Abstract
The recent increase in monkeypox (MPX) cases has attracted attention of public health authorities due to its quick spread and transmission across non-endemic regions. This outbreak, unlike previous ones, displays different epidemiological features and transmission dynamics, which appear to be largely influenced by the newly divergent MPX lineages (B.1). Yet, the genomic characteristics driving the high dispersal and diversification of these lineages remain largely unknown. Herein, we sought to explore and characterize the genomic features and phylogenetic diversity of the B.1 lineages through a comparative genomic analysis inclusive of 1900 high quality complete MPXV genomes. Our analyses indicate that the current MPXV-2022 outbreak encompasses thirteen derived lineages with ten unique non-synonymous mutations in several genes linked to immune evasion, virulence factors and host recognition. Such mutations may translate in the rapid evolution and diversification of current MPXV lineages. Moreover, our analyses uncovered signals of genomic modifications suggestive of immune-modulatory enzymatic activity, such as APOBEC3 editing, which, as previously suggested could have favored evolutionary trends leading to the rapid spread of MPXV into non-endemic countries. Genomic surveillance continues to play a major role in unveiling the genomic signatures signaling potential adaptation of this emerging MPXV lineage and how it will continue to impact public health in the near future.
Keywords: Monkeypox virus, Phylogenomic and mutational analyses, B.1 lineages, APOBEC3 activity, MPXV proteins
1. Introduction
Monkeypox (MPX), a disease whose clinical manifestations resemble smallpox, is a zoonotic disease caused by the Monkeypox virus (MPXV) [1]. This virus, a member of the Poxviridae family contains a double-stranded DNA genome (∼197 kb) with ∼200 non-overlapping coding genes [2], which are responsible for its various biological features [3]. Although MPXV is considered endemic to central and western African countries, with two major known outbreaks (1970 and 1996-97) reported to date; in more recent years (2003–2021), smaller scale outbreaks of infection have followed in non-endemic countries including the USA, United Kingdom, Israel and Singapore. These outbreaks trailed sporadic infection events associated with travel or contact with people from MPXV-endemic countries. More recently, a steady increase of MPXV cases across 104 countries worldwide, mainly in the USA, Brazil, Spain, France, Colombia and United Kingdom (https://www.cdc.gov/poxvirus/monkeypox/response/2022/world-map.html accessed on 22 January 2023) has been reported. This outbreak, unlike previous ones, has featured a rapid global spread with lower mortality, case fatality and complication rates.
The high prevalence and increasing number of MPXV cases in non-endemic countries prompted different public health organizations to rapidly implement genomic surveillance programs and monitor the epidemiology and evolution of MPXV. In terms of its genomic and evolutionary characteristics, MPXV embraces two distinct clades (I and II) [4]. Clade I (former Central African Clade) includes isolates associated with epidemiological outbreaks from the Democratic Republic of Congo (Congo Basin), which typically show increased transmissibility, severe clinical symptoms and higher mortality rates. Clade II (former West African Clade) on the other hand, reveals two distinct subgroupings (IIa and IIb) [4]. Subclade IIa, comprising the West African isolates often associated with milder course of infection, lower mortality and transmissibility [5]; and subclade IIb that only includes genomes associated with the most recent outbreaks in humans (hMPXV), including the current MPXV outbreak and whose ongoing clinical and epidemiological features remain yet to be fully characterized.
Moreover, the different outbreaks occurring in recent years have reshaped the genomic landscape of subclade IIb leading to a further divergence of lineages (A, A.1, A.1.1, A.1.1, B.1), based on a trail of molecular evolutionary events. Interestingly, circulating strains from the current outbreak actually fall into the B.1 lineage, with diverse differences in coding regions, which have been linked to immunomodulatory and host recognition antigenic determinants [[6], [7], [8]]. In addition, this lineage shows marked divergence as a product of the various epidemiological events governing previous outbreaks, which in turn, may have driven the different microevolutionary events modulating its genomic architecture [9]. To date, some characteristics of this lineage, such as its unique mutational repertoire favoring its dispersal and diversification, remain to be deciphered. Therefore, the aim of this study was to identify the unique mutational profile defining the phylogenetic diversity of the B.1 lineages genomes through a comparative genomic analysis of publicly available genomes.
2. Methodology
A total of 1900 high-quality, complete MPXV genomes available in the GISAID and NCBI Viral databases as of September 30, 2022 (Table S1) were analyzed following a similar scheme to that used by [10]. This consists of an alignment using NextClade v2.4.2 tool [11], geographic analysis of B.1 lineages, mutational analysis of coding regions, especially for those genes in which unique mutations have been found for the MPXV B.1 lineages, and analysis on the phylogenomic relationships of genome sequences publicly available. For phylogenomic analysis, the aligned dataset obtained from NextClade was used to build a maximum likelihood (ML) in IQ-TREE multicore v1.6.12 [12], using as the best substitution model and other parameters by default.
Furthermore, we estimated the potential introduction date and dispersion dynamics of the MPXV lineages using TreeTime software [13], which considers a fixed clock rate of 5 x10−5 (SD = 10.2 × 10−6) [20,29], a strict clock (SC) under a coalescent tree skyline prior and a root step to minimize residuals in a root-to-tip. The obtained tree was graphically represented in Interactive Tree Of Life (iTOL) v5 [14] where the clades were assigned using the intrataxa classification system that most accurately describes the viral diversity and the currently MPXV Clades I, IIa and IIb as described in Ref. [4].
3. Results
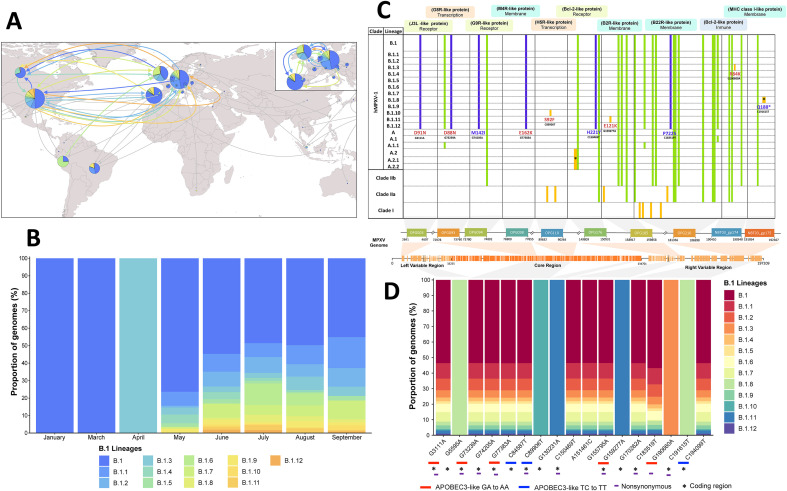
Of the 1900 genomes analyzed, 1118 (97.4%) fall into lineages from the current MPXV outbreak: B.1 (n = 974, 51.3%), B.1.1 (n = 181, 9.53%), B.1.2 (n = 136, 7.16%), B.1.3 (n = 85, 4.47%), B.1.4 (n = 44, 2.32%), B.1.5 (n = 29, 1.53%), B.1.6 (n = 98, 5.16%), B.1.7 (n = 11, 5.84%), B.1.8 (n = 45, 2.37%), B.1.9 (n = 44, 2.32%), B.1.10 (n = 14, 0.74%), B.1.11 (n = 38, 2.00%) and B.1.12 (n = 18, 0.95%). Regarding their geographic distribution, we found a wide distribution of lineages (Fig. 1 A y Fig. 1B), with Germany, USA, the United Kingdom, and Portugal being the countries exhibiting the highest number of distinct genomes. Furthermore, it was determined that these countries represented the main geographical areas that originated and amplified the dispersal of the thirteen B.1 lineages (Fig. 1A). Although most lineages appear as widely distributed, some are predominantly present only in certain countries. For instance, the majority of B.1.6 genomes originate from Peru, B.1.4 from Canada and B.1.11 mostly from the USA (Fig. 1A). At a temporal scale, we found that in early 2022 the MPXV B.1 lineage was circulating through non-endemic countries (Fig. 1B). In addition, it was determined that genomes from the first B.1-derived lineages were already present as early as mid-April and May, to later increase in July to prevail and spread across different countries.
Fig. 1.
Descriptive and mutational analyses of MPXV B.1 lineages. (A) Geographical distribution and circulation of the current MPXV outbreak 2022. The colored arrows indicate the first presumptive dispersal routes of the B.1 lineages (B) Temporal variations in the proportions of genomes of the B.1 lineages. (C) Mutational analysis of nonsynonymous changes at the amino acid level between the lineages and clades of MPXV. The purple color represents the unique substitutions shared between the genomes of B.1 lineages analyzed (N = 1118), the green color the substitutions shared between MPXV lineages/clades, and the yellow color represent the substitutions considered unique for each taxa. In red, it highlights the amino acid modifications associated with the action of the APOBCE3 enzyme. (D) Proportion of MPXV genomes that have the unique nucleotide mutations of the B.1 lineages. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Mutational analysis showed thirty-three non-synonymous amino acid substitutions belonging to ten MPXV genes (Fig. 1C). These genes were selected as they harbor mutations unique to the B.1 lineages (Fig. 1D). Twenty-three of these substitutions (69.7%) are shared, four are present between B.1 lineages and at least one lineage of the hMPXV-1 A clade (17.4%) and twenty-one substitutions are common to different lineages and clades (82.6%). When analyzing the different B.1 lineages, we observed ten unique non-synonymous mutations (Fig. 1C). Six of them derived from APOBEC3 like activity (Fig. 1D) and located in genes whose proteins exhibit different functions such as membrane proteins (OPG098 and OPG185), receptors (OPG003), transcriptional regulators (OPG093, OPG110) or modulators of the host immune system (NBT03_gp174). Of the ten B.1 distinct mutations, five were shared among the thirteen B.1 lineages, which are located in the OPG093, OPG094, OPG098, OPG176 and OPG210 genes (Fig. 1C). Likewise, we found that the B.1.3, B.1.8, B.1.10 and B.1.11 lineages have only one shared non-synonymous substitution among the ten genes analyzed (Fig. 1C and D).
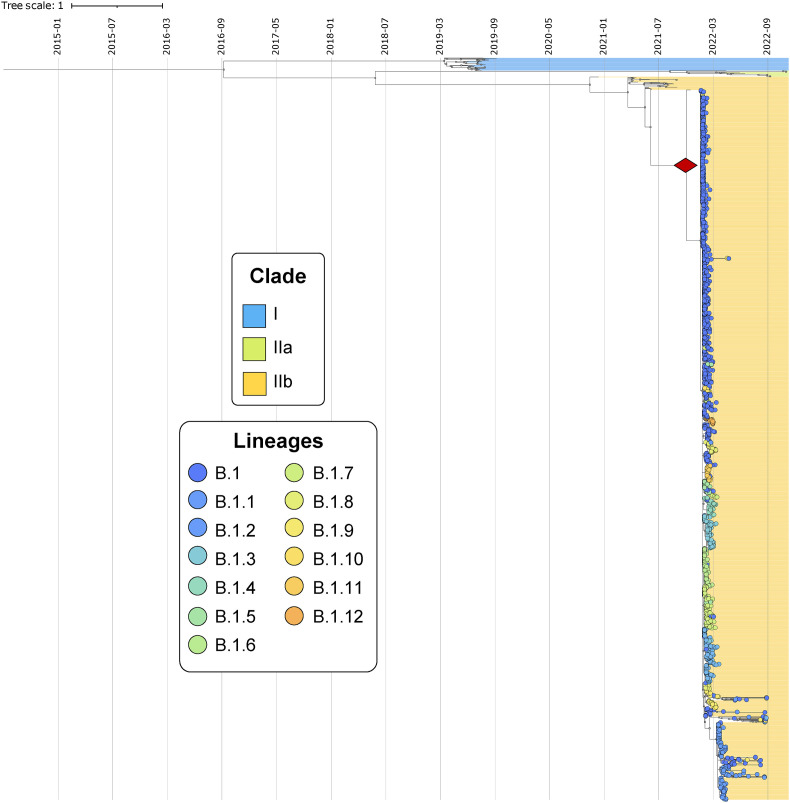
The maximum-likelihood time-scaled phylogeny (ML-TSP) obtained from Tree-Time (Fig. 2 ) depicted clustering by clade, according to the classification proposed for MPXV [4]. These clades were monophyletic and included the following number of genomes: 32 MPXV genomes associated with outbreaks in Central Africa (Clade I), 16 genomes of the outbreaks in West Africa (Clade IIa) and 1842 genomes belonged to the last two epidemiological outbreaks of MPXV (2017–2022). The latter includes the clusters identified as the MPXV lineages, where genomes closely related to the B.1 lineages were identified (Fig. 2).
Fig. 2.
Phylogenomic analysis of MPXV. The figure shows the time-scaled phylogeny with the phylogenomic relationships of B.1 MPXV genomes and the introduction date based on a node-date assignment from the Most Recent Common Ancestor (MRCA) indicated in red diamond. These genomes were grouped according to the current MPXV classification proposed by Happi et al.
The time-scaled phylogeny obtained from Tree-Time showed the introduction date based on a node-date assignment from the Most Recent Common Ancestor (MRCA) (Fig. 2). This analysis showed that clade I represents the most ancestral group, whereas lineage A.1.1 embodies the most recent amongst circulating lineages from the current outbreak (Fig. 2). Finally, the potential introduction date for the B.1 lineages was estimated 10/12/2021 [95%CI = 21/11/2021 to 31/12/2021] in Europe.
4. Discussion
The sudden increase in MPXV cases and its unprecedented spread throughout non-endemic regions has raised red flags among global public health organizations and authorities. To date, a total 73,284 MPXV-2022 cases have been reported across several countries, predominantly in Europe (Spain, France, Germany and Portugal) and North America (USA and Canada). This current outbreak, which has been associated with the emergence of the B.1 lineage, exhibits a distinct constellation of clinical manifestations, behaving mostly as a sexually transmitted infection, accompanied by myalgias, massive lymphadenitis, cutaneous (mainly monomorphic) pustular rashes, and low severity [15,16]. However, the risk for severe outcomes in high-risk populations like the immunocompromised, young children and elderly persists. Classically, MPXV transmission occurs by direct human-to-human direct contact, as well as, via respiratory droplets, or fomites [15]. Furthermore, other modes of transmission, such as transplacental or sexual contact (as for the current outbreak) [15] have been reported, highlighting the variety of mechanisms involved in virus transmission. This broad spectrum of transmission pathways, the occurrence of certain super spreading events in foci of disease clustering across specific sexual networks, coupled with an ever-increasing reactivation of international flights seems to have fueled the rapid dispersion and geographic patterns observed during the current outbreak.
Based on its evolutionary traits, MPXV-2022 presents two major evolutionary features: the diversification of clades I-II and the divergence of the B.1 lineage. The first relates to genomic modifications in coding regions involved in host recognition by antigenic determinants, such as B21R and H3L-like proteins [15]. This event led to the diversification of clade II, which comprises strains related to human-human infections and those lineages from small outbreaks in non-endemic countries (Clade IIb and lineages A, A.1, A.1.1 and A.2) [17,18]. Subsequently, a second event is thought to have caused the rapid evolution and spread of the strains of the current MPXV outbreak (B.1 lineages). Data suggests that the microevolution of the B.1 lineages may have occurred by genomic modifications in a short period of time, an unusual feature for Orthopoxviruses, which remains under investigation [15,19]. These modifications may signal a role for enzymatic activity of APOBEC3 [20], a family of deaminases that act on single-stranded DNA to mutate the complementary strand leading to viral inactivation [15], via GA→AA and TC→TT changes and thereby imprinting unique mutations across different regions of the MPXV genome. However, further evidence is needed to confirm whether the mutational bias induced by the presumed action of APOBCE3 has led to the cumulus of mutations recorded on the current 2022 MPXV outbreak and its potential role on diversification and adaptation. Therefore, elucidating the mechanisms that may be driving the evolution, diversification, and spread of current MPXV lineages is of utmost relevance.
At the genomic level, the B.1 lineage shows a predominance of APOBEC3-associated genetic modifications in different regions of its genome, which have generated 46 B.1-specific single nucleotide polymorphisms (SNPs), including four intergenic, 18 synonymous and 24 nonsynonymous mutations [15,19]. Here, we found ten genes whose amino acid mutations are linked to B.1 lineages, with six of them being associated to APOBCE3 in the OPG003, OPG093, OPG098, OPG110, OPG185 and NBT03_gp174 genes, and whose proteins correlate with MPXV functional features such as virulence factors and immune evasion. Furthermore, we also found non-APOBCE3 mutations that modify functional MPXV proteins. For instance, several amino acid changes (D209 N, P722S and M1741I) have been reported in the gene associated with the immunogenic surface glycoprotein (OPG210 or B22R-like protein) which modulates the interaction with host antibodies and virus transmissibility [15,17]. Similarly, mutations in the B2-cell lymphoma-like protein (OPG176 or BCL-2 like protein) regulate host immune activity in the presence of MPXV [15,17]. Although most of the unique mutations in B.1 are located in coding regions, homologous to highly studied sites in Variola and Vaccinia viruses, its impact on the biological characteristics of MPXV remains unknown; particularly in those genes harboring nonsynonymous substitutions in amino acids unique to or shared among viral lineages or clades. Therefore, further research is needed to evaluate the effect of these mutations on the protein and functional features of MPXV, and its role as a driver of lineage diversification.
As for the emergence of the B.1 lineages (Fig. 2), the chain of micro-evolutionary events related with these lineages is estimated to have occurred in late 2021. This result is consistent with that reported by Nextstrain [January 24, 2022] (https://nextstrain.org/monkeypox/hmpxv1?dmin=2021-07-03&label=clade:B.1 (accessed October 17, 2022)), which determined that the B.1 lineage emerged in the European continent at that time generating the first outbreaks of the virus, and subsequently spreading to other continents such as America, Oceania and Asia. In addition, during its dispersion, B.1 further diversified into the different groupings or lineages, displaying specific geographic distributions. This analysis together with the genomic data confirms that B.1 lineages emerged and dispersed recently generating a stochastic increase in MPXV cases. In addition, these lineages harbor specific mutations associated with genes involved in immune evasion, host recognition and a number of virulence factors. Given the epidemiological and genomic characteristics of B.1, genomic surveillance of this virus is necessary to design disease control and prevention mechanisms in non-endemic regions.
5. Conclusions
In summary, genomes belonging to B.1 lineages associated with the current MPXV outbreak in 2022 reveal distinct mutational and evolutionary features unique to these lineages. Our analyses suggest that the virus harbors a constellation of multiple mutations in genes with functional implications in infection and transmission, which seem to have favored the rapid spread of MPXV into non-endemic countries. On the other hand, these mutational signatures appear to be associated with the accelerated evolutionary changes that gave rise to this lineage. Therefore, we highlighted the different events that contributed to the divergence and dispersal of these lineages in context of the current outbreak.
Funding source
None.
CRediT authorship contribution statement
Nicolas Luna: Conceptualization, Methodology, Software, Data curation, Writing – original draft, Visualization, Investigation. Marina Muñoz: Conceptualization, Methodology, Software, Data curation, Writing – original draft, Visualization, Investigation. Luz H. Patiño: Conceptualization, Methodology, Software, Data curation, Writing – original draft, Visualization, Investigation. Yana Kasminskaya: Methodology, Writing – review & editing. Alberto Paniz-Mondolfi: Conceptualization, Methodology, Software, Data curation, Writing – original draft, Visualization, Investigation. Juan David Ramírez: Conceptualization, Methodology, Software, Data curation, Writing – original draft, Visualization, Investigation, Supervision.
Declaration of competing interest
Nothing to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tmaid.2023.102551.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Alakunle E., Moens U., Nchinda G., Okeke M.I. Monkeypox virus in Nigeria: infection biology, epidemiology, and evolution. Viruses. 2020;12:1257. doi: 10.3390/v12111257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shchelkunov S.N., Totmenin A.V., Safronov P.F., et al. Analysis of the monkeypox virus genome. Virology. 2002;297:172–194. doi: 10.1006/viro.2002.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kugelman J.R., Johnston S.C., Mulembakani P.M., et al. Genomic variability of monkeypox virus among humans, democratic republic of the Congo. Emerg Infect Dis. 2014;20 doi: 10.3201/eid2002.130118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Happi C., Adetifa I., Mbala P., et al. Urgent need for a non-discriminatory and non-stigmatizing nomenclature for monkeypox virus. PLoS Biol. 2022;20 doi: 10.1371/JOURNAL.PBIO.3001769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jezek Z., Szczeniowski M., Paluku K.M., Mutombo M. Human monkeypox: clinical features of 282 patients. JID (J Infect Dis) 1987;156:293–298. doi: 10.1093/infdis/156.2.293. [DOI] [PubMed] [Google Scholar]
- 6.Berthet N., Descorps-Declère S., Besombes C., et al. Genomic history of human monkey pox infections in the Central African Republic between 2001 and 2018. Sci Rep. 2021;11 doi: 10.1038/s41598-021-92315-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shchelkunov S.N., Totmenin A.V., Safronov P.F., et al. Analysis of the monkeypox virus genome. Virology. 2002;297:172–194. doi: 10.1006/viro.2002.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Toole Á., Rambaut A. 2022. Initial observations about putative APOBEC3 deaminase editing driving short-term evolution of MPXV since 2017.https://virological.org/t/initial-observations-about-putative-apobec3-deaminase-editing-driving-short-term-evolution-of-mpxv-since-2017/830 [Google Scholar]
- 9.Gomes J.P., Isidro J., Borges V., et al. 2022. Multi-country outbreak of Monkeypox virus: genetic divergence and first signs of microevolution; pp. 1–8. [Google Scholar]
- 10.Luna N., Ramírez A.L., Muñoz M., et al. Phylogenomic analysis of the monkeypox virus (MPXV) 2022 outbreak: emergence of a novel viral lineage? Trav Med Infect Dis. 2022;49 doi: 10.1016/J.TMAID.2022.102402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aksamentov I., Roemer C., Hodcroft E., Neher R. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 2021;6:3773. doi: 10.21105/joss.03773. [DOI] [Google Scholar]
- 12.Minh B.Q., Schmidt H.A., Chernomor O., et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sagulenko P., Puller V., Neher R.A. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4 doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letunic I., Bork P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/NAR/GKAB301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Zhang H., Ding K., et al. Cytokine Growth Factor Rev; 2022. The evolving epidemiology of monkeypox virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aljabali A.A.A., Obeid M.A., Nusair M.B., et al. Monkeypox virus: an emerging epidemic. Microb Pathog. 2022;173 doi: 10.1016/j.micpath.2022.105794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L., Shang J., Weng S., et al. Genomic annotation and molecular evolution of monkeypox virus outbreak in 2022. J Med Virol. 2022 doi: 10.1002/jmv.28036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riopelle J.C., Munster V.J., Port J.R. Atypical and unique transmission of monkeypox virus during the 2022 outbreak: an overview of the current state of knowledge. Viruses. 2022;14 doi: 10.3390/v14092012. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lum F.-M., Torres-Ruesta A., Tay M.Z., et al. Monkeypox: disease epidemiology, host immunity and clinical interventions. Nat Rev Immunol. 2022;22:597–613. doi: 10.1038/s41577-022-00775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isidro J., Borges V., Pinto M., et al. Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus. Nat Med. 2022 doi: 10.1038/s41591-022-01907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.