Abstract
Cancer heterogeneity has posed a great challenge to traditional cancer treatment, with the reappearance of cancer heterogeneity of inter and intra patients being especially critical. Based on this, personalized therapy has emerged as significant research focus in recent and even future years. Cancer-related therapeutic models are developing, including cell lines, patient-derived xenografts, organoids, etc. Organoids are three-dimensional in vitro models emerged in the past dozen years and are able to reproduce the cellular and molecular composition of the original tumor. These advantages demonstrate the great potential for patient-derived organoids to develop personalized anticancer therapies, including preclinical drug screening and the prediction of patient treatment response. The impact of microenvironment on cancer treatment cannot be underestimated, and the remodeling of microenvironment also allows organoids to interact with other technologies, among which organs-on-chips is a representative one. This review highlights the use of organoids and organs-on-chips as complementary reference tools in treating colorectal cancer from the perspective of clinical efficacy predictability. We also discuss the limitations of both techniques and how they complement each other well.
Subject terms: Tissue engineering, Cancer models
Facts
Tumor treatment needs personalized treatment based on standardized treatment.
Organoid can recapitulate the characteristics of parental tumor.
Several organoid studies have proved that organoid has great potential to predict drug response.
Organ-on-chip can simulate microenvironment by combining two or more cell types.
The combination of organoid and organ chip can provide broad research prospects for precise tumor treatment.
Open questions
How to carry out effective clinical transformation of drug prediction function of organoid model?
How to effectively combine organoid and organ-on-chip?
How can the interaction of these two models be used for the study of immunotherapy and antivascular therapy?
Introduction
Colorectal cancer (CRC) now ranks third in estimated new cases and emerges in the top three leading causes of death [1, 2]. There are proposals to bring forward CRC screening from 50 to 45, implying that cancer risk becomes earlier [3, 4]. Although advanced surgical techniques and modified adjuvant therapy have resulted in good colorectal cancer treatment outcomes, yet, CRC mortality remains high due to metastasis and post-treatment recurrence [5]. Colorectal cancer is a group of heterogeneous neoplastic diseases that usually originate from abnormal crypts. The heterogeneity of CRC can show distinct clinical and pathological features, leading to diverse outcomes and prognoses. Most colorectal cancers show changes in proto-oncogenes and tumor suppressor genes [6]. Generally, it is the result of an accumulation of genetic changes and epigenetic modifications over time and shows abnormalities in some of the following signaling pathways: Wnt/β-catenin, EGFR-RAS-RAF, MEK-MAPK, PI3K, p53, and TGF-β-SMADs [6, 7]. These molecular biomarkers can be used as a predictive and prognostic tool based on the application of next-generation sequencing.
The differences in clinical patient responses may be partly due to tumor heterogeneity. Thus, personalized treatment for different patients with different conditions is necessary, such as gene detection or evidence-based medicine [8]. Relying solely on diagnostic guidelines and molecular sequencing is insufficient to cover all patients. Human cancer cell lines are the most commonly used basic cancer and drug research models. However, few of them can fully recapitulate the mutation and transcriptional heterogeneity of primary tumors [9]. Another in vitro model, patient-derived xenograft (PDX), is relatively difficult to construct but with higher accuracy. The apparent limitations of its clinical application include low fluxes and long processing time. Because of the integrity of PDX microenvironment, it remains the gold standard for verifying drug sensitivity [10]. Organoids are three-dimensional cellular complex made by the self-organization of stem cells based on developmental biology principles [11], while organs-on-chips are microfluidic cell culture devices manufactured by the microchip manufacturing method [12]. The emergence of cancer organoids and organs-on-chips is currently suitable as a preclinical model for clinical transformation and precision medicine [13, 14] (Fig. 1). Significant advances in organoids and organs-on-chips technologies have facilitated the construction of in vitro near-physiological three-dimensional tissues and organs. Incorporating organoid precise recurrence of tumor features and organ-chip microenvironment integration may provide a tremendous opportunity to accelerate clinical transformation [15].
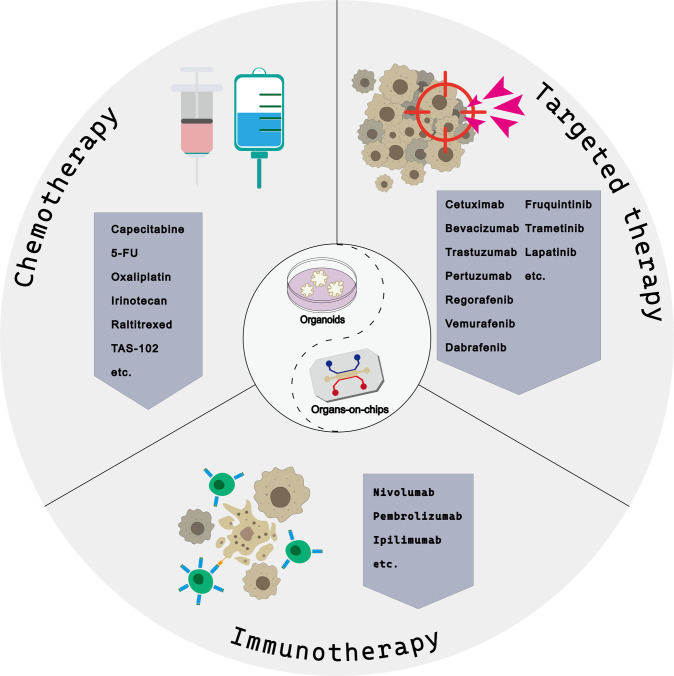
Fig. 1. Systemic therapy for colorectal cancer.
This figure depicts chemotherapy drugs, targeted drugs, and immunotherapy drugs commonly used in basic and clinical research of colorectal cancer based on organoid and organ chips.
In this review, we discuss two preclinical models of the adult stem cell-derived organoid system and organs-on-chips technology efficacy in predicting systemic treatment of colorectal cancer. We focused on the consistency of in vitro and in vivo drug responses based on these two models and the feasibility of adjusting treatment strategies in clinical patients based on this predictive effect as a supplement to the clinical treatment reference index. Finally, we discuss the two models’ limitations and look forward to organoids-on-a-chip technology.
The organoid system
Organoid is a three-dimensional cellular complex with a certain spatial structure formed by cells differentiated from stem cells cultured in vitro. Hans Clevers’s group in 2009 demonstrated that one single leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5+) intestinal stem cell could initiate 3D crypt-villus organoids. The small intestine organoids cultures were suspended in Matrigel accompanied by R- spondin 1 (a ligand of LGR5 and WNT agonist [16], epidermal growth factor (EGF), and the bone morphogenetic protein (BMP) inhibitor noggin). To date, organoids from most parts of the human body have been established, including healthy and tumor organoids from brain [17, 18], esophagus [19], lung [20, 21], breast [22], liver [23, 24], stomach [25, 26], pancreas [27, 28], kidney [29, 30], colon [21, 31], bladder [32, 33], prostate [34, 35] and so on. The culture systems for colorectal cancer organoids are summarized in Table 1, while the articles without clear reporting methods have been excluded. Adult stem cells and induced pluripotent stem cells (iPSCs) are the two main sources of organoids. Higher cell integrity and a greater diversity of cell types are frequently observed in iPSC-derived organoids. Organoids can capture the characteristics of their corresponding parental tumors [36]. The histological and genetic characteristics between organoids and parental tumors are highly consistent. Even in the course of culture for a certain period, the corresponding characteristics are not lost [37].
Table 1.
Culture systems for colorectal cancer organoids.
| Cancer lesion | Enzyme | Concentration | Digestion time | Growth factors | Required or absent | Function of growth factors in organoid culture | Ref |
|---|---|---|---|---|---|---|---|
| Colon |
Collagenase IX Dispase II |
75 U/mL 125 g/mL |
30–60 min |
Adv.DF12 + + + Wnt3a* R-spondin Noggin EGF Nicotinamide N-acetylcysteine Gastrin-1 A83-01 SB202190 Y-27632 PGE2# FGF-2# FGF-10# N2 supplement# B27 supplement |
+ +/− + + + + + + + + +/− +/− +/− +/− +/− + |
Basal medium to reduce serum use Stemness/Wnt signaling activator Stemness/Wnt signaling enhancer BMP inhibitor Mitogen Stemness/cystic phenotype Antioxidant Mitogen/Prolong survival TGF-β inhibitor p38MAPK inhibitor ROCK inhibitor /Block apoptosis Wnt activator/cystic phenotype Mitogen Mitogen Cofactor mixture Cofactor mixture based on N2 |
[21] |
| Colon and Rectum |
Collagenase II Hyaluronidase |
1.5 mg/ml 20 ug/ml |
30 min | [68] | |||
|
Colon and Rectum, Liver and ovarian (metastasis) |
Liberase TH TrypLE Express |
1x 1x |
60 min | [31] | |||
| Colon and Rectum |
Collagenase IV Trypsin-EDTA |
250 U/mL 0.05% |
unknown | [10] | |||
|
Colon and Rectum (metastasis) |
PBS/EDTA TrypLE Express |
1 mM 2x |
60 min | [67] | |||
|
Colon and Rectum, Liver and Peritoneum (metastasis) |
Collagenase II Hyaluronidase IV |
1.5 mg/mL 10 mg/mL |
15–60 min | [100] | |||
| Rectum |
Collagenase XI Dispase II TrypLE Express DNase I |
100 U/ml 125 μg/ ml 1x 3 mg |
40 + 10 min | [69] | |||
| Colon and Rectum |
Collagenase A Hyaluronidase |
0.5 mg/mL 20 mg/mL |
30 min | [85] | |||
| Rectum |
Collagenase IV Collagenase II Hyaluronidase Dispase II |
500 U/mL 1.5 mg/mL 20 mg/mL 0.1 mg/mL |
30–60 min | [70] | |||
|
Peritoneum (metastasis) |
Collagenase IV Dispase Hyaluronidase DNase I |
67.5 U/mL 0.23 U/mL 8–20 U/mL 50 units/mL |
30–60 min | [74] |
Adv.DF12 + + + is Advanced DMEM/F12-based medium supplemented with HEPES, Glutamax, penicillin and streptomycin.
*Wnt3a is usually withdrawn to reduce normal organoid contamination in colorectal tumor organoid culture.
+, Required; −, Absent; # Not strictly required.
Furthermore, organoids outline many developmental-related biological parameters, such as heterogeneous cell construction and cell-cell/cell-matrix interactions. Current applications for cancer biology in tumor organoids mainly include screening for clinical and preclinical drugs [38] and drug discovery [39], the study of the exploration microenvironment [40] and tumor heterogeneity [22], as well as response prediction and personalized medicine [10]. Peculiarly, considering time cost, economic cost, operability, and success rate, the organoid system maintains particular superiority in drug response prediction and personalized medicine. Despite the potential of organoid models in biomedicine, there are significant limitations. Lack of a vascular, nervous or immune system is a major disadvantage of organoids. Besides, matrigel and other animal-derived matrices are commonly used in organoid cultures. Uncertain protein composition and variation among batches of animal-derived matrices are ongoing issues which may result in low controllability of cell microenvironment and low reproducibility of organoids. These constraints may prevent organoids from responding to specific pathophysiological parameters and carrying out downstream conversion application.
Organs-on-chips
Organs-on-chips can be defined as a small functional unit that mimics the level of human organs in vitro [41]. Generally, it is based on an organ’s anatomy and simplified for assembly in vitro with the presence of essential elements necessary for physiological function [12]. Briefly, organ chips are microfluidic cell culture devices made of optically transparent materials, such as polydimethylsiloxane (PDMS). The main structure is the irrigation-controlled microchannel for the growth of living cells like cell lines, primary cells, and stem cells. The chips can simulate a microenvironment by combining two or more cell types. Different cells, such as organ-specific epithelial cells and stromal cells, are typically separated by ECM gel in different channels. Accordingly, it can recapitulate the multicellular structure of human organs and tissue-tissue interfaces, chemical gradients, vascular perfusion systems, and mechanical properties at the level of multicellular structure. At present, organ chips that encompass many organ types have been developed, including lung alveoli [42, 43] and bronchioles [44, 45], kidney [46, 47], liver [48], pancreas [49, 50], heart [51, 52], bone and bone marrow [53, 54] as well as the blood-brain barrier (BBB) [55, 56]. Some orthotopic cancer organ chips are developed to mimic tumor structure and physiology, such as lung adenocarcinoma chip [57], breast cancer chip [58] and multiple myeloma chip [59]. Table 2 summarizes the characteristics of CRC-related devices that have been reported in the articles since 2016 [60, 61]. The ability to capture human physiology and pathophysiology from a different perspective than organoids make them capable in vitro models for preclinical evaluation [62]. The dynamic study of microenvironment, the reduction of using animal models, novel drug development, drug efficacy, and toxicity assessment are all areas of applied research in organ chips technology that are currently in full swing [63].
Table 2.
Technological overview of organ-on-a-chip models of colorectal cancer.
| In vitro platform | Cancer cell type | Other biological components | Material | Chip configuration | Fabrication technique | Media exchange method | Ref |
|---|---|---|---|---|---|---|---|
| Vascularized micro-organs |
HCT116, SW620, SW480 |
ECs, matrix and stromal cells | PDMS | 2-layered channels | Soft lithography | Direct exchange | [77] |
| Organotypic tumor spheroids |
MC38, CT26, Primary cells from human and mouse (Organotypic Tumor Spheroids) |
Organotypic Tumor Spheroids contains immune cells and stromal cells | PDMS | 3-layered channels | Soft lithography | Perfusion | [66] |
| Colorectal tumor chip | HCT116 | HCoMEC | PDMS | 3-layered channels | Soft lithography | Perfusion | [86] |
| CRC-on-chip |
Caco2, C2BBe1, HCT116, HT29, CRC organoid |
HUVEC and human primary fibroblasts | PDMS | 2-layered channels | Soft lithography | Perfusion | [60] |
| Vascularized micro-tumors |
HCT116, SW480 |
ECs, matrix and fibroblasts | PDMS | 3-layered channels | Soft lithography | Direct exchange | [78] |
| Tumor-on-chip | SW620 | Spheroids within Matrigel | Polycarbonate | 2-layered channels | Micromachining | Perfusion | [61] |
PDMS polydimethylsiloxane, ECs endothelial cells, HCoMEC human colonic microvascular endothelial cells, HUVEC human umbilical vein endothelial cells.
Pre-clinical models for chemotherapy
Chemotherapy, surgery, and radiation are still considered the mainstream antitumor therapy for CRC in clinical setups and have improved many patients’ overall survival [5]. Despite its severe side effects and pervasive drug resistance, chemotherapy remains the most compelling part of systematic therapy. The chemotherapy backbone for CRC or mCRC treatment is drugs like 5-fluorouracil, capecitabine, oxaliplatin, and irinotecan, and several protocols that combine with them, such as FOLFOX, FOLRIRI, and CAPEOX regimen [64, 65] (Fig. 1). However, the chemotherapeutics strategies are constantly adjusted in response to changes in the patient’s actual condition. Colorectal cancer has entered the era of precision therapy, such as MMR/MSI status to predict the efficacy of adjuvant chemotherapy. Currently, preclinical models like organoids and organs-on-chips show great potential in anticancer-drug screening, personalized therapy, and drug response prediction associated with patient outcomes [66, 67]. These high-fidelity, high operational in vitro models have come to the fore in translational research.
Large-scale drug screening was performed in CRC and mCRC organoids in clinical trials and practice, including chemotherapeutic drugs and targeted agents [67, 68]. The results demonstrated the feasibility of high-throughput drug screening and forecasting the response of patients’ drug therapy. Based on these successful organoid-drug response platforms, a joint analysis of clinical patients’ therapeutic response and patient-derived organoids’ drug treatment response can sufficiently provide robust evidence [67]. For instance, they established a biorepository of 65 patient-derived rectal cancer (RC) organoids from lesions of primary, metastatic and recurrent; Ganesh and colleagues treated 21 different RC organoids with single-drug 5-FU and FOLFOX regimen (5-FU, leucovorin, and oxaliplatin). Based on the sufficient clinical follow-up of seven patients, the organoid drug responses significantly correlated with the corresponding patients’ progression-free survival [69].
Similarly, Yao and colleagues tested 80 rectal cancer organoids with single-agent 5-FU, irinotecan, or radiation and tested 23 with combinational therapies. Comparing monotherapy and combinational therapies in organoids almost remained consistent when they observed organoid drug responses with tumor regression grading (TRG) after surgical resection [70]. Although drug dosages employed during in vitro drug screening and in vivo, chemotherapeutics may remain significant, the sensitivity of the drugs could be reflected to verify the predictability of organoids. Lately, they have used liver metastatic organoids to complement their cohort and demonstrated that organoids have a good potential to predict drug sensitivity and clinical outcome of FOLFOX or FOLFIRI [71]. In addition to the single lesion, drug response on organoids from multiple lesions may explain the clinical reaction of specific patients. the test about TAS-102 (an oral combination agent) on mCRC organoid from different metastases in the same site (liver) and pre-and post-treatment metastases emphasized the correlation between heterogeneity and drug response as well as the consistency of efficacy [67]. The organoids from baseline segments were more sensitive to TAS102 than those from disease progression segments of pre-and post-treatment, consistent with the size of tumor foci from CT scan. Some prospective studies have provided the rudiment of preclinical guiding pharmacy. A successful chemotherapy response prediction was performed by mCRC organoids in contrast with CT and CEA tumor markers from the corresponding patient of treatment-refractory metastatic colon cancer along with alterations in APC and TP53. They received re-treatment of FOLFOX regimen and FOLFIRI regimen combined with panitumumab successively [72]. Before the patient benefitted from re-treatment of FOLFOX chemotherapy, organoids were confirmed sensitive to 5-FU and oxaliplatin but not to single-agent 5-FU.
Voest and colleagues reported another prospective clinical study to verify the feasibility and potential value of PDO as a predictive tool for chemotherapeutics regimens in CRC. Although there was no intervention with the patient, their PDO-based classifier indicated that it is clinically feasible to employ mCRC organoid to deliver a prediction on the outcome of irinotecan-based chemotherapy, yet, it failed on 5-FU-oxaliplatin combination therapy [73]. Still, with prospective studies that recruited 28 CRPMs patients, Woods and colleagues could not distinguish samples from 9 patients with partial response (PR) or stable disease (SD) versus progressive disease (PD) following FOLFOX treatment based on FOLFOX sensitivity of peritonoids [74]. The potential mechanism of the role of FOLFOX in mCRC organoids may be worth figuring out [75]. Impressively, they supplied a gemcitabine-capecitabine combination to a patient whose peritonoids were sensitive to multiple therapeutics. FDG PET-CT scans showed partial response after three months of treatment but followed by disease progression in further two months of treatment [74]. This variation in drug potency is prevalent in clinical settings, possibly due to multiple heterogeneous lesions or other unknown changes. Resampling organoids from altered lesions may aid in revealing changes in the tumor itself following treatment.
The organs-on-chips (OOCs) platform is another preclinical system for evaluating responses to cancer therapies. The combination of spheroid culture and microfluidic platform may be a successful strategy in the OOCs field to mimic the in vivo tumor microenvironment for drug screening of anticancer candidates [76]. Although low simulation of the tumor itself in cell lines or spheroids compared to organoids, the endotheliocyte, fibroblast, and immune cells can be assembled in organs-on-chips to mimic a relatively intact tumor microenvironment [77–79]. Earlier, the microfluidic devices combined with spheroids from human LS174T colon carcinoma cells were exposed to a continuous fluid medium to mimic microenvironment gradients in the vasculature of solid tumors [80]. A vascularized micro tumors platform was recently reported to test FOLFOX regimen and single agents such as 5-FU, oxaliplatin, and vincristine in three CRC cell lines and endothelial cells, matrix cells, stromal cells [77]. More recently, another vascularized micro tumor model (VMT) was also tested with FOLFOX regimen but compared with corresponding monolayer cultures, spheroids, and xenografts, and the tumor growth curves of VMT and xenografts were highly similar [78]. In reality, not many studies have been done to predict the anticancer agent efficacy of CRC on organs-on-chips [14]. Relatively ideal curative efficacy prediction requires primary culture spheroids or primary culture cells to ensure a degree of similarity to the patient’s parental tumor. In future studies, OOCs model may construct cellular components with primary culturing, microenvironment-related cells, and vasculature to evaluate drug response and mechanisms of drug action [66, 79]. Following that, drug concentrations similar to blood, chemical gradients caused by drug penetration, and drug response under microenvironment will be studied and adjusted as closely as possible to the drug response in vivo, based on dynamic perfusion of vasculature and integrated microenvironment. Chemotherapy efficacy can thus be predicted to some extent in this manner.
Organoids can be used as a therapeutic prediction tool and have been preliminarily used to instruct adjustment of patients’ chemotherapy strategies [71]. With the improvement of organs-on-chips technology, efficacy prediction can also be carried out from another aspect.
Targeted therapy prediction for individual
The genomic and heterogeneous similarities between patient-derived tumor organoids and corresponding parental-tumor specimens have aided in developing personalized cancer therapy [81]. Compared to chemotherapy, targeted therapy is more dependent on gene-drug associations. The well-known biologic anti-EGFR agents, such as cetuximab, and panitumumab, consistently execute poor efficacy for RAS and RAF mutant CRC tumors, leading to the indispensability of testing KRAS and NRAS, and BRAF mutations before considering the anti-EGFR therapies [5, 68] (Fig. 1). The two main signaling pathways of EGFR activation are the RAS-RAF-MAPK pathway and PI3K-PTEN/PTEN/AKT pathway. Many other targeted drugs in clinical practice have been derived from these two pathways, such as BRAF-inhibitors, MEK inhibitors, etc. Another renowned biologic agent was the anti-VEGF monoclonal antibody like bevacizumab, aflibercept, and ramucirumab. Unfortunately, resistance to targeted drugs is as common as chemotherapy drugs. Some KRAS and BRAF wild-type CRC patients also could not benefit from anti-EGFR agents because of the effect from other relevant pathways and potential pathway crosstalk [82]. The detection of molecular characteristics alone is no longer sufficient for the existing clinical adjustment of clinical therapeutic strategy [10]. Compared to potentially complex resistance mechanisms that would necessitate extensive research, combining a better phenotype and simple molecular characteristics would be a more direct and effective method.
Organoid platforms were verified that allow for genomic and functional studies at the level of individual patients [68]. The gene-drug association captured by organoids lays the foundation for investigating the molecular basis of drug response and predicting targeted therapy efficacy [83]. Despite a lack of validation in xenograft models and patients, the initial gene-related drug screening on CRC organoids shows that KRAS mutant organoids and KRAS wild-type/BRAF mutant organoids were insensitive to cetuximab, which were consistent with the response of targeted clinical therapy [68, 84]. Some KRAS-mutated organoids could be restrained by combinatorial EGFR-KRAS G12C inhibition [85]. Ulteriorly, Two CRC cases were involved in combination drug screens. KARS/TP53 mutated organoid was insensitive to most drugs, including frequently-used 5-FU, oxaliplatin, and EGFR inhibitors, consistent with clinical patient response [84]. The trametinib-based combinatorial targeted treatment, on the other hand, was highly influential on this gene-type organoid but was not validated in PDX models [10]. Another APC-mutant organoid was more insensitive to the combination of afatinib and histone deacetylase (HDAC) inhibitors than FOLFOX regimens accepted by patients at the time [10]. The result was further verified in corresponding PDX models. Five PDOs and their respective mCRC patients have recently tested the predictive value to the response of EGFR blockage cetuximab. The most impressive was the biopsy, along with EGFR amplification as well as no RAS pathway mutation, from a disease-progressive patient who initially responded to cetuximab but gradually turned to resistance. No response to cetuximab showed in its organoids consistent with the patient’s clinical response [67]. Signally, multiple tyrosine kinase inhibitors, regorafenib, have also been consistent clinical responses in PDO-xenografts due to lacking vasculature in organoids [67]. This predictive value must eventually manifest in the patient. The attempts are made by two CRC peritonoids and their respective patients. The combination of MEK inhibitors trametinib and cobimetinib and the multi-tyrosine kinase inhibitor vandetanib significantly reduced the viability of KRAS G12D-mutant organoids that were resistant to most monotherapy chemotherapeutics. No obvious response was found with adjusted four weeks of vandetanib therapy in the patient but did not include MEK inhibitors [74]. The other peritonoid showed sensitivity to adavosertib, the combination of EGFR inhibitor Osimertinib, and HDAC inhibitor vorinostat. However, adavosertib remains in phase II clinical trial for advanced colorectal cancer, and two other drugs were restricted by funding and off-label use [74]. Received gemcitabine–capecitabine combination therapy ultimately according to drug access, cost, and toxicity; this patient displayed partial response (PR) but still turned to disease progression (PD) subsequently, as described before. The general pattern of transformation from organoid to clinical is displayed in Fig. 2a. The Netherlands Cancer Institute performed a prospective clinical study (SENSOR study, NL50400.031.14 EudraCT2014-003811-13) involving colon cancer and NSCLC patients for targeted treatment. In this study, patients’ treatment strategies would be altered with screening results from organoids of these patients’ tumors [81]. Other registration numbers for other organoid clinical trials are listed in Fig. 2b. The organoids may be particularly appropriate as predictive tools for targeted therapies, considering the robust capture of genomic alterations.
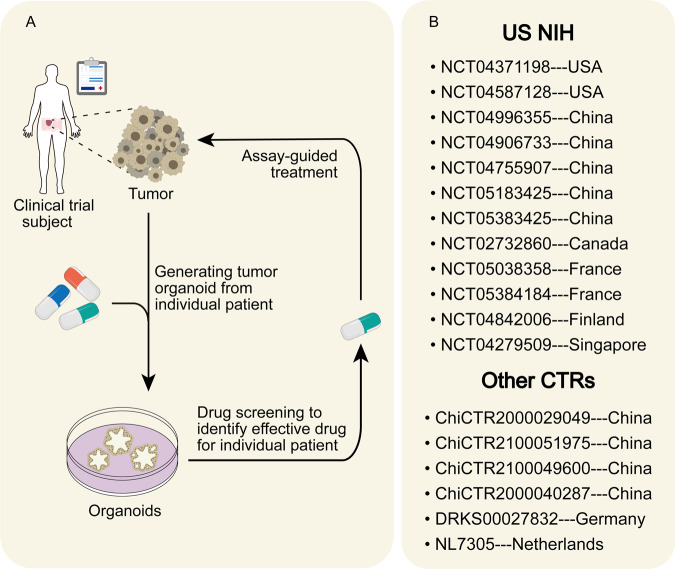
Fig. 2. Organoids as a supplement of the clinical treatment reference index.
A Patient-derived tumor organoids have the potential to choose therapy for the individual patient. It can provide feedback to the clinician to adjust the treatment decision based on the drug screening results. B This shows the registration numbers of ongoing clinical trials based on colorectal cancer organoids [129], including registered Clinical trials from US ClinicalTrials.gov, Chinese Clinical Trial Registry, etc [130, 131].
Organs-on-chips could also be used for screening specific drugs. The in vitro vascularized micro tumors (VMTs) are a relatively representative model in colorectal cancer. The vasculature derived from endothelial cells commendably complements the composition of microenvironment [77, 78]. Ten kinds of clinically-approved receptor tyrosine kinase (RTK) inhibitors targeted to VEGFR2, PDGFR, Tie2, and FGFR1, including pazopanib and sorafenib, were tested in VMTs [77]. Novel targeted drug like galunisertib, a TGF-βR1 antagonist, finished in phase II clinical trials for hepatocellular cancer, was also tested in VMT loading with SW480 and HCT116 attributed to the presence of fibroblasts in the microenvironment [78] In addition, drug delivery and penetration have been performed in similar colorectal cancer organ-chip systems [86, 87]. Though the fidelity of the tumor itself is not superior to the organoid, the integrity of microenvironment can better explore the interaction of medicine and the cell-extracellular matrix. The influence of complex microenvironments on drug efficacy cannot be ignored, such as drug targeting to the stroma, dynamic microenvironmental drug concentrations, and microenvironmental barriers [88]. These conditions can also provide a basis for predicting the efficacy of targeted therapy in organs-on-chips.
By assembling primary tumor cells, patient efficacy can be predicted to a certain extent. Similar efficacy prediction studies have been conducted on other tumor chips. Two chips were combined to test the anti-ES tumor effect and cardiotoxicity of lincitinib, a selective IGF-1R inhibitor. The results were also compared with those from clinical trials [89]. Notably, Hickman and colleagues established a reconfigurable multi-organ system to investigate anticancer drug efficacy and off-target effects in two different cancer-derived models. One was the combination of two leukemia cell lines and primary hepatocytes to study imatinib and diclofenac. Another system used to study tamoxifen was a multidrug-resistant vulva cancer line, a non–multidrug-resistant breast cancer line, primary hepatocytes, and induced pluripotent stem cell-derived cardiomyocytes [90]. The results were compared to published clinical and preclinical data. These cases provide direction and gist to explore drug efficacy prediction and evaluation in the organs-on-chips system, even in colorectal cancer.
Development of personalized immunotherapy
Immunotherapy, including immune checkpoint inhibitors and adoptive cell therapy, regulates the tumor microenvironment or immune system to identify and attack tumor cells [91]. Unfortunately, immunotherapy plays a limited role in solid tumors, while yields unusually brilliant results in hematologic tumors. For 4–5% of CRC with mismatch-repair-deficient (dMMR) or high levels of microsatellite instability (MSI-H), immune checkpoint inhibitors (ICIs), including PD-1 blockade and CTLA-4 inhibitor, have received US Food and Drug Administration’s (US-FDA) approval [92, 93] (Fig. 1). ICIs, on the other hand, are ineffective in most clinical CRC patients with mismatch-repair-proficient (pMMR) and microsatellite-stable (MSS) or low levels of microsatellite instability (MSI-L) due to low mutant neoantigen burden and a lack of immune cell infiltration [94, 95]. Several MSI CRCs with a high mutational load could not benefit from ICIs [96]. In addition, adoptive T cell therapies have limited colorectal cancer effects. Unlike pharmacotherapy, the improvement of cell therapy always directly depends on many clinical studies subjected to many restrictions [97–99]. Developing cell therapies for solid tumors may therefore be slow. Accordingly, as a predictive tool, the emerging preclinical models may effectively accelerate the development of immunotherapy in solid tumors.
Organoids generally lack immune system components, making tumor-immune interaction difficult. Exploiting the air-liquid interface (ALI) system for the immune microenvironment and organoids/immune cell co-culture system makes it possible to study immunotherapy [100, 101]. ALI system could retain the original innate immune components for a certain period. Another one was an artificial combination of organoids and immune cells. The co-culture system of peripheral blood lymphocytes and tumor organoids were validated primarily in dMMR patients. In four of eight major histocompatibility complex (MHC) class I + colorectal organoids, CD8+ T cells showed tumor-specific responses but not in MHC class I deficient organoids [100]. MHC class I deficiency was also inherited from its parental tumor. Numerous studies have found that HLA/MHC gene expression is closely related to ICI treatment response [102, 103]. The co-culture result showed that tumor-responsive T cell populations could be amplified and the capture of MHC-based immune reactivity. The author and colleagues co-cultured organoids with T cell populations loading tumor responsive with or without MHC class I and MHC class II blockages. The cytotoxic effect of T cells was observed in CRC organoids but not in healthy organoids [100]. General stratification of patient immunotherapy response includes expression of the immune checkpoint, Infiltration of immune cells, mismatch repair mechanism, tumor neoantigen burden [104], TCR clonality, and immune gene signatures. These factors have more or less been studied and recapitulated in CRC organoids, containing the expression of HLA molecular [100–103, 105].
The capture of immune-related components and co-culture system testifies to the possibility of ICIs efficacy prediction. Apparently, this treatment prediction requires validation from more prospective studies and clinical outcome correlation to screening appropriate patients for enriching clinical ICIs response. Voest and Haane reported an ongoing exploratory NICHE study about neoadjuvant immunotherapy in pMMR and dMMR early-stage colon cancer [106]. 100% and 27% responses were found in patients with dMMR and pMMR colon cancer, respectively, after the employment of CTLA-4 inhibitor and PD-1 blockage for neoadjuvant treatment. Notably, they investigate the low reactivity of pMMR tumors in clinical responders’ coculture platforms. The reactivity of autologous T cells in an organoid co-culture system was limited to colon tumors that responded clinically to ICIs. In vitro reactivity was not observed in all clinical responders [106]. The result proves the predictability of organoids in ICI therapy from the side. Otherwise, Farin and colleagues developed a quantitative co-culture platform for CAR-mediated cytotoxicity toward PDCOs. They proved that organoids could be tested for CAR toxicity in a complex and competitive microenvironment by co-culture of EGFR variant III-expressing CRC organoids, EGFRvIII-CAR NK-92 cells, and healthy colon organoids [107]. Likewise, the application of organoid-based adoptive cell therapy required further exploration in vivo experiments [100, 107]. Compared to chemotherapy and targeted therapy, the prediction of personalized immunotherapy responses in CRC organoids response suffer more challenges.
A popular organ-on-chip type is similar to the organoid-immune cell co-culture system but with a higher degree of integration. Cancer cell lines, cancer-associated fibroblasts (CAFs), immune cells from healthy donors’ PBMCs, and endothelial cells are typically involved in this system. Within the chips, dynamic changes in the tumor immune microenvironment can be quantified and visualized [108]. Accordingly, some mutual problems may be figured out. For instance, trastuzumab may promote cancer-immune cell interactions and CAFs played an inhibitory role in the immune microenvironment in breast cancer [79]. Whereas the lack of individual characteristics is insufficient for personalized precision immunotherapy. The main part of organs-on-chip construction needs to come from individual patients to accomplish the prediction of immunotherapy [66]. The murine- and patient-derived organotypic tumor spheroids (MDOTS/PDOTS) are more satisfactory subjects for forecasting immunoreaction. It retained autologous lymphoid and myeloid cell populations [66, 109]. This type of OOCs was proved that could simulate the response to PD-1 blockade and ascertain specific interventions to counteract resistance [66]. However, it was limited to tumor infiltrating lymphocytes (TILs) and could not reflect the recruitment of additional immune cells [109]. The evaluation of immune cell recruitment has also been carried out in Breast tumor-on-a-Chips [110]. The addition of autologous circulating immune cells or biomimetic lymph nodes may be a future direction [111, 112]. Even though organs-on-chips have some advantages in the integration of immune-related systems, there is still a lack of research closely related to clinical transformation.
Future prospects
Further exploration of these in vitro preclinical models has revealed many limitations. Compared to cancer cell lines, patient-derived organoids and organs-on-chips consume more time and resources in culturing. The primary limitations of tumor organoid culture are the lack of a microenvironment, the short-term expansion, contamination from normal organoids, the effects of serum and growth factors in the culture system, and the requirement for mouse-derived extracellular matrix (ECM). Meanwhile, despite simulating a relatively complete microenvironment, organs-on-chips are limited to technical robustness, time cost, raw materials for chips, and relatively lower complexity of 3d structure than organoids. Many of them have the potential to influence the prediction of curative effects. For example, except for organoids derived from colorectal cancer, some organoid drug efficacy comparisons for other cancers have not been particularly favorable [113], which may directly relate to the lack of microenvironment [114]. Likewise, the same is valid for fetal bovine serum and some mouse-derived small molecules or ECM [115, 116]. Routinely, effective predictive models require drug screening in the window of meaningful short-term clinical treatment. Long-term response to drug treatment is also critical. In terms of immunotherapy, the long-term clinical response has been unsatisfactory, attributed to immune failure related to the immune microenvironment. In the model, this may necessitate extending the duration of drug action, analyzing the internal changes of individual immune cells [117] and constantly introducing new lymphocytes or forming multi-organ organ chips containing lymph nodes to stimulate immune cell recruitment from tertiary lymphoid structure [111]. The presence of a small number of potentially healthy organoids or deliberately importing healthy organoids may make the outcome of drug response more precise [100, 107]. This demonstrates that in vitro model has great potential for improvement. Thommen and his colleagues built a patient-derived tumor fragment platform to dissect the early immunological response to ex vivo PD-1 blockade. Compared to matched clinical response data, the ex vivo immunological response clinical outcomes were in concordance with clinical outcomes in a high degree of 26 patients, including 12 melanoma and non-small cell lung cancer (NSCLC) patients who did not respond to clinical PD-1 blockade [118]. This gives inspiration to microenvironment capture. It also shows the importance of early immunotherapy intervention [106] and of tracking the long-term dynamics of the immune microenvironment from the side. Furthermore, low cost and high throughput must be considered a reference supplement for individual clinical efficacy prediction. This role could be filled by commercial organs-on-chips [88]. Therefore, without limitation to the perfection of the model itself, being consideration of pros and cons of these two models, organoids-on-a-chip may be a promising future direction.
Organoids-on-a-chip is an integration of organoids with organ-on-a-chip technology. It’s essentially an interaction between the stem cell field and the engineering field. Organoids are further designed to break the limitations by controlling stem cell behavior and cell microenvironment. Organs-on-chips are the target organ’s artificial construction and are precisely controlled artificially [119]. Organoids follow intrinsic developmental programs to self-assemble [11]. The fusion of these two distinct but complementary technologies has been accepted by most scientists [81, 120, 121]. Enhancing fidelity and reproducibility is the top priority task of organoids-on-a-chip [15]. The biophysical and biochemical microenvironment and the nutrient supply must be controlled during the assembly of organoids-on-a-chip.
Mechanical flow has been shown to promote additional physiological integrity in developing PSC-derived intestinal and islet organoids [122, 123]. Physiological flow promoting angiogenesis was demonstrated on kidney organoids-on-chip [124]. An embryonic-like malleable vasculogenic endothelial cells was reported can break through the limitations of synthetic scaffolds and semi-permeable membranes with its ability to self-organize 3D lumenized vascular networks. Co-cultures of organoids with the specialized endothelial cells can serve as a tissue-specific biological platform containing vascular niche [125]. Analogously, The co-culture system of cancer-associated fibroblasts and pancreatic cancer organoids has also been applied to niche studies [116]. Lutolf and colleagues constructed an intestinal organoids-on-a-chip to attain more physiologically relevant organ shapes, sizes, and functions [126]. The application of fluid scour, and lumen structure scaffold well simulates the biophysical microenvironment. Remarkable tissue homeostasis was shown in the self-organization of functional intestinal epithelium in confining hydrogel scaffold. Induced epithelial injury and long-term parasite infection verified the regenerative and physiologic mimicry of intestinal organoids-on-a-chip. The authors also introduced intestinal myofibroblasts and macrophages to mimic organ-level complexity [126]. Lutolf’s team recently developed a hydrogel-based microfabrication method for controlling the spatiotemporal morphogenesis of intestinal organoids with defined shape and structure [127]. Morphogenesis of other tissues may be extended based on this in the future. These strategies further improve organoid and organ-on-chip models in a visual and controllable manner, allowing the possibility of reproducing in vivo complex tissues or organs in vitro. High fidelity may be achieved by introducing a complete microenvironment, including the vasculature, and its application may take developmental medicine and regenerative medicine to a new level. We can consider the possibility of organoid-on-hips with personalized characteristics of patients based on these analogous models (Fig. 3). The patient’s primary cells are introduced into the chip in an organoid fashion, and various types of stromal cells are also delivered to further refine the physiological structure in vitro. The dynamic changes in drug response can then be detected in the flow system.On the one hand, it increases fidelity to complete the current bottleneck in basic research. On the other hand, it simplifies the technical difficulty within a controllable range to improve repeatability to serve as more versatile and predictive preclinical tools for precision medicine [15, 128]. This synergistic engineering may be a pivotal bridge to translation from lab to clinic.
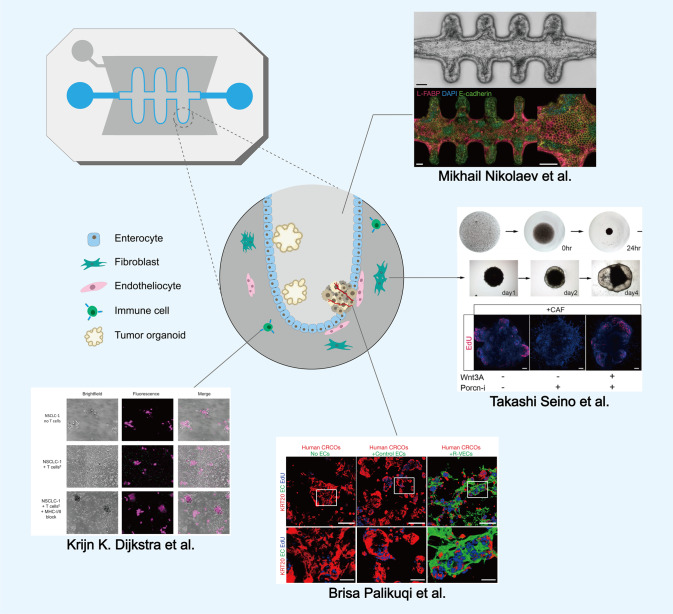
Fig. 3. Organoids meet organs-on-chips to perfect the quasi-physiological structure.
The figure depicts a possible future, more potent in vitro technology by loading organoids and other stromal cells into microfluidic chips to complement the limitation of the two models. One channel was loaded with cancer organoids and enterocytes, while the other was coated with stromal cells. The middle figure is based on Ref. [126]. The top right is adapted from Ref. [126], Springer Nature Limited. The middle right is adapted from Ref. [116], Cell Limited. The lower one is adapted from Ref. [125], Springer Nature Limited. The lower left one is adapted from Ref. [100], Cell Limited. The surrounding experimental figures do not represent the actual situation of this model.
Conclusion
Although it remains a long way from being practical and widely used, it has great potential in predicting therapeutic efficacy. A corresponding number of clinical trials based on these models are currently underway. In terms of the treatment time window, cost, drug screening efficiency, and accuracy, these in vitro models will likely be used as a clinical reference shortly to guide the optimization of treatment strategies for individual patients through technological advancement or technology fusion.
Author contributions
CXY, LZJ and FGB provided the idea of the review. ZJL, JLL and CYT wrote the manuscript. LHY, HMX, DZ, WJ and XD helped with the final revision of the review. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China 82072725 and 81872042 (to XC), 81702442 (to ZL), 81972332 to (YC), 82002583 (to MH), 82002591 (to GF), the Key Program of Jiangsu Provincial Health Commission ZD2021039 (to XC), the Natural Science Foundation of Jiangsu province BK20170623 (to ZL), BK20200273 (to GF), the China Postdoctoral Science Foundation 2020M670090ZX (to ZL), 2021MD703958 (to GF), the Postdoctoral Science Found of Jiangsu province 2018K090B (to ZL).
Competing interests
The authors declare no competing interests.
Consent for publication
All authors consent to publication.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jialong Zhu, Linlin Ji, Yitian Chen.
Contributor Information
Gongbo Fu, Email: mhksfgb@126.com.
Zengjie Lei, Email: leizengjie@163.com.
Xiaoyuan Chu, Email: chuxiaoyuan000@163.com.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin. 2018;68:250–81. doi: 10.3322/caac.21457. [DOI] [PubMed] [Google Scholar]
- 4.Patel SG, May FP, Anderson JC, Burke CA, Dominitz JA, Gross SA, et al. Updates on age to start and stop colorectal cancer screening: recommendations from the U.S. multi-society task force on colorectal cancer. Gastroenterology. 2022;162:285–99. doi: 10.1053/j.gastro.2021.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33:125–36. doi: 10.1016/j.ccell.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kyrochristos ID, Roukos DH. Comprehensive intra-individual genomic and transcriptional heterogeneity: evidence-based colorectal cancer precision medicine. Cancer Treat Rev. 2019;80:101894. doi: 10.1016/j.ctrv.2019.101894. [DOI] [PubMed] [Google Scholar]
- 9.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 10.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–77. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–97. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 12.Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Disco. 2015;14:248–60. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18:407–18. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 14.Sontheimer-Phelps A, Hassell BA, Ingber DE. Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer. 2019;19:65–81. doi: 10.1038/s41568-018-0104-6. [DOI] [PubMed] [Google Scholar]
- 15.Hofer M, Lutolf MP. Engineering organoids. Nat Rev Mater. 2021;6:402–20. doi: 10.1038/s41578-021-00279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 17.Fleck JS, Sanchis-Calleja F, He Z, Santel M, Boyle MJ, Camp JG, et al. Resolving organoid brain region identities by mapping single-cell genomic data to reference atlases. Cell Stem Cell. 2021;28:1148–59. doi: 10.1016/j.stem.2021.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, Francies HE, Secrier M, Perner J, Miremadi A, Galeano-Dalmau N, et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat Commun. 2018;9:2983. doi: 10.1038/s41467-018-05190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim M, Mun H, Sung CO, Cho EJ, Jeon HJ, Chun SM, et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun. 2019;10:3991. doi: 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 22.Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–86. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–35. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–36. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nanki K, Toshimitsu K, Takano A, Fujii M, Shimokawa M, Ohta Y, et al. Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell. 2018;174:856–69. doi: 10.1016/j.cell.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 27.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiriac H, Belleau P, Engle DD, Plenker D, Deschenes A, Somerville TDD, et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018;8:1112–29. doi: 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grassi L, Alfonsi R, Francescangeli F, Signore M, De Angelis ML, Addario A, et al. Organoids as a new model for improving regenerative medicine and cancer personalized therapy in renal diseases. Cell Death Dis. 2019;10:201. doi: 10.1038/s41419-019-1453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schutgens F, Rookmaaker MB, Margaritis T, Rios A, Ammerlaan C, Jansen J, et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat Biotechnol. 2019;37:303–13. doi: 10.1038/s41587-019-0048-8. [DOI] [PubMed] [Google Scholar]
- 31.Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–38. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, et al. Tumor evolution and drug response in patient-derived organoid models of bladder. Cancer Cell. 2018;173:515–28. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullenders J, de Jongh E, Brousali A, Roosen M, Blom JPA, Begthel H, et al. Mouse and human urothelial cancer organoids: a tool for bladder cancer research. Proc Natl Acad Sci USA. 2019;116:4567–74. doi: 10.1073/pnas.1803595116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–87. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–75. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci USA. 2015;112:13308–11. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–5. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 38.Verissimo CS, Overmeer RM, Ponsioen B, Drost J, Mertens S, Verlaan-Klink I, et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife. 2016;5:e18489. doi: 10.7554/eLife.18489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weeber F, Ooft SN, Dijkstra KK, Voest EE. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem Biol. 2017;24:1092–100. doi: 10.1016/j.chembiol.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 40.Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering stem cell organoids. Cell Stem Cell. 2016;18:25–38. doi: 10.1016/j.stem.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang B, Korolj A, Lai BFL, Radisic M. Advances in organ-on-a-chip engineering. Nat Rev Mater. 2018;3:257–78. doi: 10.1038/s41578-018-0034-7. [DOI] [Google Scholar]
- 42.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–8. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stucki AO, Stucki JD, Hall SR, Felder M, Mermoud Y, Schmid RA, et al. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip. 2015;15:1302–10. doi: 10.1039/C4LC01252F. [DOI] [PubMed] [Google Scholar]
- 44.Sellgren KL, Butala EJ, Gilmour BP, Randell SH, Grego S. A biomimetic multicellular model of the airways using primary human cells. Lab Chip. 2014;14:3349–58. doi: 10.1039/C4LC00552J. [DOI] [PubMed] [Google Scholar]
- 45.Benam KH, Villenave R, Lucchesi C, Varone A, Hubeau C, Lee HH, et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods. 2016;13:151–7. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 46.Musah S, Dimitrakakis N, Camacho DM, Church GM, Ingber DE. Directed differentiation of human induced pluripotent stem cells into mature kidney podocytes and establishment of a Glomerulus Chip. Nat Protoc. 2018;13:1662–85. doi: 10.1038/s41596-018-0007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hubner J, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15:2688–99. doi: 10.1039/C5LC00392J. [DOI] [PubMed] [Google Scholar]
- 48.Esch MB, Mahler GJ, Stokol T, Shuler ML. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip. 2014;14:3081–92. doi: 10.1039/C4LC00371C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shik Mun K, Arora K, Huang Y, Yang F, Yarlagadda S, Ramananda Y, et al. Patient-derived pancreas-on-a-chip to model cystic fibrosis-related disorders. Nat Commun. 2019;10:3124. doi: 10.1038/s41467-019-11178-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glieberman AL, Pope BD, Zimmerman JF, Liu Q, Ferrier JP, Kenty JHR, et al. Synchronized stimulation and continuous insulin sensing in a microfluidic human Islet on a Chip designed for scalable manufacturing. Lab Chip. 2019;19:2993–3010. doi: 10.1039/C9LC00253G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marsano A, Conficconi C, Lemme M, Occhetta P, Gaudiello E, Votta E, et al. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip. 2016;16:599–610. doi: 10.1039/C5LC01356A. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Wang T, Wang P, Hu N. High-throughput assessment of drug cardiac safety using a high-speed impedance detection technology-based heart-on-a-chip. Micromachines (Basel) 2016;7:122. doi: 10.3390/mi7070122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marturano-Kruik A, Nava MM, Yeager K, Chramiec A, Hao L, Robinson S, et al. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc Natl Acad Sci USA. 2018;115:1256–61. doi: 10.1073/pnas.1714282115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torisawa YS, Mammoto T, Jiang E, Jiang A, Mammoto A, Watters AL, et al. Modeling hematopoiesis and responses to radiation countermeasures in a bone marrow-on-a-chip. Tissue Eng Part C Methods. 2016;22:509–15. doi: 10.1089/ten.tec.2015.0507. [DOI] [PubMed] [Google Scholar]
- 55.Adriani G, Ma D, Pavesi A, Kamm RD, Goh EL. A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood-brain barrier. Lab Chip. 2017;17:448–59. doi: 10.1039/C6LC00638H. [DOI] [PubMed] [Google Scholar]
- 56.Wang YI, Abaci HE, Shuler ML. Microfluidic blood-brain barrier model provides in vivo-like barrier properties for drug permeability screening. Biotechnol Bioeng. 2017;114:184–94. doi: 10.1002/bit.26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS, et al. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 2017;21:508–16. doi: 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 58.Bersini S, Jeon JS, Dubini G, Arrigoni C, Chung S, Charest JL, et al. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials. 2014;35:2454–61. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang W, Lee WY, Siegel DS, Tolias P, Zilberberg J. Patient-specific 3D microfluidic tissue model for multiple myeloma. Tissue Eng Part C Methods. 2014;20:663–70. doi: 10.1089/ten.tec.2013.0490. [DOI] [PubMed] [Google Scholar]
- 60.Strelez C, Chilakala S, Ghaffarian K, Lau R, Spiller E, Ung N, et al. Human colorectal cancer-on-chip model to study the microenvironmental influence on early metastatic spread. iScience. 2021;24:102509. doi: 10.1016/j.isci.2021.102509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petreus T, Cadogan E, Hughes G, Smith A, Pilla Reddy V, Lau A, et al. Tumour-on-chip microfluidic platform for assessment of drug pharmacokinetics and treatment response. Commun Biol. 2021;4:1001. doi: 10.1038/s42003-021-02526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monteiro MV, Zhang YS, Gaspar VM, Mano JF. 3D-bioprinted cancer-on-a-chip: level-up organotypic in vitro models. Trends Biotechnol. 2022;40:432–47. doi: 10.1016/j.tibtech.2021.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vunjak-Novakovic G, Ronaldson-Bouchard K, Radisic M. Organs-on-a-chip models for biological research. Cell. 2021;184:4597–611. doi: 10.1016/j.cell.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19:329–59. doi: 10.6004/jnccn.2021.0012. [DOI] [PubMed] [Google Scholar]
- 65.Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. NCCN guidelines insights: rectal cancer, version 6.2020. J Natl Compr Canc Netw. 2020;18:806–15. doi: 10.6004/jnccn.2020.0032. [DOI] [PubMed] [Google Scholar]
- 66.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8:196–215. doi: 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–6. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–45. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ganesh K, Wu C, O’Rourke KP, Szeglin BC, Zheng Y, Sauvé CG, et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med. 2019;25:1607–14. doi: 10.1038/s41591-019-0584-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell. 2020;26:17–26. doi: 10.1016/j.stem.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 71.Mo S, Tang P, Luo W, Zhang L, Li Y, Hu X, et al. Patient-derived organoids from colorectal cancer with paired liver metastasis reveal tumor heterogeneity and predict response to chemotherapy. Adv Sci (Weinh) 2022;9:e2204097. doi: 10.1002/advs.202204097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pasch CA, Favreau PF, Yueh AE, Babiarz CP, Gillette AA, Sharick JT, et al. Patient-derived cancer organoid cultures to predict sensitivity to chemotherapy and radiation. Clin Cancer Res: Off J Am Assoc Cancer Res. 2019;25:5376–87. doi: 10.1158/1078-0432.CCR-18-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van Werkhoven E, et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med. 2019;11:eaay2574. doi: 10.1126/scitranslmed.aay2574. [DOI] [PubMed] [Google Scholar]
- 74.Narasimhan V, Wright JA, Churchill M, Wang T, Rosati R, Lannagan TRM, et al. Medium-throughput drug screening of patient-derived organoids from colorectal peritoneal metastases to direct personalized therapy. Clin Cancer Res: Off J Am Assoc Cancer Res. 2020;26:3662–70. doi: 10.1158/1078-0432.CCR-20-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bruun J, Kryeziu K, Eide PW, Moosavi SH, Eilertsen IA, Langerud J, et al. Patient-derived organoids from multiple colorectal cancer liver metastases reveal moderate intra-patient pharmacotranscriptomic heterogeneity. Clin Cancer Res: Off J Am Assoc Cancer Res. 2020;26:4107–19. doi: 10.1158/1078-0432.CCR-19-3637. [DOI] [PubMed] [Google Scholar]
- 76.Chen Y, Gao D, Liu H, Lin S, Jiang Y. Drug cytotoxicity and signaling pathway analysis with three-dimensional tumor spheroids in a microwell-based microfluidic chip for drug screening. Anal Chim Acta. 2015;898:85–92. doi: 10.1016/j.aca.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 77.Sobrino A, Phan DT, Datta R, Wang X, Hachey SJ, Romero-López M, et al. 3D microtumors in vitro supported by perfused vascular networks. Sci Rep. 2016;6:31589. doi: 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hachey SJ, Movsesyan S, Nguyen QH, Burton-Sojo G, Tankazyan A, Wu J, et al. An in vitro vascularized micro-tumor model of human colorectal cancer recapitulates in vivo responses to standard-of-care therapy. Lab Chip. 2021;21:1333–51. doi: 10.1039/D0LC01216E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nguyen M, De Ninno A, Mencattini A, Mermet-Meillon F, Fornabaio G, Evans SS, et al. Dissecting effects of anti-cancer drugs and cancer-associated fibroblasts by on-chip reconstitution of immunocompetent tumor microenvironments. Cell Rep. 2018;25:3884–93. doi: 10.1016/j.celrep.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 80.Walsh CL, Babin BM, Kasinskas RW, Foster JA, McGarry MJ, Forbes NS. A multipurpose microfluidic device designed to mimic microenvironment gradients and develop targeted cancer therapeutics. Lab Chip. 2009;9:545–54. doi: 10.1039/B810571E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aboulkheyr EsH, Montazeri L, Aref AR, Vosough M, Baharvand H. Personalized cancer medicine: an organoid approach. Trends Biotechnol. 2018;36:358–71. doi: 10.1016/j.tibtech.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 82.Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–7. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Toshimitsu K, Takano A, Fujii M, Togasaki K, Matano M, Takahashi S, et al. Organoid screening reveals epigenetic vulnerabilities in human colorectal cancer. Nat Chem Biol. 2022;18:605–14. doi: 10.1038/s41589-022-00984-x. [DOI] [PubMed] [Google Scholar]
- 84.Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325:669–85. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 85.Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, et al. EGFR blockade reverts resistance to KRAS(G12C) inhibition in colorectal cancer. Cancer Discov. 2020;10:1129–39. doi: 10.1158/2159-8290.CD-20-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carvalho MR, Barata D, Teixeira LM, Giselbrecht S, Reis RL, Oliveira JM, et al. Colorectal tumor-on-a-chip system: a 3D tool for precision onco-nanomedicine. Sci Adv. 2019;5:eaaw1317. doi: 10.1126/sciadv.aaw1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ingram N, McVeigh LE, Abou-Saleh RH, Maynard J, Peyman SA, McLaughlan JR, et al. Ultrasound-triggered therapeutic microbubbles enhance the efficacy of cytotoxic drugs by increasing circulation and tumor drug accumulation and limiting bioavailability and toxicity in normal tissues. Theranostics. 2020;10:10973–92. doi: 10.7150/thno.49670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma C, Peng Y, Li H, Chen W. Organ-on-a-Chip: a new paradigm for drug development. Trends Pharm Sci. 2021;42:119–33. doi: 10.1016/j.tips.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chramiec A, Teles D, Yeager K, Marturano-Kruik A, Pak J, Chen T, et al. Integrated human organ-on-a-chip model for predictive studies of anti-tumor drug efficacy and cardiac safety. Lab Chip. 2020;20:4357–72. doi: 10.1039/D0LC00424C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McAleer CW, Long CJ, Elbrecht D, Sasserath T, Bridges LR, Rumsey JW, et al. Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Sci Transl Med. 2019;11:eaav1386. doi: 10.1126/scitranslmed.aav1386. [DOI] [PubMed] [Google Scholar]
- 91.Egen JG, Ouyang W, Wu LC. Human anti-tumor immunity: insights from immunotherapy clinical trials. Immunity. 2020;52:36–54. doi: 10.1016/j.immuni.2019.12.010. [DOI] [PubMed] [Google Scholar]
- 92.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz H-J, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–91. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–9. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 94.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 95.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hege KM, Bergsland EK, Fisher GA, Nemunaitis JJ, Warren RS, McArthur JG, et al. Safety, tumor trafficking and immunogenicity of chimeric antigen receptor (CAR)-T cells specific for TAG-72 in colorectal cancer. J Immunother Cancer. 2017;5:22. doi: 10.1186/s40425-017-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xiao L, Cen D, Gan H, Sun Y, Huang N, Xiong H, et al. Adoptive transfer of NKG2D CAR mRNA-engineered natural killer cells in colorectal cancer patients. Mol Ther. 2019;27:1114–25. doi: 10.1016/j.ymthe.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang C, Wang Z, Yang Z, Wang M, Li S, Li Y, et al. Phase I escalating-dose trial of CAR-T therapy targeting CEA(+) metastatic colorectal cancers. Mol Ther. 2017;25:1248–58. doi: 10.1016/j.ymthe.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–98. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–88. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cho EJ, Kim M, Jo D, Kim J, Oh JH, Chung HC, et al. Immuno-genomic classification of colorectal cancer organoids reveals cancer cells with intrinsic immunogenic properties associated with patient survival. J Exp Clin Cancer Res: CR. 2021;40:230. doi: 10.1186/s13046-021-02034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Newey A, Griffiths B, Michaux J, Pak HS, Stevenson BJ, Woolston A, et al. Immunopeptidomics of colorectal cancer organoids reveals a sparse HLA class I neoantigen landscape and no increase in neoantigens with interferon or MEK-inhibitor treatment. J Immunother cancer. 2019;7:309. doi: 10.1186/s40425-019-0769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dall’Olio FG, Marabelle A, Caramella C, Garcia C, Aldea M, Chaput N, et al. Tumour burden and efficacy of immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2021. [DOI] [PubMed]
- 105.Demmers LC, Kretzschmar K, Van Hoeck A, Bar-Epraim YE, van den Toorn HWP, Koomen M, et al. Single-cell derived tumor organoids display diversity in HLA class I peptide presentation. Nat Commun. 2020;11:5338. doi: 10.1038/s41467-020-19142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chalabi M, Fanchi LF, Dijkstra KK, Van den Berg JG, Aalbers AG, Sikorska K, et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat Med. 2020;26:566–76. doi: 10.1038/s41591-020-0805-8. [DOI] [PubMed] [Google Scholar]
- 107.Schnalzger TE, de Groot MH, Zhang C, Mosa MH, Michels BE, Röder J, et al. 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 2019;38:e100928. doi: 10.15252/embj.2018100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miller CP, Shin W, Ahn EH, Kim HJ, Kim DH. Engineering microphysiological immune system responses on chips. Trends Biotechnol. 2020;38:857–72. doi: 10.1016/j.tibtech.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aref AR, Campisi M, Ivanova E, Portell A, Larios D, Piel BP, et al. 3D microfluidic ex vivo culture of organotypic tumor spheroids to model immune checkpoint blockade. Lab Chip. 2018;18:3129–43. doi: 10.1039/C8LC00322J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aung A, Kumar V, Theprungsirikul J, Davey SK, Varghese S. An engineered tumor-on-a-chip device with breast cancer-immune cell interactions for assessing T-cell recruitment. Cancer Res. 2020;80:263–75. doi: 10.1158/0008-5472.CAN-19-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shanti A, Samara B, Abdullah A, Hallfors N, Accoto D, Sapudom J, et al. Multi-compartment 3D-cultured organ-on-a-chip: towards a biomimetic lymph node for drug development. Pharmaceutics. 2020;12:464. doi: 10.3390/pharmaceutics12050464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shim S, Belanger MC, Harris AR, Munson JM, Pompano RR. Two-way communication between ex vivo tissues on a microfluidic chip: application to tumor-lymph node interaction. Lab Chip. 2019;19:1013–26. doi: 10.1039/C8LC00957K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen P, Zhang X, Ding R, Yang L, Lyu X, Zeng J, et al. Patient-derived organoids can guide personalized-therapies for patients with advanced breast cancer. Adv Sci (Weinh) 2021;8:e2101176. doi: 10.1002/advs.202101176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yuki K, Cheng N, Nakano M, Kuo CJ. Organoid models of tumor immunology. Trends Immunol. 2020;41:652–64. doi: 10.1016/j.it.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Janda CY, Dang LT, You C, Chang J, de Lau W, Zhong ZA, et al. Surrogate Wnt agonists that phenocopy canonical Wnt and beta-catenin signalling. Nature. 2017;545:234–7. doi: 10.1038/nature22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell. 2018;22:454–67. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 117.Ren X, Zhang L, Zhang Y, Li Z, Siemers N, Zhang Z. Insights gained from single-cell analysis of immune cells in the tumor microenvironment. Annu Rev Immunol. 2021;39:583–609. doi: 10.1146/annurev-immunol-110519-071134. [DOI] [PubMed] [Google Scholar]
- 118.Voabil P, de Bruijn M, Roelofsen LM, Hendriks SH, Brokamp S, van den Braber M, et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat Med. 2021;27:1250–61. doi: 10.1038/s41591-021-01398-3. [DOI] [PubMed] [Google Scholar]
- 119.Takebe T, Zhang B, Radisic M. Synergistic engineering: organoids meet organs-on-a-chip. Cell Stem Cell. 2017;21:297–300. doi: 10.1016/j.stem.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 120.Fan H, Demirci U, Chen P. Emerging organoid models: leaping forward in cancer research. J Hematol Oncol. 2019;12:142. doi: 10.1186/s13045-019-0832-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Park SE, Georgescu A, Huh D. Organoids-on-a-chip. Science. 2019;364:960–5. doi: 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tao T, Wang Y, Chen W, Li Z, Su W, Guo Y, et al. Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab Chip. 2019;19:948–58. doi: 10.1039/C8LC01298A. [DOI] [PubMed] [Google Scholar]
- 123.Workman MJ, Gleeson JP, Troisi EJ, Estrada HQ, Kerns SJ, Hinojosa CD, et al. Enhanced utilization of induced pluripotent stem cell-derived human intestinal organoids using microengineered chips. Cell Mol Gastroenterol Hepatol. 2018;5:669–77. doi: 10.1016/j.jcmgh.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat Methods. 2019;16:255–62. doi: 10.1038/s41592-019-0325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Palikuqi B, Nguyen DT, Li G, Schreiner R, Pellegata AF, Liu Y, et al. Adaptable haemodynamic endothelial cells for organogenesis and tumorigenesis. Nature. 2020;585:426–32. doi: 10.1038/s41586-020-2712-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 2020;585:574–8. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- 127.Gjorevski N, Nikolaev M, Brown TE, Mitrofanova O, Brandenberg N, DelRio FW, et al. Tissue geometry drives deterministic organoid patterning. Science. 2022;375:eaaw9021. doi: 10.1126/science.aaw9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wiedenmann S, Breunig M, Merkle J, von Toerne C, Georgiev T, Moussus M, et al. Single-cell-resolved differentiation of human induced pluripotent stem cells into pancreatic duct-like organoids on a microwell chip. Nat Biomed Eng. 2021;5:897–913. doi: 10.1038/s41551-021-00757-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vivarelli S, Candido S, Caruso G, Falzone L, Libra M. Patient-derived tumor organoids for drug repositioning in cancer care: a promising approach in the era of tailored treatment. Cancers (Basel) 2020;12:3636. doi: 10.3390/cancers12123636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.World Health Organization WHO. WHO Tral Search. Available from: https://apps.who.int/trialsearch/Default.aspx.
- 131.NIH National Institutes of Health. Clinical Trials NIH. Available from: https://clinicaltrials.gov/.