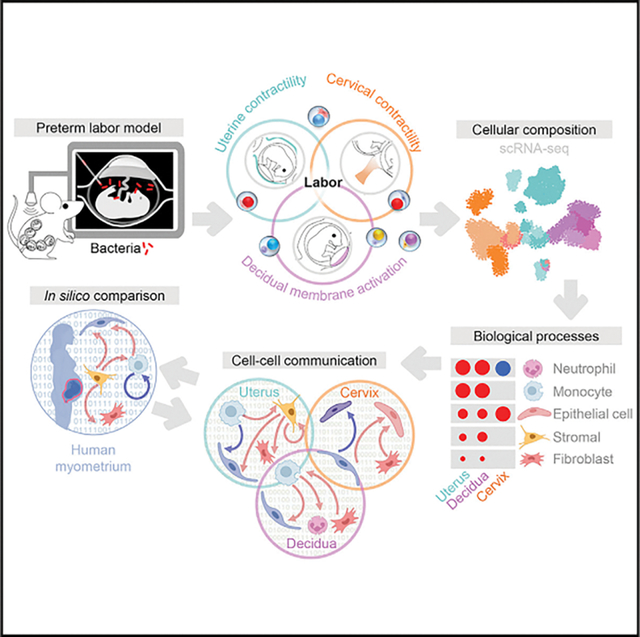
SUMMARY
Preterm birth, the leading cause of perinatal morbidity and mortality worldwide, frequently results from the syndrome of preterm labor. The best-established causal link to preterm labor is intra-amniotic infection, which involves premature activation of the parturition cascade in the reproductive tissues. Herein, we utilize single-cell RNA sequencing (scRNA-seq) to generate a single-cell atlas of the murine uterus, decidua, and cervix in a model of infection-induced preterm labor. We show that preterm labor affects the transcriptomic profiles of specific immune and non-immune cell subsets. Shared and tissue-specific gene expression signatures are identified among affected cells. Determination of intercellular communications implicates specific cell types in preterm labor-associated signaling pathways across tissues. In silico comparison of murine and human uterine cell-cell interactions reveals conserved signaling pathways implicated in labor. Thus, our scRNA-seq data provide insights into the preterm labor-driven cellular landscape and communications in reproductive tissues.
In brief
Garcia-Flores et al. generate a single-cell atlas of the murine uterus, decidua, and cervix using a model of intra-amniotic infection-induced preterm labor and demonstrate alterations in cell type composition, transcriptional profiles, and cell-cell signaling. This scRNA-seq dataset can serve as a valuable resource to be leveraged by future investigations.
Graphical Abstract
INTRODUCTION
Preterm birth, a devastating clinical condition that affects 15 million infants each year, is the leading cause of neonatal morbidity and mortality worldwide.1,2 Spontaneous preterm birth often results from preterm labor, a syndrome for which multiple etiologies have been proposed.3,4 Among them, the best-established causal link to preterm birth is intra-amniotic infection, a clinical condition resulting from invasion of microbes into the amniotic cavity.5–13 The most frequently detected bacteria in amniotic fluid of women diagnosed with intra-amniotic infection include genital mycoplasmas, Streptococcus agalactiae, Gardnerella vaginalis, and Escherichia coli, among others.14–25 Human descriptive studies have consistently shown that such microbial invasion of the amniotic cavity is accompanied by a local acute inflammatory response that includes infiltration of leukocytes into the amniotic cavity (including amniotic fluid26–39 and placental tissues40–57) as well as the reproductive tissues.58 More recently, animal models coupled with omics technologies have been utilized to strengthen this concept and establish causality between intra-amniotic infection and the inflammatory milieu observed in the reproductive tissues (e.g., uterus, decidua, and cervix) that serve to orchestrate premature activation of the common pathway of labor.59–62 However, a simultaneous investigation of the cellular landscape and interaction networks at single-cell resolution in the reproductive tissues implicated in preterm parturition has not been undertaken.
Single-cell technology has emerged as a useful tool for evaluating cellular composition, transcriptomic activity, and communication networks in gestational and reproductive tissues.63–70 We have applied single-cell RNA sequencing (scRNA-seq) to investigate the physiological and pathological processes of labor in the placenta and extraplacental membranes.68 More recently, we utilized scRNA-seq to unravel the myometrial cell types that participate in the normal process of term parturition as well as key cell-cell interactions taking place in this compartment.70 The discovery of single-cell signatures derived from the placental tissues and myometrium has translational value, as these can serve as potential non-invasive biomarkers of labor progression and/or obstetric disease.65,68,70–73
In the current study, we utilized scRNA-seq coupled with an allogeneic murine model of intra-amniotic infection to investigate the cellular landscape and cell-cell communications in the reproductive tissues (uterus, decidua, and cervix) during the process of preterm labor. We utilized a murine model of preterm labor and birth induced by the intra-amniotic inoculation of E. coli and assessed cervical shortening to establish the timing of active preterm labor. Next, using scRNA-seq and computational approaches, we generated a single-cell atlas of the uterus, decidua, and cervix during preterm labor as well as their cell type-specific transcriptomic activity. We also established the cell-cell communication networks between cell types in each tissue during preterm labor and identified key signaling pathways implicated in this process. Last, we integrated cell-cell signaling pathways derived from the murine uterus with those from the human myometrium during the processes of preterm and term labor, respectively, to demonstrate conserved labor-associated signaling.
RESULTS
A single-cell atlas of murine reproductive tissues during preterm labor induced by E. coli
Intra-amniotic infection has been documented as inducing inflammatory changes in the tissues surrounding the amniotic cavity.74–77 To investigate such changes at single-cell resolution, we established a murine model of preterm labor and birth induced by intra-amniotic inoculation with E. coli, one of the microorganisms commonly identified in the amniotic fluid of women with intra-amniotic infection.14,16,20,78 Mice with an allogeneic pregnancy underwent ultrasound-guided intra-amniotic injection of E. coli or vehicle control on 16.5 days post coitum (dpc) (Figure 1A). Intra-amniotic inoculation with E. coli reduced the gestational length in a majority of dams (Figure 1B), resulting in an 83.3% (5 of 6) rate of preterm birth (Figure 1C). We then intra-amniotically injected a second cohort of mice with E. coli or PBS to perform tissue collection for single-cell analyses. To ensure that the E. coli-injected mice were undergoing preterm labor at the time of tissue collection, we utilized ultrasound to evaluate cervical length just prior to intra-amniotic injection and again 24 h later as a readout of cervical effacement (Figure 1D). Cervical shortening was observed in all dams that received intra-amniotic E. coli 24 h after injection, indicating that these dams were in active preterm labor at the time of tissue collection, whereas no cervical shortening was observed in controls (Figure 1E). Therefore, intra-amniotic inoculation with E. coli represents a translational model that resembles the clinical scenario of intra-amniotic infection leading to preterm labor and birth.
Figure 1. Single-cell atlas of murine reproductive tissues during preterm labor induced by intra-amniotic infection.
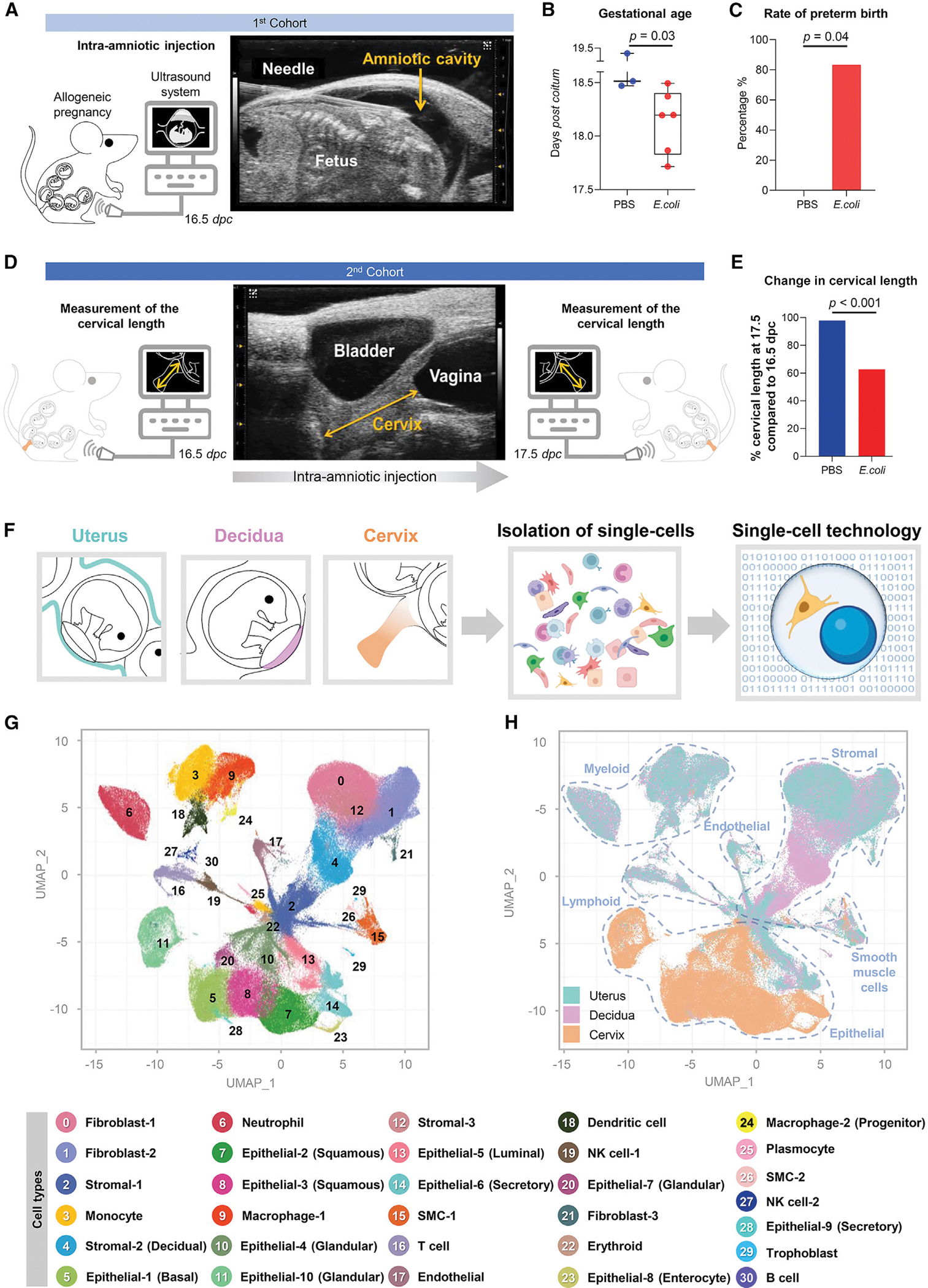
(A–C) Experimental design for the ultrasound-guided intra-amniotic injection of E. coli or PBS (vehicle control) into pregnant mice on 16.5 days post coitum (dpc) (n = 3–6 per group) (A). Mice were monitored to determine pregnancy outcomes (B and C). Shown are the gestational age and preterm birth rate of dams injected intra-amniotically with E. coli or PBS. Gestational age was compared using a 2-sided Mann-Whitney U test, and preterm birth rates were compared using a 2-sided Fisher’s exact test. p < 0.05 was considered significant.
(D) Experimental design for determination of cervical length on 16.5 dpc, prior to intra-amniotic injection of E. coli or PBS, and 24 h later (17.5 dpc) (n = 6–9 per group).
(E) Cervical length of dams injected intra-amniotically with E. coli or PBS on 16.5 and 17.5 dpc. Cervical length was compared between time points using a 2-sided Mann-Whitney U test. p < 0.05 was considered significant. The change in cervical shortening was calculated by considering the measurement at 16.5 dpc as 100%.
(F) Diagram illustrating generation of single-cell suspensions from the uterus, decidua, and cervix collected for single-cell RNA sequencing (scRNA-seq) experiments (n = 4 per group).
(G) Uniform manifold approximation and projection (UMAP) plot showing all cell types present in the uterus, decidua, and cervix.
(H) UMAP color-coded plot showing tissue-specific predominance of distinct cell types in the uterus (blue), decidua (pink), and cervix (orange). Blue dotted lines distinguish major cell types: myeloid, endothelial, stromal, smooth muscle, epithelial, and lymphoid.
SMC, smooth muscle cell; NK, natural killer. See also Figure S1.
Preterm parturition includes activation of the common pathway of labor that comprises increased uterine contractility, triggering of the local immune response in the decidual tissues, and cervical dilatation.4,79–83 Therefore, to establish a single-cell atlas of murine reproductive tissues in preterm labor, we utilized the uterus, decidua, and cervix of dams that received intra-amniotic inoculation with E. coli in the active phase of parturition (17.5 dpc) for scRNA-seq (Figure 1F). We identified 31 cell clusters across the uterus, decidua, and cervix that corresponded to multiple cell types: smooth muscle cells (SMC) (2 clusters), epithelial cells (10 clusters), fibroblasts (3 clusters), stromal cells (3 clusters), endothelial, neutrophil, monocyte, macrophages (2 clusters), dendritic cell, T cell, B cell, natural killer (NK) cells (2 clusters), erythroid, plasmocyte, and trophoblast (Figure 1G). The heterogeneous and distinct cellular composition of the uterus, decidua, and cervix was highlighted by assigning a tissue identity to each cell cluster (Figure 1H). In control dams, the uterus, decidua, and cervix each displayed a distinct basal cellular repertoire. The uterus showed a predominance of fibroblasts (clusters 0 and 1) and non-decidual stromal (clusters 2 and 12) cell types, and the decidua also included an exclusive subset of stromal cells (cluster 4) (likely corresponding to conventional decidual stromal cells) (Figure S1A). The uterus and decidua of control mice also included modest populations of innate immune cells, such as monocyte and macrophage subsets, as well as lymphocytes such as T cell, NK cell-1, NK cell-2, and B cell (Figure S1A), likely representing the resident immune populations that have been characterized in human and murine tissues.66,68,70,84–86 By contrast with the uterus and decidua, the cervix of control mice comprised a diverse compartment of epithelial subsets (clusters 5, 7, 8, 10, 11, and 14) and other major cell types (Figure S1A), as shown previously.87–89 Immune cells were scarce in the cervix although a modest macrophage-1 population was observed (Figure S1A), which is consistent with prior reports of cervical cell composition in late gestation.90–93 These data provide an overview of the single-cell composition and diversity in the murine uterus, decidua, and cervix in late murine pregnancy. By dissecting the cellular repertoire of the cervix, we demonstrated the underappreciated heterogeneity of this compartment.
Preterm labor induced by E. coli dysregulates the repertoire of immune and non-immune cell types in reproductive tissues
We then examined the effects of preterm labor on the abundance of each cell type identified across all tissues (Figure 2A) as well as in the uterus, decidua, and cervix (Figures 2B–2D and S1B; Table S1). During preterm labor, a relative increase in innate immune cell clusters, such as monocyte, macrophages, dendritic cell, and neutrophil (clusters 3, 6, 9, and 18), was observed in the uterus, decidua, and, to a lesser extent, the cervix (Figures 2B–2D and S1B). The NK cell-2 and plasmocyte subsets in the uterus and decidua also showed changes with preterm labor (Figures 2B, 2C, and S1B). Dendritic cell type was increased in the decidua (Figures 2C and S1B), with a similar tendency observed in the uterus (Figures 2B and S1B). The macrophage-1 cell type was decreased in the uterus with preterm labor (Figures 2B and S1B). The T cell population (cluster 16) also appeared to increase in the uterus and decidua with preterm labor (Figures 2B, 2C, and S1B), which is consistent with prior studies implicating T-cell infiltration and activation as a component of parturition.94–101 Although not visually apparent from the uniform manifold approximation and projection (UMAP) plots (Figure S1B), the uterus and decidua showed a substantial decrease in non-immune subsets, such as fibroblast-1, fibroblast-2, and stromal-3, with preterm labor (Figures 2B and 2C), with stromal-2 also showing modest changes in the uterus (Figure 2B). A subset of epithelial cells (cluster 11, epithelial-10) that was largely absent in the uterus and decidua of controls became apparent in preterm labor (Figure S1B), suggesting labor-induced differentiation or activation of these cells. By contrast with the uterus and decidua, the cervix only showed changes in two cell types: neutrophil and epithelial-8 were increased with preterm labor (Figure 2D), indicating that a modest cellular response to intra-amniotic infection occurs in this tissue. We also evaluated whether cells of fetal origin were represented among the populations of the uterus, decidua, and cervix during preterm labor (Figures S1C–S1E). A small population of fetal cells (Trophoblast) was detected in the uterus and decidua, which is consistent with prior single-cell studies of the human myometrium70 and may represent residual placental cells attached to the uterus and decidua.
Figure 2. Preterm labor induced by E. coli dysregulates the repertoire and gene expression of immune and non-immune cell types in reproductive tissues.
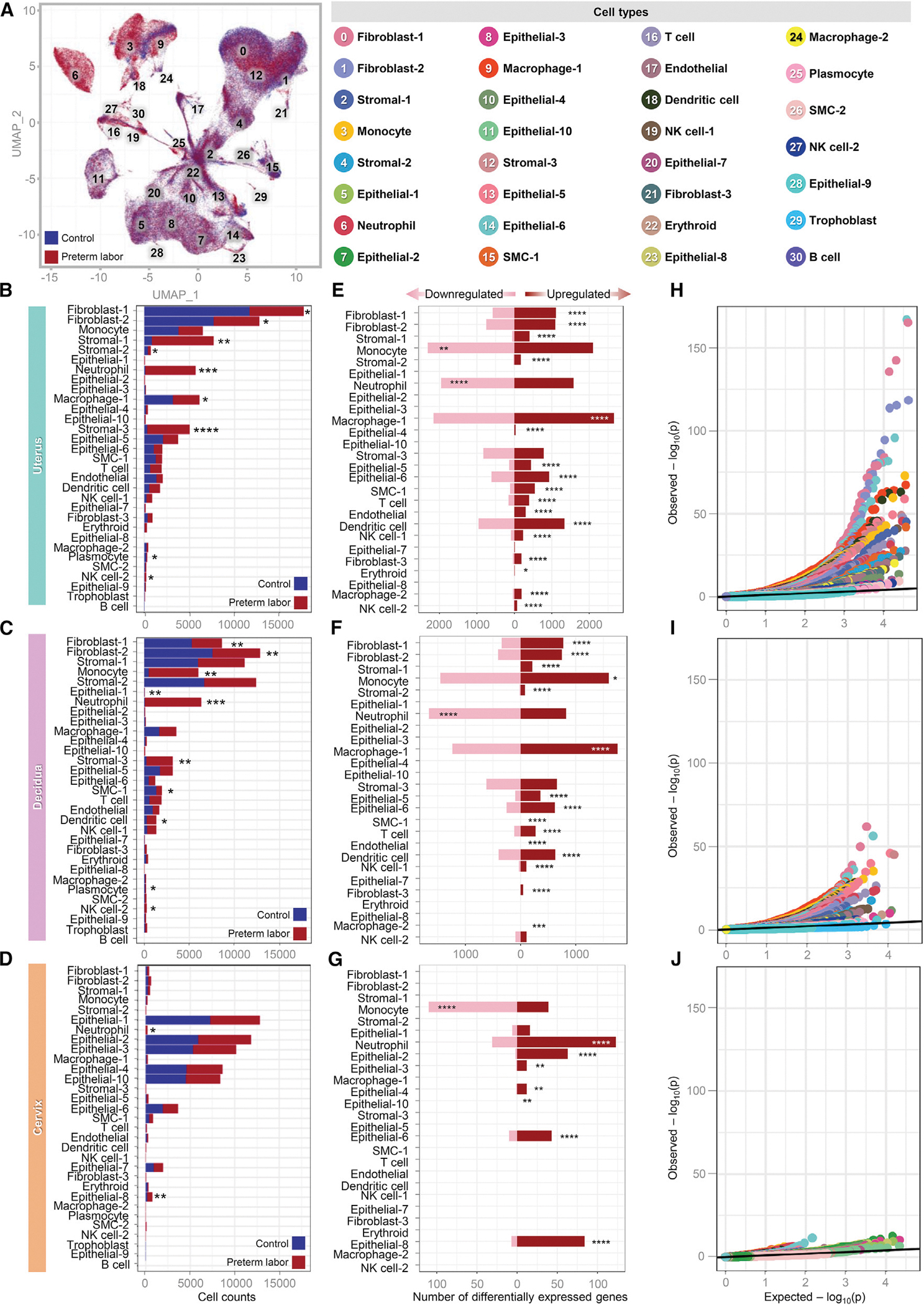
(A) Color-coded uniform manifold approximation and projection (UMAP) plot showing the effects of preterm labor on the abundance of specific cell types (red) compared to the control (blue).
(B–D) Bar plots showing the numbers of each cell type in the uterus, decidua, and cervix. The comparison of cell numbers between the two study groups for each cell type was performed using a 2-sided t test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
(E–G) Bar plots showing the numbers of differentially expressed genes (DEGs) induced by preterm labor in each cell type in the uterus, decidua, and cervix. Red and pink bars indicate upregulated and downregulated DEGs, respectively (derived from DESeq2, q < 0.1). The comparisons of the fraction of downregulated and upregulated DEGs in each cell type between the study groups were calculated using two-sided binomial tests. *q < 0.05, **q < 0.01, ***q < 0.001, ****q < 0.0001.
(H–J) Quantile-quantile plot showing differential expression of genes analyzed for selected enriched cell types from the uterus, decidua, and cervix. Deviation above the 1:1 line (solid black line) indicates enrichment.
To validate the leukocyte infiltration of the uterus, decidua, and cervix indicated by our single-cell data, we undertook a series of histological and immunohistochemical analyses (Figure S2). We observed collagen degradation in the uterine and cervical tissues with preterm labor, and mucin production by cervical cells appeared to increase compared to control tissues (Figures S2A–S2C). Histological changes in preterm labor were accompanied by increased CD45+ leukocyte infiltration in the uterus and decidua (Figures S2D–S2F). Uterine leukocytes were more evenly distributed among neutrophils, monocytes, and macrophages, whereas decidual leukocytes were predominantly neutrophils and, to a lesser extent, monocytes (Figures S2G–S2I). Similar to our scRNA-seq results, the leukocyte abundance in the cervical tissues was largely comparable between the control and preterm labor groups (Figure S2I).
Thus, our scRNA-seq data indicate a shift in the cellular composition of the murine uterus, decidua, and cervix that accompanies preterm labor.
Preterm labor induced by E. coli dysregulates gene expression of immune and non-immune cell types in reproductive tissues
Given that preterm labor altered the cellular composition of the uterus, decidua, and cervix, we next explored whether this inflammatory process would also result in transcriptomic changes to the identified cell types. Consistent with their altered abundance, multiple fibroblast, stromal, and epithelial cell types in the uterus and decidua displayed upregulated gene expression with preterm labor (Figures 2E and 2F; Table S2), whereas cervical non-immune cells with upregulated gene expression were exclusively epithelial (Figure 2G; Table S2). Innate immune cell types showed strong dysregulation of gene expression in both directions that was inconsistent among tissues; although monocyte showed more downregulated differentially expressed genes (DEGs) in the uterus (Figure 2E) and cervix (Figure 2G), this cell type showed more upregulated DEGs in the decidua (Figure 2F). Neutrophil showed stronger downregulation of DEGs in the uterus and decidua (Figures 2E and 2F), whereas DEGs were primarily upregulated in this cell type in the cervix (Figure 2G). Macrophage-1, dendritic cell, and NK cell-1 consistently displayed predominantly upregulated DEGs in the uterus and decidua (Figures 2E and 2F) and were not represented in the cervix, as noted previously. The uterine macrophage-2 and NK cell-2 populations displayed upregulated DEGs (Figure 2E), which was not observed in other tissues (Figures 2F and 2G). Although not as abundant as innate immune cells, the T cell population also displayed upregulated DEGs with preterm labor in the uterus and decidua (Figures 2E and 2F). Quantile-quantile plots of DEGs from enriched cell types indicated that the uterus is the tissue most affected by the process of labor (Figures 2H–2J). Thus, preterm labor primarily induced gene expression in the dominant cell types from each tissue; however, the substantial amount of downregulated gene expression in innate immune cells may indicate an immunological switch from one transcriptomic program to another to combat infection.
Preterm labor induced by E. coli involves conserved cell types that display distinct processes in reproductive tissues
Transcriptomic profiling of cell types suggested that specific subsets show conserved responses with preterm labor across the reproductive tissues. Therefore, we next focused on shared preterm labor-specific gene expression among the uterus, decidua, and cervix. The Venn diagram displayed in Figure 3A highlights the overlap in DEGs across tissues, particularly the uterus and decidua. Correlation analyses indicated stronger relationships between preterm labor and gene expression changes in the uterus and decidua than in the cervix (Figure S3A), which was reflective of the total preterm labor-associated DEGs in each tissue. This observation was confirmed by the correlation between the gene expression profiles of the uterus and decidua, which was stronger than the correlations between the decidua and cervix or the uterus and cervix (Figure S3B). Given that the uterus, decidua, and cervix displayed some degree of correlation for preterm labor-associated gene expression, we evaluated the cell type-specific transcriptomic changes that were conserved across all three tissues. We found that innate immune cell types (monocyte and neutrophil) as well as non-immune cell types (epithelial-3, -4, and -6 and endothelial) showed conserved gene expression changes associated with preterm labor across the uterus, decidua, and cervix (Figure 3B). We reasoned that, although the transcriptome profiles of specific cell types were affected across all tissues, such cells may display distinct biological processes according to their location. Gene Ontology (GO) analysis of the neutrophil cell type in the uterus, decidua, and cervix revealed that, although these cells shared some processes, such as “response to bacterium” and “response to lipopolysaccharide,” processes specific to neutrophil in each tissue were also observed (Figure 3C). Uterine neutrophil showed enrichment of processes related to cytokine signaling and anti-viral response, whereas decidual neutrophil showed enrichment of cellular activation-associated processes (Figure 3C). In the cervix, enriched neutrophil processes were primarily associated with response to external stimuli and bacteria (Figure 3C). Uterine and decidual monocyte and macrophage-1 cells also shared enriched processes related to cytokine production and response to bacteria/lipopolysaccharide, with decidual monocyte also showing enrichment of activation-associated processes (Figure 3D). By contrast, cervical monocyte displayed highly distinct processes related to protein synthesis and humoral immune response (Figure 3D), suggesting that such cells may functionally differ from their counterparts in the uterus and decidua. Epithelial-6, which had sufficient DEGs to perform GO analysis in all three tissues, displayed largely consistent processes across the uterus, decidua, and cervix that were related to inflammation, antibacterial response, and cytokine production (Figure 3E). The uterine epithelial-4 cell type displayed enrichment of several chemotaxis-associated processes, suggesting involvement in leukocyte recruitment to this tissue, whereas the cervical epithelial-4 showed enrichment of effector functions, such as production of NO (nitric oxide), interleukin-1 (IL-1), and interferon γ (IFNγ) (Figure 3E). Epithelial-3, which only displayed sufficient DEGs for GO analysis in the cervix, showed enrichment of multiple processes related to promotion of B cell and antibody responses (Figure 3E). Thus, the conserved cell types affected by preterm labor in the uterus, decidua, and cervix each display distinct enrichment of biological processes, suggesting that similar cell types display tissue-specific functions in the context of intra-amniotic infection leading to preterm labor. Together with the observed increase in cervical epithelial-8 cell counts with preterm labor, it is possible that the upregulation of inflammatory gene expression represents an infection-induced differentiation of cervical epithelial cells to better participate in host defense mechanisms in this compartment.
Figure 3. Preterm labor induced by E. coli involves conserved cell types that display distinct processes in reproductive tissues.
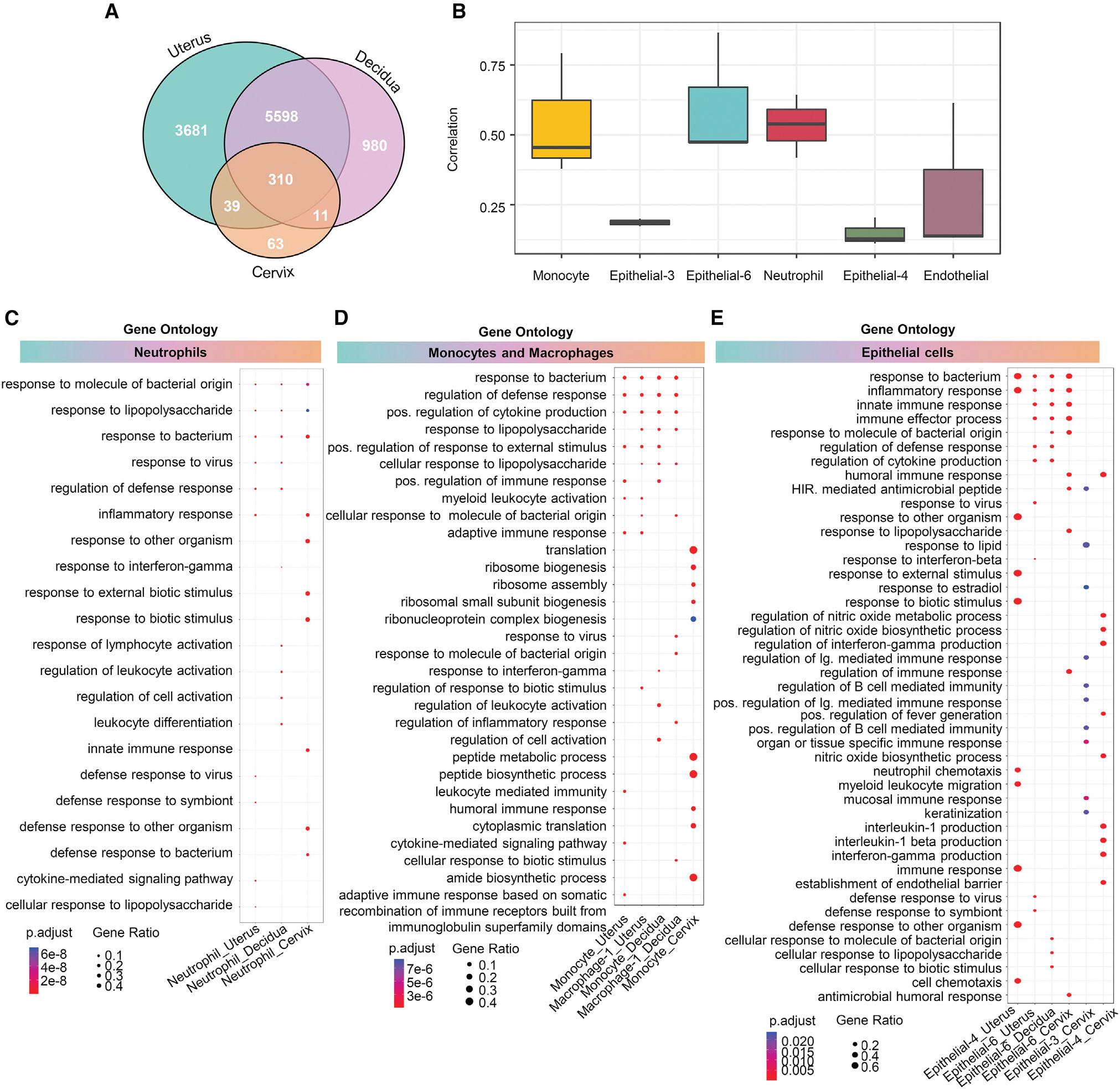
(A) Venn diagrams showing the numbers of differentially expressed genes (DEGs; q < 0.1) that are exclusive to or shared among the uterus, decidua, and cervix.
(B) Boxplots showing the correlation of specific cell types affected by preterm labor and conserved across the uterus, decidua, and cervix, using the Spearman’s method.
(C–E) Cluster profiler dot plots showing the preterm labor-associated Gene Ontology (GO) biological processes that are exclusive to or shared among
(C) neutrophil, (D) monocyte and macrophage, and (E) epithelial cell types from the uterus, decidua, and cervix. The size and color of each dot represent gene ratio and significance level, respectively. 1-sided Fisher’s exact tests were used.
See also Figures S3 and S4 and Table S2.
To infer cellular functionality in preterm labor, we utilized the Kyoto Encyclopedia of Genes and Genomes database to evaluate the pathways enriched in labor-associated DEGs in each cell type (Figure S3C). Immune and non-immune cell types with altered gene expression in preterm labor showed enrichment of immunological pathways, such as “cytokine-cytokine receptor interaction,” “NOD-like receptor signaling pathway,” and “viral protein interaction with cytokine and its receptor,” across the three tissues (Figure S3C). Such findings are consistent with previous studies showing upregulation of immune-related pathways in decidual endothelial102 and stromal cells69 from women with labor. Additional inflammatory pathways, such as “NF-κB signaling pathway” and “Toll-like receptor signaling pathway,” were also represented, to a lesser extent, by immune cells (e.g., NK cells and neutrophil) as well as non-immune cells, such as epithelial cells (Figure S3C).
We also investigated the biological processes enriched in several non-immune cell subsets that were conserved between the uterus and decidua with preterm labor (Figure S4). Stromal-1 and stromal-2 in the uterus showed largely similar enrichment of biological processes, as did the stromal-1 and stromal-2 cell types in the decidua (Figure S4A). However, these cell types differed between tissues, given that the decidual stromal cells were enriched for leukocyte migration and chemotaxis, whereas the uterine stromal cells showed enrichment for response to immune signaling (Figure S4A). The fibroblast-1 and stromal-3 cell types showed less diversity in their enriched processes when compared between the uterus and decidua, with the former associated with host defense against infection and the latter associated with immune activation, including adaptive immunity (Figure S4B). Fibroblast-2 and fibroblast-3 were also comparable between the uterus and decidua; however, the decidual fibroblast-3 showed more striking enrichment of responses to microorganisms and cytokine signaling (Figure S4C). Finally, the uterine and decidual endothelial cell types displayed similar enrichment of processes related to host defense, innate immunity, and cytokine signaling, with the decidual subset showing modestly higher enrichment for processes related to neutrophil migration (Figure S4D).
These data indicate that the uterus, decidua, and cervix contain cell types that display distinct tissue-specific gene expression profiles in preterm labor, pointing to differing functional roles for these cells in the host response to intra-amniotic infection. However, there is an overall tendency for the enrichment of similar immunological pathways in immune and non-immune cell types across tissues, likely as part of the common host response to intra-amniotic infection.
Preterm labor influences cell-cell communications in reproductive tissues
Having established that preterm labor drives distinct transcriptomic changes in specific cell types in the uterus, decidua, and cervix, we next leveraged our single-cell data to elucidate cell-cell communication networks in these tissues.
Cell-cell communications in the uterus
The uterus is a highly heterogeneous organ with multiple described regions that differ in cellular composition and function.103–112 To unravel the intercellular communications taking place in the murine uterus with preterm labor, we performed a correlation analysis across preterm labor-associated genes for each pair of identified cell types (Figure S5A). The strongest correlations were observed for non-immune cell types, such as stromal, epithelial, fibroblast, smooth muscle, and endothelial cell types (Figure S5A), suggesting that these cells exhibit similar changes in gene expression with preterm labor. Innate and adaptive immune cell types also showed moderate correlations: T cell, NK cell-1, NK cell-2, macrophage-1, macrophage-2, monocyte, and dendritic cell (Figure S5A). The cell types with the weakest correlations were primarily epithelial subsets as well as neutrophil, erythroid, stromal-3, and SMC-2 (Figure S5A), indicating that such cells show more distinct gene expression changes with preterm labor.
We next applied CellChat to infer cell-cell communications within the uterus, using our single-cell gene expression data and a database of established interactions between signaling ligands, receptors, and their cofactors.113 Signaling pathways that were enriched or diminished in preterm labor, as well as those that were unaffected by this process, are shown in Figure S5B and Table S3. The alluvial plots shown in Figures 4A and 4B display the major cell-cell communication processes taking place in the uterus as well as the cell types that participate as senders or receivers in each process. Innate and adaptive immune cell subsets (neutrophil, macrophage-1, monocyte, dendritic cell, NK cell-1, and T cell) contribute to the top signaling pathways implicated in preterm labor, such as C-C chemokine ligand (CCL), C-X-C chemokine ligand (CXCL), complement, IFN-I, IFN-II, IL-1, and IL-6 (Figures 4A and 4B). Multiple non-immune subsets also participate in these processes: fibroblast, stromal, epithelial, SMC, and endothelial cell types (Figures 4A and 4B). Although immune and non-immune cell types served as receivers of preterm labor-associated signaling, specific responders to each pathway could be distinguished (Figure 4B). For example, the signaling pathway of IL-6, which is commonly utilized as a biomarker of intra-amniotic inflammation,47 was primarily driven by immune cell types (Figure 4A); however, the receiver cells for this pathway were non-immune subsets (Figure 4B). Conversely, the primary senders for the Annexin signaling pathway were non-immune cell types (Figure 4A), with the downstream receivers being predominantly immune cells (Figure 4B). The changes in cell-cell communication occurring as a result of preterm labor were visualized using the arrow plot in Figure 4C, where the directionality of each cell type arrow reflects the propensity for increased outgoing and/or incoming interaction strength. Cell types such as macrophage-2, stromal-2, stromal-3, and fibroblast-3 showed primarily even increases in incoming and outgoing signaling with preterm labor (Figure 4C). Other cell types were more biased toward incoming interactions, such as macrophage-1, neutrophil, dendritic cell, plasmocyte, monocyte, and epithelial-6, or toward outgoing interactions, such as stromal-1, fibroblast-1, and fibroblast-2 (Figure 4C). Several cell types showed a net decrease in signaling with preterm labor: T cell and SMC-1 (Figure 4C).
Figure 4. Cellular interactions in the uterus during preterm labor.
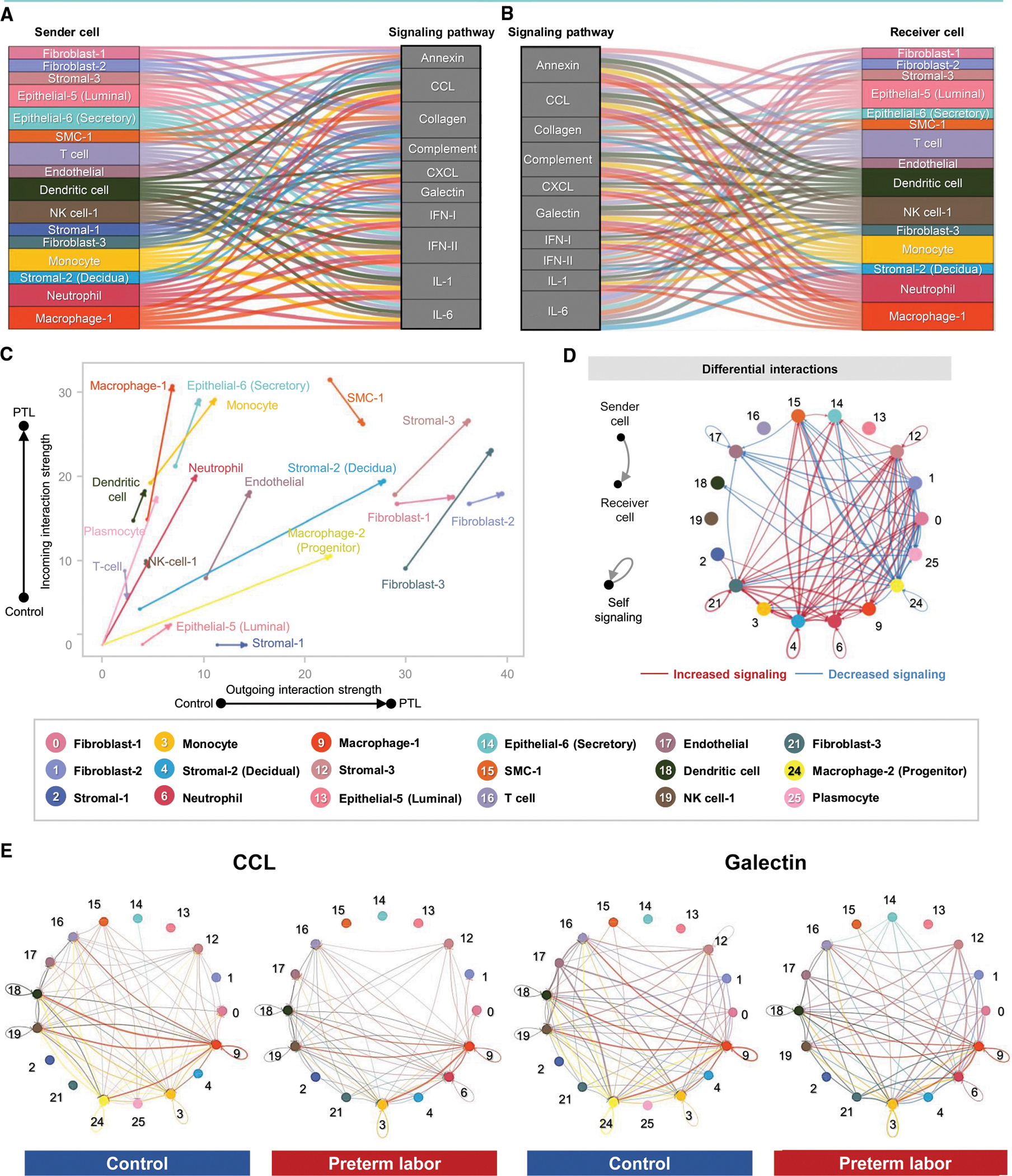
(A and B) Alluvial plots showing the roles of specific cell types as senders or receivers of preterm labor-associated signaling in the uterus based on selected enriched signaling pathways comparing the overall information flow within networks between preterm labor and control derived from CellChat (full list of pathways in Figure S5B). Connecting lines are color coded and represent the participation of each cell type as senders or receivers of the indicated signaling pathway. Line thickness is proportional to the strength of interaction.
(C) Arrow plot showing changes in outgoing and incoming interaction strength between preterm labor (point of the arrow) and control conditions (base of the arrow) for specific cell types in the uterus.
(D) Circle plots showing the top 25% increased (red) or decreased (blue) signaling interactions in the uterus for specific pathways in preterm labor compared to controls.
(E) Circle plots showing the top 25% of aggregated interactions among cell types in the uterus for control and preterm labor groups. Each node represents a cell type, and the interaction is shown by lines color coded based on the sender cell.
PTL, preterm labor. See also Figures S5 and S6 and Table S3.
The top 25% of aggregated cellular interactions in the uterus were then contrasted between the control and preterm labor groups, emphasizing the overall increase in cell-cell signaling with preterm labor as well as the incorporation of new signaling pathways from cell types that were rarely present in control tissues, such as neutrophil (Figure 4D). Although macrophage-1 signaling was increased, macrophage-2 signaling decreased, which could indicate a homeostatic role for the latter subset that is diminished in preterm labor, as reported previously.114 Next, we examined the top contributors within uterine cell-cell signaling pathways enriched with preterm labor (Figure 4E). We found that macrophage subsets and dendritic cell were primary contributors to CCL signaling between control uterine cell types, and such signaling was strengthened in preterm labor (Figure 4E). By contrast, the galectin signaling pathway, already enriched in control uterine tissues, was upregulated in new cell types in preterm labor (e.g., epithelial-6) and diminished in others (e.g., macrophage-2) (Figure 4E).
We also explored the changes in cell type-specific expression of genes related to progesterone and prostaglandin signaling in the uterus (Figures S5C and S6A). As expected, progesterone-related gene expression was consistently downregulated across uterine cell types in preterm labor (Figure S5C). Prostaglandin-related gene expression showed more activity in the uterus than in other tissues (Figure S6); however, preterm labor-associated changes in each gene were consistent across uterine cell types (Figure S6A), supporting the involvement of multiple immune and non-immune cell populations in labor-mediator signaling pathways.
These findings highlight the complex cell-cell communication network taking place in the murine uterus and how such interactions are modulated by the inflammatory process of preterm labor in immune and non-immune cell types.
Cell-cell communication in the decidua
We next examined the correlations across preterm labor-associated changes in gene expression for each pair of cell types identified in the decidua (Figure S7A). Similar to the uterine tissues, the strongest correlations were observed for non-immune cell types (e.g., stromal, epithelial, fibroblast, smooth muscle, and endothelial), followed by innate and adaptive immune cells, of which the macrophage and monocyte clusters were best correlated (Figure S7A). Similarly, the weakest correlations were observed for some epithelial cell types and neutrophil (Figure S7A). Thus, decidual cells display preterm labor-associated changes in gene expression with varying magnitudes of sharing among cell types, which resemble those observed in the uterine tissues.
The inferred cell-cell signaling pathways that were enriched or diminished in the decidua with preterm labor are shown in Figure S7B and Table S3. From among them, the top pathways are displayed, with their participating sender and receiver cell types, in Figures 5A and 5B. Similar to the uterine tissues, key cell-cell communication pathways were primarily related to immune functions, such as cytokine and chemokine signaling (Figured 5A and 5B). Among the three compared tissues, the IL-17 pathway was most prominent in the decidua (Figures 5A and 5B), which was consistent with a previous report of IL-17 signaling in endothelial cells derived from the human peripartum decidua102 and suggested that decidual T cells participate in the local inflammatory response to intra-amniotic infection. Among other identified signaling pathways, immune and non-immune cell subsets contributed as senders or receivers, including the NK cell-2 subset, which was not implicated in uterine cell-cell signaling (Figures 5A and 5B). The decidual epithelial-5 cell type appeared to be primarily functioning as a receiver of cell-cell signaling in this tissue (Figures 5A and 5B). We then visualized the preterm labor-driven changes in incoming and outgoing signaling and observed that subsets such as monocyte, macrophage-1, neutrophil, NK cells, and dendritic cell showed predominantly incoming interactions (Figure 5C). On the other hand, stromal and fibroblast subsets as well as T cell tended toward increased outgoing signaling, whereas SMC-1 and endothelial showed an overall reduction in interaction strength (Figure 5C). Outgoing interaction strength was greater in decidual T cell compared to uterine T cell (Figures 5C versus Figure 4C), which emphasizes a role of T cell-derived signals in the pathophysiology of preterm labor associated with intra-amniotic infection.115 Consistent with enhanced cell-cell signaling in preterm labor, aggregated cellular interaction plots demonstrated an overall net increase in decidual intercellular interactions compared to controls (Figure 5D). Similar to the uterine tissues, enriched signaling pathways, such as CCL and galectin, were primarily driven by macrophage-1, monocyte, and dendritic cell in preterm labor, with overall interactions among cell types increasing compared to controls (Figure 5E).
Figure 5. Cellular interactions in the decidua during preterm labor.
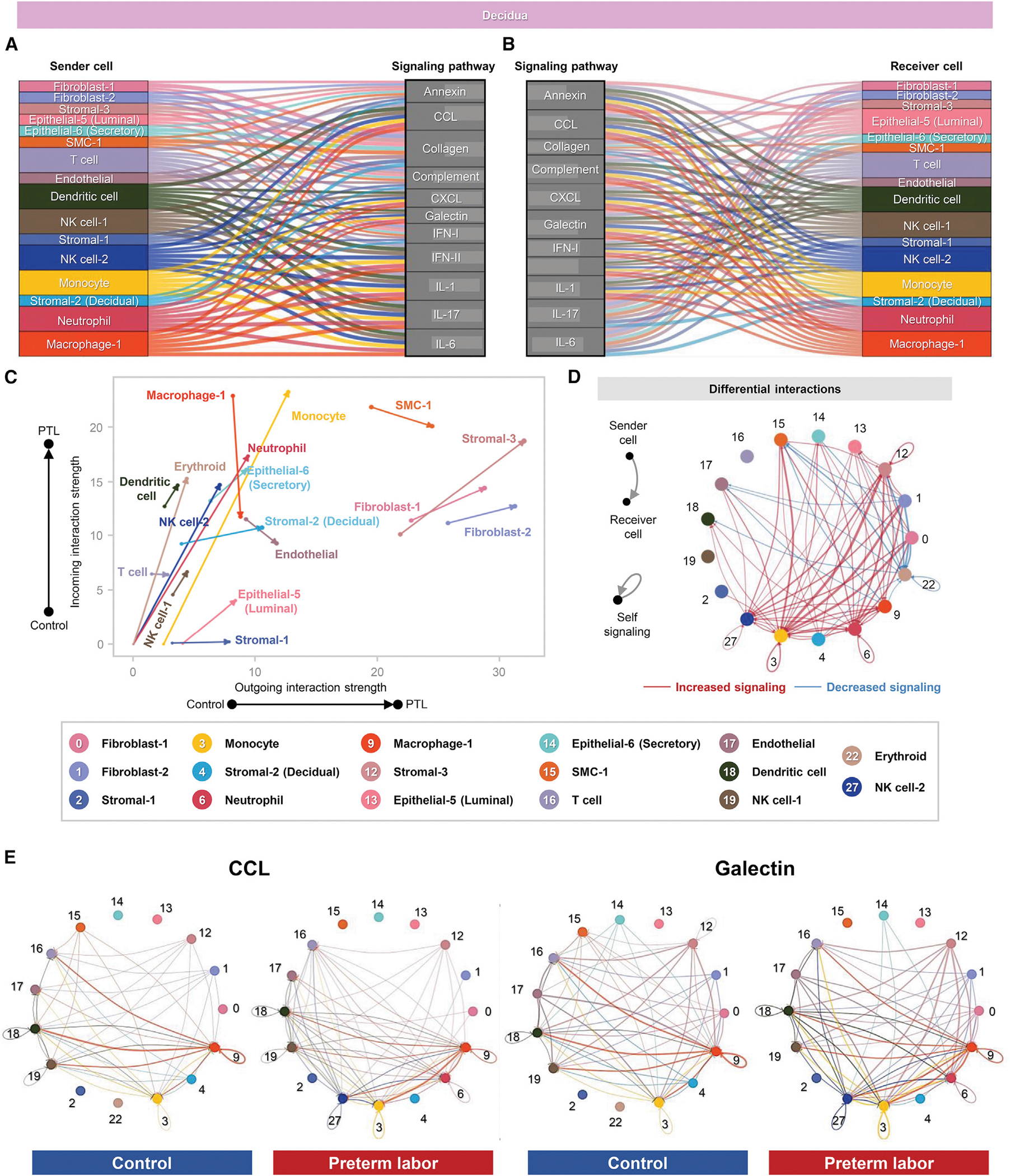
(A and B) Alluvial plots showing the roles of specific cell types as senders or receivers of preterm labor-associated signaling in the decidua based on 11 enriched signaling pathways comparing the overall information flow within networks between preterm labor and control conditions derived from CellChat (full list of pathways in Figure S6B). Connecting lines are color coded and represent the participation of each cell type as a sender or as a receiver of the indicating signaling pathway. Line thickness is proportional to the strength of interaction.
(C) Arrow plot showing changes in the outgoing and incoming interaction strength between preterm labor (point of the arrow) and control conditions (base of the arrow) for specific cell types in the decidua.
(D) Circle plots showing the top 25% increased (red) or decreased (blue) signaling interactions in the decidua for specific pathways in preterm labor compared to controls.
(E) Circle plots showing the top 25% of aggregated interactions among cell types in the decidua for control and preterm labor groups. Each node represents a cell type, and the interaction is shown by color-coded lines.
PTL, preterm labor. See also Figures S6 and S7 and Table S3.
Similar to the changes observed in the uterus, decidual expression of progesterone-related genes was consistently downregulated across cell types with preterm labor (Figure S7C). The patterns of change in prostaglandin-related gene expression were also similar between the decidua and uterus; however, some differences in the magnitude of change between compartments were observed for multiple genes, potentially indicating a stronger upregulation of preterm labor-associated prostaglandin signaling in the uterus relative to the decidua (Figure S6B).
These data provide insight into the distinct cellular interactions taking place in the decidua during the process of preterm labor, including the involvement of cell types and signaling pathways not observed in other tissues. However, the decidua and uterus also share cell type-specific communications that are affected by preterm labor.
Cell-cell communication in the cervix
Our investigation of the cell type-specific changes taking place in the cervix with preterm labor indicated that the neutrophil and monocyte subsets were most affected, in tandem with previous studies showing labor-associated infiltration of immune cells90,91,116–118 as well as multiple epithelial cell subsets (Figures 2 and 3). Correlation analysis of these cell types showed the strongest associations in gene expression changes driven by preterm labor among epithelial cell types (Figure S8A), whereas neutrophil and monocyte showed modest correlation of genes affected by preterm labor (Figure S8A). Inferred cell-cell signaling pathways were noticeably fewer compared to the other tissues and included multiple processes exclusive to the cervix (Figure S8B; Table S3), which could be attributed to the less diverse cell type composition observed in this tissue. As shown by the participating senders and receivers, signaling pathways that were strongly implicated in the uterus and decidua with preterm labor were not as enriched in cervical cell types (Figures 6A and 6B). On the other hand, cell-cell signaling pathways related to extracellular matrix were strongly represented (Figures 6A and 6B), which is consistent with the primarily connective tissue composition of the cervix.119–121 As expected, given their inferred roles as receivers, most cervical epithelial cell types showed strong incoming interactions with preterm labor, whereas the SMC-1, fibroblast-2, and stromal-1 subsets showed a tendency toward increased outgoing interactions (Figures 6B and 6C). This finding was supported by the aggregated cervical cell-cell interactions in the control and preterm labor groups showing increased receipt of signaling by epithelial-1 and epithelial-8 as well as SMC-1 and fibroblast-2 (Figure 6D). Fibroblast-2 and SMC-1 were top contributors to enriched signaling pathways such as collagen and tenascin (Figure 6E). It is possible that the fibroblast-2 and/or SMC-1 cell clusters include cervical myofibroblasts, given that a previous histological investigation indicated a pregnancy-specific accumulation of such cells, which could be interacting with the extracellular matrix to aid in supporting the mechanical stresses present during labor.122 In the last decade, a new paradigm for the role of SMCs in the human cervix has emerged, suggesting a sphincter-like function of the internal os, in which the SMCs express contractility-associated proteins that are responsive to oxytocin signaling.123 Together with our current findings, these observations support the involvement of SMC-1 and fibroblast-2 subsets in preterm labor-associated signaling in the murine cervix and emphasize the distinct cell-cell signaling pathways taking place in this tissue during preterm labor.
Figure 6. Cellular interactions in the cervix during preterm labor.
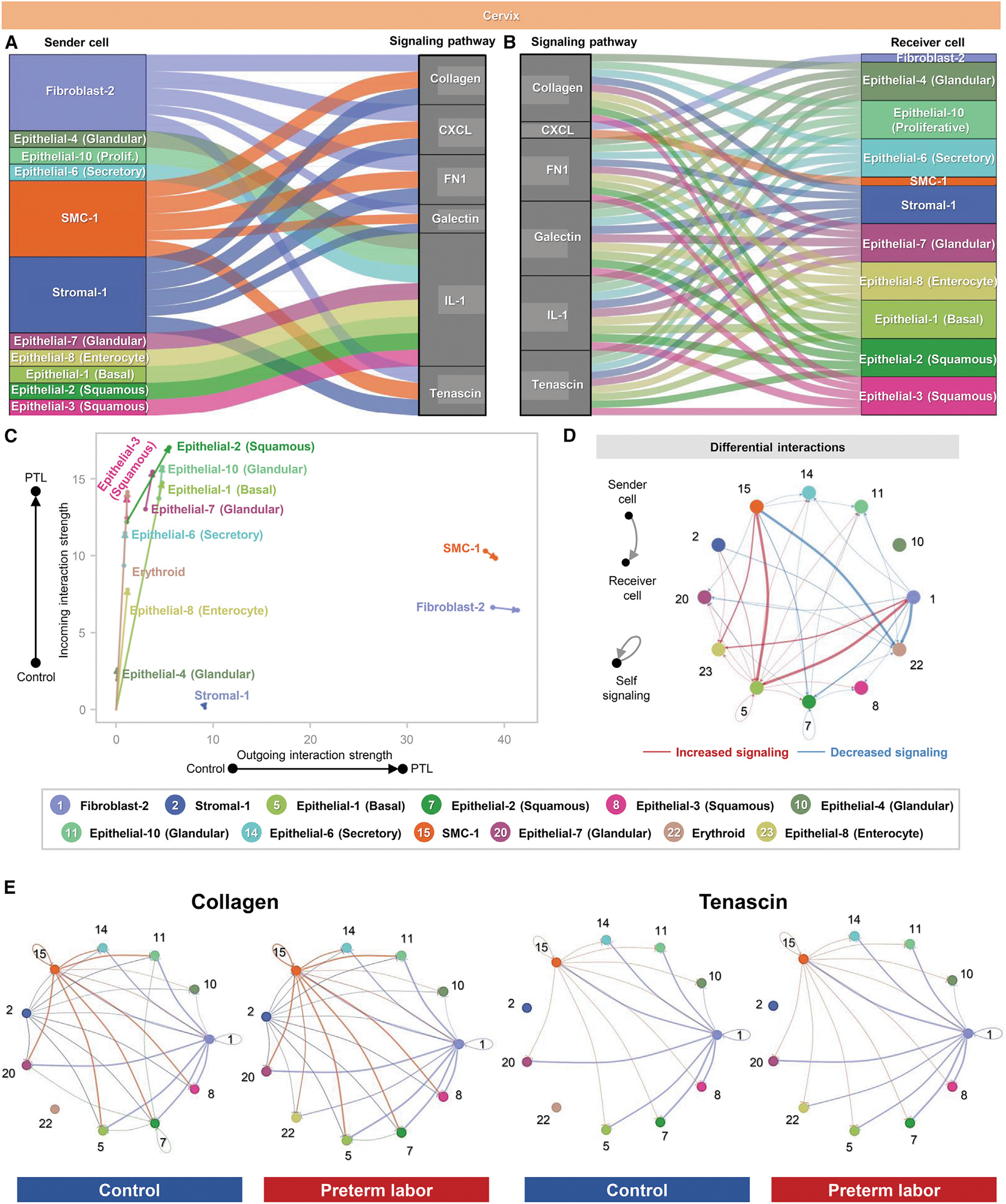
(A and B) Alluvial plots showing the roles of specific cell types as senders or receivers of preterm labor-associated signaling in the cervix based on selected enriched signaling pathways (full list of pathways in Figure S7B). Connecting lines are color coded and represent the participation of each cell type as a sender or as a receiver of the indicating signaling pathway. Line thickness is proportional to the strength of interaction.
(C) Arrow plot showing changes in the strength of outgoing and incoming interactions between preterm labor (point of the arrow) and control conditions (base of the arrow) for specific cell types in the cervix.
(D) Circle plots showing the top 25% increased (red) or decreased (blue) signaling interactions in the cervix for specific pathways in preterm labor compared to controls.
(E) Circle plots showing the top 25% of aggregated interactions among cell types in the cervix for control and preterm labor groups. Each node represents a cell type, and the interaction is shown by color-coded lines.
PTL, preterm labor. See also Figures S6 and S8 and Table S3.
Shared cellular signaling pathways in the murine uterus and human myometrium during the processes of preterm and term labor
Last, to examine the shared pathways implicated in the process of parturition in mice and humans, we utilized the differential cellular interactions in the murine uterus with preterm labor together with our previously generated single-cell atlas of the human myometrium with labor at term70 (Figure 7). We investigated the interaction strength between cell types affected by labor and prominent signaling pathways and then contrasted them between the murine and human tissues. Overall, we found that labor-associated cell-cell interactions were primarily driven by SMC, stromal, fibroblast, and innate immune cell types in the murine uterus and human myometrium, independent of differences in sender/receiver status (Figures 7A and 7B).
Figure 7. Shared cellular signaling pathways in the murine uterus and human myometrium during the processes of preterm and term labor.
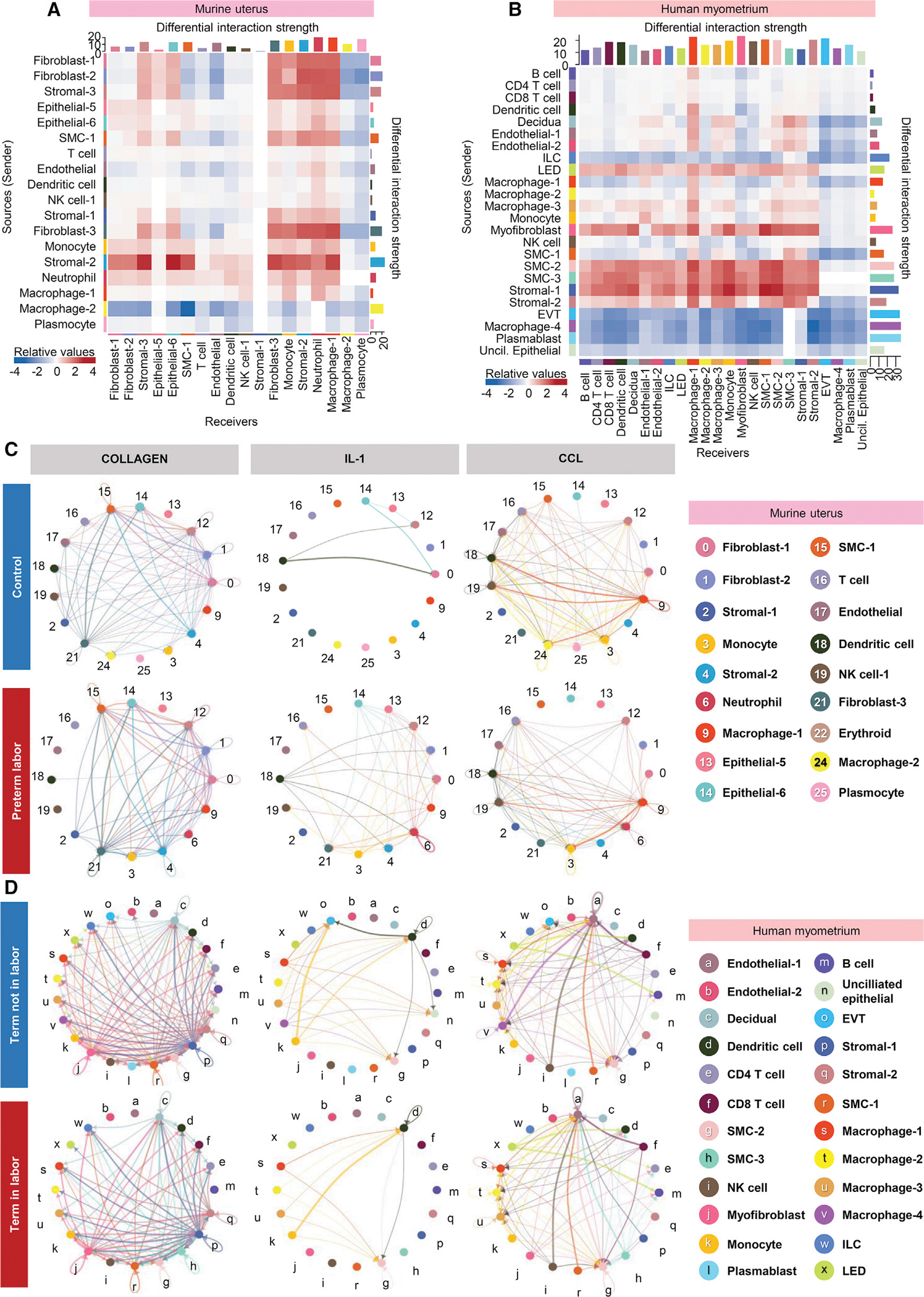
(A) Heatmap showing the differential interaction strength among cell types in the murine uterus with preterm labor. Red and blue shading indicate increased or decreased signaling, respectively, in preterm labor compared to control.
(B) Heatmap showing the differential interaction strength among cell types in the human myometrium with term labor. Red and blue shading indicate increased or decreased signaling, respectively, in term labor compared to term without labor.
(C) Circle plots representing the top 25% murine uterine cell-cell communications inferred for the collagen, IL-1, and CCL pathways for the control and preterm labor groups.
(D) Circle plots representing the top 25% human myometrial cell-cell communications inferred for the collagen, IL-1, and CCL pathways for the control and preterm labor groups.
EVT, extravillous trophoblast; ILC, innate lymphoid cell; LED, lymphoid endothelial decidual cell. See also Figure S9.
Specifically, in the murine uterus, non-immune cell types (such as fibroblast-1, -2, and -3, stromal-2 and -3, and SMC-1) showed the strongest labor-associated increase in outgoing cell-cell signaling, whereas outgoing signaling by macrophage-2 was greatly diminished (Figure 7A). The top receivers of labor-associated signaling were fibroblast-3, stromal-2 and -3, and epithelial-6 as well as innate immune cell types (monocyte, neutrophil, and macrophage-1) (Figure 7A). Endothelial, SMC-1, plasmocyte, and macrophage-2 showed diminished incoming signaling with preterm labor (Figure 7A).
In the human myometrium, labor involved increased outgoing signaling almost exclusively by SMC and stromal subsets, with myofibroblast and lymphoid endothelial decidual (LED) also contributing to this process (Figure 7B). Receivers of such outgoing interactions included macrophage subsets and monocyte, which is consistent with previous investigations indicating that myometrial cell contraction during labor is promoted by crosstalk with macrophages in a co-culture model.124 Multiple myometrial cell subsets displayed substantially reduced outgoing signaling with labor, such as extravillous trophoblast (EVT), macrophage-4, plasmablast, unciliated epithelial, and innate lymphoid cell (ILC) (Figure 7B). For incoming signaling, the majority of human myometrial cell types tended to have increased interaction only with cells that displayed greater outgoing signals, such as smooth muscle and stromal cells (Figure 7B). Some exceptions seemed to be the macrophage-1, SMC-3, and stromal-1 subsets, whose incoming signaling from the majority of other cell types was strengthened (Figure 7B).
We then examined the top 25% of aggregated cell-cell interactions in the human myometrium at term without labor and at term with labor (Figure S8A). Consistent with the correlation analysis above (Figure 7B), a clear shift in the cell types contributing to myometrial cell-cell signaling was observed between groups, with the EVT, macrophage-4, plasmablast, and unciliated epithelial clusters showing greatly diminished interactions in labor and the SMC and stromal cell subsets acquiring increased interactions (Figure S9A). This shift in interaction with labor was also reflected by the combined differential interaction analysis shown in Figure S8B. When examining the specific cell-cell signaling pathways that were affected by labor, terms such as collagen, IL-1, CCL, complement, and CXCL were found to be shared between the murine uterus and human myometrium, indicating shared labor-associated cellular signaling between species (Figures 7C, 7D, and S9C–S9G); however, closer inspection of these shared signaling pathways revealed differences in the cell types contributing to each (Figures 7C, 7D, and S9C–S9G). The relevance of immune pathways such as chemokine signaling in the process of labor is supported by previous reports demonstrating that use of a chemokine inhibitor on human myometrial cells decreased contraction and gap junction formation, disrupting intercellular communication.125,126 These data indicated that labor-associated cell-cell interactions heavily involve SMC, stromal, fibroblast, and innate immune cell types in the murine uterus and the human myometrium, providing evidence of labor-specific signaling processes between immune and non-immune cells that are shared between species. However, this interpretation should be taken with caution, given that we compared the physiologic process of labor in the human myometrium to the pathologic process of labor induced by bacteria in the murine uterus.
DISCUSSION
The current study provides a single-cell atlas of the murine uterus, decidua, and cervix that highlights the cell-type composition, transcriptional profiles, and cell-cell signaling taking place in these tissues in normal late gestation and in the context of infection-induced preterm labor and birth. scRNA-seq allowed deep characterization of immune and non-immune cells that demonstrates the underappreciated heterogeneity of cell types conventionally considered to be uniform in function, such as uterine SMCs and cervical epithelial cells. Our data can thus serve as a reference for future studies seeking to target specific subsets of these cells, which may have differing roles in pregnancy and labor, as indicated by their distinct transcriptional profiles. Shared modulation of gene expression, noted between uterine and decidual cell types, was reflected by the similar enrichment of labor-associated signaling pathways, which is consistent with the spatial proximity of these tissues. However, comparison of individual cell types across tissues indicated that the most-represented biological processes can vary according to location; therefore, the tissue of origin should be taken into consideration when inferring cellular function. Herein, we provide scRNA-seq characterization of the understudied cervical tissues, demonstrating a plethora of epithelial subsets with different potential functions as well as SMC and fibroblast cell types that indicate an unexpected level of heterogeneity in the cervix. Recent evidence has suggested a sphincter-like function of SMCs in the internal os of the human cervix,123 and our current findings support this concept. Inferred cell-cell communications provided evidence of substantial cross-talk among uterine, decidual, and cervical cell types during the process of preterm labor, highlighting key signaling pathways that could potentially be targeted in future translational studies aimed at preventing spontaneous preterm labor. This analysis demonstrated cell types with elevated or diminished interactions driven by inflammation, which can serve to identify cell types that are most and least involved in such signaling. To demonstrate the application of our single-cell dataset, we leveraged prior single-cell analyses of the human myometrium in term labor to evaluate cellular interactions compared to our murine model of preterm labor. This analysis provided useful insight into shared signaling pathways associated with the inflammatory process of labor, providing a practical demonstration of how our scRNA-seq dataset can be leveraged for in silico discovery of specific cell types, pathways, or genes that can be subsequently targeted in vitro and/or in vivo.
Limitations of the study
The current study has some limitations. First, it is important to consider that spontaneous preterm labor is a syndrome for which intra-amniotic infection represents only one known etiology.4 Herein, we focused on preterm labor triggered by intra-amniotic infection with E. coli, a Gram-negative bacterium, using an established animal model that resembles the clinical condition.127 Other known or proposed etiologies for spontaneous preterm labor may have subtle differences in the involved cell types and associated signaling pathways; thus, further characterization of the cellular atlas in each preterm labor subtype is necessary for their distinction. Second, intra-amniotic infection is often polymicrobial and/or can be induced by a variety of bacterial species,20,25 of which Ureaplasma species are the most commonly found in the amniotic cavity;16,17,76,128–130 therefore, the immune responses triggered by each bacterium or cluster of bacteria may differentially affect cellular responses in the reproductive and gestational tissues. However, in vivo standardization of a polymicrobial infection model and isolation of clinically relevant Ureaplasma species are challenging; thus, herein, we utilized intra-amniotic infection with an easily cultured bacterium, E. coli, to induce preterm birth in mice. Future investigations may utilize other bacteria detected in the amniotic cavity of women diagnosed with intra-amniotic infection.18,20 Single-cell RNA-seq is a discovery-based approach that we utilized to generate an atlas of the murine reproductive tissues, and careful interpretation is required when extrapolating specific findings to the human context. Last, RNA techniques using single-cell suspensions lose information about the spatial relationships among cell types in target tissues; therefore, such data may be complemented by using spatial transcriptomics and/or proteomics. Our data serve as a resource for targeted studies that can validate such findings using human samples.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nardhy Gomez-Lopez (nardhy.gomez-lopez@wayne.edu).
Material availability
This study did not generate new unique reagents.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication (see key resources table for accession number). Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication (see key resources table for DOI). Microscopy data reported in this paper will be shared by the lead contact upon reasonable request.
All original code has been deposited at Zenodo and is publicly available as of the date of publication (see key resources table for DOI).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| The Monoclonal Rabbit Anti-Mouse CD45 | Cell Signaling Technology | Cat#70257S; RRID: AB_2799780 |
| Rabbit FLEX Universal Negative Control | Agilent | Cat# IR60066-2 |
| Monoclonal Rabbit Anti-Mouse F4/80 | Cell Signaling Technology | Cat# 70076S; RRID: AB_2799771 |
| Monoclonal Rabbit Anti-Mouse CD3ε | Cell Signaling Technology | Cat# 78588S; RRID: AB_2889902 |
| Monoclonal Rabbit Anti-Mouse Klrb1c/CD161c | Cell Signaling Technology | Cat# 39197S; RRID: AB_2892989 |
| Polyclonal Rabbit Anti-Mouse Ly6C | HuaBio | Cat# HA500088 |
| Monoclonal Rabbit Anti-Mouse Ly6G | Cell Signaling Technology | Cat# 87048S; RRID: AB_2909808 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Escherichia coli | ATCC | ATCC 12014 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Umbilical Cord Dissociation Kit, human | Miltenyi Biotec | Cat# 130-105-737 |
| Dead Cell Removal Kit | Miltenyi Biotec | Cat# 130-090-101 |
| Chromium Next GEM Single Cell 3′ GEM, Library &Gel beads Kit | 10× Genomics | PN:1000121 |
| Chromium Next GEM Chip G Single Cell Kit | 10× Genomics | PN:1000120 |
| Single Index Kit T Set A | 10× Genomics | PN: 1000213 |
| SPRIselect Reagent | Beckman Coulter | Item Number: B23318 |
|
| ||
| Deposited data | ||
|
| ||
| scRNA-sequencing | This paper | Gene Expression Omnibus: GSE200289 |
| Scripts | This paper | Zenodo: https://doi.org/10.5281/zenodo.7335384 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6 | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mouse: BALB/cByJ | The Jackson Laboratory | RRID: IMSR_JAX:001026 |
|
| ||
| Software and algorithms | ||
|
| ||
| Cell Ranger version 4.0.0 | 10× Genomics | http://www.10xgenomics.com |
| STAR aligner | (Dobin et al.)131 | https://github.com/alexdobin/STAR |
| Demuxlet | (Kang et al.)132 | https://github.com/statgen/demuxlet |
| SoupX version 1.5.2 | (Young et al.)133 | https://github.com/constantAmateur/Soup |
| DoubletFinder 2.0.3 | (McGinnis.)134 | |
| Seurat version 4.0.3 | (Stuart et al.)135 (Hafemeister et al.)136 |
https://satijalab.org/seurat/ |
| Harmony package in R version 1.0.0 (R package from CRAN) | (Korsunsky et al.)137 | N/A |
| Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) algorithm | (McInnes et al.)138 (Becht et al.)139 |
N/A |
| SingleR package in R version 1.6.1 | (Aran et al.)140 | https://doi.org/10.18129/B9.bioc.SingleR |
| Mouse Cell Atlas and single-cell MCA (scMCA) package in R version 0.2.0 | (Han et al.)141 | http://bis.zju.edu.cn/MCA/search.html |
| DESeq2 R package version 1.32.0 | (Love et al.)142 | https://doi.org/10.18129/B9.bioc.DESeq2 |
| clusterProfiler in R version 4.0.4 | (Yu et al.)143 | https://doi.org/10.18129/B9.bioc.clusterProfiler |
| CellChat version 1.1.2 (R package from CRAN) | (Jin et al.)113 | N/A |
| ggalluvial version 0.12.3 (R package from CRAN) | N/A | |
| ggplot2 version 3.3.5 (R package from CRAN) | N/A | |
|
| ||
| Other | ||
|
| ||
| Human uterine scRNA-seq data | (Pique-Regi et al.)70 | dbGaP: phs001886.v4.pl |
EXPERIMENTAL MODEL DETAILS
Mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and bred in the animal care facility at the C.S. Mott Center for Human Growth and Development, Wayne State University (Detroit, MI, USA). Mice were under a circadian cycle of light:dark = 12:12 h. Eight- to twelve-week-old C57BL/6 (RRID:IMSR_JAX:000664) female mice were bred with BALB/cByJ male mice (RRI-D:IMSR_JAX:001026), and females were examined daily between 0800 and 0900 to check for the presence of a vaginal plug, which was considered as 0.5 days post coitum (dpc). Upon observation of a vaginal plug, females were removed from mating cages and housed separately, and their weights were monitored daily. At 12.5 dpc, a weight gain of ≥2 g was considered confirmation of pregnancy. Mice were randomized to receive intra-amniotic injection of E. coli or vehicle control (1X phosphate-buffered saline, PBS) (Thermo Fisher Scientific/Gibco, Grand Island, NY, USA), and investigators were not blinded to control or treatment assignment. No experimental mice were excluded from analysis. All procedures and experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Wayne State University under Protocol nos. 18–03-0584 and 21–04-3506.
METHOD DETAILS
Preparation of E. coli for intra-amniotic injection
Escherichia coli was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) (ATCC 12014) and was grown in Luria-Bertani (LB) broth (cat. no. L8050, Teknova, Hollister, CA, USA) at 37°C. From an overnight culture, a sub-culture was placed with fresh LB broth and grown to the logarithmic phase (OD600 0.9–1). Additional dilution was performed using sterile 1X PBS to reach a working concentration of 10 CFU/25 μL.
Measurement of cervical length by ultrasound
Dams were anesthetized on 16.5 dpc by inhalation of 2% isoflurane (Fluriso™ (Isoflurane, USP), VetOne, Boise, ID, USA) and 1 to 2 liters/min of oxygen in an induction chamber. Anesthesia was maintained with a mixture of 1.5–2% isoflurane and 1.5 to 2 liters/min of oxygen. Mice were positioned on a heating pad and stabilized with adhesive tape. Fur removal from the abdomen was performed using Nair cream (Church & Dwight Co., Inc., Ewing, NJ, USA). Sterile forceps were utilized to expose the vulva and 150 μL of Sterile Aquasonic® 100 ultrasound transmission gel (Parker laboratories, Fairfield, NJ, USA) was used to fill the vagina to create contrast and allow for clear visualization of the external limit of the uterine cervix (i.e., the external os). A trans-abdominal ultrasound approach was utilized to evaluate the cervix using the Vevo® 2100 Imaging System (VisualSonics Inc., Toronto, ON, Canada). The transducer was slowly moved toward the lower part of the abdomen and the cervix was positioned in a longitudinal view. The cervical length was measured from the internal to the external os at least three times per mouse, and its average was utilized as the final value for cervical length. This procedure was performed prior to ultrasound-guided injection with either E. coli or PBS and repeated 24 h later (on 17.5 dpc) (i.e., prior to tissue collection). The change in cervical length was determined as a percentage by considering the cervical length on 16.5 dpc as 100% and then calculating the percentage of the cervical length on 17.5 dpc.
Intra-amniotic inoculation with E. coli
Dams that underwent cervical measurement were maintained on the heating pad under anesthesia as described above. The ultrasound transducer was slowly moved toward the abdomen to localize the amniotic sacs. The syringe with E. coli suspension (10 CFU/25 μL) was stabilized by a mechanical holder (VisualSonics). Ultrasound-guided intra-amniotic inoculation with E. coli was performed in each amniotic sac using a 30G needle (BD PrecisionGlide needle; Becton Dickinson, Franklin Lakes, NJ, USA). Controls were injected with 25 μL of sterile 1X PBS into each amniotic sac. After the ultrasound injection, the dams were placed under a heat lamp for recovery (defined as when the mouse resumed normal activities such as walking and responding), which typically occurred 10 min after removal from anesthesia. After recovery, mice were video monitored to observe pregnancy outcomes.
Video monitoring
Pregnancy outcomes were recorded via video camera (Sony Corporation, Tokyo, Japan) to determine gestational length, and therefore rate of preterm birth. Preterm birth was defined as delivery occurring before 18.5 dpc, based on the earliest delivery of PBS-injected control dams, and its rate was represented by the percentage of females delivering preterm among the total number of mice injected.
Tissue collection prior to preterm birth
Dams were euthanized on 17.5 dpc and the reproductive tissues (uterus, decidua, and cervix) were collected. Tissues collected for the preparation of single-cell suspensions were placed in sterile 1X PBS, while tissues for histological analyses were fixed in 10% Neutral Buffered Formalin (Surgipath, Leyca Biosystems, Wetzlar, Germany) and embedded in paraffin. Five-μm-thick sections were cut and mounted on Superfrost® Plus microscope slides (Cat. No. 48311–703, VWR International, LLC. Radnor, PA, USA).
Histological characterization of murine reproductive tissues
Leukocyte detection using DAB immunohistochemistry
Five-μm-thick tissue sections from mice injected with PBS or E. coli were deparaffinized and rehydrated using xylene and a series of decreasing ethanol concentrations, respectively. Immunohistochemistry staining using the Monoclonal Rabbit Anti-Mouse CD45 (AB_2799780; clone D3F8Q, cat. no. 70257S, Cell Signaling Technology, Danvers, MA, USA) was performed using the Leica Bond Max Automatic Staining System in a peroxidase-mediated oxidation of 3,3’-diaminobenzidine (DAB) from the Bond™ Polymer Refine Detection Kit (both from Leica Microsystems, Wetzlar, Germany). The negative control used was the Rabbit FLEX Universal Negative Control (cat. no. IR60066–2, Agilent, Santa Clara, CA, USA). Images were scanned using the Brightfield setting of the Vectra Polaris Multispectral Imaging System (Akoya Biosciences, Marlborough, MA, USA).
Movat’s pentachrome staining
Five-mm-thick tissue sections from mice injected with PBS or E. coli were histologically characterized for the presence of collagen, elastin, muscle, and mucin using the Movat Pentachrome Stain Kit (Modified Russell-Movat; ScyTek Laboratories, Inc. Logan, UT, USA), following manufacturer’s instructions with modifications. Briefly, tissue sections were deparaffinized, stained with working Elastic Stain solution for 20 min, and rinsed in running tap water for 1 min followed by rinsing with deionized water. Then, the following reagents from the kit were sequentially applied to the entire tissue section with distilled water rinsing in between each application: 2% Ferric Chloride for 5–8 s, 5% Sodium Thiosulfate Solution for 1 min, Alcian Blue Solution (pH 2.5) for 20 min, Biebrich Scarlet-Acid Fuchsin Solution for 2 min, 5% Phosphotungstic Acid Solution for 7 min, and 1% Acetic Acid Solution for 3 min. Excess Acetic Acid Solution was drained from the slides and Yellow Stain Solution was immediately applied for 20 min. The slides were then rinsed in 100% ethanol followed by rinsing with xylene. Images were scanned using the Brightfield setting of the Vectra Polaris Multispectral Imaging System.
OPAL multiplex immunofluorescence
OPAL multiplex immunofluorescence staining was performed using the OPAL Multiplex 7-color IHC kit (Cat. no. NEL811001KT; Akoya Biosciences), according to the manufacturer’s instructions. Prior to multiplex staining, the order of antibody staining was optimized using single-plex staining paired with tyramide signal amplification (TSA)-conjugated OPAL fluorophores. The optimized detection panel includes antibody-OPAL fluorophore pairs in the following order: Monoclonal Rabbit Anti-Mouse F4/80 (AB_2799771; clone D2S9R; cat. no. 70076S, Cell Signaling Technology) with OPAL 520, Monoclonal Rabbit Anti-Mouse CD3ε (AB_2889902; clone E4T1B; cat. no. 78588S, Cell Signaling Technology) with OPAL 570, Monoclonal Rabbit Anti-Mouse Klrb1c/CD161c (AB_2892989; clone E6Y9G; cat. no. 39197S, Cell Signaling Technology) with OPAL 620, Polyclonal Rabbit Anti-Mouse Ly6C (cat. no. HA500088, HuaBio, Boston, MA, USA) with OPAL 650, and Monoclonal Rabbit Anti-Mouse Ly6G (AB_2909808; clone E6Z1T; cat. no. 87048S, Cell Signaling Technology) with OPAL 690. The Rabbit FLEX Universal Negative Control (Agilent) was used as isotype. Briefly, 5-μm-thick tissue sections from mice injected with PBS or E. coli were deparaffinized and rehydrated using xylene and a series of decreasing ethanol concentrations, respectively. The slides were rinsed in deionized water and epitope retrieval was performed by submerging the slides in appropriate antigen retrieval (AR) buffer and boiling in a microwave oven. Non-specific binding was prevented by incubating slides in OPAL antibody diluent/blocking solution prior to incubating with each primary antibody at room temperature. Next, the slides were rinsed in TBST prior to incubation with anti-mouse secondary antibody-horse radish peroxidase (HRP) conjugate followed by the selected TSA-conjugated OPAL fluorophore. Cycles of sequential epitope retrieval, target detection, and signal amplification were repeated using the optimized antibody-OPAL fluorophore pair. Once all targets were detected, the slides were incubated with DAPI (4′,6-diamidino-2-phenylindole) as a nuclear counterstain and mounted using AquaSlip™ Aqueous Permanent Mounting Medium (American MasterTech). Fluorescence image acquisition was performed using the Vectra Polaris Multispectral Imaging System at 20x magnification. Multispectral images were analyzed using the inForm software version 2.4 (Akoya Biosciences).
Tissue dissociation of murine uterus and decidua
Immediately following tissue collection, the uterus and decidua were dissociated to prepare single-cell suspensions. The tissues were mechanically dissociated and enzymatically digested by incubating at 37°C using enzymes from the Umbilical Cord Dissociation Kit (Miltenyi Biotec). A second round of mechanical dissociation was performed using the gentleMACS Dissociator (Miltenyi Biotec), and dissociated cells were rinsed with 1X PBS prior to filtration using a 100 μm cell strainer (Miltenyi Biotec). Filtered cells were pelleted by centrifugation at 300 × g for 5 min, erythrocytes were eliminated using ACK Lysing Buffer (Life Technologies), and the cells were rinsed in 0.04% Bovine Serum Albumin (BSA, Sigma Aldrich) and 0.5 mM EDTA (Sigma Aldrich) diluted in 1X PBS. Finally, the cells were filtered using a 30 μm cell strainer (Miltenyi Biotec), and the Dead Cell Removal Kit (Miltenyi Biotec) was used to remove dead cells to obtain a cell viability of ≥80%.
Tissue dissociation of the murine cervix
Immediately following the collection of the cervix, the tissue was mechanically dissociated and enzymatically digested using Collagenase A (160 mg/mL) (Sigma Aldrich) and incubated at 37°C. Then, the dissociated cells were pelleted by centrifugation at 16,000 × g for 10 min at 20°C and resuspended with 0.05% trypsin-EDTA (Thermo Fisher Scientific, Waltham, MA) prior to a second round of mincing and incubation in 0.05% trypsin-EDTA at 37°C. The enzymatic reaction was stopped by the addition of FBS (Fetal Bovine Serum, Thermo Fisher). Cells were then filtered using a 70 μm cell strainer (Miltenyi Biotec, San Diego, CA, USA) and pelleted by centrifugation at 300 × g for 10 min. Erythrocytes were removed using ACK Lysing Buffer (Life Technologies, Grand Island, NY, USA). Finally, the cells were resuspended in 0.04% BSA diluted in 1X PBS and filtered through a 30 μm cell strainer. The cell concentration and viability were determined using an automatic cell counter (Cellometer Auto 2000, Nexcelom Bioscience, Lawrence, MA, USA) and the Dead Cell Removal Kit was used to remove dead cells to obtain a cell viability of ≥80%.
Generation of gel beads-in-emulsion (GEMs) and library preparation
Generation of gel beads-in-emulsion (GEMs) and preparation of library constructs was performed on viable single-cell suspensions using the 10x Genomics Chromium Single Cell 3′ Gene Expression Version 3.1 Kit (10x Genomics, Pleasanton, CA, USA), according to the manufacturer’s instructions. Briefly, viable single cells were encapsulated in partitioning oil together with a single Gel Bead with barcoded oligonucleotides within the Chromium Controller. Reverse transcription of mRNA into complementary (c)DNA was performed using the Veriti 96-well Thermal Cycler (Thermo Fisher Scientific, Wilmington, DE, USA). Dynabeads MyOne SILANE (10x Genomics) and the SPRIselect Reagent (Beckman Coulter, Indianapolis, IN, USA) were used to purify resulting cDNA, which was optimized by enzymatic fragmentation, end-repair, and A-tailing. Next, adaptors and sample index were incorporated by ligation. The sample index PCR product was then amplified using the Veriti 96-well Thermal Cycler and double-sided size selection was performed using the SPRIselect Reagent. Following the formation of cDNA and final library construct, the Agilent Bioanalyzer High Sensitivity DNA Chip (Agilent Technologies, Wilmington, DE, USA) was used determine sample quality and concentration.
Sequencing
Prior to sequencing of post-library constructs, samples were quantified using the Kapa DNA Quantification Kit for Illumina platforms (Kapa Biosystems, Wilmington, MA, USA), following the manufacturer’s instructions. The sequencing of 10x scRNA-seq libraries was performed on the Illumina NextSeq 500 at the Genomics Services Center (GSC) of the Center for Molecular Medicine and Genetics (Wayne State University School of Medicine, Detroit, MI, USA). The Illumina 75 Cycle Sequencing Kit (Illumina, San Diego, CA, USA) was used with 58 cycles for R2, 26 for R1, and 8 for I1.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical details of all analyses are described in the figure legends or STAR Methods.
scRNA-seq data normalization and pre-processing
Sequencing data were processed using Cell Ranger version 4.0.0 (10x Genomics). The “cellranger counts” was also used to align the scRNA-seq reads by using the STAR aligner131 to produce the bam files necessary for demultiplexing the individual of origin based on genotype information using demuxlet132 and a custom vcf file. The genotype data were downloaded from ftp://ftp-mouse.sanger.ac.uk/current_snps/mgp.v5.merged.snps_all.dbSNP142.vcf.gz, the strains C57BL_6NJ and BALB_cJ were extracted, and a new synthetic vcf file was generated consisting of all the genetic variants where these two strains diverge, and containing a maternal genotype column identical to the C57BL_6NJ strain and a fetal genotype column with a “0/1” heterozygote genotype. Ambient RNA contamination and doublets were removed using SoupX version 1.5.2133 and DoubletFinder 2.0.3.134 Additionally, any cell with <200 genes or >20,000 genes detected, or that had >10% mitochondrial reads, was excluded (Table S4). All count data matrices were then normalized and combined using the Seurat package in R (Seurat version 4.0.3).135,136 The first 100 principal components were obtained, and the different libraries were integrated and harmonized using the Harmony package in R version 1.0.0137.The top 30 harmony components were then processed to embed and visualize the cells in a two-dimensional map via the Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) algorithm.138,139 A resolution of 0.5 was used to cluster the single cells.
Annotation of cell types
The SingleR140 package in R version 1.6.1 was used to annotate cell types based on their similarities to reference datasets with known labels.144,145 SingleR annotates single cells from query data by computing the Spearman’s correlation coefficient between the single-cell gene expression data and samples from the reference dataset. The correlation is measured only based on the variable genes in the reference dataset. The multiple correlation coefficients per cell type are combined according to the cell type labels of the reference dataset to assign a score per cell type. Additionally, we confirmed the cell type identities by identifying the top DEGs (see below) and the gene-cell type mapping data provided by the Mouse Cell Atlas and single-cell MCA (scMCA) package141 in R version 0.2.0. Using different annotations obtained from the reference mapping workflows, the final cell type labels were assigned based on a majority vote. If multiple clusters were assigned to the same consensus cell type, we added a sub-index to that cell type for each different original Seurat cluster: Clusters 0, 1, and 21 were annotated as Fibroblast-1, Fibroblast-2, and Fibroblast-3; clusters 2, 4, and 12 were annotated as Stromal-1, Stromal-2 (Decidua), and Stromal-3; clusters 5, 7, 8, 10, 11, 13, 14, 20, 23, and 28 were annotated as Epithelial-1 (basal), Epithelial-2 (squamous), Epithelial-3 (squamous), Epithelial-4 (glandular), Epithelial-10 (proliferative), Epithelial-5 (luminal), Epithelial-6 (secretory), Epithelial-7 (glandular), Epithelial-8 (Enterocyte), and Epithelial-9 (Secretory); clusters 9 and 24 were annotated as Macrophage-1 and Macrophage-2 (progenitor); clusters 15 and 26 were annotated as SMC-1 and SMC-2; and clusters 19 and 27 were annotated as NK-cell-1 and NK-cell-2. All remaining clusters were assigned a unique cell type identifier (Table S5).
Differential gene expression for cell type analysis
For this analysis, the differential expression of selected marker genes for each cell type/cluster was identified using the Wilcoxon Rank Sum test and the FindAllMarkers function from Seurat (Table S5). For this analysis, we compared each cluster to all cell types. We further used the top cell markers [ranked based on log2(Fold change) and requiring q < 0.1] assigned to each sub-cluster to annotate the clusters using the Mouse Cell Atlas and scMCA package.141
Differential gene expression in preterm labor
The identification of preterm labor-associated DEGs between study groups was performed using the DESeq2 R package version 1.32.0.142 A term for each library was added to the DESeq2 model to correct for technical batch effects (library identifier). For each cell type/replicate combination, we only used combinations with more than 20 cells; otherwise, it was treated as non-observed. Cell types found in <3 combinations per study group were dropped from the differential gene expression analysis (Table S2 contains all genes determined as differentially expressed). Note that these thresholds imply that clusters with <120 cells are not analyzed to ensure robust gene expression estimation. Quantile-quantile plots were used to show that p-values are well calibrated under the null hypothesis of no effect of preterm labor, and also to show which tissues and cell types are more enriched for preterm labor-associated gene expression changes (Figures 2H–2J). Multiple comparison correction was performed by controlling for false discovery rate using Benjamini-Hochberg’s method and genes with q < 0.1 were reported in Figures 2E–2G and Table S2. Statistical difference between the fraction of genes that were upregulated versus downregulated by preterm labor in each cell type was assessed with a binomial test and corrected for multiple comparisons using Benjamini-Hochberg’s method. To compare the effects of preterm labor on gene expression across different tissues and cell types, we performed Spearman’s correlation between the log2FC obtained in each DESeq2 analysis performed using genes that had been detected as differentially expressed in at least one cell-type/tissue, q < 0.1. These correlations were visualized as a heatmap in Figures S5A, S6A, and S7A and in boxplots for relevant tissue and cell-type combinations in Figures 3B–3D.
Gene ontology and pathway enrichment analysis of genes affected by preterm labor
The clusterProfiler in R version 4.0.4143 was used to perform the Over-Representation Analysis (ORA) separately for each list of genes obtained as differentially expressed for each cell type based on the Gene Ontology (GO), Kyoto Encyclopedia of Gene and Genomes (KEGG), and Reactome databases. The functions “enrichPathway”, “enrichKEGG”, and “enrichGO”, from “clusterProfiler” were used. In ORA analyses, the universe of genes for each cell type was the subset that was expressed at a level sufficient to be tested in differential gene expression analysis. When results are combined across cell types, any genes tested (with a calculated p-value) in any of the cell types are used for the universe. Only ORA results that were significant after correction were reported with q < 0.05 being considered statistically significant.
Cell-cell communication analysis
CellChat113 was used to infer the cell-cell communications using the single-cell gene expression data from preterm labor and control conditions and a database of prior knowledge of the interactions between signaling ligands, receptors, and their cofactors. The top 25% of significant cell-cell communications (p < 0.05) across different pathways were shown for the two conditions of preterm labor and control. Next, the aggregated cell-cell communication between different cell groups was calculated for the two study groups, and the interaction strength was compared among different cell types from the two study groups. The differential interaction strength was represented with circle plots with red (or blue) edges showing the increased (or decreased) signaling in preterm labor compared to controls. Additionally, the detailed differential interaction strengths were shown using heatmap representations. Major signaling sender and receiver cells were displayed using scatter plots where the changes in signaling strength from control to preterm labor were represented by arrows. The R packages CellChat version 1.1.2, ggalluvial version 0.12.3, and ggplot2 version 3.3.5 were used to visualize cell-cell communication analyses. The major sending and receiving signaling roles based on context-specific pathways across different cell groups were identified using a cut-off of 0.5 when visualizing the connection. The overall information flow [sum of the significant communication probability (p < 0.05) in the inferred cell-cell network] for each signaling network was represented using a bar plot. The comparison between the overall information flow from the two study groups (preterm labor and control) was performed using the paired Wilcoxon test with the function “rankNet” from CellChat.
Comparison between cell-cell communication in human and murine uterine tissues
We inferred cell-cell communications using the human myometrial single-cell gene expression data from term in labor and term not in labor study groups,70 and compared the inferred interactions between mouse (uterus) and human (myometrium) across the top common signaling pathways with highest numbers of DEGs.
Statistical analysis
Observational mouse data were analyzed by using SPSS v19.0 and GraphPad Prism version 8. For comparing the rates of preterm birth, the Fisher’s exact test was used. For gestational length and cervical shortening, the statistical significance of group comparisons was assessed using the Mann-Whitney U-test.
Supplementary Material
Highlights.
Single-cell atlas of the murine uterus, decidua, and cervix in preterm labor
Preterm labor alters shared and tissue-specific single-cell gene expression
Preterm labor drives communication among specific cell types across tissues
Labor-associated signaling pathways are conserved between murine and human uterus
ACKNOWLEDGMENTS
The authors would like to acknowledge Yesong Liu for assistance with performing some of the immunofluorescence imaging. This research was supported by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS) under contract HHSN275201300006C. R.R. contributed to this work as part of his official duties as an employee of the United States Federal Government. This research was also supported by the Wayne State University Perinatal Initiative in Maternal, Perinatal, and Child Health. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. Figures include art created with BioRender.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111846.
REFERENCES
- 1.Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, Cousens S, Mathers C, and Black RE (2015). Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 385, 430–440. 10.1016/s0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 2.Chawanpaiboon S, Vogel JP, Moller AB, Lumbiganon P, Petzold M, Hogan D, Landoulsi S, Jampathong N, Kongwattanakul K, Laopaiboon M, et al. (2019). Global, regional, and national estimates of levels of preterm birth in 2014: a systematic review and modelling analysis. Lancet Glob. Health 7, e37–e46. 10.1016/s2214-109x(18)30451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldenberg RL, Culhane JF, Iams JD, and Romero R (2008). Epidemiology and causes of preterm birth. Lancet 371, 75–84. 10.1016/s0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero R, Dey SK, and Fisher SJ (2014). Preterm labor: one syndrome, many causes. Science 345, 760–765. 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibbs RS, Romero R, Hillier SL, Eschenbach DA, and Sweet RL (1992). A review of premature birth and subclinical infection. Am. J. Obstet. Gynecol. 166, 1515–1528. 10.1016/0002-9378(92)91628-n. [DOI] [PubMed] [Google Scholar]
- 6.Gomez R, Ghezzi F, Romero R, Muñoz H, Tolosa JE, and Rojas I (1995). Premature labor and intra-amniotic infection. Clinical aspects and role of the cytokines in diagnosis and pathophysiology. Clin. Perinatol. 22, 281–342. [PubMed] [Google Scholar]
- 7.Gonç alves LF., Chaiworapongsa T., and Romero R. (2002). Intrauterine infection and prematurity. Ment. Retard. Dev. Disabil. Res. Rev. 8, 3–13. 10.1002/mrdd.10008. [DOI] [PubMed] [Google Scholar]
- 8.Keelan JA, Blumenstein M, Helliwell RJA, Sato TA, Marvin KW, and Mitchell MD (2003). Cytokines, prostaglandins and parturition–a review. Placenta 24, S33–S46. 10.1053/plac.2002.0948. [DOI] [PubMed] [Google Scholar]
- 9.Bastek JA, Gómez LM, and Elovitz MA (2011). The role of inflammation and infection in preterm birth. Clin. Perinatol. 38, 385–406. 10.1016/j.clp.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal V, and Hirsch E (2012). Intrauterine infection and preterm labor. Semin. Fetal Neonatal Med. 17, 12–19. 10.1016/j.siny.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Combs CA, Gravett M, Garite TJ, Hickok DE, Lapidus J, Porreco R, Rael J, Grove T, Morgan TK, Clewell W, et al. (2014). Amniotic fluid infection, inflammation, and colonization in preterm labor with intact membranes. Am. J. Obstet. Gynecol. 210, 125.e1–125.e15. 10.1016/j.ajog.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 12.Romero R, Gomez-Lopez N, Winters AD, Jung E, Shaman M, Bieda J, Panaitescu B, Pacora P, Erez O, Greenberg JM, et al. (2019). Evidence that intra-amniotic infections are often the result of an ascending invasion - a molecular microbiological study. J. Perinat. Med. 47, 915–931. 10.1515/jpm-2019-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theis KR, Romero R, Motomura K, Galaz J, Winters AD, Pacora P, Miller D, Slutsky R, Florova V, Levenson D, et al. (2020). Microbial burden and inflammasome activation in amniotic fluid of patients with preterm prelabor rupture of membranes. J. Perinat. Med. 48, 115–131. 10.1515/jpm-2019-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbs RS, Blanco JD, St Clair PJ, and Castaneda YS (1982). Quantitative bacteriology of amniotic fluid from women with clinical intraamniotic infection at term. J. Infect. Dis. 145, 1–8. 10.1093/infdis/145.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Gravett MG, Hummel D, Eschenbach DA, and Holmes KK (1986). Preterm labor associated with subclinical amniotic fluid infection and with bacterial vaginosis. Obstet. Gynecol. 67, 229–237. 10.1097/00006250-198602000-00013. [DOI] [PubMed] [Google Scholar]
- 16.Romero R, Sirtori M, Oyarzun E, Avila C, Mazor M, Callahan R, Sabo V, Athanassiadis AP, and Hobbins JC (1989). Infection and labor. V. Prevalence, microbiology, and clinical significance of intraamniotic infection in women with preterm labor and intact membranes. Am. J. Obstet. Gynecol. 161, 817–824. 10.1016/0002-9378(89)90409-2. [DOI] [PubMed] [Google Scholar]
- 17.Yoon BH, Chang JW, and Romero R (1998). Isolation of Ureaplasma urealyticum from the amniotic cavity and adverse outcome in preterm labor. Obstet. Gynecol. 92, 77–82. 10.1016/s0029-7844(98)00122-7. [DOI] [PubMed] [Google Scholar]
- 18.DiGiulio DB, Romero R, Kusanovic JP, Gómez R, Kim CJ, Seok KS, Gotsch F, Mazaki-Tovi S, Vaisbuch E, Sanders K, et al. (2010). Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am. J. Reprod. Immunol. 64, 38–57. 10.1111/j.1600-0897.2010.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, et al. (2014). A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes. Am. J. Reprod. Immunol. 71, 330–358. 10.1111/aji.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero R, Miranda J, Kusanovic JP, Chaiworapongsa T, Chaemsaithong P, Martinez A, Gotsch F, Dong Z, Ahmed AI, Shaman M, et al. (2015). Clinical chorioamnionitis at term I: microbiology of the amniotic cavity using cultivation and molecular techniques. J. Perinat. Med. 43, 19–36. 10.1515/jpm-2014-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, et al. (2015). Sterile intra-amniotic inflammation in asymptomatic patients with a sonographic short cervix: prevalence and clinical significance. J. Matern. Fetal Neonatal Med. 28, 1343–1359. 10.3109/14767058.2014.954243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero R, Miranda J, Chaemsaithong P, Chaiworapongsa T, Kusanovic JP, Dong Z, Ahmed AI, Shaman M, Lannaman K, Yoon BH, et al. (2015). Sterile and microbial-associated intra-amniotic inflammation in preterm prelabor rupture of membranes. J. Matern. Fetal Neonatal Med. 28, 1394–1409. 10.3109/14767058.2014.958463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh KJ, Romero R, Park JY, Lee J, Conde-Agudelo A, Hong JS, and Yoon BH (2019). Evidence that antibiotic administration is effective in the treatment of a subset of patients with intra-amniotic infection/inflammation presenting with cervical insufficiency. Am. J. Obstet. Gynecol. 221, 140.e1–140.e18. 10.1016/j.ajog.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon BH, Romero R, Park JY, Oh KJ, Lee J, Conde-Agudelo A, and Hong JS (2019). Antibiotic administration can eradicate intra-amniotic infection or intra-amniotic inflammation in a subset of patients with preterm labor and intact membranes. Am. J. Obstet. Gynecol. 221, 142.e1–142.e22. 10.1016/j.ajog.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnham P, Gomez-Lopez N, Heyang M, Cheng AP, Lenz JS, Dadhania DM, Lee JR, Suthanthiran M, Romero R, and De Vlaminck I (2020). Separating the signal from the noise in metagenomic cell-free DNA sequencing. Microbiome 8, 18. 10.1186/s40168-020-0793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romero R, Quintero R, Nores J, Avila C, Mazor M, Hanaoka S, Hagay Z, Merchant L, and Hobbins JC (1991). Amniotic fluid white blood cell count: a rapid and simple test to diagnose microbial invasion of the amniotic cavity and predict preterm delivery. Am. J. Obstet. Gynecol. 165, 821–830. 10.1016/0002-9378(91)90423-o. [DOI] [PubMed] [Google Scholar]
- 27.Romero R, Yoon BH, Mazor M, Gomez R, Diamond MP, Kenney JS, Ramirez M, Fidel PL, Sorokin Y, Cotton D, et al. (1993). The diagnostic and prognostic value of amniotic fluid white blood cell count, glucose, interleukin-6, and gram stain in patients with preterm labor and intact membranes. Am. J. Obstet. Gynecol. 169, 805–816. 10.1016/0002-9378(93)90009-8. [DOI] [PubMed] [Google Scholar]
- 28.Romero R, Yoon BH, Mazor M, Gomez R, Gonzalez R, Diamond MP, Baumann P, Araneda H, Kenney JS, Cotton DB, et al. (1993). A comparative study of the diagnostic performance of amniotic fluid glucose, white blood cell count, interleukin-6, and gram stain in the detection of microbial invasion in patients with preterm premature rupture of membranes. Am. J. Obstet. Gynecol. 169, 839–851. 10.1016/0002-9378(93)90014-a. [DOI] [PubMed] [Google Scholar]
- 29.Gomez R, Romero R, Galasso M, Behnke E, Insunza A, and Cotton DB (1994). The value of amniotic fluid interleukin-6, white blood cell count, and gram stain in the diagnosis of microbial invasion of the amniotic cavity in patients at term. Am. J. Reprod. Immunol. 32, 200–210. 10.1111/j.1600-0897.1994.tb01115.x. [DOI] [PubMed] [Google Scholar]
- 30.Yoon BH, Yang SH, Jun JK, Park KH, Kim CJ, and Romero R (1996). Maternal blood C-reactive protein, white blood cell count, and temperature in preterm labor: a comparison with amniotic fluid white blood cell count. Obstet. Gynecol. 87, 231–237. 10.1016/0029-7844(95)00380-0. [DOI] [PubMed] [Google Scholar]
- 31.Gomez-Lopez N, Romero R, Xu Y, Leng Y, Garcia-Flores V, Miller D, Jacques SM, Hassan SS, Faro J, Alsamsam A, et al. (2017). Are amniotic fluid neutrophils in women with intraamniotic infection and/or inflammation of fetal or maternal origin? Am. J. Obstet. Gynecol. 217, 693.e1–693.e16. 10.1016/j.ajog.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Varea A, Romero R, Xu Y, Miller D, Ahmed AI, Chaemsaithong P, Chaiyasit N, Yeo L, Shaman M, Lannaman K, et al. (2017). Clinical chorioamnionitis at term VII: the amniotic fluid cellular immune response. J. Perinat. Med. 45, 523–538. 10.1515/jpm-2016-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez-Lopez N, Romero R, Xu Y, Miller D, Leng Y, Panaitescu B, Silva P, Faro J, Alhousseini A, Gill N, et al. (2018). The immunophenotype of amniotic fluid leukocytes in normal and complicated pregnancies. Am. J. Reprod. Immunol. 79, e12827. 10.1111/aji.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomez-Lopez N, Romero R, Xu Y, Miller D, Arenas-Hernandez M, Garcia-Flores V, Panaitescu B, Galaz J, Hsu CD, Para R, and Berry SM (2019). Fetal T cell activation in the amniotic cavity during preterm labor: a potential mechanism for a subset of idiopathic preterm birth. J. Immunol. 203, 1793–1807. 10.4049/jimmunol.1900621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Lopez N, Romero R, Leng Y, Xu Y, Slutsky R, Levenson D, Pacora P, Jung E, Panaitescu B, and Hsu CD (2019). The origin of amniotic fluid monocytes/macrophages in women with intra-amniotic inflammation or infection. J. Perinat. Med. 47, 822–840. 10.1515/jpm-2019-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Lopez N, Romero R, Galaz J, Xu Y, Panaitescu B, Slutsky R, Motomura K, Gill N, Para R, Pacora P, et al. (2019). Cellular immune responses in amniotic fluid of women with preterm labor and intraamniotic infection or intra-amniotic inflammation. Am. J. Reprod. Immunol. 82, e13171. 10.1111/aji.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galaz J, Romero R, Xu Y, Miller D, Slutsky R, Levenson D, Hsu CD, and Gomez-Lopez N (2020). Cellular immune responses in amniotic fluid of women with preterm clinical chorioamnionitis. Inflamm. Res. 69, 203–216. 10.1007/s00011-019-01308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galaz J, Romero R, Slutsky R, Xu Y, Motomura K, Para R, Pacora P, Panaitescu B, Hsu CD, Kacerovsky M, and Gomez-Lopez N (2020). Cellular immune responses in amniotic fluid of women with preterm prelabor rupture of membranes. J. Perinat. Med. 48, 222–233. 10.1515/jpm-2019-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez-Lopez N, Romero R, Varrey A, Leng Y, Miller D, Done B, Xu Y, Bhatti G, Motomura K, Gershater M, et al. (2021). RNA sequencing reveals diverse functions of amniotic fluid neutrophils and monocytes/macrophages in intra-amniotic infection. J. Innate Immun. 13, 63–82. 10.1159/000509718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olding L (1970). Value of placentitis as a sign of intrauterine infection in human subjects. Acta Pathol. Microbiol. Scand. 78, 256–264. 10.1111/j.1699-0463.1970.tb03300.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang JM, Kraus FT, and Aquino TI (1985). Chorioamnionitis: a comparative histologic, bacteriologic, and clinical study. Int. J. Gynecol. Pathol. 4, 1–10. [PubMed] [Google Scholar]
- 42.Guzick DS, and Winn K (1985). The association of chorioamnionitis with preterm delivery. Obstet. Gynecol. 65, 11–16. [PubMed] [Google Scholar]
- 43.Svensson L, Ingemarsson I, and Mårdh PA (1986). Chorioamnionitis and the isolation of microorganisms from the placenta. Obstet. Gynecol. 67, 403–409. [PubMed] [Google Scholar]
- 44.Hillier SL, Martius J, Krohn M, Kiviat N, Holmes KK, and Eschenbach DA (1988). A case-control study of chorioamnionic infection and histologic chorioamnionitis in prematurity. N. Engl. J. Med. 319, 972–978. 10.1056/nejm198810133191503. [DOI] [PubMed] [Google Scholar]
- 45.van Hoeven KH, Anyaegbunam A, Hochster H, Whitty JE, Distant J, Crawford C, and Factor SM (1996). Clinical significance of increasing histologic severity of acute inflammation in the fetal membranes and umbilical cord. Pediatric pathology & laboratory medicine. Pediatr. Pathol. Lab. Med. 16, 731–744. [PubMed] [Google Scholar]
- 46.Odibo AO, Rodis JF, Sanders MM, Borgida AF, Wilson M, Egan JF, and Campbell WA (1999). Relationship of amniotic fluid markers of intra-amniotic infection with histopathology in cases of preterm labor with intact membranes. J. Perinatol. 19, 407–412. 10.1038/sj.jp.7200210. [DOI] [PubMed] [Google Scholar]
- 47.Yoon BH, Romero R, Moon JB, Shim SS, Kim M, Kim G, and Jun JK (2001). Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am. J. Obstet. Gynecol. 185, 1130–1136. 10.1067/mob.2001.117680. [DOI] [PubMed] [Google Scholar]
- 48.Pacora P, Chaiworapongsa T, Maymon E, Kim YM, Gomez R, Yoon BH, Ghezzi F, Berry SM, Qureshi F, Jacques SM, et al. (2002). Funisitis and chorionic vasculitis: the histological counterpart of the fetal inflammatory response syndrome. J. Matern. Fetal Neonatal Med. 11, 18–25. 10.1080/jmf.11.1.18.25. [DOI] [PubMed] [Google Scholar]
- 49.Redline RW, Faye-Petersen O, Heller D, Qureshi F, Savell V, and Vogler C Society for Pediatric Pathology; Perinatal Section; Amniotic Fluid Infection Nosology Committee (2003). Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns. Pediatr. Dev. Pathol. 6, 435–448. 10.1007/s10024-003-7070-y. [DOI] [PubMed] [Google Scholar]
- 50.Kim MJ, Romero R, Gervasi MT, Kim JS, Yoo W, Lee DC, Mittal P, Erez O, Kusanovic JP, Hassan SS, and Kim CJ (2009). Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intra-amniotic infection. Lab. Invest. 89, 924–936. 10.1038/labinvest.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park CW, Moon KC, Park JS, Jun JK, Romero R, and Yoon BH (2009). The involvement of human amnion in histologic chorioamnionitis is an indicator that a fetal and an intra-amniotic inflammatory response is more likely and severe: clinical implications. Placenta 30, 56–61. 10.1016/j.placenta.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oh KJ, Park KH, Kim SN, Jeong EH, Lee SY, and Yoon HY (2011). Predictive value of intra-amniotic and serum markers for inflammatory lesions of preterm placenta. Placenta 32, 732–736. 10.1016/j.placenta.2011.07.080. [DOI] [PubMed] [Google Scholar]
- 53.Redline RW (2012). Inflammatory response in acute chorioamnionitis. Semin. Fetal Neonatal Med. 17, 20–25. 10.1016/j.siny.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Kim CJ, Romero R, Chaemsaithong P, Chaiyasit N, Yoon BH, and Kim YM (2015). Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am. J. Obstet. Gynecol. 213, S29–S52. 10.1016/j.ajog.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim SM, Romero R, Park JW, Oh KJ, Jun JK, and Yoon BH (2015). The relationship between the intensity of intra-amniotic inflammation and the presence and severity of acute histologic chorioamnionitis in preterm gestation. J. Matern. Fetal Neonatal Med. 28, 1500–1509. 10.3109/14767058.2014.961009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomez-Lopez N, Romero R, Leng Y, Garcia-Flores V, Xu Y, Miller D, and Hassan SS (2017). Neutrophil extracellular traps in acute chorioamnionitis: a mechanism of host defense. Am. J. Reprod. Immunol, 77. 10.1111/aji.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oh JW, Moon KC, Park CW, Park JS, and Jun JK (2021). Acute chorioamnionitis and intra-amniotic inflammation are more severe according to outside-in neutrophil migration within the same choriodecidua. Taiwan. J. Obstet. Gynecol. 60, 639–652. 10.1016/j.tjog.2021.05.011. [DOI] [PubMed] [Google Scholar]
- 58.Keski-Nisula LT, Aalto ML, Kirkinen PP, Kosma VM, and Heinonen ST (2003). Myometrial inflammation in human delivery and its association with labor and infection. Am. J. Clin. Pathol. 120, 217–224. 10.1309/kc6k-dtx9-8lfy-b3j7. [DOI] [PubMed] [Google Scholar]
- 59.Migale R, MacIntyre DA, Cacciatore S, Lee YS, Hagberg H, Herbert BR, Johnson MR, Peebles D, Waddington SN, and Bennett PR (2016). Modeling hormonal and inflammatory contributions to preterm and term labor using uterine temporal transcriptomics. BMC Med. 14, 86. 10.1186/s12916-016-0632-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willcockson AR, Nandu T, Liu CL, Nallasamy S, Kraus WL, and Mahendroo M (2018). Transcriptome signature identifies distinct cervical pathways induced in lipopolysaccharide-mediated preterm birth. Biol. Reprod. 98, 408–421. 10.1093/biolre/iox180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Motomura K, Romero R, Tarca AL, Galaz J, Bhatti G, Done B, Arenas-Hernandez M, Levenson D, Slutsky R, Hsu CD, and Gomez-Lopez N (2020). Pregnancy-specific transcriptional changes upon endotoxin exposure in mice. J. Perinat. Med. 48, 700–722. 10.1515/jpm-2020-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toothaker JM, Presicce P, Cappelletti M, Stras SF, McCourt CC, Chougnet CA, Kallapur SG, and Konnikova L (2020). Immune cells in the placental villi contribute to intra-amniotic inflammation. Front. Immunol. 11, 866. 10.3389/fimmu.2020.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson AC, Mould AW, Bikoff EK, and Robertson EJ (2016). Single-cell RNA-seq reveals cell type-specific transcriptional signatures at the maternal-foetal interface during pregnancy. Nat. Commun. 7, 11414. 10.1038/ncomms11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pavličev M, Wagner GP, Chavan AR, Owens K, Maziarz J, Dunn-Fletcher C, Kallapur SG, Muglia L, and Jones H (2017). Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal-fetal interface. Genome Res. 27, 349–361. 10.1101/gr.207597.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang P, Lui KO, Ni YB, To KF, Cheng YKY, Chiu RWK, and Lo YMD (2017). Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc. Natl. Acad. Sci. USA 114, E7786–E7795. 10.1073/pnas.1710470114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park JE, Stephenson E, Polański K, Goncalves A, et al. (2018). Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353. 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, Kustagi M, Tuschl T, and Williams Z (2018). A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 4, eaau4788. 10.1126/sciadv.aau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pique-Regi R, Romero R, Tarca AL, Sendler ED, Xu Y, Garcia-Flores V, Leng Y, Luca F, Hassan SS, and Gomez-Lopez N (2019). Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 8, e52004. 10.7554/eLife.52004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang J, Li Q, Peng Q, Xie Y, Wang W, Pei C, Zhao Y, Liu R, Huang L, Li T, et al. (2021). Single-cell RNA sequencing reveals heterogeneity and differential expression of decidual tissues during the peripartum period. Cell Prolif 54, e12967. 10.1111/cpr.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pique-Regi R, Romero R, Garcia-Flores V, Peyvandipour A, Tarca AL, Pusod E, Galaz J, Miller D, Bhatti G, Para R, et al. (2022). A single-cell atlas of the myometrium in human parturition. JCI Insight 7, e153921. 10.1172/jci.insight.153921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tarca AL, Romero R, Xu Z, Gomez-Lopez N, Erez O, Hsu CD, Hassan SS, and Carey VJ (2019). Targeted expression profiling by RNA-Seq improves detection of cellular dynamics during pregnancy and identifies a role for T cells in term parturition. Sci. Rep. 9, 848. 10.1038/s41598-018-36649-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tarca AL, Romero R, Erez O, Gudicha DW, Than NG, Benshalom-Tirosh N, Pacora P, Hsu CD, Chaiworapongsa T, Hassan SS, and Gomez-Lopez N (2021). Maternal whole blood mRNA signatures identify women at risk of early preeclampsia: a longitudinal study. J. Matern. Fetal Neonatal Med. 34, 3463–3474. 10.1080/14767058.2019.1685964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gomez-Lopez N, Romero R, Galaz J, Bhatti G, Done B, Miller D, Ghita C, Motomura K, Farias-Jofre M, Jung E, et al. (2022). Transcriptome changes in maternal peripheral blood during term parturition mimic perturbations preceding spontaneous preterm birth. Biol. Reprod. 106, 185–199. 10.1093/biolre/ioab197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Senthamaraikannan P, Presicce P, Rueda CM, Maneenil G, Schmidt AF, Miller LA, Waites KB, Jobe AH, Kallapur SG, and Chougnet CA (2016). Intra-amniotic Ureaplasma parvum-induced maternal and fetal inflammation and immune responses in rhesus macaques. J. Infect. Dis. 214, 1597–1604. 10.1093/infdis/jiw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faro J, Romero R, Schwenkel G, Garcia-Flores V, Arenas-Hernandez M, Leng Y, Xu Y, Miller D, Hassan SS, and Gomez-Lopez N (2019). Intra-amniotic inflammation induces preterm birth by activating the NLRP3 inflammasome. Biol. Reprod. 100, 1290–1305. 10.1093/biolre/ioy261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Motomura K, Romero R, Xu Y, Theis KR, Galaz J, Winters AD, Slutsky R, Garcia-Flores V, Zou C, Levenson D, et al. (2020). Intraamniotic infection with Ureaplasma parvum causes preterm birth and neonatal mortality that are prevented by treatment with clarithromycin. mBio 11, 007977–20. 10.1128/mBio.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Motomura K, Romero R, Galaz J, Tarca AL, Done B, Xu Y, Leng Y, Garcia-Flores V, Arenas-Hernandez M, Theis KR, et al. (2021). RNA sequencing reveals distinct immune responses in the chorioamniotic membranes of women with preterm labor and microbial or sterile intra-amniotic inflammation. Infect. Immun. 89, e00819–20. 10.1128/IAI.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoneda N, Yoneda S, Niimi H, Ueno T, Hayashi S, Ito M, Shiozaki A, Urushiyama D, Hata K, Suda W, et al. (2016). Polymicrobial amniotic fluid infection with mycoplasma/ureaplasma and other bacteria induces severe intra-amniotic inflammation associated with poor perinatal prognosis in preterm labor. Am. J. Reprod. Immunol. 75, 112–125. 10.1111/aji.12456. [DOI] [PubMed] [Google Scholar]
- 79.Romero R, Mazor M, Munoz H, Gomez R, Galasso M, and Sherer DM (1994). The preterm labor syndrome. Ann. N. Y. Acad. Sci. 734, 414–429. 10.1111/j.1749-6632.1994.tb21771.x. [DOI] [PubMed] [Google Scholar]
- 80.Norwitz ER, Robinson JN, and Challis JR (1999). The control of labor. N. Engl. J. Med. 341, 660–666. 10.1056/NEJM199908263410906. [DOI] [PubMed] [Google Scholar]
- 81.López Bernal A (2003). Mechanisms of labour–biochemical aspects. BJOG 110, 39–45. 10.1046/j.1471-0528.2003.00023.x. [DOI] [PubMed] [Google Scholar]
- 82.Romero R, Espinoza J, Kusanovic JP, Gotsch F, Hassan S, Erez O, Chaiworapongsa T, and Mazor M (2006). The preterm parturition syndrome. BJOG 113, 17–42. 10.1111/j.1471-0528.2006.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Smith R (2007). Parturition. N. Engl. J. Med. 356, 271–283. 10.1056/NEJMra061360. [DOI] [PubMed] [Google Scholar]
- 84.Trundley A, and Moffett A (2004). Human uterine leukocytes and pregnancy. Tissue Antigens 63, 1–12. 10.1111/j.1399-0039.2004.00170.x. [DOI] [PubMed] [Google Scholar]
- 85.Bartmann C, Segerer SE, Rieger L, Kapp M, Sütterlin M, and Kämmerer U (2014). Quantification of the predominant immune cell populations in decidua throughout human pregnancy. Am. J. Reprod. Immunol. 71, 109–119. 10.1111/aji.12185. [DOI] [PubMed] [Google Scholar]
- 86.Li Y, Lopez GE, Vazquez J, Sun Y, Chavarria M, Lindner PN, Fredrickson S, Karst N, and Stanic AK (2018). Decidual-placental immune landscape during syngeneic murine pregnancy. Front. Immunol. 9, 2087. 10.3389/fimmu.2018.02087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koh W, Wu A, Penland L, Treutlein B, Neff NF, Mantalas GL, Blumenfeld YJ, El-Sayed YY, Stevenson DK, Shaw GM, and Quake SR (2019). Single cell transcriptomes derived from human cervical and uterine tissue during pregnancy. Adv. Biosyst. 3, e1800336. 10.1002/adbi.201800336. [DOI] [PubMed] [Google Scholar]
- 88.Chumduri C, Gurumurthy RK, Berger H, Dietrich O, Kumar N, Koster S, Brinkmann V, Hoffmann K, Drabkina M, Arampatzi P, et al. (2021). Opposing Wnt signals regulate cervical squamocolumnar homeostasis and emergence of metaplasia. Nat. Cell Biol. 23, 184–197. 10.1038/s41556-020-00619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao Z, Wang Y, Wu Y, Li D, Zhang T, Ma Y, Teng X, and Zuo W (2021). Single-cell analysis defines the lineage plasticity of stem cells in cervix epithelium. Cell Regen. 10, 36. 10.1186/s13619-021-00096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Osman I, Young A, Ledingham MA, Thomson AJ, Jordan F, Greer IA, and Norman JE (2003). Leukocyte density and pro-inflammatory cytokine expression in human fetal membranes, decidua, cervix and myometrium before and during labour at term. Mol. Hum. Reprod. 9, 41–45. 10.1093/molehr/gag001. [DOI] [PubMed] [Google Scholar]
- 91.Timmons BC, Fairhurst AM, and Mahendroo MS (2009). Temporal changes in myeloid cells in the cervix during pregnancy and parturition. J. Immunol. 182, 2700–2707. 10.4049/jimmunol.0803138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dobyns AE, Goyal R, Carpenter LG, Freeman TC, Longo LD, and Yellon SM (2015). Macrophage gene expression associated with remodeling of the prepartum rat cervix: microarray and pathway analyses. PLoS One 10, e0119782. 10.1371/journal.pone.0119782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dubicke A, Ekman-Ordeberg G, Mazurek P, Miller L, and Yellon SM (2016). Density of stromal cells and macrophages associated with collagen remodeling in the human cervix in preterm and term birth. Reprod. Sci. 23, 595–603. 10.1177/1933719115616497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vargas ML, Sántos JL, Ruiz C, Montes MJ, Alemán P, García-Tortosa C, and García-Olivares E (1993). Comparison of the proportions of leukocytes in early and term human decidua. Am. J. Reprod. Immunol. 29, 135–140. 10.1111/j.1600-0897.1993.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 95.Sindram-Trujillo A, Scherjon S, Kanhai H, Roelen D, and Claas F (2003). Increased T-cell activation in decidua parietalis compared to decidua basalis in uncomplicated human term pregnancy. Am. J. Reprod. Immunol. 49, 261–268. 10.1034/j.1600-0897.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- 96.Gomez-Lopez N, Estrada-Gutierrez G, Jimenez-Zamudio L, Vega-Sanchez R, and Vadillo-Ortega F (2009). Fetal membranes exhibit selective leukocyte chemotaxic activity during human labor. J. Reprod. Immunol. 80, 122–131. 10.1016/j.jri.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 97.Gomez-Lopez N, Vadillo-Perez L, Hernandez-Carbajal A, Godines-Enriquez M, Olson DM, and Vadillo-Ortega F (2011). Specific inflammatory microenvironments in the zones of the fetal membranes at term delivery. Am. J. Obstet. Gynecol. 205, 235.e15–235.e24. 10.1016/j.ajog.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 98.Gomez-Lopez N, Vadillo-Perez L, Nessim S, Olson DM, and Vadillo-Ortega F (2011). Choriodecidua and amnion exhibit selective leukocyte chemotaxis during term human labor. Am. J. Obstet. Gynecol. 204, 364.e9–364.16. 10.1016/j.ajog.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 99.Gomez-Lopez N, Vega-Sanchez R, Castillo-Castrejon M, Romero R, Cubeiro-Arreola K, and Vadillo-Ortega F (2013). Evidence for a role for the adaptive immune response in human term parturition. Am. J. Reprod. Immunol. 69, 212–230. 10.1111/aji.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arenas-Hernandez M, Romero R, Xu Y, Panaitescu B, Garcia-Flores V, Miller D, Ahn H, Done B, Hassan SS, Hsu CD, et al. (2019). Effector and activated T cells induce preterm labor and birth that is prevented by treatment with progesterone. J. Immunol. 202, 2585–2608. 10.4049/jimmunol.1801350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Slutsky R, Romero R, Xu Y, Galaz J, Miller D, Done B, Tarca AL, Gregor S, Hassan SS, Leng Y, and Gomez-Lopez N (2019). Exhausted and senescent T cells at the maternal-fetal interface in preterm and term labor. J. Immunol. Res. 2019, 3128010. 10.1155/2019/3128010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang JR, Nie J, Liu LJ, Zhang XW, Xie YM, Peng QZ, Wang WN, Pei CL, Zhao YH, Liu R, et al. (2020). Single-cell transcriptomics reveals the heterogeneity of the decidual endothelial cells that participate in labor onset. Eur. Rev. Med. Pharmacol. Sci. 24, 10359–10365. 10.26355/eurrev_202010_23385. [DOI] [PubMed] [Google Scholar]
- 103.Danforth DN, and Ivy AC (1949). The lower uterine segment; its derivation and physiologic behavior. Am. J. Obstet. Gynecol. 57, 831–841. 10.1016/0002-9378(49)90639-0. [DOI] [PubMed] [Google Scholar]
- 104.Schwalm H, and Dubrauszky V (1966). The structure of the musculature of the human uterus–muscles and connective tissue. Am. J. Obstet. Gynecol. 94, 391–404. 10.1016/0002-9378(66)90661-2. [DOI] [PubMed] [Google Scholar]
- 105.Wikland M, Lindblom B, Wilhelmsson L, and Wiqvist N (1982). Oxytocin, prostaglandins, and contractility of the human uterus at term pregnancy. Acta Obstet. Gynecol. Scand. 61, 467–472. 10.3109/00016348209156592. [DOI] [PubMed] [Google Scholar]
- 106.Sparey C, Robson SC, Bailey J, Lyall F, and Europe-Finner GN (1999). The differential expression of myometrial connexin-43, cyclooxygenase-1 and -2, and Gs alpha proteins in the upper and lower segments of the human uterus during pregnancy and labor. J. Clin. Endocrinol. Metab. 84, 1705–1710. 10.1210/jcem.84.5.5644. [DOI] [PubMed] [Google Scholar]
- 107.Pollard AJ, Sparey C, Robson SC, Krainer AR, and Europe-Finner GN (2000). Spatio-temporal expression of the trans-acting splicing factors SF2/ASF and heterogeneous ribonuclear proteins A1/A1B in the myometrium of the pregnant human uterus: a molecular mechanism for regulating regional protein isoform expression in vivo. J. Clin. Endocrinol. Metab. 85, 1928–1936. 10.1210/jcem.85.5.6537. [DOI] [PubMed] [Google Scholar]
- 108.Bukowski R, Hankins GDV, Saade GR, Anderson GD, and Thornton S (2006). Labor-associated gene expression in the human uterine fundus, lower segment, and cervix. PLoS Med. 3, e169. 10.1371/journal.pmed.0030169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sooranna SR, Grigsby PL, Engineer N, Liang Z, Sun K, Myatt L, and Johnson MR (2006). Myometrial prostaglandin E2 synthetic enzyme mRNA expression: spatial and temporal variations with pregnancy and labour. Mol. Hum. Reprod. 12, 625–631. 10.1093/molehr/gal061. [DOI] [PubMed] [Google Scholar]
- 110.Mosher AA, Rainey KJ, Bolstad SS, Lye SJ, Mitchell BF, Olson DM, Wood SL, and Slater DM (2013). Development and validation of primary human myometrial cell culture models to study pregnancy and labour. BMC Pregnancy Childbirth 13 (Suppl 1), S7. 10.1186/1471-2393-13-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu L, Li H, Dargahi D, Shynlova O, Slater D, Jones SJM, Lye SJ, and Dong X (2015). HoxA13 regulates phenotype regionalization of human pregnant myometrium. J. Clin. Endocrinol. Metab. 100, E1512–E1522. 10.1210/jc.2015-2815. [DOI] [PubMed] [Google Scholar]
- 112.Patwardhan M, Hernandez-Andrade E, Ahn H, Korzeniewski SJ, Schwartz A, Hassan SS, and Romero R (2015). Dynamic changes in the myometrium during the third stage of labor, evaluated using two-dimensional ultrasound, in women with normal and abnormal third stage of labor and in women with obstetric complications. Gynecol. Obstet. Invest. 80, 26–37. 10.1159/000370001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088. 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gomez-Lopez N, Garcia-Flores V, Chin PY, Groome HM, Bijland MT, Diener KR, Romero R, and Robertson SA (2021). Macrophages exert homeostatic actions in pregnancy to protect against preterm birth and fetal inflammatory injury. JCI Insight 6, e146089. 10.1172/jci.insight.146089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gershater M, Romero R, Arenas-Hernandez M, Galaz J, Motomura K, Tao L, Xu Y, Miller D, Pique-Regi R, Martinez G 3rd., et al. (2022). IL-22 plays a dual role in the amniotic cavity: tissue injury and host defense against microbes in preterm labor. J. Immunol. 10.4049/jimmunol.2100439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sakamoto Y, Moran P, Bulmer JN, Searle RF, and Robson SC (2005). Macrophages and not granulocytes are involved in cervical ripening. J. Reprod. Immunol. 66, 161–173. 10.1016/j.jri.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 117.Gonzalez JM, Dong Z, Romero R, and Girardi G (2011). Cervical remodeling/ripening at term and preterm delivery: the same mechanism initiated by different mediators and different effector cells. PLoS One 6, e26877. 10.1371/journal.pone.0026877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Payne KJ, Clyde LA, Weldon AJ, Milford TA, and Yellon SM (2012). Residency and activation of myeloid cells during remodeling of the prepartum murine cervix. Biol. Reprod. 87, 106. 10.1095/biolreprod.112.101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Krantz KE, and Phillips WP (1962). Anatomy of the human uterine cervix, gross and microscopic. Ann. N. Y. Acad. Sci. 97, 551–563. 10.1111/j.1749-6632.1962.tb34666.x. [DOI] [PubMed] [Google Scholar]
- 120.Granström L, Ekman G, Ulmsten U, and Malmström A (1989). Changes in the connective tissue of corpus and cervix uteri during ripening and labour in term pregnancy. Br. J. Obstet. Gynaecol. 96, 1198–1202. 10.1111/j.1471-0528.1989.tb03196.x. [DOI] [PubMed] [Google Scholar]
- 121.Ludmir J, and Sehdev HM (2000). Anatomy and physiology of the uterine cervix. Clin. Obstet. Gynecol. 43, 433–439. 10.1097/00003081-200009000-00003. [DOI] [PubMed] [Google Scholar]
- 122.Montes GS, Zugaib M, Joazeiro PP, Varayoud J, Ramos JG, Muñoz-de-Toro M, and Luque EH (2002). Phenotypic modulation of fibroblastic cells in the mucous layer of the human uterine cervix at term. Reproduction 124, 783–790. 10.1530/rep.0.1240783. [DOI] [PubMed] [Google Scholar]
- 123.Vink JY, Qin S, Brock CO, Zork NM, Feltovich HM, Chen X, Urie P, Myers KM, Hall TJ, Wapner R, et al. (2016). A new paradigm for the role of smooth muscle cells in the human cervix. Am. J. Obstet. Gynecol. 215, 478.e1–478.e11. 10.1016/j.ajog.2016.04.053. [DOI] [PubMed] [Google Scholar]
- 124.Wendremaire M, Hadi T, Pezze M, Barrichon M, Lopez T, Neiers F, Sagot P, Garrido C, and Lirussi F (2020). Macrophage-induced reactive oxygen species promote myometrial contraction and labor-associated mechanisms. Biol. Reprod. 102, 1326–1339. 10.1093/biolre/ioaa032. [DOI] [PubMed] [Google Scholar]
- 125.Coleman M, Orvis A, Wu TY, Dacanay M, Merillat S, Ogle J, Baldessari A, Kretzer NM, Munson J, Boros-Rausch AJ, et al. (2020). A broad spectrum chemokine inhibitor prevents preterm labor but not microbial invasion of the amniotic cavity or neonatal morbidity in a non-human primate model. Front. Immunol. 11, 770. 10.3389/fimmu.2020.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Boros-Rausch A, Shynlova O, and Lye SJ (2021). A broad-spectrum chemokine inhibitor blocks inflammation-induced myometrial myocytemacrophage crosstalk and myometrial contraction. Cells 11, 128. 10.3390/cells11010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gomez-Lopez N, Romero R, Arenas-Hernandez M, Panaitescu B, Garcia-Flores V, Mial TN, Sahi A, and Hassan SS (2018). Intra-amniotic administration of lipopolysaccharide induces spontaneous preterm labor and birth in the absence of a body temperature change. J. Matern. Fetal Neonatal Med. 31, 439–446. 10.1080/14767058.2017.1287894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Viscardi RM (2010). Ureaplasma species: role in diseases of prematurity. Clin. Perinatol. 37, 393–409. 10.1016/j.clp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sweeney EL, Kallapur SG, Gisslen T, Lambers DS, Chougnet CA, Stephenson SA, Jobe AH, and Knox CL (2016). Placental infection with Ureaplasma species is associated with histologic chorioamnionitis and adverse outcomes in moderately preterm and late-preterm infants. J. Infect. Dis. 213, 1340–1347. 10.1093/infdis/jiv587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yoneda S, Shiozaki A, Yoneda N, Ito M, Shima T, Fukuda K, Ueno T, Niimi H, Kitajima I, Kigawa M, and Saito S (2016). Antibiotic therapy increases the risk of preterm birth in preterm labor without intraamniotic microbes, but may prolong the gestation period in preterm labor with microbes, evaluated by rapid and high-sensitive PCR system. Am. J. Reprod. Immunol. 75, 440–450. 10.1111/aji.12484. [DOI] [PubMed] [Google Scholar]
- 131.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, et al. (2018). Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94. 10.1038/nbt.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Young MD, and Behjati S (2020). SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience 9, giaa151. 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4. 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296. 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, and Raychaudhuri S (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.McInnes L, Healy J, and Melville J (2018). UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at arXiv. 10.48550/arXiv.1802.03426v3. [DOI] [Google Scholar]
- 139.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 140.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al. (2019). Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172. 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. (2018). Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091–1107.e17. 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 142.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tabula Muris Consortium; Supplemental Text Writing Group; Principal Investigators; Overall Coordination; Logistical Coordination; Organ Collection and Processing; Library Preparation and Sequencing; Computational Data Analysis; Cell Type Annotation; Writing Group (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372. 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Buechler MB, Pradhan RN, Krishnamurty AT, Cox C, Calviello AK, Wang AW, Yang YA, Tam L, Caothien R, Roose-Girma M, et al. (2021). Cross-tissue organization of the fibroblast lineage. Nature 593, 575–579. 10.1038/s41586-021-03549-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication (see key resources table for accession number). Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication (see key resources table for DOI). Microscopy data reported in this paper will be shared by the lead contact upon reasonable request.
All original code has been deposited at Zenodo and is publicly available as of the date of publication (see key resources table for DOI).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| The Monoclonal Rabbit Anti-Mouse CD45 | Cell Signaling Technology | Cat#70257S; RRID: AB_2799780 |
| Rabbit FLEX Universal Negative Control | Agilent | Cat# IR60066-2 |
| Monoclonal Rabbit Anti-Mouse F4/80 | Cell Signaling Technology | Cat# 70076S; RRID: AB_2799771 |
| Monoclonal Rabbit Anti-Mouse CD3ε | Cell Signaling Technology | Cat# 78588S; RRID: AB_2889902 |
| Monoclonal Rabbit Anti-Mouse Klrb1c/CD161c | Cell Signaling Technology | Cat# 39197S; RRID: AB_2892989 |
| Polyclonal Rabbit Anti-Mouse Ly6C | HuaBio | Cat# HA500088 |
| Monoclonal Rabbit Anti-Mouse Ly6G | Cell Signaling Technology | Cat# 87048S; RRID: AB_2909808 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Escherichia coli | ATCC | ATCC 12014 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Umbilical Cord Dissociation Kit, human | Miltenyi Biotec | Cat# 130-105-737 |
| Dead Cell Removal Kit | Miltenyi Biotec | Cat# 130-090-101 |
| Chromium Next GEM Single Cell 3′ GEM, Library &Gel beads Kit | 10× Genomics | PN:1000121 |
| Chromium Next GEM Chip G Single Cell Kit | 10× Genomics | PN:1000120 |
| Single Index Kit T Set A | 10× Genomics | PN: 1000213 |
| SPRIselect Reagent | Beckman Coulter | Item Number: B23318 |
|
| ||
| Deposited data | ||
|
| ||
| scRNA-sequencing | This paper | Gene Expression Omnibus: GSE200289 |
| Scripts | This paper | Zenodo: https://doi.org/10.5281/zenodo.7335384 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: C57BL/6 | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mouse: BALB/cByJ | The Jackson Laboratory | RRID: IMSR_JAX:001026 |
|
| ||
| Software and algorithms | ||
|
| ||
| Cell Ranger version 4.0.0 | 10× Genomics | http://www.10xgenomics.com |
| STAR aligner | (Dobin et al.)131 | https://github.com/alexdobin/STAR |
| Demuxlet | (Kang et al.)132 | https://github.com/statgen/demuxlet |
| SoupX version 1.5.2 | (Young et al.)133 | https://github.com/constantAmateur/Soup |
| DoubletFinder 2.0.3 | (McGinnis.)134 | |
| Seurat version 4.0.3 | (Stuart et al.)135 (Hafemeister et al.)136 |
https://satijalab.org/seurat/ |
| Harmony package in R version 1.0.0 (R package from CRAN) | (Korsunsky et al.)137 | N/A |
| Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) algorithm | (McInnes et al.)138 (Becht et al.)139 |
N/A |
| SingleR package in R version 1.6.1 | (Aran et al.)140 | https://doi.org/10.18129/B9.bioc.SingleR |
| Mouse Cell Atlas and single-cell MCA (scMCA) package in R version 0.2.0 | (Han et al.)141 | http://bis.zju.edu.cn/MCA/search.html |
| DESeq2 R package version 1.32.0 | (Love et al.)142 | https://doi.org/10.18129/B9.bioc.DESeq2 |
| clusterProfiler in R version 4.0.4 | (Yu et al.)143 | https://doi.org/10.18129/B9.bioc.clusterProfiler |
| CellChat version 1.1.2 (R package from CRAN) | (Jin et al.)113 | N/A |
| ggalluvial version 0.12.3 (R package from CRAN) | N/A | |
| ggplot2 version 3.3.5 (R package from CRAN) | N/A | |
|
| ||
| Other | ||
|
| ||
| Human uterine scRNA-seq data | (Pique-Regi et al.)70 | dbGaP: phs001886.v4.pl |


