Abstract
Salivary gland (SG) development, maturation, and homeostasis require coordinated roles of transcription factors (TFs) that dictate specific cell identities and fate. The ETS family of proteins are important transcriptional drivers of diverse cell lineages, tissue development, and differentiation programs and hence are also likely to play an important role in the SG. Here we have leveraged genomic and epigenomic data of the SG to examine the expression profile of ETS genes and identified 2 closely related paralogs, Elf5 and Ehf, that are highly expressed in distinct epithelial subpopulations. By using a well-defined mouse knockout model of Elf5, we show that Elf5, despite its enriched expression in the acinar cells, is functionally dispensable for maintaining the homeostatic state of the adult SG epithelium. The lack of a discernible phenotype of the Elf5-null SG might be due to possible functional redundancy with Ehf or other ETS factors. To probe this possibility and to examine the specific consequences of Ehf loss in the SG, we used CRISPR-Cas9 to generate mice in which the DNA-binding ETS domain of Ehf is disrupted due to an insertion mutation. We demonstrate that the Ehf mutant (EhfMut) mice exhibit a distinct cellular phenotype with decreased granular convoluted tubules that are accompanied by an increased accumulation of the intercalated Sox9-positive ductal cell population. Interestingly, the ductal phenotype of the EhfMut animals is highly pronounced in males, reaffirming the established sexual dimorphism of the SG that exists in rodents. Our results show that unlike Elf5, Ehf plays a nonredundant role in directing ductal cell differentiation of the SG and highlights the phenotypic subtlety in mutant mice of closely related TFs and the importance of careful consideration of cell type–specific studies.
Keywords: salivary glands, ETS, gene expression, cell fate
Introduction
The salivary gland (SG) is an exocrine organ that consists of 2 major epithelial components: a distal acinus, which produces and secretes saliva into the lumen, and a proximal labyrinth of ducts that serves as a conduit and also actively modifies the composition of the saliva (Amano et al. 2012). The submandibular gland (SMG) and the parotid gland are 2 of the largest major salivary glands that generate the bulk of the saliva, which helps in food digestion, surface lubrication, and immune defense (Maruyama et al. 2019). Additional specialized cells are also crucial components of the SG, as well illustrated by myoepithelial cells that provide contractile forces to extrude the saliva.
The cellular diversity of the SG is perhaps best typified by the various ductal subtypes that are categorized into the intercalated ducts (IDs), granular convoluted tubules (GCTs), striated ducts (SDs), and excretory ducts (EDs). Intercalated ducts, which are the smallest ducts, are directly connected to the acinus and are surrounded by basal cells. Granular convoluted tubules contain growth factor–loaded secretory granules that are well developed in male mice compared to females (Mudd and White 1975; Gresik et al. 1996). Striated ducts modify saliva and lead to the largest excretory duct that is connected to the oral cavity. The intricate process of tissue morphogenesis of the SG requires precise spatiotemporal control of gene expression that is governed by coordinated morphogenic signaling (Lombaert and Hoffman 2010; Musselmann et al. 2011; Patel and Hoffman 2014). Expression of the crucial genes encoding many of such morphogens and their receptors is controlled by a bevy of transcription factors (TFs) that are often highly regulated themselves.
Over the past several years, genetic deletions of TFs have provided important insight into their function in the SG. This includes, for instance, members of the Sox family of TFs such as Sox2, which regulates the acinar cell populations and is essential for their survival (Emmerson et al. 2017, 2018). Sox9 and Sox10, on the other hand, play a more prominent role in controlling progenitor cell fate during embryonic SG development (Chatzeli et al. 2017; Athwal et al. 2019). Similar regulatory functions are also attributed to ΔNp63, which specifies epithelial cell lineages of the embryonic and adult SG (Song et al. 2018). Some TFs have more restricted roles, as evident in the blocked maturation of the ducts observed in Tfcp2l1-null mice or Ascl3 knockout (KO) mice, which develop smaller salivary glands but secrete saliva normally (Yamaguchi et al. 2006; Arany et al. 2011).
We have recently leveraged RNA sequencing (RNA-seq) data sets to identify Elf5 and Ehf as 2 members of the ETS-domain TF family, whose expression is enriched in the SMG compared to other organs and tissues of the mouse (Gluck et al. 2016); however, their potential roles in SG biology, if any, have not been explored. Elf5 plays an important role in cell fate specification in various tissues, prominent examples being the trophectoderm of the blastocyst, embryonic kidney, and the pregnant mammary gland (Choi et al. 2009; Lee and Ormandy 2012; Latos et al. 2015; Grassmeyer et al. 2017). Conversely, studies on Ehf have, until recently, skewed more toward its role in various cancer types rather than tissue developmental programs (Reehorst et al. 2021; Oyelakin et al. 2022). Interestingly, we found both the Elf5 and Ehf genes to be associated with super-enhancers in the SG, suggestive of their likely role in defining cell fate and identity (Whyte et al. 2013; Gluck et al. 2021). Prompted by these findings, here we have carefully examined well-characterized conditional Elf5 knockouts (Elf5cKO) and a newly generated Ehf mutant (EhfMut) mouse model. We confirm that Elf5 and Ehf are the most highly enriched ETS family members in the adult mouse SMG and that they exhibit distinct expression in specific cell types. While Elf5cKO mice surprisingly exhibit no discernible SMG phenotype, loss of Ehf is associated with a distinct ductal phenotype that shows an interesting sex bias. Our studies highlight the complex TF-based gene regulatory network in the SMG and reaffirms the need for careful characterization and interpretation of loss-of-function phenotypes.
Materials and Methods
Animal studies were performed in accordance with the Roswell Park Institutional Animal Care and Use Committee (IACUC) regulations. This study conformed to the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines. Specific details of animal studies and other pertinent materials and methods can be found in the Appendix.
Results
Identification of ETS Family Members Likely to Play a Functional Role in the SMG
Recent epigenomic analyses from our laboratory have revealed that the ETS motif is particularly prevalent in the H3K27Ac-marked regular enhancers and super-enhancers, suggesting that the ETS family of TFs is likely to play an important role in SMG-specific gene expression (GSE145753) (Gluck et al. 2021). To identify specific members of the ETS family that are likely to be functionally relevant, we first examined the expression profile of the mouse ETS genes by probing RNA-seq data sets from adult mouse SMG and a large number of other mouse tissues and organs (GSE81097) (Gluck et al. 2016). Hierarchical cluster analysis revealed not only the expression profile of various ETS family members across different organs but importantly also identified Ehf and Elf5 as the 2 top-ranked factors that are highly expressed in the SMG compared to other tissues (Appendix Fig. 1A). This was not surprising given that the genomic locus surrounding the Ehf and Elf5 genes consists of a super-enhancer (Appendix Fig. 1B), in agreement with the prevailing notion of super-enhancer associated genes having significantly higher expression levels than genes controlled by regular enhancers (Wang et al. 2019).
Single-Cell Analysis of ETS Family Members in the Mouse Submandibular Gland and Investigation of the Expression Patterns of Elf5 and Ehf
To examine the cell type–specific expression of the various ETS factors, we next probed single-cell RNA-seq (scRNA-seq) data sets from embryonic and adult female mouse SMG (GSE150327 and GSE145268, respectively) (Hauser et al. 2020; Min et al. 2020). Dot plot visualization of the data sets revealed very diverse expression patterns of various ETS factors (Fig. 1A and Appendix Fig. 2). We found Elf5 and Ehf to share a somewhat similar expression profile during early development, particularly at embryonic day 12 (E12) and E14, when Ehf and Elf5 are coexpressed in the Krt19+ ducts and end buds. At E16, Elf5 and Ehf expression patterns begin to diverge, although they remain coexpressed in the Krt19+ ducts and end buds. Interestingly, at postnatal day 1 (P1), in addition to the Krt19+ ducts, both of these TFs are expressed in the Smgc+ and Bpifa2+ proacinar cells. However, in adult glands, while Elf5 is expressed in acinar cells and a subset of ductal cells, including intercalated and Ascl3+ cells, Ehf is weakly expressed in acinar cells and rather widely distributed across all the ductal cell types, with prominent expression in the intercalated and Smgc+ ducts (Fig. 1A). Interestingly, both Elf5 and Ehf showed enriched expression in the IDs (Fig. 1A).
Figure 1.
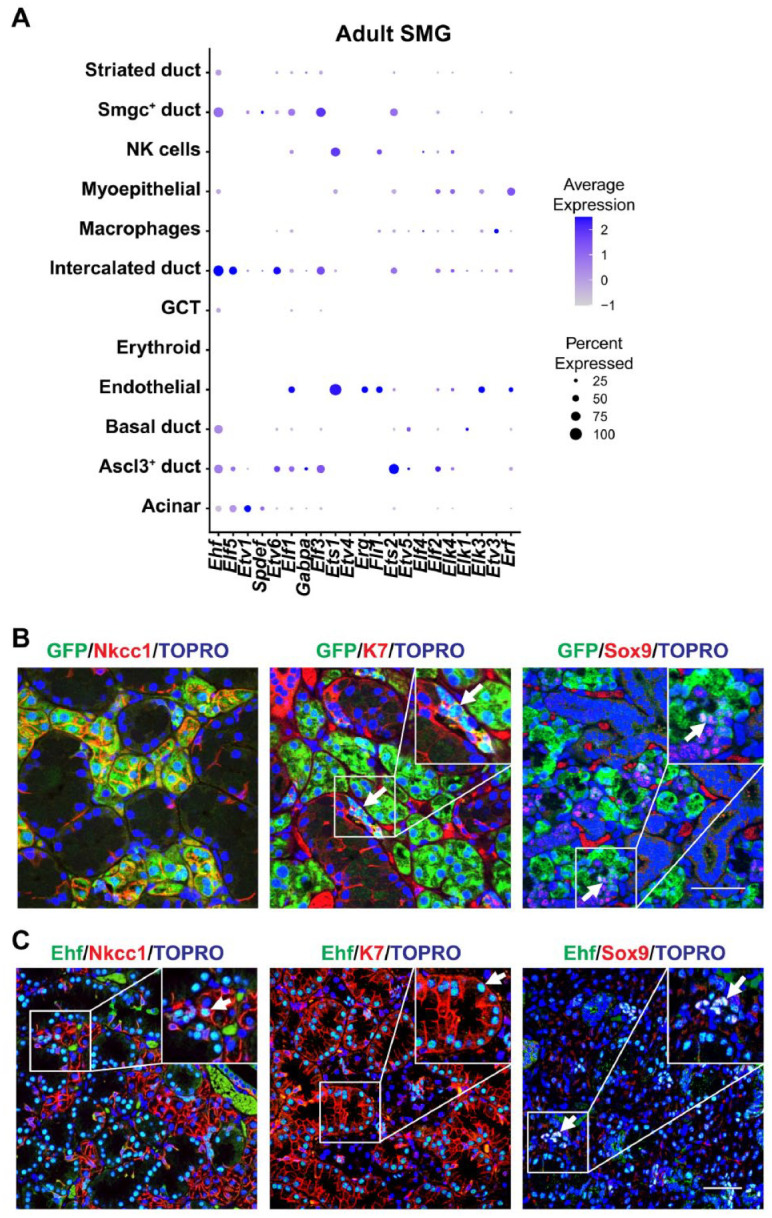
Single-cell RNA sequencing analysis and protein expression profile of Elf5 and Ehf in mouse submandibular glands (SMGs). (A) Dot plot showing the scaled expression (dot color) and percent expression (dot size) of the ETS gene family members in each cell type in adult female SMG (GSE145268) (Min et al. 2020). (B) Immunofluorescence staining of Elf5–green fluorescent protein (GFP) (Pearton et al. 2011) transgenic mouse SMGs shows coexpression of GFP with acinar (Nkcc1), ductal (K7), and intercalated ducts (Sox9). Arrows highlight double-positive cells. (C) Ehf protein expression profile in 10-wk-old adult male mouse SMG showing colocalization of Ehf with the broad ductal (K7) and specific intercalated ductal marker (Sox9). Arrows indicate Ehf colocalization with indicated cell markers. Yellow: green and red colocalization; pink: red and blue (nuclei) colocalization; turquoise: green and blue colocalization; white: green, red, and blue colocalization. Scale bar: 50 µm. GCT, granular convoluted tubule.
We next sought to validate the expression patterns of both Elf5 and Ehf in murine SMGs. For this purpose, we took advantage of Elf5 reporter mouse models expressing either green fluorescent protein (GFP) or nuclear LacZ (nLacZ), each of which serves as a surrogate marker for endogenous Elf5 expression (Choi et al. 2008; Pearton et al. 2011). Indeed, immunofluorescence staining of adult SMGs revealed colocalization of GFP with the acinar cell marker Na+/K+/2Cl− cotransporter (Nkcc1) (Fig. 1B). In addition, we also found a small population of GFP+/K7+ double-positive cells that likely represent the IDs as demonstrated by colocalization of GFP and the ID cell marker, Sox9 (Fig. 1B). Similar results were obtained from X-gal staining of adult Elf5-nLacZ SMGs, which showed β-galactosidase activity in the nucleus of acinar cells (Appendix Fig. 3A) and confirmed by immunofluorescence staining of β-galactosidase and Nkcc1 coexpression (Appendix Fig. 3B). Interestingly, we also observed modest colocalization of β-galactosidase and Sox9 in the IDs, confirming our GFP/Sox9 costaining results (Appendix Fig. 3B). Ehf, unlike Elf5, was modestly expressed in acinar cells and instead showed preferential expression in various ductal cells as evident by costaining with K7 and Sox9, which mark the various ductal cell populations and IDs, respectively (Fig. 1C). Overall, our results show that Elf5 and Ehf have some overlapping but mainly distinct expression profiles with Elf5 expression skewed to the acinar cells, while Ehf is robustly expressed in various ductal cell populations.
Dispensable Role of Elf5 in Adult Mouse Submandibular Gland
Given the distinct expression profiles of Elf5 and Ehf in the SMG, we next sought to determine the role of these 2 members of the ETS family of TFs in SG biology. First, to examine the role of Elf5 in the adult salivary gland, we crossed Elf5fl/fl mice to transgenic animals that constitutively express Cre-recombinase (Cre) under the control of either the K14 (K14-Cre;Elf5fl/fl (K14;Elf5cKO)) or Sox2 (Sox2-Cre;Elf5fl/fl (Sox2;Elf5cKO)) regulatory elements, resulting in ablation of Elf5 as confirmed by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis (Appendix Fig. 4A). Histological analysis of hematoxylin and eosin (H&E)–stained paraffin-embedded SMGs of both male and female control and K14;Elf5cKO mice revealed no significant alterations to either the ducts or acinar structures or any difference in SMG weight (Appendix Fig. 4B, C and Appendix Fig. 5A). One possibility for the surprising lack of any phenotype in Elf5-null SMGs might be in part due to an altered expression pattern of Ehf resulting from the loss of Elf5. To rule this out, we examined Ehf expression by both immunofluorescence and qRT-PCR analysis in control and K14;Elf5cKO SMGs, which revealed no appreciable differences (Appendix Fig. 6).
Next, to ascertain any possible subtle phenotypic changes resulting from the loss of Elf5, we performed immunofluorescence studies and examined both the male and female K14;Elf5cKO SMGs using a battery of well-established epithelial cell markers. Evaluation of the progenitor cell marker p63, which is restricted to the basal and myoepithelial cell populations, revealed no appreciable differences in protein expression pattern between control and K14;Elf5cKO (Appendix Figs. 4C and 5A). Similarly, we failed to observe any differences in expression of α–smooth muscle actin (SMA), a marker of the myoepithelial cells (Appendix Figs. 4C and 5A). In agreement with histological findings, immunofluorescence staining for the ductal marker, keratin 7 (K7), showed no discernible alterations in the K14;Elf5cKO SMGs when compared to control counterparts (Appendix Figs. 4C and 5A). Importantly, no obvious changes were found in Elf5-high acinar cells, as revealed by aquaporin 5 (Aqp5) staining, which showed consistent Aqp5 localization to the apical surface in both control and K14;Elf5cKO SMGs (Appendix Figs. 4C and 5A). This finding was in contrast to the observations in mammary glands, where apical–basal polarity has been shown to be affected in the absence of Elf5 (Choi et al. 2009). To account for the possibility that the normal state of the K14;Elf5cKO SMGs might reflect a partially penetrant phenotype, we next used the Sox2-Cre driver to delete Elf5. Parallel analyses of SMGs of both male and female control and Sox2;Elf5cKO mice led to similar findings, confirming that loss of Elf5 does not result in any discernable adult SMG phenotype, prompting us to thus perform follow-up studies on Ehf (Appendix Figs. 4D and 5B).
Generation of Ehf Mutant Animals and Gross Phenotypic Characterization
We employed the CRISPR-Cas9 system to introduce a mutation in the mouse Ehf locus that disrupted the coding region. Confirmation of the genotype of the animals was achieved by PCR amplification of the targeted Ehf genomic region, followed by Sanger sequencing analysis. As shown in a representative sequencing chromatogram, insertion of a T residue in exon 7 of the Ehf gene resulted in a frameshift, effectively truncating the C-terminal portion of the Ehf protein (Appendix Fig. 7). The sequence-specific recognition of the GGA core motif by ETS family members is mediated primarily by the α3 recognition helix in the ETS domain (Wang et al. 2005; Cooper et al. 2015). The in-frame mutation of Ehf truncates the α3 recognition helix and with the loss of one of the critical residues, Arg-268, and Tyr-269 makes it highly likely that the mutant Ehf protein is unable to bind to ETS-responsive elements. Importantly, adult Ehf mutant (EhfMut) mice displayed some of the phenotypes such as inflammation and swelling of the preputial glands as described recently with an independent EhfMut model (Reehorst et al. 2021).
Alterations of the GCT and Intercalated Ducts in EhfMut SMG
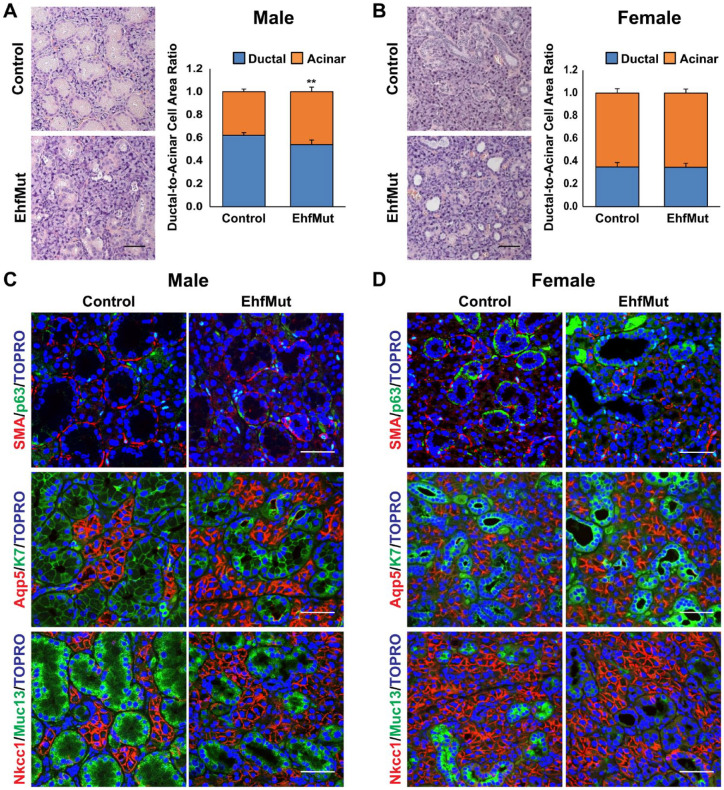
To evaluate the impact of the Ehf mutation, we first examined the SMG in adult EhfMut male and female mice. Loss of Ehf expression in both the SMG and the lacrimal gland, another tissue with high levels of Ehf, was confirmed by immunofluorescence staining with anti-Ehf antibodies that are specific to the C-termini of wild-type Ehf but not EhfMut, based on Western blot results (Appendix Fig. 8). Interestingly, we found that Ehf mutant glands were smaller and weighed less compared to the controls (Appendix Fig. 9). Closer histological examination of SMGs by H&E revealed a dramatic reduction in the GCT size and number of ducts in male EhfMut glands (Fig. 2A), while the EhfMut females presented larger ductal lumens when compared to the control animals (Fig. 2B). Indeed, this phenotype was more pronounced in the male SMGs, which have larger, well-developed GCTs compared to female mice, as illustrated by quantification analyses comparing the ductal and acinar areas (Fig. 2A, B).
Figure 2.
Phenotypic, histological, and immunochemical analysis of mouse submandibular glands (SMGs) with targeted mutation of Ehf. (A) Hematoxylin and eosin (H&E) staining of the male control and EhfMut SMGs (left panel). Compared to control, mutant SMGs show a reduction in granular convoluted tubule (GCT) size. Quantification analysis of the ratio of ductal cell area compared to acini cell area in male control and EhfMut SMG (right panel) (n = 5). (B) H&E staining of control and EhfMut female SMGs shows dilated GCTs due to functional loss of Ehf (left panel). Quantification of the ratio of ductal cell area compared to acini cell area in female control and EhfMut SMG (right panel) (n = 5). (C) Immunofluorescence staining of male control and EhfMut SMGs shows widened lumens in the EhfMut mice compared to control. (D) Immunofluorescence staining of female control and EhfMut SMGs reveals loss of Muc13 expressing GCTs and widened lumens in the EhfMut mice (n = 5). Data are represented as mean ± standard deviation (SD). **P < 0.01. Scale bar: 50 µm.
To better appreciate the cellular alterations associated with the Ehf mutation, we costained male and female glands with basal and myoepithelial markers, p63 and SMA, which revealed a decrease in SMA expression in the male EhfMut glands compared to the control but no significant difference in the female glands (Fig. 2C, D). Interestingly, expression of the ductal marker K7 and the acinar marker Aqp5 showed no appreciable differences in expression pattern between mutant and control glands (Fig. 2C, D). Given the changes in the GCTs in both male and female glands, we next focused our attention on the GCT-specific marker mucin 13 (Muc13) to probe further the observed ductal phenotype. Interestingly, while costaining with Muc13 and Nkcc1 did not reveal dramatic differences in Nkcc1 protein expression between control and EhfMut glands, we observed a reduction of Muc13 expression in male glands with more pronounced changes in female glands (Fig. 2C, D). To confirm these findings, we performed qRT-PCR for a select panel of genes that mark various SMG cell populations (Appendix Fig. 10). Results from such experiments showed that while some genes such as Krt18 were downregulated at modest levels, others such as Muc13 were significantly reduced in the EhfMut SMGs. Interestingly, downregulation of 2 of the myoepithelial markers, Krt14 and Acta2, in the EhfMut glands was more pronounced in male glands (Appendix Fig. 10A).
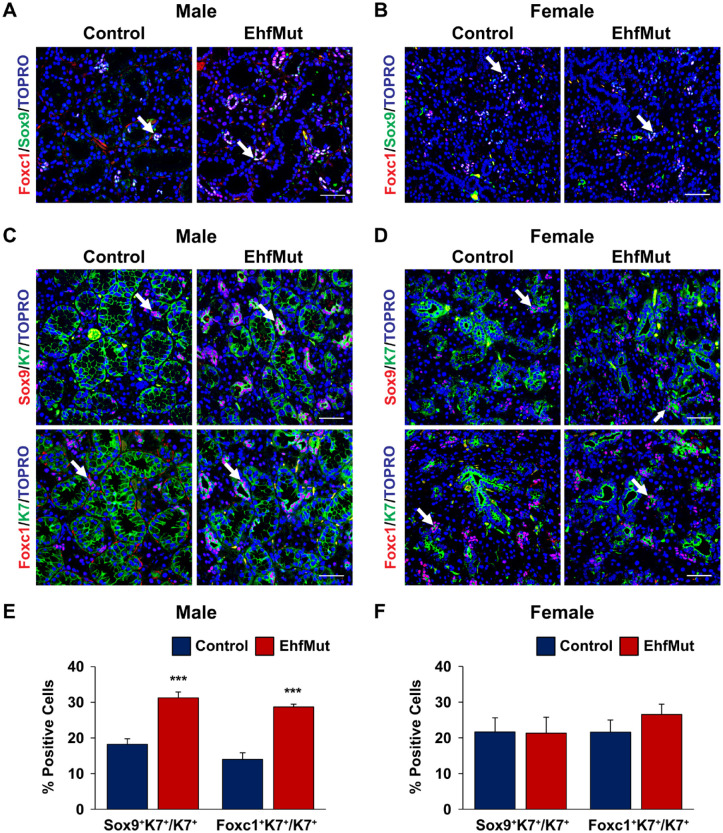
Given the enriched expression of Ehf messenger RNA (mRNA) in the IDs (Fig. 1A), we next examined if there were any alterations to this specific cell type in EhfMut glands. Interestingly, evaluation of the ID markers Sox9 and Foxc1 (Chatzeli et al. 2017; Gluck et al. 2021) revealed elevated numbers of Sox9 and Foxc1 double-positive (Sox9+/Foxc1+) cells in the male EhfMut glands, but not the female EhfMut glands, when compared to control mice (Fig. 3A, B). To further confirm our findings, we costained control and EhfMut glands with Sox9 and K7, which we reasoned would mark the Sox9-expressing ID cells. Indeed, quantification of the percentage of Sox9+ cells, which coexpressed K7, revealed a significant increase in ID cells of the EhfMut male mice compared to control animals (Fig. 3C, E). This was not the case for the female EhfMut mice, which showed no differences when compared to the control (Fig. 3D, F). Additional costaining and quantification studies using Foxc1 and K7 revealed a significant increase in the percentage of Foxc1+ cells, which coexpressed K7 in the male EhfMut and not the females when compared to the controls (Fig. 3C–F).
Figure 3.
Increased Sox9+ and Foxc1+ intercalated ductal cell populations in EhfMut submandibular glands (SMGs). (A) Immunostaining of adult male control and EhfMut SMGs with the intercalated ductal markers Sox9 and Foxc1 reveals increased intercalated ductal cells (white arrows) in the mutant glands. (B) Immunostaining of adult control and EhfMut female SMGs with the intercalated ductal markers Sox9 and Foxc1 reveals no significant difference in the intercalated ductal cells (white arrow). (C) Ducts costained with Sox9/K7 and Foxc1/K7 (white arrows) reveals increased numbers of intercalated ductal cells in the male EhfMut glands compared to the controls while no changes are detected in female mutant glands (D). Quantification of the Sox9+K7+/K7+ and Foxc1+K7+/K7+ intercalated ducts of the (E) male and (F) female control and EhfMut SMGs. Data are represented as mean ± SD (n = 5). ***P < 0.001. Scale bar: 50 µm.
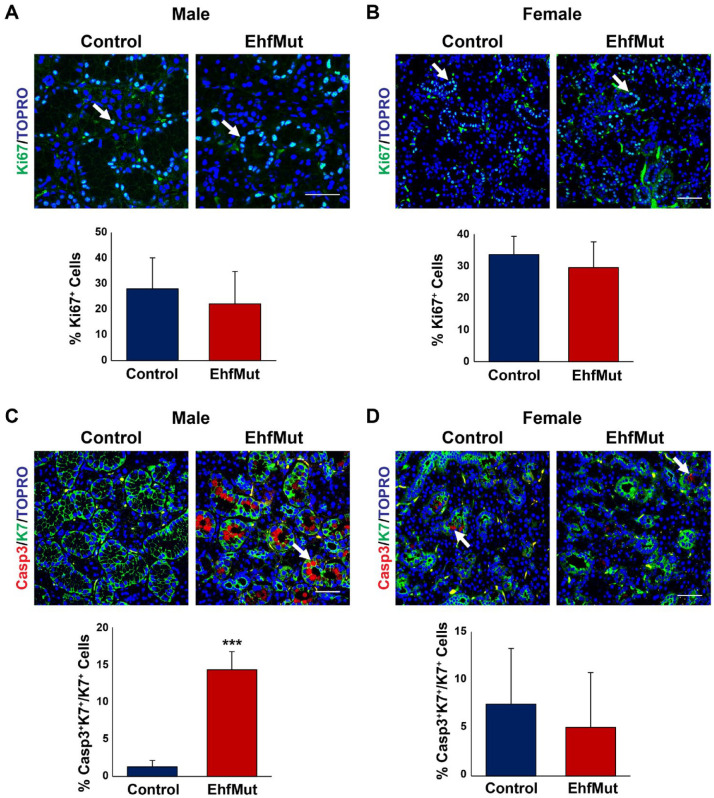
Since our immunofluorescence analysis did not reveal any mechanisms that could account for the overall decrease in the size of the GCTs, we assessed whether these changes were driven by proliferation defects. While the EhfMut glands did not reveal any significant changes in proliferation based on expression of the proliferation marker Ki67 in either the male or female SMGs compared to control (Fig. 4A, B), we did observe a significant increase in apoptosis in the male SMGs as demonstrated by the elevated numbers of K7+ ductal cells that stained positive for the apoptotic marker cleaved caspase-3 (Casp3) as compared to control glands (Fig. 4C). Interestingly, we did not observe any differences in the corresponding female mutant SMGs (Fig. 4D). Taken together, these results revealed a loss of the GCTs as well as changes in the number of ID cells in EhfMut male glands as depicted in our proposed model (Fig. 5) and in agreement with the enriched expression of Ehf in these cell types.
Figure 4.
Ductal cells undergo apoptosis in EhfMut mice. (A) Expression and quantification analysis of cell proliferation based on Ki67 expression (white arrows) show no differences between control and male EhfMut glands and (B) female control and mutant glands. (C) Expression and quantification analysis of cleaved caspase-3 (Casp3) reveals increased apoptosis in the K7+ ducts of the male EhfMut submandibular gland (SMG) compared to control mice. Arrows indicate the K7+Casp3+ double-positive cells. (D) Expression and quantification analysis of cleaved Casp3 reveals similar levels of apoptosis in the ducts of the female EhfMut SMG compared to control mice. Arrows indicate ductal cells undergoing apoptosis. Data are represented as mean ± standard deviation (SD) (n = 5). ***P < 0.001. Scale bar: 50 µm.
Figure 5.
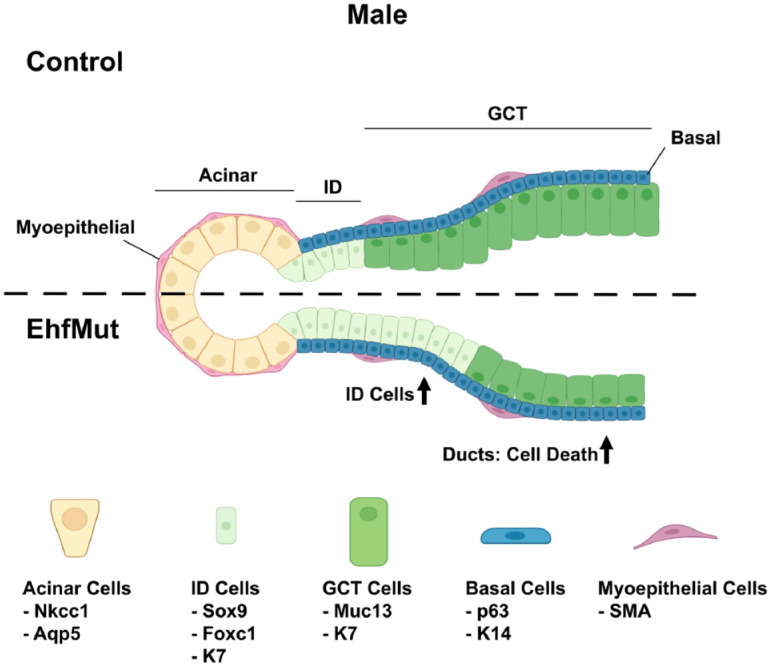
Model outlining the role of Ehf in maintaining the cell differentiation program of the adult salivary gland. Loss of Ehf results in increased apoptosis in the male EhfMut. This is accompanied by elevated numbers of Sox9+K7+ and Foxc1+K7+ intercalated ductal cells (ID cells) and widened lumens. Cell population markers are shown below the individual cell types. Created with BioRender.com. GCT, granular convoluted tubule.
Discussion
A careful analysis of the existing bulk and scRNA-seq data sets has demonstrated Elf5 and Ehf as 2 ETS factors that exhibit highly enriched expression in the SMG and associated with super-enhancers. Of these 2 TFs, Elf5 has been studied in more detail primarily in the context of the pregnant and lactating mammary gland, where it is highly expressed. Indeed, we and others have demonstrated that Elf5-null mouse mammary glands fail to initiate alveologenesis and accumulate stem and luminal progenitor cells (Chakrabarti, Hwang, et al. 2012, Chakrabarti, Wei, et al. 2012). Given the mammary gland phenotype of Elf5cKO mice, it is surprising that Elf5 loss in the SMG has no discernible phenotype—we speculate compensatory mechanisms are likely at play.
Unlike Elf5, mouse KO models for Ehf have been unavailable until only recently with the publication of 2 studies that focused on its role in intestinal stem cell differentiation (Zhu et al. 2018; Reehorst et al. 2021). The overall gross phenotype of our EhfMut mouse model has significant overlap with the published EhfMut studies by Reehorst et al. (2021) and thus provide an independent validation of the functional role of Ehf. One caveat worth stressing is that since the ETS DNA-binding domain was targeted for deletion (Reehorst et al. 2021) or truncation (our study), the potential for expression of a C-terminal deleted Ehf protein that can retain some functionality or instead harbor a gain-of-function effect cannot be excluded. These shortcomings notwithstanding, it is clear that the loss of the DNA-binding function of Ehf is sufficient to induce specific phenotypes in multiple tissues, including the SMG.
It was interesting to find that the males and females present different ductal phenotypes with an increased intercalated ductal cell population and increased cell death in male SMGs only (Fig. 5). We suspect that Ehf is required for maintaining the delicate balance of different ductal subpopulations. However, a careful examination of various ductal subtypes by scRNA-seq analysis is challenging at this point due to lack of specific markers. A case in point is the newly annotated Smgc+, basal, and Ascl3+ ductal subtypes, which might not be functionally and morphologically distinct from the intercalated ducts (Hauser et al. 2020). In contrast, acinar cells, which express more Elf5, seem to be unaffected by the loss of either Elf5 or Ehf. Although Elf5 and Ehf are 2 top-ranked genes in the SMG, it is worth noting that other ETS factors might compensate for the loss of either Elf5 or Ehf.
Our study has some limitations, the primary one being that we have only characterized the SMG of adult Elf5cKOs and EhfMut animals of a narrow range of age. We suspect that under different physiological and/or pathological conditions, such as aging or injury, loss of Elf5 and Ehf might reveal more striking phenotypes other than what we have described here. Another possibility is the likelihood of functional redundancy between Elf5 and Ehf and other ETS proteins that are structurally very similar and bind to similar DNA regulatory motifs (Wei et al. 2010). Future studies with compound knockout mouse models will likely reveal interesting molecular clues regarding the role of various ETS factors in shaping the gene regulatory networks of the diverse cell populations of the SMGs.
Author Contributions
E.A.C. Song, K. Smalley, A. Oyelakin, E. Horeth, M. Che, T. Wrynn, J. Osinski, contributed to data analysis, critically revised the manuscript; R.A. Romano, contributed to conception and design, data analysis, drafted and critically revised the manuscript; S. Sinha, contributed to conception and design, data analysis, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-docx-1-jdr-10.1177_00220345221130258 for Genetic Study of Elf5 and Ehf in the Mouse Salivary Gland by E.A.C. Song, K. Smalley, A. Oyelakin, E. Horeth, M. Che, T. Wrynn, J. Osinski, R.A. Romano and S. Sinha in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) training grant (NIH/NIDCR) DE023526 to the State University of New York at Buffalo, School of Dental Medicine, Department of Oral Biology for E.A.C. Song, A. Oyelakin, E. Horeth, and M. Che.
ORCID iDs: A. Oyelakin  https://orcid.org/0000-0003-0607-6631
https://orcid.org/0000-0003-0607-6631
E. Horeth  https://orcid.org/0000-0002-1376-2042
https://orcid.org/0000-0002-1376-2042
R.A. Romano  https://orcid.org/0000-0001-9652-9555
https://orcid.org/0000-0001-9652-9555
References
- Amano O, Mizobe K, Bando Y, Sakiyama K. 2012. Anatomy and histology of rodent and human major salivary glands: overview of the Japan Salivary Gland Society–sponsored workshop. Acta Histochem Cytoc. 45(5):241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany S, Catalan MA, Roztocil E, Ovitt CE. 2011. Ascl3 knockout and cell ablation models reveal complexity of salivary gland maintenance and regeneration. Dev Biol. 353(2):186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athwal HK, Murphy G, 3rd, Tibbs E, Cornett A, Hill E, Yeoh K, Berenstein E, Hoffman MP, Lombaert IMA. 2019. Sox10 regulates plasticity of epithelial progenitors toward secretory units of exocrine glands. Stem Cell Reports. 12(2):366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Hwang J, Andres Blanco M, Wei Y, Lukacisin M, Romano RA, Smalley K, Liu S, Yang Q, Ibrahim T, et al. 2012. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing snail2. Nat Cell Biol. 14(11):1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Wei Y, Romano RA, DeCoste C, Kang Y, Sinha S. 2012. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells. 30(7):1496–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzeli L, Gaete M, Tucker AS. 2017. Fgf10 and sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Development. 144(12):2294–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Chakrabarti R, Escamilla-Hernandez R, Sinha S. 2009. Elf5 conditional knockout mice reveal its role as a master regulator in mammary alveolar development: failure of stat5 activation and functional differentiation in the absence of Elf5. Dev Biol. 329(2):227–241. [DOI] [PubMed] [Google Scholar]
- Choi YS, Cheng J, Segre J, Sinha S. 2008. Generation and analysis of Elf5-LacZ mouse: unique and dynamic expression of Elf5 (ESE-2) in the inner root sheath of cycling hair follicles. Histochem Cell Biol. 129(1):85–94. [DOI] [PubMed] [Google Scholar]
- Cooper CD, Newman JA, Aitkenhead H, Allerston CK, Gileadi O. 2015. Structures of the Ets protein DNA-binding domains of transcription factors Etv1, Etv4, Etv5, and Fev: determinants of DNA binding and redox regulation by disulfide bond formation. J Biol Chem. 290(22):13692–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson E, May AJ, Berthoin L, Cruz-Pacheco N, Nathan S, Mattingly AJ, Chang JL, Ryan WR, Tward AD, Knox SM. 2018. Salivary glands regenerate after radiation injury through sox2-mediated secretory cell replacement. EMBO Mol Med. 10(3):e8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson E, May AJ, Nathan S, Cruz-Pacheco N, Lizama CO, Maliskova L, Zovein AC, Shen Y, Muench MO, Knox SM. 2017. Sox2 regulates acinar cell development in the salivary gland. Elife. 6:e26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck C, Min S, Oyelakin A, Che M, Horeth E, Song EAC, Bard J, Lamb N, Sinha S, Romano RA. 2021. A global vista of the epigenomic state of the mouse submandibular gland. J Dent Res. 100(13):1492–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck C, Min S, Oyelakin A, Smalley K, Sinha S, Romano RA. 2016. RNA-seq based transcriptomic map reveals new insights into mouse salivary gland development and maturation. BMC Genomics. 17(1):923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassmeyer J, Mukherjee M, deRiso J, Hettinger C, Bailey M, Sinha S, Visvader JE, Zhao H, Fogarty E, Surendran K. 2017. Elf5 is a principal cell lineage specific transcription factor in the kidney that contributes to Aqp2 and Avpr2 gene expression. Dev Biol. 424(1):77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresik EW, Hosoi K, Kurihara K, Maruyama S, Ueha T. 1996. The rodent granular convoluted tubule cell—an update. Eur J Morphol. 34(3):221–224. [DOI] [PubMed] [Google Scholar]
- Hauser BR, Aure MH, Kelly MC, Genomics and Computational Biology Core, Hoffman MP, Chibly AM. 2020. Generation of a single-cell RNAseq atlas of murine salivary gland development. iScience. 23(12):101838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos PA, Sienerth AR, Murray A, Senner CE, Muto M, Ikawa M, Oxley D, Burge S, Cox BJ, Hemberger M. 2015. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev. 29(23):2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Ormandy CJ. 2012. Elf5, hormones and cell fate. Trends Endocrinol Metab. 23(6):292–298. [DOI] [PubMed] [Google Scholar]
- Lombaert IM, Hoffman MP. 2010. Epithelial stem/progenitor cells in the embryonic mouse submandibular gland. Front Oral Biol. 14:90–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama CL, Monroe MM, Hunt JP, Buchmann L, Baker OJ. 2019. Comparing human and mouse salivary glands: a practice guide for salivary researchers. Oral Dis. 25(2):403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Oyelakin A, Gluck C, Bard JE, Song EC, Smalley K, Che M, Flores E, Sinha S, Romano RA. 2020. P63 and its target follistatin maintain salivary gland stem/progenitor cell function through TGF-β/activin signaling. iScience. 23(9):101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd BD, White SC. 1975. Sexual dimorphism in the rat submandibular gland. J Dent Res. 54(1):193. [DOI] [PubMed] [Google Scholar]
- Musselmann K, Green JA, Sone K, Hsu JC, Bothwell IR, Johnson SA, Harunaga JS, Wei Z, Yamada KM. 2011. Salivary gland gene expression atlas identifies a new regulator of branching morphogenesis. J Dent Res. 90(9):1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyelakin A, Nayak KB, Glathar AR, Gluck C, Wrynn T, Tugores A, Romano RA, Sinha S. 2022. Ehf is a novel regulator of cellular redox metabolism and predicts patient prognosis in HNSCC. NAR Cancer. 4(2):zcac017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VN, Hoffman MP. 2014. Salivary gland development: a template for regeneration. Semin Cell Dev Biol. 25–26:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearton DJ, Broadhurst R, Donnison M, Pfeffer PL. 2011. Elf5 regulation in the trophectoderm. Dev Biol. 360(2):343–350. [DOI] [PubMed] [Google Scholar]
- Reehorst CM, Nightingale R, Luk IY, Jenkins L, Koentgen F, Williams DS, Darido C, Tan F, Anderton H, Chopin M, et al. 2021. Ehf is essential for epidermal and colonic epithelial homeostasis, and suppresses Apc-initiated colonic tumorigenesis. Development. 148(12):dev199542. [DOI] [PubMed] [Google Scholar]
- Song EC, Min S, Oyelakin A, Smalley K, Bard JE, Liao L, Xu J, Romano RA. 2018. Genetic and scRNA-seq analysis reveals distinct cell populations that contribute to salivary gland development and maintenance. Sci Rep. 8(1):14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cairns MJ, Yan J. 2019. Super-enhancers in transcriptional regulation and genome organization. Nucleic Acids Res. 47(22):11481–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Feng L, Said M, Balderman S, Fayazi Z, Liu Y, Ghosh D, Gulick AM. 2005. Analysis of the 2.0 a crystal structure of the protein-DNA complex of the human PDEF Ets domain bound to the prostate specific antigen regulatory site. Biochemistry. 44(19):7095–7106. [DOI] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. 2010. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29(13):2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153(2):307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Yonemura S, Takada S. 2006. Grainyhead-related transcription factor is required for duct maturation in the salivary gland and the kidney of the mouse. Development. 133(23):4737–4748. [DOI] [PubMed] [Google Scholar]
- Zhu P, Wu J, Wang Y, Zhu X, Lu T, Liu B, He L, Ye B, Wang S, Meng S, et al. 2018. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat Cell Biol. 20(10):1134–1144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-jdr-10.1177_00220345221130258 for Genetic Study of Elf5 and Ehf in the Mouse Salivary Gland by E.A.C. Song, K. Smalley, A. Oyelakin, E. Horeth, M. Che, T. Wrynn, J. Osinski, R.A. Romano and S. Sinha in Journal of Dental Research