in liquid fermentation [35]. Researchers have also found that different camphor plant varieties could promote the growth of the hyphae of A. cinnamomea to varying degrees [36]. Lin et al. [22] reported that the metabolites of A. cinnamomea cultured on C. kanehirae wood substrates were more abundant and distinctly different from those cultured on other wood substrates, such as ergostane and lanostane triterpenes. As the only natural host of A. cinnamomea, camphor trees contain special metabolite components not possessed by other camphor plants. However, the camphor plants that can be cultured on wood substrates should contain one or more common A. cinnamomea growth-promoting substances.
In this study, the differential expression of genes of A. cinnamomea mycelium and the dynamic changes in the terpene metabolite contents under different camphor culture conditions were explored, and a total of 78,729 transcripts were obtained by assembling on the Illumina HiSeq 2 000 Transcriptome High-Throughput Sequencing Platform. Unigenes had 18,391, and N50 had 4,463 bp. The Unigenes obtained from the assembly of A. cinnamomea transcriptome with the NR, GO, KEGG, and eggNOG databases were annotated and classified using the BLAST algorithm. GO function was divided into 67 branches and three categories: biological processes, cell composition, and molecular function; of which, metabolic process, cellular process, cell, cell part, catalytic activity, and binding were more concentrated. Lee et al. [37] identified the differentially expressed genes related to terpenoid metabolism in Polyporus brumalis, and the GO enrichment analysis results showed that the Unigenes of mycelium were divided into 46 functional groups, mainly biological processes, cellular components, and molecular functions, which were significantly enriched in the membranes, organelles, catalytic activities, and cellular metabolic processes. These results were consistent with the present study results. A total of 5,994 Unigenes were obtained through the KEGG metabolic classification analysis; of which, the metabolism-related pathways accounted for the largest proportion (41.83%). Similarly, in the studies of Chaga Bacterium, the Unigenes and the biosynthesis of secondary metabolites pathways were significantly enriched through the KEGG pathway [38], suggesting that the related genes in both pathways could maintain the mycelial metabolic activity.
The analysis of transcriptome-related genes of A.cinnamomea cultured on different wood substrates showed that the expression of OCS, SQS, and SE was higher on NZM compared to XZM and YZM. Squalene is the common precursor of all triterpenoids. SQS catalyzes the synthesis of squalene from two molecules of farnesyl pyrophosphate. It is the first key enzyme in the triterpenoid synthesis pathway. The concentration and activity in vivo determine the yield of subsequent products [39,40]. Kim et al. [41] reported that the upregulated expression of SQS in Panax ginseng significantly improved the biosynthesis of triterpenoids and sterols. The expression of SQS2 during the synthesis of terpenes in I. obliquus significantly increased with the addition of substrates, indicating its involvement in triterpenoid production [42]. The expressions of OCS, SQS, and SE in A. cinnamomea were higher under the culture conditions of NZM, indicating that the NZM culture method positively regulated triterpene biosynthesis. This result verified that camphor plants, as the only natural host of A. cinnamomea, were more suitable for the growth of A. cinnamomea than other species.
In this study, the mycelium of A. cinnamomea angustifolia was selected as the research object, and the dynamic changes of terpene metabolite contents in A. cinnamomea under different wood cultures conditions were analyzed using transcriptome sequencing and qRT-PCR. The results demonstrated that different wood substrates showed different terpenoid gene expressions, thus affecting the content of triterpenoids. This result indicates that different culture conditions have a significant impact on the growth of fungi and secondary metabolites synthesis. Additionally, the key genes involved in the terpene metabolism of A. cinnamomea angustifolia sinensis were discovered, which provided a reference for the genome research of A. cinnamomea angustifolia and other medicinal fungi. In the future, we will use the technology of compound separation and purification to separate and purify the compounds induced by A. cinnamomea under different wood cultures conditions, and further study the composition and content differences of terpenoid metabolites.
Supplementary Material
Funding Statement
The authors are grateful for the support of the National Natural Science Foundation of China [No. 32160736, 31860177]; Major Project of the Agricultural Basic Research Program in Yunnan Province [202101BD070001-020]; General Project of the Basic Research Program in Yunnan Province [202101AT070218, 202101AT070044]; the Reserve Talents for Young and Middle-aged Academic and Technical Leaders of the Yunnan Province [202205AC160044]; Yunnan Key Laboratory for Fungal Diversity and Green Development [E03A311261-3].
Author contributions
Jiaojiao Chen, Zhang zhang and Yi Wang completed the experiment, analyzed the experimental results, and wrote the main part of the manuscript. Zhang Zhang wrote a smaller part and revised it. Xiaolong Yuan, Juan Wang, and Yuming Yang revised and proofread the manuscript. Yi Wang and Yuan Zheng conceived the experiment and were responsible for the implementation of the project. All authors have read and approved the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Zhang BB, Guan YY, Hu PF, et al. Production of bioactive metabolites by submerged fermentation of the medicinal mushroom Antrodia cinnamomea: recent advances and future development. Crit Rev Biotechnol. 2019;39(4):541–554. [DOI] [PubMed] [Google Scholar]
- 2.Chen CY, Chien SC, Tsao NW, et al. Metabolite Profiling and comparison of bioactivity in Antrodia cinnamomea and Antrodia salmonea fruiting bodies. Planta Med. 2016;82(3):244–249. [DOI] [PubMed] [Google Scholar]
- 3.Cha WS, Ding JL, Choi DB.. Comparative evaluation of antioxidant, nitrite scavenging, and antitumor effects of Antrodia camphorata extract. Biotechnol Bioproc E. 2009;14(2):232–237. [Google Scholar]
- 4.Mau JL, Huang PN, Huang SJ, et al. Antioxidant properties of methanolic extracts from two kinds of Antrodia camphorata mycelia. Food Chem. 2004;86(1):25–31. [Google Scholar]
- 5.Geethangili M, Tzeng YM.. Review of pharmacological effects of Antrodia camphorata and its bioactive compounds. Evid Based Complement Alternative Med. 2011;2011:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganesan N, Baskaran R, Velmurugan BK, et al. Antrodia cinnamomea—an updated minireview of its bioactive components and biological activity. J Food Biochem. 2019;43(8):e12936. [DOI] [PubMed] [Google Scholar]
- 7.Yeh CT, Rao YK, Yao CJ, et al. Cytotoxic triterpenes from Antrodia camphorata and their mode of action in HT-29 human Colon cancer cells. Cancer Lett. 2009;285(1):73–79. [DOI] [PubMed] [Google Scholar]
- 8.Toyomasu T. Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci Biotechnol Biochem. 2008;72(5):1168–1175. [DOI] [PubMed] [Google Scholar]
- 9.Hsiao G, Shen MY, Lin KH, et al. Antioxidative and hepatoprotective effects of Antrodia camphorata extract. J Agric Food Chem. 2003;51(11):3302–3308. [DOI] [PubMed] [Google Scholar]
- 10.Joshi RA. Antrodia camphorata with potential anticancerous activities: a review. Journal of Medicinal Plants Studies. 2017;5(1):284–291. [Google Scholar]
- 11.Qiao X, Wang Q, Ji S, et al. Metabolites identification and multi-component pharmacokinetics of ergostane and lanostane triterpenoids in the anticancer mushroom Antrodia cinnamomea. J Pharm Biomed Anal. 2015;111:266–276. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Wang Y, Yuan XL, et al. Effects of culture mechanism of Cinnamomum kanehirae and C. camphora on the expression of genes related to terpene biosynthesis in Antrodia cinnamomea. Mycobiology. 2022;50(2):121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang LP, Liang J, Xie X, et al. Direct formation of the sesquiterpeonid ether liguloxide by a terpene synthase in senecio scandens. Plant Mol Biol. 2021;105(1-2):55–64. [DOI] [PubMed] [Google Scholar]
- 14.Mitsuhashi T, Abe I.. Chimeric terpene synthases possessing both terpene cyclization and prenyltransfer activities. Chem Bio Chem. 2018;19(11):1106–1114. [DOI] [PubMed] [Google Scholar]
- 15.Lu MYJ, Fan WL, Wang WF, et al. Genomic and transcriptomic analyses of the medicinal fungus Antrodia cinnamomea for its metabolite biosynthesis and sexual development. Proc Natl Acad Sci U S A. 2014;111(44):E4743–E4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu CJ, Chiang CC, Chiang BH.. The elicited two-stage submerged cultivation of Antrodia cinnamomea for enhancing triterpenoids production and antitumor activity. Biochem Eng J. 2012;64:48–54. [Google Scholar]
- 17.Ma TW, Lai Y, Yang FC.. Enhanced production of triterpenoid in submerged cultures of Antrodia cinnamomea with the addition of citrus peel extract. Bioprocess Biosyst Eng. 2014;37(11):2251–2261. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, Jia W, Zhang J, et al. GC-MS and GC-Olfactometry analysis of aroma compounds extracted from culture fluids of Antrodia camphorate. World J Microbiol Biotechnol. 2008;24(8):1599–1602. [Google Scholar]
- 19.Wang X, Jia LH, Wang MD, et al. The complete mitochondrial genome of medicinal fungus Taiwanofungus camphoratus reveals gene rearrangements and intron dynamics of polyporales. Sci Rep. 2020;10(1):16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang FC, Yang YH, Lu HC.. Enhanced antioxidant and antitumor activities of Antrodia cinnamomea cultured with cereal substrates in solid state fermentation. Biochem Eng J. 2013;78:108–113. [Google Scholar]
- 21.Lin LT, Tai CJ, Su CH, et al. The Ethanolic extract of Taiwanofungus camphoratus (Antrodia camphorata) induces cell cycle arrest and enhances cytotoxicity of cisplatin and doxorubicin on human hepatocellular carcinoma cells. Biomed Res Int. 2015;2015:415269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin TY, Chen CY, Chien SC, et al. Metabolite profiles for Antrodia cinnamomea fruiting bodies harvested at different culture ages and from different wood substrates. J Agric Food Chem. 2011;59(14):7626–7635. [DOI] [PubMed] [Google Scholar]
- 23.Bastos DZL, Pimentel IC, De Jesus DA, et al. Biotransformation of betulinic and betulonic acids by fungi. Phytochemistry. 2007;68(6):834–839. [DOI] [PubMed] [Google Scholar]
- 24.Ohm RA, De Jong JF, Lugones LG, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28(9):957–963. [DOI] [PubMed] [Google Scholar]
- 25.Riley R, Salamov AA, Brown DW, et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A. 2014;111(27):9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castanera R, Lopez-Varas L, Borgognone A, et al. Transposable elements versus the fungalgenome: impact on whole-genome architecture and transcriptional profiles. PLoS Genet. 2016;12(6):el006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen SL, Xu J, Liu C, et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat Commun. 2012;3(3):913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen LF, Gong YH, Cai YL, et al. Genome sequence of the edible cultivated mushroom Lentinula edodes (shiitake) reveals insights into lignocellulose degradation. PLoS One. 2016;11(8):e0160336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, Liu F, Jiang YJ, et al. The multigene family of fungal laccases and their expression in the white rot basidiomycete Flammulina velutipes. Gene. 2015;563(2):142–149. [DOI] [PubMed] [Google Scholar]
- 30.Yu GJ, Wang M, Huang J, et al. Deep insight into the Ganoderma lucidum by comprehensive analysis of its transcriptome. PLoS One. 2012;7(8):e44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y, Wu XQ, Jian D, et al. De novo transcriptome analysis of a medicinal fungi Phellinus linteus and identification of SSR markers. Biotechnol Biotechnol Equip. 2015;29(2):395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plaza DF, Lin CW, van der Velden NSJ, et al. Comparative transcriptomics of the model mushroom Coprinopsis cinerea reveals tissue-specific armories and a conserved circuitry for sexual development. BMC Genomics. 2014;15(1):492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li ZW, Kuang Y, Tang SN, et al. Hepatoprotective activities of Antrodia camphorata and its triterpenoid compounds against CCl4-induced liver injury in mice. J Ethnopharmacol. 2017;206:31–39. [DOI] [PubMed] [Google Scholar]
- 34.Meng LH, Luo BB, Yang Y, et al. Addition of vegetable oil to improve triterpenoids production in liquid fermentation of medicinal fungus Antrodia cinnamomea. JoF. 2021;7(11):926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu YD, Zhang H, Lu RQ, et al. Enabling the biosynthesis of antroquinonol in submerged fermentation of Antrodia camphorate. Biochem Eng J. 2014;91:157–162. [Google Scholar]
- 36.Lu MC, El-Shazly M, Wu TY, et al. Recent research and development of Antrodia cinnamomea. Pharmacol Ther. 2013;139(2):124–156. [DOI] [PubMed] [Google Scholar]
- 37.Lee SY, Kim M, Kim SH, et al. Transcriptomic analysis of the white rot fungus Polyporus brumalis provides insight into sesquiterpene biosynthesis. Microbiol Res. 2016;182:141–149. [DOI] [PubMed] [Google Scholar]
- 38.Zou L, Sun TT, Li TL, et al. De novo transcriptome analysis of inonotus baumii by RNA-seq. J Biosci Bioeng. 2016;121(4):380–384. [DOI] [PubMed] [Google Scholar]
- 39.Haralampidis K, Trojanowska M, Osbourn AE.. Biosynthesis of triterpenoid saponins in plants. Advances in Biochemical Engineering. 2002;75(2):31–49. [DOI] [PubMed] [Google Scholar]
- 40.Lee MH, Jeong JH, Seo JW, et al. Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol. 2004;45(8):976–984. [DOI] [PubMed] [Google Scholar]
- 41.Kim TD, Han JY, Huh GH, et al. Expression and functional characterization of three squalene synthase genes associated with saponin biosynthesis in Panax ginseng. Plant Cell Physiol. 2011;52(1):125–137. [DOI] [PubMed] [Google Scholar]
- 42.Fradj N, Santos KCGD, Montigny ND, et al. RNA-Seq de novo assembly and differential transcriptome analysis of chaga (Inonotus obliquus) cultured with different betulin sources and the regulation of genes involved in terpenoid biosynthesis. IJMS. 2019;20(18):4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.