Abstract
Surface levels of membrane proteins are determined by a dynamic balance between exocytosis-mediated surface delivery and endocytosis-dependent retrieval from the cell surface. Imbalances in surface protein levels perturb surface protein homeostasis and cause major forms of human disease such as type 2 diabetes and neurological disorders. Here, we found a Reps1-Ralbp1-RalA module in the exocytic pathway broadly regulating surface protein levels. Reps1 and Ralbp1 form a binary complex that recognizes RalA, a vesicle-bound small guanosine triphosphatases (GTPase) promoting exocytosis through interacting with the exocyst complex. RalA binding results in Reps1 release and formation of a Ralbp1-RalA binary complex. Ralbp1 selectively recognizes GTP-bound RalA but is not a RalA effector. Instead, Ralbp1 binding maintains RalA in an active GTP-bound state. These studies uncovered a segment in the exocytic pathway and, more broadly, revealed a previously unrecognized regulatory mechanism for small GTPases, GTP state stabilization.
A Reps1-Ralbp1-RalA module promotes cargo exocytosis, in which Ralbp1 regulates RalA through GTP-state stabilization.
INTRODUCTION
Integral and peripheral membrane proteins on the cell surface enable the cell to constantly communicate with the environment (1, 2). Surface levels of membrane proteins must be precisely maintained as imbalances in surface protein levels disrupt surface protein homeostasis, impair cell physiology, and cause major forms of human disease such as cancer, type 2 diabetes, and neurodegeneration (3). The surface level of a protein is determined by the dynamic balance of its delivery to the plasma membrane through exocytic pathways (including recycling exocytosis) and removal from the plasma membrane by endocytic pathways (4–6).
Central players in exocytic pathways include vesicle-anchored small guanosine triphosphatases (GTPases) from the Ral and Rab families, which promote vesicle tethering and fusion by interacting with the exocyst complex localized to the plasma membrane (7–9). Similar to other small GTPases, Ral and Rab cycle between a guanosine diphosphate (GDP)–bound inactive state and a GTP-bound active state (10–12). Guanine nucleotide exchange factors (GEFs) recognize the GDP-bound form of small GTPases and accelerate GDP release, leading to GTP binding and activation. GTPase-activating proteins (GAPs), on the other hand, recognize the GTP-bound form of small GTPases and stimulate guanosine 5′-triphosphate (GTP) hydrolysis, inactivating the small GTPases (12).
A major endocytic route is clathrin-mediated endocytosis (CME), a vesicle budding event on the plasma membrane driven by the coat protein clathrin (13, 14). To recruit cargo proteins to endocytic vesicles, CME requires the heterotetrameric adaptor protein complex AP2 complex that acts as an adaptor between cargo proteins and clathrin coats (5, 15–18).
In this study, we performed a genome-scale CRISPR screen to identify genes required for surface protein homeostasis. The screen isolated genes encoding known exocytic and endocytic mediators, as well as many genes not previously linked to surface protein homeostasis. A high-ranking gene isolated in the screen is Reps1 (Ralbp1-associated eps domain containing 1), which encodes a ubiquitously expressed soluble protein lacking an annotated function. Reps1 forms a binary complex with Ralbp1 (RalA-binding protein 1; also known as Rlip76), another factor isolated in our CRISPR screen. Loss of the Reps1-Ralbp1 complex inhibits cargo exocytosis without affecting endocytosis, altering the surface levels of numerous proteins. The Reps1-Ralbp1 binary complex recognizes RalA, a small GTPase involved in exocytosis, followed by the release of Reps1 and formation of a Ralbp1-RalA binary complex. Ralbp1 binding maintains RalA in a GTP-bound active state, enabling RalA to interact with exocyst to promote cargo exocytosis. In neurons, mutations of the Reps1-Ralbp1 complex severely impair exocytosis-driven neurite outgrowth in neuronal morphogenesis. Together, these studies uncovered a Reps1-Ralbp1-RalA module in the exocytic pathway, in which Ralbp1 acts as a GTP-state stabilizing factor (GSF) for RalA. The GSF function represents a previously unrecognized regulatory mechanism that is likely involved in the activation cycles of other small GTPases.
RESULTS
Regulators of surface protein homeostasis identified in a genome-scale CRISPR genetic screen
To identify genes required for the maintenance of surface protein homeostasis, we carried out a genome-wide CRISPR genetic screen using a fluorescent surface protein reporter. The reporter is based on the glucose transporter GLUT4 (Fig. 1A), which depends on general exocytic and endocytic pathways to regulate its surface levels (3, 19–21). Thus, we anticipate that the glucose transporter–based surface protein reporter (GLUT-SPR) would enable us to identify factors broadly regulating surface protein homeostasis.
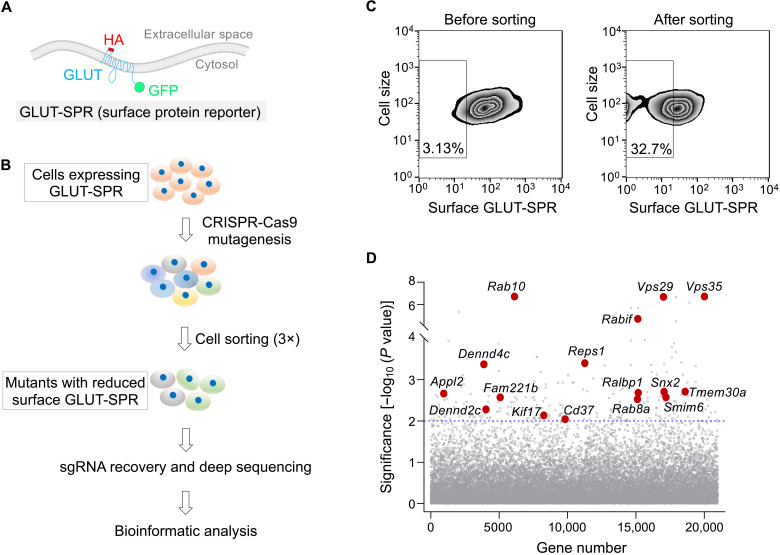
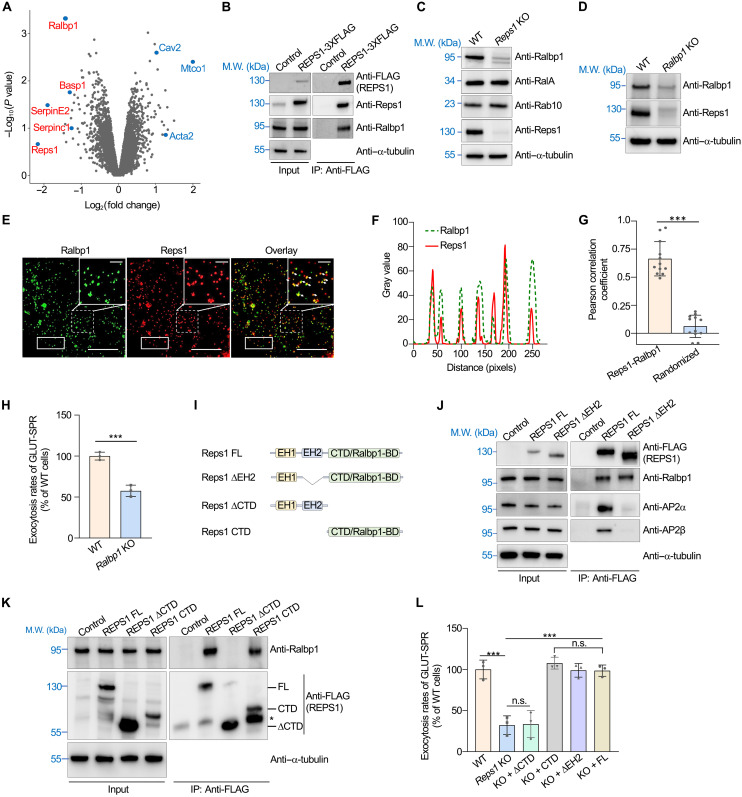
Fig. 1. Identification of genes involved in surface protein homeostasis using a genome-wide CRISPR genetic screen.
(A) Diagram of the surface protein reporter GLUT-SPR (HA-GLUT4-GFP) used in the CRISPR screen. The reporter is based on the multitransmembrane glucose transporter GLUT4. (B) Illustration of the genome-wide genetic screen to identify genes required for maintaining the surface levels of GLUT-SPR. (C) Flow cytometry analysis of the starting CRISPR library of preadipocyte fibroblasts (left) and the final population after three rounds of sorting (right) in the genome-wide CRISPR screen. GLUT-SPR molecules on the cell surface were labeled using anti-HA antibodies and allophycocyanin (APC)–conjugated secondary antibodies. APC and GFP fluorescence was measured by flow cytometry and used to calculate relative surface levels of the reporter. (D) Ranking of genes in the CRISPR screen based on their P values. Each dot represents a gene. A P value of 0.01 is set as the cutoff (shown as a horizontal dotted line). Selected genes are labeled. Full datasets of the CRISPR screen are included in tables S1 and S2. sgRNA, single-guide RNA.
Mouse preadipocyte fibroblasts expressing GLUT-SPR were mutagenized using a pooled genome-wide CRISPR single-guide RNA (sgRNA) library (22). The reporter bears an exoplasmic hemagglutinin (HA) epitope and a cytoplasmic green fluorescent protein (GFP) tag (Fig. 1A), which enabled us to simultaneously monitor both surface and total levels of the reporter. Fluorescence-activated cell sorting was used to identify and collect mutant cells with reduced surface but normal total levels of the reporter (Fig. 1, B and C). After three rounds of sorting, sgRNAs were recovered from the sorted and unsorted populations and analyzed using deep sequencing. Genes were ranked on the basis of enrichment of their corresponding sgRNAs using the Model-based Analysis of Genome-wide CRISPR-Cas9 knockout (MAGeCK) algorithm (Fig. 1D and tables S1 and S2) (19, 21, 23). Hits from the screen included genes encoding known exocytic and endocytic mediators (Fig. 1D and table S2). However, a large fraction of the hits have not been previously linked to surface protein homeostasis.
Mutations of Reps1 alter surface levels of numerous membrane proteins
A high-ranking hit from the screen is Reps1, which encodes a ubiquitously expressed soluble protein of ~125 kDa (Fig. 1D). REPS1 mutations in humans cause neurodegeneration with brain iron accumulation (NBIA), a progressive neurodegenerative disease (24–26). The biological function of Reps1, however, was unknown. To begin unraveling its function, we deleted the Reps1 gene in adipocytes (Fig. 2A and fig. S1A), a cell type demanding strict surface protein homeostasis (27–30). However, in accordance with our screen, Reps1 knockout (KO) adipocytes exhibited a strong reduction in surface levels of GLUT-SPR (Fig. 2, B and C). Further analysis showed that a large portion of GLUT-SPR molecules were retained in intracellular compartments in Reps1 KO cells, while the total levels of the reporter remained unchanged (Fig. 2, B and C, and fig. S1, B to D). Thus, Reps1 regulates the subcellular localization but not the overall expression of GLUT-SPR.
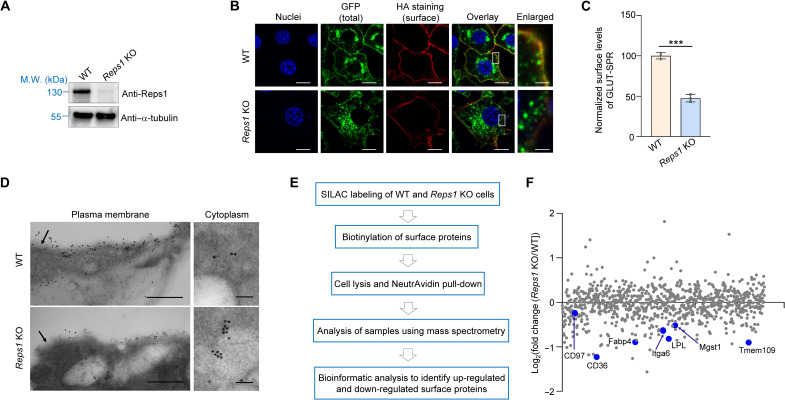
Fig. 2. Reps1 plays a critical role in maintaining surface protein homeostasis.
(A) Representative immunoblots showing the expression of the indicated proteins in WT and Reps1 KO adipocytes. M.W., molecular weight. (B) Representative confocal images showing the localization of GLUT-SPR (depicted in Fig. 1A) in unpermeabilized WT and Reps1 KO adipocytes. Before harvesting, cells were switched to culture media containing 20 nM insulin to mimic a physiological fed state. After 20 min, surface reporters were labeled using anti-HA antibodies and Alexa Fluor 568–conjugated secondary antibodies. Scale bars, 10 μm (1 μm for the enlarged images on the right). (C) Flow cytometry measurements of surface GLUT-SPR in cells described in (B). Data of mutant cells were normalized to those of WT cells. Data are presented as means ± SD of three biological replicates. ***P < 0.001 (calculated using Student’s t test). In all figures, data normalization was performed by setting the mean value of WT data points as 100 or 1, and all data points including WT ones were normalized to that mean value. (E) Electron micrographs of GLUT-SPR distribution in WT and Reps1 KO adipocytes. Cells were cultured as in (B) and frozen in acetone containing 0.25% glutaraldehyde and 0.1% uranyl acetate. Cell sections were stained with anti-GFP antibodies and gold-conjugated secondary antibodies. Arrows point to the plasma membrane. Direct magnification: ×49,000. Scale bars, 500 (left) and 100 nm (right). (E) Procedure of mass spectrometry–based surface proteomic analysis of WT and Reps1 KO cells. (F) Scatter plot showing adjusted fold changes of surface protein levels (Reps1 KO/WT) in mouse adipocytes. To calculate the adjusted values, fold changes of surface protein levels were divided by those of total protein levels based on whole-cell proteomic data shown in table S5.Selected down-regulated surface proteins are labeled. Full datasets are shown in table S3. In addition, see figs. S1 to S3.
To detect its subcellular localization at a higher resolution, GLUT-SPR was labeled using immunogold and detected using electron microscopy (EM). Consistent with the findings of confocal imaging and flow cytometry, GLUT-SPR was markedly redistributed to intracellular compartments in Reps1 KO cells compared to its predominant surface localization in wild-type (WT) cells (Fig. 2D and fig. S1E). KO of Reps1 did not affect the differentiation or insulin signaling of adipocytes (fig. S2). We also generated mutant HeLa cells lacking REPS1 (figs. S1 and S3A). Similar to the results of adipocyte experiments, surface levels of GLUT-SPR were strongly reduced in REPS1-deficient HeLa cells (fig. S3, B and C). Together, these data confirmed the findings of the fibroblast-based CRISPR screen and demonstrate that the function of Reps1 in surface protein homeostasis is not restricted to a specific cell type.
To globally assess the role of Reps1 in surface protein homeostasis, we carried out mass spectrometry–based proteomic analysis to compare the surface proteomes of WT and Reps1 KO adipocytes (Fig. 2E). Surface proteins were biotinylated, purified using NeutrAvidin, and analyzed by mass spectrometry. The total number of surface proteins identified in mass spectrometry was similar in WT and Reps1 KO cells (table S3). However, surface levels of many integral and peripheral membrane proteins were substantially down-regulated in Reps1 KO cells (Fig. 2F and table S3). Thus, mutations of Reps1 substantially alter the surface proteome, validating the use of GLUT-SPR as a reporter to identify genes broadly regulating surface protein homeostasis.
Reps1 is dispensable for CME-dependent cargo internalization
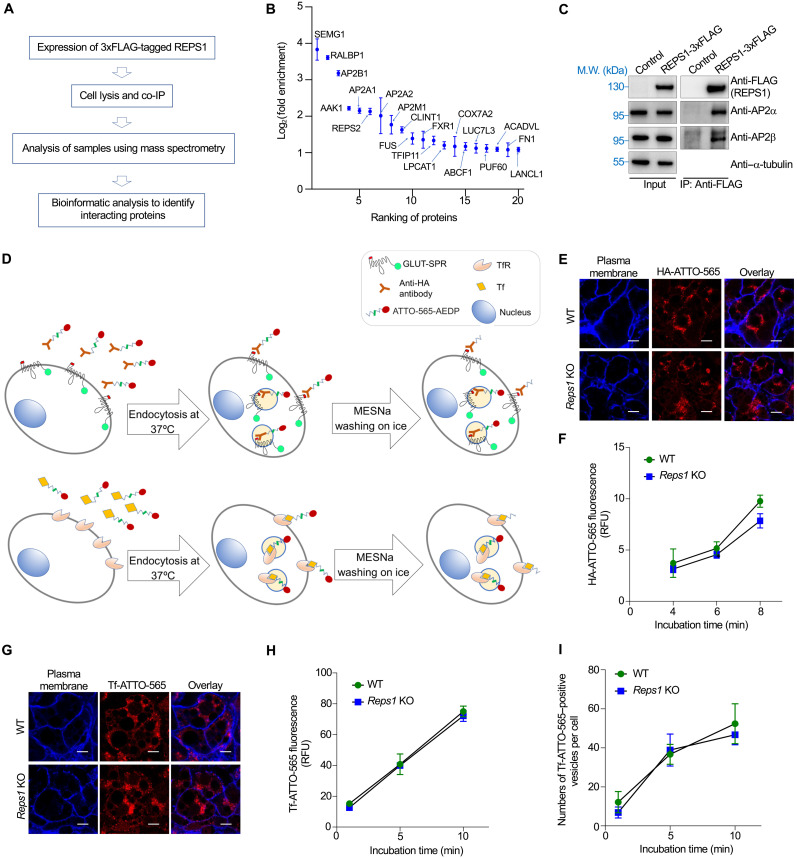
Next, we sought to determine how Reps1 regulates surface protein levels. We began by identifying the binding partners of Reps1. Reps1-bound proteins were isolated using co-immunoprecipitation (co-IP) and analyzed using mass spectrometry (Fig. 3A). Top-ranking binding proteins of Reps1 included subunits of the AP2 adaptor complex (Fig. 3B and table S4), a key regulator of CME required for GLUT-SPR internalization (21). Reps1-AP2 interaction was confirmed using co-IP and immunoblotting (Fig. 3C). Because cargo endocytosis is a key determinant of surface protein levels, we tested whether Reps1 influences surface levels of GLUT-SPR through regulating CME. To accurately measure CME, we developed cargo endocytosis assays using the cleavable fluorescent dye ATTO-565-3-[(2-aminoethyl)dithio]propionic acid N-hydroxysuccinimide ester (ATTO-565-AEDP-NHS-ester) (31–33). After conjugation, the ATTO-565 dye moiety is linked to a cargo protein through a disulfide bond (Fig. 3D). Surface-exposed ATTO-565 dyes can be rapidly removed by breaking the disulfide bond using membrane-impermeable reducing agents such as sodium 2-mercaptoethane sulfonate (MESNa). By contrast, dyes on internalized cargo proteins are resistant to MESNa cleavage (Fig. 3D).
Fig. 3. Reps1 is dispensable for CME-mediated cargo internalization.
(A) Proteomic analysis procedure of Reps1-interacting proteins in HeLa cells. (B) Scatter plot showing the top proteins enriched in 3xFLAG-tagged REPS1 immunoprecipitates compared to the control sample. Error bars indicate SD of biological duplicates. Full datasets are included in table S4. (C) Representative immunoblots showing the interaction of REPS1 with AP2 adaptor. The 3xFLAG-tagged REPS1 protein was stably expressed in preadipocytes and immunoprecipitated using anti-FLAG antibodies. Control cells were transfected with an empty vector. IP, immunoprecipitation. (D) Diagrams of assays measuring the endocytosis of GLUT-SPR (top) and Tf (bottom). (E) Representative confocal images showing GLUT-SPR endocytosis. Cells were incubated with ATTO-565–conjugated anti-HA antibodies (HA-ATTO-565; 1 μg/ml) for 8 min at 37°C. After MESNa treatment on ice, cells were fixed and labeled with CF405M-conjugated concanavalin A (50 μg/ml). Scale bars, 10 μm. (F) Flow cytometry analysis of HA-ATTO-565 endocytosis. Cells were incubated with ATTO-565–conjugated anti-HA antibodies (HA-ATTO-565; 1 μg/ml) at 37°C for the indicated durations. After washing with MESNa, cells were disassociated using Accutase and analyzed using flow cytometry. Error bars indicates SD, n = 3. RFU, relative fluorescence unit. (G) Representative confocal images showing the endocytosis of ATTO-565–conjugated Tf (Tf-ATTO-565) into WT or Reps1 KO adipocytes. Cells were incubated with Tf-ATTO-565 (5 μg/ml) at 37°C for 10 min before the cells were harvested and processed as in (E). Scale bars, 10 μm. (H) Time course of Tf-ATTO-565 endocytosis into WT or Reps1 KO adipocytes measured by flow cytometry. Error bars indicate SD, n = 3. (I) Tf-ATTO-565 endocytosis was performed as in (G), and Tf-ATTO-565–positive endocytic vesicles were quantified on the basis of confocal images. About 100 cells from seven fields were analyzed for each sample. Error bars indicate SD of different fields.
We labeled anti-HA antibodies using the cleavable dye and measured the endocytosis of GLUT-SPR. The rate of GLUT-SPR endocytosis remained largely unchanged in Reps1 KO cells (Fig. 4, E and F). The moderate reduction of GLUT-SPR endocytosis during a longer incubation was likely due to decreased surface GLUT-SPR available for internalization. We also examined the endocytosis of transferrin (Tf), a classic CME cargo internalized through binding to the Tf receptor (TfR) (Fig. 3D). We observed that Tf endocytosis remained intact in Reps1 KO cells (Fig. 3, G and H), consistent with the finding that TfR surface levels were unchanged in the KO cells (fig. S1F). Moreover, the number of Tf-positive CME vesicles was comparable in WT and Reps1 KO cells (Fig. 3I). These results demonstrate that despite its interaction with AP2, Reps1 is dispensable for the internalization of CME cargoes including GLUT-SPR.
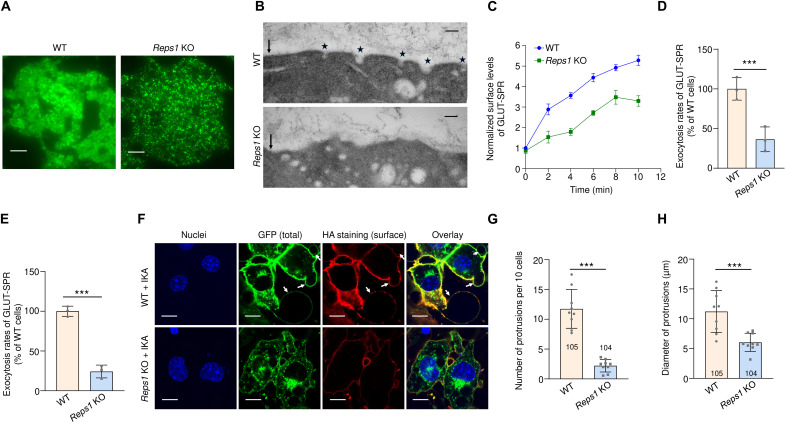
Fig. 4. Reps1 promotes cargo exocytosis.
(A) Representative TIRFM images of GLUT-SPR in WT and Reps1 KO adipocytes. Cells were cultured as in Fig. 2B, and GLUT-SPR on and near the plasma membrane was visualized using TIRFM. Scale bars, 10 μm. (B) Representative transmission electron microscopy (TEM) images showing vesicles near the plasma membrane in WT and Reps1 KO adipocytes. Cells were cultured as in Fig. 2B, and 80-nm sections of cells were visualized using TEM without staining. Arrows point to the plasma membrane, whereas stars point to vesicles fusing with the plasma membrane. Direct magnification: ×68,000. Scale bars, 100 nm. (C) Time course of GLUT-SPR surface levels in WT and Reps1 KO adipocytes. Cells were cultured as in (B), and surface reporter levels were measured using flow cytometry after the indicated periods in insulin-containing media. (D) Normalized initial exocytosis rates of the reporter based on data in (C). Data are presented as means ± SD of three biological replicates. ***P < 0.001 (calculated using Student’s t test). (E) Normalized initial exocytosis rates of GLUT-SPR in the presence of IKA. Experiments were carried out and analyzed as in (C) and (D) except that 4 μM IKA was included. (F) Representative confocal images showing the GLUT-SPR reporter in WT and Reps1 KO adipocytes. Cells were cultured as in Fig. 2B and incubated with 4 μM IKA for 20 min before cell harvesting. Arrows point to protrusions emanating from the plasma membrane. Scale bars, 25 μm. (G and H) Quantification of the number and diameter of protrusions in WT and Reps1 KO adipocytes based on nine confocal images from three independent experiments. Numbers of cells analyzed are indicated on the graphs. Error bars indicate SD. ***P < 0.001 (calculated using Student’s t test). Also, see fig. S4.
Reps1 promotes cargo exocytosis
Next, we examined whether Reps1 regulates GLUT-SPR exocytosis—the fusion of GLUT-SPR–containing vesicles with the plasma membrane. Total internal reflection fluorescence microscopy (TIRFM) was used to selectively visualize GLUT-SPR molecules on and near the plasma membrane. We observed substantial GLUT-SPR localization to the cell surface in WT cells (Fig. 4A). However, in Reps1 KO cells, GLUT-SPR signal was confined to punctate structures near the plasma membrane (Fig. 4A), suggesting that the molecules were sequestered in intracellular vesicles. To confirm this finding, we used EM to detect vesicles near the plasma membrane. In WT cells, vesicles were frequently observed in the process of fusing with the plasma membrane, an event rarely detected in Reps1 KO cells (Fig. 4B). Instead, vesicles accumulated near the plasma membrane in Reps1 KO cells (Fig. 4B), consistent with the TIRFM data. Together, these data suggest that Reps1 promotes GLUT-SPR exocytosis.
Next, we measured surface levels of GLUT-SPR over time, which enabled us to calculate exocytosis rates of the reporter (Fig. 2, C and D). We observed that the exocytosis rate of GLUT-SPR was strongly decreased in Reps1 KO cells (Fig. 2D). We also measured exocytosis rates in the presence of ikarugamycin (IKA), a selective inhibitor of CME (34). Again, we observed a large reduction in GLUT-SPR exocytosis in Reps1 KO cells (Fig. 4E). To confirm the role of Reps1 in exocytosis, we took advantage of the observation that after prolonged inhibition of CME using IKA, protrusions began to emanate from the cell surface due to accumulation of excess membranes delivered by ongoing exocytosis and a lack of compensatory endocytosis (Fig. 4F). Strikingly, both the number and size of these protrusions were strongly reduced in Reps1 KO cells (Fig. 4, F to H), confirming an exocytosis defect in the KO cells. F-actin was absent in the protrusions (fig. S4), suggesting that the protrusions were not generated by internal actin polymerization. Notably, the membrane protrusion phenotype reflects global alterations in exocytic addition of membranes to the cell surface, in agreement with our conclusion that Reps1 broadly regulates surface protein homeostasis. Together, these results demonstrate that Reps1 plays a key role in cargo exocytosis.
Reps1 acts in concert with Ralbp1 to regulate exocytosis
To gain insights into the mechanism of Reps1 in exocytosis, we compared the whole-cell proteomes of WT and Reps1 KO cells. We found that Ralbp1, a soluble factor of 76 kDa, was among the most depleted proteins in Reps1 KO cells (Fig. 5A and table S5). Ralbp1 was also identified as a significant hit in our CRISPR screen (Fig. 1D) and is an interacting factor of Reps1 (Figs. 3B and 5B) (35), suggesting that Ralbp1 and Reps1 are involved in the same biological pathway.
Fig. 5. Reps1 acts in concert with Ralbp1 to regulate exocytosis.
(A) Whole-cell proteomic analysis to identify proteins down-regulated or up-regulated in Reps1 KO cells. Full datasets are shown in table S5. (B) Representative immunoblots showing the interaction of Reps1 with Ralbp1. The 3xFLAG-tagged REPS1 protein was stably expressed in preadipocytes and immunoprecipitated using anti-FLAG antibodies. Control cells were transfected with an empty vector. (C and D) Representative immunoblots showing the expression of the indicated proteins. (E) Representative SIM images showing the subcellular localization of Reps1 and Ralbp1. The 3xFLAG-tagged Reps1 protein was stably expressed in HeLa cells and stained using anti-FLAG antibodies, whereas endogenous Ralbp1 was stained using anti-Ralbp1 antibodies. Scale bars, 2 μm for main images and 1 μm for enlarged images. (F) Profile analysis plot comparing the distributions of Ralbp1 and Reps1 within the rectangular areas of (E). (G) Quantification of Ralbp1-Reps1 colocalization using the Pearson correlation coefficient. Images were captured as in (E) and analyzed using Image J. Each dot represents data of an individual cell. In randomized images, Reps1 images were rotated 90° clockwise, whereas Ralbp1 images were not rotated. (H) Normalized initial exocytosis rates of GLUT-SPR. Error bars indicate SD, n = 3. ***P < 0.001 (calculated using Student’s t test). (I) Diagrams of FL and mutant Reps1. (J and K) Representative immunoblots showing protein expression and the interactions of REPS1 with Ralbp1 and AP2 adaptor. The asterisk denotes a possible degradation product of Reps1 CTD. (L) Normalized initial exocytosis rates of GLUT-SPR. Error bars represent SD, n = 3. ***P < 0.001; not significant (n.s.), P > 0.05 [calculated using one-way analysis of variance (ANOVA)]. In addition, see fig. S5.
Ralbp1 is implicated in a range of physiological and pathological processes, but its function and mechanism were unclear (35–37). Using immunoblotting, we confirmed that Ralbp1 expression was strongly reduced in Reps1 KO cells, while other exocytic regulators we examined remained unchanged (Fig. 5C and figs. S3A and S7B). Reciprocally, Reps1 expression was diminished in Ralbp1 KO cells (Fig. 5D). These results are reminiscent of the observations that elimination of one subunit of a multimeric protein complex often leads to loss of other subunits (38, 39). Super-resolution imaging using structured illumination microscopy (SIM) showed that Ralbp1 and Reps1 exhibited substantial colocalization in the cell (Fig. 5, E to G). KO of Ralbp1 in adipocytes markedly reduced the exocytosis rate of GLUT-SPR (Fig. 5H), similar to the phenotype of Reps1 KO cells. These biochemical and genetic data suggest that Reps1 and Ralbp1 are physically and functionally linked.
Next, we examined whether the functions of Reps1 and Ralbp1 in exocytosis require their interaction. Reps1 has two EH (Eps15 homology) domains, protein modules involved in protein-protein interactions (40), and an evolutionarily conserved C-terminal domain (CTD) with no sequence homology with other proteins (Fig. 5I). We deleted these domains individually and examined how the deletions affect Ralbp1 binding and Reps1 function. While the Reps1 ΔEH1 mutant could not be expressed, the Reps1 ΔEH2 mutant was expressed to a similar level as full-length (FL) Reps1 and still interacted with Ralbp1 (Fig. 5J). The Reps1 ΔEH2 mutant, however, was unable to interact with AP2 adaptor (Fig. 5J). The Reps1 ΔCTD mutant, on the other hand, did not bind Ralbp1, whereas CTD itself interacted with Ralbp1 (Fig. 5K). Thus, we renamed CTD as Ralbp1-binding domain (Ralbp1-BD). In functional assays, the Ralbp1-BD of Reps1 fully restored the exocytosis rate of GLUT-SPR when expressed in Reps1 KO cells, whereas the Reps1 ΔRalbp1-BD mutant did not (Fig. 5L). Expression of the ΔEH2 mutant in Reps1 KO cells fully restored the exocytosis rate of GLUT-SPR (Fig. 5L), indicating that AP2 binding is dispensable for the function of Reps1 in exocytosis. Using a similar approach, we mapped the Reps1-binding domain of Ralbp1 to its CTD (fig. S5). Thus, Reps1 and Ralbp1 interact through their CTDs. Together, these data demonstrate that Reps1 and Ralbp1 form a binary complex and act in concert to promote exocytosis.
Reps1 and Ralbp1 promote exocytosis by regulating RalA activation
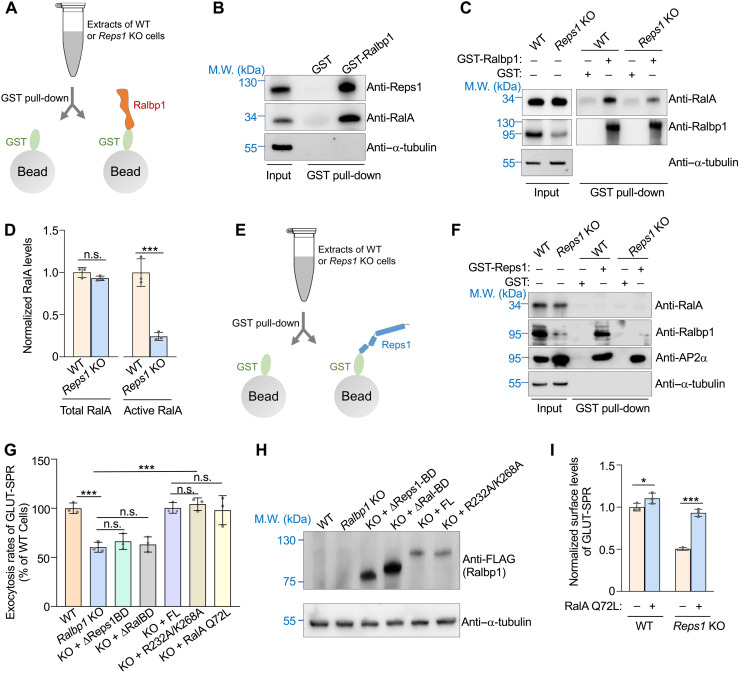
Ralbp1 binds to RalA, a small GTPase regulating vesicle tethering and fusion through interacting with exocyst (11, 20, 41–44). Next, we sought to determine whether Ralbp1 and Reps1 promote exocytosis by regulating RalA activity. In a glutathione S-transferase (GST) pull-down assay, recombinant Ralbp1 interacted with both RalA and Reps1 from adipocyte extracts (Fig. 6, A and B). Ralbp1 selectively binds to the active GTP-bound form of Ral GTPases, offering a reliable approach to quantify active RalA proteins (37, 45, 46). We observed that active RalA was reduced to a near-background level in Reps1 KO cells, whereas the total level of RalA remained unchanged (Fig. 6, C and D). These data suggest that the exocytosis defects observed in Reps1-Ralbp1–deficient cells are due to diminished activities of RalA.
Fig. 6. Reps1 and Ralbp1 maintain RalA in an active state.
(A) Illustration of GST pull-down assays measuring interactions of Ralbp1 with Reps1 and RalA. (B) Representative immunoblots detecting proteins pulled down from WT adipocytes using GST or GST-Ralbp1. (C) Representative immunoblots detecting proteins pulled down from WT or Reps1 KO adipocytes using GST or GST-Ralbp1. (D) Quantification of total and active RalA proteins in WT and Reps1 KO adipocytes. Ralbp1, which selectively binds active GTP-bound RalA, was used to pull down active RalA proteins from cell extracts. Intensities of proteins on immunoblots were quantified using ImageJ and normalized to those of α-tubulin. Data of Reps1 KO samples were normalized to those of WT cells. Error bars represent SD, n = 3. ***P < 0.001; n.s., P > 0.05 (calculated using Student’s t test). (E) Illustration of GST pull-down assays measuring interactions of Reps1 with Ralbp1 and RalA. (F) Representative immunoblots detecting proteins pulled down from WT or Reps1 KO adipocytes using GST or GST-Reps1. (G) Normalized initial exocytosis rates of GLUT-SPR in WT and Ralbp1 KO adipocytes. Error bars indicate SD, n = 3. ***P < 0.001 (calculated using one-way ANOVA). (H) Representative immunoblots showing the expression of the indicated proteins. (I) Normalized surface levels of GLUT-SPR in WT or Reps1 KO adipocytes stably expressing the constitutively active RalA Q72L mutant. The cells were cultured and labeled as in Fig. 2C. Error bars indicate SD, n = 3. *P < 0.05 and ***P < 0.001 (calculated using two-way ANOVA).
Next, we performed GST pull-down assays using recombinant GST-Reps1 (Fig. 6E). We observed that GST-Reps1 did not pull down RalA from either WT or Reps1 KO cells (Fig. 6F). GST-Reps1, however, readily pulled down Ralbp1 and AP2 subunits (Fig. 6F), consistent with the co-IP results (Fig. 3C). Although the Reps1-AP2 interaction is dispensable for Reps1 function in exocytosis, detection of the interaction further suggests that recombinant GST-Reps1 proteins were properly folded. These GST pull-down results are consistent with the Reps1 interactome data (Fig. 3B and table S4) and indicate that Ralbp1 does not simultaneously interact with Reps1 and RalA. Instead, when Ralbp1 binds to RalA, Reps1 is released from Ralbp1 without continuing to associate with the newly formed Ralbp1-RalA binary complex.
Because Ralbp1 selectively binds to active GTP-bound RalA, and its mutations diminish active RalA, we postulate that Ralbp1 is required for maintaining RalA in its active state. To test this possibility, we expressed a RalA Q72L mutant, in which RalA was locked in a GTP-bound state (47). When expressed in Ralbp1 KO cells, the RalA Q72L mutant fully restored the exocytosis rate of GLUT-SPR (Fig. 6G). Likewise, the expression of the RalA Q72L mutant also fully rescued the exocytosis phenotype of Reps1 KO cells (Fig. 6I). Expression of RalA Q72L had limited effect on the surface level of GLUT-SPR in WT cells (Fig. 6I), suggesting that RalA Q72L selectively rescued a Ralbp1-Reps1–dependent activity rather than stimulating exocytosis through a different pathway. In WT cells, there were likely sufficient amounts of active RalA to drive efficient exocytosis such that introduction of additional active RalA did not substantially enhance the exocytosis rate. Thus, the functions of Reps1 and Ralbp1 in exocytosis can be bypassed by constitutive activation of RalA, supporting the notion that Reps1 and Ralbp1 promote exocytosis by regulating RalA activation rather than being RalA effectors.
Ralbp1 contains a Ral-binding domain and an Reps1-binding domain (CTD) (fig. S4A), both of which are essential to the function of Ralbp1 in exocytosis (Fig. 6, G and H). In addition, Ralbp1 has a putative RhoGAP domain, although its catalytic activity is very low (48, 49). The activity of the RhoGAP domain is abolished by mutations of a conserved arginine finger (R232) and a stabilizing lysine residue (K268) (50). However, when expressed in Ralbp1 KO cells, the Ralbp1 R232A/K268A mutant fully restored GLUT-SPR exocytosis (Fig. 6, G and H). Thus, the function of Ralbp1 in exocytosis does not require the activity of RhoGAP, a major recognizable functional domain in Ralbp1. These data further support our conclusion that Ralbp1 does not act as a RalA effector in the exocytic pathway. Together, these results revealed a Reps1-Ralbp1-RalA module in the exocytic pathway, in which Ralbp1 regulates RalA through a previously unrecognized mechanism of GTP state stabilization.
The Reps1-Ralbp1-RalA module is required for neurite outgrowth in neurons
Because REPS1 mutations cause neurodegenerative diseases, next, we examined the functional role of the Reps1-Ralbp1-RalA module in neurons. Neurons are highly polarized and compartmentalized cells (51). During neuronal morphogenesis, neuronal progenitor cells send out processes that become neurites including a single axon and multiple dendrites (52). Neurite outgrowth depends on exocytosis to deliver membrane materials to specific surface regions, a process requiring RalA and exocyst (53–55). However, the roles of Reps1 and Ralbp1 in neuronal morphogenesis remained unknown.
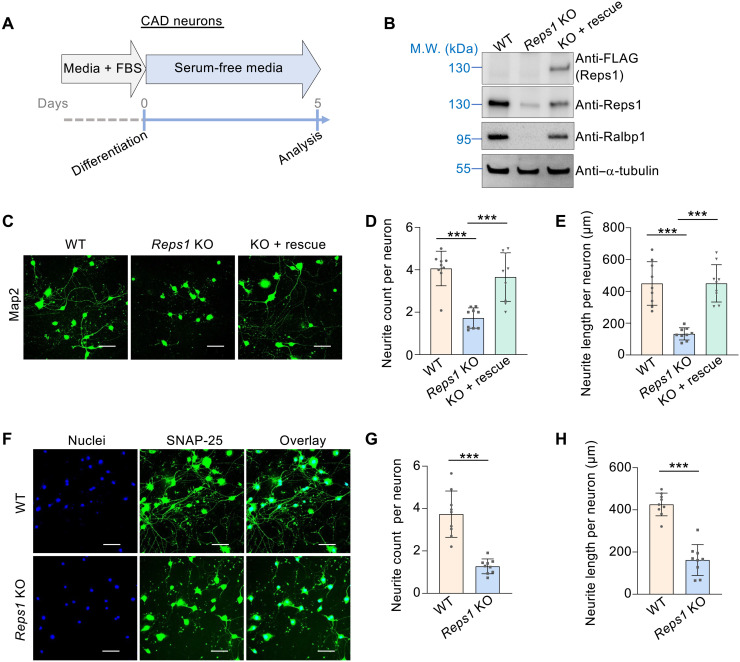
We first examined Cath.a-differentiated (CAD) cells, a mouse catecholaminergic neuronal cell line (56). After serum withdrawal, CAD progenitor cells differentiated into neurons with characteristic neurites (Fig. 7, A to C). KO of Reps1 in CAD cells led to loss of both Reps1 and Ralbp1 proteins (Fig. 7B). CAD neurons were labeled using antibodies against Microtubule associated protein 2 (Map2), a cytosolic neuronal maker (57). We observed that the total cell number did not change in Reps1 KO CAD neurons, but neurite number and length were strongly reduced (Fig. 7, C to E). The KO phenotype was fully rescued by expression of a WT Reps1 gene (Fig. 7, C to E). We also labeled Synaptosome associated protein 25 (SNAP-25), a membrane-bound neuronal protein (58). Again, both neurite number and length visualized with SNAP-25 staining were markedly decreased in Reps1 KO CAD neurons (Fig. 7, F to H). When the Ralbp1 gene was deleted in CAD cells, both Ralbp1 and Reps1 proteins were diminished (fig. S6A). In agreement with the Reps1 KO data, neurite outgrowth was substantially reduced in Ralbp1 KO CAD neurons (fig. S6, B to D). Thus, Reps1 and Ralbp1 are essential to the morphogenesis of CAD neurons.
Fig. 7. Reps1 is essential to neurite outgrowth in CAD neurons.
(A) Diagram illustrating the differentiation of CAD neurons. (B) Representative immunoblots showing the expression of the indicated proteins. (C) Staining of Map2 in the indicated CAD neurons. Scale bars, 100 μm. (D and E) Quantification of neurite number and total neurite length per CAD neuron. Neurites were traced on the basis of Map2 staining and processed using the NeuronJ plugin for Fiji. A total of 80 WT cells, 93 Reps1 KO cells, and 95 cells of Reps1 KO with a rescue gene were analyzed from nine fields each. Error bars indicate SD of different fields. ***P < 0.001 (calculated using one-way ANOVA). (F) Staining of SNAP-25 in the indicated CAD neurons. Scale bars, 100 μm. (G and H) Quantification of neurite number and total neurite length per cell in WT and Reps1 KO CAD neurons. Neurites were traced and analyzed on the basis of SNAP-25 staining. A total of 106 WT cells and 103 Reps1 KO cells were analyzed from nine fields each. Error bars indicate SD of different fields. ***P < 0.001 (calculated using Student’s t test). In addition, see fig. S6.
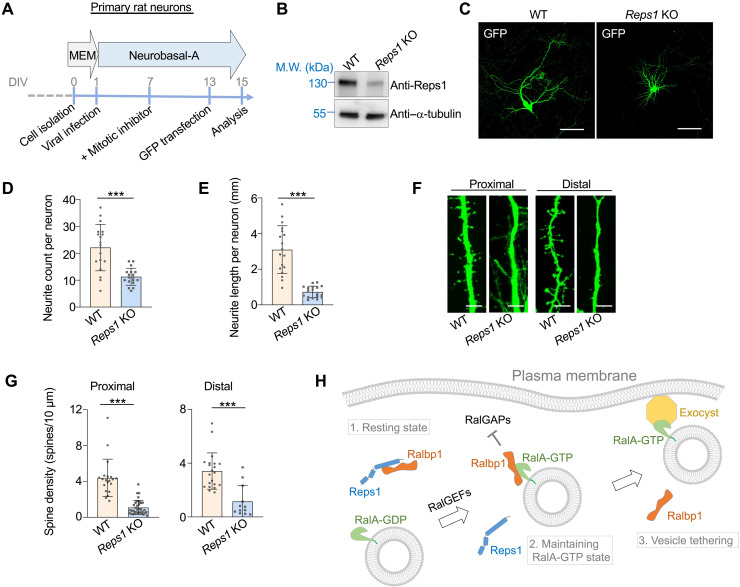
Next, we examined the function of the Reps1-Ralbp1 complex in cultured primary hippocampal neurons isolated from newborn rats. Lentiviral CRISPR particles were introduced into the primary neurons to delete the Reps1 gene (Fig. 8, A and B). Despite the delay between cell seeding and Reps1 depletion, the number of neurites in Reps1 KO primary neurons was markedly decreased, while the total cell number did not change (Fig. 8, C and D). Likewise, the length of neurites was markedly reduced in Reps1 KO primary neurons (Fig. 8, C and E), consistent with the results of the CAD experiments. We also quantified dendritic spines, micro-sized dendritic protrusions contacted by axons to form synaptic connections (51). The formation of dendritic spines also depends on Ral-mediated exocytosis (55). We observed that dendritic spines were largely eliminated in Reps1 KO neurons at both proximal and distal regions of dendrites (Fig. 8, F and G). Thus, similar to RalA, Reps1 and Ralbp1 are required for neurite outgrowth. These findings demonstrate that the Reps1-Ralbp1-RalA module plays an essential role in exocytosis-driven neurite outgrowth during neuronal morphogenesis.
Fig. 8. Reps1 is essential to neurite outgrowth in primary neurons.
(A) Schematic illustration of culturing primary rat hippocampal neurons. DIV: days in vitro. (B) Representative immunoblots showing the expression of the indicated proteins. (C) Representative images showing GFP-labeled WT and Reps1 KO rat hippocampal neurons. Cells were transiently transfected using a GFP-encoding plasmid. After 48 hours, cells were fixed and visualized using a 20× objective. Only a small number of neurons were GFP+ such that the morphologies of individual neurons were readily discerned. (D and E) Quantification of neurite number and total neurite length per cell in WT or Reps1 KO rat hippocampal neurons based on GFP labeling. A total of 23 WT neurons and 21 Reps1 KO primary neurons were analyzed from 20 fields each. Error bars indicate SD of different fields. ***P < 0.001 (calculated using Student’s t test). (F) Representative images showing reconstructed dendrites of rat hippocampal neurons. Z-stacking was performed at 1 μm per step. Scale bars, 5 μm. (G) Quantification of spine densities located at proximal or distal dendrites of WT and Reps1 KO primary neurons. Error bars indicate SD of different dendritic segments from 12 WT neurons from 12 fields and 11 Reps1 KO neurons from 11 fields. ***P < 0.001 (calculated using Student’s t test). (H) Model depicting the function of the Reps1-Ralbp1-RalA module in exocytosis. For simplicity, other effectors of RalA such as motor proteins are not shown (75).
DISCUSSION
In this work, we found a Reps1-Ralbp1-RalA module that regulates exocytosis through sequential, dynamic molecular interactions. Reps1 and Ralbp1 form a binary complex at the resting state. When the Reps1-Ralbp1 binary complex recognizes the vesicle-bound small GTPase RalA, Reps1 dissociates from Ralbp1, allowing for the formation of a new binary complex of Ralbp1 and RalA. Ralbp1 binding maintains RalA in its GTP-bound active state, enabling the latter to interact with the Sec5 and Exo84 subunits of the plasma membrane-localized exocyst complex to facilitate vesicle tethering and fusion. RalA-exocyst association is accompanied by the release of Ralbp1 from RalA (Fig. 8H).
Reps1 is an essential component of the Reps1-Ralbp1-RalA module, although it does not directly associate with RalA. Our findings suggest that Reps1 not only stabilizes Ralbp1 but also presents binding-competent Ralbp1 to RalA. The functions of Reps1 and Ralbp1 are tightly integrated such that deletion of either one leads to the loss of both proteins. Thus, Reps1 and Ralbp1 form an obligate protein complex in the cytosol. Because Reps1 and RalA recognize distinct regions on Ralbp1, the dissociation of Reps1 from Ralbp1 is likely triggered by a spatial hindrance or conformational change in Ralbp1 upon RalA binding. Subsequent dissociation of Ralbp1 from RalA, on the other hand, is expected to result from direct competition from exocyst as Ralbp1 and exocyst recognize the same domain of GTP-bound Ral GTPases (4, 9, 45). The Reps1-Ralbp1-RalA module constitutes an integral segment of the Ral-dependent exocytic pathway, which operates in parallel with the Rab-dependent pathway in mediating cargo exocytosis and maintaining surface protein homeostasis (11, 59).
Ralbp1 represents a previously unrecognized type of regulator for small GTPases, a GSF. The GSF function of Ralbp1 is supported by three lines of evidence: (i) Ralbp1 selectively recognizes the active GTP-bound form of RalA but is not a RalA effector; (ii) its binding stabilizes the GTP-bound state of RalA; and (iii) its function in exocytosis can be bypassed by artificially locking RalA in a GTP-bound state. A key conclusion of these findings is that while GEFs directly activate RalA through nucleotide exchange, a dedicated GSF is required to maintain the active state of RalA till the arrival of RalA effectors. Ralbp1 interacts with switches 1 and 2 of GTP-bound Rals, the same region recognized by RalGAPs (12, 45, 60). Thus, Ralbp1 may stabilize the GTP-bound state of RalA by preventing RalGAPs from stimulating GTP hydrolysis. Since other small GTPases face the same challenges in their activation cycles, they may also require a similar GSF-dependent regulatory mechanism.
Reps1, Ralbp1, and RalA are all implicated in a range of physiological and pathological processes (10, 61, 62). We anticipate that these processes are mediated primarily by the role of the proteins in exocytosis. The Reps1-Ralbp1-RalA module regulates distinct cargo proteins across cell types, but its molecular mechanism of action is the same. We note that surface levels of many proteins were up-regulated in Reps1 KO cells. The reason for their up-regulation is unclear. We posit that mutations of Reps1 inhibit exocytic delivery of factors involved in retrieval of certain surface proteins, leading to increases in their surface levels. Besides their primary function in exocytosis, the components of the Reps1-Ralbp1-RalA module may play additional roles in cell physiology. Reps1 and Ralbp1 were speculated to regulate CME-dependent receptor down-regulation (40, 62, 63). Previous studies of Reps1 and Ralbp1 in endocytosis, however, were based on overexpression of dominant-negative mutants in WT cells, precluding direct assessment of their roles in CME (63, 64). Our data show that Reps1 and Ralbp1 do not represent core mediators of CME despite the binding of Reps1 to the AP2 adaptor. It is possible that the Reps1-AP2 binding mode does not play a significant role in the cell types we examined. Alternatively, the Reps1-AP2 interaction may regulate the internalization of a restricted group of cargoes such that its function becomes evident only under certain conditions. Moreover, RalA and Ralbp1 have been implicated in mitochondrial dynamics (65). It would be interesting to determine whether Reps1 is also involved and whether this mitochondrial function is connected to the role of the Reps1-Ralbp1-RalA module in exocytosis. Last, it remains possible that in another biological pathway, Ralbp1 acts as a RalA effector rather than a GSF. Further studies are needed to address these questions.
NBIA is a heterogenous group of rare monogenetic neurodegenerative diseases exhibiting focal accumulation of intracellular iron (25). Patients bearing compound heterozygous REPS1 point mutations show progressive cerebral and cerebellar atrophy (24, 25). Mutations in RALBP1 and RALA are also linked to neurodegeneration (66, 67). A complete loss of Reps1 abolishes neuronal morphogenesis and is expected to cause early lethality during mammalian development. Thus, NBIA-causing REPS1 mutations are likely hypomorphic in nature with mutant proteins retaining partial functionality, selectively affecting neurite outgrowth at specific brain regions most sensitive to reduction of its function. Most of NBIA-causing genes including REPS1 have no obvious connection to iron metabolism or transport. TfR trafficking is slightly altered in fibroblasts bearing NBIA-causing mutations (24). However, KO of Reps1 or Ralbp1 in CAD cells had little effect on the surface and total levels of TfR or Tf endocytosis (fig. S7), raising the possibility that REPS1-linked NBIA pathology involves other cargo molecule(s). Iron accumulation is also observed in other neurodegenerative disorders including Alzheimer’s disease and Parkinson’s disease (68, 69), although a causal link between iron overload and neurodegeneration has not been established. With the discovery of the Reps1-Ralbp1-RalA module in exocytosis and neuronal morphogenesis, an important future direction is to determine how disease-causing REPS1 mutations affect exocytosis kinetics and neuronal functions using animal models, as well as neurons and organoids derived from human pluripotent stem cells.
MATERIALS AND METHODS
Culture of cell lines
Mouse preadipocytes, 293T cells and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% Fetal Bovine Essence (FBE; VWR, #10803-034) and penicillin/streptomycin (Thermo Fisher Scientific, #15140122). CAD cells were cultured in DMEM/nutrient mixture F-12 (DMEM/F12) (Thermo Fisher Scientific, #11320033) supplemented with 8% fetal bovine serum (FBS; Hyclone, # SH30088.03HI). All cell lines were maintained in a humidified incubator at 37°C with 5% CO2.
To differentiate preadipocytes into mature adipocytes, preadipocytes were grown to ~95% confluence before a differentiation cocktail was added at the following final concentrations: insulin (5 μg/ml; Sigma-Aldrich, #I0516), 1 nM triiodo-l-thyronine (T3; Sigma-Aldrich, #T2877), 125 μM indomethacin (Sigma-Aldrich, #I-7378), 5 μM dexamethasone (Sigma-Aldrich, #D1756), and 0.5 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich, #I5879). After 2 days, the cells were switched to DMEM supplemented with 10% FBE, insulin (5 μg/ml), and 1 nM T3. After another 2 days, fresh DMEM media supplemented with 10% FBE and 1 nM T3 were added to the cells. Differentiated adipocytes were usually analyzed 6 days after addition of the differentiation cocktail.
Genome-wide CRISPR mutagenesis of preadipocyte fibroblasts
The surface protein reporter GLUT-SPR was stably expressed in preadipocytes (19, 21). The cells were mutagenized using a mouse CRISPR knockout pooled library (GeCKO v2) (Addgene, #1000000052) (22). Lentiviruses were produced by transfecting GeCKO v2 library plasmids into 293T cells along with pAdVAntage (Promega, #E1711), pCMV-VSVG (Addgene, #8454), and psPax2 (Addgene, #12260) as previously described (19, 21). Starting at 48 hours after transfection, cell culture supernatants containing lentiviruses were collected every 24 hours, and a total of four collections were made. Lentiviral particles were pelleted in a Beckman SW28 rotor at 25,000 rpm for 1.5 hours. The lentiviral pellets were resuspended in phosphate-buffered saline (PBS), snap-frozen, and stored at −70°C. Preadipocytes expressing GLUT-SPR were seeded into 12-well plates at 0.5 million cells per well and were infected by CRISPR lentiviruses to achieve a multiplicity of infection of ~0.4. Transduced cells were seeded onto 10-cm dishes and treated with puromycin for 48 hours. The resulted CRISPR mutant cell library was harvested and frozen 6 days after lentiviral infection.
Genome-wide CRISPR genetic screen
Forty million mutagenized mouse preadipocytes were seeded at a density of 1 million cells per 10-cm dish. On the following day, the cells were incubated in Krebs-Ringer bicarbonate HEPES (KRH) buffer [121 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4, 0.33 mM CaCl2, and 12 mM Hepes (pH 7.4)] for 2 hours. Subsequently, the cells were switched to KRH buffer containing 100 nM insulin to mimic the physiological fed state. After 30 min, the dishes were chilled on an ice bath, incubated with KRH buffer containing 5% FBE, and labeled with anti-HA antibodies (BioLegend, #901501, RRID: AB_2565006) and allophycocyanin (APC)–conjugated secondary antibodies (eBioscience, #17-4015-82). After dissociation from the plates using Accutase, the cells were sorted on a MoFlo XDP Flow Cytometer (Beckman Counter, Inc.) to collect 3% GFP+ cells with the lowest APC fluorescence. The collected cells were expanded and sorted for another two rounds using the same fluorescence gating set for the first round of sorting. In the passage control population, the mutant CRISPR cell library was cultured continuously for 3 weeks without sorting.
Genomic DNA was isolated from the final sorted population using a GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific, #K0721). The unsorted control population contained ~40 million cells, whereas the sorted population contained about 5 million cells. The isolated genomic DNA was used as templates to amplify guide sequences using NEBNext Ultra II Q5 Master Mix (New England BioLabs Inc., #M0544). In the first round of polymerase chain reaction (PCR), each reaction was performed in a total volume of 100 μl containing 3 μg of genomic DNA in the presence of sequencing adaptors (forward and reverse). PCR products were purified using SPRIselect beads (Beckman Coulter Inc., #B23318) and used as templates for the second round of PCR reactions. Oligo sequences are shown in the reagent/resource table, in which stagger sequences are shown to the 5′ of the barcode in upper case, whereas the priming sites are shown to the 3′ of the barcode in lower case. PCR products were pooled and purified using SPRIselect beads and sequenced on a NovaSeq 6000 sequencing system (Illumina Inc., San Diego, CA, USA). Sequencing reads were demultiplexed and processed to contain only the 20-bp unique guide sequences using the FAXTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Bioinformatic analysis was performed using the MAGeCK algorithm to identify significant hits (https://sourceforge.net/projects/mageck/) (23).
Gene KO using CRISPR-Cas9
The guide RNAs (gRNAs) targeting a gene were subcloned into the pLenti-CRISPR-V2 vector (Addgene, #52961) or a modified version of the pLentiGuide-Puro vector (Addgene, #52963) in which the puromycin selection marker was replaced with a hygromycin selection marker (19). CRISPR plasmids were transfected into 293T cells along with pAdVAntage, pCMV-VSVG, and psPax2. The 293T cell culture media containing lentiviral particles were harvested daily for 4 days and centrifuged at 25,000 rpm for 1.5 hours using a Beckman SW28 rotor. Viral pellets were resuspended in PBS and used to infect target cells. After lentiviral infection, cells were selected using puromycin (3.5 μg/ml; Sigma-Aldrich, #3101118) for 2 days, followed by selection using hygromycin B (500 μg/ml; Thermo Fisher Scientific, #10687010) for another 2 days. All KO cells used in this work were pooled populations. Guide sequences are listed in the reagent/resource table.
Gene expression in mammalian cells
Human REPS1 and RALBP1 genes were subcloned into the SHC003BSD-GFPD vector (Addgene, #133301) with a 3xFLAG-encoding sequence at the 3′ ends of the genes. The protospacer adjacent motif of human REPS1 was mutated to prevent targeting by gRNAs used for gene KO. The human RALA gene was subcloned into the SHC003BSD-GFPD vector with a 3xFLAG-encoding sequence at the 5′ end. The constitutively active RALA Q72L mutant was generated using a site-directed mutagenesis kit (Agilent, #210518). The constructs were transfected into 293T cells to produce lentiviral particles using a similar procedure as CRISPR lentiviral production. The lentiviruses were used to infect target cells, followed by selection using blasticidin (10 μg/ml; Thermo Fisher Scientific, #BP2647).
Flow cytometry analysis
Cells were washed with the KRH buffer and blocked with 5% FBE in the KRH buffer at 4°C. Next, the cells were stained using primary antibodies and APC-conjugated secondary antibodies. The cells were disassociated using Accutase and analyzed on a CyAN ADP analyzer (Beckman Coulter) or MACSQuant Analyzer (Miltenyi Biotec). Data were analyzed using the FlowJo software (FlowJo, LLC) based on experiments run in biological triplicates. To measure surface levels of GLUT-SPR, cells were washed three times with KRH buffer, followed by starvation in KRH buffer for 2 hours. The cells were cultured in media containing insulin for various durations before being chilled on ice. Surface GLUT-SPR molecules were stained using anti-HA antibodies and APC-conjugated secondary antibodies. The cells were dislodged using Accutase, and GFP and APC fluorescence was measured. To calculate normalized surface levels of GLUT-SPR, mean APC fluorescence was divided by mean GFP fluorescence. For surface TfR measurements, cells were blocked with 5% FBE, labeled with APC-conjugated anti-CD71 antibodies (BioLegend, #334108, RRID: AB_10915138), and dislodged using Accutase. To measure total TfR levels, cells were dislodged using Accutase, fixed using 2% paraformaldehyde (PFA), permeabilized using 0.2% saponin (Sigma-Aldrich, #84510), and stained with APC-conjugated anti-CD71 antibodies.
Endocytosis assays
Anti–HA-ATTO-565 and Tf-ATTO-565 were generated by labeling anti-HA antibodies (Sigma-Aldrich, #901501) and human Tf (Sigma-Aldrich, #616397) with ATTO-565-AEDP-NHS-ester (Atto-tec, Siegen, Germany). For anti-HA antibody conjugation, ATTO-565-AEDP-NHS-ester was added to anti-HA antibodies such that the molar ratio of dye to antibody was approximately 10:1. The mixture was incubated at room temperature (RT) for 1 hour. For Tf conjugation, ATTO-565-AEDP-NHS-ester was added to Tf (5 mg/ml) such that the molar ratio of dye to Tf was approximately 10:1. The mixture was incubated at RT for 1 hour, followed by incubation for 18 hours at 4°C. After centrifugation to remove precipitates, the samples were transferred to dialysis tubing and dialyzed for 24 hours at 4°C against 1 liter of PBS to remove free dyes. Protein concentrations were measured using Coomassie protein assay reagents (Thermo Fisher Scientific, #1856209). After the addition of the preservative Kathon CG/ICP (Supelco Analytical, #5-00119), the samples were stored at 4°C protected from light.
On the day of endocytosis assays, cells were washed three times with KRH buffer and starved in KRH buffer for 30 min before being chilled on ice. Subsequently, the cells were incubated with prewarmed HA-ATTO-565 (1 μg/ml) in the presence of 20 nM insulin or Tf-ATTO-565 (5 μg/ml) in a 37°C incubator for desired durations. The cells were chilled on ice and washed three times with 150 mM ice-cold MESNa (Sigma-Aldrich, #M1511) in PBS and once with ice-cold PBS. Subsequently, the cells were disassociated using Accutase, and ATTO-565 fluorescence was measured using the Phycoerythrin (PE) channel (filter of 586/15 nm) on a MACSQuant Analyzer. About 10,000 cells of each sample were analyzed. To detect endocytosis by confocal imaging, cells were grown on micro cover glasses with a diameter of 12 mm (VWR, #89015-725) in 24-well plates. Endocytosis assays were carried out as described above. After washing with MESNa three times and once with PBS, the cells were fixed with 4% PFA. After washing three times with PBS, the cells were incubated with CF405M-conjugated concanavalin A (50 μg/ml; Biotium, #29074) for 20 min at RT to stain the cell membrane. The 405-nm laser was used to detect CF405M-conjugated concanavalin A (cell membrane staining), and the 561-nm laser was used to detect ATTO-565 fluorescence using a 100× oil immerse objective on a Nikon A1 laser-scanning confocal microscope. Tf-ATTO-565–positive endocytic vesicles were analyzed using Fiji.
Immunostaining and imaging
Cells grown on coverslips were fixed using 4% PFA and permeabilized in PBS containing 5% FBE and 0.2% saponin. Stably expressed 3xFLAG-tagged Reps1 was labeled using anti-FLAG M2 antibodies (Sigma-Aldrich, #F1804, RRID: AB_262044) and Alexa Fluor 647–conjugated anti-mouse secondary antibodies (Invitrogen, #A32733, RRID: AB_2633282). Endogenous Ralbp1 was labeled using anti-Ralbp1 primary antibodies (Cell Signaling Technology, #3630, RRID: AB_2176169) and Alexa Fluor 488–conjugated secondary antibodies (Thermo Fisher Scientific, #A11008, RRID: AB_142165). To visualize surface GLUT-SPR, preadipocytes cultured in four-chamber glass bottom dishes were differentiated into mature adipocytes, fixed with 4% paraformaldehyde, and blocked with 2% bovine serum albumin (BSA) without permeabilization. Surface reporters were stained with anti-HA antibodies and Alexa Fluor 568–conjugated secondary antibodies (Thermo Fisher Scientific, #A11004, RRID: AB_2534072). Nuclei were stained with Hoechst 33342 (Sigma-Aldrich, #D9642). To visualize F-actin, adipocytes were fixed, permeabilized, and stained with CF647-conjugated phalloidin (5 U/ml; Biotium, #00041) for 20 min at room temperature. Confocal images were captured using a 100× oil immersion objective on a Nikon A1 laser scanning confocal microscope. TIRFM was carried out using a 100× oil immerse objective on a Nikon N-STORM TIRF microscope. In SIM, cells were grown on glass slides, labeled as in confocal microscopy, and imaged using a 100× oil immersion objective on a Nikon SIM microscope.
Electron microscopy
Cells were seeded onto 3-mm sapphire discs in 24-well plates. The sapphire discs were carefully picked from the plates and frozen using a Wohlwend Compact 02 high-pressure freezer (Technotrade International, Manchester, NH). Frozen specimens were freeze-substituted in anhydrous acetone containing 0.25% glutaraldehyde and 0.1% uranyl acetate and embedded in HM20 resin. Serial thin sections (80 nm) were cut using a Leica UCT ultramicrotome and collected onto formvar-coated nickel slot grids. For immunogold labeling of GLUT-SPR, the cell sections were sequentially incubated with rabbit anti-GFP antibodies (made in house) and gold-conjugated anti-rabbit secondary antibodies (TED PEKKA Inc., #15727). Electron micrographs were captured on a Tecnai T12 Spirit transmission electron microscopy operating at 100 kV with an AMT charge-coupled device system (Advanced Microscopy Techniques Corp., Danvers, MA).
Mass spectrometry
For whole-cell proteomics, WT and mutant cells were harvested in lysis buffer [100 mM triethyl ammonium bicarbonate (Millipore-Sigma, #18597), 5 mM tris (2-carboxyethyl) phosphine, 20 mM chloroacetamide (Millipore-Sigma, #C0267), and 1% SDS]. The samples were labeled using the TMTsixplex Isobaric Label Reagent Set (Thermo Fisher Scientific, #90061). For surface proteomics, WT and mutant preadipocytes were grown in regular cell culture media and differentiated into mature adipocytes. On the day of harvest, the cells were serum-starved for 2 hours followed by incubation in cell culture media containing 20 nM insulin. After 20 min, surface proteins were biotinylated using Sulfo-NHS-Biotin (Thermo Fisher Scientific, #A39256). The reactions were quenched using 100 mM glycine in PBS before the cells were lysed in a lysis buffer [20 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, and a protease inhibitor cocktail]. Equal amounts of cell extracts were mixed, and biotinylated proteins were isolated using NeutrAvidin agarose resins (Thermo Fisher scientific, #29201). Proteins were eluted from resins using an elution buffer containing 5% SDS, 50 mM tris (pH 8.5), 10 mM tris-(2-carboxyethyl) phosphine, 40 mM 2-chloroacetamide, 2 mM biotin, and 1 mM dithiothreitol (DTT).
Samples were prepared for mass spectrometry using the protein aggregation capture method (70). Peptides were prefractionated using high-pH fractionation and concatenated into 10 fractions before they were resolved on the Thermo Ultimate 3000 RSLCnano System in a direct injection mode (70). Raw data files from mass spectrometry were processed using MaxQuant/Andromeda (version 1.6.2.10) (71) and searched against the Uniprot database of protein sequences. False discovery rates were set to 0.01 for both protein and peptide identifications with a minimum peptide length of four residues and a minimum peptide number of two.
Immunoblotting and IP
Cells grown in 24-well plates were lysed in a SDS protein sample buffer. Cell lysates were resolved on 8% bis-tris SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and probed using primary antibodies and horseradish peroxidase (HRP)–conjugated secondary antibodies. Primary antibodies used in immunoblotting included anti–phospho-Akt (Ser473) antibodies (Cell Signaling Technology, #9271, RRID: AB_2716452), anti-Akt antibodies (Cell Signaling Technology, #9272, RRID: AB_329827), anti-Reps1 (Cell Signaling Technology, #6404, RRID: AB_11220228), anti-Ralbp1, anti–α-adaptin (BD Biosciences, #610502, RRID: AB_397868), anti-RalA (Cell Signaling Technology, #4799, RRID: AB_10612771), anti-peroxisome proliferator–activated receptor γ (Santa Cruz Biotechnology, #sc-7273, RRID: AB_628115), anti–α-tubulin (DSHB, clone: #12G10, RRID: AB_1210456), anti-AP2β (Thermo Fisher Scientific, #A304-719A, RRID: AB_2620914), anti-Rab10 antibodies (Cell Signaling Technology, #8127, RRID: AB_10828219), mouse monoclonal HRP-conjugated anti-FLAG M2 antibodies (Sigma-Aldrich, #A8592, RRID: AB_439702), and HRP-conjugated anti-HA antibodies (Roche, #12013819001, RRID: AB_390917). Secondary antibodies used in immunoblotting included HRP-conjugated anti-rabbit immunoglobulin G (IgG; Sigma-Aldrich, #A6154, RRID: AB_258284) and HRP-conjugated anti-mouse IgG (Sigma-Aldrich, #A6782, RRID: AB_258315).
In IP experiments, cells were lysed in IP buffer [25 mM Hepes (pH 7.4), 138 mM NaCl, 10 mM Na3PO4, 2.7 mM KCl, 0.5% CHAPS, 1 mM DTT, and a protease inhibitor cocktail]. After centrifugation, proteins were immunoprecipitated from cell extracts using anti-FLAG M2 antibodies and protein A/G agarose beads (Thermo Fisher Scientific, #WF324079). Immunoprecipitates were resolved on 8% bis-tris SDS-PAGE, and proteins were detected using primary antibodies and HRP-conjugated secondary antibodies. All experiments were run in biological triplicates.
Quantitative reverse transcription PCR
The RNeasy Mini Kit (Qiagen, #74104) was used to purify total RNAs. After treatment with ezDNase (Thermo Fisher Scientific, #8091150), first-strand complementary DNA was synthesized using the SuperScript IV Kit (Thermo Fisher Scientific, #18091050). Gene expression levels were measured by quantitative reverse transcription PCR on the Applied Biosystems 7500 Fast Real-Time PCR System using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, #172-5272) with gene-specific primer sets. The cycle threshold values of a gene were normalized to those of GAPDH, a reference gene, and the Δcycle threshold values were calculated. The results were plotted as fold changes relative to the WT sample. The PCR primers for mouse Reps1 were as follows: 5′-CTCCCACGATTTGTTGCTTC-3′ (forward) and 5′-GGATCACACCCGAGTAAGAG-3′ (reverse). Primers for human REPS1 were as follows: 5’′-CCAGTAGTTTCACCACAGCA-3′ (forward) and 5′-AGGCCTCTCACTATTCCCA-3′ (reverse). Primers for human GAPDH were as follows: 5′-GACAGTCAGCCGCATCTTCT-3′ (forward) and 5′-GCGCCCAATACGACCAAATC-3′ (reverse). Primers for mouse Gapdh were as follows: 5′-AGGTCGGTGTGAACGGATTTG-3′ (forward) and 5′-TGTAGACCATGTAGTTGAGGTCA-3′ (reverse).
Recombinant protein expression and GST pull-down assays
REPS1 and RALBP1 were subcloned into the pGEX-4T-3 vector (GE Healthcare). The empty pGEX-4T-3 vector was used to express the GST control protein. Recombinant proteins were expressed in BL21 (DE3) Escherichia coli (Stratagene, #230132) as previously described (19, 72). When optical density at 600 nm of E. coli cultured in 2xYT media reached ~0.6, 1 mM isopropyl β-d-1-thiogalactopyranoside was added to induce protein expression. After 2 hours, bacterial cells were harvested, lysed, and centrifuged to remove debris. Recombinant proteins were purified from the supernatant using glutathione beads (Thermo Fisher Scientific, #PI16101). GST-tagged proteins on glutathione beads were added to mammalian cell extracts prepared using a similar way as IP. After incubation and centrifugation, proteins bound to the beads were resolved on 8% bis-tris SDS-PAGE and detected using primary antibodies and HRP-conjugated secondary antibodies. All experiments were run in biological triplicates.
Lipid droplet staining and analysis
Preadipocytes were grown in clear-bottom 96-well plates. After differentiation, adipocytes were fixed using 4% PFA, washed three times with PBS, and stained with 0.3% Oil Red O (Sigma-Aldrich, #O0625). To quantify neutral lipids within lipid droplets, adipocytes were fixed using 4% PFA and stained with Oil Red O. Images of stained lipid droplets were captured on a bright-field microscope. In parallel, the stained cells were air-dried at RT for 2 hours. Methanol was added to solubilize the cells, and Oil Red O absorbance in the supernatant was measured at the wavelength of 520 nm on a plate reader. The absorbance of KO cells was normalized to that of WT cells. The experiments were performed in biological triplicates.
Differentiation and analysis of CAD neurons
CAD neuronal progenitors were seeded on 12-mm coverslips coated with poly-d-lysine in a 24-well plate at a density of 15,000 cells per well. The next day, the cells were switched to serum-free DMEM/F12 media to initiate differentiation (56). After 5 days, CAD neurons were washed with PBS, fixed using 4% PFA, permeabilized using 0.2% saponin, and blocked using 2% BSA. Map2 of CAD neurons was labeled with mouse anti-Map2 (Santa Cruz Biotechnology Inc., #sc-74421, RRID: AB_112621) and Alexa Fluor 488–conjugated secondary antibodies. SNAP-25 was labeled using rabbit anti–SNAP-25 antibodies (Sigma-Aldrich, #S9684, RRID: AB_261576) and Alexa Fluor 488–conjugated secondary antibodies. Cell nuclei were stained with Hoechst 33342 (10 μg/ml). The images of CAD neurons were captured using a 20× objective on a Nikon A1 confocal fluorescent microscope. Neurite number and length per neuron were calculated using a NeuronJ Plugin in the Fiji software.
Primary neuronal culture and analysis
All animal procedures were conducted in accordance with the Institutional Animal Care and Use Committee at University of Colorado. Primary hippocampal neurons were prepared from neonatal Sprague-Dawley rats following procedures reported previously (73, 74). Hippocampi were dissected from the brains of mixed sex neonatal rat pups (postnatal days 0 to 1) and dissociated by papain digestion. Neurons were plated at 125,000 cells per well in glutamine-free minimum essential medium (Thermo Fisher Scientific, #11090081) supplemented with 10% FBS and penicillin/streptomycin on poly-d-lysine–coated 12-well plates. For imaging, 18-mm coverslips were placed into the plates before coating. Neurons were maintained at 37°C in a humidified incubator with 5% CO2. On day in vitro (DIV) 1, the cell culture media were replaced with Neurobasal-A media (Life Technologies, #10888022) supplemented with B27 (Life Technologies, #17504044) and GlutaMAX (Life Technologies, #35050061). CRISPR lentiviruses were added to the neurons on DIV 1. To limit the growth of actively dividing cells, mitotic inhibitors (uridine and 5-fluoro-2′deoxyuridine) were added by replacing half of the media on DIV 7. Neurons were transfected with a GFP-encoding plasmid using Lipofectamine 2000 (Thermo Fisher Scientific, #11668027) on DIV 13. On DIV 15, cells were harvested into a SDS protein sample buffer for immunoblotting or fixed with 4% PFA for imaging.
Oligonucleotide sequences
The oligonucleotide sequences are as follows: SequencingAdaptor_F, AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG; SequencingAdaptor_R, TCTACTATTCTTTCCCCTGCACTGTtgtgggcgatgtgcgctctg; SequencingF01 ,AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTAAGTAGAGtcttgtggaaaggacgaaacaccg; SequencingF02, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATACACGATCtcttgtggaaaggacgaaacaccg; SequencingF03, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGATCGCGCGGTtcttgtggaaaggacgaaacaccg; SequencingF04, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCGATCATGATCGtcttgtggaaaggacgaaacaccg; SequencingF05, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTCGATCGTTACCAtcttgtggaaaggacgaaacaccg; SequencingF06, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATCGATTCCTTGGTtcttgtggaaaggacgaaacaccg; SequencingR01, CAAGCAGAAGACGGCATACGAGATAAGTAGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTttctactattctttcccctgcactgt; mouse Reps1 sgRNA1, GAAGCAACAAATCGTGGGAG; mouse Reps1 sgRNA2, GAAGTTGGTGGGGACTGCTG; human REPS1 sgRNA1, ACAGGTAACGTCGCGTCCCG; human REPS1 sgRNA2, AGAGTGGAAAGTATAAATAC; rat Reps1 sgRNA1, CTTTGCGGTGCAACAAGACT; rat Reps1 sgRNA2, TCACACCCGCGTAAGCGCTC; mouse Ralbp1 sgRNA1, GGCTCATTGTGCACCTGGAC; and mouse Ralbp1 sgRNA1, GCGGAGATCCAGAGGTACAG.
Acknowledgments
We thank Y. Ye, M. Stowell, H. Yu, Y. Tian, S. Kajimura, D. James, A.-C. Tan, and T. Wang for advice or reagents. We thank Y. Ouyang, T. L. Antwine, J. Orth, J. Dragavon, C. C. Ebmeier, E. T. O’Toole, G. P. Morgan, C. R. Ozzello, and K. Diener for technical assistance.
Funding: This work was supported by the National Institutes of Health (NIH) grants GM126960 (to J.S.), DK124431 (to J.S.), GM138685 (to Q.Y.), and NS116879 (to M.J.K.); a fellowship from Beijing University of Chinese Medicine 2232022JYBXJSJJ013 to (S.W.); and an NIH institutional predoctoral training grant GM088759 (to L.C.). Publication of this article was partially funded by the University of Colorado Boulder Libraries Open Access Fund.
Author contributions: S.W. and J.S. conceived the project. S.W., X.C., L.C., X.D., C.Wi., C.Wa., and H.P. performed the experiments. S.W., X.C., L.C., M.J.K., Q.Y., and J.S. analyzed the data. S.W. and J.S. wrote the manuscript with inputs from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S5
Other Supplementary Material for this : manuscript includes the following:
Tables S1 to S5
REFERENCES AND NOTES
- 1.J. S. Bonifacino, B. S. Glick, The mechanisms of vesicle budding and fusion. Cell 116, 153–166 (2004). [DOI] [PubMed] [Google Scholar]
- 2.D. Bausch-Fluck, U. Goldmann, S. Müller, M. van Oostrum, M. Müller, O. T. Schubert, B. Wollscheid, The in silico human surfaceome. Proc. Natl. Acad. Sci. U.S.A. 115, E10988–E10997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.N. J. Bryant, R. Govers, D. E. James, Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell Biol. 3, 267–277 (2002). [DOI] [PubMed] [Google Scholar]
- 4.S. Fukai, H. T. Matern, J. R. Jagath, R. H. Scheller, A. T. Brunger, Structural basis of the interaction between RalA and Sec5, a subunit of the sec6/8 complex. EMBO J. 22, 3267–3278 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.K. Briant, L. Redlingshofer, F. M. Brodsky, Clathrin’s life beyond 40: Connecting biochemistry with physiology and disease. Curr. Opin. Cell Biol. 65, 141–149 (2020). [DOI] [PubMed] [Google Scholar]
- 6.M. Mettlen, P. H. Chen, S. Srinivasan, G. Danuser, S. L. Schmid, Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 87, 871–896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.B. Wu, W. Guo, The exocyst at a glance. J. Cell Sci. 128, 2957–2964 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D. M. Lepore, L. Martinez-Nunez, M. Munson, Exposing the elusive exocyst structure. Trends Biochem. Sci. 43, 714–725 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R. Jin, J. R. Junutula, H. T. Matern, K. E. Ervin, R. H. Scheller, A. T. Brunger, Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J. 24, 2064–2074 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. Yan, D. Theodorescu, RAL GTPases: Biology and potential as therapeutic targets in cancer. Pharmacol. Rev. 70, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R. Shirakawa, H. Horiuchi, Ral GTPases: Crucial mediators of exocytosis and tumourigenesis. J. Biochem. 157, 285–299 (2015). [DOI] [PubMed] [Google Scholar]
- 12.J. Cherfils, M. Zeghouf, Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 93, 269–309 (2013). [DOI] [PubMed] [Google Scholar]
- 13.A. Fotin, Y. Cheng, P. Sliz, N. Grigorieff, S. C. Harrison, T. Kirchhausen, T. Walz, Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature 432, 573–579 (2004). [DOI] [PubMed] [Google Scholar]
- 14.M. Kaksonen, C. P. Toret, D. G. Drubin, Harnessing actin dynamics for clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 7, 404–414 (2006). [DOI] [PubMed] [Google Scholar]
- 15.B. M. Pearse, M. S. Robinson, Purification and properties of 100-kd proteins from coated vesicles and their reconstitution with clathrin. EMBO J. 3, 1951–1957 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. D. Conner, S. L. Schmid, Differential requirements for AP-2 in clathrin-mediated endocytosis. J. Cell Biol. 162, 773–780 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.B. M. Collins, A. J. McCoy, H. M. Kent, P. R. Evans, D. J. Owen, Molecular architecture and functional model of the endocytic AP2 complex. Cell 109, 523–535 (2002). [DOI] [PubMed] [Google Scholar]
- 18.M. Kaksonen, A. Roux, Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 19, 313–326 (2018). [DOI] [PubMed] [Google Scholar]
- 19.D. R. Gulbranson, E. M. Davis, B. A. Demmitt, Y. Ouyang, Y. Ye, H. Yu, J. Shen, RABIF/MSS4 is a Rab-stabilizing holdase chaperone required for GLUT4 exocytosis. Proc. Natl. Acad. Sci. U.S.A. 114, E8224–E8233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Y. Skorobogatko, M. Dragan, C. Cordon, S. M. Reilly, C. W. Hung, W. Xia, P. Zhao, M. Wallace, D. E. Lackey, X. W. Chen, O. Osborn, J. G. Bogner-Strauss, D. Theodorescu, C. M. Metallo, J. M. Olefsky, A. R. Saltiel, RalA controls glucose homeostasis by regulating glucose uptake in brown fat. Proc. Natl. Acad. Sci. U.S.A. 115, 7819–7824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D. R. Gulbranson, L. Crisman, M. S. Lee, Y. Ouyang, B. L. Menasche, B. A. Demmitt, C. Wan, T. Nomura, Y. Ye, H. Yu, J. Shen, AAGAB controls AP2 adaptor assembly in clathrin-mediated endocytosis. Dev. Cell 50, 436–446.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.N. E. Sanjana, O. Shalem, F. Zhang, Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.W. Li, H. Xu, T. Xiao, L. Cong, M. I. Love, F. Zhang, R. A. Irizarry, J. S. Liu, M. Brown, X. S. Liu, MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A. Drecourt, J. Babdor, M. Dussiot, F. Petit, N. Goudin, M. Garfa-Traoré, F. Habarou, C. Bole-Feysot, P. Nitschké, C. Ottolenghi, M. D. Metodiev, V. Serre, I. Desguerre, N. Boddaert, O. Hermine, A. Munnich, A. Rötig, Impaired transferrin receptor palmitoylation and recycling in neurodegeneration with brain iron accumulation. Am. J. Hum. Genet. 102, 266–277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Levi, V. Tiranti, Neurodegeneration with brain iron accumulation disorders: Valuable models aimed at understanding the pathogenesis of iron deposition. Pharmaceuticals (Basel) 12, 27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. Levi, D. Finazzi, Neurodegeneration with brain iron accumulation: Update on pathogenic mechanisms. Front. Pharmacol. 5, 99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. P. Czech, Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 23, 804–814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C. N. Antonescu, T. E. McGraw, A. Klip, Reciprocal regulation of endocytosis and metabolism. Cold Spring Harb. Perspect. Biol. 6, a016964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. S. Bogan, K. V. Kandror, Biogenesis and regulation of insulin-responsive vesicles containing GLUT4. Curr. Opin. Cell Biol. 22, 506–512 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A. Aslamy, D. C. Thurmond, Exocytosis proteins as novel targets for diabetes prevention and/or remediation? Am. J. Physiol. Regul. Integr. Comp. Physiol. 312, R739–R752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D. Tobys, L. M. Kowalski, E. Cziudaj, S. Müller, P. Zentis, E. Pach, P. Zigrino, T. Blaeske, S. Höning, Inhibition of clathrin-mediated endocytosis by knockdown of AP-2 leads to alterations in the plasma membrane proteome. Traffic 22, 6–22 (2021). [DOI] [PubMed] [Google Scholar]
- 32.V. Bitsikas, I. R. Correa Jr., B. J. Nichols, Clathrin-independent pathways do not contribute significantly to endocytic flux. eLife 3, e03970 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.S. Wang, C. Wan, G. T. Squiers, J. Shen, Endocytosis assays using cleavable fluorescent dyes. Methods Mol. Biol. 2473, 181–194 (2022). [DOI] [PubMed] [Google Scholar]
- 34.S. R. Elkin, N. W. Oswald, D. K. Reed, M. Mettlen, J. B. MacMillan, S. L. Schmid, Ikarugamycin: A natural product inhibitor of clathrin-mediated endocytosis. Traffic 17, 1139–1149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. Yamaguchi, T. Urano, T. Goi, L. A. Feig, An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J. Biol. Chem. 272, 31230–31234 (1997). [DOI] [PubMed] [Google Scholar]
- 36.M. Ikeda, O. Ishida, T. Hinoi, S. Kishida, A. Kikuchi, Identification and characterization of a novel protein interacting with Ral-binding protein 1, a putative effector protein of Ral. J. Biol. Chem. 273, 814–821 (1998). [DOI] [PubMed] [Google Scholar]
- 37.S. B. Cantor, T. Urano, L. A. Feig, Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol. Cell. Biol. 15, 4578–4584 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.C. Wan, L. Crisman, B. Wang, Y. Tian, S. Wang, R. Yang, I. Datta, T. Nomura, S. Li, H. Yu, Q. Yin, J. Shen, AAGAB is an assembly chaperone regulating AP1 and AP2 clathrin adaptors. J. Cell Sci. 134, jcs258587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Motley, N. A. Bright, M. N. Seaman, M. S. Robinson, Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 162, 909–918 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. E. Salcini, S. Confalonieri, M. Doria, E. Santolini, E. Tassi, O. Minenkova, G. Cesareni, P. G. Pelicci, P. P. di Fiore, Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 11, 2239–2249 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.P. Chardin, A. Tavitian, The ral gene: A new ras related gene isolated by the use of a synthetic probe. EMBO J. 5, 2203–2208 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.L. Wang, G. Li, S. Sugita, RalA-exocyst interaction mediates GTP-dependent exocytosis. J. Biol. Chem. 279, 19875–19881 (2004). [DOI] [PubMed] [Google Scholar]
- 43.X. W. Chen, D. Leto, J. Xiao, J. Goss, Q. Wang, J. A. Shavit, T. Xiong, G. Yu, D. Ginsburg, D. Toomre, Z. Xu, A. R. Saltiel, Exocyst function regulated by effector phosphorylation. Nat. Cell Biol. 13, 580–588 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.M. Kawato, R. Shirakawa, H. Kondo, T. Higashi, T. Ikeda, K. Okawa, S. Fukai, O. Nureki, T. Kita, H. Horiuchi, Regulation of platelet dense granule secretion by the Ral GTPase-exocyst pathway. J. Biol. Chem. 283, 166–174 (2008). [DOI] [PubMed] [Google Scholar]
- 45.R. B. Fenwick, L. J. Campbell, K. Rajasekar, S. Prasannan, D. Nietlispach, J. Camonis, D. Owen, H. R. Mott, The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction. Structure 18, 985–995 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.V. Jullien-Flores, O. Dorseuil, F. Romero, F. Letourneur, S. Saragosti, R. Berger, A. Tavitian, G. Gacon, J. H. Camonis, Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J. Biol. Chem. 270, 22473–22477 (1995). [DOI] [PubMed] [Google Scholar]
- 47.T. Maehama, M. Tanaka, H. Nishina, M. Murakami, Y. Kanaho, K. Hanada, RalA functions as an indispensable signal mediator for the nutrient-sensing system. J. Biol. Chem. 283, 35053–35059 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S. H. Park, R. A. Weinberg, A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene 11, 2349–2355 (1995). [PubMed] [Google Scholar]
- 49.K. V. Rajasekar, L. J. Campbell, D. Nietlispach, D. Owen, H. R. Mott, The Structure of the RLIP76 RhoGAP-Ral binding domain dyad: Fixed position of the domains leads to dual engagement of small G proteins at the membrane. Structure 21, 2131–2142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.N. F. Neel, K. L. Rossman, T. D. Martin, T. K. Hayes, J. J. Yeh, C. J. Der, The RalB small GTPase mediates formation of invadopodia through a GTPase-activating protein-independent function of the RalBP1/RLIP76 effector. Mol. Cell. Biol. 32, 1374–1386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.M. J. Kennedy, C. Hanus, Architecture and dynamics of the neuronal secretory network. Annu. Rev. Cell Dev. Biol. 35, 543–566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.F. L. Urbina, S. L. Gupton, SNARE-mediated exocytosis in neuronal development. Front. Mol. Neurosci. 13, 133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.G. Lalli, RalA and the exocyst complex influence neuronal polarity through PAR-3 and aPKC. J. Cell Sci. 122, 1499–1506 (2009). [DOI] [PubMed] [Google Scholar]
- 54.G. Lalli, A. Hall, Ral GTPases regulate neurite branching through GAP-43 and the exocyst complex. J. Cell Biol. 171, 857–869 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R. O. Teodoro, G. Pekkurnaz, A. Nasser, M. E. Higashi-Kovtun, M. Balakireva, I. G. McLachlan, J. Camonis, T. L. Schwarz, Ral mediates activity-dependent growth of postsynaptic membranes via recruitment of the exocyst. EMBO J. 32, 2039–2055 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. Qi, J. K. Wang, M. McMillian, D. M. Chikaraishi, Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J. Neurosci. 17, 1217–1225 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.C. Shen, Y. Liu, H. Yu, D. R. Gulbranson, I. Kogut, G. Bilousova, C. Zhang, M. H. B. Stowell, J. Shen, The N-peptide-binding mode is critical to Munc18-1 function in synaptic exocytosis. J. Biol. Chem. 293, 18309–18317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.G. Grosse, J. Grosse, R. Tapp, J. Kuchinke, M. Gorsleben, I. Fetter, B. Höhne-Zell, M. Gratzl, M. Bergmann, SNAP-25 requirement for dendritic growth of hippocampal neurons. J. Neurosci. Res. 56, 539–546 (1999). [DOI] [PubMed] [Google Scholar]
- 59.D. Leto, A. R. Saltiel, Regulation of glucose transport by insulin: Traffic control of GLUT4. Nat. Rev. Mol. Cell Biol. 13, 383–396 (2012). [DOI] [PubMed] [Google Scholar]
- 60.O. Daumke, M. Weyand, P. P. Chakrabarti, I. R. Vetter, A. Wittinghofer, The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 429, 197–201 (2004). [DOI] [PubMed] [Google Scholar]
- 61.K. H. Lim, K. O’Hayer, S. J. Adam, S. D. S. Kendall, P. M. Campbell, C. J. Der, C. M. Counter, Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr. Biol. 16, 2385–2394 (2006). [DOI] [PubMed] [Google Scholar]
- 62.J. Cornish, D. Owen, H. R. Mott, RLIP76: A structural and functional triumvirate. Cancers (Basel) 13, 2206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.V. Jullien-Flores, Y. Mahe, G. Mirey, C. Leprince, B. Meunier-Bisceuil, A. Sorkin, J. H. Camonis, RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: Involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 113 (Pt 16), 2837–2844 (2000). [DOI] [PubMed] [Google Scholar]
- 64.S. Nakashima, K. Morinaka, S. Koyama, M. Ikeda, M. Kishida, K. Okawa, A. Iwamatsu, S. Kishida, A. Kikuchi, Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 18, 3629–3642 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.D. F. Kashatus, K. H. Lim, D. C. Brady, N. L. K. Pershing, A. D. Cox, C. M. Counter, RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 13, 1108–1115 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.A. Hindle, S. P. Singh, J. A. Pradeepkiran, C. Bose, M. Vijayan, S. Kshirsagar, N. A. Sawant, P. H. Reddy, Rlip76: An unexplored player in neurodegeneration and Alzheimer’s disease? Int. J. Mol. Sci. 23, 6098 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.M. Wagner, Y. Skorobogatko, B. Pode-Shakked, C. M. Powell, B. Alhaddad, A. Seibt, O. Barel, G. Heimer, C. Hoffmann, L. A. Demmer, Y. Perilla-Young, M. Remke, D. Wieczorek, T. Navaratnarajah, P. Lichtner, D. Klee, H. E. Shamseldin, F. al Mutairi, E. Mayatepek, T. Strom, T. Meitinger, F. S. Alkuraya, Y. Anikster, A. R. Saltiel, F. Distelmaier, Bi-allelic variants in RALGAPA1 cause profound neurodevelopmental disability, muscular hypotonia, infantile spasms, and feeding abnormalities. Am. J. Hum. Genet. 106, 246–255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.R. J. Ward, F. A. Zucca, J. H. Duyn, R. R. Crichton, L. Zecca, The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 13, 1045–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.L. Zecca, M. B. Youdim, P. Riederer, J. R. Connor, R. R. Crichton, Iron, brain ageing and neurodegenerative disorders. Nat. Rev. Neurosci. 5, 863–873 (2004). [DOI] [PubMed] [Google Scholar]
- 70.T. S. Batth, M. A. X. Tollenaere, P. Rüther, A. Gonzalez-Franquesa, B. S. Prabhakar, S. Bekker-Jensen, A. S. Deshmukh, J. V. Olsen, Protein aggregation capture on microparticles enables multipurpose proteomics sample preparation. Mol. Cell. Proteomics 18, 1027–1035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.J. Cox, N. Neuhauser, A. Michalski, R. A. Scheltema, J. V. Olsen, M. Mann, Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 72.H. Yu, L. Crisman, M. H. B. Stowell, J. Shen, Functional reconstitution of intracellular vesicle fusion using purified SNAREs and Sec1/Munc18 (SM) proteins. Methods Mol. Biol. 1860, 237–249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Q. Liu, B. L. Sinnen, E. E. Boxer, M. W. Schneider, M. J. Grybko, W. C. Buchta, E. S. Gibson, C. L. Wysoczynski, C. P. Ford, A. Gottschalk, J. Aoto, C. L. Tucker, M. J. Kennedy, A photoactivatable botulinum neurotoxin for inducible control of neurotransmission. Neuron 101, 863–875.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.J. D. Garcia, S. E. Gookin, K. C. Crosby, S. L. Schwartz, E. Tiemeier, M. J. Kennedy, M. L. Dell’Acqua, P. S. Herson, N. Quillinan, K. R. Smith, Stepwise disassembly of GABAergic synapses during pathogenic excitotoxicity. Cell Rep. 37, 110142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.X. W. Chen, D. Leto, S. H. Chiang, Q. Wang, A. R. Saltiel, Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev. Cell 13, 391–404 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S5
Tables S1 to S5