Abstract
109Tissue-engineered scaffolds are more commonly used to construct three-dimensional (3D) tumor models for in vitro studies when compared to the conventional two-dimensional (2D) cell culture because the microenvironments provided by the 3D tumor models closely resemble the in vivo system and could achieve higher success rate when the scaffolds are translated for use in pre-clinical animal model. Physical properties, heterogeneity, and cell behaviors of the model could be regulated to simulate different tumors by changing the components and concentrations of materials. In this study, a novel 3D breast tumor model was fabricated by bioprinting using a bioink that consists of porcine liver-derived decellularized extracellular matrix (dECM) with different concentrations of gelatin and sodium alginate. Primary cells were removed while extracellular matrix components of porcine liver were preserved. The rheological properties of biomimetic bioinks and the physical properties of hybrid scaffolds were investigated, and we found that the addition of gelatin increased hydrophilia and viscoelasticity, while the addition of alginate increased mechanical properties and porosity. The swelling ratio, compression modulus, and porosity could reach 835.43 ± 130.61%, 9.64 ± 0.41 kPa, and 76.62 ± 4.43%, respectively. L929 cells and the mouse breast tumor cells 4T1 were subsequently inoculated to evaluate biocompatibility of the scaffolds and to form the 3D models. The results showed that all scaffolds exhibited good biocompatibility, and the average diameter of tumor spheres could reach 148.52 ± 8.02 μm on 7 d. These findings suggest that the 3D breast tumor model could serve as an effective platform for anticancer drug screening and cancer research in vitro.
Keywords: Tumor model, Decellularized extracellular matrix, Gelatin, Sodium alginate, Three-dimensional bioprinting
1. Introduction
110Cancer is a disease with extremely high morbidity and mortality. According to a report of the World Health Organization (WHO), the mortality rate of cancer has surpassed that of coronary heart disease and stroke, and cancer is recognized for contributing to the highest mortality worldwide[1]. The latest survey by the International Agency for Research on Cancer (IARC) in 2020 showed that breast cancer is the most common cancer in women, and also one of the cancers with the highest mortality worldwide. Therefore, research on breast cancer has become extensive in the research community[2]. Using in vitro tumor models constructed by tissue engineering method to study the mechanism of cancer and perform anti-cancer drug screening is very common. However, the development of traditional two-dimensional (2D) tumor models is limited by the growth environment they confer that cannot accurately mimic the heterogeneity and complexity of tumor tissues in vivo. In addition to tumor cells, multiple types of stromal cells, such as fibroblasts, endothelial cells, and immune cells, as well as complex network of blood vessels and extracellular matrix structure, exist within tumor microenvironment[3,4]. Therefore, threedimensional (3D) tumor models and their construction have been widely studied in recent years. 3D scaffolds fabricated with various desired materials can simulate the complex extracellular matrix components and structures of tumor microenvironment that be unattainable for 2D counterparts, and they are often used as a bridge between 2D-level researches and animal experiments[4,5].
In the process of constructing 3D tumor models, it is very important to choose appropriate methods and materials to prepare scaffolds for cell growth. 3D bioprinting is an effective additive manufacturing technology. Living cells, biochemical factors, proteins, drugs, and other biological materials were deposited on solid or gel surfaces or liquid collectors through layer-by-layer stacking of bioink to construct functional biomaterials[6-8]. By writing and controlling the computer program, this technology can achieve precise control of cell distribution and material structure, thereby printing tissue engineering scaffold, which is flexible and adjustable for different models and applications[9,10]. Due to its advantages in accurate reproducibility, high throughput, and high efficiency, 3D bioprinting has been gradually applied in tissue engineering in recent years[11]. 3D bioprinting technologies include material extrusion[12], material jetting,[13] and vat polymerization[14], among which material extrusion is the most widely used printing method on the grounds of simplicity and low cost. Nevertheless, since extrusion printing involves the squeezing of the bioink out of the nozzle through a pressure system, the structure of the material can be destroyed by high pressure; therefore, the bioink must have a high viscosity in order to maintain the structure of extruded material intact[15].
Extracellular matrix (ECM) is an important component in biological tissues, which supports cell growth and is involved in various complex cell-cell and cell-matrix interactions, as well as the release and reception of biochemical signals. ECM is a highly dynamic structure which is constantly reshaped by cells in tissues through synthesizing and degrading various chemical components and reorganizing their 3D structure. These complex processes require strict regulation to maintain tissue homeostasis, and imbalance of ECM remodeling may lead to disease[16]. In tumor tissues, the composition, content, and structure of ECM are related to tumor type, degree of malignancy, and stage of development. In clinical treatment, ECM molecules can be used as indicators to predict tumor prognosis[17]. Studies have shown that remodeling of ECM plays an important role in the formation and development of tumors, during which the composition and content of ECM in tumor tissues may change. Therefore, the study of in vitro ECM and its remodeling is conducive to further investigation of the pathogenesis of tumors[18]. Currently, the materials commonly used to construct in vitro ECM as tissue engineering scaffold can be divided into natural materials and synthetic materials. Natural materials include gelatin[19], alginate[20], chitosan[21], collagen[22], decellularized extracellular matrix (dECM)[23], and hyaluronic acid (HA)[24], etc. Synthetic materials include polylactic acid glycolic acid (PLGA)[25], polycaprolactone (PCL)[26] and polyethylene glycol (PEG)[27], etc. Natural materials exhibit good biocompatibility and can provide effective support for cell growth but often have limited physical properties, while synthetic materials show good physical properties but lack the sites for cell attachment.
dECM is a natural material commonly used to construct tissue engineering models. It is a biological macromolecular network obtained after the removal of cell components in biological tissues. It retains the structure, various chemical components, and vascular network of ECM, which could promote cell proliferation and tumor progression[28]. dECM had been reported to increase normal cell migration and invasion of cancer cells, and regulate cell behavior to maintain tissue integrity[23,29]. According to statistics, more than 50% of the patients diagnosed with breast cancer develop liver metastasis[30]. Owing to the association of the liver with metabolism of xenobiotics and drugs in the body, the liver proves to be a very challenging site for antimetastatic chemotherapies[30]. Therefore, it is imperative to establish a tumor model simulating liver microenvironment for continuous adhesion, proliferation, and invasion of breast cancer cells for the screening of antimetastatic drugs of 111breast cancer to liver. The liver-derived dECM contains collagen, fibronectin, laminin, glycosaminoglycan (GAG), proteoglycan (PG), and a variety of insoluble growth factors, which can mimic the liver environment, provide support and anchorage, and regulate intercellular communication for liver metastatic breast cancer cells[31]. However, although dECM has good biocompatibility, it is difficult to effectively fabricate tissue-engineered scaffolds by 3D printing using dECM alone due to its low viscosity. Therefore, it needs to be modified to make dECM more suitable for 3D printing[32]. Gelatin is a hydrolytic product of collagen, which has good biocompatibility and temperature sensitivity. The viscosity of dECM solution would increase after combining with gelatin under warm bath, and gelation can be formed at lower temperature, which is conducive to fabricate scaffolds with high-resolution 3D printing[11]. Sodium alginate is an easily available natural polymer with good biocompatibility. It can be crosslinked with divalent cations to form hydrogels under mild conditions. When combine with other biomolecules, it can significantly change the properties of the gel and make it suitable for different applications.
In this study, dECM derived from porcine liver, gelatin, and sodium alginate were used as materials to prepare tissue-engineered scaffolds. Using appropriate decellularization methods, the extracellular matrix components in the tissue and its original structure could be preserved while completely removing the cells at the same time. dECM was resolved and mixed with different concentration of gelatin and sodium alginate to prepare bioinks, among which the printable bioinks were selected to fabricate tissue-engineered scaffolds. The porosity, swelling, mechanical properties, and biocompatibility of scaffolds were tested, and metastatic mouse breast cancer cells 4T1 were seeded on scaffolds. The current study aimed to construct a metastatic tumor model to provide a platform simulating the in vivo environment of tumor tissues for anti-cancer drug screening and the delineation of the mechanism underlying tumor progression.
2. Materials and methods
2.1. Materials
Fresh porcine liver was purchased from the supermarket. L929 cell line and 4T1 cell line were donated by Zhengzhou Institute of Emerging Industrial Technology (Zhengzhou, China) and Cancer Hospital of China Medical University (Shenyang, China), respectively. NaCl, KCl, KH2PO4, and Na2HPO4·12H2O were provided by Tianjin Kemiou Chemical Reagent Co., Ltd. (Tianjin, China). Triton X-100, sodium dodecyl sulfate (SDS), and trypsin were provided by Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China). NH3·H2O was provided by Xilong Scientific Co., Ltd. (Shantou, China). EDTA, glutaraldehyde, and ethyl alcohol were provided by Damao Chemical Reagent Factory (Tianjin, China). Hematoxylin and eosin (H&E) dye, Masson dye, and pepsin were provided by Beijing Coolaber Science & Technology Co., Ltd. (Beijing, China). Hoechst 33324, Calcein-AM, and propidium iodide (PI) were provided by Shanghai Beyotime Biotechnology Co., Ltd. (Shanghai, China). α-DMEM and penicillin-streptomycin were provided by Sigma-Aldrich Inc. (St. Louis, MO, USA). RPMI 1640 was provided by Procell Life Science & Technology Co., Ltd. (Wuhan, China). CCK-8 kit was provided by Dojindo Laboratories (Kumamoto, Japan).
2.2. Cell culture
Mouse fibroblasts L929 were cultured in Dulbecco’s modified eagle medium (DMEM), supplemented with 10% fetal calf serum and 1% penicillin-streptomycin at 37°C with 5% CO2. The medium was changed once every 2 or 3 d, and when cells covered 90% of culture flask, they were digested by 2 mL 0.25% trypsin, and divided into 3–5 new culture flasks. Mouse breast tumor cells 4T1 were cultured in Roswell Park Memorial Institute medium (RPMI) supplemented with 10% fetal calf serum and 1% penicillin-streptomycin at 37°C with 5% CO2. The medium was changed every day, and when cells covered 80% of culture flask, they were digested by 2 mL 0.25% trypsin and divided into 3–5 new culture flasks.
2.3. Preparation of dECM and decellularization efficiency
Fresh porcine liver purchased from the supermarket was firstly washed with distilled water until most blood stains on the tissue had been removed. After that, it was cut into pieces with the size of 10 mm × 10 mm × 5 mm and stirred in distilled water for 2 h. The water was replaced every 0.5 h. The porcine liver pieces were then subjected to the first step of decellularization treatment with 2.5% trypsin- EDTA solution and stirred at 37°C for 6 h. The liver pieces were then placed into a 2% Triton X-100 solution with 1.25% NH3∙H2O and stirred for 72 h. Finally, the liver pieces were decellularized by stirring in 0.1% SDS solution until they turned white, and the dECM was obtained after washing with distilled water for 24 h to remove the residual reagents.
The microstructures of the dECM were observed by scanning electron microscopy (SEM; QUANTA 450, FEI, USA) to evaluate the decellularization efficiency. Briefly, the dECM (native tissue) was cut into the size of 3 mm × 3 mm × 1 mm and fixed with 2.5% glutaraldehyde solution for 3 h. Gradient dehydration was carried out with 50%, 70%, 90%, and 100% alcohol successively for 0.5 h. Then the specimens were observed with SEM after metal spraying. The specimens were also stained with H&E 112staining kit and Masson staining kit and observed under optical microscope (IX83, OLYMPUS, Japan) to evaluate the decellularization efficiency.
The decellularization efficiency was further evaluated to determine the DNA, GAGs, and collagen contents of dECM compared to the native tissue[33-36]. For quantification, 1 mg/mL of lyophilized dECM was digested in a papain solution (125 μg∕mL papain in 0.1 M Na3PO4 with 5 mM Na2EDTA and 5 mM cysteine at pH 6.5) for 16 h at 60°C. Native tissue of similar weight was also digested in a same manner as the control. The DNA content was determined using Hoechst 33324 assay. Briefly, 200 μL sample solution and 10 μL Hoechst dye were added to a 96-well plate, and incubated away from light for 30 min to measure the fluorescence intensity (excitation wavelength: 360 nm, emission wavelength: 450 nm). The standard curve for DNA was generated using calf thymus DNA and used for quantifying the DNA in samples. The GAGs content was estimated via quantifying the amount of sulphated GAGs using 1, 9-dimethylmethylene blue (DMMB) solution. Briefly, 20 μL sample solution and 200 μL DMMB were added to a 96-well plate, and incubated for 4–6 min to measure the absorbance at wavelength of 520 nm. The standard curve was made using chondroitin sulphate A and used for estimating the sulphated GAGs in samples. The collagen content was determined via a conventional hydroxyproline assay. Briefly, 1 mL 0.01mol/L CuSO4 solution, 1 mL 2.5 M NaOH solution, and 0.2 mL 3.6% H2O2 were added to 1 mL sample solution and shaken for 5 min. Then, it was left to stand for 30 min, bathed in water at 30°C for 10 min, and shaken violently again for 5 min. 4 mL H2SO4 and 2 mL 5% p-dimethylaminobenzaldehyde (P-DMAB) solution were added after water bath at 65°C for 20 min. Finally, 100 μL solution was added to a 96-well plate to measure the absorbance at 560 nm and quantified by referring to a standard curve made with hydroxyproline.
The surface of specimen was gently smoothed with a blade and the water contact angle of specimen was investigated by a contact angle meter with a high-speed camera (OCAH200, Data Physics, Germany). The videos were recorded from the moment the droplets touched the materials, until the droplets completely penetrated the scaffolds or became stable on the scaffolds. The photos of droplet at 0 s, 0.5 s, and 1 s were captured, and the water contact angles were measured by Image Pro Plus 6.0 (IPP, Media Cybernetics, USA). The results were averaged in each group by performing three parallel experiments.
2.4. Preparation and characterization of bioinks
The dECM solution was prepared by dissolving the lyophilized dECM pieces with pepsin. Briefly, 1 g dECM pieces were added and stirred in 100 mL 0.5 M acetic acid solution containing 10 mg pepsin at room temperature for 3 d until dECM pieces were completely dissolved. Then, the pH of dECM solution was adjusted to 7 with 1 M NaOH solution. The gelatin/sodium alginate/dECM (Gel/SA/ dECM) bioinks were prepared by mixing the dECM with different weight ratio of Gel and SA. The concentrations of gelatin were 4%, 5%, 6%, and 7%, while the concentrations of sodium alginate were 2% and 3%. The prepared bioinks were placed in 4°C refrigerator for later use.
The rheological properties of bioinks were tested by a rheometer (Anton-Paar, Austria)[37]. To determine the viscoelasticity of the bioinks, the measuring position was set to 0.1 mm, and the angular frequency range was 0.01–100 rad∙s−1. During the test, the temperature was set as 20°C, and the shear strain remained at 1%. The temperature sensitivity (“G’-Temp” model) of the bioinks was tested by serious reduction of temperature from 40 to 10°C (5°C / min), and the frequency was set to 1 Hz. To measure the steady viscosity, the relationship between shear rate and viscosity was tested at the shear rate ranged of 0.01 to 1000 (1∙s−1) at 20°C, and then the test was performed in the same manner again at the shear rate range of 1000 to 0.01 s−1 to determine thixotropy of the boinks. The thixotropic curves were graphed by fitting these two inverse shear rate-shear stress curves.
2.5. Manufacture of Gel/SA/dECM hybrid scaffold
The bioinks were pre-printed to select printable inks for the construction of 3D hybrid scaffolds by a 3D bioprinter (Pro, Regenovo, Hangzhou). Bioinks with appropriate concentrations were selected to construct 3D hybrid scaffolds, and the printed scaffolds were crosslinked with CaCl2 solution and EDC/NHS successively. Briefly, the hybrid scaffolds were soaked in 3% CaCl2 solution for 2 h and washed with distilled water for 3 times to remove the residual crosslinking agent. Then, the hybrid scaffolds were immersed in EDC/NHS crosslinking agent (50 mM EDC, 50mM NHS, 50mM MES, 60% ethyl alcohol) for 24 h and were washed three times with distilled water (0.5 h each time) to remove the residual crosslinking agent. The Gel/ SA/dECM (GSd) scaffolds were obtained after lyophilizing for 24 h.
2.6. Scaffolds characterization
2.6.1. Macro- and micro-structure of scaffolds
The 3D-printed scaffolds were cut into the size of 5 mm × 5 mm × 1 mm and were pasted on the conductive adhesive of the sample table. The surfaces of specimens were gently blown with nitrogen, and specimens were placed on the metal spraying instrument and sprayed with metal evenly for 10 min. Then, the microstructure of the scaffold was observed by SEM. The pore size data of the scaffolds determined by IPP software were sorted into pore size 113distribution graph using Origin Pro 9.5 software to analyze the pore structure of the scaffolds.
2.6.2. Infrared spectroscopic analysis
The chemical structure and the status of chemical bonds in the scaffolds were investigated by Fourier transform infrared spectroscopy (FTIR). Briefly, the scaffolds were milled into powder with KBr particles and presented to the Fourier transform infrared spectrometer (EQUINOX55, Bruker, Germany) to generate infrared (IR) spectra. The FTIR spectrometer was performed over the wavenumber range of 4000–500 cm−1 with a resolution of 4 cm−1.
2.6.3. Swelling ratio
The swelling behavior was characterized by gravimetric analysis[38]. The dried scaffold (m1) was placed in a 24-well plate, and completely immersed in distilled water. Then, the plate was placed in an incubator set at 37°C and taken out every 2 h to be weighed as (m2). Excessive water was gently absorbed with filter paper before test. The swelling ratio was calculated using Equation I:
| (I) |
The results were averaged in each group by performing three parallel experiments.
2.6.4. Mechanical properties
The surface of the scaffold was ground flat with a blade and make sure that its thickness was no less than 3 mm. The compression performance of the scaffold was tested using a universal testing machine (SANS, Shenzhen). Briefly, the length, width, and height of the sample were measured with a vernier caliper and marked as a1, b1, and l1, respectively. The compression was performed with 0.5 mm/min of crosshead speed until the specimen height had decreased by 50%. Then, the length, width, and height of the sample were measured again as a2, b2, and l2, respectively. The compression modulus was calculated using Equation II:
| (II) |
The results were averaged in each group by performing three parallel experiments.
2.6.5. Porosity
The porosity of the scaffolds was measured by pycnometer method, and ethanol was used as a substitution liquid because it is not a solvent for the scaffolds[39]. The dry scaffold was weighed as m0, and the weight of pycnometer filled with ethanol was noted as m1. The scaffold was immersed in the pycnometer and placed in a vacuum oven until the air bubbles were completely removed. The pycnometer was taken out and filled with ethanol again and weighed it as m2. Then, the scaffold was slowly taken out, and the pycnometer with the remaining ethanol was weighed as m3. The porosity was calculated using Equation III:
| (III) |
The results were averaged in each group by performing three parallel experiments.
2.6.6. Hemolytic properties
The scaffold was placed in normal saline. The negative control group was given normal saline without the scaffold, while the positive control group was given distilled water. 2 mL fresh rabbit blood was added to each group and incubated at 37°C for 1 h before taken out. The solutions were centrifuged with a centrifugation rate of 1000 rpm for 5 min to observe and compare whether blood cells were lysed.
2.6.7. Water contact angle
Water contact angles were measured with the method as described in section 2.3. The surface of scaffold was gently smoothed with a blade, and the water contact angle of the scaffold was tested with a contact angle meter with a highspeed camera. The videos were recorded from the moment the droplets touched the materials, until the droplets completely penetrated the scaffolds or became stable on the scaffolds. The photos of droplet at 0 s, 0.5 s, and 1 s were captured, and the water contact angles were measured by IPP software. The results were averaged in each group by performing three parallel experiments.
2.7. Evaluation of biocompatibility and skin model construction
The scaffolds were cut into the size of 5 mm × 5 mm × 1 mm and immersed in 75% ethanol under ultraviolet light overnight for sterilization purposes. The scaffolds were then slaked twice with phosphate-buffered saline (PBS) to remove residual ethanol as well as UV irradiated for 1 h each time. A cell suspension of fibroblast L929 with the concentration of 2×106 cells/mL was prepared and 10 μL of the cell suspension was inoculated on one side of the scaffold. After 0.5 h, 300 μL medium was added and the plate was put into a 37°C incubator with 5% CO2. After 3 h, the same amount of L929 were inoculated on the other side of the scaffold and supplemented with 200 μL of medium. 2D control group was directly inoculated with 20 μL of the cell suspension supplemented with 1 mL medium on the well plate.
The scaffolds inoculated with the cells for 1 d, 4 d, and 7 d, respectively, were taken out for fluorescence staining. Briefly, 300 μL staining solution, which was prepared by mixing 1 mL PBS with 2 μL Calcein-AM and 1141 μL propidium iodide (PI) was added to each well and incubated at 37°C with 5% CO2 for 15–20 min. The live/ dead staining images of cell-scaffolds were observed under single-photon laser confocal microscopy (IX83, Olympus, Japan).
The proliferation of L929 fibroblasts on the scaffolds was assessed by CCK-8 assay. The medium was removed followed by addition of 500 μL CCK-8 solution (CCK: α-DMEM = 1:10) to the cell-scaffolds cultured for 1 d, 4 d, and 7 d in each well, and placed in an incubator at 37°C with 5% CO2 for 3 h. The optical density (OD) values were tested with an enzyme-linked immunoassay at 450 nm. The results were averaged in each group by performing three parallel experiments. Moreover, the morphology of the cells cultured on the scaffolds for 1 d and 7 d were observed by SEM. The procedure was similar to that described in section 2.3.
2.8. Construction of breast tumor model
Briefly, a 4T1 cell suspension with the concentration of 1×106 cells/mL was prepared and 10 μL of the cell suspension was inoculated on one side of the scaffold. After 0.5 h, 300 μL medium was added and the plate was put into a 37°C incubator with 5% CO2. After 3 h, the same amount of 4T1 were inoculated on the other side of the scaffold and supplemented with 200 μL of medium. 2D control group was directly inoculated with 20 μL of the cell suspension supplemented with 1 mL medium on the well plate.
The scaffolds inoculated with the cells for 1 d, 4 d, and 7 d, respectively, were taken out for fluorescence staining. Briefly, 300 μL staining solution, which was prepared by mixing 1 mL PBS with 2 μL Calcein-AM, 1 μL PI and 5 μL Hoechst 33342, was added to each well and incubated at 37°C with 5% CO2 for 15–20 min. The staining images of cell-scaffolds were observed under single-photon laser confocal microscopy. The diameters of tumor spheres formed on 4 d and 7 d were determined by IPP software and sorted into a column graph. 4T1 grown on 2D plates were observed by inverted microscope and pictures were also taken.
The proliferation of L929 fibroblasts on the scaffolds was assessed by CCK-8 assay. The medium was removed followed by addition of 500 μL CCK-8 solution (CCK: α-DMEM = 1:10) to the cell-scaffolds cultured for 1 d, 4 d, and 7 d in each well, and placed in an incubator at 37°C with 5% CO2 for 3 h. The OD values were tested with an enzyme-linked immunoassay at 450 nm. The results were averaged in each group by performing three parallel experiments. Moreover, the morphology of the cells cultured on the scaffolds for 1 d, 4 d, and 7 d were observed by SEM. The procedure was similar to that described in section 2.3.
2.9. Statistical analysis
All data were presented as mean ± standard deviation (SD). One-way analysis of variance (ANOVA) was used for analyzing significant difference of data in each group, and P<0.05 indicated that there was significant difference between groups. All statistical analyses were performed using Origin Pro 9.5 software (Origin Lab, MA, USA).
3. Results and discussion
3.1. Evaluation of decellularization efficiency
If the process of decellularization is too intense, the microstructure in tissues may be destroyed, and some bioactive components, such as proteins, glycoproteins, and other chemicals that are important for cell proliferation, may be lost in large quantities, which could inhibit cell attachment and migration[40]. If the decellularization process is too mild, the original cells in the tissue may not be completely removed, so the growth and proliferation of subsequent inoculated cells may also be inhibited as a result of immune activity[41]. Therefore, it is necessary to evaluate the efficiency of decellularization. Figure 1A showed the macroscopic morphology of porcine liver tissues before and after decellularization. It was obvious that fresh porcine liver tissue was red before decellularization, while pale and soft after being washed by distilled water. dECM obtained after decellularization was white, slightly transparent, and softer, with visible pore structures on the surface. After freeze-drying, the dECM turned into white slices, and the internal tissue was found to be white floccules. This result was identical to the dECM of pig liver obtained by Sellaro et al.[42] In order to confirm that the decellularization process was complete and the original structure of the tissue was not damaged, SEM (Figure 1B), H&E staining (Figure 1C), and Masson staining (Figure 1D) were used to observe the tissues before and after decellularization, and the results were similar to those obtained by Saleh et al.[43] and Wu et al.[44] SEM results showed that there were no cells in dECM and its surface showed irregular pore structure. In addition, a large number of fiber networks could be seen in dECM under high magnification, which may be collagen or other components in extracellular matrix that provided sites for cells to attach and grow. A large number of cells could be seen in H&E staining and Masson staining images before decellularization, and actin was more than collagen in porcine liver tissue. ECM and collagen in liver tissue were preserved in dECM, while actin was removed. The complex network structure was not damaged, which was consistent with the results of SEM.
Figure 1.
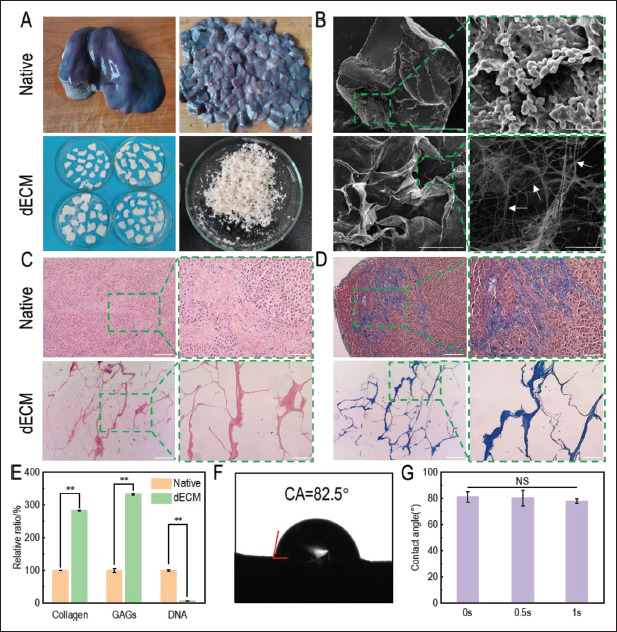
Characterization of porcine liver tissues before and after decellularization. (A) Macrostructure of tissues before and after decellularization. (B) Microstructure of tissues observed by scanning electron microscope. Images on the left (scale bar: 100 μm) were enlarged and shown on the right (scale bar: 50 μm). White arrows: fibers. (C) H&E staining (red: nuclear; blue: extracellular matrix) and (D) Masson staining (red: actin; blue: collagen) of tissues before and after decellularization were performed. Images on the left (scale bar: 400 μm) were enlarged and shown on the right (scale bar: 100 μm). (E) The relative contents of collagen, GAGs, and DNA after decellularization were investigated by standard curve method. **P<0.01, represents significant difference between two groups. (F) Water contact angle of dECM at 0 s. (G) Water contact angles at 0 s, 0.5 s, and 1 s. NS indicated that no significant difference existed between different groups.
To further evaluate the efficiency of decellularization, collagen, GAGs, and DNA in the tissues before and after decellularization were quantitatively detected. GAGs are important component of ECM. Due to their hydrophilic 115structure, GAGs can bind to growth factors and retain water in tissues[45]. The results showed that collagen and GAGs significantly increased while DNA significantly decreased after decellularization, which turned out to be 283.17 ± 0.47%, 333.31 ± 2.18%, and 6.45 ± 0.68% compared with those before decellularization, respectively (Figure 1G). Since the total weight of the decellularized tissue decreased, while collagen and GAGs were mainly retained, the relative ratios increased compared with those before decellularization. The contents of collagen and GAGs in dECM were 353.85 ± 13.60 μg∕mg and 468.82 ± 16.23 μg∕mg, respectively, calculated by the fitting formula of the standard curve. Compared with 52.19 ± 11.18 μg∕mg and 5.62 ± 0.65 μg∕mg obtained by Struecker et al.[46], the content of collagen and GAGs retained in dECM in this study were higher. DNA content in dECM was found to be 42.16 ± 4.06 ng/mg calculated by the fitting formula of the standard curve. It is generally believed that the process of decellularization is complete when DNA content is less than 50 ng/mg[16]. Therefore, the decellularization process in this study met the requirements. Sun et al.[47] prepared microbeads based on porcine liver dECM and sodium alginate, in which the DNA content of dECM was 290.67 ± 54.31 ng/mg. Compared with their result, the decellularization in this study was more thorough. In conclusion, the decellularization method used in this study exhibited a good efficiency.
The adhesion of cells to scaffolds is affected by the hydrophilicity of scaffolds[48]. As the exchange of substances between cells and medium takes place in liquid environment, scaffolds should be highly hydrophilic so that the cells inoculated on them can fully contact with the medium. The water contact angle of the dECM tissue after 116decellularization was evaluated, and it was found that when the water droplets contacted the dECM surface at 0 s, 0.5 s, and 1 s, the water contact angles were 81.16 ± 4.09°, 80.35 ± 2.47°, and 78.05 ± 1.34° (Figure 1F). The water angle decreased with time, indicating that the material showed strong hydrophobicity. The dECM water contact angle obtained by Li et al.[49] from porcine lung was 82°, which was similar to that of pig liver dECM in this study. In order to enable cells to better exchange nutrients and metabolites with liquid medium, modification was required to make dECM more hydrophilic.
3.2. Printability analysis
Different concentrations of bioinks were prepared, and in order to evaluate whether the bioinks meet the requirements of 3D printing, these bioinks were prebioprinted. The concentrations of bioinks and printing conditions were shown in Table 1. With the increase of bioink concentration, the required printing temperature and pressure also had to increase accordingly, otherwise it may lead to disconnection during bioprinting, failing to meet the requirements of scaffolds for tissue engineered models. In addition, the higher the concentration of gelatin and sodium alginate in the bioink, the higher the resolution of the printed scaffold under suitable conditions; this was attributed to the high viscosity of these two materials. When the gelatin concentration in the bioink was less than 5%, it was difficult to mold under similar printing conditions, and the resolution of scaffold was low and easy to collapse, which was consistent with the results of rheological properties.
Table 1.
Parameter of 3D bioprinting with different bioinks
| Gelatin (wt%) | Alginate (wt%) | dECM (wt%) | Thickness (mm) | Space (mm) | Temperature (°C) | Pressure (MPa) | Speed (mm/s) | |
|---|---|---|---|---|---|---|---|---|
| 4G2S1d | 4 | 2 | 1 | 0.2 | 0.7 | 10 | 0.17 | 5 |
| 4G3S1d | 4 | 3 | 1 | 0.2 | 0.7 | 15 | 0.15 | 10 |
| 5G3S1d | 5 | 3 | 1 | 0.2 | 1.0 | 20 | 0.20 | 8 |
| 6G3S1d | 6 | 3 | 1 | 0.2 | 1.0 | 20 | 0.20 | 8 |
| 7G2S1d | 7 | 2 | 1 | 0.2 | 1.0 | 24 | 0.24 | 8 |
In addition to pre-bioprinting, rheological properties of bioinks were also tested for further investigation of printability, and the results were shown in Figure 2. Temperature sensitivity is the property that storage modulus (G’) of fluid changes with temperature. As shown in Figure 2A, the gel points of several bioinks were all around 20°C, which was mainly attributed to the temperature sensitivity of gelatin. The change trend was the same as the temperature sensitivity of the gelatin/ sodium alginate/Matrigel bioinks prepared by Mao et al.[50]
Figure 2.
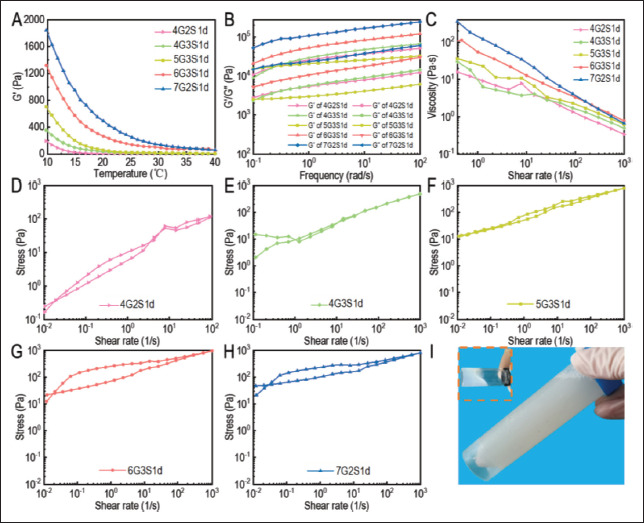
Rheology characterization of Gel/SA/dECM bioinks. (A) Thermosensitivity. (B) Viscoelasticity. (C) Viscosity. Thixotropy of 4G2S1d (D), 4G3S1d (E), 5G3S1d (F), 6G3S1d (G), and 7G2S1d (H) were evaluated. (I) After gelatin and sodium alginate were added, the Gel/SA/dECM bioink turned more viscous as well as a little yellow compared to the dECM solution (inset), a white viscous liquid.
Due to the addition of high concentration of Matrigel, the latter showed higher energy storage and loss modulus at the same temperature. The structure and properties of collagen fiber are determined by the electrostatic force and hydrophobic interaction between adjacent collagen molecules. With the increase of temperature or pH, the hydrophobic interaction and electrostatic attraction of amino acid side chains in collagen fiber are strengthened, so the collagen fiber stiffens and the material changes from fluid state to gel state[51]. The storage modulus (G’) of the three bioinks changed more obvious with the higher concentration of gelatin from 40 to 10°C, among which 7G2S1d increased from 56.2 Pa to 1844.2 Pa, and 6G3S1d and 5G3S1d increased from 52.4 Pa and 6.3 Pa to 1321.0 Pa and 704.0 Pa, respectively. The G’ of the two bioinks with 4% gelatin increased the least, which increased from 1.5 Pa and 3.1 Pa to 194.4 and 356.7 Pa, respectively, so the G’ change is related to the gelatin concentration. The higher the gelatin concentration, the greater the storage modulus change.
Figure 2B showed the viscoelasticity of several bioinks, which was the comprehensive change of elasticity and viscosity of materials during the fluidization process. Storage modulus (G’) represented elasticity and loss modulus (G”) represented viscosity. When the two curves intersect, it indicates that the material has undergone fluidization or gelation. The energy storage modulus of the materials in the figure was always higher than the loss modulus because the externally applied energy of the fluid was stored and converted into repulsive force during the process of the fluid changing from gel state to solid state[52]. The storage modulus of the material with high concentrations could reach more than 10000 Pa and the highest could reach 66907 Pa, while the storage modulus of 4G2S1d was 1036.3 Pa. No intersection point was seen between the two curves, indicating that the elasticity of the material was greater than viscosity within the test range, and no fluidization phenomenon occurred during the process. The higher the elasticity of the system was, the more difficult the material was to fluidize, and the less possible the printed scaffold would deform.
117The steady viscosity of several bioinks was shown in Figure 2C. Generally speaking, the viscosity of each material decreased with the increase of shear frequency, indicating that they were pseudoplastic fluids and shear thinning fluids[53]. The higher the material concentration, the higher the fluid viscosity. In the test range shown in the figure, the maximum viscosity of 7G2S1d was 182.93 Pa, and the maximum viscosity of 4G2S1d was 15.91 Pa. It was found that the higher the concentration was, the more stable the decrease rate of viscosity with the increase of shear frequency, indicating that scaffold constructed by the material with high concentration had a lower risk of deformation or collapse after 3D printing.
Thixotropy is a reversible sol rheological phenomenon in which the viscosity and shear stress of the gel change with time as a result of external forces such as oscillation, agitation, and compression, reflecting the time dependence of fluid viscosity. The area enclosed by the two curves formed by the change of fluid shear stress as a result of the increase and decrease of shear rate is called thixotropic ring. The larger the area of the thixotropic ring is, the stronger the time stability of the fluid is. As shown in Figure 2D-H, the higher the bioink concentration was, the stronger the thixotropy was in general. The thixotropy of 6G3S1d was stronger than that of 7G2S1d, possibly because sodium alginate has a greater effect on improving the viscosity of the material than gelatin. Therefore, 5G3S1d, 6G3S1d, and 7G2S1d were selected as bioinks for printing scaffolds with consideration of both rheological properties test results and pre-printing results.
Han et al.[54] prepared porcine liver dECM solution and collagen solution with concentrations of 1.5% and 3%, respectively, for 3D-printed tissue-engineered scaffolds. In the shear frequency range from 1 to 100 rad·s−1, the maximum storage modulus of the two bioinks were no more than 1000 Pa. Compared with their bioinks, the viscoelasticity of bioinks in our study exhibited higher viscoelasticity. In addition, during the shear rate range from 0 to 1000 s−1, the steady viscosity of the bioink with sodium alginate in our study was also higher than that of pure collagen solution. It is suggested that the addition of gelatin and sodium alginate, two high-viscosity materials, can improve the stability and printability of bioink. Compared with the dECM bioinks derived from porcine fat, cartilage, and heart prepared by Pati et al.[33], the groups of bioinks prepared in our study had higher viscoelasticity and steady viscosity. After gelatin and sodium alginate were added, Gel/SA/dECM bioink turned more viscous as well as a little yellow (Figure 2I) compared to the dECM solution (Figure 2I inset), a white viscous liquid.
3.3. Structure and pore size of scaffolds
118As shown in Figure 3A, the three printed scaffolds were translucent with high resolution and clear pore structure, but the structure was loose and easy to deform with external forces. In order to further improve the stability of the scaffolds, CaCl2 and chemical crosslinking agent EDC/NHS were both used to crosslink the scaffolds. After crosslinking, the stiffness and resolution of the scaffolds were further improved. After freeze-drying, the scaffolds presented a white slice with evenly distributed pore structure. In order to further observe the microstructure of the scaffolds, they were observed by SEM. Figure 3B showed a series of uniformly distributed macropores which were formed from printing gaps and many interconnected micropores on the beam which were created by lyophilization after crystallization. However, there were no pore structures in some areas, while the surface exhibited rough ravine, similar to the results of Chaji et al.[55]. The complex network of small pores nested with large holes within the scaffold increased the specific surface area of the scaffold and reduced the risk of dissolution in liquid environment[56]. The large pore sizes of the three groups of scaffolds were 262.62 ± 49.78 μm, 202.57 ± 14.23 μm and 533.58 ± 52.41 μm, respectively. The pore size distribution was shown in Figure 3C. The small pore sizes of the three groups of scaffolds ranged from 15.97 μm to 140.32 μm, 22.55 μm to 173.67 μm, and 5.10 μm to 50.21 μm, respectively. The complex porous structure with these pore sizes could promote the growth, adhesion, and invasion of cells on the scaffold, and the large pore size was beneficial to improve the exchange efficiency of metabolic waste and nutrients in the tumor model.
Figure 3.
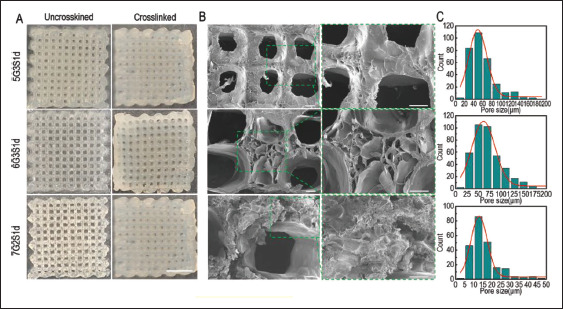
Structure of 3D-printed Gel/SA/dECM scaffolds. 5G3S1d and 6G3S1d were printed with the temperature of 20° and pressure of 0.20 Mpa, while 7G2S1d was printed with the temperature of 24° and pressure of 0.24 Mpa. (A) Macrostructure of the scaffolds before and after crosslinking by 3% CaCl2 and EDC/NHS. Scale bar: 5 mm. (B) Microstructure of crosslinked scaffolds observed by scanning electron microscope. The left images (scale bar: 100 μm) were enlarged and shown on the right (scale bar: 100 μm). (C) Distribution of small pore size in the scaffolds.
Jeong et al.[57] prepared porous scaffolds by 3D printing using pure porcine liver dECM as bioink. The small pore size in the scaffold turned out to be 20–60 μm, while the large pore size was 600–1000 μm, and the small pore size increased as large pore size increased. In contrast, the three groups of 3D-printed scaffolds in our study had smaller large pores but larger small pores, indicating that the addition of gelatin and sodium alginate improved the porosity of the material and was conducive to providing support for the growth of more cells.
3.4. Physical properties and hemolytic properties of scaffolds
IR spectra of the scaffold before and after crosslinking were shown in Figure 4A and B. It could be seen that the main absorption peaks before and after crosslinking showed no significant difference, but the transmittances of some absorption peaks were changed. This was due to CaCl2 is a physical crosslinking agent, combining alginate and divalent cation to form a stable network structure by ionic bond, thus improving the stability of the system. This process does not change the chemical composition of the material[58]. In addition, EDC/NHS is a chemical crosslinking agent, whose crosslinking mechanism is to transform the amino and carboxyl groups in the system into amide bonds, which can repair collagen broken in the process of decellularization[49]. Therefore, in the infrared spectrogram after crosslinking, the transmittance of absorption peaks C=O and N-H changed greatly, and the two absorption peaks at 1653 cm−1 and 1553 cm−1 were the same as those of gelatin, corresponding to the C=O stretching vibration of amide I and N-H deformation vibration of amide II and C=N in protein, respectively[59,60]. In addition, N-H stretching vibration peaks of amide I and amide II were observed at 3294 cm−1 and 3086 cm−1, respectively. N-deformation vibration peak of amide III was observed at 1238 cm−1, and absorption peak of GAG 119was also observed at 1048 cm−1[59,61]. The absorption peak at 1390 cm−1 was the stretching vibration of C-N, and the wide band at 3000–3500 cm−1 corresponding to the -OH group in the system[60]. In general, the IR spectra of scaffold before and after crosslinking were similar to those of porcine lung dECM obtained by Li et al.[49], indicating that the ECM components of porcine lung and liver were similar. It had been reported that the broken N-H-O hydrogen bond between the triple helix chains of collagen leaded to the destruction of its triple structure, and the absorption peaks in the infrared spectrogram were found to migrate to a lower wavenumber, indicating protein denaturation[62]. It can be seen from Figure 4A and B that some absorption peaks migrated to higher wavenumbers after the scaffold was crosslinked, which may be caused by the restoration of the triple structure of collagen under the effect of EDC/NHS during the crosslinking process. Therefore, the damaged collagen structure in dECM was effectively repaired during the crosslinking process[45].
Figure 4.
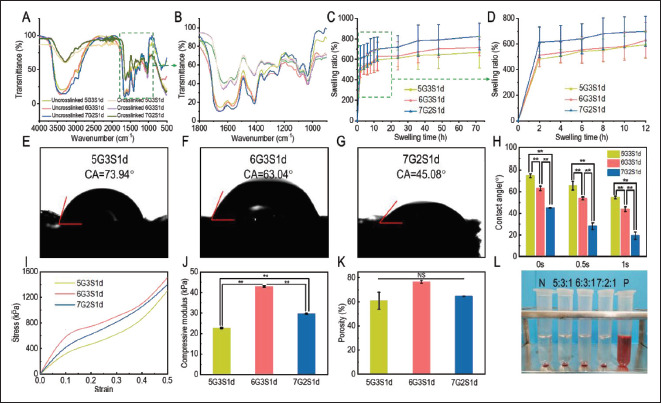
Physical characteristics of the 3D-printed Gel/SA/dECM scaffolds. (A) FITR spectra of the scaffolds before and after crosslinking. (B) FITR spectra at 1000–1800cm−1. (C) Swelling ratio of the scaffolds. (D) Swelling ratio during the initial 6 h. (E-G) Water contact angles of the scaffolds at 0 s. (H) Water contact angles of the scaffolds at 0 s, 0.5 s, and 1 s. **P<0.01 represents significant difference between two groups. (I) Stress-strain curve of the scaffolds. (J) Compressive modulus of the scaffolds within elastic deformation stage. **P<0.01 represents significant difference between two groups. (K) Porosity of the scaffolds was investigated. NS represents no significant difference between two groups. Hemolytic test was performed on scaffolds and result is shown in (L).
Since tumor tissues in vivo grow in a liquid environment, in vitro tumor models have to exhibit good hydrophilicity to simulate the effects of tumor microenvironment on tumor cell behaviors. In order to evaluate the hydrophilicity of the scaffolds, the swelling ratios of the three groups of crosslinked scaffolds in water were tested, and the results were shown in Figure 4C and D. Obviously, in the initial 2 h, the swelling ratios of scaffolds increased rapidly, while increased slowly after 2 h, and gradually stabilized after 48 h. After 72 h, the swelling ratios of 5G3S1d, 6G3S1d, and 7G2S1d reached 727.99 ± 162.49%, 752.62 ± 82.22%, and 835.43 ± 130.61%, respectively. The results showed that the hydrophilicity of the scaffold could be improved by increasing the concentration of gelatin or sodium alginate, but with the same total concentration, the swelling ratio of 7G2S1d was significantly higher than that of 6G3S1d, indicating that the hydrophilicity of gelatin was stronger than that of sodium alginate. Addition of gelatin into the system improved the hydrophilicity more obviously. The poly (n-propenyl l-lysine)/ hyaluronic acid scaffold prepared by Xu et al.[63] generally stabilized after immersion for 120 h, increasing from 8.9 ± 0.6 to 11.8 ± 0.5 as the degree of methacrylate increased. The swelling degree of the scaffolds prepared in our study was similar to that of 20% methacrylate. Figure 4E-H showed the test results of water contact angles of the three groups of scaffolds. Compared with dECM alone, the water contact angles of the scaffolds added with gelatin and sodium alginate decreased, indicating that their hydrophilicity was enhanced. The water contact angles of 5G3S1d, 6G3S1d, and 7G2S1d at 0s were 45.08 ± 0.41°, 63.04 ± 1.99°, and 74.65 ± 1.77°, respectively. The correlation between water contact angle and bioink concentration was consistent with the results of swelling ratio test. In addition, compared with the pure porcine liver dECM, the water droplets on these three scaffolds completely penetrated the material 120quickly, and the water contact angles after 1 s were 54.55 ± 1.21°, 43.78 ± 2.34°, and 19.30 ± 3.39°, respectively, indicating that the hydrophilicity of the scaffolds was greatly improved. The water contact angles of a series of porcine skin dECM/ gelatin/chitosan scaffolds prepared by Xu et al.[34] ranged from 84.84° to 126.25° at 0 s. Moreover, with the addition of dECM, water droplets penetrated the scaffolds at a slower rate, remaining between 52.75° and 70.25° after 3–6 s. The scaffolds prepared in our study were more hydrophilic.
In order to support the growth of cells and the formation of tumor spheres, the scaffold should have a certain mechanical strength, so that it can stand the weight of cells filling the entire scaffold. Therefore, the compression performance of the scaffold was tested, and the results were shown in Figure 4I. It can be seen that when the strain reached 0.5, the stress of the three scaffolds could reach more than 1000 Pa, among which 6G3S1d showed the best compression performance. When the scaffold was compressed, the stress-strain curves was nearly a straight line at first, which is called the elastic deformation stage, and the relationship between stress and strain follows Hooke’s law. Then, the scaffold enters the micro-plastic strain stage and yield stage as it continues to be compressed. Therefore, a tangent line was taken for the part at the beginning of the compression curve, which was nearly straight, and its slope was the compression modulus. The results were shown in Figure 4J, turning out to be 22.68 ± 0.28 kPa, 29.64 ± 0.41 kPa, and 42.90 ± 0.19 kPa, respectively. The 3D printed scaffold based on hydroxyethyl cellulose/sodium alginate/gelatin (HCSG) composite biomaterial prepared by Li et al.[64] was no more than 50 kPa at a strain of 0.6, and the GelMA hydrogel prepared by Peela et al.[65] for the construction of breast cancer model had a compression modulus of 748 ± 90 Pa. The three groups of 3D-printed scaffolds prepared in our study exhibited better compression performance, and the stress could all reach more than 1000 Pa when the strain was only 0.5, and the compression modulus of 5G3S1d was also more than 20 kPa, indicating that the scaffolds prepared in our study were not easy to deform and could meet the requirements for inoculating cells. It had been reported that the behavior of tumor cells was sensitive, and dependent on the mechanical forces in the extracellular matrix, so scaffolds with high mechanical strength were conducive to the adhesion and migration of tumor cells[66]. The high mechanical strength of these scaffolds was attributed to the double crosslinking of Ca2+ and EDC/ NHS crosslinkers, which also restricted the degradation ratio of these scaffolds by serum enzymes (lysozyme and collagenase) to some extent (Figure S1). The appropriate mechanical strength and degradation rate of these scaffolds offered stout “houses” for tumor cell proliferation and further development of tumor sphere.
The porous structure of scaffolds could promote cell adhesion and proliferation. High porosity and close connection between pores can ensure effective mass transfer between high-density cells and improve water absorption[67]. To evaluate whether the scaffolds were suitable for cell inoculation, the porosity of the three groups of scaffolds was investigated. The results were shown in Figure 4K, which turned out to be 61.02 ± 6.95%, 76.62 ± 4.43%, and 64.68 ± 1.21%, respectively. It had been reported that with the increase of hydrogel material concentration, the average pore size of the scaffold decreased and porosity increased after lyophilization, which was due to the influence of solution viscosity, ice crystal size, and nucleation rate during lyophilization. It can be seen in the results that although the total concentration was the same, the porosity of 6G3S1d was higher than that of 7G2S1d, possibly because sodium alginate forms a more stable network after crosslinking by Ca2+[21,68].
Hemolysis test results of scaffolds were shown in Figure 4L. It could be observed that in the positive control group, red blood cells were lysed as a result of osmotic pressure inside and outside the cells, while in the solutions of the three groups of scaffolds, red blood cells precipitated at the bottom of the tubes were not lysed and the solutions were clear and transparent, which was similar to the results displayed in the negative control group. It could be preliminary evidence that substances harmful to the cell were not contained in the scaffolds.
3.5. Biocompatibility
In order to further evaluate the biocompatibility of the scaffolds, mouse fibroblasts L929 were inoculated on the scaffolds and observed by confocal laser microscope on 1 d, 4 d, and 7 d, respectively, as shown in Figure 5A-C. Calcein-AM contains methyl acetate with high lipophilic properties, which can pass through the cell membrane, and go through the action of esterase inside living cells. The remaining Calcein can emit strong green fluorescence at the excitation wavelength of 490 nm. PI is commonly used as a nuclear stain. It cannot pass through the membrane of living cells, but it can pass through the membrane of dead cells, and reach the nucleus, embedding into the DNA double helix structure and emitting red fluorescence at the excitation wavelength of 535 nm. Therefore, Calcein-AM and PI are usually used as fluorescent staining agents to label living cells and dead cells, respectively. From Figure 5A-C, it can be seen that with the increase of incubation time, quantity of living cells on the scaffolds also gradually increased. On 4 d, cell clusters had appeared on the scaffold (white arrows), and the diameter of cell clusters increased 121on 7 d. During the whole culture process, no dead cells were observed, showing that the biocompatibility of the scaffolds was good. In general, 6G3S1d had the highest number of living cells, indicating the best biocompatibility.
Figure 5.
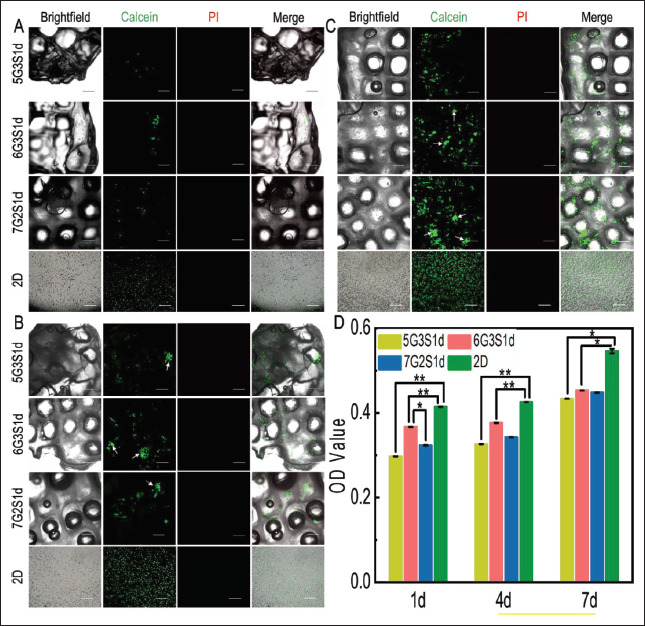
Cell viability, distribution, and growth of L929 cells in the Gel/SA/dECM hybrid scaffolds and 2D culture condition were both investigated. Live/ dead staining performed at 1 d (A), 4 d (B), and 7 d (C), respectively. Green: live; red: dead; white arrow: cell colonies (scale bar: 500 μm). (D) Viability of L929 grown on scaffolds evaluated by CCK-8 kit. *P<0.05, **P<0.01 represent significant difference between two groups.
In order to further determine the biocompatibility of the scaffolds, CCK-8 kit was used for quantitative detection of cell viability, and the results were shown in Figure 5D. It could be seen that CCK-8 results were consistent with those of fluorescence staining. No significant improvement of cell viability could be seen from 1 d to 4 d, but it significantly increased on 7 d. Overall, cells on 6G3S1d showed the highest viability, which was related to the higher porosity, hydrophilicity, and mechanical properties of the hybrid scaffold. In the whole process of culture, the cell viability in 2D environment was always higher than that in 3D environment, which may be due to hypoxia and less nutrients in the tumor microenvironment compared to the 2D culture[69]. In addition, on 7 d of culture, no significant difference in cell viability among the three scaffolds was shown, indicating that the number of cells inside the scaffolds reached the maximum. It had been reported that cells grew faster on harder materials than on softer materials[70]. The in vivo tissue microenvironment of organisms is softer than materials of plates, so our scaffolds could simulate the in vivo microenvironment more closely than 2D counterparts.
Figure 6A showed the change of L929 distribution on the scaffolds with the culture time. As could be seen from the 3D view of Calcein-AM staining, the proliferation cells in the scaffolds increased with the culture time both horizontally and vertically at the same time. On 7 d, most of the field of vision on the flat was filled with cells, and infiltration in the vertical direction was deeper. Figure 6B showed the distribution of L929 on three different specific heights on 6G3S1d scaffold. It can be seen that the number of the cells on the scaffold surface was the largest, which was the depth of 0 μm, and with the increase of depth, cells began to appear in different areas, while the total fluorescence intensity was reduced, which is due to the harder exchange of nutrients, and metabolic 122wastes between internal scaffold, and culture medium. In addition, cells penetrated the interior from the surface, so fewer cells could be found at the deeper site of the scaffold. The number of cells at the same depth on 4 d culture increased compared with that on 1 d, indicating that L929 gradually penetrated the scaffold, which was consistent with the change of 3D view with the days of culture.
Figure 6.
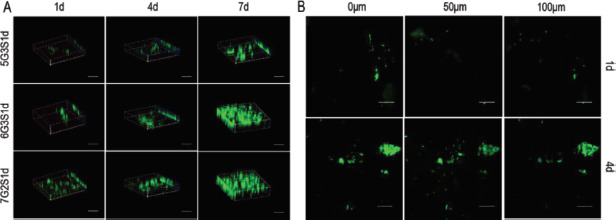
Infiltration of L929 within the Gel/SA/dECM scaffolds was observed by laser scanning confocal microscope. (A) 3D views of cells grown in the scaffolds at 1 d, 4 d, and 7 d (scale bar: 500 μm). (B) Distribution of cells on different depth in 6G3S1d scaffolds (scale bar: 500 μm).
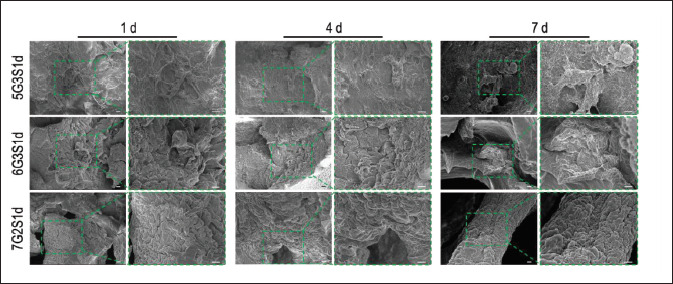
The growth morphology of cells on the scaffolds was observed by SEM, and the results were shown in Figure 7. It can be seen that the cells were mostly attached to the corner of the scaffold, the ravine or the edge of the large pores. On 1 d, several cells, which were almost spherical, were observed on the scaffolds. A few cells were distributed together while most of them were randomly scattered in the same area. On 7 d, the number of cells on the scaffolds increased significantly, and cell clusters appeared in some areas. The cell morphology became more flat, which resembled the long spindle shape observed by Kwak et al.[71]. Under high magnification, the pseudopodia of cells attached to the scaffolds could be clearly observed. This result was consistent with the fluorescence staining results and CCK-8 results, indicating that our scaffolds exhibited good biocompatibility, and cell clusters with high viability could be formed after 7 d of inoculation.
Figure 7.
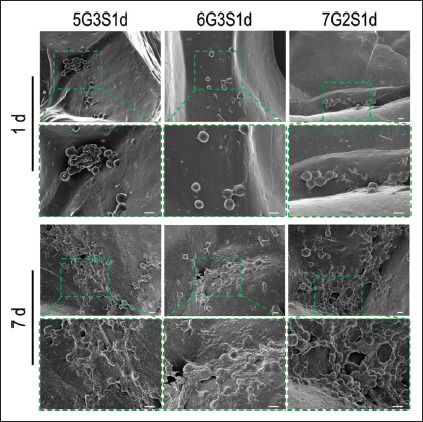
Obvious growth of L929 cells on Gel/SA/dECM scaffolds at 1 d and 7 d as observed by a scanning electron microscope. Cell colonies were observed at 7 d. The images in upper panel (scale bar: 100 μm) were enlarged and shown in the lower panel (scale bar: 10 μm) to show the antennae binding cells with the scaffolds.
3.6. Viability of 4T1 on 3D scaffolds and 2D plates
4T1 is a tumor cell line extracted by Fred Miller and others from mice with portability as well as high tumorigenicity and invasion, and could metastasize from primary breast tumors to distant tissues and organs spontaneously. The invasion and metastasis of 4T1 are identical to metastatic breast tumor cells of human[72]. Therefore, 4T1 was utilized in the present study as seed cells to construct the metastatic tumor model. Mice metastatic breast cancer cells 4T1 were inoculated on three groups of scaffolds and 2D plates, respectively, and cultured in an incubator with fluid exchange every 2 d to establish the model of metastatic breast cancer. Figure 8A-C showed the growth of cells on the scaffold on 1 d, 4 d, and 7 d of culture. Hoechst could travel across the cell membrane and bind to small grooves on the DNA double strands in living cells, and emit blue fluorescence at an excitation wavelength of 461 nm. So, it is often used to label living cells. However, at the same time, it can also bind with gelatins and other biocompatible scaffolds to fluoresce, so the results of Hoechst staining should be compared with those of Calcein-AM staining to draw a conclusion.
Figure 8.
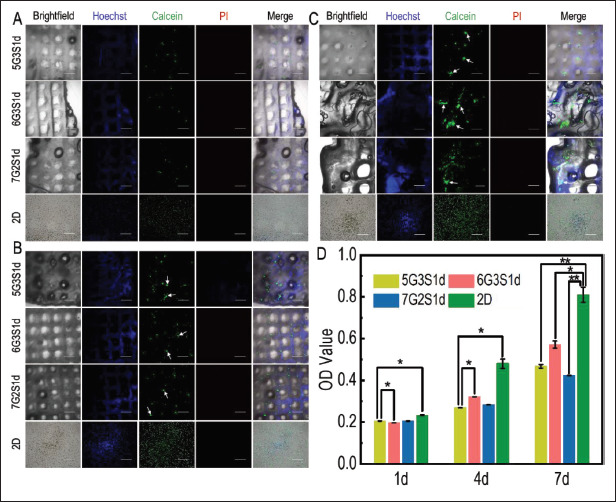
Cell viability, distribution, and growth of 4T1 cells in the Gel/SA/dECM hybrid scaffolds and 2D culture condition were both investigated. Fluorescence staining was performed on the scaffolds and cells at 1 d (A), 4 d (B), and 7 d (C), and the growth of 4T1 on the scaffold was observed with confocal laser microscope. Green: live; red: dead; white arrows: cell colonies or tumor spheres (scale bars: 500 μm). (D) Viability of 4T1 grown on scaffolds evaluated by CCK-8 kit. *P<0.05, **P<0.01 represent significant difference between two groups.
Fluorescence staining results of cells on the scaffolds showed that the number of cells increased gradually with time. Compared with mouse fibroblast L929, the distribution of 4T1 on the scaffold was more dispersed, forming cell clusters and tumor spheres clearly visible at low magnification on 7 d. Compared with the previous fluorescence staining results of L929, the fluorescence intensity of 4T1 was lower, resulting from the fewer 4T1 123inoculated in this experiment. In general, the number of cells on 6G3S1d scaffold was the highest, and the proliferation was the fastest, which was consistent with the growth of L929.
Figure 8D presented the results of quantitative test of 4T1 viability evaluated by CCK-8 kit. On 1 d, the viability of three groups was similar, but the difference of viability also became more significant with time. Similar to the L929 cells, 4T1 cells on 6G3S1d also showed the highest viability in three groups, which was consistent with fluorescence staining results. Compared with the CCK-8 results of L929, 4T1 showed higher viability as culture time increased, because 4T1 is a highly aggressive tumor cell and therefore shows higher viability than normal cells. In addition, compared with L929, the difference of 4T1 activity in each group was still very significant on 7 d, indicating that the scaffold was not full of cells at this time, which was also consistent with the fluorescence staining results of the two cell lines. Similar to L929, the proliferation rate of cells on the scaffold was slower than that in the 2D environment, which was similarly reported by Mahmoudzadeh et al.[69]. Cells in the 3D environment were not able to get as much oxygen and nutrients as those in the 2D environment and formed vascular networks before they could proliferate rapidly, leading to a slower proliferation rate than in the 2D group.
In order to explore the morphology of 4T1 on the scaffold, SEM was used to observe 4T1, and the results were shown in Figure 9. It could be found that on 1 d, no large-scale cell cluster was formed on the scaffolds, and only a few cells were scattered at the edges and corners of the scaffolds. The long spindle shape of cells could be clearly observed. On 4 d, the cells proliferated rapidly, and a large number of cells gathered and grew. The cells shrank a little, but the boundary of cell was clear. At this point, tumor spheres have appeared on the scaffolds. The cells cultured for 7 d had better proliferation rate than the cells cultured in the first few days. Besides the corners of the scaffolds, a large number of cells could also be observed on the edge of the main parts of the scaffolds, which were connected together and distributed in sheets. In addition, the size of tumor spheres formed in some areas on the scaffolds ranged from 100 μm to 200 μm. The results revealed the effective adhesion, invasion, proliferation, and tumorigenicity of the 4T1 cells on these hybrid scaffolds, which also could maintain good cell morphology.
Figure 9.
Morphology of 4T1 cells cultured on the Gel/SA/dECM scaffolds for several days. Pseudopodia were observed clearly on 1 d, and cell colonies appeared on the scaffolds after 4 d of culture (4 d and 7 d). The images on the right panel (scale bar: 10 μm) show magnified images of those on the left panel for each day category (scale bar: 100 μm).
3.7. Infiltration of 4T1 on scaffolds
In order to explore the distribution of 4T1 cells in the scaffolds, laser confocal microscope was used to scan the scaffolds along the Z-axis to generate 3D fluorescence 124staining images, as shown in Figure 10. It could be seen that during cell growth, all the cells in the three groups infiltrated the scaffold vertically, and the number of cells also increased gradually. On 4 d and 7d, 4T1 on scaffolds formed larger cell clusters and tumor spheres, or proliferated at different areas of the same depth, but the total fluorescence intensities on the same day of different scaffolds were similar, indicating that cells exhibited both trends of forming into tumor spheres at primary sites and infiltrating other parts of the scaffolds. The characteristic was consistent with the development of metastatic cancer cells in native organisms. Migration of cells to different parts of the scaffolds may be achieved by travelling in liquid medium.
Figure 10.
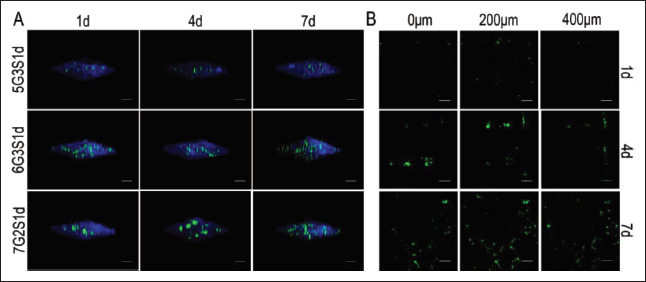
Infiltration and invasion of 4T1 cells within the Gel/SA/dECM scaffolds were investigated under a confocal laser microscope. (A) 3D views of cells grown in scaffolds at 1 d, 4 d, and 7 d (scale bar: 500 μm). (B) Distribution of cells on different depth in the 5G3S1d scaffolds (scale bar: 500 μm).
It had been reported that the tumorigenicity and metastasis of 4T1 cells were positively and negatively correlated with their inoculation density, respectively. Gregorio et al.[73] injected different amounts of 4T1 cells into mice, and it turned out that 100% lung metastasis rate could be achieved when the number of cells was 500, while only 45% when the cell density increased to 1×106. Tumorigenicity rates at the same density were 87% and 95%, respectively. In this study, a compromise scheme was adopted as 20,000 cells, in order to keep both the tumorigenicity and metastasis at a high level and simulate the tumor progression in vivo as precisely as possible. Although the cell densities of 4T1 onto superficial layer of the scaffolds were lower than that of L929 due to half concentration of cell inoculation compared to L929, 4T1 cells penetrated and developed availably into the deeper layer of the scaffolds (Figure 10B). 4T1 invaded 400 μm beneath the surface of the scaffold on 4 d, and the fluorescence intensity was higher at the same depth. On 7 d, 4T1 further proliferated horizontally on the plane of the same depth, indicating that the cells could still maintain high viability at the interior of the scaffolds where it was difficult to exchange nutrients and metabolites with the medium. The tumor model had successfully stimulated the infiltration of tumor cells into surrounding tissues over time in vivo. No significant difference in the invasion of cells among the 125three groups of scaffolds was observed, indicating that the change of gelatin and sodium alginate concentration in a small range in this study had no significant effect on the invasion of 4T1. If the concentration difference was more significant, the results may turn out to be different.
3.8. Formation and growth of tumor spheres
In native tumor microenvironment, tumor cells can gradually develop into tumor spheres. In order to determine the progression of 4T1 cells on the scaffolds, laser confocal microscopy was used to observe the change of tumor sphere diameter over the time of culture. As shown in Figure 11A, scattered tumor spheres began to form on the scaffolds on 3 d, but the size was small. The tumor spheres in different areas of the same group were photographed and their diameters were measured by IPP software. As shown in Figure 11C, the average diameters of tumor spheres of the three groups were 78.81 ± 6.06 μm, 88.91 ± 12.61 μm, and 84.87 ± 10.93 μm, respectively, which indicating there were no significant difference between the groups. On 5 d, the diameter of tumor spheres increased and more tumor spheres developed in the same area. The average diameters of tumor spheres on 5 d were 96.99 ± 6.55 μm, 115.18 ± 4.29 μm, and 100.06 ± 14.43 μm, respectively. The diameters of some tumor spheres were still small, similar to that on 3 d. It was speculated that these spheres may be formed by newly proliferating cells during culture. On 7 d, the average diameters of tumor spheres increased to 116.19 ± 4.63 μm, 148.52 ± 8.02 μm, and 122.25 ± 7.63 μm, respectively. There were significant differences between the groups, indicating that the tumor spheres became mature at this time, showing the influence of scaffold materials on tumor cell viability and tumorigenicity.
Figure 11.
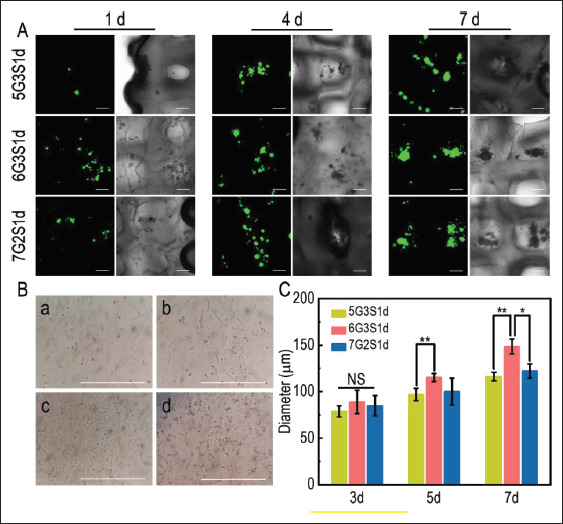
Progression of 4T1 tumor spheres on the Gel/SA/dECM scaffolds. (A) Confocal laser microscopy was used to observe the progression of tumor spheres on the three groups of scaffolds for 1 d, 4 d, and 7 d (scale bar: 200 μm). (B) Growth and morphology of 4T1 in 2D environment (scale bar: 200 μm). (a), (b), (c), and (d) show the images of 4T1 under optical microscope on 1 d, 3 d, 5 d, and 7 d, respectively. (C) Quantitative detection of the change of tumor sphere diameter on the scaffolds on 3 d, 5 d, and 7 d. The diameters were evaluated by IPP software. *P<0.05, **P<0.01 represent significant difference between two groups.
Overall, tumor spheres on the 6G3S1d scaffolds were the largest in diameter and grew at a faster rate than those on the other two groups, consistent with the results of viability. However, from 3 d to 7 d, the growth rate of tumor spheres on the same group of scaffolds remained almost the same, indicating that the proliferation rate of tumor spheres on the scaffolds was relatively stable. The shape and size of spheres formed by 4T1 cells on the scaffolds were similar to the results obtained by Keklikoglou et al.[74]. Lv et al.[56] established a 3D-printed chitosan/gelatin scaffold, which was inoculated with human breast cancer cells MCF-7, and the diameter of spheres was about 150 μm after 7 d, which was close to that of 6G3S1d in the present study. Bright field images showed that tumor spheres were attached at the corner of the scaffolds and 126along the edge of scaffolds, which gradually developed into a larger cell cluster. Figure 11B showed the morphology of 4T1 grown in a 2D environment. Cells exhibited fully extended and long fusiform shape. With the increase of cells, the cells shrank in size, and no tumor spheres formed during the whole process until the cells gradually detached from the plate as a result of insufficient space. Therefore, compared with 2D environment, in vitro tumor model constructed in the present study could simulate tumor microenvironment in vivo better.
4. Conclusion
In this study, a metastatic breast tumor model was fabricated by 3D printing using a bioink consisting of porcine liver- derived dECM with different concentrations of gelatin and sodium alginate. The process of decellularization was appropriate as cells were removed while the microstructure and biochemical components that are important for cell adhesion and tumor progression of ECM were retained. The concentration of bioinks played an important role in stability and biocompatibility of scaffolds, and the addition of gelatin and alginate increased hydrophilicity and mechanical properties, respectively. Distribution of cells and physical properties could be regulated by changing the concentration of bioinks. These tumor models exhibited good compatibilities and stimulated tumor infiltration as well as formation of tumor spheres in native tumor microenvironment. Compared to 2D models, the proliferation rate of 3D models in the present study was more similar to that of tumor tissues in vivo, indicating that our model could serve as a flexible platform for cancer research and anti-tumor drug screening.
Acknowledgments
Not applicable.
Funding
This research was funded by the National Natural Science Foundation of China (31670978), the Fok Ying Tung Education Foundation (132027), the State Key Laboratory of Fine Chemicals (KF1111), and the Fundamental Research Funds for the Central Universities (DUT21YG113/DUT22YG213/DUT22YG116).
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Conceptualization: Kedong Song, Jie Xu, Shuangjia Yang
Data curation: Jie Xu, Shuangjia Yang
Formal analysis: Jie Xu, Shuangjia Yang, Ya Su, Xueyan Hu
Investigation: Shuangjia Yang, Jie Xu, Yue Xi
Methodology: Kedong Song, Shuangjia Yang, Jie Xu, Ya Su, Xueyan Hu
Project administration: Kedong Song, Yi Nie, Yue Kang, Bo Pan
Resources: Kedong Song, Yi Nie, Yue Kang, Bo Pan
Supervision: Kedong Song, Yi Nie
Validation: Kedong Song, Yi Nie, Yue Kang, Bo Pan
Visualization: Jie Xu, Shuangjia Yang, Kedong Song
Writing – original draft: Shuangjia Yang, Jie Xu, Yue Xi
Writing – review & editing: Kedong Song, Yuen Yee Cheng, Yi Nie, Jie Xu
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data
Not applicable.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer . 2015;136(5):E359–86. doi: 10.1002/ijc.29210. https://doi.org/10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: An overview. Int J Cancer . 2021;149(8):778, 789. doi: 10.1002/ijc.33588. https://doi.org/10.1002/ijc.33588. [DOI] [PubMed] [Google Scholar]
- 3.Xin X, Yang H, Zhang F, et al. 3D cell coculture tumor model: A promising approach for future cancer drug discovery. Process Biochem . 2019;78:148, 160. https://doi.org/10.1016/j.procbio.2018.12.028. [Google Scholar]
- 4.Fong E, Harrington D, Farach-Carson MC, et al. Heralding a new paradigm in 3D tumor modeling. Biomaterials . 2016;108:197. doi: 10.1016/j.biomaterials.2016.08.052. https://doi.org/10.1016/j.biomaterials.2016.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang C, Yang Z, Dong DL, et al. 3D culture technologies of cancer stem cells: promising ex vivo tumor models. J Tissue Eng . 2020;11:1–7. doi: 10.1177/2041731420933407. https://doi.org/10.1177/2041731420933407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shapira A, Dvir T. 3D tissue and organ printing-hope and reality. Adv Sci . 2021;8(10):2003751. doi: 10.1002/advs.202003751. https://doi.org/10.1002/advs.202003751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.127Ng W, Chua C, Shen Y-F. Print me an organ! Why we are not there yet. Prog Polym Sci . 2019;97: 101145. https://doi.Org/10.1016/j.progpolymsci.2019.101145. [Google Scholar]
- 8.Ma L, Li Y, Wu Y, et al. 'The construction of in vitro tumor models based on 3D bioprinting. Bio-Des Manuf . 2020;3(3):227, 236. https://doi.org/10.1007/s42242-020-00068-6. [Google Scholar]
- 9.Mao S, Pang Y, Liu T, et al. Bioprinting of in vitro tumor models for personalized cancer treatment: a review. Biofabrication . 2020;12(4):042001. doi: 10.1088/1758-5090/ab97c0. https://doi.org/10.1088/1758-5090/ab97c0. [DOI] [PubMed] [Google Scholar]
- 10.Xie F, Sun L, Pang Y, et al. Three-dimensional bioprinting of primary human hepatocellular carcinoma for personalized medicine. Biomaterials . 2021;265: 120416. doi: 10.1016/j.biomaterials.2020.120416. https://doi.org/10.1016/j.biomaterials.2020.120416. [DOI] [PubMed] [Google Scholar]
- 11.Bahcecioglu G, Basara G, Ellis BW, et al. Breast cancer models: Engineering the tumor microenvironment. Acta Biomater . 2020;106:1, 21. doi: 10.1016/j.actbio.2020.02.006. https://doi.org/10.1016/j.actbio.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang T, Munguia-Lopez J, Flores-Torres S, et al. Extrusion bioprinting of soft materials: An emerging technique for biological model fabrication. Appl Phys Rev . 2019;6(1):011310. https://doi.org/10.1063/L5059393. [Google Scholar]
- 13.Li X, Liu B, Pei B, et al. Inkjet bioprinting of biomaterials. Chem Rev . 2020;120(19):10793–10833. doi: 10.1021/acs.chemrev.0c00008. https://doi.org/10.1021/acs.chemrev.0c00008. [DOI] [PubMed] [Google Scholar]
- 14.Ng W, Lee J, Zhou M, et al. Vat polymerizationbased bioprinting-process, materials, applications and regulatory challenges. Biofabrication . 2020;12(2):022001. doi: 10.1088/1758-5090/ab6034. https://doi.org/10.1088/1758-5090/ab6034. [DOI] [PubMed] [Google Scholar]
- 15.Oztan Y, Nawafleh N, Zhou Y, et al. Recent advances on utilization of bioprinting for tumor modeling. Bioprinting . 2020;18:e00079. doi: 10.1016/j.bprint.2020.e00079. https://doi.org/10.1016/j.bprint.2020.e00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazza G, Telese A, Al-Akkad W, et al. Cirrhotic human liver extracellular matrix 3d scaffolds promote smad- dependent tgf-beta1 epithelial mesenchymal transition. Cells . 2019;9(1):83. doi: 10.3390/cells9010083. https://doi.org/10.3390/cells9010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoshiba T. Decellularized extracellular matrix for cancer research. Materials (Basel) . 2019;12(8):1311. doi: 10.3390/ma12081311. https://doi.org/10.3390/ma12081311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabirian F, Mozafari M. Decellularized ECM-derived bioinks: Prospects for the future. Methods . 2020;171:108, 118. doi: 10.1016/j.ymeth.2019.04.019. https://doi.org/10.1016/j.ymeth.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 19.Su J, Satchell S, Wertheim JA, et al. Poly(ethylene glycol)-crosslinked gelatin hydrogel substrates with conjugated bioactive peptides influence endothelial cell behavior. Biomaterials . 2019;201:99. doi: 10.1016/j.biomaterials.2019.02.001. https://doi.org/10.1016/j.biomaterials.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estrada M, Rebelo S, Davies EJ, et al. Modelling the tumour microenvironment in long-term microencapsulated 3D co-cultures recapitulates phenotypic features of disease progression. Biomaterials . 2016;78:50. doi: 10.1016/j.biomaterials.2015.11.030. https://doi.org/10.1016/j.biomaterials.2015.11.030. [DOI] [PubMed] [Google Scholar]
- 21.Xu K, Wang Z, Copland JA, et al. 3D porous chitosan-chondroitin sulfate scaffolds promote epithelial to mesenchymal transition in prostate cancer cells. Biomaterials . 2020;254:120126. doi: 10.1016/j.biomaterials.2020.120126. https://doi.org/10.1016/j.biomaterials.2020.120126. [DOI] [PubMed] [Google Scholar]
- 22.Choi S, Friedrichs J, Song YH, et al. Intrafibrillar, bone-mimetic collagen mineralization regulates breast cancer cell adhesion and migration. Biomaterials . 2019;198:95. doi: 10.1016/j.biomaterials.2018.05.002. https://doi.org/10.1016/j.biomaterials.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alabi B, Laranger R, Shay J. Decellularized mice colons as models to study the contribution of the extracellular matrix to cell behavior and colon cancer progression. Acta Biomater . 2019;100:213. doi: 10.1016/j.actbio.2019.09.033. https://doi.org/10.1016/j.actbio.2019.09.033. [DOI] [PubMed] [Google Scholar]
- 24.Suo A, Xu W, Wang Y, et al. Dual-degradable and injectable hyaluronic acid hydrogel mimicking extracellular matrix for 3D culture of breast cancer MCF-7 cells. Carbohydr Polym . 2019;211:336, 348. doi: 10.1016/j.carbpol.2019.01.115. https://doi.org/10.1016/j.carbpol.2019.01.115. [DOI] [PubMed] [Google Scholar]
- 25.Zhang T, Zhang Q, Chen J, et al. The controllable preparation of porous PLGA microspheres by the oil/ water emulsion method and its application in 3D culture of ovarian cancer cells. Colloids Surf Physicochem Eng Aspects . 2014;452:115, 124. https://doi.org/10.1016/j.colsurfa.2014.03.085. [Google Scholar]
- 26.Ariadna G-P, Marc R, Teresa P, et al. Optimization of poli(ε-caprolactone) scaffolds suitable for 3D cancer cell culture. Procedia CIRP . 2016;49:61. https://doi.org/10.1016/j.procir.2015.07.031. [Google Scholar]
- 27.Wang C, Li J, Sinha S, et al. Mimicking brain tumorvasculature microanatomical architecture via co-culture of brain tumor and endothelial cells in 3D hydrogels. Biomaterials . 2019;202:35. doi: 10.1016/j.biomaterials.2019.02.024. https://doi.org/10.1016/j.biomaterials.2019.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira L, Gaspar V, Mano J. Decellularized extracellular matrix for bioengineering physiomimetic 3D in vitro tumor models. Trends Biotechnol . 2020;38(12):13971414. doi: 10.1016/j.tibtech.2020.04.006. https://doi.org/10.1016/j.tibtech.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 29.128Zhao L, Huang L, Yu S, et al. Decellularized tongue tissue as an in vitro model for studying tongue cancer and tongue regeneration. Acta Biomater . 2017;58:122, 135. doi: 10.1016/j.actbio.2017.05.062. https://doi.org/10.1016/j.actbio.2017.05.062. [DOI] [PubMed] [Google Scholar]
- 30.Narkhede A, Shevde L, Rao S. Biomimetic strategies to recapitulate organ specific microenvironments for studying breast cancer metastasis. Int J Cancer . 2017;141(6):1091, 1109. doi: 10.1002/ijc.30748. https://doi.org/10.1002/ijc.30748. [DOI] [PubMed] [Google Scholar]
- 31.Lang R, Stern M, Smith L, et al. Three-dimensional culture of hepatocytes on porcine liver tissue-derived extracellular matrix. Biomaterials . 2011;32(29):7042, 7052. doi: 10.1016/j.biomaterials.2011.06.005. https://doi.org/10.1016/j.biomaterials.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Zhao F, Cheng J, Sun M, et al. Digestion degree is a key factor to regulate the printability of pure tendon decellularized extracellular matrix bio-ink in extrusionbased 3D cell printing. Biofabrication . 2020;12(4):045011. doi: 10.1088/1758-5090/aba411. https://doi.org/10.1088/1758-5090/aba411. [DOI] [PubMed] [Google Scholar]
- 33.Pati F, Jang J, Ha DH, et al. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat Commun . 2014;5:3935. doi: 10.1038/ncomms4935. https://doi.org/10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Fang H, Zheng S, et al. A biological functional hybrid scaffold based on decellularized extracellular matrix/ gelatin/chitosan with high biocompatibility and antibacterial activity for skin tissue engineering. Int J Biol Macromol . 2021;187:840, 849. doi: 10.1016/j.ijbiomac.2021.07.162. https://doi.org/10.1016/j.ijbiomac.2021.07.162. [DOI] [PubMed] [Google Scholar]
- 35.Kim B, Kwon Y, Kong JS, et al. 3D cell printing ofin vitro stabilized skin model and in vivo pre-vascularized skin patch using tissue-specific extracellular matrix bioink: A step towards advanced skin tissue engineering. Biomaterials . 2018;168:38. doi: 10.1016/j.biomaterials.2018.03.040. https://doi.org/10.1016/j.biomaterials.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 36.Kim J, Shim I, Hwang DG, et al. 3D cell printing of islet-laden pancreatic tissue-derived extracellular matrix bioink constructs for enhancing pancreatic functions. J Mater Chem B . 2019;7(10):1773, 1781. doi: 10.1039/c8tb02787k. https://doi.org/10.1039/c8tb02787k. [DOI] [PubMed] [Google Scholar]
- 37.Xu J, Fang H, Su Y, et al. A 3D bioprinted decellularized extracellular matrix/gelatin/quaternized chitosan scaffold assembling with poly(ionic liquid)s for skin tissue engineering. Int JBiol Macromol . 2022;220:1253, 1266. doi: 10.1016/j.ijbiomac.2022.08.149. https://doi.org/10.1016/j.ijbiomac.2022.08.149. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Yuan B, Zhang Y, et al. Biomimetic lignin/ poly(ionic liquids) composite hydrogel dressing with excellent mechanical strength, self-healing properties, and reusability. Chem Eng J . 2020;400:125984. https://doi.org/10.1016/j.cej.2020.125984. [Google Scholar]
- 39.Du J, Hu X, Su Y, et al. Gelatin/sodium alginate hydrogel-coated decellularized porcine coronary artery to construct bilayer tissue engineered blood vessels. Int J Biol Macromol . 2022;209:2070. doi: 10.1016/j.ijbiomac.2022.04.188. https://doi.org/10.1016/j.ijbiomac.2022.04.188. [DOI] [PubMed] [Google Scholar]
- 40.Coronado R, Somaraki-Cormier M, Natesan S, et al. Decellularization and solubilization of porcine liver for use as a substrate for porcine hepatocyte culture. Cell Transplant . 2018;26(12):1840, 1854. doi: 10.1177/0963689717742157. https://doi.org/10.1177/0963689717742157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abaci A, Guvendiren M. Designing decellularized extracellular matrix-based bioinks for 3D bioprinting. Adv Healthc Mater . 2020;9(24):e2000734. doi: 10.1002/adhm.202000734. https://doi.org/10.1002/adhm.202000734. [DOI] [PubMed] [Google Scholar]
- 42.Sellaro T, Ranade A, Faulk DM, et al. Maintenance of human hepatocyte function in vitro by liver-derived extracellular matrix gels. Tissue Eng Part A . 2010;16(3):10751082. doi: 10.1089/ten.tea.2008.0587. https://doi.org/10.1089/ten.tea.2008.0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saleh T, Ahmed E, Yu L, et al. Silver nanoparticles improve structural stability and biocompatibility of decellularized porcine liver. Artif Cells Nanomed Biotechnol . 2018;46(sup2):273–284. doi: 10.1080/21691401.2018.1457037. https://doi.org/10.1080/21691401.2018.1457037. [DOI] [PubMed] [Google Scholar]
- 44.Wu Q, Bao J, Zhou YJ, et al. Optimizing perfusion- decellularization methods of porcine livers for clinical-scale whole-organ bioengineering. Biomed Res Int . 2015;2015:785474. doi: 10.1155/2015/785474. https://doi.org/10.1155/2015/785474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poornejad N, Nielsen J, Morris RJ, et al. Comparison of four decontamination treatments on porcine renal decellularized extracellular matrix structure, composition, and support of human renal cortical tubular epithelium cells. JBiomater Appl . 2016;30(8):1154, 1167. doi: 10.1177/0885328215615760. https://doi.org/10.1177/0885328215615760. [DOI] [PubMed] [Google Scholar]
- 46.Struecker B, Hillebrandt K, Voitl R, et al. Porcine liver decellularization under oscillating pressure conditions: A technical refinement to improve the homogeneity of the decellularization process. Tissue Eng Part C Methods . 2015;21(3):303, 313. doi: 10.1089/ten.TEC.2014.0321. https://doi.org/10.1089/ten.TEC.2014.0321. [DOI] [PubMed] [Google Scholar]
- 47.Sun D, Liu Y, Wang H, et al. Novel decellularized liver matrix-alginate hybrid gel beads for the 3D culture of hepatocellular carcinoma cells. Int J Biol Macromol . 2018;109:1154, 1163. doi: 10.1016/j.ijbiomac.2017.11.103. https://doi.org/10.1016/j.ijbiomac.2017.11.103. [DOI] [PubMed] [Google Scholar]
- 48.Hu X, Li W, Li L, et al. A biomimetic cartilage gradient hybrid scaffold for functional tissue engineering of cartilage. Tissue Cell . 2019;58:84. doi: 10.1016/j.tice.2019.05.001. https://doi.org/10.1016/j.tice.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 49.129Li W, Hu X, Yang S, et al. A novel tissue-engineered 3D tumor model for anti-cancer drug discovery. Biofabrication . 2018;11(1):015004. doi: 10.1088/1758-5090/aae270. https://doi.org/10.1088/1758-5090/aae270. [DOI] [PubMed] [Google Scholar]
- 50.Mao S, He J, Zhao Y, et al. Bioprinting of patient- derived in vitro intrahepatic cholangiocarcinoma tumor model: Establishment, evaluation and anti-cancer drug testing. Biofabrication . 2020;12(4):045014. doi: 10.1088/1758-5090/aba0c3. https://doi.org/10.1088/1758-5090/aba0c3. [DOI] [PubMed] [Google Scholar]
- 51.Song K, Li L, Yan X, et al. Characterization of human adipose tissue-derived stem cells in vitro culture and in vivo differentiation in a temperature-sensitive chitosan/beta- glycerophosphate/collagen hybrid hydrogel. Mater Sci Eng C Mater Biol Appl . 2017;70(Pt 1):231–240. doi: 10.1016/j.msec.2016.08.085. https://doi.org/10.1016/j.msec.2016.08.085. [DOI] [PubMed] [Google Scholar]
- 52.Ijima H, Nakamura S, Bual R, et al. Physical properties of the extracellular matrix of decellularized porcine liver. Gels . 2018;4(2):39. doi: 10.3390/gels4020039. https://doi.org/10.3390/gels4020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polley C, Mau R, Lieberwirth C, et al. Bioprinting of three dimensional tumor models: a preliminary study using a low cost 3D printer. Curr Dir Biomed Eng . 2017;3(2):135, 138. https://doi.org/10.1515/cdbme-2017-0028. [Google Scholar]
- 54.Lee H, Han W, Kim H, et al. Development of liver decellularized extracellular matrix bioink for threedimensional cell printing-based liver tissue engineering. Biomacromolecules . 2017;18(4):1229, 1237. doi: 10.1021/acs.biomac.6b01908. https://doi.org/10.1021/acs.biomac.6b01908. [DOI] [PubMed] [Google Scholar]
- 55.Chaji S, Al-Saleh J, Gomillion C. Bioprinted threedimensional cell-laden hydrogels to evaluate adipocytebreast cancer cell interactions. Gels . 2020;6(1):10. doi: 10.3390/gels6010010. https://doi.org/10.3390/gels6010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lv K, Zhu J, Zheng S, et al. Evaluation of inhibitory effects of geniposide on a tumor model of human breast cancer based on 3D printed Cs/Gel hybrid scaffold. Mater Sci Eng C . 2021;119:111509. doi: 10.1016/j.msec.2020.111509. https://doi.org/10.1016/j.msec.2020.111509. [DOI] [PubMed] [Google Scholar]
- 57.Jeong W, Kim M, Kang H. Effect of detergent type on the performance of liver decellularized extracellular matrix-based bio-inks. J Tissue Eng . 2021;12:1, 14. doi: 10.1177/2041731421997091. https://doi.org/10.1177/2041731421997091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bordoni M, Karabulut E, Kuzmenko V, et al. 3D printed conductive nanocellulose scaffolds for the differentiation of human neuroblastoma cells. Cells . 2020;9(3):682. doi: 10.3390/cells9030682. https://doi.org/10.3390/cells9030682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao C, Li Y, Peng G, et al. Decellularized liver matrix-modified chitosan fibrous scaffold as a substrate for C3A hepatocyte culture. J Biomater Sci, Polym Ed . 2020;31(8):1041, 1056. doi: 10.1080/09205063.2020.1738690. https://doi.org/10.1080/09205063.2020.1738690. [DOI] [PubMed] [Google Scholar]
- 60.Lu S, Cuzzucoli F, Jiang J, et al. Development of a biomimetic liver tumor-on-a-chip model based on decellularized liver matrix for toxicity testing. Lab Chip . 2018;18(22):3379–3392. doi: 10.1039/c8lc00852c. https://doi.org/10.1039/c8lc00852c. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Zhu Y, Ma HC, et al. Developing multicellular tumor spheroid model (MCTS) in the chitosan/ collagen/alginate (CCA) fibrous scaffold for anticancer drug screening. Mater Sci Eng C Mater Biol Appl . 2016;62:215. doi: 10.1016/j.msec.2016.01.045. https://doi.org/10.1016/j.msec.2016.01.045. [DOI] [PubMed] [Google Scholar]
- 62.Sachlos E, Reis N, Ainsley C, et al. Novel collagen scaffolds with predefined internal morphology made by solid freeform fabrication. Biomaterials . 2003;24(8):1487, 1497. doi: 10.1016/s0142-9612(02)00528-8. https://doi.org/10.1016/S0142-9612(02)00528-8. [DOI] [PubMed] [Google Scholar]
- 63.Xu W, Qian J, Zhang Y, et al. A double-network poly(Nvarepsilon-acryloyl L-lysine)/hyaluronic acid hydrogel as a mimic of the breast tumor microenvironment. Acta Biomater . 2016;33:131, 141. doi: 10.1016/j.actbio.2016.01.027. https://doi.org/10.1016/j.actbio.2016.01.027. [DOI] [PubMed] [Google Scholar]
- 64.Li X, Deng Q, Zhuang T, et al. 3D bioprinted breast tumor model for structure-activity relationship study. Bio- Des Manuf . 2020;3(4):361, 372. https://doi.org/10.1007/s42242-020-00085-5. [Google Scholar]
- 65.Peela N, Sam F, Christenson W, et al. A three dimensional micropatterned tumor model for breast cancer cell migration studies. Biomaterials . 2016;81:72, 83. doi: 10.1016/j.biomaterials.2015.11.039. https://doi.org/10.1016/j.biomaterials.2015.11.039. [DOI] [PubMed] [Google Scholar]
- 66.Antunes J, Gaspar V, Ferreira L, et al. In-air production of 3D co-culture tumor spheroid hydrogels for expedited drug screening. Acta Biomater . 2019;94:392. doi: 10.1016/j.actbio.2019.06.012. https://doi.org/10.1016/j.actbio.2019.06.012. [DOI] [PubMed] [Google Scholar]
- 67.Li L, Qin S, Peng J, et al. Engineering gelatin-based alginate/carbon nanotubes blend bioink for direct 3D printing of vessel constructs. Int J Biol Macromol . 2020;145:262. doi: 10.1016/j.ijbiomac.2019.12.174. https://doi.org/10.1016/j.ijbiomac.2019.12.174. [DOI] [PubMed] [Google Scholar]
- 68.Xu K, Ganapathy K, Andl T, et al. 3D porous chitosanalginate scaffold stiffness promotes differential responses in prostate cancer cell lines. Biomaterials . 2019;217:119311. doi: 10.1016/j.biomaterials.2019.119311. https://doi.org/10.1016/j.biomaterials.2019.119311. [DOI] [PubMed] [Google Scholar]
- 69.Mahmoudzadeh A, Mohammadpour H. Tumor cell culture on collagen-chitosan scaffolds as three-dimensional tumor model: A suitable model for tumor studies. J Food Drug Anal . 2016;24(3):620, 626. doi: 10.1016/j.jfda.2016.02.008. https://doi.org/10.1016/j.jfda.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.130Ananthanarayanan B, Kim Y, Kumar S. Elucidating the mechanobiology of malignant brain tumors using a brain matrix-mimetic hyaluronic acid hydrogel platform. Biomaterials . 2011;32(31):7913, 7923. doi: 10.1016/j.biomaterials.2011.07.005. https://doi.org/10.1016/j.biomaterials.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwak B, Choi W, Jeon J-W, et al. In vitro 3D skin model using gelatin methacrylate hydrogel. J Ind Eng Chem . 2018;66:254, 261. https://doi.org/10.1016/j.jiec.2018.05.037. [Google Scholar]
- 72.Pulaski B, S. O-R. Mouse 4T1 breast tumor model. Curr Protoc Immunol . 2000;39(1):1, 16. doi: 10.1002/0471142735.im2002s39. https://doi.org/10.1002/0471142735.im2002s39. [DOI] [PubMed] [Google Scholar]
- 73.Gregorio A, Fonseca N, Moura V, et al. Inoculated cell density as a determinant factor of the growth dynamics and metastatic efficiency of a breast cancer murine model. PLoS One . 2016;11(11):e0165817. doi: 10.1371/journal.pone.0165817. https://doi.org/10.1371/journal.pone.0165817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keklikoglou I, Cianciaruso C, Guc E, et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat Cell Biol . 2019;21(2):190, 202. doi: 10.1038/s41556-018-0256-3. https://doi.org/10.1038/s41556-018-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


