Abstract
Although rare, infection and vaccination can result in antibodies to human leukocyte antigens (HLA). We analyzed the effect of SARS-CoV-2 infection or vaccination on HLA antibodies in waitlisted renal transplant candidates. Specificities were collected and adjudicated if the calculated panel reactive antibodies (cPRA) changed after exposure. Of 409 patients, 285 (69.7 %) had an initial cPRA of 0 %, and 56 (13.7 %) had an initial cPRA > 80 %. The cPRA changed in 26 patients (6.4 %), 16 (3.9 %) increased, and 10 (2.4 %) decreased. Based on cPRA adjudication, cPRA differences generally resulted from a small number of specificities with subtle fluctuations around the borderline of the participating centers’ cutoff for unacceptable antigen listing. All five COVID recovered patients with an increased cPRA were female (p = 0.02). In summary, exposure to this virus or vaccine does not increase HLA antibody specificities and their MFI in approximately 99 % of cases and 97 % of sensitized patients. These results have implications for virtual crossmatching at the time of organ offer after SARS-CoV-2 infection or vaccination, and these events of unclear clinical significance should not influence vaccination programs.
Keywords: Kidney transplantation, Organ allocation, Virtual crossmatch, COVID-19 vaccination, SARS-CoV-2 infection
1. Introduction
Organ transplantation, pregnancy, and blood transfusion trigger new Human Leukocyte Antigen (HLA) antibody production or increase existing HLA antibody levels, which are particularly interesting to patients awaiting renal transplantation. [1], [2], [3], [4], [5] Exposure to pathogens can result in antibody formation by inducing alloreactivity (termed heterologous immunity) and via costimulatory factors that activate bystander alloreactive leukocytes.[1] Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic, and new and recurrent infections with this virus are expected to continue for the foreseeable future. Vaccination efforts are widespread, and patients with renal failure awaiting kidney transplants are strongly encouraged to get vaccinated because the risk of mortality is high in this population prior to transplant [6], [7]. Vaccination after transplant is less protective due to immunosuppression. [8], [9] SARS-CoV-2 infects cells expressing angiotensin-converting enzyme 2 and Transmembrane Serine Protease 2 surface proteins, and activates both an innate and adaptive immune response, resulting in cytokine storm in some patients.[10] A recent report describes the presence of HLA antibodies in the convalescent serum of male patients without any known allosensitizing events who recovered from COVID-19, suggesting that infection with this virus could trigger the generation of antibodies that recognize HLA antigens.[11].
Confidence in the most recent antibody profile is critical at the time of organ offer[12], and there is a lack of consensus among experts about the need to repeat single antigen testing after a recent COVID infection or vaccination at the time of an organ offer. Also, vaccination after transplant in the setting of immunosuppression leaves many high-risk patients unprotected from SARS-CoV-2[8], but the granular effects of this vaccine on HLA antibody profiles are not described.
We previously reported that SARS-CoV-2 infection did not result in HLA antibody formation in a small group of patients awaiting kidney transplant at a single center [13], but a small cohort of patients cannot adequately address this question. The current study brought together 8 transplant programs aiming to directly address whether patients with renal failure who are waitlisted for kidney transplant develop HLA antibodies after being 1) infected with SARS-CoV-2, or 2) receiving the vaccination against COVID-19.
2. Materials & methods
2.1. Waitlisted Renal Transplant Candidates
This is a retrospective cohort study of prospectively maintained databases of adult renal transplant candidates at 8 transplant centers in the US, performed with the approval of the institutional review board (IRB Number: 21–33936). Data were collected about patients exposed between 4/19/2020 to 1/20/2022. Centers routinely performed HLA antibody testing of waitlisted patients approaching the top of the deceased donor waiting list at some interval. The inclusion criteria were waitlisted patients with exposures to either 1) COVID infection or 2) complete vaccination while on the waitinglist, that also had HLA single antigen testing within 3 months prior to the exposure and within 3 months after the exposure. The close proximity of single-antigen testing to the COVID infection or vaccination event is critical for a granular understanding of the HLA antibody profile, or confounders become limiting (antibody levels fade, other sensitizing events occur, etc.). Chart review was used at each center to collect patient demographics, including historic sensitizing events, pre- and post-exposure cPRA, as well as clinical details surrounding COVID infection or vaccination, and the exact process for collection of these variables was center specific. Many waitlisted patients are treated for infections at non-transplant hospitals, and vaccination is also generally not performed at transplant centers, so most of the clinical details of infection and vaccination were obtained via chart review of waitlisted patients records at neighboring medical centers by the participating center. Patients with both vaccination and infection during the study period were excluded before data was submitted to the coordinating center, so no patients were included in both cohort. Patients with a sensitizing event between the single antigen tests were excluded before data was sent to the coordinating center. Data collection posed unique challenges at each center and therefore took a variable amount of time. Once completed, data was sent as a single batch to the coordinating center, and centers were not expected to collect data on additional patients.
2.2. cPRA and specificities
cPRA used in this study were the cPRA used for UNOS listing by the center. Specifically, local MFI thresholds were used for cPRA calculations prior to and after exposure by the center per local protocol for UNOS listing. If the second cPRA was different from the first cPRA, the local HLA lab director identified and reported the specificities and MFI responsible for the cPRA change. Deidentified data was sent to the coordinating center. HLA lab directors adjudicated specificities for each case in which the cPRA changed.
2.3. SARS‐CoV‐2 RNA testing
Nasopharyngeal and oropharyngeal testing samples were collected based on clinical indication and analyzed locally as previously described.[14].
2.4. Vaccination
Vaccination occurred per the center protocol, and patients were vaccinated if they completed the primary vaccination series defined by CDC during the study period (received two doses of an mRNA vaccine or 1 dose of the DNA vaccine).
2.5. HLA antibody testing
Quantification of antibodies to HLA class I and class II was performed at the centers with the Luminex-based SAB assay as previously described (One Lambda Inc., Canoga Park, CA).[12] One center used the supplement kit to cover more single antigen beads. Center protocol determined the need for pre-treatment with dithiothreitol (DTT) or ethylenediaminetetraacetic acid (EDTA) of all serum samples to prevent aggregation of high titer antibodies and improve the sensitivity of antibody detection. One center used a secondary biotinylated anti-IgG with streptavidin-conjugated phycoerythrin (SAPE) to reduce the need for dilution of the sera [15]. Specific criteria for HLA antibody interpretation, MFI cutoffs, and listing unacceptable antigens for cPRA calculations and match run were previously established by the center and not adjusted based on participation in this study, and such center-specific criteria are summarized below. We compared each patient's cPRA results and antibody specificities before and after COVID-19.
2.5.1. Site#1, Emory
HLA antibody assignments are based on reaction patterns to epitopes/eplets identifying unique (private) or shared (cross-reactive) sequences of amino acids HLA class I and class II molecules. MFI values are a semi-quantifiable metric to assess antibody quantity. If multiple beads have allelic variants of the same antigen (e.g., HLA-A*02:01 vs *02:03 vs *02:06 are each unique representations of the HLA-A2 antigen), each has different MFI values, the MFIs of all three beads are averaged to report the HLA-A2 antibody strength. While many laboratories use EDTA to abrogate the “prozone effect” seen with some patient samples, the Emory HLA Laboratory uses a method that avoids EDTA treatment. Targets of HLA antibodies identified are listed as unacceptable antigens in UNOS according to the following criterion: HLA-A/B/DRB1/DRB3/DRB4/DRB5/DQA1 and DQB1 allotypes are listed as unacceptable antigens when the patient displays antibodies with ≥ 3,000 MFI on a Luminex 3D instrument (2000 MFI if using a Luminex 200). For HLA-C, HLA-DPA1, and HLA-DPB1 locus antibodies, the MFI cutoffs are > 5000 MFI.
2.5.2. Site#2, Georgetown University
All sera are EDTA-treated. HLA antibody assignments are based on reaction patterns to epitopes/eplets that identify unique (private) or shared (cross-reactive) sequences of amino acids HLA class I and II molecules. MFI values are a semi-quantifiable metric to assess antibody quantity. The SAB-detected antibody specificities are confirmed by the phenotype (multi-antigen) bead assay (specificities that are not confirmed are not called positive and not listed as unacceptable antigens regardless of the MFI values). When beads carrying allelic variants of the same antigen (e.g., HLA-A*02:01, *02:03, *02:06 - variants of HLA-A2 antigen) are all positive, then the average MFI is used to quantify the HLA-A2 antibody levels; however, when allele-specific reactions are detected (e.g., DRB1*04:02 vs all other DR4 alleles), then only the positive allele (or alleles) is reported if MFI > 1,000, or at a lower MFI if it is a part of a shared epitope or eplet pattern. Targets of the identified HLA antibodies are listed as unacceptable antigens in UNOS if they are thought to increase the risk of graft rejection. The following are the criteria for listing unacceptable antigens: 1) HLA-A/B/C/DR/DR51/DR52/DR53/DQB1/DQA1 allotypes are listed as unacceptable antigens if the patient displays antibodies > 1,000–2,000 MFI (depending on the pattern); 2) Bw4 or Bw6 are listed as unacceptable antigens at any MFI; 3) DPA1 is not listed; 4) DPB1 are listed if titers are > 1:64.
2.5.3. Site#3, UCSF
All sera are DTT-treated before being used for HLA antibody testing to remove interfering substances. HLA antibody specificity is determined based on known cross-reactivity patterns. The MFI is used as an arbitrary unit of HLA antibody quantity. If multiple beads have allelic variants of the same antigen (e.g., HLA-A*02:01, *02:03, *02:06 – the variants of HLA-A2 antigen), then the average MFI of all positive beads is used to quantify HLA-A2 antibody MFI strength. Targets of HLA antibodies identified are listed as unacceptable antigens in UNOS if they are thought to increase the risk of graft rejection. The following are the criterion for listing unacceptable antigens: 1).HLA-A/B/C/DR/DR51/DR52/DR53/DQB1 allotypes are listed as unacceptable antigens if the patient displays antibodies with ≥ 2,000 MFI reactivity against these allotypes; 2) Bw4 or Bw6 are listed as unacceptable antigens if the patient displayed antibodies to these epitopes at any MFI; 3) none of the DQA1, DPA1, and DPB1 allotypes and HLA-A/B/C/DR/DR51/DR52/DR53/DQB1 alleles are listed as unacceptable antigens despite targeted antibodies of any MFI to maximize donor offers.
2.5.4. Site#4, MGH
HLA antibody specificity is determined based on a combination of MFI cutoff and known cross-reactivity patterns. The serum is pre-treated with EDTA. The following are the general criteria for listing unacceptable antigens: 1) antigens with all solid phase beads exhibiting > 3,000–4,000 MFI for HLA-A, B, DRB1, DRB3/4/5, DQB1 - on occasion, alleles, rather than antigens, are listed as unacceptable; 2) Bw4 or Bw6, if there is a clear pattern of reactivity > 1,000 MFI; 3) HLA-C, if there is a clear epitope pattern and/or if associated with prior positive cross matches, and MFI > 10,000; 4) mismatched antigens present on prior grafts have a lower cutoff of > 1,000 MFI; 4) DPB1 antigens, if they match common epitope patterns and are associated with prior positive cross matches; 5) DQA1 or DPA1 antigens are rarely listed as unacceptable, but may be entered if there is clearly clustering of all single antigen beads associated with that allele.
2.5.5. Site#5, UAB
Sera are pre-treated using a Melon IgG spin column to reduce interference similar to EDTA treatment. HLA antibody assignments are based on reactions greater than the cutoff of 1500 MFI, displaying known reaction patterns identifying shared (cross-reactive) amino acid sequences (epitopes) on HLA class I and II molecules. All antibody assignments between 1500 MFI and 5000 MFI are confirmed with a second single antigen bead assay from a different vendor. HLA antigens listed as unacceptable antigens in UNOS will vary with patient and assessment of risk (repeated mismatch, shared epitope, known sensitization through pregnancy) but generally include all clearly defined specificities greater than > 2000 MFI. If the patient is highly sensitized (CPRA = 100), specificities, particularly C and DP, may be left off the unacceptable antigens up to an MFI of 5,000 to allow for more offers.
2.5.6. Site#6, Upenn
Sera are treated with DTT to abrogate the “prozone effect” seen with some patient samples. The following are the criterion for listing unacceptable antigens: HLA antibody assignments are based on reaction patterns to epitopes/eplets identifying unique (private) or shared (cross-reactive) sequences of amino acids HLA class I and class II molecules. Targets of HLA antibodies identified are listed as unacceptable antigens in UNOS according to the following criterion: HLA-A/B/DRB1/DRB3/DRB4/DRB5/DQA1 and DQB1 specificities are listed as unacceptable antigens when the patient displays antibodies with ≥ 3,000 MFI on a Luminex 3D instrument except for HLA-C which has 500 MFI cutoff. If multiple beads have allelic variants of the same antigen (e.g., HLA-A*02:01 vs *02:03 vs *02:06 are each unique representation of the HLA-A2 antigen), each has different MFI values, only the alleles with ≥ 3,000 MFI are listed.
2.5.7. Site#7, UTSA
All sera are EDTA-treated prior to HLA antibody testing. HLA antibody specificity is determined based on known cross-reactivity patterns. MFI is used as an arbitrary, semi-quantitative, unit of HLA antibody detection. If multiple beads have allelic variants of the same antigen (e.g., HLA-A*02:01, *02:03, *02:06 – the variants of HLA-A2 antigen), the average MFI of all positive beads is used to estimate HLA-A2 antibody MFI strength. Targets of HLA antibodies identified are listed as unacceptable antigens in UNOS if they are thought to increase the risk of graft rejection. The following are the criterion for listing unacceptable antigens: 1).HLA-A/B/DR/DR51/DR52/DR53 allotypes are listed as unacceptable antigens if the patient displays antibodies with ≥ 4,000 MFI reactivity against these allotypes; 2) HLA-C allotypes are listed as unacceptable antigens if the patient displays antibodies with ≥ 5,000 MFI reactivity against these allotypes; 3) HLA-DPB allotypes are listed as unacceptable antigens if the patient displays antibodies with ≥8,000 MFI reactivity against these allotypes; 4) Bw4 or Bw6 are listed as unacceptable if a patient displays antibodies against all alleles horboring these epitopes in a tight stacked pattern at any MFI and is homozygous for the alternative epitope; 5) DQA1 and DPA1 allotypes are not listed as unacceptable to maximize donor offers.
2.5.8. Site#8, USC:
HLA antibody assignments are based on reaction patterns to epitopes/eplets identifying unique (private) or shared (cross-reactive) sequences of amino acids HLA class I and class II molecules. MFI values are a semi-quantifiable metric to assess antibody quantity. If multiple beads have allelic variants of the same antigen (e.g., HLA-A*02:01 vs *02:03 vs *02:06 are each unique representations of the HLA-A2 antigen), each has different MFI values, the MFIs of all three beads is averaged to report the HLA-A2 antibody strength. If the HLA-A*02:03 bead is reactive and the other HLA-A2 variants are not, we will list in UNOS as HLA-A2 antigen due to UNOS current match run system. We used EDTA to eradicate the inhibitory effects of “prozone”. Targets of HLA antibodies identified are listed as unacceptable antigens in UNOS according to the following criterion: HLA-A/B/DRB1/DRB3/DRB4/DRB5/DQA1 and DQB1 allotypes are listed as unacceptable antigens when the patient displays antibodies with ≥ 5,000 MFI on a Luminex 3D instrument. For HLA-C, HLA-DPA1, and HLA-DPB1 locus antibodies, the MFI cutoffs are > 5000 MFI.
2.6. Statistical analysis
Descriptive baseline characteristics were quantified as percentages or median values with interquartile range when appropriate. Mean with standard deviation was used to compare cPRA between groups. A 2-tailed T-test was used to evaluate the significance between cPRA groups (P < 0.05). The distribution of gender, retransplant status, and other demographic characteristics between the study groups were estimated by Pearson chi-square (P < 0.05).
3. Results
Seven centers contributed data for 409 patients with SAB HLA antibody class I and class II testing meeting the strict testing period around the exposure, including 149 patients with SARS-CoV-2 infection and 260 that were vaccinated. Baseline patient characteristics are shown in Table 1 , and as expected, 85.3 % of the patients had hypertension and 45.6 % had diabetes. Of the patients with SARS-CoV-2 infection, 34.2 % required hospitalization, 4.7 % ICU level care and 3.4 % mechanical ventilation. Of the 260 vaccinated patients, the vast majority received an mRNA vaccine, and 4 (1.5 %) received a DNA vaccine. Two hundred eighty-five patients (69.7 %) of the entire study population had an initial cPRA of 0 %, and 56 (13.7 %) had an initial cPRA > 80 %. Our cohort resembles the national UNOS waitlist in terms of sensitization (approximately 80 % of the waitlist is ‘unsensitized” with a cPRA of < 10 %, and 20 % with a cPRA > 80 %), gender (57.7 % of males in our cohort vs 62 % UNOS cohort) and the history of previously undergoing a transplant (14.7 % of our cohort vs 13.8 % UNOS cohort) (https://WWW.SRTR.org).
Table 1.
Patient baseline characteristics.
|
Entire cohort |
Infected |
Vaccinated |
|
|---|---|---|---|
| (n = 409) | (n = 149) | (n = 260) | |
| Age (yrs) | 55.0 (17.0) | 53.0 (19.0) | 57.0 (15.5) |
| Sex male | 57.70 % | 55.0(%) | 59.20 % |
| BMI | 28.6 (7.2) | 28.5 (7.3) | 28.6 (7.5) |
| History of prior to transplant | 14.70 % | 16.10 % | 13.80 % |
| Hypertension | 85.30 % | 83.90 % | 86.00 % |
| Diabetes | 45.60 % | 45.00 % | 45.90 % |
| History of transfusion | 37.90 % | 50.80 % | 31.30 % |
| Multiparous | 25.10 % | 28.40 % | 23.20 % |
| On immunosuppressing medication | 13.70 % | 17.40 % | 11.60 % |
| Hospitalized for COVID | NA | 34.20 % | NA |
| Required ICU care for COVID | NA | 4.70 % | NA |
| Required mechanical ventilation for COVID | NA | 3.40 % | NA |
| Required only oxygen supplementation for COVID | NA | 16.10 % | NA |
| Time from exposure to single antigen test (days) | 55.0 (49.0) | 64.0 (43.0) | 51.5 (49.0) |
| Initial cPRA | |||
| 0 % | 285 (69.7 %) | 98 (65.8 %) | 187 (71.9) |
| 1–20 % | 31 (7.6 %) | 18 (12.1 %) | 13 (5.0 %) |
| 21–80 % | 37 (9.0 %) | 14 (9.4 %) | 23 (8.8 %) |
| 81–97 % | 26 (6.4 %) | 9 (6.0 %) | 17 (6.5 %) |
| 98 % | 5 (1.2 %) | 2 (1.3 %) | 3 (1.2 %) |
| 99 % | 5 (1.2 %) | 2 (1.3 %) | 3 (1.2 %) |
| 100 % | 20 (4.9 %) | 6 (4.0 %) | 14 (5.4 %) |
| Patient distribution by participating centers | |||
| Center ID_1 | 20 | 1 | 19 |
| Center ID_2 | 71 | 19 | 52 |
| Center ID_3 | 43 | 15 | 28 |
| Center ID_4 | 36 | 35 | 1 |
| Center ID_5 | 36 | 20 | 16 |
| Center ID_6 | 46 | 11 | 35 |
| Center ID_7 | 72 | 40 | 32 |
| Center ID_8 | 85 | 8 | 77 |
Categorical values are stated as number (percentage), or median (interquartile range). Initial cPRA values are stated as the number of patients (percentage). Not applicable (NA).
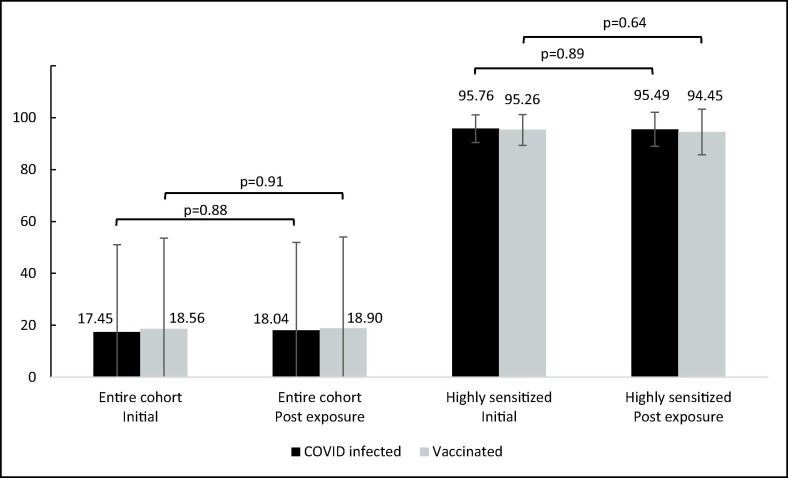
After exposure, 26 patients (6.4 %) had a change in cPRA value (Table 2 ), 16 (3.9 %) had an increase, and 10 (2.4 %) had a decrease. Of the 16 patients with a cPRA increase, 5 patients (3.4 %) were from the COVID-recovered cohort and 11 (4.2 %) were from the vaccinated cohort. Notably, all five COVID recovered patients with an increased cPRA were female (p = 0.02), although gender did not influence the likelihood of an increased cPRA among the vaccinated group (supplement table 1). The cPRA change following infection or vaccination was not associated with any other demographic characteristic, including the history of undergoing a previous transplant. Fig. 1 shows the cPRA change in the entire cohort, and in the highly sensitized patients (cPRA ≥ 80 %).
Table 2.
Mean cPRA Change in Patients who Experienced a Change Post-Exposure (n = 26).
| All Pts | Infected | Vaccinated | |
|---|---|---|---|
| Mean cPRA Change (absolute) | 17.42 ± 16.85 (n = 26) |
17.98 ± 12.19 (n = 6) |
17.25 ± 18.29 (n = 20) |
| Mean cPRA Increase | 19.60 ± 18.01 (n = 16) |
19.52 ± 12.96 (n = 5) |
19.64 ± 20.48 (n = 11) |
| Mean cPRA Decrease | 13.94 ± 15.04 (n = 10) |
10.31 (n = 1) |
14.34 ± 15.89 (n = 9) |
| Categorical values stated as mean ± standard deviation. | |||
Fig. 1.
Initial cPRA and cPRA after exposure.
Each patient with a cPRA change was adjudicated (Table 3 ). No patients with an initial cPRA of 0 % that were infected with COVID had a cPRA > 0 % after exposure, although 5 vaccinated patients with an initial cPRA of 0 % that were vaccinated had a cPRA > 0 % after vaccination. Adjudication revealed the cPRA differences were almost all the result of local cPRA practices and specificities with subtle fluctuations around the borderline of the participating centers’ mean fluorescence intensity (MFI) cutoff for unacceptable antigen listing rather than the appearance of a new antibody or a memory response. None of the new or increased antibodies detected in the candidates awaiting retransplant were directed to the previous donor. The spouse HLA types are unavailable to determine if the new or increased antibodies are due to potential memory response to the spouse HLA allotypes. Four of the 5 cases with a significantly increased antibodies were sensitized patients (4 of the 124 subjects with cPRA > 0 %).
Table 3.
Patients with cPRA Change following COVID-19 infection or vaccination.
| Center #; Patient # | Re-transplantation (yes/no) | Pregnancies (yes/no/NA) | Initial cPRA | CPRA after exposure | Actual cPRA change | Direction of change | HLA antibody specificities (MFI) influencing this cPRA change | Comment |
|---|---|---|---|---|---|---|---|---|
| COVID Infected patients | ||||||||
| 5;2 | no | yes | 8 | 39 | 31 | ↑ | A23 (<1000 to 6949), A24 (<1000 to 9516), A29 (<1000 to 3932) | High MFI antibodies to A3 (A23, A24) and A29 |
| 7;2 | no | yes | 42 | 72 | 30 | ↑ | B*51:02 (<1000 to 1449), B*14:01 (<1000 to 1187), C*15:02 (<1000 to 1624), C*17:01 (<1000 to 1683), C*18:02 (<1000 to 1272), C*02:02 (<1000 to 1201), C*05:01 (<1000 to 1705), C*06:02 (<1000 to 1172) | Subtle increase over MFI cut-off |
| 4;2 | no | no | 22 | 47 | 25 | ↑ | new antibodies DQ2 (MFI range 1774–4556) | Subtle increase over MFI cut-off |
| 5;1 | yes | yes | 11 | 17 | 6 | ↑ | B38 (<1000 to 1696), B39 (<1000 to 1648) | Subtle increase over MFI cut-off |
| 7;1 | no | NA | 95 | 100 | 5 | ↑ | B*15:13 (<1000 to 1717), C*01:02 (<1000 to 1346), C*03:02 (<1000 to 1924), C*02:02 (<1000 to 1050), C*03:03 (<1000 to 2396), DRB1*01:03 (<1000 to 2602), DQA1*05:01 (<1000 to 1720). | Subtle increase over MFI cut-off |
| 4;1 | no | yes | 89 | 78 | 11 | ↓ | B61 (3773 to 2194), B60 (4309 to 1948), B48 (3755 to 1978), B57 (3616 to 1919), B78 (3530 to 1315), B39 (3236 to 1130), B8 (3112 to 1050) | Subtle decrease under MFI cut-off |
| COVID Vaccinated patients | ||||||||
| 7;8 | no | yes | 0 | 68 | 68 | ↑ | B*82:01 (<1000 to 1502), DQB1*05:02 (<1000 to 1562), DRB4*01:01 (<1000 to 1379) | Subtle increase over MFI cut-off |
| 8;6 | no | NA | 0 | 37 | 37 | ↑ | DQA1*01:02&DQB1*06:04 (3972 to 5090), DQA1*01:02&DQB1*06:02 (3752 to 4985) | Subtle increase over MFI cut-off |
| 2;1 | yes | NA | 57 | 90 | 33 | ↑ | DQ8 (0 to 3000–5000), DQ9 (0 to 3000–5000) DQ7 (0 to 1700) | Subtle increase over MFI cut-off |
| 8;7 | no | NA | 0 | 23 | 23 | ↑ | C*03:03 (133 to 17396), C*15:02 (44 to 16521), C*03:04 (66 to 16368), C*03:02 (37 to 14868), C*15:05 (0 to 13497), A*80:01 (180 to 7101), A*25:01 (267 to 5061) | High MFI antibodies to certain Cw, A25 and A80 |
| 8;9 | no | yes | 0 | 19 | 19 | ↑ | B*15:17 (2383 to 5888), B*49:01 (2712 to 5457), B*51:02 (2150 to 4352), A*32:01 (2235 to 4261), B*51:01 (2157 to 4091), B*59:01 (1704 to 4048), B*15:16 (1961 to 4043) | Subtle increase over MFI cut-off |
| 7;5 | no | yes | 81 | 99 | 18 | ↑ | B*15:12 (1277 to 1438), B*15:13 (1277 to 2311), B*82:01 (1277 to 2968) C*15:02 (1277 to 3068), DQA1*05:01 (1277 to 1323), DPB1*28:01 (1277 to 2634), DRB4*01:03 (1277 to 1028) | Subtle increase over MFI cut-off |
| 3;1 | no | no | 0 | 11 | 11 | ↑ | B57 (<1000 to 5600) B58 (<1000 to 3500) | Subtle increase over MFI cut-off |
| 8;3 | yes | NA | 95 | 99 | 4 | ↑ | B*15:17 (3600 to 6939), B*15:16 (2203 to 4933), A*29:01 (2167 to 4707), A*29:02 (2216 to 4425), DRB4*01:03 (2267 to 13822), DRB4*01:01 (1484 to 12326), DRB1*09:01 (2362 to 7750), DRB1*01:01 (0 to 5468), DRB1*07:01 (0 to 5022), DRB1*01:03 (0 to 4561), DRB1*01:02 (0 to 4451) | Increase in DR7, DR9 and DR53 antibodies |
| 7;3 | yes | NA | 98 | 99 | 1 | ↑ | B46 (<1000 to 1251), B49 (<1000 to 1295), B50 (<1000 to 1246), B52 (<1000 to 1159), B60 (<1000 to 1087), B61 (<1000 to 1117), B*15:11 (<1000 to 1147), B77 (<1000 to 1121) | Subtle increase over MFI cut-off |
| 8;2 | no | NA | 97 | 98 | 1 | ↑ | B*14:01 (3174 to 4357), DRB4*01:01 (4671 to 5102), DRB4*01:03 (3797 to 4122) | Subtle increase over MFI cut-off |
| 8;4 | no | yes | 61 | 62 | 1 | ↑ | B*15:10 (3975 to 5016), B*48:01 (3813 to 4574) | Subtle increase over MFI cut-off |
| 8;5 | no | no | 49 | 0 | 49 | ↓ | C*07:01 (9012 to 7352), C*07:02 (8464 to 7173), C*07:04 (5465 to 4112), C*17:03 (5060 to 3767), B*73:01 (4272 to 3271) | Subtle decrease under MFI cut-off |
| 7;4 | no | yes | 95 | 67 | 28 | ↓ | A*23:01 (1813 to < 1000), A*25:01 (2881 to < 1000), A*69:01 (1027 to < 1000), A*80:01 (1499 to < 1000), B*48:01 (1878 to < 1000), B*40:01 (2193 to < 1000), B*40:02 (2214 to < 1000), B*40:06 (1801 to < 1000), B*07:02 (2735 to < 1000), B*15:02 (1030 to < 1000), B*15:12 (1425 to < 1000), B*81:01 (2847 to < 1000), C*18:02 (2766 to < 1000), C*02:02 (2240 to < 1000), C*04:01 (1116 to < 1000), C*05:01 (3636 to < 1000), C*06:02 (3667 to < 1000). | Subtle decrease under MFI cut-off |
| 7;6 | no | NA | 81 | 63 | 18 | ↓ | A66 (3302 to < 1000), B48 (1714 to < 1000), B60 (2797 to < 1000), B61 (1922 to < 1000), B73 (1678 to < 1000), B81 (3018 to < 1000), DQA1*05:03 (1059 to < 1000), DQA1*05:05 (1102 to < 1000) | Subtle decrease under MFI cut-off |
| 7;7 | no | NA | 54 | 37 | 17 | ↓ | DQA1*05:05 (1080 to < 1000), DQA1*06:01 (1069 to < 1000), DP1 (1911 to < 1000) | Subtle decrease under MFI cut-off |
| 4;5 | no | no | 28 | 25 | 3 | ↓ | B58 (3541 to 2575) | Subtle decrease under MFI cut-off |
| 4;6 | no | NA | 5 | 0 | 5 | ↓ | DRB1*14:02 (747 to 596) | Subtle decrease under MFI cut-off |
| 4;4 | yes | NA | 91 | 86 | 5 | ↓ | A*66:01 (1277 to 1046), B47 (3270 to 2987), B54 (3171 to 2759). | Subtle decrease under MFI cut-off |
| 4;3 | yes | no | 96 | 94 | 2 | ↓ | A*66:01 (1277 to 1046), B47 (3270 to 2987), B54 (3171 to 2759) | Subtle decrease under MFI cut-off |
| 8;1 | no | yes | 100 | 99 | 1 | ↓ | A*24:03 (4340 to 3817) | Subtle decrease under MFI cut-off |
CPRA - calculated panel reactive antibody, MFI - mean fluorescence intensity, NA - not available.
4. Discussion.
The SARS‐CoV‐2 pandemic has evolved into a global epidemic that will not disappear. Many aspects of this infection hold specific interest for patients with chronic kidney disease awaiting kidney transplantation. Transplant providers are learning to work around this pathogen to perform kidney transplants in a safe and efficient manner.
Virtual crossmatching is increasingly used in place of a physical crossmatch at the time of organ offer. Virtual crossmatching depends on the presence of a reliable HLA antibody profile determined by the single antigen bead assay at the time of an organ offer. Infections and vaccinations against this pathogen have become routine, therefore, an understanding of the effects of exposure to this virus and/or vaccination against it is essential for patients atop the waiting list. If SARS‐CoV‐2 infection or vaccination causes the HLA antibody profile to change, the risk of rejection after transplant may be increased. Currently, guidelines regarding the need for HLA antibody testing prior to moving forward with kidney transplant do not exist for candidates following infection with SARS‐CoV‐2, or COVID vaccination.
Infection with SARS‐CoV‐2 causes a somewhat unique immune dysregulation.[16] Additionally, infection with several other viruses has been shown to cause HLA antibodies via T-cell cross-reactivity (termed heterologous immunity) [17], [18], [19], [20], [21]. Also, male convalescent plasma donors with no known sensitizing events after COVID-19 infection have displayed HLA antibodies.[11] Lastly, it is largely unknown if SARS‐CoV‐2 infection or COVID vaccination induces a memory response which could cause an increase in the MFI of an existing HLA antibody, termed a memory response.
Anecdotal expert consensus is that the risk of HLA antibody formation after one of these exposures is low, but since the actual risk is unknown as well as the absence of data, opinions differ about the safety of proceeding directly to transplant after a recent SARS‐CoV‐2 infection or COVID vaccination. HLA antibody formation after this type of exposure is likely to be a rare event so a large cohort of patients is required to address this question. Individual centers lack the volume of patients with single antigen testing at the proper time points around exposure, so a multicenter study is required. Additionally, highly sensitized patients are needed to identify activation of a memory response, and a single-center cohort of highly sensitized patients is even rarer.
There are reasons for the paucity of data published on this topic. First, reliable data collection, able to precisely capture the HLA antibody formation in this population, poses unique challenges. The data must capture a large number of patients because the frequency of the event is low. Capturing HLA antibody formation after any exposure requires single antigen testing performed in a relatively narrow time window because antibodies may wane and the possibility of non-identified exposures in patients receiving care at non-transplant hospitals increases over time. For these reasons a strict single antigen testing window is critical. Given the need for data precision a single-center experience would seem to provide the best study environment, but the number of patients required is too large for a single-center study. Another challenge is center variation in single antigen testing protocols for waitlisted patients, and interpretation of the results, limiting options for study design and data analysis. The cPRA is derived from HLA antibody specificities which fluctuate (up and down) a small amount between tests [22] so granularity of the data and thoughtful interpretation within the context of the center MFI cutoffs is required.
Patients with no called antibodies prior to exposure are an optimal cohort to determine if SARS‐CoV‐2 infection or COVID vaccination results in new antibody development. No patients with an initial cPRA of 0 % had an increase in the cPRA after infection. After vaccination a very similar number of patients had a decrease in cPRA or an increase in cPRA, including 5 patients that had an initial cPRA of 0 % and a final cPRA > 0 %. Most of these patients exhibited a slight increase in 2–7 HLA antibody specificities with MFI of around 2000, just above their center’s cutoffs for unacceptable antigen listing. Despite the cPRA increase, these changes presumably do not impact the outcome of flow cytometry crossmatching. Patient 7 from center 7 is a very good example of the difficulty interpreting cPRA across centers with varied practices, an issue that is rarely described. Some centers acknowledge the antibodies displayed in this post exposure sample (C-locus and A*80-locus) could reflect artifactual reactivity in the solid phase SAB assay, and further testing could confirm donors with these antibodies should not be excluded, while other centers tentatively list them as avoids. In any event, it was reassuring to see the overwhelming majority of patients had no identifiable increase in risk of rejection after exposure to infection or vaccination. Consistent with our findings, reports on kidney and heart transplant recipients revealed that neither SARS-CoV-2 infection nor Covid-19 vaccination was associated with significant changes in the donor-specific HLA antibodies (DSA) [23], [24], [25], [26], [27]. However, a case report revealed a positive B cell flow cytometry crossmatch in a patient waiting for second kidney transplantation after receiving the COVID-19 vaccine, presumably due to the bystander activation of memory response by the COVID-19 vaccination[28]. Another case report of ABO incompatible living donor kidney transplant revealed appearance of new DSA following COVID-19 vaccination[29].
The strengths of this study are the multi-institutional nature allowing an adequate real-world study population, including a large number of both unsensitized and highly sensitized patients, with a strict study window for single antigen testing around the exposure, and the granularity of the antibody specificity data provided by the centers. The study is a retrospective cohort study of patients on the national kidney transplant waiting list exposed to infection or vaccination available at the time of the study. The SAB interpretation and cPRA calculation used for UNOS listing vary between centers, since various centers may accept different levels of immunological risk for diverse patient populations. As it may not be practical to standardize the cPRA calculations based on the common cutoffs, this study aimed to determine whether the patient’s immunological assessment within each transplant center may be affected by COVID infection/vaccination. Our data show that the risk of increasing an immunological risk is insignificant; however, the study design captures very granular data for all cases with cPRA change. The weaknesses of the study primarily revolve around the fluid nature of this virus and our efforts to vaccinate against it, as the variants continue to evolve, and we did not capture booster vaccinations that became more common after the study period. The fact that many patients are treated at hospitals that are not their transplant center increases the risk of confounding events occurring between single antigen tests and makes data collection challenging. Centers captured as much data about other sensitizing events between single antigen tests as possible from other institutions and excluded these patients, but it is likely we did not capture all confounding exposures. The retrospective cohort selection could introduce bias. Immunosuppression in retransplant candidates may play a role in suppressing HLA antibody production following COVID-19 infection and vaccination. Also, center-specific protocols, such as the use of DTT, EDTA or SAPE to prevent complement interference, could have a subtle effect on specificities, but the number of centers in the study helps overcome confounders such as these. Center level decisions about UNOS listing cPRA vary, and bias may have been introduced by the number of patients a center enrolled, possibly underestimating the risk of antibody formation (note- center 7 enrolled a large number of vaccinated patients). Lastly, the UNOS cPRA calculator does not include DQA1 and DP antibodies, and therefore one of the limitations is that the cPRA values provided in this study do not consider changes in DQA1 and DP antibodies. However, only a few subtle DQA1/DP antibody changes were observed only in 5 patients, and thus the effect of this limitation is minimal.
In conclusion, SARS-CoV2 infection and vaccination did not meaningfully alter anti HLA antibody specificities in 99 % of this cohort. Changes displayed in the vast majority of patients were very small fluctuations in specificities. Data from the small number of patients with a significant cPRA change can be interpreted as showing there is a minimal increased immunological risk to transplant candidates based on a patient's prior infection with or vaccination to SARS-CoV-2. This has implications for virtual crossmatching at the time of organ offer after SARS-CoV-2 infection or vaccination. Vaccination does not lead to sensitization and is safe for waitlisted patients from that perspective, even if they are highly sensitized. This is important in light of the limited response to vaccination seen in immunosuppressed recipients after transplant.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.humimm.2023.02.005.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Masson E., Vidal C., Deschamps M., et al. Incidence and risk factors of anti-HLA immunization after pregnancy. Hum. Immunol. 2013;74(8):946–951. doi: 10.1016/j.humimm.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 2.Hyun J., Park K.D., Yoo Y., et al. Effects of different sensitization events on HLA alloimmunization in solid organ transplantation patients. Transplant Proc. 2012;44(1):222–225. doi: 10.1016/j.transproceed.2011.12.049. [DOI] [PubMed] [Google Scholar]
- 3.Lopes D., Barra T., Malheiro J., et al. Effect of different sensitization events on HLA alloimmunization in kidney transplantation candidates. Transplant Proc. 2015;47(4):894–897. doi: 10.1016/j.transproceed.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Picascia A., Grimaldi V., Sabia C., Napoli C. Comprehensive assessment of sensitizing events and anti-HLA antibody development in women awaiting kidney transplantation. Transpl. Immunol. 2016;36:14–19. doi: 10.1016/j.trim.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Akgul S.U., Ciftci H.S., Temurhan S., et al. Association Between HLA antibodies and different sensitization events in renal transplant candidates. Transplant Proc. 2017;49(3):425–429. doi: 10.1016/j.transproceed.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Azzi Y., Bartash R., Scalea J., Loarte-Campos P., Akalin E. COVID-19 and solid organ transplantation: a review article. Transplantation. 2021;105(1):37–55. doi: 10.1097/TP.0000000000003523. [DOI] [PubMed] [Google Scholar]
- 7.Cravedi P., Mothi S.S., Azzi Y., et al. COVID-19 and kidney transplantation: results from the TANGO international transplant consortium. Am. J. Transplant. 2020;20(11):3140–3148. doi: 10.1111/ajt.16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyarsky B.J., Werbel W.A., Avery R.K., et al. Antibody Response to 2-Dose SARS-CoV-2 mRNA vaccine series in solid organ transplant recipients. JAMA. 2021;325(21):2204–2206. doi: 10.1001/jama.2021.7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartzell S., Bin S., Benedetti C., et al. Evidence of potent humoral immune activity in COVID-19-infected kidney transplant recipients. Am. J. Transplant. 2020;20(11):3149–3161. doi: 10.1111/ajt.16261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKechnie J.L., Blish C.A. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27(6):863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juskewitch J.E., Stubbs J.R., Gandhi M.J. Elevated Rate of HLA Antibodies in Male COVID-19 convalescent plasma donors: a risk factor for transfusion-related acute lung injury. Mayo Clin. Proc. 2021;96(2):500–502. doi: 10.1016/j.mayocp.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roll G.R., Webber A.B., Gae D.H., et al. A virtual crossmatch-based strategy facilitates sharing of deceased donor kidneys for highly sensitized recipients. Transplantation. 2020;104(6):1239–1245. doi: 10.1097/TP.0000000000002924. [DOI] [PubMed] [Google Scholar]
- 13.Roll G.R., Lunow-Luke T., Braun H.J., et al. COVID-19 does not impact HLA antibody profile in a series of waitlisted renal transplant candidates. Hum. Immunol. 2021;82(8):568–573. doi: 10.1016/j.humimm.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X., Wang L., Sakthivel S.K., et al. US CDC Real-Time Reverse Transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(8) doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullivan H.C., Gebel H.M., Bray R.A. Understanding solid-phase HLA antibody assays and the value of MFI. Hum. Immunol. 2017;78(7–8):471–480. doi: 10.1016/j.humimm.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., et al. Complex Immune Dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000 e1003. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Orsogna L., van den Heuvel H., van Kooten C., Heidt S., Claas F.H.J. Infectious pathogens may trigger specific allo-HLA reactivity via multiple mechanisms. Immunogenetics. 2017;69(8–9):631–641. doi: 10.1007/s00251-017-0989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katerinis I., Hadaya K., Duquesnoy R., et al. De novo anti-HLA antibody after pandemic H1N1 and seasonal influenza immunization in kidney transplant recipients. Am. J. Transplant. 2011;11(8):1727–1733. doi: 10.1111/j.1600-6143.2011.03604.x. [DOI] [PubMed] [Google Scholar]
- 19.van den Heuvel H., Heutinck K.M., van der Meer-Prins E.M.W., et al. Allo-HLA Cross-Reactivities of Cytomegalovirus-, Influenza-, and Varicella Zoster Virus-Specific Memory T Cells are shared by different healthy individuals. Am. J. Transplant. 2017;17(8):2033–2044. doi: 10.1111/ajt.14279. [DOI] [PubMed] [Google Scholar]
- 20.Amir A.L., D'Orsogna L.J., Roelen D.L., et al. Allo-HLA reactivity of virus-specific memory T cells is common. Blood. 2010;115(15):3146–3157. doi: 10.1182/blood-2009-07-234906. [DOI] [PubMed] [Google Scholar]
- 21.Morice A., Charreau B., Neveu B., et al. Cross-reactivity of herpesvirus-specific CD8 T cell lines toward allogeneic class I MHC molecules. PLoS One. 2010;5(8):e12120. doi: 10.1371/journal.pone.0012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed E.F., Rao P., Zhang Z., et al. Comprehensive assessment and standardization of solid phase multiplex-bead arrays for the detection of antibodies to HLA-drilling down on key sources of variation. Am. J. Transplant. 2013;13(11):3050–3051. doi: 10.1111/ajt.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCune T.R.B.R., Baran D.A., Toepp A.J., Forte S.J., Gilgannon L.T., Williams T., Chen S., Sadr H., Gebel H.M., Herre J.M. Development of donor specific antibodies after SARS-CoV-2 vaccination in kidney and heart transplant recipients. Transplant Immunoi. 2022;75 doi: 10.1016/j.trim.2022.101722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kueht M.K.K., Scott Lea A., Stevenson H.L., Fair J., Kathleen Gamilla-Crudo A., Hussain S., Mujtaba M. Donor-directed immunologic safety of COVID-19 vaccination in renal transplant recipients. Hum. Immunol. 2022;83:607–612. doi: 10.1016/j.humimm.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.V.P.W.M.V.S. Wijtvliet, B. Depreter, C. Heylen, E. Coeman, S. Abrams, B.Y. De Winter, A. Massart, R. Hellemans, L. Pipeleers, F.H.J. Claas, K.K. Ariën, K.M. Wissing, D. Abramowicz, K.J. Ledeganck, SARS-CoV-2 mRNA vaccination is not associated with the induction of anti-HLA or non-HLA antibodies, Transpl. Immunol. 74 (2022) 101670. [DOI] [PMC free article] [PubMed]
- 26.Cassaniti I.G.M., Bergami F., Arena F., Sammartino J.C., Percivalle E., Soleymaninejadian E., Abelli M., Ticozzelli E., Nocco A., Minero F., Pattonieri E.F., Lilleri D., Rampino T., Baldanti F. Effect of a third dose of SARS-CoV-2 mRNA BNT162b2 vaccine on humoral and cellular responses and Serum Anti-HLA antibodies in kidney transplant recipients. Vaccines (Basel). 2022;10:921. doi: 10.3390/vaccines10060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abu-Khader A.W.W., Berka M., Galaszkiewicz I., Khan F., Berka N. SARS Cov-2 vaccination induces de novo donor-specific HLA antibodies in a renal transplant patient on waiting list: a case report. HLA. 2022;99:25–30. doi: 10.1111/tan.14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Q.S.P., Helmick D., Lomago J.S., Tevar A.D., Zeevi A. Positive flow cytometry crossmatch with discrepant antibody testing results following COVID-19 vaccination. Am. J. Transplant. 2021;21:3785–3789. doi: 10.1111/ajt.16753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J.K.B.H., Ko G.Y., Lee J., Jung J., Jekarl D.W., Choi A.R., Lee S., Chung B.H., Yang C.W., Park S.C., Oh E.J. Successful ABO-incompatible living donor kidney transplantation in a recipient who developed flow cytometry crossmatch-positive donor-specific class I HLA antibodies following COVID-19 vaccination. HLA. 2022;100:152–158. doi: 10.1111/tan.14649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.