Abstract
Cardiac fibroblasts are one of the major constituents of a healthy heart. Cultured cardiac fibroblasts are a crucial resource for conducting studies on cardiac fibrosis. The existing methods for culturing cardiac fibroblasts involve complicated steps and require special reagents and instruments. The major problems faced with primary cardiac fibroblast culture are the low yield and viability of the cultured cells and contamination with other heart cell types, including cardiomyocytes, endothelial cells, and immune cells. Numerous parameters, including the quality of the reagents used for the culture, conditions maintained during digestion of the cardiac tissue, composition of the digestion mixture used, and age of the pups used for culture determine the yield and purity of the cultured cardiac fibroblasts. The present study describes a detailed and simplified protocol to isolate and culture primary cardiac fibroblasts from neonatal murine pups. We demonstrate the transdifferentiation of fibroblasts into myofibroblasts through transforming growth factor (TGF)-β1 treatment, representing the changes in fibroblasts during cardiac fibrosis. These cells can be used to study the various aspects of cardiac fibrosis, inflammation, fibroblast proliferation, and growth.
Keywords: Cardiac fibroblasts, Fibrosis, TGF-β1, Primary culture, Cardiac fibrosis
Background
Cardiac fibroblasts or fibrocytes are one of several cells constituting a healthy heart. These are non-myocyte cells involved in developing and maintaining the cardiac architecture (Snider et al., 2009). In a physiological condition, they interact with other heart cells and maintain cardiac homeostasis (Kurose, 2021). Cardiac fibroblasts preserve the structural integrity of the heart by producing and depositing collagen I, III, V, and VI and fibronectin as part of the extracellular matrix (ECM) (Snider et al., 2009). Fibroblasts are the major players in modulating ECM by degrading its proteins through the secretion of matrix metalloproteinases (Eghbali and Weber, 1990; Chacar et al., 2017).
In a healthy heart, one of the largest populations of cells is the fibroblasts (Camelliti et al., 2005). During cardiac stress or injury, fibroblasts divide and differentiate into myofibroblasts or activated fibroblasts with an increase in their activity (Rohr, 2011). To compensate for the loss of cardiomyocytes during a pathophysiological condition, activated fibroblasts deposit ECM proteins, such as collagen, to sustain the structural integrity of the heart (Kurose, 2021). However, if deposited in excess, the ECM expands the cardiac interstitium, a characteristic of cardiac fibrosis that leads to heart dysfunction (N. G. Frangogiannis, 2020) and premature death. Since cardiac fibroblasts hold great significance in the functioning of a heart, it is essential to characterize the various roles played by fibroblasts (Rohr, 2011).
To investigate various cellular aspects of cardiac fibroblasts in detail, including changes in cellular morphology, regulatory molecular pathways, gene-expression levels, and changes in the expression level of proteins, cultured neonatal primary cardiac fibroblasts can prove to be a valuable model system. Cultured neonatal cardiac fibroblasts mimic various characteristics of fibroblasts in vitro. These cells have the propensity to phenotypically differentiate into myofibroblasts, thus recapitulating the conditions of a stressed or injured heart (Rohr, 2011). Obtaining pure primary cardiac fibroblasts and procuring homogeneity in the cells is indeed very advantageous. It allows for studies to be carried out on these specific cells without any interference from other heart cell types or other regulatory paracrine signaling pathways that can affect their activity. Cultured cardiac fibroblasts can also be subjected to treatments stimulating these cells to mimic cardiac pathologic conditions, such as cardiac fibrosis, in vitro.
Transforming growth factor β (TGF-β) plays a significant role in fibrogenesis in the fibroblasts (N. Frangogiannis, 2020). There are three isoforms of TGF-β: TGF-β1, TGF-β2, and TGF-β3 (Schiller et al., 2004). All three have been shown to contribute to fibrogenesis. However, TGF-β1 has been reported to be the most important isoform involved in fibrosis (Yu et al., 2003). The signaling pathways involved in driving fibrosis are either Smad3-dependent or Smad3-independent pathways, involving various co-receptors and regulators (N. Frangogiannis, 2020). Primary cardiac fibroblasts can be used to study various signaling pathways involved in cardiac fibrosis, including the TGF- β/SMAD3 signaling pathway. Overall, primary cardiac fibroblasts act as a reliable in vitro model that can be used to induce and mimic fibrosis that takes place in vivo.
Isolation and culture of primary cardiac fibroblasts from neonatal rat and mouse pups
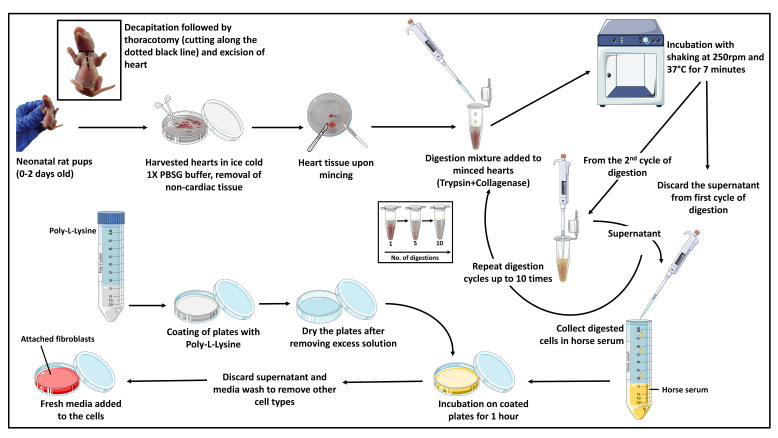
This protocol describes an easy procedure for isolating and culturing cardiac fibroblasts from neonatal rat and mouse pups. The described procedure involves the use of enzymes such as collagenase type II and trypsin for the digestion of the heart tissue (to digest the ECM and dissociate the cells). No additional growth supplements are required for the culture of primary cardiac fibroblasts. Tissue culture plates are coated with poly-L-lysine to enhance the attachment of cardiac fibroblasts. The procedure includes dissecting the rat/mouse pups and taking out the hearts. This is followed by making small chunks of the heart and digesting the heart tissue with the prepared digestion mixture (trypsin + collagenase). Eventually, collection of the digested cells is done in horse serum; the collected cells are plated in tissue culture plates and incubated in an incubator for further subculturing and experimentation. The whole procedure takes approximately 4–5 h (Figure 1).
Figure 1. Diagrammatic representation of the major steps involved in the isolation and culture of primary cardiac fibroblasts from murine pup hearts.
The figure was designed using images adapted from Servier Medical Art by Servier. Original images are licensed under a Creative Commons Attribution 3.0 Unported License.
It is advised for the users to first consult the institutional guidelines on animal usage for experiments and to get the necessary permissions from the institute’s animal ethics committees. Gloves and lab coats should always be worn when handling animals, and there should be no direct contact of bare hands with animals and/or tissues. Carcasses and tissue wastes generated while harvesting the pups should be collected and sealed in double bags and disposed of in an incinerator, following institutional guidelines.
Basic protocol: Isolation and culture of primary cardiac fibroblasts from neonatal murine pups
Materials and Reagents
Sterile cotton
Sterile 10 cm Petri plates (Eppendorf, catalog number: EP0030702115)
Sterile 1.5 mL microcentrifuge tubes (Eppendorf, catalog number: 0030121872)
50 mL conical centrifuge tubes (Eppendorf, catalog number: 0030122178)
Rat pups or mouse pups (0–2 days old)
Distilled water
70% ethanol (HIMEDIA, catalog number: MB228) (v/v) in double distilled water
Horse serum (Gibco, catalog number: 16050122)
Trypsin 0.25% solution 1× without phenol red (HIMEDIA, catalog number: TCL006)
Complete media: Dulbecco’s Modified Eagle Medium (DMEM) (HIMEDIA, catalog number: AL007A), supplemented with 10% fetal bovine serum (FBS) (HIMEDIA, catalog number: RM-10432) and 1× antibiotic-antimycotic mix (Gibco, catalog number: 15640-055) (see Recipes)
Phosphate-buffered saline (PBS), 1× (see Recipes)
Phosphate-buffered saline buffer (1×) with D-glucose (PBSG) (see Recipes)
Digestion mixture (DM) (see Recipes)
Equipment
Sterile surgical scissors (Harvard Apparatus, catalog number: ST2 72-8438)
Forceps (Harvard Apparatus, catalog number: ST2 72-8949)
Blades (Harvard Apparatus, catalog number: ST2 72-8366)
Tissue culture laminar hood (vertical flow) (Thermo Scientific, model: 1338)
Vortex mixer (SPINIX by Tarsons, 3020)
Shaker incubator (Thermo, model: MAXQ 4450)
CO2 incubator for cell culture (maintained at 37 °C and 5% CO2)
Procedure
-
Heart tissue harvesting
-
Clean each mouse/rat pup with lukewarm distilled water followed by 70% ethanol, gently using a sterile absorbent cotton ball.
Notes:
This step is important to remove any filth or contaminants due to environmental exposure and is done to avoid any contamination in the later stages of cell culture.
The number of pups required should be decided based on the planned experiments.
-
Anesthetize pups and then decapitate them with the help of a pair of sterile scissors in a sterile dissection hood.
Note: Pups should be 0–2 days old (P0–P2) as the yield of viable cells decreases with older pups. Culling of the pups should be done one by one. Make sure to wipe off the scissors with 70% ethanol before and after using them.
-
Hold the animals in supine position, allowing access to the head, neck, and thoracic region of the animal. Make a horizontal incision in the neck region and then a vertical incision, cutting through thoracic ribs with the help of a pair of sterile scissors (the cut is shaped like a T).
Note: Make the vertical cut inside the rib cage until it exposes the heart. Press a little with fingers from where the animal is held until the heart protrudes.
-
Cut out the heart with the help of sterile scissors and quickly put the excised heart in ice-cold sterile PBSG.
Note: Putting the heart in ice-cold PBSG is essential because it will decrease the rate of metabolism in the heart, which will lessen stress on the heart.
-
After washing the hearts, transfer them to another dish containing fresh, ice-cold PBSG in a tissue culture laminar hood. With the help of sterile forceps, try to squeeze out any excess blood inside the heart and stop squeezing once the blood stops oozing out.
Note: Get rid of parts of any other organs/tissues attached to the heart during its excision, like lungs, diaphragm, and any blood clots. Also, make sure to remove the top part of the heart that consists of the atria and large blood vessels.
-
-
Digesting the heart tissue
-
Cut a heart into 4–5 pieces (pieces should be around 1 mm3) with the help of a sterile surgical blade. Collect the pieces into a sterile 1.5 mL microcentrifuge tube containing ice-cold DM (approximately 130 μL per rat heart or two mice hearts).
Notes:
Try not to make very small pieces as it may be difficult to avoid taking them while collecting cells with a micropipette after rounds of digestion.
One 1.5 mL microcentrifuge tube can accommodate up to three rat hearts and 5–6 mouse hearts. The volume of the DM should be adjusted according to the number of hearts in each tube.
Crushing or grinding should not be included while cutting the heart pieces as it may lead to cell death.
Start the digestion by placing the microcentrifuge tubes containing DM and the heart chunks into a shaker incubator, which should be maintained at 37 °C and set at 250 rpm for 7 min for each digestion cycle.
-
After the first digestion cycle, vortex the tubes and then spin them for no more than 10 s at approximately 600 × g in a tabletop centrifuge to let the heart chunks settle at the bottom of the tube. Discard the supernatant.
Note: Supernatant from the first digestion cycle is discarded as it contains parts of the ECM, RBCs, and other debris.
-
Add fresh DM to these tubes in the tissue culture laminar hood and vortex briefly before placing them back in the shaker incubator, maintained at the same optimum conditions for 7 min.
Note: Prolonged digestion or vortexing should be avoided, as excessive stress can cause damage and death of the cells.
-
Once the digestion is complete, vortex and spin the tubes for less than 10 s at approximately 600 × g in a tabletop centrifuge. Inside the tissue culture laminar hood, collect the supernatant from the microcentrifuge tubes in a 50 mL conical centrifuge tube containing pre-warmed horse serum (at 37 °C). Keep this tube containing horse serum with cells in an incubator maintained at 37 °C and 5% CO2 saturation until the next digestion cycle is complete. Collect the supernatant from each digestion in the same tube.
Note: To avoid contamination, a completely sterile environment should be maintained for the cells collected from this step onwards. Keep the screw cap of the 50 mL conical centrifuge tube partially open (not completely closed) to allow aeration.
-
Continue the digestion process (add DM, keep in shaker incubator for 7 min, vortex and spin, collect the supernatant in horse serum in the 50 mL conical centrifuge tube, and place in the CO2 incubator) until the heart chunks are completely digested. It takes approximately 9–10 cycles to digest the mouse/rat heart tissue completely.
Notes:
The amount of horse serum in the 50 mL conical centrifuge tube used to collect the digested cells after each digestion cycle should be approximately double the total volume of supernatant collected upon all digestions.
The enzymes present in the DM get neutralized by horse serum upon collection of the digested cells. This is important to prevent over-digestion, or the cells may be damaged.
Sterile conditions should be maintained throughout the procedure. The number of digestions may be increased to get a higher cellular yield, but longer digestions may lead to damage to the cells, thus reducing the overall yield of viable fibroblasts.
-
-
Plating of cells
-
Mix the cells and horse serum to a near homogenous mixture before seeding the cells in poly-L-lysine–coated 10 cm culture dishes (Refer to the Support protocol for coating the cell culture dishes before seeding the fibroblasts). Incubate these dishes for 1 h in a CO2 incubator to allow the attachment of the fibroblasts.
Notes:
The number of dishes used will be based on the number of rat/mouse pups used according to the experiment. One 10 cm culture plate can accommodate fibroblasts from around 4–5 and 9–10 hearts from rat and mouse pups, respectively. The expected yield is approximately 2 × 105 cells per rat heart and approximately 1 × 105 cells per mouse heart.
Fibroblasts attach to the plate because of the differential attachment properties of cardiac fibroblasts and cardiomyocytes. Cardiac fibroblasts get attached to the dishes before cardiomyocytes (within 1 h) in the presence of poly-L-lysine. Cardiomyocytes adhere poorly to the coated surface of the dishes within this short period and, therefore, can easily be washed and collected along with the supernatant. If the incubation time is increased, separating cardiac fibroblasts from cardiomyocytes may be challenging as the cardiomyocytes will also adhere to the coated surface of the 10 cm culture plates. Hence, the differential attachment properties of the two cell types have proved beneficial for the specific isolation of cardiac fibroblasts.
After 1 h of incubation, remove the supernatant containing the unattached cardiomyocytes. The cardiomyocytes can either be discarded along with the serum or collected through centrifugation and can further be resuspended in fresh media to be used for experiments if needed.
-
Wash the plates containing attached fibroblasts with fresh complete media by adding media to each plate (approximately 2 mL) and gently swirling it around before collecting it back. The collected media can be added to the collected serum supernatant from the last step before centrifugation.
Note: Media wash is given to wash any remaining cardiomyocytes still there on the surface of the plate and obtain a pure culture of fibroblast cells.
Add 10 mL of fresh complete media to each plate and keep in a CO2 incubator maintained at 37 °C and 5% CO2 saturation level.
-
-
Propagation and seeding of fibroblasts
-
Discard the media from the culture dishes in which the cells were seeded.
Note: After removing used media using a serrated pipette and a pipette aid, keep the dishes in a slanting position to drain and collect the remaining media.
-
Wash the cells (attached to the surface of the plate) with 1× PBS (4 mL per dish). Gently swirl the PBS and discard it in the same way as the used media in the previous step.
Note: Proper washing of cells and removal of the used media is important because serum present in complete media may hamper the activity of trypsin. The serum contains protease inhibitors that inhibit trypsin activity.
-
Add 1 mL of 0.25% trypsin-EDTA (1×) to each 10 cm culture dish containing primary cardiac fibroblasts and swirl the dish gently to cover all cells with the solution.
Note: Keep the dishes with trypsin-EDTA in a CO2 incubator to activate trypsin’s activity for one to two minutes while constantly checking for the detachment of the cells (slight tapping can help the cells detach from the base).
-
Add 4 mL of complete media to the trypsinized cells in the 10 cm culture dishes making the total volume as 5 mL (1 mL trypsin-EDTA + 4 mL complete media) in each dish.
Note: Media is added as it contains serum that helps to prevent over-trypsinization of cells. If trypsin activity is not inhibited, prolonged trypsinization may lead to cell damage.
-
Collect the cells + media + trypsin in a 15 mL conical centrifuge tube and spin at 100 × g for 5 min at room temperature in a centrifuge.
Note: The media + trypsin solution in the plates can be used to flush the cells from the surface of the plates by pipetting them up and down several times in the culture dish. This step will help detach the maximum number of cells from the surface of the culture dish.
-
After centrifugation, discard the supernatant and resuspend the cell pellet with fresh complete media.
Note: Make sure to resuspend the cells to a near-homogenous mixture so that the seeding can be done appropriately for experiments.
-
The resuspended cells can then be seeded in suitably labeled tissue culture dishes or other plates as per the requirements of the designed experiments (Refer ‘Support protocol’ for coating the cell culture dishes before seeding the fibroblasts).
Note: Primary neonatal cardiac fibroblasts can be trypsinized and sub-cultured up to two times.
-
Support protocol: Coating the tissue culture plates with poly-L-lysine for effective attachment of cardiac fibroblasts to the surface of the dishes
Materials and Reagents
10 cm tissue culture plates (Eppendorf, catalog number: EP0030702115)
0.22 μm syringe filter (WhatmanTM Uniflo 25 syringe filter, Merck, catalog number: WHA9913-2502)
0.45 µm PVDF membrane (GE Healthcare, catalog number: 10600023)
Poly-L-lysine (Sigma-Aldrich, catalog number: P8920)
Trypsin 0.25% solution 1× without phenol red (HIMEDIA, catalog number: TCL006)
Collagenase type II (Gibco Collagenase, Type II, powder, catalog number: 17101015)
DMEM, high glucose (HIMEDIA, catalog number: AL007A)
Fetal bovine serum (FBS) (Gibco, catalog number: RM-10432)
100× antibiotic-antimycotic solution (Gibco, catalog number: 15640-055)
Formaldehyde solution (Sigma-Aldrich, catalog number: F8775)
Triton X-100 (Sigma-Aldrich, catalog number: 9002-93-1)
Bovine Serum Albumin (HIMEDIA, catalog number: MB-083)
Tween-20 (Sigma-Aldrich, catalog number: P1379)
Fluoromount (Sigma-Aldrich, catalog number: F4680)
2× Laemmli Sample Buffer (Bio-Rad, catalog number: 161-0737)
β-Mercaptoethanol (Sigma-Aldrich, catalog number: M3148)
Skimmed Milk Powder (HIMEDIA, catalog number: GRM-1254)
Clarity Western ECL Substrate (Bio-Rad, catalog number: 1705061)
RNAiso Plus Reagent (TAKARA, catalog number: 9108)
PrimeScript 1st Strand cDNA Synthesis kit (TAKARA, catalog number: RR014B)
SYBR-Green PCR master mix (Bio-Rad, catalog number: 1725124)
TGF-β1 (Sigma, catalog number: T7039)
KCl
NaCl
KH2PO4
Na2HPO4
Double-distilled water (DDW)
HCl
Glucose (Qualigens, catalog number: 15405)
Antibodies (Table 1)
Digestion mixture (see Recipes)
DMEM with 10% FBS and antibiotic-antimycotic mix (complete media) (see Recipes)
0.01% (w/v) Poly-L-lysine (see Recipes)
Phosphate-buffered saline (PBS), 1× (see Recipes)
Phosphate-buffered saline-glucose (PBSG), 1× (see Recipes)
Phosphate-buffered saline-Tween-20 (PBST), 1× (see Recipes)
Tris-buffered saline-Tween-20 (TBST), 1× (see Recipes)
Transfer Buffer (see Recipes)
Table 1. List of antibodies used in the study.
| S. No. | Antibody | Company | Catalog no. | Application |
|---|---|---|---|---|
| 1 | Anti-Actin, α-Smooth Muscle antibody, Mouse monoclonal | Sigma | A5228 |
Western Blotting (1:1,000), Immunofluorescence (1:500) |
| 2 | Fibronectin (EP5) mouse monoclonal | Santa-Cruz | sc-8422 |
Western Blotting (1:1,000), Immunofluorescence (1:250) |
| 3 | COL1A1 (C-18) goat polyclonal | Santa-Cruz | sc-8784 |
Western Blotting (1:1,000), Immunofluorescence (1:250) |
| 4 | COL3A1 (B-10) mouse monoclonal | Santa-Cruz | sc-271249 |
Western Blotting (1:1,000), Immunofluorescence (1:250) |
| 5 | Ms mAB to beta Actin | Abcam | ab8226 | Western Blotting (1:2,000) |
Equipment
-20 °C freezer
Confocal Laser Scanning Microscope (LSM 880, Zeiss, USA)
ChemiDoc Imaging System, Bio-Rad (Version 2.2.0.08)
QuantStudio 6 Flex, Life Technologies
Procedure
Poly-L-lysine is an effective agent that promotes adherence of cells and ECM to coated solid surfaces.
Polycationic poly-L-lysine molecules are strongly adsorbed onto the solid surfaces. The anionic sites on the cells recognize the exposed polycationic sites on these molecules, therefore enhancing the attachment of the cells to the surface of the coated tissue culture plates, forming a monolayer of cells (Mazia et al., 1975; Macieira-Coelho and Avrameas, 1972). For the seeding of fibroblasts, coating of tissue culture ware is done by covering the surface briefly with a 0.01% working solution of poly-L-lysine.
Tissue culture plates coating
Prepare sterile 0.01% working solution (in deionized water) from commercially available 0.1% stock solution of poly-L-lysine.
-
Coat 10 cm tissue culture dishes by covering the entire surface with 0.01% poly-L-lysine.
Note: 5 mL of working poly-L-lysine solution is sufficient for covering the surface of a 10 cm culture plate.
-
Incubate the plates for half an hour in a CO2 incubator maintained at 37 °C and a 5% CO2 saturation level.
Notes:
Take the plates from the incubator and remove the excess poly-L-lysine solution.
Collect any excess solution from the plates. Avoid putting any scratch on the coated surface with the pipette tips.
-
Keep the plates open under UV for 30 min at room temperature inside a sterile tissue culture hood.
Note: Avoid over-drying the coated plates, as it might compromise the attachment of cardiac fibroblasts because of coating disruption.
These plates are now ready for seeding the cardiac fibroblasts after completing the digestion steps.
Methodology
Immunofluorescence protocol:
Immunofluorescence experiments were performed with primary neonatal rat cardiac fibroblasts as described previously (Maity et al., 2020). 3.7% formaldehyde was used for the fixation of the cells for 10 min at room temperature, and permeabilization was done using 0.2% Triton X-100 for 5 min at room temperature. Three washes with 1× PBS for 3 min each were given to the cells before each step. Blocking the cells was done using 5% BSA in PBST buffer for 1 h at room temperature. Primary antibodies were prepared in 1% BSA in PBST and incubated with the cells overnight at 4 °C. After washing the cells thrice with 1× PBS for 3 min each, they were incubated with the secondary antibody prepared in 1% BSA in PBST for 1 h at room temperature. Nuclei were stained using Hoechst 33258 (1:2,000) for 10 min. Fluoromount aqueous mounting medium was used to mount the coverslips on clean glass slides, and imaging was done with a confocal microscope. The antibodies used have been listed in Table 1.
Western Blotting:
Western blotting was performed for primary neonatal rat cardiac fibroblasts as previously described (Maity et al., 2020). In short, total protein lysates were prepared using RIPA buffer supplemented with phosphatase inhibitors and protease inhibitors. Samples were prepared by mixing the lysates with 2× Laemmli sample buffer with 5% β-mercaptoethanol. The prepared samples were heated at 95 °C for 5 min and resolved on a 10% SDS-PAGE gel at 80 V. PVDF membrane (0.45 µm) was utilized for transferring the proteins through overnight wet transfer at 20 V, using transfer buffer. Blocking of the membrane after the transfer was done using 5% skimmed milk in TBST for 1 h at room temperature. Upon washing the blocked membrane with TBST, it was incubated with primary antibody (prepared in 5% BSA in TBST) overnight at 4 °C, followed by incubation with secondary antibody (prepared in 1% skimmed milk in TBST) for 1 h at room temperature. Antibodies bound to the PVDF membrane were visualized using a western ECL reagent in a chemiluminescence imager. The antibodies used have been listed in Table 1.
Real-time PCR:
qRT-PCR analysis was performed as described previously (Maity et al., 2020). RNAiso Plus reagent was used for the isolation of total RNA from the cells following the manufacturer’s protocol. Purity and integrity of the isolated RNA were confirmed, and cDNA synthesis was done using 1 μg of the RNA with the PrimeScript 1st Strand cDNA Synthesis kit. RT-PCR was performed with the SYBR-Green PCR master mix in a real-time PCR machine. The primers used in the study are listed in Table 2.
Table 2. List of primers used in the study.
| S. No. | Name | Sequence (5′-3′) |
|---|---|---|
| 1 | α-SMA Forward | GGAGATGGCGTGACTCACAA |
| 2 | α-SMA Reverse | CGCTCAGCAGTAGTCACGAA |
| 3 | FN1 Forward | CCACCATCACTGGTCTGGAG |
| 4 | FN1 Reverse | GGGTGTGGAAGGGTAACCAG |
| 5 | Col1a Forward | CAATGGCACGGCTGTGTGCG |
| 6 | Col1a Reverse | CACTCGCCCTCCCGTCTTTGG |
| 7 | Col3a Forward | TGGCACAGCAGTCCAACGTA |
| 8 | Col3a Reverse | AAGGACAGATCCTGAGTCACAGA |
| 9 | Actin Forward | CACTGTCGAGTCGCGTCC |
| 10 | Actin Reverse | TCATCCATGGCGAACTGGTG |
TGF-β1 treatment:
Used media was removed from the cells and they were washed with PBS; 10 ng/mL TGF-β1 in serum-free DMEM was added to the cells, followed by incubation for 24 h at 37 °C and 5% CO2 saturation. Upon completion of the treatment, cells were either harvested for lysate preparation (western blot analysis) or RNA isolation (qRT-PCR analysis) according to the planned experiments. The treated cells could also be fixed using 4% formaldehyde for immunofluorescence experiments.
Commentary:
All major organs have resident fibroblasts, the most widely present active cells of the connective tissue (Kendall and Feghali-Bostwick, 2014). They play a crucial role in the proper functioning of the organs. Fibroblasts are essential for regulating gene expression affecting the site-specific differentiation of cells in different organs (Rinn et al., 2006). Interactions of fibroblasts with different cell types have been proven to play a significant role in the patterning of various organs, including the lung, skin, and gastrointestinal tract. Fibroblasts express specific genes, which help provide identity to cells in different organs, and, therefore, can be used to study organ-specific functions and the process of organogenesis (Rinn et al., 2006). Fibrosis, the excess production of ECM proteins, including collagen, to form scar tissue, is the underlying process for all fibrotic-related diseases. The production of scar tissue is important for sealing open wounds. However, if unregulated, it may act as the basis of numerous diseases such as idiopathic pulmonary fibrosis, systemic sclerosis, kidney fibrosis, liver cirrhosis, and cardiac fibrosis (Zhang and Zhang, 2020). Other than fibrosis, fibroblasts have also been shown to play a role in inflammation and cancer progression. In a published study, fibroblasts have been found to maintain inflammation, eventually leading to rheumatoid arthritis and supporting the growth of tumor mass (Mizoguchi et al., 2018; Winkler et al., 2020).
In a healthy heart, fibroblasts can regenerate functional tissue. They are known to be involved in wound healing, inflammation, the proliferation of cells, deposition of ECM, and eventually remodeling (desJardins-Park et al., 2018). Cardiac fibroblasts play an important role in maintaining homeostasis and normal heart function, as well as in pathological conditions involving cardiac remodeling, such as hypertension and myocardial infarction. The functions that fibroblasts are involved in include the production and deposition of ECM proteins, the interaction and communication with other cell types including myocytes, and the intercellular signaling with endothelial cells and other fibroblasts. These interactions have been shown to regulate the electrophysiological properties of the cells, secretion of cytokines and growth factors, and angiogenesis. Several studies have been conducted to elucidate the role of fibroblasts in all these processes, except angiogenesis (Souders et al., 2009). Cardiac fibroblasts form a scaffold by secreting ECM, in which the cardiomyocytes are embedded to create the 3D structure of a healthy heart. This leads to dense packaging of cardiomyocytes and cardiac fibroblasts with each other, making it difficult to study each cell type separately. Hence, gaining information about these cells becomes difficult while they are in a three-dimensional arrangement in vitro.
Cell culture in two-dimension has proven to be a very proficient approach for studying conceptual molecular biology. Though the cultured immortalized cell lines can be maintained for many passages, they do not accurately represent the in vitro conditions. Therefore, they are not the best model to study the physiological and pathophysiological aspects of the cell. Consequently, it is of utmost importance to be able to culture cardiac fibroblasts and cardiomyocytes separately in two-dimensional culture in vitro to study their functions individually. Most of what is already known about these cells, including their function and morphology in the heart and their behavior upon stimulation, has been possible by conducting experiments in two-dimensional primary tissue culture. In the present study, cultured primary cardiac fibroblasts proved to be of better biological relevance than immortalized cell lines. Upon stimulation, these cells in culture reflect various molecular changes in vitro (Landry et al., 2021). Therefore, these cells can be used to investigate multiple cellular responses under different conditions as these are the closest mimic of the processes that occur in vitro (Eghbali, 1992). However, upon passaging the cells 2–3 times, the cardiac fibroblasts tend to transdifferentiate into myofibroblasts, which are characterized by an increased α-SMA secretion (Rohr, 2011).
Since fibroblasts are involved in the pathogenesis of various diseases, cultured fibroblasts can act as a suitable model for studying several diseases and pathological conditions. Moreover, cultured fibroblasts can be used to study drug toxicology and even drug screening. Fibroblasts cultured from human skin have been used in a published study to elucidate the cytotoxic effects and skin irritation of surfactants (Lee et al., 2000). In another study, fibroblasts isolated from lungs have been utilized to study the toxicity of antimicrobial drugs (Krempaska et al., 2020).
Although cultured cardiac fibroblasts cannot completely mimic the in vitro phenotype, they prove to be an advantageous model in several aspects for both physiological and pathophysiological studies. For example, they can be selectively manipulated with numerous treatments without interference from cardiomyocytes as they are cultured as a distinct homogenous population (Ravi et al., 2021). These cells can be used for various genetic and/or drug screening purposes, which may be completed in comparatively less time than in vitro studies. Cultured cardiac fibroblasts are also a beneficial tool for studying the factors responsible for their transformation and regulating the production and degradation of the ECM. Moreover, they can be used to elucidate cardiac fibroblasts’ role in the endocrine activity (Grupp and Müller, 1999).
Mouse and rats are used for culturing the primary cardiac fibroblasts due to several reasons. Rat and mice models are easily available without the requirement of high cost and skills. Additionally, these murine models are very well studied, and most of the studies on signaling pathways and metabolics that have been published have been carried out in these models (Zaragoza et al., 2011).
Cultured primary fibroblasts act as a better model system in biological research when compared to immortal cancer cell lines. These cells lack mutations in the tumor repressor genes and the oncogenes. Therefore, they make for a preferred model system for studies on DNA repair, cell cycle regulation, and apoptosis (Alikhani et al., 2005; Schauble et al., 2012; Marthandan et al., 2016). Moreover, most existing protocols use sophisticated and specialized techniques like density gradient centrifugation. Our protocol, however, is a set of simple steps involving commonly available machines and chemicals that can be easily found in all laboratories.
Although cultured primary fibroblasts are an excellent model for studying fibrosis, they have their limitations. The primary fibroblasts in culture may spontaneously transdifferentiate into myofibroblasts due to the contact with solid surfaces of the culture dishes. Additionally, since these cells are grown in isolation from other cell types present around them in a living organism, it is impossible to fully comprehend the interactions and signaling pathways regulated by those interactions between fibroblasts and the various other cell types in different organs.
Results
Using the described protocol, we successfully cultured primary cardiac fibroblasts from neonatal murine pups (P0-P2). Further, the cultured cells were treated with TGF-β1, a major regulator of fibrosis. Overall, our results demonstrate the expected outcome of the protocol as described. Moreover, we show that these cells can be transdifferentiated into activated myofibroblasts upon TGF-β1 treatment.
Visualization and characterization of cultured cardiac fibroblasts
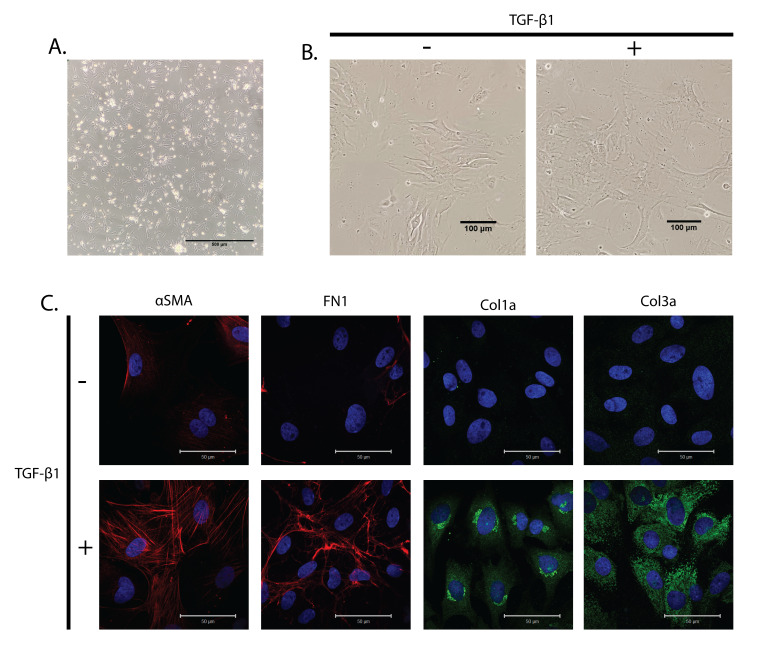
Fibroblasts appear round during the initial stages of pre-plating and the beginning of attachment to the coated surface of the cell culture dishes. With time, the fibroblasts spread and attach firmly to the coated culture dishes. Eventually, they form a monolayer of plump, spindle-shaped cells (Figure 2A). Some cells remain rounded and do not attach even after a day; these cells can be washed off using PBS while passaging the cells or changing the medium. On keeping the fibroblasts in a confluent state, starving the cells of serum, or treating the cells with TGF-β1, the spindle-shaped fibroblasts transdifferentiate into myofibroblasts, which have a characteristic large surface area with various protrusions of the plasma membrane (Figure 2B).
Figure 2. Cultured primary cardiac fibroblasts treated with TGF-β1.
(A) Bright-field image of attached neonatal rat cardiac fibroblasts on poly-L-lysine-coated dishes. Scale bar: 500 μm. (B) Bright-field image of primary cardiac fibroblasts with and without TGF-β1 treatment for 24 h. Scale bar: 100 μm. (C) Primary cardiac fibroblasts were treated with TGF-β1 for 24 h and stained with α-smooth muscle actin (α-SMA) as a marker for myofibroblasts, and fibronectin 1 (FN1), collagen 1a (Col1a), and Collagen 3a (Col3a) as fibrotic markers. Hoechst 33342 was used to stain the nuclei blue. Scale bar: 50 μm.
Treatment of cardiac fibroblasts with TGF-β1
TGF-β1 is one of the major regulators of fibrosis. TGF-β1 treatment is a well-established model for studying the induction of fibrosis in mice and cultured cells. Upon TGF-β1 treatment, the fibroblasts get activated and transdifferentiate into myofibroblasts. This can be characterized by enhanced production of α-smooth muscle actin (α-SMA), a myofibroblast-specific marker. α-SMA is a cytoskeletal protein responsible for activated myofibroblasts' mobility. Additionally, since TGF-β1 treatment results in the induction of fibrosis, it is accompanied by an increase in the production and deposition of the ECM proteins, including fibronectin 1 (FN1), collagen 1a (Col1a), and collagen 3a (Col3a). These act as fibrotic markers, and their levels can indicate whether fibrosis has set in upon the treatment.
To show that the isolated cells are indeed cardiac fibroblasts and can be differentiated into myofibroblasts, we treated the cells with TGF-β1 (10 ng/mL) for 24 h to induce fibrosis. The cells with and without TGF-β1 treatment were stained with myofibroblast specific markers (α-SMA) and fibrotic markers (FN1, Col1a, and Col3a). We observed an increase in the protein levels of α-SMA and a difference in its arrangement, confirming the transdifferentiation of fibroblasts into myofibroblasts upon TGF-β1 treatment (Figure 2C). Moreover, we could note an increase in the protein levels of FN1, Col1a, and Col3a, all of which are fibrotic markers (Figure 2C). This implies that fibrosis is enhanced in the cells treated with TGF-β1 compared to the controls.
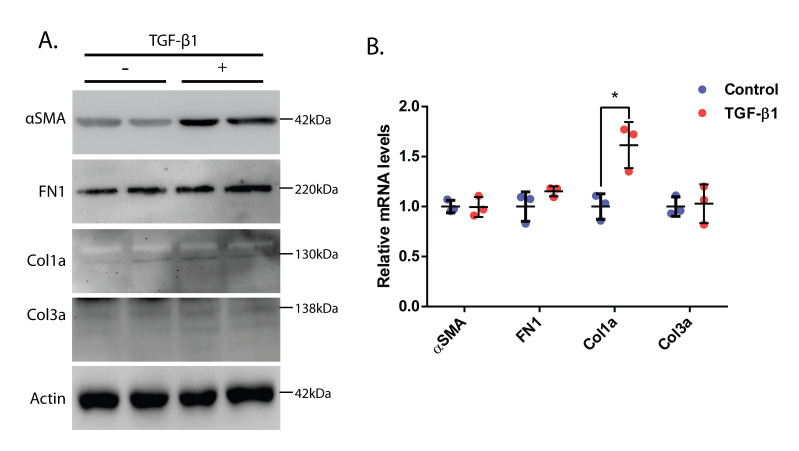
To further solidify our claims, we performed a western blot analysis to check the protein levels of α-SMA, FN1, Col1a, and Col3a. The cells were treated with TGF-β1 (10 ng/mL) for 24 h in serum-free media, and the lysates were prepared and run in an SDS-PAGE gel. We found an increase in the levels of α-SMA, FN1, Col1a, and Col3a, thus confirming that there was an increased population of myofibroblasts than fibroblasts in the TGF-β1-treated samples as compared to control samples (Figure 3A). Further, we performed qRT-PCR analysis to check if there was any change in the mRNA levels of α-SMA, FN1, Col1a, and Col3a. Upon analysis of the qRT-PCR results, we found that the mRNA levels of Col1a were elevated in TGF-β1-treated samples compared to controls (Figure 3B). However, we could not see a significant difference in the mRNA levels of other genes.
Figure 3. Molecular analysis of cardiac fibroblasts treated with TGF-β1.
(A) Western blot analysis of neonatal rat cardiac fibroblasts upon 24 h of TGF-β1 treatment. Α-SMA was used as a myofibroblast marker, and FN1, Col1a, and Col3a were used as fibrotic markers. Actin was used as the loading control. (B) Relative mRNA levels of α-SMA, FN1, Col1a, and Col3a in neonatal rat primary cardiac fibroblasts treated with TGF-β1 for 24 h. Actin has been used for normalization (as an internal control). Student’s t-test used for statistical analysis (*p ≤ 0.05). Data presented as mean ± S.D.
Our results prove that the cells isolated using the protocol are indeed cardiac fibroblasts and can be differentiated into myofibroblasts using TGF-β1 treatment. The cultured cardiac fibroblasts can be used for similar studies to evaluate the toxicity of various drugs and phenotypic and subcellular changes resulting from such treatments.
Time required
The total amount of time required to carry out the primary cardiac fibroblast culture from neonatal rat pups according to the described protocol has been mentioned in Table 3.
Table 3. Time requirement.
| Procedure | Time (h:min) | Notes |
|---|---|---|
| Basic protocol | ||
| Washing and sterilization of pups with water and 70% ethanol, respectively | 0:10 | The indicated time periods are for 5–7 rat pups. Timings can be scaled up and down according to the number of animals being used |
| Euthanasia of pups | 0:15 | |
| Harvesting the hearts through dissection | 0:15 | |
| Mincing the hearts into smaller pieces | 0:10 | |
| Digestion of the cardiac tissue and collection of cells in horse serum | 1:30 | |
| Seeding cells on culture dishes (Pre-plating) | 1:30 | |
| Washing and resubstituting the cells with media (to remove unattached cardiomyocytes) | 0:10 | |
| Total | 4:00 | The total time may vary depending upon the number of animals being used for the experiment and the expertise of the user. |
| Support protocol | ||
| Adding poly-L-lysine coating solution and incubating | 0:30 | |
| Removing the solution and keeping for air-drying | 0:30 | The coating should be done either before or during the digestion of tissues. |
Notes
Important parameters and troubleshooting
The critical points to be noted while culturing primary cardiac fibroblasts using our protocol have been listed. Wherever possible, we have included tips along with the protocol to avoid potential issues for each step. We have also compiled a table listing all the possible problems that may be faced and their potential solutions in Table 4.
Table 4. Troubleshooting for common problems.
| Problem | Possible reasons for problems | Solution |
|---|---|---|
|
Low yield of cardiac fibroblasts Less viability |
Pieces of mouse/rat heart tissues are too big for proper digestion The digestion mixture used is insufficient Optimum conditions such as speed (rpm) or temperature not maintained Improper digestion due to expired or substandard digestion reagents Not enough cycles of digestion Too much digestion Excess mechanical stress Optimum conditions not maintained during harvesting and digestion of heart Improper coating of tissue culture plates |
Taking large chunks would cause incomplete digestion of heart tissue. One heart should be cut into pieces of ~1 mm3 (4–5 pieces for rat pups and 2–3 pieces for mouse pups), for better yield. For better digestion, sufficient DM should be added (130 μL per four mice or one rat heart). The shaker incubator should be set at 250 rpm and 37 °C for digestion. Ensure that the reagents used are fresh and new and stored appropriately at the recommended temperatures. Digestion should be done for an adequate number of cycles (9–10 cycles). Digestion of the heart tissue should not exceed 10 cycles. Digestion time for one cycle should not be longer than 7 min. Excessive mechanical stress should be avoided such as excessive mincing of the heart tissue and prolonged vortexing. The speed of the shaker incubator should not exceed 280 rpm. The excised heart should be placed immediately in ice-cold PBSG buffer upon harvesting. After each cycle of digestion, cells should be collected in horse serum and the 50 mL tube should be kept inside a CO2 incubator maintained at 37 °C and a 5% CO2 saturation level. Proper coating of tissue culture plates with appropriate adhesion reagent should be done. |
| Contamination of cells by bacteria/fungi |
Use of unsterilized dissection tools and/or reagents Contamination in CO2 incubator or tissue culture hood Use of expired or bad antibiotics |
All the steps should be performed under sterile conditions. All tools and reagents should be sterilized including scissors and forceps. Fresh blades and a new sterile 50 mL conical centrifuge should be used. Buffers should be made fresh and autoclaved. DM, horse serum, complete media, and the reagent used for coating the plates should be filter-sterilized. Heat-sterilize the incubator to decontaminate it. Fumigation or UV sterilization of the tissue culture hood should be practiced frequently. Before starting the digestion process, expose the tissue culture hood to UV for a minimum of 30 min. Antibiotics should be added to the media right before usage. Repeated freeze-thaw cycles should be avoided for the media with antibiotics. |
| Contamination by non-fibroblast cells |
Excessive pre-plating Tissues from other organs not removed completely during harvesting of the hearts |
Pre-plating should not exceed 1 h as other cells (cardiomyocytes) may also start to get attached to the coated surface of the plate. Non-cardiac tissues such as pieces of lungs, diaphragm, or any other connective tissue stuck to the heart should be completely removed before starting the digestion process. |
Recipes
-
Digestion mixture (DM)
DM with a composition of 0.4 mg per ml collagenase type II and 0.25% trypsin is prepared by mixing 8 mL trypsin 0.25% solution 1× without phenol red with 4 mg of collagenase type II and making up the volume to 10 ml by adding 1× sterile PBSG buffer. Sterilize this mixture by passing it through a 0.22 μm syringe filter. Prepared DM can be aliquoted into vials and stored in a -20 °C freezer for up to three months.
-
DMEM with 10% FBS and antibiotic-antimycotic mix (complete media)
445 mL of DMEM, high glucose is supplemented with 50 mL of FBS (sterilized through filtration using a 0.22 μm syringe filter) and 5 mL of 100× antibiotic-antimycotic solution to prepare complete media. This media can be used for seeding the primary cardiac fibroblasts after the steps of digestion and for subculturing the cells. The media should be prepared fresh as the antibiotics and antimycotics may degrade upon prolonged storage in the fridge at 4 °C.
-
0.01% (w/v) Poly-L-lysine
0.1% (w/v) poly-L-lysine is diluted to a working concentration of 0.01% with deionized water before use. This diluted working solution can be stored in a refrigerator at 4 °C. The stored solution can be used for up to three months if no turbidity or bacterial growth is present (discard if contaminated).
-
Phosphate-buffered saline (PBS) (1×)
Prepare PBS buffer (1×) by dissolving 2.7 mM KCl, 137 mM NaCl, 1.8 mM KH2PO4, and 10 mM Na2HPO4 in DDW. The pH has to be set at 7.4 with the help of HCl. It should be autoclaved before use for cell culture and stored in the refrigerator at 4 °C (discard if contamination can be seen).
-
Phosphate-buffered saline-glucose (PBSG) (1×)
PBS is prepared as per the recipe and autoclaved. 0.01 M glucose in PBS is prepared by adding 1 mL of sterile 1 M D-glucose (in water) to 100 mL of autoclaved 1× PBS. PBSG cannot be autoclaved as glucose degrades upon autoclaving. The prepared buffer can be stored at 4 °C for up to one month.
-
Phosphate-buffered saline with Tween-20 (PBST) (1×)
PBS is prepared as per the recipe and autoclaved. 0.05% Tween-20 in PBS is prepared by adding 0.05 mL of Tween-20 to 100 mL of autoclaved 1× PBS. The prepared buffer can be stored at 4 °C for up to one month.
-
Tris-buffered saline with Tween-20 (TBST) (1×)
Tris-buffered saline with 0.05% Tween-20 is prepared by dissolving 20 M Tris, 160 mM NaCl and 0.05% Tween-20 in DDW. The pH has to be set at 7.4 with the help of HCl. The prepared buffer can be stored at 4 °C for up to one month.
-
Transfer Buffer (1×)
Transfer buffer is prepared by dissolving 12.5 mM Tris base, 230 mM glycine, 0.125% SDS, and 20% methanol in DDW. It can be stored at 4 °C and should be used up within a week.
Acknowledgments
The central animal facility, confocal facility (Department of Microbiology and Cell Biology), and Indian Institute of Science, Bengaluru, are acknowledged for their services and technical help. This work is supported by funding from the Department of Science and Technology, the Department of Biotechnology, the Indian Council of Medical Research, and the Department of Biotechnology-Indian Institute of Science partnership program for advanced research. N.R.S. is a recipient of the Innovative Young Biotechnologist Award (IYBA), the National Bioscience Award for Career Development, and the Ramalingaswami Re-entry Fellowship from the Department of Biotechnology, Government of India. The protocol has been modified and improved from the one used in a previous publication (Maity et al., 2020).
Author Contributions
Shweta Kumar: data curation, analysis, investigation, methodology, writing original draft, review, and editing, Dimple Nagesh: investigation, methodology, writing original draft, Venketsubbu Ramasubbu: data curation, investigation, analysis, methodology, Arathi Bangalore Prabhashankar: data curation, review, and editing, Ravi Sundaresan: funding acquisition, project administration, resources, supervision, review, and editing.
Competing interests
The authors declare no conflict of interest.
Data Availability Statement
The data supporting the study are available to the corresponding author upon request.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1.Alikhani Z., Alikhani M., Boyd C. M., Nagao K., Trackman P. C. and Graves D. T.(2005). Advanced glycation end products enhance expression of pro-apoptotic genes and stimulate fibroblast apoptosis through cytoplasmic and mitochondrial pathways. J Biol Chem 280(13): 12087-12095. [DOI] [PubMed] [Google Scholar]
- 2.Camelliti P., Borg T. K. and Kohl P.(2005). Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res 65(1): 40-51. [DOI] [PubMed] [Google Scholar]
- 3.Chacar S., Farès N., Bois P. and Faivre J. F.(2017). Basic Signaling in Cardiac Fibroblasts. J Cell Physiol 232(4): 725-730. [DOI] [PubMed] [Google Scholar]
- 4.desJardins-Park H. E., Foster D. S. and Longaker M. T.(2018). Fibroblasts and wound healing: an update. Regenerative Medicine 13(5): 491-495. [DOI] [PubMed] [Google Scholar]
- 5.Eghbali M.(1992). Cardiac fibroblasts: function, regulation of gene expression, and phenotypic modulation. Basic Res Cardiol 2: 183–189.. [DOI] [PubMed] [Google Scholar]
- 6.Eghbali M. and Weber K. T.(1990). Collagen and the myocardium: fibrillar structure, biosynthesis and degradation in relation to hypertrophy and its regression. Mol Cell Biochem 96(1): 1-14. [DOI] [PubMed] [Google Scholar]
- 7.Frangogiannis N.(2020). Transforming growth factor-β in tissue fibrosis. J Exp Med 217(3): e20190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frangogiannis N. G.(2020). Cardiac fibrosis. Cardiovasc Res 117(6): 1450-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grupp C. and Müller G. A.(1999). Renal fibroblast culture. Exp Nephrol 7(5-6): 377-385. [DOI] [PubMed] [Google Scholar]
- 10.Kendall R. T. and Feghali-Bostwick C. A.(2014). Fibroblasts in fibrosis: novel roles and mediators. Front Pharmacol 5: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krempaska K., Barnowski S., Gavini J., Hobi N., Ebener S., Simillion C., Stokes A., Schliep R., Knudsen L., Geiser T. K., et al.(2020). Azithromycin has enhanced effects on lung fibroblasts from idiopathic pulmonary fibrosis(IPF) patients compared to controls.(vol 21, 25, 2019). Respir Res 21(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurose H.(2021). Cardiac Fibrosis and Fibroblasts. Cells 10(7): 1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landry N. M., Rattan S. G. and Dixon I. M. C.(2021). Soft Substrate Culture to Mechanically Control Cardiac Myofibroblast Activation. Methods Mol Biol 2299: 171-179. [DOI] [PubMed] [Google Scholar]
- 14.Lee J. K., Kim D. B., Kim J. I. and Kim P. Y.(2000). In vitro cytotoxicity tests on cultured human skin fibroblasts to predict skin irritation potential of surfactants. Toxicol In Vitro 14(4): 345-349. [DOI] [PubMed] [Google Scholar]
- 15.Macieira-Coelho A. and Avrameas S.(1972). Modulation of cell behavior in vitro by the substratum in fibroblastic and leukemic mouse cell lines. Proc Natl Acad Sci U S A 69(9): 2469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maity S., Muhamed J., Sarikhani M., Kumar S., Ahamed F., Spurthi K. M., Ravi V., Jain A., Khan D., Arathi B. P., et al.(2020). Sirtuin 6 deficiency transcriptionally up-regulates TGF-beta signaling and induces fibrosis in mice. J Biol Chem 295(2): 415-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marthandan S., Menzel U., Priebe S., Groth M., Guthke R., Platzer M., Hemmerich P., Kaether C. and Diekmann S.(2016). Conserved genes and pathways in primary human fibroblast strains undergoing replicative and radiation induced senescence. Biol Res 49(1): 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazia D., Schatten G. and Sale W.(1975). Adhesion of cells to surfaces coated with polylysine. Applications to electron microscopy. J Cell Biol 66(1): 198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizoguchi F., Slowikowski K., Wei K., Marshall J. L., Rao D. A., Chang S. K., Nguyen H. N., Noss E. H., Turner J. D., Earp B. E., et al.(2018). Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun 9(1): 789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravi V., Jain A., Taneja A., Chatterjee K. and Sundaresan N. R.(2021). Isolation and Culture of Neonatal Murine Primary Cardiomyocytes. Curr Protoc 1(7): e196. [DOI] [PubMed] [Google Scholar]
- 21.Rinn J. L., Bondre C., Gladstone H. B., Brown P. O. and Chang H. Y.(2006). Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet 2(7): e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohr S.(2011). Cardiac fibroblasts in cell culture systems: myofibroblasts all along? J Cardiovasc Pharmacol 57(4): 389-399. [DOI] [PubMed] [Google Scholar]
- 23.Schauble S., Klement K., Marthandan S., Munch S., Heiland I., Schuster S., Hemmerich P. and Diekmann S.(2012). Quantitative model of cell cycle arrest and cellular senescence in primary human fibroblasts. PLoS One 7(8): e42150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiller M., Javelaud D. and Mauviel A.(2004). TGF-beta-induced SMAD signaling and gene regulation: consequences for extracellular matrix remodeling and wound healing. J Dermatol Sci 35(2): 83-92. [DOI] [PubMed] [Google Scholar]
- 25.Snider P., Standley K. N., Wang J., Azhar M., Doetschman T. and Conway S. J.(2009). Origin of cardiac fibroblasts and the role of periostin. Circ Res 105(10): 934-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Souders C. A., Bowers S. L. K. and Baudino T. A.(2009). Cardiac Fibroblast: The Renaissance Cell. Circulation Research 105(12): 1164-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkler J., Abisoye-Ogunniyan A., Metcalf K. J. and Werb Z.(2020). Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun 11(1): 5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu L., Border W. A., Huang Y. and Noble N. A.(2003). TGF-β isoforms in renal fibrogenesis. Kidney Int 64(3): 844-856. [DOI] [PubMed] [Google Scholar]
- 29.Zaragoza C., Gomez-Guerrero C., Martin-Ventura J. L., Blanco-Colio L., Lavin B., Mallavia B., Tarin C., Mas S., Ortiz A. and Egido J.(2011). Animal models of cardiovascular diseases. J Biomed Biotechnol 2011: 497841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M. and Zhang S.(2020). T Cells in Fibrosis and Fibrotic Diseases. Front Immunol 11: 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]