Abstract
The advent of multimodal brain atlases promises to accelerate progress in neuroscience by allowing in silico queries of neuron morphology, connectivity, and gene expression. We used multiplexed fluorescent in situ RNA hybridization chain reaction (HCR) technology to generate expression maps across the larval zebrafish brain for a growing set of marker genes. The data were registered to the Max Planck Zebrafish Brain (mapzebrain) atlas, thus allowing covisualization of gene expression, single-neuron tracings, and expertly curated anatomical segmentations. Using post hoc HCR labeling of the immediate early gene cfos, we mapped responses to prey stimuli and food ingestion across the brain of freely swimming larvae. This unbiased approach revealed, in addition to previously described visual and motor areas, a cluster of neurons in the secondary gustatory nucleus, which express the marker calb2a, as well as a specific neuropeptide Y receptor, and project to the hypothalamus. This discovery exemplifies the power of this new atlas resource for zebrafish neurobiology.
A gene expression atlas links development, structure, and function in the larval zebrafish brain.
INTRODUCTION
Development and function of neural circuits depend on the spatiotemporal dynamics of differential gene expression across the brain (1, 2). Particular classes of genes play prominent roles in organizing the layout of the nervous system and may serve as markers of cell-type identity and/or function. These include genes whose protein products directly influence cell-cell interactions and local properties of the synaptic network and those that regulate the expression of other genes to determine neuronal cell fate, diversification, morphology, and connectivity (3–6). Often the corresponding RNAs are expressed in a region- and cell type-specific fashion. Another class of important marker genes encode functional determinants, e.g., neurotransmitter-synthesizing enzymes, ion channels, synaptic machinery components, and neuropeptides; the presence of the corresponding proteins provides information about physiological properties of the expressing neurons and tends to be not cell-type specific (except for neuropeptides). Visualizing the expression of these markers in an atlas format offers a glimpse into the cellular constituents of neural circuitry and their molecular endowment (5, 6). Such an atlas then allows researchers to select markers for labeling of neurons of interest and genetic handles for their manipulation. Moreover, covisualizing many of these markers offers a holistic view of the genetic architecture of the brain and may reveal general organizing principles.
The most prominent attempt at visualizing brain-wide transcript expression, while preserving spatial layout of the tissue, is probably the Allen Brain Atlas project, which published the results of RNA in situ hybridizations (ISHs) for thousands of genes in tissue sections, first in mouse (5) and later in human (6). These pioneering studies, together with both prior and more recent work with similar aims [e.g., (7–9)], are of immense value for the research community. The resulting resources are routinely consulted by experimenters seeking to corroborate hypotheses and identify genetic markers for neuronal subpopulations of interest.
Atlases of the nervous system have also been devised for other model organisms, although, to our knowledge, not yet for RNA ISH patterns to faithfully map the endogenous gene expression. The nervous system of the nematode worm Caenorhabditis elegans has been charted in unsurpassed detail (10). Most recently, a strain of C. elegans has been developed by genetic engineering in which each of its 302 neurons is identifiable in one animal by a combination of fluorescent proteins (11). This multicolor atlas enables in vivo interrogation of the spatial arrangement of identifiable neurons and serves as a convenient readout of the effects of developmental perturbations on cell fate and cell positioning (11). In the fruitfly Drosophila melanogaster, the “virtual fly brain browser” (12) and the “fruit fly brain observatory” (13) offer standardized neuroanatomical frames in which thousands of transgene expression patterns are coregistered, generating a light-microscopic mesoscale connectome.
In the zebrafish Danio rerio, histological brain atlases have been available for some time [adult: (14, 15); embryo/larva: (16)], as well as atlas databases of hundreds of transgenic lines and immunostainings, such as “ViBE-Z” (17), “Z-brain” (18), and the “Zebrafish Brain Browser” (19). The Max Planck Zebrafish Brain (“mapzebrain”) atlas (20) at https://mapzebrain.org is a multimodal digital atlas of the 6-day postfertilization (dpf) larval brain, integrating data portals for transgenic lines, histological stains, and expertly curated neuroanatomical regions, as well as a growing number (currently ca. 4300) of single-neuron morphologies. These datasets are all registered into the same spatial coordinate system, the so-called standard brain (20). The mapzebrain atlas has also been cross-linked to Z-brain and the Zebrafish Brain Browser to enable data transfer between the different resources (20). Its Web interface offers refined visualization and animation tools, as well as options for downloading original or processed data for offline analyses.
Here, we set out to add RNA fluorescent in situ hybridization (FISH) patterns to the mapzebrain atlas. Single-cell resolution was achieved by hybridization chain reaction (HCR), a spatial transcriptomic method that is highly sensitive, specific, and multiplexable, allowing visualization of several RNA species in one specimen with different probe-specific fluorophores (21). Moreover, as a powerful tool for recording activity hotspots across the brain, we have devised an HCR FISH protocol for detecting cfos RNA following various sensory stimulations and behavioral tasks. All the data are publicly available and free to download through the mapzebrain website (https://mapzebrain.org/). This new resource, which can be straightforwardly expanded in the future by community contributions, offers convenient Web-based access to markers of neuronal subpopulations and paves the way for a genetic analysis of circuit development and structure in this widely used vertebrate model.
RESULTS
Pipeline for HCR in situ labeling and registration to the mapzebrain atlas
Recent developments of FISH techniques, such as RNAscope, MERFISH, and HCR (9, 21, 22), offer single RNA molecule detection and multiplexing capabilities. We chose HCR, because the staining protocol is easily implemented in larval and juvenile fish (21, 23) and is suitable for registration into a common three-dimensional (3D) reference (23, 24). HCR technology enables multiplexed FISH of up to five probe sets per round of staining with different fluorophores (21). To scale up the number of RNA species mapped, we tested two approaches: either multiround HCR FISH by stripping the HCR signal by deoxyribonuclease I treatment and restaining of the same sample (24, 25) or labeling of different genes in different specimens and computational registering into a common coordinate system. Although the former strategy gives theoretically unlimited information about coexpression in individual cells, we decided against it mainly for two reasons: First, we noticed that iterative rounds of stripping and relabeling, together with repeated agarose embedding for imaging, lead to a reduced fluorescence of the reference channel [Tg(elavl3:H2b-GCaMP6s); see below] and a weaker in situ staining. Second, the initial set of markers is, of course, not final, as new markers or better probes are certain to become available in the future. Thus, to allow for seamless addition of new data to the atlas, we chose to generate expression maps from single rounds of HCR and computational coregistration into a standard brain.
To enable coalignment of different brains, we carried out HCR FISH stainings in the background of the Tg(elavl3:H2b-GCaMP6s) transgenic line, in which nuclear-localized GCaMP6s is expressed in almost all neurons. We generated an averaged brain template of the Tg(elavl3:H2b-GCaMP6s) line using the Advanced Normalization Tools (ANTs) toolkit (fig. S1) (26) and calibrated a registration protocol (see Methods; movie S1). We then acquired FISH data for 290 genes (table S1), which were previously identified as distinct, often cell type–specific, markers in retina, tectum, and diencephalon (27–29). This initial set of markers includes genes encoding transcription factors, cell surface and secreted molecules, enzymes involved in transmitter synthesis and transport, neuropeptides, calcium-binding proteins, and other factors (Fig. 1 and table S1).
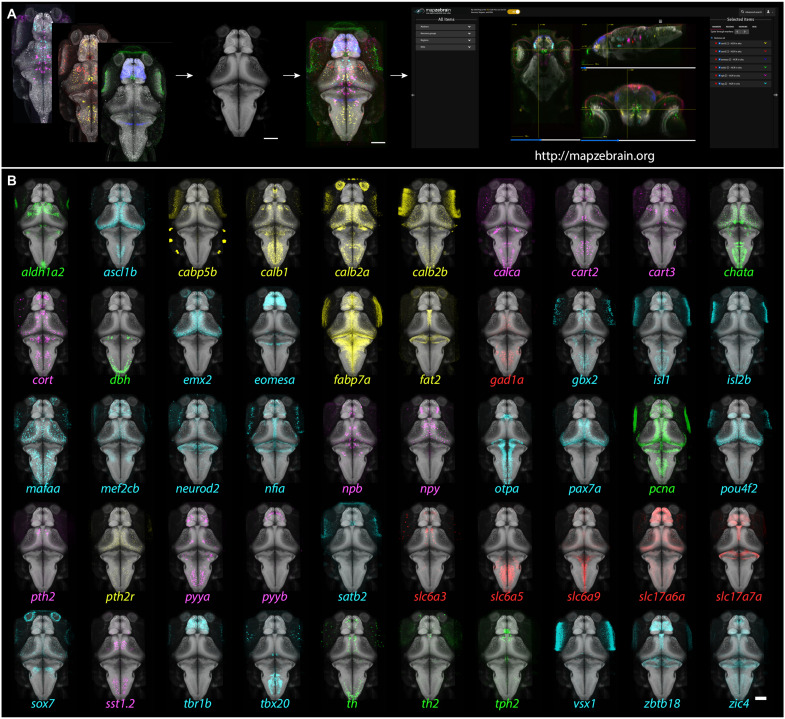
Fig. 1. Gene expression atlas of the larval zebrafish brain: Pipeline and example patterns.
(A) Multiplexed FISH was performed on 6-dpf Tg(elavl3:H2b-GCaMP6s) fish. The GCaMP6s signal was used to align the images to the reference brain, and the aligned images were then uploaded to the mapzebrain atlas. Scale bar, 100 μm. (B) 3D projections of 50 selected FISH images, registered into the standard brain. Color coding: green, enzymes; cyan, transcription factors; red, neurotransmitter synthesizing enzyme (GABAergic) and transmitter transporters (glutamatergic); magenta, neuropeptides; yellow, miscellaneous other. Scale bar, 100 μm.
For each marker gene, we labeled and imaged three 6-dpf fish, which we registered to the standard brain and averaged to ensure we are mapping a consistent expression signal among animals (Fig. 1; see Methods). We multiplexed two genes per sample (table S1) and acquired the confocal data at single-cell resolution, revealing the number of cells expressing a particular gene per region of interest (ROI), as well as any overlapping expression (Fig. 2). The confocal images were acquired from dorsal to ventral; this resulted in a reduced signal in the deep ventral parts due to light scattering. We therefore implemented a signal enhancement visualization option at mapzebrain.org to better explore the ventral expression patterns. All the expression patterns of individual marker genes, including those in individual and averaged brains, are publicly accessible through the latest version of mapzebrain at http://mapzebrain.org/ (Fig. 1).
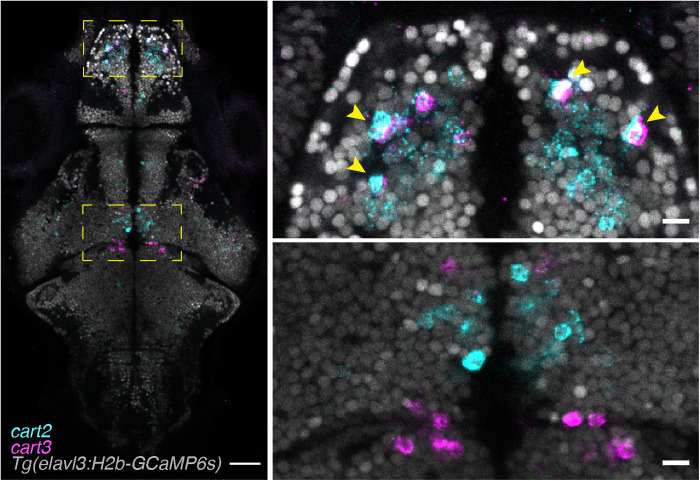
Fig. 2. Multiplexed HCR in situ labeling of single cells.
Example of a single focal plane of a multiplexed HCR labeled fish. The endogenous expression of Tg(elavl3:H2b-GCaMP6s) labels neuronal nuclei and can be used to segment single neurons and to identify whether genes (here cart2 and cart3) are coexpressed (top right panel, arrowheads) or not (bottom right panel). Scale bars, 100 μm (left) and 10 μm (right).
FISH image coregistration with other mapzebrain data modalities
Coregistration of the new FISH dataset with existing mapzebrain datasets frequently revealed spatial overlaps. For example, after registration, the expression of aldh1a2 partially overlapped with the pattern of the transgenic gene trap line mpn321Gt in the pretectum (fig. S2). To verify this in silico overlap of registered data acquired by different procedures (i.e., live imaging, immunostaining, and FISH), we carried out aldh1a2 FISH labeling in larvae expressing the mpn321Gt transgene and indeed found coexpression of the two markers in individual cells in the pretectum (fig. S2). This finding highlights the potential to identify and confirm marker genes for cell populations of interest via rapid FISH labeling and demonstrates the robustness and reproducibility of our digital image registration pipeline across samples and data acquisition procedures.
Generating post hoc cfos brain activity maps of freely swimming fish
To expand the capabilities of our gene expression resource, we challenged the fish larvae in sensory or behavioral paradigms, for a total of 40 min, and carried out post hoc FISH staining of RNA from the immediate early gene cfos (Fig. 3, fig. S3, and movie S2). The cfos gene, also known as fosab, is expressed in neurons after prolonged depolarization and is commonly used as a marker integrating recent neuronal activity (30, 31).
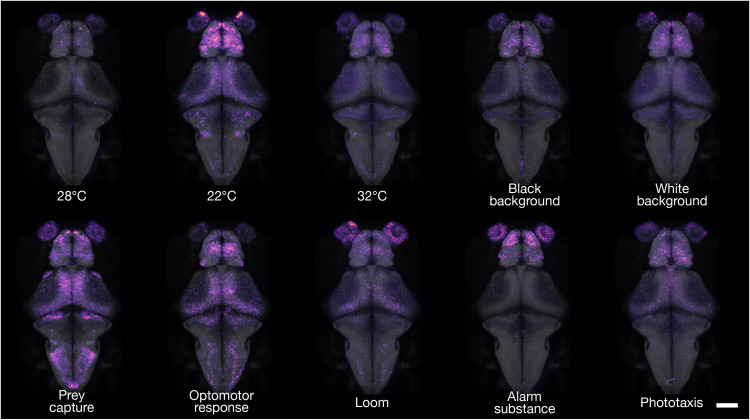
Fig. 3. cfos brain activity maps of freely swimming fish.
3D projections of cfos FISH-labeled areas of neuronal activity during behavior. Dorsal views are shown for each experimental condition. For interactive viewing, spanning the full depth of the brain, see mapzebrain atlas at https://mapzebrain.org/. Scale bar, 100 μm.
We exposed two groups of larval zebrafish to high temperature (32°C) or low temperature (22°C). Another group was allowed to hunt prey. Other larvae were held on a black, or a white, background. Still, others were subjected to a drifting, black-and-white grating, which induced an optomotor response (32, 33), or to a repeatedly looming black disk on a white background, thus evoking a series of visual escape responses (34). Two groups were exposed either to an asymmetric light source, which evoked phototaxis (35, 36), or to the alarm substance, “Schreckstoff,” which was extracted from fish skin (see Methods). For each condition, we imaged, registered, and analyzed six larvae. Qualitative inspection of the cfos FISH-labeled areas revealed specific activity hotspots for different behavioral conditions (Fig. 3, fig. S3, and movie S2). Such activity maps not only largely confirmed previously published calcium imaging studies of embedded animals (34, 37–40) but also revealed previously unidentified hotspots. For example, in larvae that were hunting prey, we observed strong cfos labeling in the anterior optic tectum and retinal arborization field 7 (AF7; Fig. 3, fig. S3, and movie S2), which have been implicated in prey detection and prey capture initiation in both embedded and freely swimming fish (40–44), as well as in cells of the secondary gustatory nucleus (SGN), which is located adjacent to the cerebellar commissure (Fig. 3 and movie S2) (45–47).
Molecular and cellular analysis of a brain area associated with hunting
Since activity in the SGN had not been reported in brain-wide imaging studies in embedded animals, we suspect that it is contingent on the animal to freely swim and hunt. Since it is likely driven by gustatory inputs, we propose that its activation is linked to the ingestion of food rather than the detection of prey or hunting-related movement (42, 44). To further explore how cfos maps can be used for circuit analysis, we interrogated the mapzebrain atlas for marker genes overlapping with the cfos signal and found that this region contains a group of neurons labeled by calb2a (calretinin), a known marker for the SGN (46) and its corresponding anti-Calretinin immunostaining. We verified coexpression of cfos and calb2a by multiplexed HCR FISH (Fig. 4, A to C).
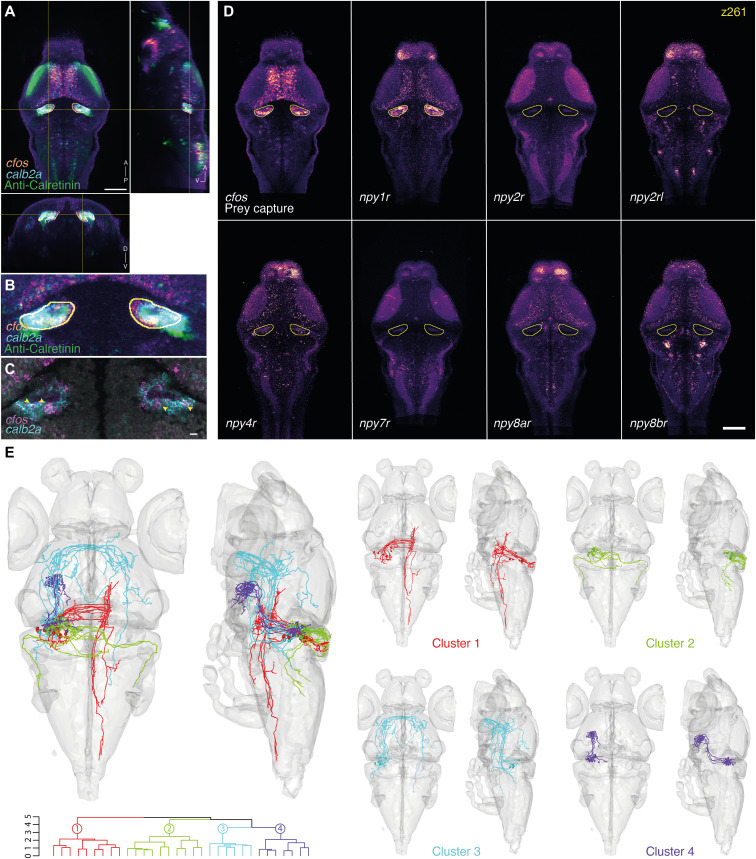
Fig. 4. Marker gene expression and hypothetical connectivity of SGN neurons involved in prey capture.
(A) Orthogonal views of the cfos prey capture brain activity map with the registered calb2a FISH and anti-Calretinin immunostaining. An ROI is drawn around the cfos-positive SGN area (yellow). Scale bar, 100 μm. A, anterior; D, dorsal; P, posterior; V, ventral. (B) Enlargement of the cfos-positive SGN area. Possible colabeling was identified between the registered cfos and calb2a data. (C) Multiplexed FISH of calb2a and cfos verified the suggested colabeling (arrowheads). A single focal plane of an individual larva is shown. Scale bar, 10 μm. (D) Expression patterns of several NPY receptors in the same single z plane (z261). The yellow line labels the cfos + SGN area. npy1r shows strong expression within the SGN, which is not seen for the other tested NPY receptors. Scale bar, 100 μm. (E) The ROI drawn in (A) was used to search for single neurons whose somas are within the ROI. Thirty-two neurons were identified, mirrored onto the left hemisphere [see (20)] and hierarchically clustered according to their morphology. One cluster of Purkinje cells (cluster 2) projecting to the octavolateralis nucleus was identified, as well as three clusters of SGN neurons, with projections to the contralateral hindbrain (cluster 1), to the contralateral retinal AF7 and tectum through the postoptic commissure (cluster 3), and to the hypothalamus (cluster 4).
We scanned other gene expression maps and discovered that npy1r, which encodes a neuropeptide Y (NPY) receptor, also overlaps with the SGN-associated cfos pattern (Fig. 4D). NPY has been implicated in the regulation of food consumption and foraging (48–50), and the expression map of npy1r suggests that NPY could function as a modulator of the gustatory pathway through direct signaling to SGN neurons. Other NPY receptors that were examined did not express within the SGN area (Fig. 4D) but showed various, complementary expression patterns in other brain areas. Nonoverlapping expression of the NPY receptor repertoire in distinct neuronal subpopulations may underlie local specificity of modulation by this neuropeptide.
Next, we drew a 3D ROI encompassing the cfos-labeled region near the SGN to query the database for individual, morphologically reconstructed neurons (see movie S3 for a detailed tutorial). We used the search feature of mapzebrain and identified 32 neurons that had their somata within the 3D ROI (Fig. 4E). SGN neurons were hierarchically clustered into four morphological types using the NBLAST algorithm (51). One of these clusters is located at the edge of the 3D ROI mask, evidently in the cerebellum, and is composed of Purkinje cells (cluster 2; Fig. 4E). Other groups project to the contralateral hindbrain (cluster 1) or to the tectum and the contralateral AF7 through the postoptic commissure (cluster 3). Notably, one cluster of SGN neurons (cluster 4) extends axons to the dorsal zone of the lateral hypothalamus (Fig. 4E), an area that is activated in response to both prey stimuli and satiety (39, 52) and which also shows strong cfos expression following hunting behavior (movie S2). Thus, exploring cfos activity maps with morphologically reconstructed neurons revealed connections between SGN, AF7-associated pretectum, tectum, and hindbrain, brain areas that are jointly engaged during pursuit and consummation of prey.
DISCUSSION
Here, we present an essential addition to the neuroscience resources for larval zebrafish: whole-brain, single-cell resolution gene expression maps for an initial set of 290 markers, coregistered with thousands of single-neuron tracings, hundreds of transgenic lines, and more than 120 expertly curated anatomical segmentations in a common atlas space. For more than 20 years, the main zebrafish RNA expression resource has been the one produced by Thisse and Thisse (53, 54). It provides colorimetric whole-mount ISH images in 2D, which are accessible through the Zebrafish Information Network (54, 55). While this resource contributed greatly to zebrafish research in many fields, its utility for neuroscience is limited: Most of the data were collected for early developmental stages (up to 72 hours postfertilization), do not offer cellular resolution or spatial accuracy due to the colorimetric ISH method used, and cannot be registered to the newly emerging zebrafish brain atlases. Our new dataset closes this gap.
As an added feature, we provide cfos brain activity maps for several commonly studied behaviors of larval zebrafish. The cfos method has at least two experimental advantages over other post hoc activity labeling methods: (i) cfos expression is localized to the soma, without neuropil labeling, which improves cell detection and segmentation compared to phosphorylated extracellular-signal related kinase (pERK) detection (14) and CAMPARI (52). (ii) cfos labeling can be combined with cell marker probes by iterative (20) or multiplexed HCR stains.
In proof-of-concept experiments, we have succeeded in linking cfos maps with other data modalities, such as gene expression and single-cell connectivity. This has enabled the discovery of a behaviorally relevant pathway. The calb2a+ SGN neurons are activated during hunting by gustatory cues (Fig. 4) (38, 40). By using the atlas query tool (20) and gene expression maps, we identified their projection patterns, which likely convey this information to an area in the lateral hypothalamus (39). In addition, SGN neurons also express npy1r and are thus targets of modulation by the orexigenic peptide NPY. While an experimental test of these hypotheses is beyond the scope of the current work, these findings illustrate the power of integrating gene expression, functional cfos labeling, transgene, and single-cell reconstructions in a multimodal digital atlas.
The Web browser interface of the mapzebrain atlas offers advanced computational, visualization, and downloading options. Bridging registrations between mapzebrain and other atlases have been made available (20). Moreover, standardized protocols for staining and imaging enable coregistration of data acquired in different laboratories, supporting different microscopes and imaging modalities (56, 57). The mapzebrain atlas thus offers a platform for covisualization of gene expression and cellular architecture and a starting point for explorations of circuit function and structure. Future work will be directed at integrating brain-wide functional imaging data (e.g., from calcium or voltage recordings) and electron microscopic reconstructions (58) into the atlas.
METHODS
Zebrafish husbandry and maintenance
Adult zebrafish of the TLN strain were kept at 28°C with a day/night cycle of 14/10 hours, pH 7 to 7.5, and a conductivity of 600 μS. All the experiments were performed on Tg(elavl3:H2b-GCaMP6s) larvae produced by natural matings. Larvae were raised until 6 dpf at 28°C in Danieau’s solution in petri dishes. The larvae were treated with phenylthiocarbamide (phenylthiourea, PTU) starting 24 hours postfertilization to prevent pigmentation. For cfos experiments, Tg(elavl3:H2b-GCaMP6s) larvae in the TLN background, carrying the mitfa mutation and therefore lacking melanin pigment, were used without PTU treatment. The animal experiments were performed under the regulations of the Max Planck Society and the regional government of Upper Bavaria (Regierung von Oberbayern) approved protocols: ROB-55.2-2532.Vet_02-20-11, ROB-55.2-2532.Vet_02-21-93, 55.2-ROB-2532.Vet_02-20-183, and ROB-55.2-2532.Vet_02-19-16.
HCR FISH staining
HCR reagents, including probes, hairpins, and buffers, were purchased from Molecular Instruments (Los Angeles, CA, USA). The staining was performed according to a modified protocol of the “HCR RNA-FISH protocol for whole-mount zebrafish embryos and larvae” provided by Molecular Instruments (21). Tg(elavl3:H2b-GCaMP6s)-positive larvae were anesthetized in 1.5 mM tricaine and fixed with ice-cold 4% paraformaldehyde (PFA)/Dulbecco’s phosphate-buffered saline (DPBS) overnight at 4°C with gentle shaking. The following day, larvae were washed three times for 5 min with DPBST (1× DPBS + 0.1% Tween 20) to stop fixation, followed by a short 10-min treatment with ice-cold 100% methanol (MeOH) at −20°C to dehydrate and permeabilize the tissue samples. Next, rehydration was performed by serial washing of 50% MeOH/50% DPBST and 25% MeOH/75% DPBST for 5 min each and lastly 5 × 5 min in DPBST. Ten to 12 larvae were transferred into a 1.5-ml Eppendorf tube and prehybridized with prewarmed hybridization buffer for 30 min at 37°C. Probe solution was prepared by transferring 2 pmol of each HCR probe set (2 μl of 1 μM stock) to 500 μl of hybridization buffer at 37°C. For cfos experiments, 4 μl of 1 μM probe stock was used, while for genes with relatively weak expression levels (i.e., receptors), 8 μl of 1 μM probe stock was used. The hybridization buffer was replaced with probe solution, and the samples were incubated for 12 to 16 hours at 37°C with gentle shaking. To remove excess probe, larvae were washed 4 × 15 min with 500 μl of prewarmed probe wash buffer at 37°C. Subsequently, larvae were washed 2 × 5 min with 5× SSCT (5× sodium chloride sodium citrate + 0.1% Tween 20) buffer at room temperature. Next, preamplification was performed by incubating the samples in 500 μl of amplification buffer for 30 min at room temperature. Separately, 30 pmol of hairpin h1 and 30 pmol of hairpin h2 were prepared by snap-cooling 10 μl of 3 μM stock by incubating the hairpins in 95°C for 90 s and cooling down to room temperature in a dark environment. After cooling down for 30 min, hairpin solution was prepared by transferring the h1 and h2 hairpins to 500 μl of amplification buffer. The preamplification buffer was removed, and the samples were incubated in the hairpin solution for 12 to 16 hours in the dark at room temperature. Excess hairpins were washed the next day 3 × 20 min using 5× SSCT at room temperature. Larvae were then long-term stored at 4°C in 5× SSCT until imaging.
Imaging
Samples were embedded in 2% low-melting agarose in 1× DPBS and imaged with a Zeiss LSM700 confocal scanning microscope (upright), equipped with a 20× water immersion objective (W “Plan-Apochromat” 20×/1.0 DIC M27 75 mm; Zeiss, catalog no. 421452-9800). We consistently used HCR hairpins that were fluorescently labeled with Alexa Fluor 546 and Alexa Fluor 647, and therefore, imaging was performed with the following lasers: Diode 488 (10.0 mW), Diode 555 (10.0 mW), and Diode 639 (5.0 mW). Although we did not experience photobleaching for the majority of samples, some reduction in the intensity of the Alexa Fluor 647 occasionally occurred. To overcome the reduced signal in the ventral parts of the brain due to scattering, we applied z brightness correction, by increasing the laser power in the deep parts of the sample. Z-stacks, comprising two tiles, were scanned in an upward direction, with a dwell time of 1.58 μs, and stitched to produce a final image with a size of 1039 × 1931 pixels (463.97 × 862.29 μm, 1 μm in z).
Generation of reference brain and image registration and averaging
All the image registrations and averaging procedures were performed using ANTs (26), which were previously calibrated and used for the generation of zebrafish average brain images (20, 59). Twelve transgenic fish expressing the transgenes Tg(elavl3:H2b-GCaMP6s) and Tg(elavl3:lynTag-RFP) were fixed and underwent the entire HCR buffer procedure, but without the usage of any probes or hairpins, and imaged as described. The images were used to generate an unbiased average brain by running the following ANTs command on the GWDG computing cluster:
antsMultivariateTemplateConstruction2.sh -d 3 -o ${outputPath}/T_ -i 4 -g 0.2 -j 32 -v 500 -c 2 -k 2 -w 1x1 -f 12x8x4x2 -s 4x3x2x1 -q 200x200x200x200 -n 0 -r 1 -l 1 -z ${target1} -z ${target2} -m CC[2] -t SyN[0.05,6,0.5] ${inputPath}
To register the newly generated reference to the mapzebrain coordinate system, we first registered the older Tg(elavl3:H2b-GCaMP6s) average image that existed in mapzebrain (20) onto the newly generated reference and applied the inverse transformation on the newly generated reference image to align to the mapzebrain coordinate system. First, the following ANTs command was used to transform the older mapzebrain Tg(elavl3:H2b-GCaMP6s) average to a new reference:
antsRegistration -d 3 --float 1 -o [${output1},${output2}] --interpolation WelchWindowedSinc --use-histogram-matching 0 -r [${template},${input1},1] -t rigid[0.1] -m MI[${template},${input1},1,32,Regular,0.25] -c [200x200x200x0,1e-8,10] --shrink-factors 12x8x4x2 --smoothing-sigmas 4x3x2x1vox -t Affine[0.1] -m MI[${template},${input1},1,32,Regular,0.25] -c [200x200x200x0,1e-8,10] --shrink-factors 12x8x4x2 --smoothing-sigmas 4x3x2x1 -t SyN[0.05,6,0.5] -m CC[${template},${input1},1,2] -c [200x200x200x200x10,1e-7,10] --shrink-factors 12x8x4x2x1 --smoothing-sigmas 4x3x2x1x0
Second, inverse transformation was applied to the newly generated reference using the inverse Warp transformation file by running the following ANTs command:
antsApplyTransforms -d 3 -v 0 -- float -n WelchWindowedSinc -i ${input3} -r ${template} -o ${output4} -t ${output1}1InverseWarp.nii.gz -t ${output1}0GenericAffine.mat
The newly registered reference brain (fig. S1 and movie S1) is available to download through https://mapzebrain.org. All the imaged HCR in situ images were registered onto this new reference by registering the Tg(elavl3:H2b-GCaMP6s) image channel using the following ANTs command:
antsRegistration -d 3 --float 1 -o [${output1},${output2}] --interpolation WelchWindowedSinc --use-histogram-matching 0 -r [${template},${input1},1] -t rigid[0.1] -m MI[${template},${input1},1,32,Regular,0.25] -c [200x200x200x0,1e-8,10] --shrink-factors 12x8x4x2 --smoothing-sigmas 4x3x2x1vox -t Affine[0.1] -m MI[${template},${input1},1,32,Regular,0.25] -c [200x200x200x0,1e-8,10] --shrink-factors 12x8x4x2 --smoothing-sigmas 4x3x2x1 -t SyN[0.1,6,0.0] -m CC[${template},${input1},1,2] -c [200x200x200x200x10,1e-7,10] --shrink-factors 12x8x4x2x1 --smoothing-sigmas 4x3x2x1x0,
followed by applying the transformation files on the HCR image channels using the following ANTs command:
antsApplyTransforms -d 3 -v 0 -- float -n WelchWindowedSinc -i ${input3} -r ${template} -o ${output4} -t ${output1}1Warp.nii.gz -t ${output1}0GenericAffine.mat
Each HCR-labeled RNA was imaged from three different fish and registered as described. The three images were then arithmetically averaged using ImageJ (60). Both the average image and the individual registered images are accessible through https://mapzebrain.org.
cfos FISH experiments
For cfos experiments, larvae were raised until 6 dpf under optimal conditions (14-hour light/10-hour dark cycle, 28°C) in Danieau’s solution. At 6 dpf, the larvae were transferred into a six-well plate, 10 larvae per well until 7 dpf, when they were exposed to different stimuli for 40 min. The larvae were then euthanized in an overdose of Tricaine for 2 min and immediately fixed in ice-cold 4% PFA. Six fish of each condition were imaged and registered as described before, followed by arithmetical averaging with ImageJ (60). For visualization purposes of the averaged images, the contrast was linearly enhanced. Movie S2 anatomical legend was generated using QuPath (61).
Temperature adaptation
The six-well plate containing the fish was placed into a water bath set to either 22° or 32°C with a similar background color for 40 min.
Background adaptation
The six-well plate containing the fish was placed on either a white or a black background at 28°C for 40 min.
Prey capture
Paramecia were added into the six-well plate containing the larvae. Food was not limited: Many paramecia were still uneaten after 40 min.
Phototaxis
The six-well plate containing the fish was placed on a half-white and half-black background (splitting the well background color equally), at 28°C for 40 min.
Looming
Larvae were exposed to black looming disc stimuli using a setup previously described (62, 63). Briefly, groups of 10 larvae were placed into a 10-cm watch glass containing Danieau’s solution and left for 2 hours to acclimate to room temperature and white projector illumination from below. For stimulation, expanding black discs were projected to the center of the dish from below. Discs expanded linearly from 0 to 10 cm diameter within 500 ms, once every 60 s. This cycle was repeated over 40 min.
Optomotor response
Larvae were exposed to moving gratings using a setup previously described (55, 56). Briefly, groups of 10 larvae were placed into a 10-cm watch glass containing Danieau’s solution and left for 2 hours to acclimate to a background of a stationary grating (black/white step grating, 10-mm spatial frequency) projected from below. After 2 hours, gratings were moved in alternating directions at 1 cycle/s in the following sequence: 15 s in one direction, 5 s pause, and 15 s in the opposite direction. This sequence was repeated over 40 min. Visual inspection confirmed a strong behavioral optomotor response under these stimulus conditions. A control group of larvae remained on stationary gratings the entire time.
Alarm substance response
Alarm substance is stored in the club cells of zebrafish epidermis and released upon skin damage. For the experiment, five adult wild-type donor fish were euthanized, and scales from both flanks were collected with the blunt side of a sterile razor blade. The scales were washed off with sterile-filtered Danieau’s solution and collected in a Falcon tube, avoiding any contamination with blood. The total 20 ml of isolate was vortexed, filtered through a Whatman paper, and placed on ice until use (64). Five hundred microliters of the isolate was distributed evenly over the wells of the six-well plate containing the larvae.
Single neuron morphology clustering
An ROI surrounding the SGN cfos ISH-labeled area was labeled using the ImageJ segmentation editor plugin and saved as a binary mask .tif file. The .tif file was then uploaded as an ROI to mapzebrain.org and used to search the mapzebrain atlas for neurons whose somas were located within it (to enable this feature in mapzebrain.org, a free user account is required. See movie S3 for a detailed tutorial). Thirty-two neurons were identified and flipped to the left [as described in (20)], and a morphology similarity score between them was calculated using the NBLAST algorithm (51) by applying the nblast_allbyall function implemented in the R package NeuroAnatomy Toolbox (nat) (65) with default parameters. Next, unsupervised hierarchical clustering was performed using the nhclust function of the nat package with default parameters, and the results were plotted using the R packages dendroextras and rgl. The analysis and plotting R code can be found at https://zenodo.org/record/7308624.
Acknowledgments
We thank R. Kasper for microscopy consultation and support, and K. Slanchev, M. Stemmer, and M. Wullimann for advice and support with experiments. We thank all members of the Baier laboratory for beta testing of atlas features and fruitful discussions.
Funding: This work was supported by the Max Planck Society (all authors), an Alexander von Humboldt Foundation research fellowship for postdoctoral researchers (I.S.), and a NARSAD Young Investigator Award (J.L.).
Author contributions: Conceptualization: I.S., E.K., M.K., and H.B. Methodology: I.S., E.K., E.L., M.A.K., S.S., J.L., and M.K. Investigation: I.S. and E.K. Data analysis: I.S., E.K., M.A.K., and N.M. Software and website development and design: M.A.K., N.M., and H.B. Supervision: H.B. Writing—original draft: I.S., E.K., and H.B. Writing—review and editing: I.S., E.K., E.L., S.S., J.L., M.K., and H.B.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials, or available through the mapzebrain.org website.
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
Table S1
Legends for movies S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S3
REFERENCES AND NOTES
- 1.T. E. Bakken, J. A. Miller, S.-L. Ding, S. M. Sunkin, K. A. Smith, L. Ng, A. Szafer, R. A. Dalley, J. J. Royall, T. Lemon, S. Shapouri, K. Aiona, J. Arnold, J. L. Bennett, D. Bertagnolli, K. Bickley, A. Boe, K. Brouner, S. Butler, E. Byrnes, S. Caldejon, A. Carey, S. Cate, M. Chapin, J. Chen, N. Dee, T. Desta, T. A. Dolbeare, N. Dotson, A. Ebbert, E. Fulfs, G. Gee, T. L. Gilbert, J. Goldy, L. Gourley, B. Gregor, G. Gu, J. Hall, Z. Haradon, D. R. Haynor, N. Hejazinia, A. Hoerder-Suabedissen, R. Howard, J. Jochim, M. Kinnunen, A. Kriedberg, C. L. Kuan, C. Lau, C.-K. Lee, F. Lee, L. Luong, N. Mastan, R. May, J. Melchor, N. Mosqueda, E. Mott, K. Ngo, J. Nyhus, A. Oldre, E. Olson, J. Parente, P. D. Parker, S. Parry, J. Pendergraft, L. Potekhina, M. Reding, Z. L. Riley, T. Roberts, B. Rogers, K. Roll, D. Rosen, D. Sandman, M. Sarreal, N. Shapovalova, S. Shi, N. Sjoquist, A. J. Sodt, R. Townsend, L. Velasquez, U. Wagley, W. B. Wakeman, C. White, C. Bennett, J. Wu, R. Young, B. L. Youngstrom, P. Wohnoutka, R. A. Gibbs, J. Rogers, J. G. Hohmann, M. J. Hawrylycz, R. F. Hevner, Z. Molnár, J. W. Phillips, C. Dang, A. R. Jones, D. G. Amaral, A. Bernard, E. S. Lein, A comprehensive transcriptional map of primate brain development. Nature 535, 367–375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.C. L. Thompson, L. Ng, V. Menon, S. Martinez, C.-K. Lee, K. Glattfelder, S. M. Sunkin, A. Henry, C. Lau, C. Dang, R. Garcia-Lopez, A. Martinez-Ferre, A. Pombero, J. L. R. Rubenstein, W. B. Wakeman, J. Hohmann, N. Dee, A. J. Sodt, R. Young, K. Smith, T.-N. Nguyen, J. Kidney, L. Kuan, A. Jeromin, A. Kaykas, J. Miller, D. Page, G. Orta, A. Bernard, Z. Riley, S. Smith, P. Wohnoutka, M. J. Hawrylycz, L. Puelles, A. R. Jones, A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron 83, 309–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Y. Ko, S. A. Ament, J. A. Eddy, J. Caballero, J. C. Earls, L. Hood, N. D. Price, Cell type-specific genes show striking and distinct patterns of spatial expression in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 110, 3095–3100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L. Ng, A. Bernard, C. Lau, C. C. Overly, H.-W. Dong, C. Kuan, S. Pathak, S. M. Sunkin, C. Dang, J. W. Bohland, H. Bokil, P. P. Mitra, L. Puelles, J. Hohmann, D. J. Anderson, E. S. Lein, A. R. Jones, M. Hawrylycz, An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 12, 356–362 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., et al. , Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Hawrylycz M. J., Lein E. S., Guillozet-Bongaarts A. L., Shen E. H., Ng L., Miller J. A., Lagemaat L. N. V. D., Smith K. A., Ebbert A., Riley Z. L., et al. , An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong S., Zheng C., Doughty M. L., Losos K., Didkovsky N., Schambra U. B., Nowak N. J., Joyner A., Leblanc G., Hatten M. E., Heintz N., A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 (2003). [DOI] [PubMed] [Google Scholar]
- 8.C. Ortiz, J. F. Navarro, A. Jurek, A. Märtin, J. Lundeberg, K. Meletis, Molecular atlas of the adult mouse brain. Sci. Adv. 6, eabb3446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Zhang, S. W. Eichhorn, B. Zingg, Z. Yao, K. Cotter, H. Zeng, H. Dong, X. Zhuang, Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598, 137–143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. J. Cook, T. A. Jarrell, C. A. Brittin, Y. Wang, A. E. Bloniarz, M. A. Yakovlev, K. C. Q. Nguyen, L. T.-H. Tang, E. A. Bayer, J. S. Duerr, H. E. Bülow, O. Hobert, D. H. Hall, S. W. Emmons, Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.E. Yemini, A. Lin, A. Nejatbakhsh, E. Varol, R. Sun, G. E. Mena, A. D. T. Samuel, L. Paninski, V. Venkatachalam, O. Hobert, NeuroPAL: A multicolor atlas for Whole-Brain neuronal identification in C. elegans. Cell 184, 272–288.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.N. Milyaev, D. Osumi-Sutherland, S. Reeve, N. Burton, R. A. Baldock, J. D. Armstrong, The Virtual Fly Brain browser and query interface. Bioinformatics 28, 411–415 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Ukani N. H., Yeh C.-H., Tomkins A., Zhou Y., Florescu D., Ortiz C. L., Huang Y.-C., Wang C.-T., Turkcan M. K., Liu T., Richmond P., Lo C., Coca D., Chiang L., Lazar A. A., The fruit fly brain observatory: From structure to function. bioRxiv, 580290 (2019). [Google Scholar]
- 14.J. W. Kenney, P. E. Steadman, O. Young, M. T. Shi, M. Polanco, S. Dubaishi, K. Covert, T. Mueller, P. W. Frankland, A 3D adult zebrafish brain atlas (AZBA) for the digital age. eLife 10, e69988 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. F. Wullimann, B. Rupp, H. Reichert, Neuroanatomy of the Zebrafish Brain (Birkhäuser, 1996). [DOI] [PubMed]
- 16.T. Mueller, M. Wullimann, Atlas of Early Zebrafish Brain Development: A Tool for Molecular Neurogenetics (Academic Press, 2015). [Google Scholar]
- 17.Ronneberger O., Liu K., Rath M., Rueβ D., Mueller T., Skibbe H., Drayer B., Schmidt T., Filippi A., Nitschke R., Brox T., Burkhardt H., Driever W., ViBE-Z: A framework for 3D virtual colocalization analysis in zebrafish larval brains. Nat. Methods 9, 735–742 (2012). [DOI] [PubMed] [Google Scholar]
- 18.O. Randlett, C. L. Wee, E. A. Naumann, O. Nnaemeka, D. Schoppik, J. E. Fitzgerald, R. Portugues, A. M. B. Lacoste, C. Riegler, F. Engert, A. F. Schier, Whole-brain activity mapping onto a zebrafish brain atlas. Nat. Methods 12, 1039–1046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.K. M. Tabor, G. D. Marquart, C. Hurt, T. S. Smith, A. K. Geoca, A. A. Bhandiwad, A. Subedi, J. L. Sinclair, H. M. Rose, N. F. Polys, H. A. Burgess, Brain-wide cellular resolution imaging of Cre transgenic zebrafish lines for functional circuit-mapping. eLife 8, e42687 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. Kunst, E. Laurell, N. Mokayes, A. Kramer, F. Kubo, A. M. Fernandes, D. Förster, M. D. Maschio, H. Baier, A cellular-resolution atlas of the larval zebrafish brain. Neuron 103, 21–38.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 21.H. M. T. Choi, M. Schwarzkopf, M. E. Fornace, A. Acharya, G. Artavanis, J. Stegmaier, A. Cunha, N. A. Pierce, Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.F. Wang, J. Flanagan, N. Su, L.-C. Wang, S. Bui, A. Nielson, X. Wu, H.-T. Vo, X.-J. Ma, Y. Luo, RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.J. M. Kappel, D. Förster, K. Slangewal, I. Shainer, F. Svara, J. C. Donovan, S. Sherman, M. Januszewski, H. Baier, J. Larsch, Visual recognition of social signals by a tectothalamic neural circuit. Nature 608, 146–152 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Lovett-Barron, R. Chen, S. Bradbury, A. S. Andalman, M. Wagle, S. Guo, K. Deisseroth, Multiple convergent hypothalamus–brainstem circuits drive defensive behavior. Nat. Neurosci. 23, 959–967 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Xu, H. Yang, V. Menon, A. L. Lemire, L. Wang, F. E. Henry, S. C. Turaga, S. M. Sternson, Behavioral state coding by molecularly defined paraventricular hypothalamic cell type ensembles. Science 370, eabb2494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.B. B. Avants, N. J. Tustison, G. Song, P. A. Cook, A. Klein, J. C. Gee, A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S. Sherman, K. Kawakami, H. Baier, Retinal input influences pace of neurogenesis but not cell-type configuration of the visual forebrain. bioRxiv, 468630 (2021).
- 28.Y. Kölsch, J. Hahn, A. Sappington, M. Stemmer, A. M. Fernandes, T. O. Helmbrecht, S. Lele, S. Butrus, E. Laurell, I. Arnold-Ammer, K. Shekhar, J. R. Sanes, H. Baier, Molecular classification of zebrafish retinal ganglion cells links genes to cell types to behavior. Neuron 109, 645–662.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.B. Raj, J. A. Farrell, J. Liu, J. E. Kholtei, A. N. Carte, J. N. Acedo, L. Y. Du, A. McKenna, Đ. Relić, J. M. Leslie, A. F. Schier, Emergence of neuronal diversity during vertebrate brain development. Neuron 108, 1058–1074.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D. G. Herrera, H. A. Robertson, Activation of c-fos in the brain. Prog. Neurobiol. 50, 83–107 (1996). [DOI] [PubMed] [Google Scholar]
- 31.K. J. Kovacs, Measurement of immediate-early gene activation-c-fos and beyond. J. Neuroendocrinol. 20, 665–672 (2008). [DOI] [PubMed] [Google Scholar]
- 32.S. C. F. Neuhauss, O. Biehlmaier, M. W. Seeliger, T. Das, K. Kohler, W. A. Harris, H. Baier, Genetic disorders of vision revealed by a behavioral screen of 400 essential loci in zebrafish. J. Neurosci. 19, 8603–8615 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. B. Orger, M. C. Smear, S. M. Anstis, H. Baier, Perception of Fourier and non-Fourier motion by larval zebrafish. Nat. Neurosci. 3, 1128–1133 (2000). [DOI] [PubMed] [Google Scholar]
- 34.I. Temizer, J. C. Donovan, H. Baier, J. L. Semmelhack, A visual pathway for looming-evoked escape in larval zebrafish. Curr. Biol. 25, 1823–1834 (2015). [DOI] [PubMed] [Google Scholar]
- 35.S. E. Brockerhoff, J. B. Hurley, U. Janssen-Bienhold, S. C. Neuhauss, W. Driever, J. E. Dowling, A behavioral screen for isolating zebrafish mutants with visual system defects. Proc. Natl. Acad. Sci. U.S.A. 92, 10545–10549 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.M. B. Orger, E. Gahtan, A. Muto, P. Page-McCaw, M. C. Smear, H. Baier, Behavioral screening assays in zebrafish. Methods Cell Biol. 77, 53–68 (2004). [DOI] [PubMed] [Google Scholar]
- 37.X. Chen, Y. Mu, Y. Hu, A. T. Kuan, M. Nikitchenko, O. Randlett, A. B. Chen, J. P. Gavornik, H. Sompolinsky, F. Engert, M. B. Ahrens, Brain-wide organization of neuronal activity and convergent sensorimotor transformations in larval zebrafish. Neuron 100, 876–890.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.J. S. M. Chia, E. S. Wall, C. L. Wee, T. A. J. Rowland, R.-K. Cheng, K. Cheow, K. Guillemin, S. Jesuthasan, Bacteria evoke alarm behaviour in zebrafish. Nat. Commun. 10, 3831 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Muto, P. Lal, D. Ailani, G. Abe, M. Itoh, K. Kawakami, Activation of the hypothalamic feeding centre upon visual prey detection. Nat. Commun. 8, 15029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J. L. Semmelhack, J. C. Donovan, T. R. Thiele, E. Kuehn, E. Laurell, H. Baier, A dedicated visual pathway for prey detection in larval zebrafish. eLife 3, e04878 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.P. Antinucci, M. Folgueira, I. H. Bianco, Pretectal neurons control hunting behaviour. eLife 8, e48114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L. Cong, Z. Wang, Y. Chai, W. Hang, C. Shang, W. Yang, L. Bai, J. Du, K. Wang, Q. Wen, Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio). eLife 6, e28158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.E. Gahtan, P. Tanger, H. Baier, Visual prey capture in larval zebrafish is controlled by identified reticulospinal neurons downstream of the tectum. J. Neurosci. 25, 9294–9303 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Z. Zhang, L. Bai, L. Cong, P. Yu, T. Zhang, W. Shi, F. Li, J. Du, K. Wang, Imaging volumetric dynamics at high speed in mouse and zebrafish brain with confocal light field microscopy. Nat. Biotechnol. 39, 74–83 (2021). [DOI] [PubMed] [Google Scholar]
- 45.E. Rink, M. F. Wullimann, Some forebrain connections of the gustatory system in the goldfish Carassius auratus visualized by separate DiI application to the hypothalamic inferior lobe and the torus lateralis. J. Comp. Neurol. 394, 152–170 (1998). [DOI] [PubMed] [Google Scholar]
- 46.K. Volkmann, Y. Chen, M. P. Harris, M. F. Wullimann, R. W. Köster, The zebrafish cerebellar upper rhombic lip generates tegmental hindbrain nuclei by long-distance migration in an evolutionary conserved manner. J. Comp. Neurol. 518, 2794–2817 (2010). [DOI] [PubMed] [Google Scholar]
- 47.J. Yáñez, Y. Souto, L. Piñeiro, M. Folgueira, R. Anadón, Gustatory and general visceral centers and their connections in the brain of adult zebrafish: A carbocyanine dye tract-tracing study. J. Comp. Neurol. 525, 333–362 (2017). [DOI] [PubMed] [Google Scholar]
- 48.M. B. Sokolowski, NPY and the regulation of behavioral development. Neuron 39, 6–8 (2003). [DOI] [PubMed] [Google Scholar]
- 49.H. Volkoff, L. F. Canosa, S. Unniappan, J. M. Cerdá-Reverter, N. J. Bernier, S. P. Kelly, R. E. Peter, Neuropeptides and the control of food intake in fish. Gen. Comp. Endocrinol. 142, 3–19 (2005). [DOI] [PubMed] [Google Scholar]
- 50.G. J. Morton, M. W. Schwartz, The NPY/AgRP neuron and energy homeostasis. Int. J. Obes. 25, S56–S62 (2001). [DOI] [PubMed] [Google Scholar]
- 51.M. Costa, J. D. Manton, A. D. Ostrovsky, S. Prohaska, G. S. X. E. Jefferis, NBLAST: Rapid, sensitive comparison of neuronal structure and construction of neuron family databases. Neuron 91, 293–311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wee C. L., Song E. Y., Johnson R. E., Ailani D., Randlett O., Kim J.-Y., Nikitchenko M., Bahl A., Yang C.-T., Ahrens M. B., Kawakami K., Engert F., Kunes S., A bidirectional network for appetite control in larval zebrafish. eLife 8, e43775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thisse C., Thisse B., High resolution whole-mount in situ hybridization. Zebrafish Sci. Monit. 5 ( 1998). [Google Scholar]
- 54.Thisse B., Thisse C., Fast release clones: A high throughput expression analysis. ZFIN Direct Data Submiss. (2004). [Google Scholar]
- 55.Y. M. Bradford, C. E. Van Slyke, L. Ruzicka, A. Singer, A. Eagle, D. Fashena, D. G. Howe, K. Frazer, R. Martin, H. Paddock, C. Pich, S. Ramachandran, M. Westerfield, Zebrafish Information Network, the knowledgebase for Danio rerio research. Genetics 220, iyac016 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.L. Corradi, M. Bruzzone, M. D. Maschio, S. Sawamiphak, A. Filosa, Hypothalamic Galanin-producing neurons regulate stress in zebrafish through a peptidergic, self-inhibitory loop. Curr. Biol. 32, 1497–1510.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 57.Y. Wu, M. Dal Maschio, F. Kubo, H. Baier, An optical illusion pinpoints an essential circuit node for global motion processing. Neuron 108, 722–734.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 58.F. Svara, D. Förster, F. Kubo, M. Januszewski, M. Dal Maschio, P. J. Schubert, J. Kornfeld, A. A. Wanner, E. Laurell, W. Denk, H. Baier, Automated synapse-level reconstruction of neural circuits in the larval zebrafish brain. Nat. Methods 19, 1357–1366 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.G. D. Marquart, K. M. Tabor, E. J. Horstick, M. Brown, A. K. Geoca, N. F. Polys, D. D. Nogare, H. A. Burgess, High-precision registration between zebrafish brain atlases using symmetric diffeomorphic normalization. GigaScience 6, gix056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.C. A. Schneider, W. S. Rasband, K. W. Eliceiri, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bankhead P., Loughrey M. B., Fernández J. A., Dombrowski Y., McArt D. G., Dunne P. D., McQuaid S., Gray R. T., Murray L. J., Coleman H. G., James J.A., Salto-Tellez M., Hamilton P.W., QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.J. Larsch, H. Baier, Biological motion as an innate perceptual mechanism driving social affiliation. Curr. Biol. 28, 3523–3532.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 63.C. Pantoja, J. Larsch, E. Laurell, G. Marquart, M. Kunst, H. Baier, Rapid effects of selection on brain-wide activity and behavior. Curr. Biol. 30, 3647–3656.e3 (2020). [DOI] [PubMed] [Google Scholar]
- 64.B. Waldman, Quantitative and developmental analyses of the alarm reaction in the zebra danio, Brachydanio rerio. Copeia , 1–9 (1982). [Google Scholar]
- 65.A. S. Bates, J. D. Manton, S. R. Jagannathan, M. Costa, P. Schlegel, T. Rohlfing, G. S. X. E. Jefferis, The natverse, a versatile toolbox for combining and analysing neuroanatomical data. eLife 9, e53350 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Table S1
Legends for movies S1 to S3
Movies S1 to S3