Abstract
The predatory deltaproteobacterium Myxococcus xanthus uses a helically-trafficked motor at bacterial focal-adhesion (bFA) sites to power gliding motility. Using total internal reflection fluorescence and force microscopies, we identify the von Willebrand A domain-containing outer-membrane (OM) lipoprotein CglB as an essential substratum-coupling adhesin of the gliding transducer (Glt) machinery at bFAs. Biochemical and genetic analyses reveal that CglB localizes to the cell surface independently of the Glt apparatus; once there, it is recruited by the OM module of the gliding machinery, a heteroligomeric complex containing the integral OM β barrels GltA, GltB, and GltH, as well as the OM protein GltC and OM lipoprotein GltK. This Glt OM platform mediates the cell-surface accessibility and retention of CglB by the Glt apparatus. Together, these data suggest that the gliding complex promotes regulated surface exposure of CglB at bFAs, thus explaining the manner by which contractile forces exerted by inner-membrane motors are transduced across the cell envelope to the substratum.
An outer-membrane complex exposes the cell-surface CglB adhesin at bacterial focal-adhesion sites to mediate gliding motility.
INTRODUCTION
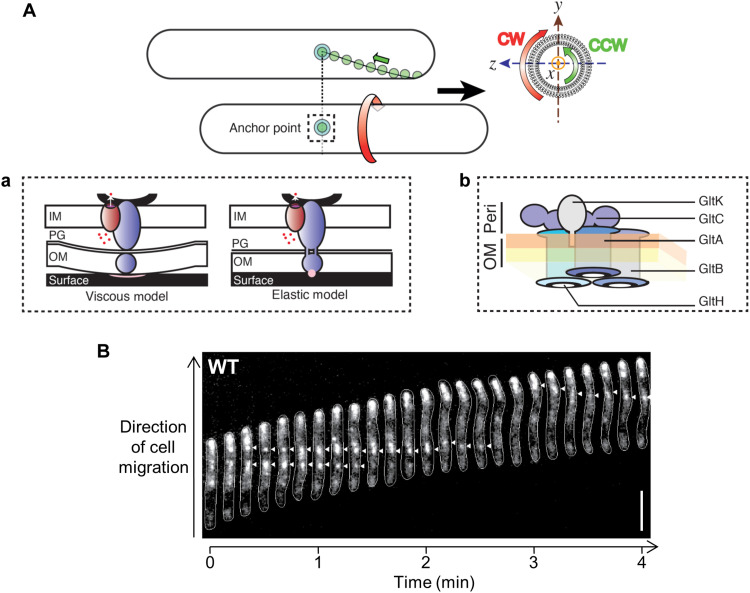
Directed surface motility of cells from all biological kingdoms involves highly dynamic cell-substratum interactions. In eukaryotic cells, this process involves the engagement and activation of surface-exposed integrin(-like) adhesins, directionally transported by molecular motors (myosin) via integrin coupling to the internal cytoskeleton (actin) (1). For metazoan organisms, nascent integrin adhesions to the extracellular matrix (ECM) lead to integrin nucleation and the formation of large eukaryotic focal-adhesion (eFA) sites; these assemblies remain fixed in space relative to a translocating cell, promoting local traction, transduction of motor forces, and cell translocation (2). Such surface motility is not, however, restricted to eukaryotic cells. Although known to move in groups on softer substrata via type IV pilus (T4P) extension/retraction (in concert with exo- and biosurfactant-polysaccharide secretion) (3–5), individual cells of the Gram-negative predatory deltaproteobacterium Myxococcus xanthus use gliding motility on harder substrata. Gliding occurs in the absence of outward appendages (e.g., flagella or T4P) (6), instead using a trans-envelope multiprotein Agl–gliding transducer (Glt) complex (Fig. 1A) to power cell locomotion (7, 8). In gliding cells, Agl–Glt complexes associate at the leading pole and move directionally in the bacterial inner membrane (IM) toward the lagging cell pole, following a right-handed helical trajectory (Fig. 1A) (9–11). These rotational movements likely probe the substratum beneath gliding cells, leading to immobilization of the Agl–Glt complex at fixed bacterial focal-adhesion (bFA) sites (Fig. 1B) and cell translocation via left-handed rotation of the bacterium around its long axis (Fig. 1A) (11).
Fig. 1. Concept of bFA-mediated gliding motility in M. xanthus.
(A) Following assembly at the leading pole, motility complexes move toward the lagging cell pole in a counterclockwise (CCW) rotational trajectory. Clockwise (CW) and CCW directionalities are defined by observing the cell cylinder from the leading pole (yz plane). When the complexes interact with the substratum, bacterial focal adhesions (bFAs) form and propel rotational cell movements. (a) In the “viscous interaction” model for bFA formation, the periplasmic complex accumulates at bFAs and pushes against the elastic peptidoglycan (PG) to create cell-envelope deformations at bFAs, creating viscous substratum interactions. Outer-membrane (OM) complex function is not accounted for in this model. In the “elastic” model, the periplasmic complex transiently interacts (through the PG) with the OM complex, which itself interacts with the substratum via an unknown adhesive molecule (pink circle). IM, inner membrane. Blue, trans-envelope Glt complex; dark red, IM AglRQS H+ motor; dots, protons; black curve, MreB. (b) OM localization of the proposed GltA/B/H/C/K OM platform is based on structural bioinformatic and fractionation analyses presented here and elsewhere (7, 17, 21). The integral association of GltABH is based on bioinformatic and proteinase K accessibility assays herein and elsewhere (17). GltA–GltB, GltA–GltC, and GltB–GltC interactions were demonstrated by pulldown assays (17). Connection with GltH is indicated from results reported in this study. The OM periplasmic (orange) and outer (yellow) leaflets are indicated. Peri, periplasm. (B) Position of a bFA (arrowheads) revealed via fluorescence microscopy of wild-type (WT) M. xanthus expressing AglZ-YFP (yellow fluorescent protein). Scale bar, 5 μm.
Direct imaging of bFAs is possible via live-cell microscopy (12), where they appear as bright fluorescent clusters that retain fixed positions relative to the substratum in a gliding cell (Fig. 1B). At the molecular level, polar activation of bFAs is driven by a cytoplasmic scaffold formed by bacterial actin MreB, the Ras-like protein MglA, and the coiled-coil protein AglZ (13, 14). This scaffold recruits the IM components of the gliding complex by as-yet-undefined interactions, activating the molecular motor within (14). The motor itself is constituted by the proteins AglR, Q, and S, which form a TolQR/ExbBD/MotAB-like H+-gated channel that uses the proton gradient formed across the bacterial IM to energize long-range movements of the IM complex in the bacterial envelope (15). However, the manner in which these intracellular motions are coupled to the substratum to propel the cell is unknown. One hypothesis states that trafficking motor units deform the peptidoglycan (PG) meshwork in the periplasm, propagating surface-wave deformations and viscous interactions between the outer membrane (OM) and the substratum (Fig. 1Aa). However, observations and mechanical modeling of cell-cell collision events suggest that interactions between a gliding cell and the substratum are elastic in nature, consistent with localized adhesion points and the existence of an anchored adhesin (Fig. 1Aa) (16). Also, when gliding on glass surfaces, cells occasionally abandon patches of OM motility complex proteins on the substratum at positions formerly occupied by bFA sites (detected with a GltC-mCherry protein) (11). In the cell envelope, the IM motor moves by establishing transient contacts with a group of Glt proteins localized in the OM (herein called the OM platform, see below) (Fig. 1Ab), linked via periplasmic domains of putatively contractile proteins that traverse the PG meshwork (Fig. 1A) (11). When in contact with the substratum, these motions tether the OM platform at bFAs (17), which is proposed to create local adhesions and movement of the cell (Fig. 1A).
A putative protein platform at the cell surface could be constituted by multiple integral OM Glt proteins, including predicted β barrels GltA, GltB, and GltH (formerly CglE/AgmV), as well as the OM-lipoprotein GltK (formerly CglC/AgmO) and OM-associated periplasmic protein GltC (Fig. 1A) (6, 7, 17). While little is known about GltH function [except that it is required for gliding motility (7, 18)], GltK/B/A/C are all encoded by the same gene cluster (7) and there is evidence that they form a functional complex (17). Specifically, GltA interacts with GltB and each protein cannot be stably expressed in the absence of the other (17). The periplasmic protein GltC also interacts with both GltA and GltB, and its expression can only be detected if these two proteins are also expressed (17). Lastly, GltK appears to be required for the proper insertion of GltA and GltB into the OM (17). During gliding, GltA, GltB, and GltC are all recruited at bFAs (17), suggesting that they are important for contact with the substratum. However, a precise adhesion function could not be established because single deletions of gltA/B/C all abolish bFA formation (11, 17), revealing that this OM complex is also essential for the assembled structure at bFAs.
In this study, we identify a hetero-oligomeric OM complex formed by GltABCHK. Furthermore, we demonstrate that the OM platform regulates recruitment, exposure, and retention of the OM lipoprotein CglB at the cell surface. In turn, CglB is shown to function as a principal adhesin essential for coupling the trans-envelope Glt apparatus to the substratum, thus mediating gliding. These results support a gliding model in which a CglB-loaded OM platform selectively unmasks the adhesin upon stimulation by the motorized IM complex, thus coupling the gliding machinery to the substratum and creating a bFA.
RESULTS
CglB, a predicted von Willebrand A domain-containing protein, is a candidate motility adhesin
We first searched for a candidate adhesin that might interact with the OM platform. The CglB protein is an ideal candidate because it is essential for single-cell gliding motility (7, 18–20) (analogous to Glt OM-platform constituents) (fig. S1A), and it localizes to the M. xanthus OM as a lipoprotein (21, 22). Homology searching across diverse bacterial genomes revealed that cglB co-occurs with genes encoding the complete Agl–Glt machinery in bacterial genomes, supporting a functional link (fig. S2).
CglB has been proposed to contain a von Willebrand A (VWA) domain (17, 23). Fold-recognition analysis of CglB indicated structural analogies with numerous metazoan α integrins (table S1). Of the 18 identified human α-subunit integrin variants, half have an intervening module (termed αI or αA) containing a VWA domain (24), characterized by a Rossmann fold with multiple α helices shielding an interior β sheet (25). Structurally similar Apicomplexan parasite gliding motility adhesins micronemal protein 2 (MIC2) and thrombospondin-related anonymous protein (TRAP) (from Toxoplasma and Plasmodium, respectively) (26, 27) containing VWA domains (Pfam: PF00092) typically involved in adhesion (25) were also matched to CglB (table S1). The top bacterial match was the αI/αA domain-like GBS104 adhesive tip pilin (28) from Streptococcus agalactiae (table S1). In bacteria, VWA domains have been much less studied but they are also involved in adhesion as it has recently been shown that type IV pili in Streptococcus sanguinis adhere via the VWA module in two-domain pilins inserted in the pilus fiber (29).
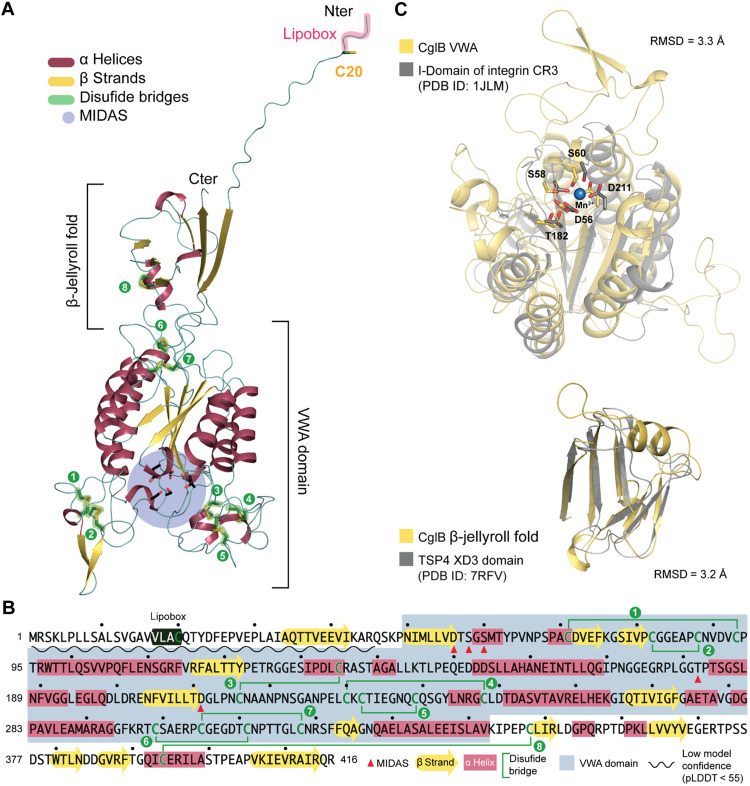
Generation of a CglB tertiary-structure model using AlphaFold2 (30, 31) confirmed that CglB likely contains a VWA domain (Fig. 2, A and B, and fig. S3). In turn, the VWA domain contains a predicted MIDAS (metal ion-dependent adhesion site) motif, a discontinuous structural feature (Asp-x-Ser-x-Ser…Thr…Asp). In general, the coordination of a divalent metal ion (e.g., Ca2+/Mg2+/Mn2+) at this site induces structural changes in VWA domains upon ligand binding that stabilize this adhesive domain in a high-affinity state for the ligand (32). For CglB, highly-conserved putative MIDAS residues map to D56, S58, S60, T182, and D211 (Fig. 2, B and C). An N-terminal β-jellyroll domain was also predicted for CglB (Fig. 2, A and B). While the function of such a domain in CglB is unknown, these domains promote oligomerization in viral capsid proteins (33). Lastly, CglB contains a high number of Cys residues (17 of 416 amino acids = 4.1%) (fig. S1B) that are predicted to stabilize structural loops of the protein by forming disulfide bonds (Fig. 2A and fig. S3) at the necks of these loops; this notion was supported by the observation that titration of reducing agent resulted in a migration shift from faster- to slower-moving CglB-specific bands via SDS–polyacrylamide gel electrophoresis (PAGE) and α-CglB Western immunoblot (Fig. 3A).
Fig. 2. CglB is a cell-surface protein with a potential integrin αI-domain-like VWA fold.
(A) AlphaFold model of CglB. CglB is predicted to contain a VWA domain and a smaller domain adopting a β-jellyroll fold. Within the CglB VWA domain, conserved residues previously described in VWA domains to coordinate divalent cations and constituting the metal ion-dependent adhesion site (MIDAS) are also present (blue circle). CglB also contains a lipobox motif with a conserved cysteine (C20). The other 16 cysteines are likely involved in disulfide bridges that stabilize the CglB structure. (B) Amino acid sequence of CglB. Secondary structures are reported as well as the cysteines potentially forming disulfide bonds (green lines). The limits of the VWA domain are also reported in gray, and the MIDAS residues are highlighted with red triangles. (C) The Dali server was used to scan the CglB structural model against the Protein Data Bank (PDB). Top: Structural alignment of CglB VWA model (yellow) to the αI domain of integrin CR3 (gray, PDB ID: 1JLM) (80). Predicted CglB MIDAS residues superimpose with the MIDAS residues (coordinating Mn2+) of the integrin CR3. Bottom: Structural alignment of CglB smaller domain adopting a β-jellyroll fold (yellow) to the XD3 domain of the bacteriophage tailspike protein 4 (TSP4 in gray, PDB ID: 7RFV) (81). RMSD, root-mean-square deviation; pLDDT, predicted local distance difference test.
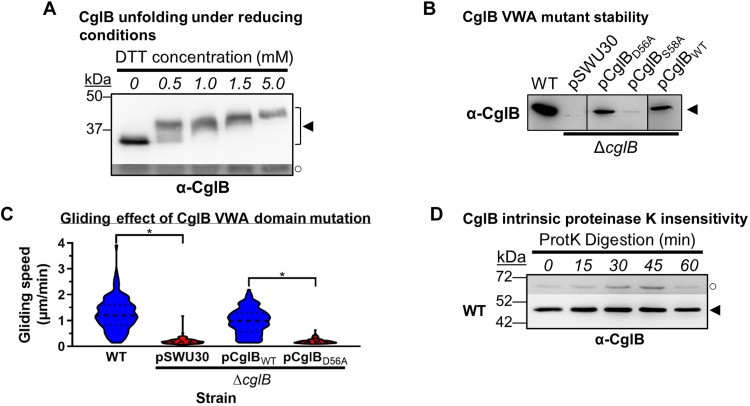
Fig. 3. The VWA domain is important for CglB stability and function.
(A) α-CglB Western blot of WT cell lysates treated with increasing dithiothreitol (DTT) concentrations to break disulfide bonds. The lower/darker zone on the blot corresponds to the same blot image section shown with higher contrast to highlight lower-intensity bands. Legend: ◄, full-length CglB; ○, loading control (nonspecific band labeled by α-CglB polyclonal antibody). (B) α-CglB Western immunoblot of CglB MIDAS motif mutant lysates. Nonadjacent lanes from the same blot are separated by vertical black lines. (C) Violin plots of single-cell gliding speeds on hard (1.5%) agar for M. xanthus DZ2 ΔcglB (n = 120 cells) complemented with CglBWT or CglBD56A. The median (dashed line) as well as lower and upper quartiles (dotted lines) are indicated. Asterisks denote datasets displaying statistically significant dataset differences (P < 0.0001) compared to strains harboring either the pSWU30 empty-vector control or the pCglBD56A VWA domain mutant CglB complementation construct, as determined via two-tailed Mann-Whitney U tests. (D) Protein samples from WT cells resuspended in tris-phosphate-magnesium (TPM) buffer and digested with exogenous proteinase K. Aliquots of the digestion mixture were removed at 15-min intervals and trichloroacetic acid (TCA)-precipitated to stop digestion. The higher, darker zone on the blot corresponds to a section of the same blot image for which the contrast has been increased to highlight lower-intensity protein bands. The lack of CglB degradation was not due to lack of proteinase K activity (see below). Legend: ◄, full-length CglB; ○, loading control (nonspecific protein band labeled by α-CglB antibody).
We thus tested the potential in vivo function of the MIDAS motif by complementing a ΔcglB strain via ectopic expression of a CglBD56A mutant construct. Contrary to ΔcglB cells in which CglBWT expression restored motility, CglBD56A was stably expressed (unlike CglBS58A) but failed to complement gliding deficiency (Fig. 3, B and C). These data are consistent with the MIDAS motif being functionally important, suggesting overall that the predicted VWA domain and intraprotein disulfide bonds are important structural determinants of CglB.
CglB is exposed at the cell surface at bFA sites in gliding cells
If CglB functions as an adhesin to anchor the Glt complex to the substratum, then it would be expected for CglB to be detectable at the cell surface. This would be an important determination as OM lipoproteins are generally considered to localize to the periplasmic leaflet of the OM. However, this does not preclude surface localization, with a growing list of OM lipoproteins having been shown to be surface-exposed in bacteria (34, 35). To probe cell-surface exposure of CglB, its susceptibility to nonspecific digestion by proteinase K was tested on intact liquid-grown cells (17). In wild-type (WT) cells, the cellular pool of CglB was not substantially depleted by this treatment (Fig. 3D), suggesting that CglB is either (i) present at the cell surface but protected from digestion or (ii) located in the periplasmic leaflet of the OM and is thus protected from extracellular digestion by proteinase K.
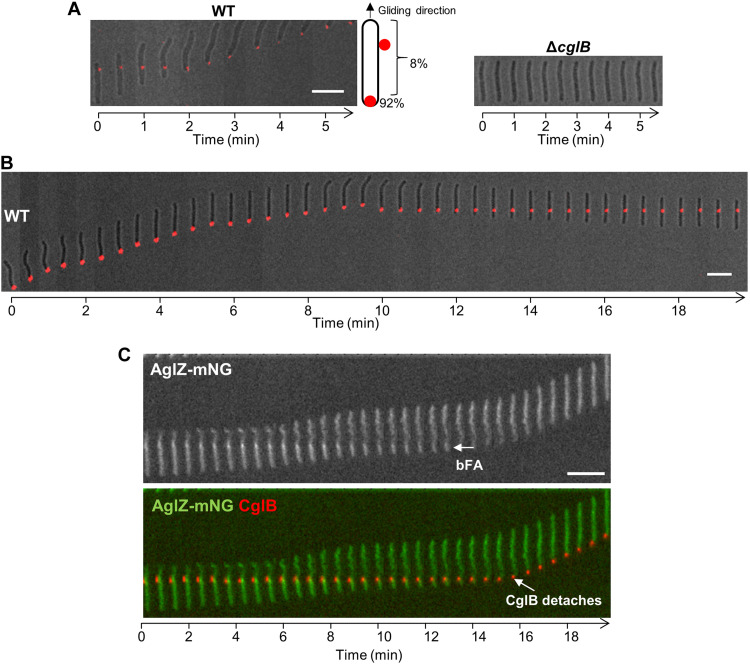
To visualize CglB localization, we first attempted to express the protein with a C-terminal translational fusion to either the fluorescent mNeonGreen reporter or the self-labeling Halo tag; however, both approaches were unsuccessful because of loss of CglB functionality. To circumvent this problem, we instead used an immunofluorescence approach, treating WT and ΔcglB cells with α-CglB primary antibodies and fluorescently labeled secondary antibodies, followed by live-cell fluorescence microscopy. As expected, 100% of individual ΔcglB cells (361 in total) imaged on agar pads did not exhibit any immunofluorescence (Fig. 4A). Conversely, 53% of single-gliding WT cells (i.e., 295 of 552) across four independent experiments displayed a fluorescent signal, manifesting as a single fluorescent cluster in each (Fig. 4A). Consistent with localization at bFAs, α-CglB immunofluorescent clusters detected along the length of the motile cell body [~8% (25 of 295) of labeled cells] remained stationary relative to the substratum, while the cell glided forward relative to the fixed fluorescent signal (Fig. 4A). Once the rear of a gliding cell arrived at the position of the fluorescent cluster, the cell began to drag the cluster behind it at the lagging pole (Fig. 4A), demonstrating that the cluster is attached to the cell (and not transiently associated). Dragged clusters accounted for ~92% (i.e., 270 of 295) of fluorescent foci in motile cells, suggesting that CglB does not adhere to the substratum at the back of the cells. Incidentally, only ~1% of cells (i.e., 3 of 270) that dragged a cluster later left this cluster behind on the substratum. However, upon reversal of gliding direction, it was possible for a “dragged” fluorescent cluster to become immobilized once again relative to the substratum, while the gliding cell moved relative to the fixed cluster (Fig. 4B).
Fig. 4. CglB is a cell-surface protein that localizes to bFA sites.
(A) Montage of live WT and ΔcglB cells immunolabeled with α-CglB 1° antibody, followed by goat α-rabbit 2° antibody conjugated to Alexa Fluor 647 on agar pads at 32°C. Images were acquired at 30-s intervals. Scale bar, 5 μm. (B) Montage of a live immunolabeled WT cell [labeled as in (A)] in which a dragged fluorescent cluster becomes immobilized relative to the substratum upon reversal of gliding direction. Scale bar, 5 μm. (C) Montage of a live immunolabeled cell expressing AglZ-mNeonGreen (82) in which an α-CglB antibody colocalizes with a fixed AglZ-mNeonGreen cluster at a bFA site. Note that the CglB cluster detaches from the surface when it reaches the lagging cell pole. Scale bar, 5 μm.
The above-described results are consistent with CglB being associated with bFAs that become active at the leading cell pole and maintain a fixed position relative to the substratum until they are disassembled at the lagging cell pole (Fig. 1B). To probe for signal overlap between CglB and bFA sites, we immunolabeled WT cells expressing fluorescently tagged AglZ for simultaneous detection of CglB and imaged via live-cell fluorescence microscopy. This analysis revealed that CglB is detected at bFA sites (Fig. 4C). It is intriguing that only single CglB clusters were observed at bFAs, given that multiple bFAs can sometimes be detected in the same cell (Fig. 1B). However, it is unclear how each of these bFAs contributes to locomotion. For example, a specific mutant strain (MglAQ82A/L) that only assembles one bFA per cell moves as fast as cells that assemble several bFAs (14), so there may be distinct features of these bFAs that need to be clarified. It is also possible that the immunolabeling procedure, which requires prelabeling of CglB before imaging, is also limiting the detection of additional clusters. Thus, together with the lack of intrinsic proteinase K susceptibility for CglB in WT cells, these results suggest that CglB is either (i) selectively transported from the periplasmic face of the OM to the cell surface at bFAs or (ii) masked at the cell surface until it is exposed at bFA sites during cell-gliding events.
CglB is essential for substratum coupling of the Agl–Glt machinery
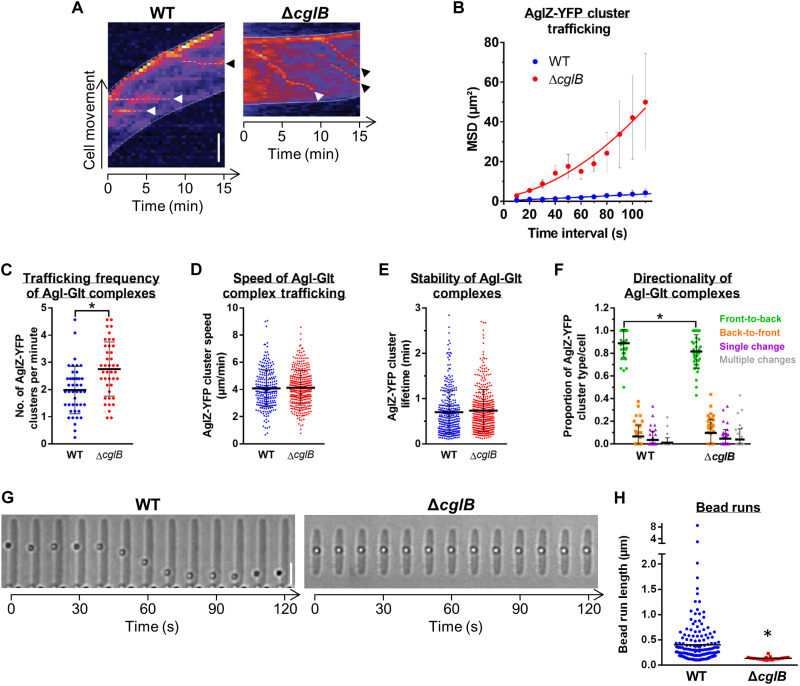
We subsequently investigated the contribution of CglB to surface coupling of the Agl–Glt complex. To probe the role of CglB in bFA formation, we analyzed the dynamics of AglZ-YFP (yellow fluorescent protein) clusters in cells on hard agar for the ΔcglB mutant (which stably expresses AglZ-YFP; fig. S4A). AglZ-YFP clusters still appeared in ΔcglB cells; however, in marked contrast to WT cells (Fig. 1B), AglZ-YFP clusters in ΔcglB cells were not stationary relative to the substratum but rather moved directionally from one pole to the other (Fig. 5, A and B, and fig. S4B). This behavior was consistent with that observed previously in nonadhered motility complexes (11). CglB is therefore required to immobilize trafficked AglZ-YFP clusters (relative to the substratum) and assemble bFAs on hard agar surfaces. The function of CglB is clearly distinct from the OM-platform β-barrel proteins because trafficking AglZ-YFP clusters are not formed in any of the ΔgltA/B/H mutant backgrounds (11).
Fig. 5. CglB is essential for gliding-complex substratum adhesion.
(A) Temporal AglZ-YFP cluster position (dashed lines) kymographs (cells on agar pads). Scale bar, 2 μm. White arrowheads: Clusters followed for entire lifetimes. Black arrowheads: Clusters followed for incomplete lifetimes. (B) Mean square displacement (MSD) of AglZ-YFP cluster position tracking in WT (n = 48 clusters) and ΔcglB (n = 23 clusters) cells. Mean MSD (± SEM) at each time interval is displayed, with a second-order polynomial fit to each dataset. For (C) to (F), experiments were performed via TIRFM on chitosan-coated glass in polydimethylsiloxane microfluidic chambers; mean values are indicated by a black line ± SEM. Distributions of the WT–ΔcglB datasets were compared via unpaired two-tailed Mann-Whitney U tests; those with significant differences (P < 0.05) are indicated (*). (C) AglZ-YFP complex trafficking frequency: WT (n = 44 cells), ΔcglB (n = 41 cells). (D) Agl-Glt complex trafficking speed: WT (n = 260 clusters), ΔcglB (n = 371 clusters). (E) Trafficking Agl–Glt complex stability: WT (n = 333 clusters), ΔcglB (n = 409 clusters). (F) Directionality of trafficked Agl–Glt complexes: WT (n = 44 cells), ΔcglB (n = 41 cells). Front and back are defined as cell poles with high/low AglZ-YFP fluorescence intensity, respectively. (G) Trafficking phenotypes of surface-deposited polystyrene beads. Scale bar, 3 μm. (H) Lengths of tracked bead runs >0.1 μm. Images from 10-s intervals were analyzed. The distributions of the two datasets are significantly different (*) as determined via unpaired two-tailed Mann-Whitney U test (P < 0.05).
In ΔcglB cells, trafficking AglZ-YFP clusters move in and out of the epifluorescence focal plane as they rotate counterclockwise around the cell envelope (Fig. 1B and fig. S4B), making it difficult to precisely track the nonadhered foci and thus accurately study their dynamic properties. To resolve these difficulties, we recently developed a total internal reflection fluorescence microscopy (TIRFM) assay in which M. xanthus cells glide in chitosan-coated microfluidic chambers (fig. S4C); in this system, trafficking AglZ-YFP clusters are also observable in WT cells because of the suboptimal nature of the chitosan surface for M. xanthus bFA adhesion (11, 36). Because the depth of field in TIRFM is restricted to near the cell-substratum interface, photobleaching is reduced and thus tracking of the trafficking AglZ-YFP clusters near the ventral face of the cell can be performed at high spatiotemporal resolution (11). On chitosan, ΔcglB cells were also nonmotile, and again, immobilized AglZ-YFP clusters could not be detected (fig. S4C). Trafficking AglZ-YFP clusters in ΔcglB cells behaved similarly to those in WT cells; although slight effects were observed via TIRFM on the trafficking frequency of AglZ-YFP clusters (from the leading to the lagging cell poles), the trafficking speed and lifetime of AglZ-YFP clusters were unchanged in the absence of CglB (Fig. 5, C to F). Because AglZ-YFP trafficking reflects the activity of the motility engine (11), we conclude that CglB does not affect the activity of the motor, but rather its adhesion to the underlying substratum at bFAs.
To test the contribution of adhesive properties by CglB to the tip of the motility complex, we adopted a force microscopy approach; here, force generation by the motility complex can be directly monitored in live M. xanthus cells immobilized atop a semisolid agarose matrix deposited on glass slides (15). In this environment, the motility complex cannot propel cells (likely because it cannot adhere to the substratum), but its activity can transport polystyrene beads that are nonspecifically adsorbed to the cell surface after being deposited using an optical trap (Fig. 5G). Trafficking gliding machinery units that collide with and recruit these beads move them directionally over long distances (fig. S4D) (15, 16). We therefore tested whether bead transport requires the CglB adhesin. While beads were transported multiple times, at lengths up to ~8 μm, along the surface of WT cells, these events were nonexistent in ΔcglB cells (Fig. 5, G and H). This demonstrates that bead recruitment and trafficking require CglB, consistent with adhesion and force-transduction functions for CglB.
Together, we conclude that CglB is required for tethering the gliding motility complex to an engaged extracellular motif, be it a solid surface for cell gliding or cargo for transport in immobilized cells. Contrary to the OM-platform proteins GltA/B/H (11) (see below), CglB is not required for Agl–Glt complex assembly and trafficking, suggesting that it functions to couple trafficking units to the substratum, as would be expected for an adhesin essential for gliding motility.
The Glt OM platform regulates CglB exposure and retention at the cell surface
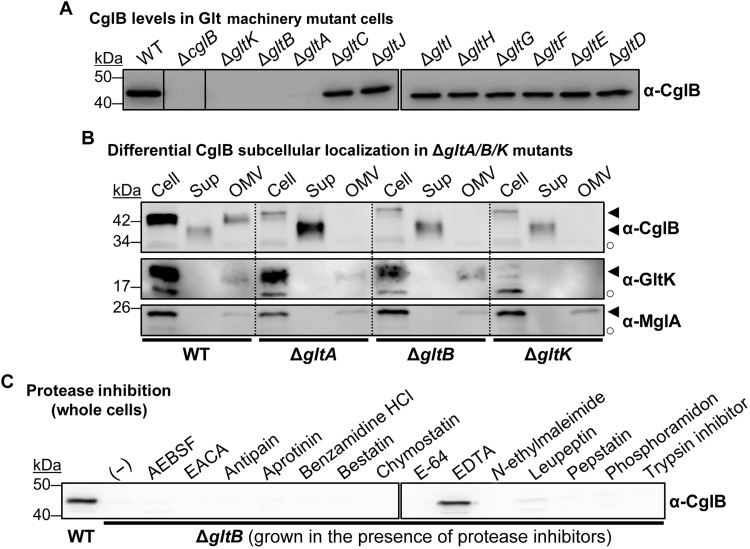
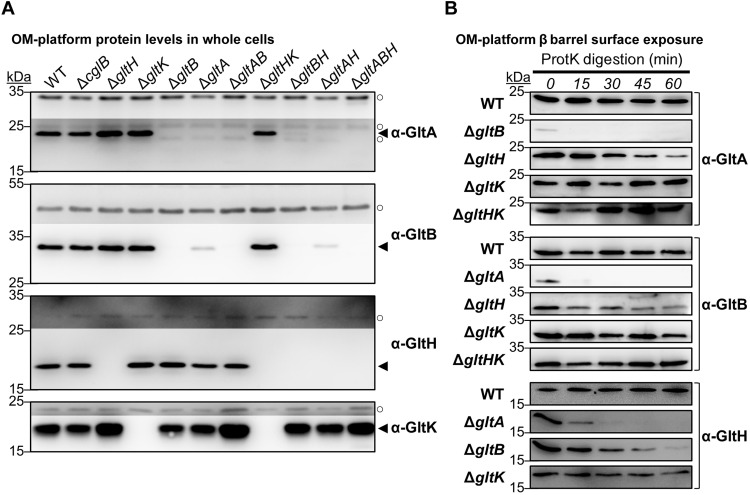
We next set out to examine the factors that regulate the exposure of the CglB adhesin at the cell surface. Because the surface dynamics of CglB suggest a direct connection between CglB and the Agl–Glt complex, we first compared CglB levels in whole-cell samples of each respective glt mutant strain. While present at comparable levels in ΔgltC/D/E/F/G/H/I/J backgrounds, cell-associated CglB was severely depleted in OM-platform mutants ΔgltA, ΔgltB, and ΔgltK (but not ΔgltH) (Fig. 6A). Given that ΔgltK cells are deficient in OM-inserted GltA and GltB (17), the nature of the CglB deficiency in ΔgltK cells may be the same as that in both ΔgltA and ΔgltB cells, namely, an absence of OM-integrated GltA and GltB β barrels.
Fig. 6. Glt OM-platform constituents mediate cellular retention of CglB.
(A) Whole-cell extracts from different Δglt mutants. Nonadjacent lanes on the same blot are separated by vertical black lines. White space separates two distinct blots processed at the same time. (B) Fractionated samples containing whole cells (Cell), supernatants (Sup), and OM vesicles (OMV) from various genetic backgrounds. Detection of the gliding motility OM lipoprotein GltK was added as a control, with the protein only detected in Cell and OMV samples, showing that the various mutations do not affect OM integrity, with the supernatant localization in this instance being specific to CglB. MglA is a cytoplasmic protein added as a control to show that cell lysis is negligible and does not account for the presence of CglB in supernatants. Legend: ◄, full-length protein; ○, loading control (nonspecific protein band labeled by the respective polyclonal antibody). (C) Whole-cell extracts from ΔgltB cells grown in the presence of different protease inhibitors. White space separates two distinct blots from the same experiment. AEBSF, 4-(2-aminoethyl benzenesulfonyl fluoride HCl; EACA, ε-aminocaproic acid.
Fractionation analysis revealed that CglB was still produced by the ΔgltA/B/K mutants (Fig. 6B); however, unlike in WT cells—where CglB was detected in whole-cell and OM vesicle (OMV) fractions—CglB in these three mutant backgrounds was only recovered in culture supernatants (Fig. 6B and fig. S5A). In the ΔgltA and ΔgltB mutants, such shedding to the supernatant was not observed for (i) GltK (Fig. 6B) which remained OMV-associated nor for (ii) the cytoplasmic protein MglA, which was detected at levels comparable to the WT strain (Fig. 6B). Therefore, cell association (and thus OM localization) of CglB depends on GltA, GltB, and GltK.
Supernatant-localized CglB from ΔgltA/B/K cultures was found to migrate faster than cell-associated CglB via SDS-PAGE (in both whole-cell and OMV samples) under equivalent denaturing conditions (Fig. 6B), suggesting that supernatant CglB is of reduced molecular weight and may have been proteolytically cleaved. Further support for proteolytic processing of CglB was provided via mass spectrometry analysis of tryptic peptides obtained from CglB immunoprecipitated from supernatant, which revealed that the first 76 N-terminal residues were unaccounted for (fig. S5B). Our efforts at N-terminal sequencing of supernatant-isolated CglB were inconclusive, and hence, we were unable to identify the initial amino acids of the truncated protein. Nonetheless, these data suggest that CglB may be cleaved by a protease in ΔgltA/B/K cells before its release into the supernatant. To test this hypothesis, we screened the effect of various protease inhibitors for their capacity at restoring CglB localization to the cell envelope in ΔgltB cells. Growth in the presence of EDTA restored cell-associated CglB in this background (Fig. 6C). Similarly, EDTA also restored cell-associated CglB in the ΔgltK and ΔgltA mutants (Fig. 7A). Under these conditions, CglB was detected as a doublet band, the relative ratio of which varied depending on the mutant background (Fig. 7A) (discussed below). Nevertheless, resuspension of EDTA-grown ΔgltB cells in EDTA-free buffer resulted in the resumption of CglB release from the cells, indicating that CglB restoration is not permanent, consistent with a protease-inhibition effect rather than a nonspecific effect of EDTA on the cell envelope (fig. S5C). Because EDTA chelates divalent cations, CglB release from ΔgltA/B/K cells would be consistent with the activity of a metalloprotease, the identification of which was not within the scope of this paper and which will require downstream experimentation.
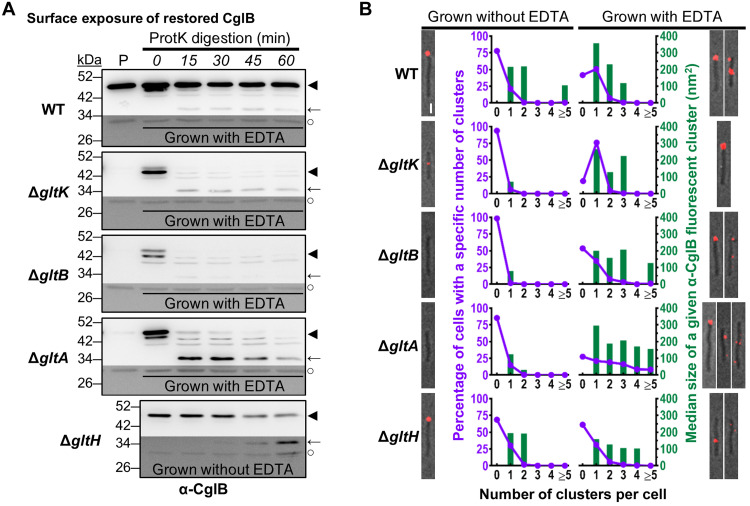
Fig. 7. CglB surface exposure is mediated by the Glt OM platform.
(A) Protein samples from intact cells digested with exogenous proteinase K. Digestion mixture aliquots were removed at 15-min intervals and TCA-precipitated to stop digestion. “P”: Lanes containing the untreated parent strain grown without EDTA. Lower/darker zones on each blot correspond to sections of the same blot image for which the contrast has been increased to highlight lower-intensity protein bands. The samples and blot for ΔgltH were obtained at the same time as that for WT (Fig. 3D), indicating that the proteinase K was active during treatment of the latter. Legend: ◄, full-length CglB; ←, CglB degradation band; ○, loading control (nonspecific band labeled by α-CglB antibody). (B) Fluorescence micrographs of live immunolabeled WT, ΔgltK/B/A/H cells grown with(out) EDTA (labeled with α-CglB 1° antibody, followed by goat α-rabbit 2° antibody conjugated to Alexa Fluor 647) on agar pads at 32°C. Representative images are provided for cluster-labeling patterns observed on ~20% or more of analyzed cells for a given strain/treatment. Scale bar, 1 μm. For each strain grown with(out) EDTA, the number of fluorescent clusters detected per cell was counted (x axis) and compared against the proportion of cells with such a labeling phenotype (purple left-side y axis). The size of each cluster was also measured, with the median area (dark green right-side y axis) given for each labeling phenotype. The number of cells analyzed for each treatment is as follows (±EDTA): WT 304/347, ΔgltK 306/306, ΔgltB 437/199, ΔgltA 424/251, and ΔgltH 505/471.
The CglB doublet band described above was only detected in all EDTA-grown WT and ΔgltA/B/K samples that were subjected to protein denaturation and precipitation via treatment with trichloroacetic acid (TCA) and acetone (Figs. 7A and 8B). Hence, the doublet is very likely an artifact of altered refolding (following TCA precipitation) in samples initially depleted of divalent cations by the effect of EDTA. These data are consistent with the proposed coordination of a divalent cation by CglB (Figs. 2C and 3B). Altered CglB refolding is further supported by the observation that no doublet band was detected in EDTA-grown samples that were not subjected to TCA precipitation before sample resolution via SDS-PAGE (Fig. 6C and fig. S5C).
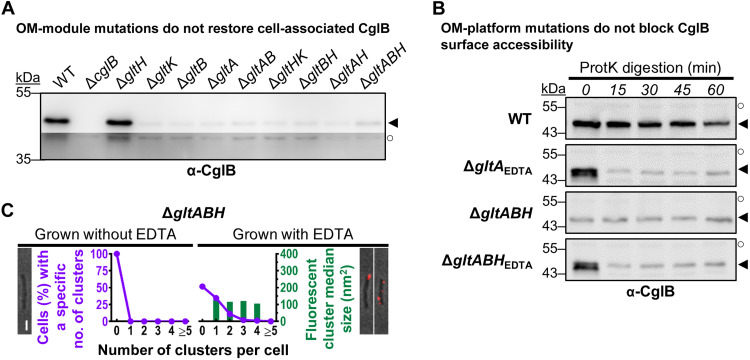
Fig. 8. CglB secretion to the cell surface is not mediated by the Glt OM platform.
(A) α-CglB Western immunoblots for whole-cell extracts from different combinations of Δglt OM-module mutations in the same strain. Legend: ◄, full-length CglB; ○, loading control (nonspecific protein band labeled by α-CglB polyclonal antibody). (B) α-CglB Western immunoblots for protein samples from cells resuspended in TPM buffer and digested with exogenous proteinase K. Aliquots of the digestion mixture were removed at 15-min intervals and TCA-precipitated to stop digestion. Legend: ◄, full-length CglB; ○, loading control (nonspecific protein band labeled by α-CglB polyclonal antibody). (C) Fluorescence micrographs of live immunolabeled ΔgltABH cells grown with(out) EDTA (labeled with α-CglB 1° antibody, followed by goat α-rabbit 2° antibody conjugated to Alexa Fluor 647) on agar pads at 32°C. Representative images are provided for cluster-labeling patterns observed on ~20% or more of analyzed cells for a given treatment. Scale bar, 1 μm. For cultures grown with(out) EDTA, the number of fluorescent clusters detected per cell was counted (x axis) and compared against the proportion of cells with such a labeling phenotype (purple left-side y axis). The size of each cluster was also measured, with the median area (dark green right-side y axis) given for each labeling phenotype. The number of cells analyzed for each treatment is as follows (±EDTA): ΔgltABH 370/344.
To determine the subcellular localization of restored CglB in EDTA-grown ΔgltA, ΔgltB, and ΔgltK mutants, we probed CglB sensitivity to exogenous proteinase K. In WT cells grown with EDTA, CglB was again protected from proteinase K attack (Fig. 7A), analogous to results in the absence of the chelator (Fig. 3D). In stark contrast, cell-associated restored CglB was immediately digested by proteinase K in the EDTA-grown ΔgltK, ΔgltB, and ΔgltA backgrounds (Fig. 7A). Upon digestion of full-length CglB in ΔgltA cells, there was an immediate appearance of an ~34-kDa CglB degradation product that was detected throughout the time course, suggesting that it was partially protected from further digestion. This protection required GltK and GltB as the ~34-kDa product was almost undetectable in the respective mutant backgrounds (Fig. 7A). As a predicted β barrel gliding-motility protein, GltH could also be a component of the OM platform. If so, its connection to CglB may not be as central as GltA and GltB given that CglB remained cell-associated in the ΔgltH mutant (Fig. 6A). Nevertheless, the cellular pool of full-length CglB decreased steadily throughout the proteinase K digestion time course in the absence of GltH, with a concurrent appearance and steady accumulation of an ~34-kDa proteinase K–resistant band (Fig. 7A).
In these putative OM-platform mutants, to assure that differences in proteinase K (a 28,900-Da protein) susceptibility of CglB were not somehow due to increased OM permeability in the various EDTA-grown mutant strains, we first tested the proteinase K susceptibility of an IMss-mCherry construct expressed in these backgrounds; this is a modified fluorescent mCherry reporter that localizes to the periplasmic space but which remains tethered to the IM (37). No degradation of the mCherry signal was detected in this experiment (fig. S5D). Next, we probed the sensitivity of these mutants to killing with vancomycin (a 1449-Da molecule), an antibiotic that disrupts periplasmic PG biosynthesis and more effectively so if the OM is permeable (38). This analysis revealed no increased drug susceptibility of the various mutants relative to WT (irrespective of growth in the absence/presence of EDTA) (fig. S5, E and F). Thus, EDTA treatment and mutations in the OM platform do not make cells more permeable than WT cells for entry of bulky molecules such as proteinase K (or antibodies; see below).
To directly demonstrate that the proteinase K sensitivity of CglB in ΔgltA, ΔgltB, and ΔgltK cells (with EDTA) and ΔgltH cells (without EDTA) reflects exposure of CglB at the cell surface, we probed these cells with α-CglB antibodies for immunolabeling as described above. In each of ΔgltA/B/KEDTA, CglB was detected in typically one to three clusters (Fig. 7B, purple lines) spread around the cell periphery, confirming cell-surface localization of CglB in these cells. Scattered foci were also 26 to 65% smaller in median size than those in WTEDTA cells (Fig. 7B, green bars); these data are consistent with compromised gliding motor-mediated CglB transport and clustering, which in WT cells would typically result in bFA formation (11). Similar data were obtained for ΔgltH cells, in which α-CglB signal was detected around the periphery in dispersed foci (Fig. 7B, purple lines) of smaller median size (Fig. 7B, green bars) than those in WT cells.
Together, the abovementioned results further support the notion that interactions between GltA, GltB, GltH, and GltK regulate CglB exposure at the cell surface. Specifically, these results indicate that CglB becomes surface-exposed in the absence of individual OM-platform components GltA, GltB, GltK, and, to a lesser extent, GltH. The differential effects observed in the various OM-platform mutant strains for both proteinase K susceptibility and immunofluorescence point to complex interaction schemes between these proteins that will need to be further explored (see Discussion).
The Glt OM β-barrel proteins are not required for CglB secretion to the cell surface
Two hypotheses could explain the cell-surface protease sensitivity of CglB in ΔgltA/B/K/H cells: (i) CglB accesses the cell surface via an as-yet unknown system and subsequently interacts with the Glt OM-platform proteins, which shield the adhesin from the action of the putative surface metalloprotease. Alternatively, (ii) the Glt OM-platform proteins are directly responsible for CglB cell-surface exposure through a regulated pore-like function that becomes constitutive as soon as one of its components (i.e., GltA, GltB, GltK, and, to a lesser extent, GltH) is removed.
To examine whether the OM-platform β barrel proteins form a pore through which CglB is exported across the OM and reaches the cell surface, we first probed cell association of CglB in double- and triple-mutant cells lacking various combinations of the OM-platform β barrel proteins. The rationale herein was that by removing all potential β-barrel pore components, this would prevent the secretion of CglB to the cell surface (and its downstream release from the cell). However, in all tested β-barrel mutant combinations, the level of cell-associated CglB remained depleted relative to WT cells, even in ΔgltABH triple-mutant cells lacking any of the β-barrel components of the OM platform (Fig. 8A). Expectedly, CglB was instead found to be enriched in the supernatants of the various β-barrel double- and triple-mutant backgrounds (fig. S5A). Akin to restored CglB in the single-mutant ΔgltA and ΔgltB strains (Fig. 7A), restored CglB in ΔgltABH triple-mutant cells (grown in the presence of EDTA) was also rapidly degraded by treatment with proteinase K (Fig. 8B), indicating that localization of the gliding adhesin to the M. xanthus cell surface is independent of the Glt OM platform.
Surface localization of CglB independent of the Glt OM platform was also probed via live-cell α-CglB immunolabeling in various strains with different combinations of OM Glt component deficiencies, grown in the absence/presence of EDTA. Most convincingly, even ΔgltABH triple-knockout cells [analogous to double-knockout combinations (fig. S6, purple lines)] grown in the presence of EDTA displayed extensive surface decoration with the α-CglB antibody, which was lacking in non-EDTA–grown cells (Fig. 8C, purple lines). It is interesting that fluorescent clusters were detected even in ΔgltABHEDTA cells (albeit of a smaller size); this could be due to CglB–CglB interactions or interaction with an as-yet unidentified partner at the surface (Fig. 8C).
Together, these proteinase K susceptibility and α-CglB immunolabeling data indicate that in the absence of all OM-platform β-barrel components, the lipoprotein CglB is still secreted to the cell surface. Therefore, CglB does not access the cell surface via a pore formed by GltABH; instead, the adhesin must become surface-localized via an as-yet unknown mechanism after which it becomes shielded by members of the Glt OM platform.
CglB directly interacts with a GltABCHK heteroligomeric OM protein complex
Lastly, we set out to characterize the nature of the proposed Glt OM platform. We began by probing for any interdependencies within the OM-platform proteins given that GltA and GltB were already known to be reciprocally depleted in ΔgltB and ΔgltA single-mutant cells (respectively), and that insertion of GltA and GltB into the OM requires the function of GltK (17). The levels of GltA, GltB, GltH, and GltK in single-, double-, and triple-mutant backgrounds corresponding to various constituents of the OM module were thus tested. Consistent with previous data (17), both GltA and GltB were stably expressed at equivalent levels across all mutant combinations, except in instances where the gltA and/or gltB genes were deleted; neither GltH nor GltK had an impact on the levels of GltA or GltB. Conversely, GltH and GltK were stably expressed in all mutant backgrounds, except for strains carrying the respective deletion (Fig. 9A). Therefore, while GltA affects the stability of GltB (and vice versa), neither GltH nor GltK affects the levels of any OM-module constituent.
Fig. 9. Glt OM-platform constituents exhibit interdependencies.
(A) Western immunoblots of GltA, GltB, GltH, and GltK in various single-, double-, and triple-mutant combinations of OM-platform constituents. Legend: ◄, full-length protein; ○, loading control (nonspecific protein band labeled by the respective α-GltA/α-GltB/α-GltH/α-GltK polyclonal antibody). (B) Western immunoblots for Glt OM-platform β-barrel constituent susceptibility to digestion by proteinase K in Glt OM-module mutant strains. Digestion aliquots were removed at 15-min intervals and TCA-precipitated to stop digestion.
To support the notion of a bonafide OM-platform protein complex, the proteinase K susceptibility of a given constituent was also tested in the absence of a different OM-platform protein. The rationale was that cell-surface topology for various OM-platform proteins could be altered because of a disrupted interaction network resulting from the missing platform component. As previously detected (17), neither GltA nor GltB was stable in a mutant background lacking the other (Fig. 9, A and B). The absence of GltH rendered GltA and GltB more proteinase K sensitive (Fig. 9B). Similarly, GltH was more sensitive to proteinase K digestion in the absence of GltA or GltB (Fig. 9B). The absence of GltK did not alter the proteinase K susceptibility of any of the three integral OM β barrels (Fig. 9B).
Previously, the insertion of GltA and GltB into the OM was proposed to be compromised in the absence of GltK because GltA and GltB were not efficiently packaged into OMV samples from ΔgltK cells (17). Here, GltA and GltB were not sensitive to proteinase K digestion in the absence of GltK but were indeed sensitive in the absence of GltH (Fig. 9B), suggesting that the former two β barrel proteins were accessible to the cell surface in the latter scenario. We thus tested the proteinase K sensitivity of GltA and GltB in a ΔgltHK strain in which both of these β barrel proteins are expressed at WT levels (Fig. 9A). In this double-mutant background, both GltA and GltB remained insensitive to digestion by proteinase K located outside the cell (Fig. 9B), supporting the notion that GltA and GltB were not correctly inserted into the OM and were thus not surface-exposed in the absence of GltK and GltH. Therefore, GltK exerts its function on the OM module before that of GltH.
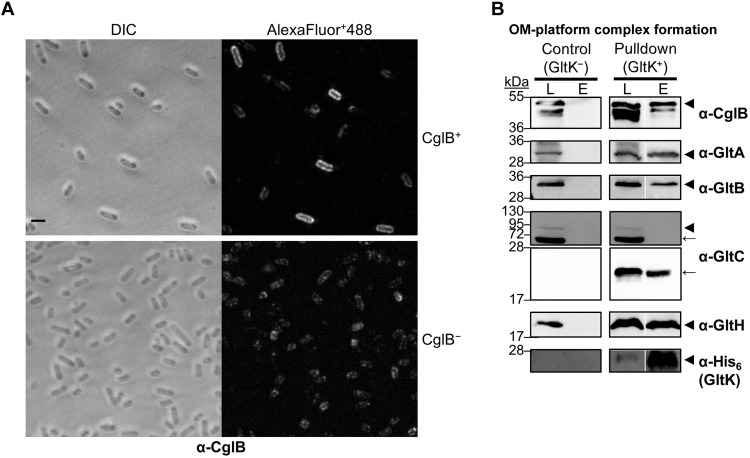
Lastly, given the evidence for functional interplay between Glt OM-platform members, we directly tested complex formation between the Glt OM-platform proteins GltA, GltB, GltC, GltH, GltK, and CglB using a biochemical pulldown approach. For this, we constructed two plasmids allowing for expression of all six proteins in Escherichia coli cells. The first plasmid contained gltK as a monocistronic sequence (encoded with a C-terminal hexa-histidine tag for pulldown experiments) and a synthetic operon of gltB, gltA, and gltC-StrepII, both downstream from T7 promoters (pCDF-Duet) (fig. S7A). This organization was chosen to mimic the genetic organization of these genes on the M. xanthus chromosome (7, 39) (a control version of this plasmid, pACYC-Duet, was created lacking gltK-His6). The second plasmid encoded gltH and cglB as a synthetic operon also downstream from a T7 promoter (pET-Duet) (fig. S7A). All genes were cloned in their entirety, including any signal sequences and lipoprotein-processing motifs, an approach previously shown to maintain targeting of M. xanthus OM proteins to the OM of E. coli (40). Following isopropyl-β-d-thiogalactopyranoside (IPTG)–induced expression in doubly transformed cells, we first tested whether CglB is also exposed at the cell surface in E. coli BL21(DE3) cells, probing intact cells by immunofluorescence with α-CglB 1º antibodies followed by Alexa Fluor Plus 488-labeled 2º antibodies. CglB was also detected at the E. coli BL21(DE3) surface in CglB+ cells as revealed by strong peripheral fluorescence around cells, which was not observed in CglB− control cells (Fig. 10A). This staining was not due to permeabilization during the labeling process because lysozyme treatment (which permeabilizes the cells) led to intense whole-cell fluorescence revealing the pool of CglB that had yet to be trafficked to the cell surface (fig. S7B).
Fig. 10. CglB directly interacts with the Glt OM-platform heteroligomeric complex.
(A) Fluorescence micrographs of E. coli BL21(DE3) cells immunolabeled with α-CglB 1° antibody, followed by goat α-rabbit 2° antibody conjugated to Alexa Fluor Plus 488 (AlexaFluor+488). Cells had been transformed with the following plasmid combinations: “pCDF-Duet-GltK6H+GltBACS & pET-Duet-GltH-CglB” (for coexpression of GltA, GltB, GltC-StrepII, GltK-His6, GltH, and CglB) or “pCDF-Duet and pET-Duet” (as empty-vector controls). Cells were induced overnight with 1.0 mM IPTG and then fixed with paraformaldehyde before immunolabeling. Scale bar, 2 μm. DIC, differential interference contrast. (B) Western immunoblotting of purified OM-platform proteins from the pulldown assay (right side) or negative control (left side) using α-CglB, α-GltA, α-GltB, α-GltH, α-His (GltK), and α-GltC 1º antibodies. Calculated molecular weights for monomeric forms of each protein construct (lacking signal peptide): CglB (42.3 kDa), GltA (25.4 kDa), GltB (27.5 kDa), GltC-StrepII (74.4 kDa), GltH (20.0 kDa), and GltK-His6 (17.5 kDa). Nonadjacent lanes from the same blot are separated by white spaces. Lane legend: L, column loading fraction; E, column elution fraction. Blot legend: ◄, full-length protein; ←, degradation product of the protein of interest.
We next performed pulldown assays on these induced cells using GltK-His6 as bait. To preserve the integrity of any OM complexes, we gently solubilized membrane fractions from lysed cells using a combination of mild detergents [n-dodecyl-β-d-maltopyranoside (DDM) and decylmaltose neopentyl glycol (DM-NPG)], followed by passage down a His-trap column, with retained proteins eluted with imidazole. As expected, GltK-His6 was specifically retained on the column; full-length GltA, GltB, GltH, and CglB were also retained on the column (Fig. 10B, triangle bands). These co-elution profiles reflect the formation of intact protein complexes, as when the experiment was repeated with plasmids encoding all proteins except GltK-His6 (fig. S7A), these proteins were similarly expressed but not retained on the column (Fig. 10B). Notably in our setup, a signal peptidase I-processed monomeric GltC (amino acids 25 to 673) with an eight-residue StrepII tag is predicted to have a molecular weight of ~74.4 kDa, similar to a previous construct cloned with a His6 tag (17). In the latter, the monomeric form of this construct was found to migrate near 100 kDa (17). However, in our experiments, full-length monomeric GltC-StrepII could not be stably maintained, though specific smaller–molecular weight degradation bands were clearly enriched and consistently detected in control and pulldown membrane fractions, with these bands also co-eluting from the column in samples containing GltK-His6 (Fig. 10B, arrow bands). Degradation bands were also detected for GltA, GltB, and CglB, but the relative signal intensity of these bands was minor in comparison (fig. S7C, arrow bands). In further support of complex formation, bands for GltA, GltB, and GltK-His6 were also observed migrating higher in the gel, representing likely oligomeric assemblies incorporating each respective protein (fig. S7C, asterisk bands). Together, these data conclusively demonstrate that the OM proteins GltA, GltB, GltC, GltK, and GltH form a heteroligomeric complex together with the adhesin CglB. This complex is stable as it could be purified in a single-step pulldown assay without the addition of cross-linking agents.
DISCUSSION
Previously, we demonstrated that, on hard surfaces, Myxococcus cells are propelled by directionally transported Agl–Glt complexes that become tethered at bFAs where they exert traction forces against the underlying substratum (11). However, the manner in which OM Glt proteins interact with the substratum and the possible implication of specific adhesins remained unclear. The characterization of CglB, a protein first studied >40 years ago (18), and its functional interactions with the OM Glt proteins (Figs. 9B and 10B) provides a potential solution to these questions.
Four main lines of evidence suggest that CglB is a cell-surface adhesin of the Agl-Glt complex:
1) CglB contains a VWA domain, which is typically found in proteins that interact with the ECM.
2) CglB becomes cell surface-exposed at bFA sites, where it forms fixed clusters that detach from the substratum (and remain cell-associated) when a bFA site reaches the lagging cell pole.
3) In the absence of CglB, trafficking motility complexes do not become immobilized, which would be expected if they fail to adhere to the surface. In addition, these trafficking complexes cannot transport surface-associated cargo in the absence of CglB.
4) CglB localizes to the cell surface where it is shielded by direct interaction with the Glt OM platform.
Below, we discuss possible adhesion mechanisms and outstanding questions for the future.
Mechanism of CglB secretion
While OM lipoproteins are generally thought to be exposed on the periplasmic leaflet of the OM, surface-exposed lipoproteins have recently come to the fore in bacterial cell biology, though the surface-exposure mechanisms for most have yet to be solved (35, 41, 42). An open question thus remains as to how exactly CglB is able to access the cell surface and subsequently associate with the integral OM β-barrel platform. Organizational parallels exist between CglB-containing Agl–Glt gliding machinery and bipartite iron-scavenging systems. In species of Neisseria and other bacterial pathogens, the cell-surface lipoproteins TbpB/LbpB (parallel: CglB) interact with the integral OM β barrel TbpA/LbpA (parallel: GltABH) to bind human transferrin/lactoferrin (respectively) and abstract iron. The TbpA/LbpA β barrels contain a Ton box and are TonB-dependent transporters (parallel: GltA/B), which can then import the abstracted iron into the cell upon activation of the integral IM ExbBD-TonB (parallel: AglRQS) motor (43, 44). Recently, a previously unknown class of transporter (corresponding to DUF560) has been shown to mediate secretion of lipoproteins such as TbpB/LbpB (and soluble proteins) across the OM (45–48). However, such a proposed “Type 11 secretion system” is not encoded in the M. xanthus genome, suggesting that this is not the mechanism for CglB trans-OM export. Considering heterologously expressed CglB can also be detected on the surface of E. coli BL21(DE3) cells (Fig. 10A), the targeting mechanism for the adhesin may depend on a more general pathway rather than an organism-specific secretion system. For instance, one possibility would be for CglB to piggyback to the cell surface during Bam-mediated OM insertion of a “host” β barrel [akin to surface lipoprotein stress sensor RcsF and OmpA (49)]. In M. xanthus, however, none of the Glt OM-platform β barrels are involved as CglB is still able to access the cell surface even in the absence of all three OM proteins.
Regulation of CglB exposure at the cell surface
CglB interacts with the OM platform, and in vivo experiments suggest that surface exposure of CglB is regulated by this interaction. Unless CglB becomes associated with the OM platform, it is rapidly cleaved from the cell surface, presumably by the action of a metalloprotease (of which there are >150 encoded in the M. xanthus genome, as identified via the MEROPS database) (50). With yet another parallel to iron-scavenging systems, the LbpB surface lipoprotein can be released into the extracellular milieu by a cell-surface serine protease (NalP, an autotransporter) (51). The functional significance of CglB cleavage is unclear as it is not detected in WT cells and is likely a by-product of the sensitized genetic backgrounds. One possibility could be that excessive cell-surface adhesin presence is detrimental to M. xanthus physiology by way of undesired cell–cell/substratum connections, and cells thus have a cleanup mechanism with which to remove free adhesin from among the cell-surface lipopolysaccharide molecules (52) of the OM. Nevertheless, the data suggest that the OM platform selectively regulates the exposure of CglB because:
1) Despite the abundance of CglB (as detected by Western blot) (Fig. 6A), only discrete foci are detected by immunofluorescence on gliding cells (mainly at the lagging cell pole) (Figs. 4 and 7B).
2) In addition, many more foci are detected in ΔgltA, ΔgltB, ΔgltK (+EDTA), and ΔgltH mutant cells, especially along the length of the cell body (Fig. 7B), suggesting that many CglB–OM platform complexes are formed that are not detected by immunofluorescence.
3) CglB localizes all around the surface of E. coli BL21(DE3) cells when it is expressed together with GltABCHK (Fig. 10A), suggesting that an additional clustering mechanism is present in M. xanthus cells.
In WT cells, CglB exposure may be coupled to adhesion via its recruitment by the Agl–Glt apparatus, which would lead to the major grouping of surface CglB clusters. Such coupling may be lost in the OM-platform mutants, leading to the formation of multiple (smaller) clusters. This hypothesis is consistent with the protection of CglB (from digestion by proteinase K) afforded by its interaction(s) with the Glt OM-platform proteins. Structure determination will be needed to identify the precise interaction network of CglB with the OM-platform proteins and elucidate the manner in which interactions between three distinct β-barrel proteins (GltA, GltB, and GltH) regulate CglB surface exposure.
Mechanism of adhesion
Surface-exposed CglB clusters become inert when they reach the lagging cell pole, where bFAs are inactivated. We cannot currently infer the nature of any interactions that CglB maintains at the lagging cell pole and it is possible that our immunolabeling procedure affects its dynamics at this location. Nevertheless, the CglB clusters can be recruited again to bFAs when cells reverse, which suggests that CglB may only be adhesive when coupled with active motor units, perhaps because it is selectively exposed, or because its adhesive properties become engaged through the mechanical action of the AglRQS motor, or both. Interactions between the OM platform and the underlying Agl–Glt machinery could trigger CglB exposure at bFAs, thus ensuring just-in-time adhesion and force transduction. We propose that when the IM AglRQS motor recruits the OM platform at bFAs (11), CglB becomes concentrated at Agl–Glt sites directly coupling adhesion to contractile forces exerted by the motor. This could occur following dynamic interactions between the IM motor and the OM platform in a manner similar to that proposed for the homologous Tol–Pal complex in E. coli. The TolQR–TolA motor localizes at division septa where it is proposed to function as a conveyor belt to concentrate the OM lipoprotein Pal locally (53). In this process, the proton flow through the channel assembled by TolQR energizes conformational changes on the TolA protein, which can stretch through pores in the PG layer to interact with Pal at the inner face of the OM (53). Although it remains to be shown, the predicted structures of the putative AglRQS motor-associated proteins suggest that they could operate like the TolA protein. Specifically, the IM proteins GltG and GltJ are both predicted to adopt TolA-like folds (6, 11), and both OM β barrel proteins GltA and GltB contain extended, unstructured N-terminal domains, with a potential TonB-box consensus sequence in the latter (11). Conceptually, such a mechanism could be compared to the firing of a gunlock cannon: A projectile (CglB) is loaded into the front-most opening of a cannon barrel (GltABH), after which an arm (GltG/J) exerts mechanical force on a lanyard (TonB box on the Glt OM module), resulting in firing of the loaded projectile (CglB) through the barrel of the cannon (GltABH).
Lastly, the extracellular CglB ligand remains to be found. Eukaryotic VWA domains, such as those found in integrins, are predominantly found in adhesins and ECM proteins (25). Prokaryotic VWA domains have been less characterized, but it was recently shown that bacterial pathogens use pilus adhesins, in type I and type IV pili, that adhere to host cell proteins via the VWA domains of tip proteins (29, 54, 55). Because gliding motility implicates a complex ECM (37, 56), the CglB VWA domain might bind to protein and/or polysaccharide components of the ECM. The recruitment of CglB at bFAs further highlights functional parallels between bFA and eFA mechanisms. In eukaryotic cells, the migration of surface-adhered cells via eFA-based locomotion involves the coordinated actions of a trans-envelope suite of proteins to transduce integrin-mediated cell–substratum adhesion to mechanical force and movement to propel the cell forward. In integrins, interaction with VWA ligands provokes large conformational changes that reinforce adhesion and trigger signal transduction (24), thus probing the biophysical properties of the ECM. It would be interesting to test whether these properties are also present in CglB, perhaps to adjust bFA activity and composition in response to the extracellular environment. These properties might be essential for multicellular behaviors (3): Similar to eukaryotic integrins, CglB could act as a sensor, regulating cell–cell interactions during development and predation.
MATERIALS AND METHODS
Bacterial cell culture and phenotypic analysis
M. xanthus strains were cultured in CYE [1% (w/v) Bacto Casitone peptone, 0.5% (w/v) yeast extract, 0.1% (w/v) MgCl2, and 10 mM Mops (pH 7.4)] broth with shaking (220 rpm) or on CYE solidified with 1.5% agar, at 32°C. To examine the effects of protease inhibition on CglB liberation, cells were grown in the presence of individual protease inhibitor panel constituents (Sigma-Aldrich, catalog no. INHIB1) at the recommended concentration: 4-(2-aminoethyl benzenesulfonyl fluoride HCl (1 mM), ε-aminocaproic acid (5 mg/ml), antipain HCl (100 μM), aprotinin (300 nM), benzamidine HCl hydrate (2 mM), bestatin HCl (40 μM), chymostatin (50 μg/ml), E-64 (10 μM), EDTA (1 mM), N-ethylmaleimide (500 μM), leupeptin hemisulfate (75 μM), pepstatin A (1 μM), phosphoramidon disodium salt (10 μM), and soybean trypsin inhibitor (1 μM). Cell resuspensions were done in TPM buffer [10 mM tris-HCl (pH 7.6), 8 mM MgSO4, and 1 mM KH2PO4]. All M. xanthus and E. coli strains used are listed in tables S2 and S3 (respectively). All plasmids used are listed in table S4.
For vancomycin-susceptibility testing, 3 ml of CYE broth (in sterile 10-ml glass tubes) were inoculated to a starting OD600 (optical density at 600 nm) of 0.05 in the absence/presence of EDTA (1 mM), with vancomycin added at increasing concentrations (0 to 100 μg/ml). Tubes were incubated with shaking (220 rpm) at 32°C for 26 hours, followed by mixing via vortex and aspiration and ejection using a pipette to break up aggregates; 1 ml of culture was then used to read the OD600 via spectrophotometer in a disposable cuvette.
Mutagenesis of cglB
The upstream region of cglB [from −213 base pairs (bp)], including a promoter region (from −190 to −141 bp) predicted by the BDGP (Berkeley Drosophila Genome Project) tool on “prokaryote” mode (57), and cglB itself was amplified via polymerase chain reaction (PCR) using Q5 high-fidelity DNA polymerase, followed by digestion of the product and plasmid pSWU30 with HindIII-HF (5′) and SacI-HF (3′), then ligation via T4 DNA ligase (all enzymes from New England Biolabs) to yield pCglBWT. Oligonucleotide primers for QuikChange site-directed mutagenesis were generated using PrimerX (http://bioinformatics.org/primerx/). Sequencing results were analyzed by Sequencher and/or ApE software.
Construction of CglB–OM platform interaction constructs
To generate plasmids for expression of OM-platform proteins and CglB in E. coli, PCRs of template genes were performed using Q5 DNA polymerase, with restriction enzymes (New England Biolabs) used according to the manufacturer’s instructions. Custom oligonucleotides were synthesized by Eurogentec; sequences are available upon request.
The pCDF-GltK6H intermediate plasmid was constructed by restriction cloning (hot fusion technique). Briefly, the sequence encoding the full-length gltK gene (MXAN_2538, residues 1 to 555) was PCR-amplified using M. xanthus DZ2 chromosomal DNA as a template with forward and reverse primers (CDF-K6H_Fw and CDF-K6H_Rev) and Q5 DNA polymerase. The PCR product introduced a 5′ NcoI–truncated site and a 3′ HindIII restriction site and a C-terminal His6 extension. The gltK6H PCR product was subcloned into the multiple cloning site (MCS) 1 pCDF-Duet1 (Novagen) corresponding restriction sites. To obtain the final construction pCDF-GltK6H _ BACS, encoding for the operon structure gltB, gltA, and gltC, a second step of restriction cloning was done by hot fusion technique. The full-length gltB (MXAN_2539, residues 1 to 828), gltA (MXAN_2540, residues 1 to 771) and gltC (MXAN_2541, residues 1 to 2022) genes were PCR-amplified using the primer pairs CDF-BACS_Fw (1)/CDF-BACS_Rev (1); CDF-BACS_Fw (2)/CDF-BACS_Rev (2), and CDF-BACS_Fw (3)/CDF-BACS_Rev (3). The PCR introduced a C-terminal streptavidin extension on gltC. The three PCR products were synthesized with 20-bp overhangs, from both 5′ and 3′ ends, corresponding to the designed overhangs genes regions and integration sites into the pCDF-GltK6H MCS2 plasmid.
The pACYC-BACS plasmid was constructed, as previously described, as pCDF-BACS but leaving the MCS1 polylinker site empty (the same primers were used). This plasmid was used as a negative control for the purification of the OM proteins.
The pET-GltH intermediate plasmid was also constructed by the restriction-cloning hot fusion technique. The full-length gltH was PCR-amplified with the primers (pET-GltH_Fw and pET-GltH_Rev) and introduced a 5′ Nco I–truncated restriction site. The PCR product was then subcloned into the pET-Duet (Novagen) MCS1 at corresponding restriction sites. Lastly, the cglB gene was amplified with the primers (pET-CglB_Fw and pET-CglB_Rev) by Q5 polymerase, with the PCR introducing a shine dalgarno sequence in the 5′ and overlapping regions from both 5′ and 3′ ends corresponding to the integration sites into pET-GltH plasmid. All constructs were verified by DNA sequencing (Eurofins) and plasmids generated were preserved in transformed E. coli DH5α cells.
Generation of α-CglB and α-GltC polyclonal antibodies
CglB (lacking signal peptide) elaborating a C-terminal hexa-histidine tag (CglB21–416-His6) was purified under denaturing conditions. Fractions were collected in 50 mM tris (pH 8.0), 300 mM NaCl, 250 mM imidazole, and 6 M urea and used to immunize rabbits (Eurogentec). The α-CglB 1° polyclonal antibody (pAb) produced was then tested for specificity by using the WT and the ΩcglB strains. For GltC, a peptide corresponding to GltC54–67 was synthesized and then used to immunize rabbits to generate pAb (GenScript). The α-GltA, α-GltB, and α-GltH 1° pAbs were raised previously (7, 17). The α-His6 antibody was commercially purchased (Sigma-Aldrich, #SAB4301134).
Immunofluorescence labeling of live M. xanthus cells
Specific volumes of overnight culture were sedimented via centrifugation (5000g, 5 min) such that pellet resuspension in 600 μl of TPM yielded an OD600 of 2.5. Following this wash, cell resuspensions were sedimented (5000g, 5 min) and then resuspended in 600 μl of TPM + BSA (bovine serum albumin) (5%, w/v), with α-CglB antiserum (1 μl for 3 ml of TPM + BSA). The solution was agitated for 1 hour at 20°C on a Nutator platform and then sedimented (5000g, 5 min). The pellet was washed twice with TPM (600 μl) and then resuspended in 600 μl of TPM + BSA with goat α-rabbit immunoglobulin G (IgG) Fab2 conjugated to Alexa Fluor 647 (Cell Signaling Technology) [1 μl of monoclonal antibody (mAb) for 3 ml of TPM + BSA]. The suspension was agitated for 1 hour at 20°C on a Nutator platform (covered with aluminum foil), washed twice with TPM as described above, and then resuspended in 600 μl of TPM. Resuspensions of immunolabeled cells were spotted (2 μl) on glass-bottomed microscopy fluorodishes (World Precision Instruments) and overlaid with TPM 1.5% (w/v) agar pads. Cells were imaged using an Axio Observer 7 (Zeiss) inverted fluorescence microscope with a heated chamber (32°C), with an alpha Plan-Apochromat 100× oil immersion objective, captured with 10% light-emitting diode illumination intensity on an Axiocam 512 camera, at 15-s intervals (binning mode: 5 × 5). Phase-contrast imaging was carried out without filters (100-ms acquisition time). Alexa Fluor 647 fluorescence was detected using the BP640/30 excitation filter and BP690/50 emission filter (200-ms acquisition time). AglZ-mNeonGreen fluorescence was detected using the BP470/40 excitation filter and BP525/50 emission filter (150-ms acquisition time).
Immunofluorescence labeling of fixed E. coli cells
Cells of E. coli BL21(DE3), cotransformed with either pCDF-Duet-GltK6H + GltBACS and pET-Duet-GltH-CglB or pCDF-Duet and pET-Duet, were induced overnight in lysogeny broth (LB) (1.0 mM IPTG, 16°C). The next day, cells (500 μl) from the induced cultures were fixed with 100 μl of 16% paraformaldehyde, 0.2 μl of 25% glutaraldehyde, and 20 μl of NaPO4 (pH 7.4). Then, 10 μl of each mix was applied to wells of a commercial 2× nine-well μ-Slide (Ibidi). Following incubation at room temperature (RT) (20 min), the wells were washed three times with 1× phosphate-buffered saline (PBS). For the relevant samples, wells were then treated with GTE (glucose-tris-EDTA) buffer containing lysozyme (1 μg/ml) at RT (4 min), followed by three washes with 1× PBS. Irrespective of treatment (or not) with lysozyme, wells were then left to air dry. For antibody labeling, all samples were incubated for 20 min at RT with 1× PBS containing 2% BSA. This buffer was then replaced by one of the same composition containing also α-CglB antibody (1:1000) and left to incubate overnight at 4°C without agitation. After 10 washes with 1× PBS, the samples were incubated with α-rabbit IgG Alexa Fluor Plus 488 (1:200; Invitrogen) for 4 hours at 4°C in the dark (without agitation) followed by 10 washes with 1× PBS. Cells were imaged by epifluorescence with an inverted Eclipse TiE microscope with Perfect Focus (Nikon), using a 100× numerical aperture (NA) = 1.45 phase-contrast objective and an ORCA-Flash4.0 digital complementary metal-oxide semiconductor camera (Hamamatsu) at RT. A mercury fluorescent lamp with a green optical filter was used when necessary. Image stacks were prepared for publication using Fiji, with fluorescence micrographs subjected to background subtraction (rolling ball radius: 10 pixels).
Phylogeny and gene co-occurrence
This study explored 61 myxobacterial genomes, distributed within three suborders and nine families (58–71), in addition to 59 outgroup genomes [members from 32 non-Myxococcales Deltaproteobacteria, 4 α-, 6 β-, 9 γ-, 4 £-proteobacteria, 2 Firmicutes, 1 Actinobacteria, and 1 FCB (Fibrobacteres, Chlorobi, and Bacteroidetes) group organism]. Highly conserved gapless concatenated alignment of 26 housekeeping protein sequences (68, 72) was subjected to RAxML to build a maximum likelihood phylogenetic tree using JTT Substitution Matrix and 100 bootstrap values (73). Sequential distribution of gliding motility genes, i.e., agl, glt (M1, G1, and G2 clusters) (7), and cglB (19, 23, 74) was identified within all 120 genomes under study using two iterations of homology searching via JackHMMER (HMMER 3.3.2 suite released in November 2020) (75) with an E value cutoff of 1 × 10−5 and other default parameters. The relative distribution of gliding motility proteins was mapped to the multiprotein phylogeny using iTol v6.5.3 (76). The strip to the right of the phylogeny depicts the taxonomic classes (from top to bottom: Myxococcales; non-Myxococcales δ-proteobacteria; α-, β-, γ-, and £-proteobacteria; Actinobacteria; Firmicutes; and Fibrobacteres, respectively).
Tertiary structure homology detection and protein modeling
Identification of structural homologs to CglB was carried out using fold-recognition searches of the Protein Data Bank using HHpred (77). Deep learning-based relaxed tertiary structure modeling of CglB was carried out via AlphaFold using the ColabFold pipeline with default settings (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb) (30, 31). The highest-confidence CglB model was used to generate structural alignments and figures with PyMol (The PyMol Molecular Graphics System, Version 2.0, Schrödinger, LLC).
SDS-PAGE, in-gel fluorescence, and Western immunoblotting
For detection of proteins from whole cells via Western immunoblot, TPM-washed cells were sedimented and resuspended at OD600 1.0 in 1× Laemmli sample buffer containing 5% β-mercaptoethanol for reducing SDS-PAGE (unless otherwise indicated). For analysis of CglB unfolding in the presence of reducing agent, TPM-washed cells were instead resuspended in 2× Laemmli (lacking reducing agent) and diluted to 1× with double-distilled H2O (ddH2O) containing increasing concentrations of dithiothreitol (DTT) (0 to 5 mM). Samples were boiled (10 min), loaded (20 μl) on 10-well 1-mm-thick gels, resolved on 10% acrylamide gels (80 V for 45 min for stacking and 120 V for 75 min for resolving), and then electroblotted (100 V for 60 min) to nitrocellulose membranes. Blots were rinsed with tris-buffered saline (TBS) buffer, blocked for 30 min at RT with 5% (w/v) milk in TBS, and then incubated rocking overnight in the 4°C cold room in the presence of primary antibodies. Primary antisera were all used at a concentration of 1:10,000 in TBS with 0.05% Tween 20 (TBS-T). The next day, blots were rinsed twice (5 min) with TBS-T, incubated with goat α-rabbit 2° antibody conjugated to horseradish peroxidase (HRP) (1:5000) (Bio-Rad) in TBS-T at RT (1 hour), and then rinsed twice (5 min) again with TBS-T. All immunoblots were developed using the SuperSignal West Pico (Thermo Fisher Scientific) chemiluminescence substrate, captured on either a GE Imager with ImageQuant software or an Amersham Imager 600 machine.
To monitor the resumption of CglB release from EDTA-grown cells, cells were sedimented via centrifuge (5000g, 5 min) and resuspended at an OD600 of 1.0 in 12.5 ml of fresh CYE or TPM and incubated at 20°C at an OD600 of 1. Each hour, the OD600 was read to calculate the volume of culture to take out to get a final suspension of 100 μl at an OD600 of 2. The volume was removed and sedimented (5000g, 5 min) and resuspended in 100 μl of Laemmli buffer before use for Western immunoblotting.
For fractionated whole cell–supernatant–OMV samples in 1× Laemmli buffer, samples were boiled (10 min) and loaded (20 μl) on 15-well 4 to 20% acrylamide precast gradient gels (Bio-Rad). Supernatant-alone samples were similarly boiled and loaded on a cast 10% acrylamide gel. Gels were resolved at 120 V, followed by electroblotting to nitrocellulose membranes at 100 V. Immunodetection was performed with diluted polyclonal antisera as follows: α-CglB (1:10,000), α-MglA (1:5000), and α-GltK (1:5000). Detection via secondary antibody was done with goat α-rabbit mAb (1:5000) conjugated to HRP (Bio-Rad). Immunoblots were developed using the SuperSignal West Femto (Thermo Fisher Scientific) chemiluminescence substrate, captured on GE Imager with ImageQuant software.
For analysis of AglZ-YFP in-gel fluorescence, TPM-washed cells were resuspended in 1× nonreducing Laemmli sample buffer to an OD600 of 4.0. Cell resuspensions were heated for 30 min (65°C), loaded (20 μl) on an 8% SDS-PAGE gel, and resolved for 45 min at 80 V, then 75 min at 120 V. Cultures, cell resuspensions, and SDS-PAGE gels (before, during, and after resolution) were all shielded from ambient light to reduce photobleaching of the YFP moiety. Resolved gels were scanned on a Typhoon FLA9500 flat-bed imager (GE Healthcare). AglZ-YFP was excited with a 473-nm laser, with fluorescence captured using the BPB1 filter (PMT 800). Prestained protein ladder bands were detected via excitation with a 635-nm laser and captured using the LPR filter (PMT 800). Quantification of band fluorescence intensity was performed using ImageJ via the “plot lanes” function, followed by determination of the area under the curve. AglZ-YFP signal for each lane was normalized to the faster-migrating autofluorescent band in the same lane; these values were then expressed as a percentage of the signal in WT cells for a given biological replicate.
For analysis of IMss-mCherry in-gel fluorescence (see below), SDS-PAGE-resolved (10% acrylamide; 80 V for 45 min and then 120 V for 75 min) samples (20 μl) were shielded from light and then scanned on a Typhoon imager. The mCherry was excited with a 532-nm laser, with fluorescence capture using the LPR filter (PMT 800).
Sample fractionation
To separate supernatant and OMV fractions, WT, ΔgltA, ΔgltB, and ΔgltK vegetative cells were grown in CYE medium to OD600 0.7. Intact cells were first eliminated by sedimentation at 7830 rpm (10 min, RT). After addition of 1 mM phenylmethylsulfonyl fluoride, supernatants were sedimented at 125,000g (2 hours, 4°C). The resulting pellets (OMV fraction) and supernatants (soluble fractions) were then treated separately. The OMV pellets were washed with TPM, sedimented again at 125,000g (2 hours, 4 °C), and then resuspended directly in 500 μl of 1× Laemmli protein sample buffer. The soluble supernatant fractions were treated with TCA (10% final concentration) for 30 min on ice and then sedimented at 11,000 rpm (1 hour, 4°C). The resulting pellets (precipitated proteins) were washed with 100% acetone, sedimented at 7830 rpm (10 min, 4°C), and dried overnight at RT. Dried pellets were then resuspended in 1.5 ml of TPM, sedimented at 15,000 rpm (30 min, 4°C), and lastly resuspended in 500 μl of 1× Laemmli protein sample buffer.
For isolation of supernatant-alone samples, 10 ml of CYE cultures (inoculated at OD600 0.02) were grown overnight with shaking (220 rpm, 32°C) to OD600 0.6 to 1.0 and then sedimented (5000g, 10 min, 20°C). Supernatants were then sedimented in an ultracentrifuge (Beckman, SW 41 Ti rotor, 120,000g, 75 min, 4°C) to remove any remaining membrane material. Clarified 10 ml of supernatant samples was treated with 1 ml of 100% TCA to precipitate the proteins. Tubes were heated at 65°C for 5 min and then spun in a centrifuge (16,300g, 20 min, RT) to sediment precipitate in 2-ml microtubes. TCA-precipitated pellets were washed with 1 ml of acetone and sedimented (16,300g, 20 min, RT), followed by supernatant aspiration. Protein pellets were left uncapped in the chemical hood overnight to ensure evaporation of acetone. Pellets were resuspended in 500 μl of 2× Laemmli sample buffer lacking reducing agent and then diluted to 1× with ddH2O.
Immunoprecipitation and mass spectrometry
For analysis of cleaved CglB in culture supernatants, cells of ΔgltB from 100 ml of CYE cultures were first sedimented (4000g, 24°C, 15 min), after which supernatants were decanted, pooled, and passed through a 0.2-μm syringe filter. Filtered supernatant was then concentrated using four Vivaspin20 columns (10-kDa cutoff) (Sartorius), spun at 8000g (20°C) in a fixed-angle centrifuge, with the supernatant concentrated to the dead volume limit of each column. Concentrated supernatants (~80 μl each) were subsequently pooled and diluted 1:2 with filter-sterilized 1× PBS (binding buffer) to equilibrate sample pH. Separately, a single 1-ml Pierce Protein A column (Thermo Fisher Scientific) per pooled supernatant was equilibrated in filter-sterilized binding buffer at RT as per the manufacturer’s instructions. Filtered α-CglB antiserum (1 ml) was sedimented in a microfuge to remove remnant cells and/or debris (4000g, 5 min), diluted 1:1 with binding buffer, and then sedimented at 12,000g to clarify the sample as binding buffer addition may have resulted in lipoprotein precipitation. The Protein A column was primed by passage of 5 ml of binding buffer. To bind antibody to the column, the 2 ml of diluted antiserum was added to the top of the column and allowed to drip through, followed by washing with 15 ml of binding buffer to remove unbound pAb. The ~960 μl of supernatant concentrate was added to the top of the column and allowed to distribute throughout the resin bed at RT (60 min). The column was then again washed with 15 ml of binding buffer. To elute bound pAb (and any associated proteins) from the column, 5 ml of elution buffer (0.1 M glycine, pH to 2.5 with HCl) was added.
To analyze the protein content of the pulldown, 500 μl of column eluate was concentrated in a microfuge using a Vivaspin500 column (10-kDa cutoff) to a dead volume of ~20 μl, then diluted 1:1 with 2× reducing Laemmli sample buffer. Samples (20 μl) were run into the stacking gel via SDS-PAGE (80 V, 13 min). Gel bands stained with SimplyBlue Safestain were excised from the stacking portion of the gel and the proteins digested by trypsin or Endoproteinase Glu-C. Liquid chromatography coupled to tandem mass spectrometry analyses were performed on a Q-Exactive plus mass spectrometer (Thermo Fisher Scientific) by staff at the Proteomics Platform of the Mediterranean Institute of Microbiology (Marseille). Processing of the spectra for protein identification was performed with Proteome Discoverer software (Thermo Fisher Scientific, versions 1.4.0.288 and 2.1.0.81).
Expression, purifications, and detection of the OM-platform proteins from E. coli cells
The pCDF-GltK6H-GltBACS and pET-GltH-CglB plasmids were used to transform E. coli BL21(DE3) (Invitrogen). Cells were grown at 37°C in LB (BD, Difco), with streptomycin and ampicillin antibiotics (100 μg/ml) (Sigma-Aldrich), to OD600 0.8 to 0.9. Expression of the gltKBACH and cglB genes was induced with 1.0 mM IPTG overnight at 16°C. The following day, cell pellets were resuspended in 50 mM tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, and 10 mM MgCl2, supplemented with deoxyribonuclease I (10 μg/ml) and lysozyme (10 μg/ml). The cell suspension was further broken using an Emulsiflex-C5 (Avestin). The broken cell suspension was clarified via centrifugation (26,000g, 15 min, 4°C). The membrane fraction was then collected via high-speed centrifugation (195,000g, 45 min, 4°C). Sedimented membranes were mechanically homogenized and solubilized in 50 mM tris-HCl (pH 8.0), 50 mM NaCl, EDTA-free protease inhibitor (Roche), 0.5% (w/v) DDM (Anatrace), 0.75% (w/v) DM-NPG (Anatrace), and 1 mM EDTA at 4°C overnight. The suspension was then clarified by high-speed centrifugation (126,000g, 35 min, 4°C). The clarified supernatant (supplemented with 20 mM imidazole) was loaded onto a 1-ml HisTrap HP (Cytivia) column and then washed with 50 mM tris-HCl (pH 8.0), 50 mM NaCl, and 0.05% (w/v) DM-NPG (Affinity buffer) with 50 mM imidazole at 4°C. The OM-platform proteins were eluted in the same buffer supplemented instead with 250 mM imidazole. Peak fractions were pooled with 1× Laemmli buffer containing β-mercaptoethanol and 1 mM DTT to be used for Western blotting.
For the negative control, we used the same protocols as previously described except that we transformed E. coli BL21(DE3) with the pACYC-BACS and pET-GltH-CglB plasmids. Cells were grown at 37°C in LB with ampicillin (100 μg/ml) and chloramphenicol (30 μg/ml) antibiotics (Sigma-Aldrich).
For detection, SDS-PAGE was performed on Bio-Rad Mini-PROTEAN systems using a standard protocol. For detection of proteins from pulldown assay via Western immunoblot, the load and eluted fractions were resuspended in 1× Laemmli sample buffer containing β-mercaptoethanol and 1 mM DTT. Samples were boiled (10 min), loaded (10 μl for load and 20 μl for eluted fractions) on 10-well 1-mm-thick gels, resolved on 12% acrylamide gels (200 V during 45 min), and then electroblotted (100 V for 60 min) to nitrocellulose membranes. Blots were blocked for 1 hour at RT with 5% milk in PBS with 0.05% Tween 20 (PBS-T; for α-GltC only) and TBS-T (for other antibodies), and then incubated rocking overnight in the 4°C cold room in 1:5000 α-CglB, 1:5000 α-GltB, 1:5000 α-GltA, 1:5000 α-His (i.e., GltK), and 1:5000 α-GltH mixture in milk 5% TBS-T, except for GltC-StrepII detection which was carried out with 1:500 α-GltC mixture in milk 5% PBS-T. The next day, blots were rinsed three times (10 min) with TBS-T or PBS-T, incubated with goat α-rabbit 2° antibody conjugated to HRP (1:5000) (Bio-Rad) in milk 5% TBS-T or PBS-T at RT (1 hour), and then rinsed three times (10 min) again with TBS-T or PBS-T. All immunoblots were developed using the SuperSignal West FEMTO (Thermo Fisher Scientific) chemiluminescence substrate, captured on an Image Quant LAS4000 Mini Imager with Image Quant software.
Proteinase K surface digestion
Cells were resuspended in TPM at OD600 2.0, followed by addition of proteinase K (200 μg/ml) and a brief vortex pulse to mix. An aliquot (50 μl) was immediately removed at t = 0 and placed into a tube containing 5 μl of 100% TCA. Digestion mixtures were incubated at RT on a rocker platform, with aliquots removed every 15 min and placed into respective pre-aliquoted tubes of TCA. Upon removal of digestion reaction aliquots, TCA-containing sample tubes were heated at 65°C for 5 min, chilled on ice, and then sedimented at 14,000g (5 min). Following supernatant removal, precipitated protein pellets were washed via resuspension in 500 μl of 100% acetone. Samples were then sedimented as before (14,000g, 5 min), followed by careful aspiration of the supernatants. Tubes were left uncapped overnight in the fume hood to promote evaporation of residual acetone, followed by storage at −80°C until needed. Precipitated protein pellets were resuspended in 50 μl of 1× Laemmli sample buffer (with reducing agent as indicated) and analyzed via SDS-PAGE and Western immunoblot.
For EDTA-grown cells expressing the IMss-mCherry construct (37), the above protocol was modified so as to not denature the fluorophore. Briefly, cells were similarly resuspended in TPM and digested with proteinase K, with 50-μl aliquots removed at the same intervals. However, upon removal, each aliquot was immediately transferred to a PCR tube, incubated at 95°C for 15 min in a thermocycler to inactivate the proteinase K, and then mixed with 50 μl of 2× Laemmli buffer lacking reducing agent. Samples were then resolved via SDS-PAGE and scanned for in-gel fluorescence (see below).
Motility and fluorescence analysis
For phase-contrast and fluorescence microscopy on agar pads, cells from exponentially growing cultures were sedimented and resuspended in TPM buffer to OD600 5.0, spotted (5 μl) on a glass coverslip, and then overlaid with a pad of 1.5% agar prepared with TPM. For motility analysis, cells were left to adhere for 5 min before imaging at 32°C using a TE2000-E-PFS microscope (Nikon) with a 40× objective and a CoolSNAP HQ2 camera (Photometrics) with Metamorph software (Molecular Devices). AglZ-YFP fluorescence was imaged using a monolithic aluminum microscope (homemade) equipped with a 1.49-NA/100× objective (Nikon Instruments) and imaged on an iXon DU 897 electron-multiplying charge coupled device (EMCCD) camera (Andor Technology). Illumination was provided by a 488-nm diode-pumped solid-state (DPSS) laser (Vortran Stradus), and sample positioning was performed using a P611 three-axis nanopositioner (Physik Instrument). Instrument control was programmed in LabView (National Instruments) providing integrated control of all components. Cell-gliding speeds were calculated using the MicrobeJ module for Fiji (78). Gliding cell montages were generated using Fiji. Kymograph panels were generated using the Fiji Kymograph Builder function. AglZ-YFP clusters were detected manually and tracked with the MTrackJ Fiji plugin. Using an R software script, the points of the AglZ-YFP cluster trajectories (x0, x1, ..., xn; y0, y1, ..., yn) were used to calculate the mean square displacement (MSD) at time
For TIRFM, imaging of real-time AglZ-YFP trafficking was performed as previously detailed in chitosan-coated polydimethylsiloxane microfluidic channels (11). Briefly, cells were injected into the chamber and left to adhere (30 min) without flow, with unadhered cells then removed via manual injection with TPM. TIRFM was performed on attached cells with active autofocus using an inverted microscope with 100× oil-immersion Plan-Achromat objective, atop a closed-loop piezoelectric stage. AglZ-YFP was excited with a 488-nm laser, with emission collected by the objective, through a dichroic mirror and band-pass filters, and captured by an EMCCD camera. For imaging of the YFP channel in real time, 500 images were captured at 20 Hz (11).
Flow chamber construction and bead assay
Before experiments, 1 ml of M. xanthus DZ2 WT + AglZ-YFP and mutant DZ2 ΔcglB + AglZ-YFP overnight culture was grown to OD600 ~ 0.6, sedimented (8000 rpm, 5 min), and resuspended in 400 ml of TPM buffer. Flow chambers were made by combining two layers of double-sided tape, a 1-mm-thick glass microscope slide, and a 100-μm-thick glass cover slip (#1.5) as previously described (79). The tape was separated to allow a final volume of approximately 60 μl. Agarose (40 μl at 0.7%) dissolved in 6 M dimethyl sulfoxide was injected into the chamber and allowed to sit at RT for 15 min. The chamber was washed with 400 μl of TPM, then injected with M. xanthus cells (60 μl), and left at RT to facilitate cell attachment to the agarose-coated surface for 30 min. Unattached cells were then thoroughly washed away with a total of 2 ml TPM media containing 10 mM glucose. The flow chamber was then mounted onto the microscope for imaging. For bead experiments, 1 μl of uncoated polystyrene beads (diameter, 520 nm) (Bangs Laboratories) was washed and diluted in 1 ml of TPM containing 10 mM glucose and injected into the flow chamber. Beads were optically trapped and placed about a third of the cell length away from the pole of the immobilized cell of interest.
Bead tracking and video analysis
For a chosen M. xanthus cell (WT or mutant), 3-min movies were recorded and analyzed using a custom MATLAB tracking code. The code uses filtering mechanisms to subtract the image background from that of the cell-attached bead. First, an internal MATLAB centroid function identified the x, y pixel values of the center of the bead for each frame in the video, followed by pixel value conversion to micrometers. This was then used to compute motor-driven bead runs and velocities for each cell. The threshold value for a run was previously determined by disabling molecular motors and decreasing bead motion in WT cells by carefully injecting 20 μM of nigericin, a pH-gradient/proton motive force-inhibitory drug, into the mounted flow chamber. This drug concentration decreased bead velocity but not motor force production translated to the beads. In these previous experiments, 40 μM nigericin was used, leading to negligible bead motion (16).
Statistical analysis
For all comparisons of dataset distributions (Figs. 3C and 5, C to F and H, and fig. S1A), analyses of statistical significance were carried out via unpaired two-tailed Mann-Whitney U test. Differences in mean values for AglZ-YFP fluorescence levels in WT versus ΔcglB were evaluated for statistical significance using a Wilcoxon signed-rank test performed relative to the reference value of “100” for WT samples (fig. S4A). Differences in mean relative culture density values for vancomycin-sensitivity testing were compared for each mutant strain against WT for each antibiotic concentration tested; this analysis was carried out via two-way analysis of variance (ANOVA) and Dunnett’s multiple comparisons test with a single-pooled variance (fig. S5, E and F). All statistical analyses were carried out in GraphPad Prism (version 8) at a confidence interval of 95% (P < 0.05).
Acknowledgments
We would like to thank J. Selkrig and N. Nickerson for valuable suggestions and troubleshooting regarding protease accessibility; A. Konovalova for constructive input on surface lipoproteins; R. Fieldhouse for insightful discussions on protein modeling and evolutionary couplings; S. Gauthier for cloning pSWU30-pCglBWT; L. Søgaard-Andersen for the gift of α-GltK, α-GltB, α-GltA, and α-GltC polyclonal antibodies; A. Roussel and R. Vincentelli (CNRS–Aix-Marseille University, Architecture et function des macromolecules biologiques) for CglB protein used to generate pAb; R. Lebrun, P. Mansuelle, and R. Puppo of the Mediterranean Institute of Microbiology’s Proteomics Platform for insightful discussions and sample processing for mass spectrometry; and E. Mauriello for invaluable assistance with E. coli immunofluorescence labeling experiments.
Funding: This work was funded by the Natural Sciences and Engineering Research Council of Canada, discovery grant RGPIN-2016-06637 (to S.T.I., N.Y.J., and F.S.); the Banting Research Foundation, Discovery Award 2018-1400 (to S.T.I.); the Canadian Institutes of Health Research fellowship (to S.T.I.); the Aix-Marseille University AMIDEX Excellence Program (to S.T.I.); PROTEO, The Quebec Network for Research on Protein Function, Engineering, and Applications studentship (to N.Y.J. and F.S.); the European Research Council, advanced grant JAWS (to T.M.); the Bettencourt-Schueller Foundation, Coup d’élan pour la recherche française 2011 (to T.M.); the National Science Foundation, grant no. CAREER PHY-0844466 (to J.W.S.); the Fondation ARC studentship (to L.M.F.); the Glenn Centers for Aging Research award (to B.P.B.); and the NIH award P50 GM071508 (to B.P.B.).
Author contributions: Conceptualization: S.T.I., N.Y.J., C.C., E.D., and T.M. Methodology: S.T.I., N.Y.J., C.C., A.M.B., L.M., J.H., E.D., and T.M. Investigation: S.T.I., N.Y.J., C.C., E.D., A.M.B., L.M., U.M., B.F., L.M.F., G.S., A.A.K., F.S., J.-B.F., B.P.B., and J.H. Visualization: S.T.I., N.Y.J., G.S., J.H., and T.M. Supervision: S.T.I., T.M., J.W.S., M.N., E.D., and G.S. Writing—original draft: S.T.I., N.Y.J., and T.M. Writing—review and editing: S.T.I., T.M., N.Y.J., J.W.S., G.S., and M.N.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S4
REFERENCES AND NOTES
- 1.Z. Sun, S. S. Guo, R. Fässler, Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445–456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.P. Kanchanawong, G. Shtengel, A. M. Pasapera, E. B. Ramko, M. W. Davidson, H. F. Hess, C. M. Waterman, Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S. T. Islam, I. Vergara Alvarez, F. Saïdi, A. Guiseppi, E. Vinogradov, G. Sharma, L. Espinosa, C. Morrone, G. Brasseur, J. F. Guillemot, A. Benarouche, J. L. Bridot, G. Ravicoularamin, A. Cagna, C. Gauthier, M. Singer, H. P. Fierobe, T. Mignot, E. M. F. Mauriello, Modulation of bacterial multicellularity via spatio-specific polysaccharide secretion. PLOS Biol. 18, e3000728 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.F. Saïdi, N. Y. Jolivet, D. J. Lemon, A. Nakamura, A. M. Belgrave, A. G. Garza, F. J. Veyrier, S. T. Islam, Bacterial glycocalyx integrity drives multicellular swarm biofilm dynamism. Mol. Microbiol. 116, 1151–1172 (2021). [DOI] [PubMed] [Google Scholar]
- 5.F. Saïdi, U. Mahanta, A. Panda, A. A. Kezzo, N. Y. Jolivet, R. Bitazar, G. John, M. Martinez, A. Mellouk, C. Calmettes, Y. W. Chang, G. Sharma, S. T. Islam, Bacterial outer membrane polysaccharide export (OPX) proteins occupy three structural classes with selective β-barrel porin requirements for polymer secretion. Microbiol. Spectr. 10, e01290-22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.S. T. Islam, T. Mignot, The mysterious nature of bacterial surface (gliding) motility: A focal adhesion-based mechanism in Myxococcus xanthus. Semin. Cell Dev. Biol. 46, 143–154 (2015). [DOI] [PubMed] [Google Scholar]
- 7.J. Luciano, R. Agrebi, A. V. le Gall, M. Wartel, F. Fiegna, A. Ducret, C. Brochier-Armanet, T. Mignot, Emergence and modular evolution of a novel motility machinery in bacteria. PLOS Genet. 7, e1002268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.B. Nan, E. M. F. Mauriello, I.-H. Sun, A. Wong, D. R. Zusman, A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol. Microbiol. 76, 1539–1554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.B. Nan, J. N. Bandaria, A. Moghtaderi, I. H. Sun, A. Yildiz, D. R. Zusman, Flagella stator homologs function as motors for myxobacterial gliding motility by moving in helical trajectories. Proc. Natl. Acad. Sci. U.S.A. 110, E1508–E1513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.G. Fu, J. N. Bandaria, A. V. le Gall, X. Fan, A. Yildiz, T. Mignot, D. R. Zusman, B. Nan, MotAB-like machinery drives the movement of MreB filaments during bacterial gliding motility. Proc. Natl. Acad. Sci. U.S.A. 115, 2484–2489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.L. M. Faure, J. B. Fiche, L. Espinosa, A. Ducret, V. Anantharaman, J. Luciano, S. Lhospice, S. T. Islam, J. Tréguier, M. Sotes, E. Kuru, M. S. van Nieuwenhze, Y. V. Brun, O. Théodoly, L. Aravind, M. Nollmann, T. Mignot, The mechanism of force transmission at bacterial focal adhesion complexes. Nature 539, 530–535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.T. Mignot, J. W. Shaevitz, P. L. Hartzell, D. R. Zusman, Evidence that focal adhesion complexes power bacterial gliding motility. Science 315, 853–856 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.E. M. F. Mauriello, F. Mouhamar, B. Nan, A. Ducret, D. Dai, D. R. Zusman, T. Mignot, Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J. 29, 315–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. Treuner-Lange, E. Macia, M. Guzzo, E. Hot, L. M. Faure, B. Jakobczak, L. Espinosa, D. Alcor, A. Ducret, D. Keilberg, J. P. Castaing, S. Lacas Gervais, M. Franco, L. Søgaard-Andersen, T. Mignot, The small G-protein MglA connects to the MreB actin cytoskeleton at bacterial focal adhesions. J. Cell Biol. 210, 243–256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Sun, M. Wartel, E. Cascales, J. W. Shaevitz, T. Mignot, Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl. Acad. Sci. U.S.A. 108, 7559–7564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R. Balagam, D. B. Litwin, F. Czerwinski, M. Sun, H. B. Kaplan, J. W. Shaevitz, O. A. Igoshin, Myxococcus xanthus gliding motors are elastically coupled to the substrate as predicted by the focal adhesion model of gliding motility. PLOS Comput. Biol. 10, e1003619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.B. Jakobczak, D. Keilberg, K. Wuichet, L. Søgaard-Andersen, Contact- and protein transfer-dependent stimulation of assembly of the gliding motility machinery in Myxococcus xanthus. PLOS Genet. 11, e1005341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.J. Hodgkin, D. Kaiser, Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U.S.A. 74, 2938–2942 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. M. Rodriguez, A. M. Spormann, Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J. Bacteriol. 181, 4381–4390 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.P. Youderian, N. Burke, D. J. White, P. L. Hartzell, Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol. Microbiol. 49, 555–570 (2003). [DOI] [PubMed] [Google Scholar]
- 21.J. Kahnt, K. Aguiluz, J. Koch, A. Treuner-Lange, A. Konovalova, S. Huntley, M. Hoppert, L. Søgaard-Andersen, R. Hedderich, Profiling the outer membrane proteome during growth and development of the social bacterium Myxococcus xanthus by selective biotinylation and analyses of outer membrane vesicles. J. Proteome Res. 9, 5197–5208 (2010). [DOI] [PubMed] [Google Scholar]
- 22.S. Bhat, X. Zhu, R. P. Patel, R. Orlando, L. J. Shimkets, Identification and localization of Myxococcus xanthus porins and lipoproteins. PLOS ONE 6, e27475 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D. T. Pathak, D. Wall, Identification of the cglC, cglD, cglE, and cglF genes and their role in cell contact-dependent gliding motility in Myxococcus xanthus. J. Bacteriol. 194, 1940–1949 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Shimaoka, T. A. Springer, Therapeutic antagonists and conformational regulation of integrin function. Nat. Rev. Drug Discov. 2, 703–716 (2003). [DOI] [PubMed] [Google Scholar]
- 25.C. A. Whittaker, R. O. Hynes, Distribution and evolution of von Willebrand/integrin A domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13, 3369–3387 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.G. Song, A. C. Koksal, C. Lu, T. A. Springer, Shape change in the receptor for gliding motility in Plasmodium sporozoites. Proc. Natl. Acad. Sci. U.S.A. 109, 21420–21425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.G. Song, T. A. Springer, Structures of the Toxoplasma gliding motility adhesin. Proc. Natl. Acad. Sci. U.S.A. 111, 4862–4867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.V. Krishnan, P. Dwivedi, B. J. Kim, A. Samal, K. Macon, X. Ma, A. Mishra, K. S. Doran, H. Ton-That, S. V. L. Narayana, Structure of Streptococcus agalactiae tip pilin GBS104: A model for GBS pili assembly and host interactions. Acta Crystallogr. D Biol. Crystallogr. 69, 1073–1089 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. Raynaud, D. Sheppard, J.-L. Berry, I. Gurung, V. Pelicic, PilB from Streptococcus sanguinis is a bimodular type IV pilin with a direct role in adhesion. Proc. Natl. Acad. Sci. U.S.A. 118, e2102092118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli, D. Hassabis, Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov, M. Steinegger, ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.M. Shimaoka, J. Takagi, T. A. Springer, Conformational regulation of integrin structure and function. Annu. Rev. Biophys. Biomol. Struct. 31, 485–516 (2002). [DOI] [PubMed] [Google Scholar]
- 33.S. Cheng, C. L. Brooks III, Viral capsid proteins are segregated in structural fold space. PLOS Comput. Biol. 9, e1002905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.G. B. Cole, T. J. Bateman, T. F. Moraes, The surface lipoproteins of gram-negative bacteria: Protectors and foragers in harsh environments. J. Biol. Chem. 296, 100147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. M. Wilson, H. D. Bernstein, Surface-exposed lipoproteins: An emerging secretion phenomenon in Gram-negative bacteria. Trends Microbiol. 24, 198–208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. Tréguier, L. Bugnicourt, G. Gay, M. Diallo, S. T. Islam, A. Toro, L. David, O. Théodoly, G. Sudre, T. Mignot, Chitosan films for microfluidic studies of single bacteria and perspectives for antibiotic susceptibility testing. MBio 10, e01375-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A. Ducret, B. Fleuchot, P. Bergam, T. Mignot, Direct live imaging of cell–Cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. eLife 2, e00868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.N. Ruiz, B. Falcone, D. Kahne, T. J. Silhavy, Chemical conditionality: A genetic strategy to probe organelle assembly. Cell 121, 307–317 (2005). [DOI] [PubMed] [Google Scholar]
- 39.R. Jain, B. H. Habermann, T. Mignot, Complete genome assembly of Myxococcus xanthus strain DZ2 using long high-fidelity (hifi) reads generated with PacBio technology. Microbiol. Resour. Announc. 10, e0053021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.C. Holkenbrink, E. Hoiczyk, J. Kahnt, P. I. Higgs, Synthesis and assembly of a novel glycan layer in Myxococcus xanthus spores. J. Biol. Chem. 289, 32364–32378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. Konovalova, T. J. Silhavy, Outer membrane lipoprotein biogenesis: Lol is not the end. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Y. Hooda, T. F. Moraes, Translocation of lipoproteins to the surface of gram negative bacteria. Curr. Opin. Struct. Biol. 51, 73–79 (2018). [DOI] [PubMed] [Google Scholar]
- 43.A. K. Pogoutse, T. F. Moraes, Iron acquisition through the bacterial transferrin receptor. Crit. Rev. Biochem. Mol. Biol. 52, 314–326 (2017). [DOI] [PubMed] [Google Scholar]
- 44.A. J. Beddek, A. B. Schryvers, The lactoferrin receptor complex in gram negative bacteria. Biometals 23, 377–386 (2010). [DOI] [PubMed] [Google Scholar]
- 45.A. S. Grossman, T. J. Mauer, K. T. Forest, H. Goodrich-Blair, L. E. Comstock, A widespread bacterial secretion system with diverse substrates. MBio 12, e01956–e01921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Y. Hooda, C. C. L. Lai, A. Judd, C. M. Buckwalter, H. E. Shin, S. D. Gray-Owen, T. F. Moraes, Slam is an outer membrane protein that is required for the surface display of lipidated virulence factors in Neisseria. Nat. Microbiol. 1, 16009 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Y. Hooda, C. C. L. Lai, T. F. Moraes, Identification of a large family of Slam-dependent surface lipoproteins in Gram-negative bacteria. Front. Cell. Infect. Microbiol. 7, 207 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.M. S. Huynh, Y. Hooda, Y. R. Li, M. Jagielnicki, C. C. L. Lai, T. F. Moraes, Reconstitution of surface lipoprotein translocation through the Slam translocon. eLife 11, e72822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. Tata, S. Kumar, S. R. Lach, S. Saha, E. M. Hart, A. Konovalova, High-throughput suppressor screen demonstrates that RcsF monitors outer membrane integrity and not Bam complex function. Proc. Natl. Acad. Sci. U.S.A. 118, e2100369118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.N. D. Rawlings, A. J. Barrett, P. D. Thomas, X. Huang, A. Bateman, R. D. Finn, The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucl. Acids Res. 46, D624–D632 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.V. Roussel-Jazédé, I. Jongerius, M. P. Bos, J. Tommassen, P. van Ulsen, NalP-mediated proteolytic release of lactoferrin-binding protein B from the meningococcal cell surface. Infect. Immun. 78, 3083–3089 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.F. Saïdi, O. J. Gamboa Marin, J. I. Veytia-Bucheli, E. Vinogradov, G. Ravicoularamin, N. Y. Jolivet, A. A. Kezzo, E. Ramirez Esquivel, A. Panda, G. Sharma, S. P. Vincent, C. Gauthier, S. T. Islam, Evaluation of azido 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) analogues for click chemistry-mediated metabolic labeling of Myxococcus xanthus DZ2 lipopolysaccharide. ACS Omega 7, 34997–35013 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. N. Webby, D. P. Williams-Jones, C. Press, C. Kleanthous, Force-generation by the trans-envelope Tol-Pal system. Front. Microbiol. 13, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.T. Izoré, C. Contreras-Martel, L. el Mortaji, C. Manzano, R. Terrasse, T. Vernet, A. M. di Guilmi, A. Dessen, Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure 18, 106–115 (2010). [DOI] [PubMed] [Google Scholar]
- 55.A. Treuner-Lange, Y. W. Chang, T. Glatter, M. Herfurth, S. Lindow, G. Chreifi, G. J. Jensen, L. Søgaard-Andersen, PilY1 and minor pilins form a complex priming the type IVa pilus in Myxococcus xanthus. Nat. Commun. 11, 5054 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.A. Ducret, M.-P. Valignat, F. Mouhamar, T. Mignot, O. Theodoly, Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc. Natl. Acad. Sci. U.S.A. 109, 10036–10041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M. G. Reese, Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26, 51–56 (2001). [DOI] [PubMed] [Google Scholar]
- 58.B. S. Goldman, W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, H. B. Kaplan, Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. U.S.A. 103, 15200–15205 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.K. Han, Z. F. Li, R. Peng, L. P. Zhu, T. Zhou, L. G. Wang, S. G. Li, X. B. Zhang, W. Hu, Z. H. Wu, N. Qin, Y. Z. Li, Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci. Rep. 3, 2101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.S. Huntley, N. Hamann, S. Wegener-Feldbrugge, A. Treuner-Lange, M. Kube, R. Reinhardt, S. Klages, R. Muller, C. M. Ronning, W. C. Nierman, L. Sogaard-Andersen, Comparative genomic analysis of fruiting body formation in Myxococcales. Mol. Biol. Evol. 28, 1083–1097 (2011). [DOI] [PubMed] [Google Scholar]
- 61.S. Huntley, S. Kneip, A. Treuner-Lange, L. Søgaard-Andersen, Complete genome sequence of Myxococcus stipitatus strain DSM 14675, a fruiting myxobacterium. Genome Announc. 1, e0010013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.S. Huntley, Y. Zhang, A. Treuner-Lange, S. Kneip, C. W. Sensen, L. Søgaard-Andersen, Complete genome sequence of the fruiting myxobacterium Corallococcus coralloides DSM 2259. J. Bacteriol. 194, 3012–3013 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.N. Ivanova, C. Daum, E. Lang, B. Abt, M. Kopitz, E. Saunders, A. Lapidus, S. Lucas, T. Glavina del Rio, M. Nolan, H. Tice, A. Copeland, J. F. Cheng, F. Chen, D. Bruce, L. Goodwin, S. Pitluck, K. Mavromatis, A. Pati, N. Mikhailova, A. Chen, K. Palaniappan, M. Land, L. Hauser, Y. J. Chang, C. D. Jeffries, J. C. Detter, T. Brettin, M. Rohde, M. Göker, J. Bristow, V. Markowitz, J. A. Eisen, P. Hugenholtz, N. C. Kyrpides, H. P. Klenk, Complete genome sequence of Haliangium ochraceum type strain (SMP-2T). Stand Genomic Sci. 2, 96–106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Z.-F. Li, X. Li, H. Liu, X. Liu, K. Han, Z. H. Wu, W. Hu, F. F. Li, Y. Z. Li, Genome sequence of the halotolerant marine bacterium Myxococcus fulvus HW-1. J. Bacteriol. 193, 5015–5016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. Müller, J. W. Willett, S. M. Bahr, C. L. Darnell, K. R. Hummels, C. K. Dong, H. C. Vlamakis, J. R. Kirby, Draft genome sequence of Myxococcus xanthus wild-type strain DZ2, a model organism for predation and development. Genome Announc. 1, e00217-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S. Schneiker, O. Perlova, O. Kaiser, K. Gerth, A. Alici, M. O. Altmeyer, D. Bartels, T. Bekel, S. Beyer, E. Bode, H. B. Bode, C. J. Bolten, J. V. Choudhuri, S. Doss, Y. A. Elnakady, B. Frank, L. Gaigalat, A. Goesmann, C. Groeger, F. Gross, L. Jelsbak, L. Jelsbak, J. Kalinowski, C. Kegler, T. Knauber, S. Konietzny, M. Kopp, L. Krause, D. Krug, B. Linke, T. Mahmud, R. Martinez-Arias, A. C. McHardy, M. Merai, F. Meyer, S. Mormann, J. Muñoz-Dorado, J. Perez, S. Pradella, S. Rachid, G. Raddatz, F. Rosenau, C. Rückert, F. Sasse, M. Scharfe, S. C. Schuster, G. Suen, A. Treuner-Lange, G. J. Velicer, F. J. Vorhölter, K. J. Weissman, R. D. Welch, S. C. Wenzel, D. E. Whitworth, S. Wilhelm, C. Wittmann, H. Blöcker, A. Pühler, R. Müller, Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotech. 25, 1281–1289 (2007). [DOI] [PubMed] [Google Scholar]
- 67.G. Sharma, I. Khatri, S. Subramanian, Complete genome of the starch-degrading myxobacteria Sandaracinus amylolyticus DSM 53668T. Genome Biol. Evol. 8, 2520–2529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.G. Sharma, T. Narwani, S. Subramanian, Complete genome sequence and comparative genomics of a novel myxobacterium Myxococcus hansupus. PLOS ONE 11, e0148593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.D. C. Stevens, J. Young, R. Carmichael, J. Tan, R. E. Taylor, Draft genome sequence of gephyronic acid producer Cystobacter violaceus strain Cb vi76. Genome Announc. 2, e01299-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.G. Sharma, S. Subramanian, Unravelling the complete genome of Archangium gephyra DSM 2261T and evolutionary insights into myxobacterial chitinases. Genome Biol. Evol. 9, 1304–1311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.G. Sharma, I. Khatri, S. Subramanian, Comparative genomics of myxobacterial chemosensory systems. J. Bacteriol. 200, e00620-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.M. Wu, J. A. Eisen, A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9, R151 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A. Stamatakis, RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- 74.E. Nudleman, D. Wall, D. Kaiser, Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309, 125–127 (2005). [DOI] [PubMed] [Google Scholar]
- 75.L. S. Johnson, S. R. Eddy, E. Portugaly, Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11, 431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.I. Letunic, P. Bork, Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.J. Söding, A. Biegert, A. N. Lupas, The HHpred interactive server for protein homology detection and structure prediction. Nucl. Acids Res. 33, W244–W248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.A. Ducret, E. M. Quardokus, Y. V. Brun, MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.S. Wang, H. Arellano-Santoyo, P. A. Combs, J. W. Shaevitz, Measuring the bending stiffness of bacterial cells using an optical trap. J. Vis. Exp. , e2012 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.J.-O. Lee, L. A. Bankston, M. A. Arnaout, R. C. Liddington, Two conformations of the integrin A-domain (I-domain): A pathway for activation? Structure 3, 1333–1340 (1995). [DOI] [PubMed] [Google Scholar]
- 81.K. L. Chao, X. Shang, J. Greenfield, S. B. Linden, A. B. Alreja, D. C. Nelson, O. Herzberg, Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N). Sci. Rep. 12, 2061 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.S. Seef, J. Herrou, P. de Boissier, L. My, G. Brasseur, D. Robert, R. Jain, R. Mercier, E. Cascales, B. H. Habermann, T. Mignot, A Tad-like apparatus is required for contact-dependent prey killing in predatory social bacteria. eLife 10, e72409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S4