Abstract
Abstract
Nucleoprotein (NP) functions crucially in the replicative cycle of influenza A virus (IAV) via forming the ribonucleoprotein complex together with PB2, PB1, and PA proteins. As its high conservation, NP ranks one of the hot targets for design of universal diagnostic reagents and antiviral drugs for IAV. Here, we report an anti-NP murine monoclonal antibody (mAb) 5F10 prepared from traditional lymphocyte hybridoma technique with the immunogen of a clade 2.3.4.4 H5N1 subtype avian influenza virus. The specificity of mAb 5F10 to NP protein was confirmed by immunofluorescence assay and western blotting, and the mAb 5F10 could be used in immunoprecipitation and immunohistochemistry assays. Importantly, mAb 5F10 possessed broad-spectrum reactivity against H1~H11 subtypes of avian influenza viruses, including various HA clades of H5Nx subtype. In addition, mAb 5F10 also showed good affinity with H1N1 and H3N2 subtype influenza viruses of swine and human origin. Furthermore, the recognized antigenic epitope of mAb 5F10 was identified to consist of the conserved amino acid motif 81EHPSA85 in the second flexible loop region of NP protein through screening the phage display peptide library. Collectively, the mAb 5F10 which recognizes the novel universal NP linear B-cell epitope of IAV with diverse origins and subtypes will be a powerful tool for NP protein-based structural, functional, and mechanistic studies, as well as the development of detection methods and universal vaccines for IAV.
Key points
• A broad-spectrum mAb against various subtypes and sources of IAV was developed
• The mAb possessed good reactivity in IFA, western blot, IP, and IHC assays
• The mAb targeted a novel conserved linear B-cell epitope involving 81EHPSA85 on NP protein
Keywords: Influenza virus, Linear B-cell epitope, Nucleoprotein, Monoclonal antibody, Universal reactivity
Introduction
Influenza viruses are negative-sense and single-stranded RNA viruses of the Orthomyxoviridae family, in which the viral genome is divided into different fragments (Hutchinson 2018). According to the antigenic differences of nucleocapsid protein (NP) and matrix protein (M), four types of influenza viruses have been designated including A, B, C, and D (Hutchinson 2018). Specifically, influenza A virus (IAV) possesses by far the widest host range; it can infect humans, poultry, pigs, horses, dogs, and many other animals. Moreover, IAV poses a severe threat to public health as it has caused global annual epidemics and occasional pandemics in the human population (Joseph et al. 2017; Long et al. 2019). Genomically, 8 RNA segments are packaged per IAV particle. And due to the discrepancy of surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) that are respectively encoded by gene segment 4 and 6, IAV could be further categorized to 18 HA subtypes (H1~H18) and 11 NA subtypes (N1~N11) (Hutchinson 2018).
In avian species, H9N2, H5Nx, and H7N9 are the major subtypes that seriously affect poultry industry worldwide (Carnaccini and Perez 2020; Lee et al. 2017; Lee et al. 2021). Noteworthy, various HA clades such as clade 2.3.4.4 and clade 2.3.2.1, plus multiple NA subtypes such as H5N1, H5N6, and H5N8, have additionally increased the complexity of the highly pathogenic avian influenza (HPAI) H5Nx viruses (Antigua et al. 2019; Ge et al. 2021; Ge et al. 2022). As for the novel reassortant H7N9 virus which emerged in 2013, it has evolved from low pathogenicity avian influenza (LPAI) to HPAI form and exhibited obvious antigenic variation to challenge the vaccine efficiency (Liu et al. 2021; Wu et al. 2021; Yin et al. 2021). Despite being listed as a LPAI virus, H9N2 could also stimulate serious respiratory symptoms and egg drop to fowl production, and has derived into numerous genotypes in which the S or G57 genotype even tends to donate the whole internal genes for generation of novel influenza reassortants (Gu et al. 2017; Pu et al. 2015). Not only that, such H5, H7, and H9 avian influenza viruses also have caused a zoonotic concern on account of many human infections and fatalities (Joseph et al. 2017).
While in human and swine populations, H1N1 and H3N2 are the mainly prevalent IAV subtypes, both of which have aroused human pandemics and become the stable constituents to induce seasonal influenza outbreaks (Anderson et al. 2021; Petrova and Russell 2018; Sun et al. 2020). Besides, the dominantly circulating Eurasian avian-like (EA) H1N1 swine influenza virus even has been regarded as the most potential animal influenza virus to cause the next flu pandemic (Yang et al. 2016). And human infections with EA H1N1 and H3N2 swine influenza viruses have sporadically occurred (Anderson et al. 2021; Deng et al. 2020; Parys et al. 2021). Researchers also noted that H10N8, H6N1, H10N3, and other LPAI viruses have gained the capacity of direct host jump from bird to human beings, so it is reasonable to speculate that any of the avian influenza viruses might become a possibly risky zoonotic pathogen (Li et al. 2019; Qi et al. 2022). Therefore, high-efficiency and broad-spectrum rapid IAV diagnostic methods are urgently needed for timely prevention and control of human and animal influenza, as well as early warning of possible influenza pandemics.
Traditionally, the etiological tests to identify IAV through inoculation of chicken embryos or cells followed by hemagglutination (HA) and hemagglutination inhibition (HI) are usually labor-intensive and time-consuming. Routine PCR and agar gel immunodiffusion (AGID) are yet not appropriate for rapid detection due to the relatively low sensitivities (Spackman et al. 2009). In contrast, molecular biology methods such as real-time quantitative PCR or serological methods like immunofluorescence assay (IFA), enzyme linked immunosorbent assay (ELISA), and immune colloidal gold technique are more and more applied in practice because of easier operation, higher sensitivity, and faster results (Luo et al. 2020; Nutter et al. 2012; Sun et al. 2018; Zhang et al. 2017). However, the universal recognition of detection antibody to various different IAVs directly determines the efficiency and reliability of those serological tests.
As one of the internal structural proteins, NP is deeply conserved in different strains or subtypes of IAV with over 90% amino acid similarity (Tan et al. 2021; Yu et al. 2020). Though not presented on the virion surface, NP protein is expressed at high levels in infected cells with strong immunogenicity to serve as a potential target for universal vaccine, detection reagents, and antiviral drugs development (Santak and Matic 2022; Tan et al. 2021). In the present study, we characterized an mAb (5F10) targeting a conserved linear B-cell epitope on NP protein and showing broad-spectrum reactivity against different subtypes ranging from H1 to H11 of avian, swine, or human origin IAVs, with different HA clades of H5N1, H5N2, H5N6, and H5N8 avian influenza viruses included. Moreover, the mAb also exhibited good applications in immunoprecipitation (IP) and immunohistochemistry (IHC) assays. The universal NP mAb would certainly benefit the in-depth research of NP protein structure, function, and mechanism.
Materials and methods
Viruses, plasmids, and cells
The clade 2.3.4.4 H5N1 subtype avian influenza virus A/chicken/Anhui/QD1/2014(QD1), as we described previously (Li et al. 2020), was served as the immunogen to prepare the mAb. As shown in Table 1, 20 strains of IAV involving different origins, subtypes, and HA clades, plus 2 strains of influenza B virus (FluB), were used to determine the universal reactivity of the mAb. The viruses were preserved in our laboratory and propagated in 10-day-old specific-pathogen-free (SPF) chicken embryos to acquire virus stocks. And virus titration was performed in Madin-Darby canine kidney (MDCK) cells (ATCC, CCL-34) for determination of 50% tissue culture infectious dose. Experiments related to HPAI viruses were conducted in the animal biosafety level 3 laboratory of Yangzhou University. The plasmid pHW2000-QD1-NP constructed by Li J et al. as previously reported (Li et al. 2020) was applied to validate the target protein of the mAb. Both human embryonic kidney 293T (ATCC, CRL-3216) cells and MDCK cells were routinely maintained in Dulbecco’s modified Eagle medium (DMEM) (#PM150210, Procell, China) supplemented with 10% fetal bovine serum (FBS) (#10099-141, Gibco, Australia). The murine myeloma Sp2/0 (ATCC, CRL-158) cells were cultured in the above-mentioned medium additionally containing hypoxanthine and thymidine (HT) while chicken embryo fibroblast (CEF) cells were prepared from 9-day-old SPF chicken embryos and grown in M199 medium (#PM150610, Procell, China) supplemented with 4% FBS.
Table 1.
Background information of different influenza viruses for evaluation of the universal reactivity of mAb 5F10
| Influenza virus type | Host | Subtype (lineage/clade) | Virus name |
|---|---|---|---|
| Type A (IAV) | Avian | H1N1 | A/duck/Shandong/SDd11/2013 |
| H2N3 | A/duck/Hubei/1115/2019 | ||
| H3N2 | A/duck/Jiangsu/YZD3/2013 | ||
| H4N6 | A/duck/Anhui/AHd38/2014 | ||
| H5N1 (clade 2.3.2.1) | A/chicken/EasternChina/1404/2014 | ||
| H5N2 (clade 7.2) | A/duck/Beijing/BJ7/2014 | ||
| H5N6 (clade 2.3.4.4) | A/chicken/Yangzhou/YJD/2014 | ||
| H5N8 (clade 2.3.4.4) | A/goose/Yangzhou/0420/2014 | ||
| H6N2 | A/duck/Jiangsu/119/2015 | ||
| H7N9 | A/chicken/Zhejiang/JX164/2015 | ||
| H8N4 | A/duck/Yangzhou/02/2005 | ||
| H9N2 | A/chicken/Eastern China/0923/2015 | ||
| H10N7 | A/duck/Jiangsu/XZD53/2014 | ||
| H11N9 | A/duck/Jiangsu/YZD1/2013 | ||
| Swine | H1N1 (EA H1N1) | A/swine/Shandong/SG03/2019 | |
| H1N1 (2009 Pandemic H1N1) | A/swine/Jiangsu/JS48/2010 | ||
| H3N2 | A/swine/Anhui/SZ14/2019 | ||
| Human | H1N1 (2009 Pandemic H1N1) | A/California/04/2009 | |
| H1N1 (Seasonal) | A/Brisbane/59/2007 | ||
| H3N2 (Seasonal) | A/Jiangsu/202/2010 | ||
| Type B (FluB) | Human | Yamagata | B/Jiangsu/3221/BY/2019 |
| Victoria | B/Jiangsu/02/BV/2019 |
Experimental animals
We chose 6-week-old female BALB/c mice for virus immunization and inoculation while 10-week-old multiparous BALB/c mice for ascites extraction. Eight-week-old ICR mice were devoted to preparation of feeder cells. Those mice were supplied by the Experimental Animal Center of Yangzhou University. All of the mice studies were allowed by the Jiangsu Administrative Committee for Laboratory Animals (permission number: SYXKSU-2017-0044), and complied with the principles for laboratory animal welfare and the ethical guidelines of Jiangsu Administrative Committee for Laboratory Animals.
Immunization of mice and preparation of mAbs
The QD1 virus inactivated with β-propiolactone (BPL) (#P816096, Macklin, China) was purified by sucrose density gradient centrifugation before emulsion with complete (#F5881) or incomplete (#F5506) Freund’s adjuvant (Sigma-Aldrich, USA). Subsequently, 6-week-old female BALB/c mice were immunized through multi-site subcutaneously injection with the mixture thrice at 2-week interval. Three days after ictus immunisatorius via the tail vein, the spleen lymphocytes of the vaccinated mice with high antibody titers were fused with Sp2/0 cells using a polyethylene glycol (PEG) solution (#P7181, Sigma-Aldrich, USA). The generated hybridoma cells were screened using ELISA and subcloned for three consecutive rounds using limited dilution to acquire monoclonal cell lines. Ten-week-old multiparous BALB/c mice were injected intraperitoneally with the hybridoma cells to prepare ascites fluid, while the corresponding mAbs were obtained via purification with a HiTrap Protein G HP column (#17040501, GE Healthcare, USA). Isotyping of the mAbs was determined with the PierceTM Rapid Isotyping Kit-Mouse (#26178, Thermo Fisher Scientific, USA) according to the manual.
Indirect immunofluorescence assay
293T cells were transfected with recombinant plasmid pHW2000-QD1-NP and its empty vector control pHW2000 to ensure the mAb specificity to NP protein. Thirty-six hours (h) post-transfection, the cells were fixed with pre-cooling 4% paraformaldehyde (#P0099, Beyotime, China) for 20 min at 4 °C. The 1:500 diluted mAb 5F10 or mouse (G3A1) IgG1 isotype control (#5415, Cell Signaling Technology, USA) was incubated as a primary antibody with the fixed cells overnight at 4 °C. After washing 3 times with phosphate-buffered saline (PBS) containing 0.1% Tween-20 (PBST), the cells were treated in shade with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (#1030-02, SouthernBiotech, USA) at a 1:500 dilution for 1 h at room temperature. Afterwards, the cell nuclei were stained by 4,6-diamidino-2-phenylindole (DAPI) staining solution (#C1006, Beyotime, China) for 10 min at 37 °C. The cells were finally washed thrice with PBS and observed under a Leica DM IL LED fluorescence microscope to take typical images. Furthermore, to determine the broad-spectrum reactivity of the mAb to influenza viruses of various host origins, subtypes, lineages, or clades, MDCK cells were infected with different viruses at a multiplicity of infection (MOI) of 1 and fixed at 24 h post-infection. Following the same way of incubation with primary and secondary antibodies as described above, infected cells with fluorescence or not were observed under microscopy to capture representative images.
Western blot analysis
Apart from IFA, western blot assay was also performed to validate the specific and universal reactivity of mAb 5F10 to NP protein of IAVs. Similarly, 293T cells were transfected with plasmids pHW2000-QD1-NP and pHW2000, while MDCK cells were infected with different influenza viruses that were simultaneously tested in the IFA. The lysates of plasmid-transfected 293T cells or virus-infected MDCK cells were separated by SDS-PAGE and then transferred onto a polyvinylidene fluoride (PVDF) membrane (#1620177, Bio-Rad, USA). The membrane was blocked with 5% skim milk for 2 h at room temperature and incubated overnight at 4 °C with 1:1000 diluted mAb 5F10, mouse IgG1 isotype control (#5415, Cell Signaling Technology, USA), or anti-β-Actin mouse mAb (#HC201-01, TransGen Biotech, China) as the primary antibody, in which the housekeeping protein β-Actin was served as the internal reference. Subsequently, the horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (#CW0102, CWBIO, China) was added onto the membrane that had been washed 3 times with Tris-buffered saline (TBS) containing 0.1% Tween-20 (TBST). After incubation for 1 h at 37 °C and then washed thrice, the immunoreactive bands on the PVDF membrane were developed through an enhanced chemiluminescence (ECL) detection kit BeyoECL Plus (#P0018, Beyotime, China).
Immunoprecipitation (IP) assay
To determine the application of mAb 5F10 in IP assay, an avian H5N6 virus from group 1 subtypes and an avian H3N2 virus from group 2 subtypes of IAV (Sangesland and Lingwood 2021) were selected as representatives for the experiment. MDCK cells were infected with IAV H3N2 or H5N6 at 1 MOI. At 24 h post-infection, the cells were lysed in IP buffer (#P0013, Beyotime, China) and then incubated with 3 µg mAb 5F10 or IgG1 isotype (#5415, Cell Signaling Technology, USA) for 30 min at 37 °C. After addition of the protein A+G magnetic beads (#P2108, Beyotime, China), the lysates were incubated at 4 °C overnight. Subsequently, the protein complex bound to the beads was then eluted into 5× sodium dodecyl sulfate (SDS) loading buffer (#P0015, Beyotime, China). Finally, the immunoprecipitated proteins were analyzed by western blot with mAb 5F10 as the primary antibody and HRP-labeled goat anti-mouse IgG (#CW0102, CWBIO, China) as the secondary antibody.
Confocal fluorescence microscopy and immunohistochemistry (IHC) assay
CEF cells were seeded into 24-well plates containing glass coverslips and cultured overnight. Upon reaching ~80% confluency, cells were infected with IAV H3N2 or H5N6 at 1 MOI. At 12 h post-infection, the cells were fixed and then permeabilized with 0.1% Triton X-100 (#P0096, Beyotime, China) for 15 min. After blocking with QuickBlock™ Blocking Buffer for Immunol Staining (#P0260, Beyotime, China) at room temperature for 30 min, the cells were incubated with anti-NP mAb 5F10 or IgG1 isotype (#5415, Cell Signaling Technology, USA) at 4 °C overnight. Next, the cells were washed with PBS for three times, and stained with Alexa Fluor 594-conjugated goat anti-mouse IgG secondary antibody (#A-11005, Thermo Fisher Scientific, USA) for 1 h at 37 °C. After trice washing with PBS, the cell nuclei were labeled with DAPI (#C1006, Beyotime, China). The fluorescence in cells was then visualized with a Leica TCS SP8 confocal microscope.
In the IHC assay, lung tissues from 6-week-old BALB/c mice that intranasally infected with 106.0EID50 of IAV H3N2 and H5N6 or mock-infected with PBS were collected on day 3 post-infection. After fixation in 10% formalin solution for 48 h at room temperature, the tissues were embedded in paraffin and sectioned at 3~4 µm. On one hand, hematoxylin and eosin (HE) staining was conducted on the sections for histopathological examination. On the other hand, the sections were overnight incubated with mAb 5F10 as the primary antibody at 4 °C before incubation with the HRP-coupled goat anti-mouse secondary antibody (#CW0102, CWBIO, China) at 37 °C for 40 min. Those prepared sections were then scanned by the Pannoramic DESK slide scanner (3DHISTECH, Hungary). Finally, the images of pathological changes via HE staining and viral staining after coloration with diaminobenzidine (DAB) (#P0203, Beyotime, China) in the lung tissues were observed and collected by the CaseViewer 2.4 software (3DHISTECH, Hungary).
Epitope identification with phage-displayed peptide library
We used the Ph.D.-7TM Phage Display Peptide library kit (#E8100S, New England Biolabs, USA) including random heptapeptides to map the antigenic epitope of the mAb according to the manufacturer’s instructions with slight modification. Briefly, 60-mm Petri dishes were coated with 0.3 µg mAb 5F10 which was diluted in 0.1 mol/L NaHCO3 (pH 8.6) at 4 °C overnight. Following blocked with 5 mg/mL BSA in 0.1 mol/L NaHCO3 (pH 8.6) for 1 h at 4 °C and then washed with TBST for 6 times, the dishes were incubated with 1×1011 plaque forming unit (PFU) of the phages from the Ph.D.-7TM phage display library in 1 mL of TBST at room temperature for 30 min. After 10 times of washing with TBST at room temperature, the unbound phages in the phage-antibody mixture were washed away. Subsequently, 1 mL Glycine-HCl buffer (0.2 mol/L, pH 2.2, 1 mg/mL BSA) was added to elute the bound phages for 15 min at room temperature before neutralization with 150 µL of 1 mol/L Tris-HCl (pH 9.1). A small amount of eluted phage solution was used for titration and the rest eluate was used for amplification in the host bacterial strain E. coli ER2738. The required phage supernatants were further purified by precipitation and centrifugation before the next round of screening. After at least three consecutive rounds of such biopanning, 18 individual positive phage clones were randomly selected and characterized by DNA sequencing. The phage heptapeptide-gIII fusion gene was sequenced with the primer-96 gIII (5′-CCC TCA TAG TTA GCG TAA CG-3′) according to the manufacturer’s protocol. And the deduced amino acid sequences of those DNA inserts were aligned with the IAV NP protein using MEGA software version 10.0 (freely available from https://www.megasoftware.net/) (Kumar et al. 2018) to analyze the mimotopes. The location of the identified epitope on NP monomer was visualized and analyzed using PyMOL software version 2.5 (Schrödinger, Inc., New York, USA). The conservation of the identified epitope was intuitively displayed by using the WebLogo tool (freely accessible at https://weblogo.threeplusone.com/) (Crooks et al. 2004).
Results
Generation and characterization of the mAb
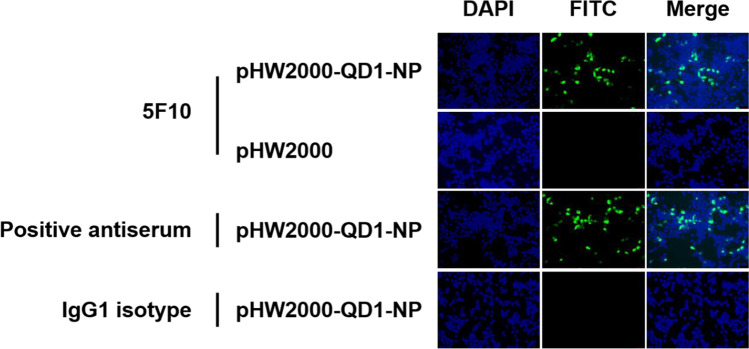
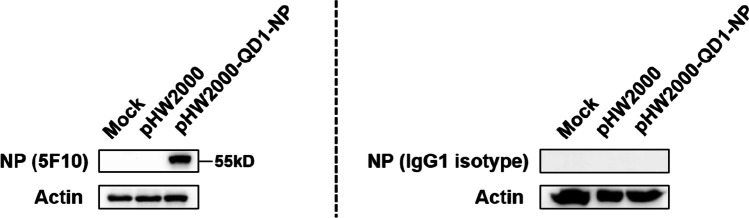
The hybridoma specifically secreting the anti-NP protein mAb 5F10 was generated through fusion of Sp2/0 myeloma cells with the spleen cells from mice immunized with the clade 2.3.4.4 H5N1 virus QD1 after the final boost. As classified using the mouse mAb isotyping kit, 5F10 belonged to the IgG1 isotype. And the NP specificity of 5F10 was validated by the binding to the transiently expressed NP protein in 293T cells through IFA (Fig. 1) and western blot (Fig. 2). Specifically, the pHW2000-QD1-NP-transfected 293T cells which had treated with mAb 5F10 as the primary antibody exhibited comparable yellowish-green fluorescence to that of the polyvalent antiserum from QD1 immunized mice (Fig. 1). In contrast, no positive fluorescent signal was detected for the cells mock-transfected with the empty pHW2000 vector or the cells transfected with pHW2000-QD1-NP but incubated with the control IgG1 isotype antibody (Fig. 1). Meanwhile, an immunopositive band localized at ~55 kD molecular mass of NP protein was detected using western blot analysis in the 293T cells that had transfected with pHW2000-QD1-NP and incubated with mAb 5F10 (Fig. 2). Similar as that found in above IFA, the result from transfected cells using IgG1 isotype as the primary antibody remained negative. Therefore, the mAb 5F10 possessed the ability to recognize NP protein. Also, since the cell samples in western blot analysis were treated under traditional reducing conditions (DTT+), it was reasonable to speculate mAb 5F10 of a linear antigenic epitope.
Fig. 1.
IFA determination of mAb 5F10 targeting NP protein in 293T cells. At 36 h post-transfection with plasmids pHW2000-QD1-NP and pHW2000, 293T cells were fixed and then cultured with primary antibody of mAb 5F10, QD1 virus-positive mouse antiserum, or the mouse (G3A1) mAb IgG1 isotype control. After incubation with the goat anti-mouse FITC-conjugated secondary antibody and then the 4′,6-diamidino-2-phenylindole (DAPI) for nucleus staining, the 293T cells were observed under fluorescence microscopy. The cells treated with the mouse antisera were used for positive control, while the cells transfected with pHW2000 empty vector or incubated with the commercial IgG1 isotype were used for negative control. Representative pictures were taken with the scale bar of 100 µm
Fig. 2.
Western blot analysis of the mAb 5F10 against NP protein. 293T cells transfected with empty pHW2000 vector or recombinant pHW2000-QD1-NP plasmid were lysed at 36 h post-transfection. The mAb 5F10 or IgG1 isotype and HRP-coupled goat anti-mouse IgG were served as primary and secondary antibodies, respectively. Incubation with mAb 5F10 produced a specific immunoreactive band in the lane marked with pHW2000-QD1-NP. The band was localized at about 55 kD, corresponding to the size of IAV NP protein. Treatment with an IgG1 isotype control antibody had no effect in generating positive bands. The housekeeping protein β-Actin was chosen as the internal reference
Broad-spectrum reactivity of mAb 5F10
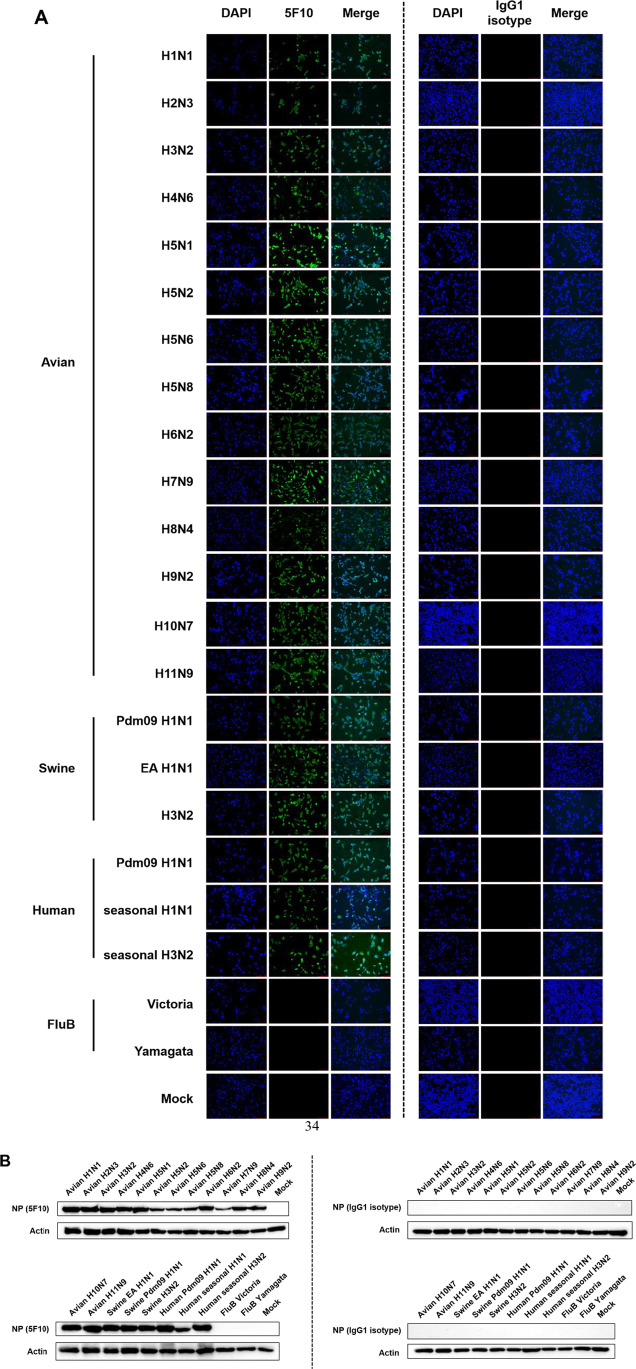
As NP is a highly conserved internal protein of IAV, we then evaluated the broad-spectrum reactivity of the anti-NP mAb 5F10 to different virus strains by IFA and western blot. MDCK cells were initially infected with different subtypes of avian IAV strains from H1 to H11, including the subtypes H1N1, H2N3, H3N2, H4N6, H5Nx, H6N2, H7N9, H8N4, H9N2, H10N7, and H11N9. Particularly, various HA clades and NA subtypes of clade 2.3.2.1 H5N1, clade 7.2 H5N2, clade 2.3.4.4 H5N6, and clade 2.3.4.4 H5N8 were contained in the tested H5Nx viruses. The infected cells were collected at 24 h post-infection for IFA analysis, with mAb 5F10 or IgG1 isotype serving as the primary antibody. As shown in Fig. 3A, all those infected MDCK cells that had incubated with mAb 5F10 emit bright green fluorescence except the negative control (mock-infected with PBS) and the isotype control. Therefore, mAb 5F10 exhibited positive reactivity to all the selected IAV subtypes of avian origin.
Fig. 3.
The broad-spectrum reactivity of mAb 5F10 against different hosts and subtypes of IAV-infected MDCK cells analyzed through IFA (A) and western blotting (B). MDCK cells were infected with avian H1~H11 IAV subtypes including H5Nx strains of multiple HA clades and NA subtypes, swine and human H1N1 and H3N2 IAV subtypes, and human type B influenza (FluB) viruses of Victoria and Yamagata lineages, respectively. Cells mock-inoculated with PBS were served as negative control. At 24 h post-infection, the infected cells were all subjected to incubation with mAb 5F10 as the primary antibody. An IgG1 isotype control antibody was simultaneously used for comparison. In the IFA analysis, typical images were taken with the scale bar of 100 µm after treatment with the FITC-conjugated goat anti-mouse IgG secondary antibody. In the western blot analysis, the HRP-conjugated goat anti-mouse IgG secondary antibody was applied to detect the immunoreactive protein bands
To further check whether mAb 5F10 recognizes more IAV strains of other host sources, MDCK cells infected with prevalent swine and human IAV strains were also subjected to IFA test. The investigated viruses involved EA H1N1, 2009 pandemic H1N1, and H3N2 subtype swine IAV strains plus 2009 pandemic H1N1, seasonal H1N1, and H3N2 subtype human IAV strains, as well as human type B influenza (FluB) viruses of Victoria and Yamagata lineages. As revealed in Fig. 3A, mAb 5F10 exhibited positive fluorescence to all the selected swine and human IAV strains but not to the FluB viruses of neither lineages. Hence, the anti-NP mAb 5F10 owned a wide-spectrum binding property to distinct IAV strains, and the reactivity was specifically limited to type A influenza viruses.
Moreover, we measured all the tested viruses in infected MDCK cells to characterize their reaction to mAb 5F10 in western blot analysis. Consistent with the results from IFA, either the avian or the mammalian IAV strains of different subtypes but not the FluB viruses were well recognized by mAb 5F10 (Fig. 3B). Each of the immunoreactive bands was approximately 55 kD in size, in accordance with the imprinting signal of IAV NP protein.
Taken together, the above results of IFA and western bot demonstrated that mAb 5F10 had a universal reactivity to IAV strains of different subtypes and hosts, at least including avian H1~H11 subtypes plus swine or human H1N1 and H3N2 subtypes. However, it was noteworthy that the broad spectrum was exclusively confined to IAV strains, but not expanded to FluB viruses.
mAb 5F10 targets a conserved B-cell linear epitope of NP protein
To identify the antigenic epitope of mAb 5F10, a commercially available M13 phage display 7-mer random peptide library was screened for biopanning. Following three rounds of successive affinity panning, 18 positive phage clones were independently selected and amplified for DNA sequencing. Through multiple alignment, we found that 18 phage clones therein harbor an identical consensus sequence containing a 3-amino acid motif “HPS.” In addition, extended consecutive amino acid patterns of “EHPS” (n=7), “HPSA” (n=6), and “EHPSA” (n=2) were also identified. As aligned to the clade 2.3.4.4 avian H5N1 virus QD1 which was treated as the immunogen, “EHPSA” exactly matched to residues E81-H82-P83-S84-A85 in the NP gene sequence (Table 2).
Table 2.
Identification of the mimotope peptides recognized by the anti-NP mAb 5F10
| Positive phage clones | Displayed random sequences | Matched peptides in NP protein of the clade 2.3.4.4 QD1(H5N1) virus | |
|---|---|---|---|
| Nucleotide sequences | Amino acid sequences | ||
| 1 | TTTCAGTTGCAGCATCCGTCT | FQLQHPS | 82HPS84 |
| 2 | AAGCTGTTTGAGCATCCTAGT | KLFEHPS | 81EHPS84 |
| 3 | TGGCAGGTTCATCCGTCTTCT | WQVHPSS | 82HPS84 |
| 4 | CTTTTGCAGCATCCTTCGGGG | LLQHPSG | 82HPS84 |
| 5 | TATTGGTTTGAGCATCCGTCT | YWFEHPS | 81EHPS84 |
| 6 | TGGACGGCGGAGCATCCTTCG | WTAEHPS | 81EHPS84 |
| 7 | AATAGTTTTGAGCATCCTTCT | NSFEHPS | 81EHPS84 |
| 8 | CAGTATCTTCATCCGTCGGCG | QYLHPSA | 82HPSA85 |
| 9 | GGTACGATTATGCATCCTTCG | GTIMHPS | 82HPS84 |
| 10 | CTTATGAATCATCCGTCTGCT | LMNHPSA | 82HPSA85 |
| 11 | TATATTGAGCATCCGAGTGCT | YIEHPSA | 81EHPSA85 |
| 12 | AATATTGAGCATCCGAGTGCT | NIEHPSA | 81EHPSA85 |
| 13 | TTTCCTCAGCATCCTTCGGCT | FPQHPSA | 82HPSA85 |
| 14 | AATCTTTGGCAGCATCCTTCG | NLWQHPS | 82HPS84 |
| 15 | AATAGTTTTGAGCATCCTTCT | NSFEHPS | 81EHPS84 |
| 16 | AATTTGATGCATCCTAGTGCG | NLMHPSA | 82HPSA85 |
| 17 | AGTACTTTTCAGCATCCGAGT | STFQHPS | 82HPS84 |
| 18 | TCTGTTATTCGTCATCCGTCT | SVIRHPS | 82HPS84 |
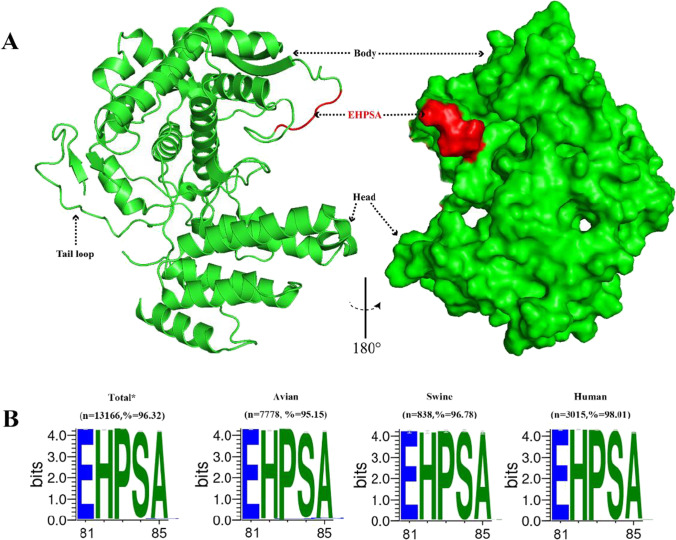
Also, such motif was consistently predicted to be involved in a potential B-cell epitope by bioinformatics analytical tools of ABCpred (Saha and Raghava 2007), Bepipred (Jespersen et al. 2017), and iBCE-EL (Manavalan et al. 2018) (data not shown). Structurally, the mimotope 81EHPSA85 was contained in the second basic flexible loop region (residues 72~92) of NP protein (Cui et al. 2019; Lo et al. 2018). To better characterize the spatial position of 81EHPSA85, the NP protein structure of IAV (PDB: 7NT8) was selected to be three-dimensionally displayed using PyMOL software. And we observed that the motif of “EHPSA” was located in a protruding loop and exposed to the protein surface (Fig. 4A), suggesting its accessibility. Consequently, we recognize the novel linear motif 81EHPSA85 as the core component of the target antigenic epitope of mAb 5F10.
Fig. 4.
Conformation and conservation analysis of the epitope recognized by the anti-NP mAb 5F10. A Location of the identified epitope on NP monomer. The NP monomer structure (PDB: 7NT8) was visualized and analyzed with PyMOL software. The epitope recognized by 5F10 was labeled in red, and it locates in a protruding loop region and on the protein surface. B Sequence conservation analysis of the identified epitope. The IAV NP sequences of all sources (n=13,166) and specifically of avian (n=7778), swine (n=838), and human (n=3015) origins were retrieved from GISAID EpiFlu database with the keywords “Type”= A and “Location”= China. The dataset was collected on 7 September 2022. The variation of the epitope was analyzed with WebLogo program, and the conservation degree of each amino acid was reflected by the size of corresponding character. *Total means of the analyzed NP sequences were of all host origins including avian, swine, human, canine, and equine
In order to further assess the epitope conservation, complete coding regions of NP genes from various IAV strains were retrieved from the Global Initiative on Sharing Avian Influenza Data (GISAID) database for multiple sequence alignment and then the WebLogo analysis. As demonstrated in Fig. 4B, the motif 81EHPSA85 targeted by anti-NP mAb 5F10 was highly conserved among IAV strains of all sources (96.32%), and specifically of avian origin (95.15%), swine origin (96.78%), and human origin (98.01%) with various subtypes.
Application of mAb 5F10 in the detection of IAV and the NP protein
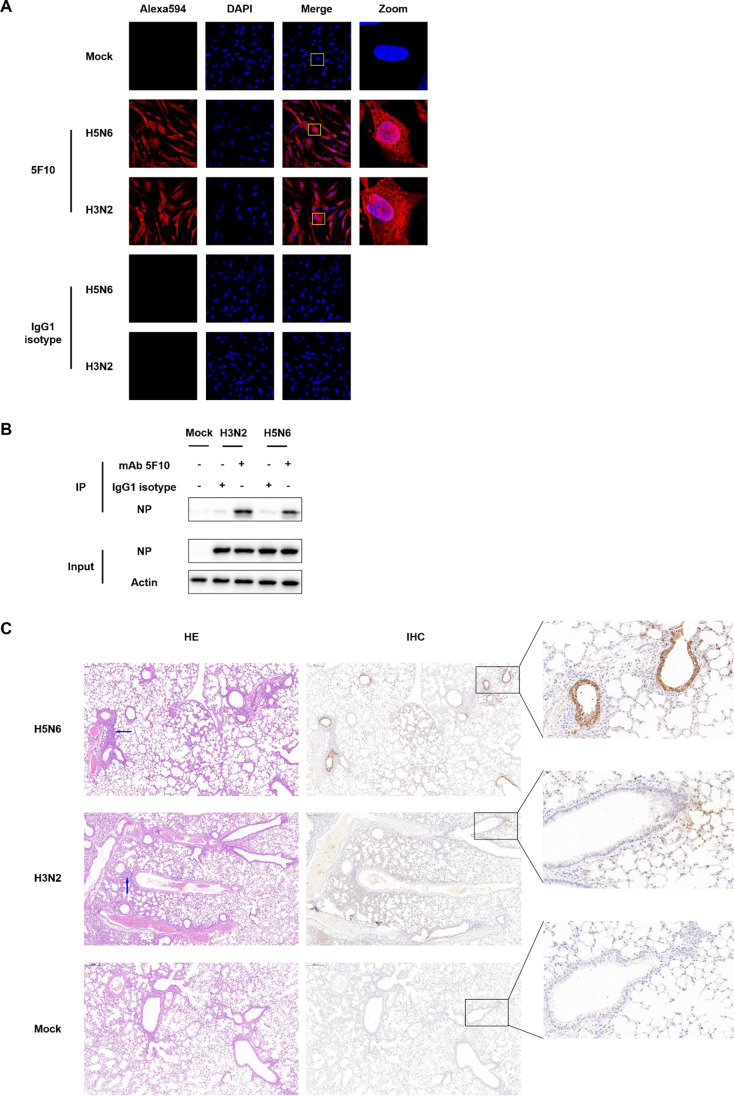
We chose H5N6 and H3N2 avian IAV isolates to representatively validate the application of mAb 5F10 in confocal microscopy, IP, and IHC assays, respectively. CEF cells infected with 1 MOI IAV at 12 h post-infection were fixed and incubated with mAb 5F10 as the primary antibody to microscopically detect the intracellular localization of NP protein. As shown in Fig. 5A, both H5N6 and H3N2 harbored the NP proteins not only in cytoplasm but also in nucleus of virus-infected MDCK cells. Thus, mAb 5F10 could be utilized to study the nucleocytoplasmic shuttling of IAV NP protein.
Fig. 5.
Application of mAb 5F10. A Intracellular localization of IAV NP protein analyzed via confocal microscopy. The CEF cells were infected with a H5N6 or a H3N2 subtype IAV at 1 MOI for 12 h. B Reaction of mAb 5F10 with IAV NP protein by IP assay. MDCK cells were infected with IAV subtypes H3N2 and H5N6 at 1 MOI, and were harvested and lysed after culturing for 24 h. The lysates were then subjected to the IP assay with mAb 5F10. C Detection of IAV antigen in the lung tissue of infected mice through IHC analysis. Six-week-old female BALB/c mice were intranasally infected with a H5N6 or a H3N2 subtype IAV at 106.0EID50, with mock-treated (with PBS) mice served as the negative control. The mice lungs were collected on day 3 post-infection for IHC staining, with mAb 5F10 serving as the primary antibody. Histopathological examination of the HE-stained sections was correspondingly performed, with the H5N6 virus induced severe lung tissue injury and atrophy of the alveoli whereas the H3N2 virus caused a relatively mild lung tissue destruction. Blue arrows indicated representative inflammatory cell infiltration, while brown signals indicated positive IHC staining. Scale bars were shown in the upper-left corner of microscopic images, with 200 µm for core images and 20 µm for zoom-in images
To further verify whether the anti-NP mAb 5F10 possesses IP activity, 24-h infected MDCK cells with 1 MOI H3N2 or H5N6 IAV were extracted for preparation of protein samples. After incubation with mAb 5F10 or IgG1 isotype and then the protein A+G magnetic beads, the formed antigen-antibody-beads complex were washed with PBS and boiled in the SDS loading buffer for western blot assay. As shown in Fig. 5B, the mAb 5F10 efficiently immunoprecipitated the NP protein in MDCK cells infected with H3N2 and H5N6 subtype IAVs. Thus, the anti-NP mAb 5F10 would provide a powerful tool for identification of the interaction between NP and host proteins.
Additionally, lung tissues of BALB/c mice intranasally inoculated with 106.0EID50 H5N6 or H3N2 IAV were collected for pathological characterization. The IHC results revealed that IAV antigen stained by the primary antibody of mAb 5F10 was obviously noted in both H5N6-infected and H3N2-infected mice lungs, while the histopathological changes by HE staining were evidently severe or somewhat mild (Fig. 5C). That is, the anti-NP mAb 5F10 could also be applied in IHC analysis of IAV distribution in infected tissues.
Discussion
The so-called mAb is a kind of lab-grown antibodies that possess a high mono-specificity and homogeneity for the target antigen or epitope. Accompanied with the rapid development of molecular biology, structural biology, and bioinformatics, mAb has experienced a series of technical breakthrough over decades to make itself a shining star for both immunodiagnostics and immunotherapy (Buss et al. 2012; Ecker et al. 2015; Gao et al. 2018). For example, over 100 patent applications concerning the preparation and application of mAbs against SARS-CoV-2, the causative agent of the ongoing global coronavirus disease 2019 (COVID-19) pandemic, have been registered according to Asdaq SMB et al. (Asdaq et al. 2021).
As for IAV, a zoonotic pathogen notorious for quick and frequent genetic variability, the most common antigen target for diagnostics via mAb assessment is NP (Phuong et al. 2018). Though it is well known that the surface antigens like HA and NA could elicit neutralizing antibodies to block viral cell entry or progeny virion release, the periodic antigenic variation of IAV has dramatically prevented the HA- and NA-specific mAbs from being applicable for the development of broad-spectrum influenza diagnostic reagents (Bhat et al. 2013; Phuong et al. 2018). However, anti-NP mAbs have been intensively developed for that purpose since the target viral protein is highly conserved among different subtypes of IAV and abundantly expressed in the virion (Bhat et al. 2013; Fujimoto et al. 2016).
In this study, we developed a mAb (5F10) specifically targeting the NP protein of IAV. Through IFA and western blot analyses, we demonstrated the universal reactivity of 5F10 to different IAV strains. 5F10 responded well not only to H5Nx avian viruses of distinct HA clades and NA subtypes, but also to various avian IAV subtypes ranging H1~H11. In addition, immune-positive signals were also evident against representative lineages of swine IAV such as EA H1N1, Pdm09 H1N1, and H3N2 plus human IAV such as Pdm09 H1N1, seasonal H1N1, and seasonal H3N2. Definitely, 5F10 did not interact with human FluB viruses either of Yamagata or Victoria lineages. Therefore, the anti-NP mAb 5F10 possessed good specificity and versatility among IAVs.
According to the phage display assay using a commercial randomized heptapeptide library, B-cell epitope prediction, and residue surface-exposure presentation, a consecutive amino acid motif of EHPSA at sites 81~85 in NP protein was recognized as the epitope of mAb 5F10. And through WebLogo analysis, this novel identified linear B-cell epitope was further revealed highly conservative across diverse subtypes of IAV strains from different hosts of avian, swine, and human. Thus, it is reasonable to speculate that the anti-NP mAb 5F10 would be extremely serviceable for recognizing a wide range of IAV isolates, including the emerging variants.
As one of the indispensable elements for ribonucleoprotein (RNP) that else involving the trimeric RNA-dependent RNA polymerase complex, NP protein interacts with viral RNA, other viral protein components, or specific macromolecules on infected cells to function critically at different stages of IAV life cycle such as transcription, replication, and assembly (Lo et al. 2018). Besides, it has been frequently documented that specific amino acid sites of S, T, and Y, on NP protein, could be phosphorylated to dynamically regulate the NP homo-oligomerization activity, NP nuclear import/export (nucleocytoplasmic shuttling), NP ubiquitination, virus polymerase activity, virus replicability, etc. (Cui et al. 2019; Li et al. 2015; Li et al. 2018; Turrell et al. 2015). Therefore, our anti-NP mAb 5F10 will be a useful tool for studying the function of IAV NP protein.
Briefly in structure, the NP molecule forms a crescent shape which comprises three evident domains of the head, body, and tail-loop. Particularly, the R-rich groove, crucially important for RNA binding, is involved in the region between the head and the body domain (Ng et al. 2009). In addition, a basic flexible loop spanning residues 73 to 91 which situated at the groove of NP monomer also plays critical roles in RNA binding activity (Ng et al. 2008). However, this structurally flexible loop is reported to be poorly resolved (Ng et al. 2009). Since the epitope of mAb 5F10 (E81-H82-P83-S84-A85) in the current study just located in such flexible loop, it would be great helpful to characterize the structure of IAV NP protein more deeply.
Despite not presenting on the IAV envelope, NP proteins are of high expression levels and able to be processed for antigen presentation on major histocompatibility complex (MHC) molecules to T cells (Doucet et al. 2011). On one hand, cytotoxic T lymphocytes (CTL) responses play an important role in the destruction of influenza virus-infected cells through cell lysis and apoptosis, thus leading to the inhibition of virion release and elimination of viral infection (Tan et al. 2021). On the other hand, antibody responses could also be stimulated on the basis of the Th epitope of NP protein to hinder viral RNA transcription and genome assembly, thus interfering the process of influenza virus infection (Tan et al. 2021). Additionally, NP has been reported on the surface of experimentally infected respiratory epithelial cells in vitro, suggesting that antibody-mediated immunity of virus-infected cells through antibody-dependent cellular cytotoxicity (ADCC), complement-dependent cell cytotoxicity (CDCC), or other mechanisms may help viral clearance in vivo (Bodewes et al. 2013; Tan et al. 2021; Vanderven et al. 2022). Therefore, provided the high homology of NP protein throughout different subtypes of IAV, the CTL and binding antibodies with cross-reactivity can be directed against pan-IAV isolates, and NP has been explored as a promising target for design of the universal vaccine (Nachbagauer and Palese 2020). In particular, since our mAb 5F10 recognizes a novel conserved linear epitope instead of a conformational epitope, it would be beneficial to promote the study of NP-induced heterosubtypic immunity and virus clearance.
We further evaluated the application of the mAb 5F10 in recognition of the universal IAV NP epitope; the mAb was used to characterize the cellular location of NP by confocal laser scanning microscopy, IP, and IHC assays. These results revealed that the mAb 5F10 could apparently locate the IAV NP not only in the cytoplasm but also in the nucleus of the host cells, consistent with the protein nature of nucleocytoplasmic transportation. In the IP assay, the mAb 5F10 not merely captured the viral NP protein but simultaneously identified the interacted polymerase (PB2 protein for example). Additionally, the mAb 5F10 stained the lung tissues of mice infected with either tested IAV strains. Although just one H3N2 subtype and one H5N6 subtype avian IAVs were selected for the determination, our findings again suggest that 5F10 can be broadly applicable in the detection of NP protein or IAV in vitro and in vivo.
Although many anti-NP mAbs have been reported by colleagues home and abroad (Liu et al. 2006; Phuong et al. 2018; Vanderven et al. 2022; Xiao et al. 2018), some of them lacked characteristic information such as precise epitope location or pan-reactivity evaluation, which may probably limited further application. In contrast to some other NP-specific mAbs with identified antigenic epitopes (Bodewes et al. 2013; Fujimoto et al. 2016; Gui et al. 2014; Huang et al. 2022), the mAb 5F10 developed in this study recognized a novel conservative linear B-cell epitope of 81EHPSA85 which located in a basic flexible loop at the N-terminal of NP protein. What is more, mAb 5F10 responded broadly to the NP protein across different IAVs of avian H1-H11, swine, and human H1 and H3 subtypes, and this anti-NP mAb was successfully applied in IFA, IP, and IHC assays for detection of IAV. The results in the present study collectively indicated that mAb 5F10 targeting the universal NP epitope of IAVs will be a powerful biomedical tool for studying the structure and function of IAV NP protein, as well as the development of methods for diagnosis of IAV infection and the design of universal flu vaccines.
Author contribution
MG and XFL conceived and designed the project. JJ, SHL, WCZ, ZCG, KRC, LJX, DCH, and XYZ carried out the experiments. MG, XQ, WMJ, PHZ, XQW, SLH, and XFL analyzed the data. MG wrote the manuscript. XFL supervised all the experiments and data analyses. All authors have read and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (32072892), the Earmarked Fund for China Agriculture Research System (CARS-40), the Jiangsu Provincial Postdoctoral Science Foundation (1501075C), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Jiangsu Qinglan Project, and the High-end Talent Support Program of Yangzhou University.
Data availability
The datasets used and analyzed in the present study are available from the corresponding author on reasonable request.
Declarations
Ethical approval
The animal experiment was approved by the Institutional Animal Ethics Committee of Yangzhou University, China (permission number: SYXK (SU) 2017-0044), and complied with the animal care and ethics guidelines.
Conflict of interest
The authors declare no competing interests
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Min Gu, Jun Jiao, and Suhan Liu contributed equally to the article.
Contributor Information
Min Gu, Email: gumin@yzu.edu.cn.
Jun Jiao, Email: 348249667@qq.com.
Suhan Liu, Email: 592603210@qq.com.
Wanchen Zhao, Email: 781240968@qq.com.
Zhichuang Ge, Email: 1411485893@qq.com.
Kairui Cai, Email: 2280953460@qq.com.
Lijun Xu, Email: 510766334@qq.com.
Dongchang He, Email: 605231753@qq.com.
Xinyu Zhang, Email: 873612832@qq.com.
Xian Qi, Email: qixiansyc@163.com.
Wenming Jiang, Email: jiangwenming@cahec.cn.
Pinghu Zhang, Email: zhangpinghu@yzu.edu.cn.
Xiaoquan Wang, Email: wxq@yzu.edu.cn.
Shunlin Hu, Email: slhu@yzu.edu.cn.
Xiufan Liu, Email: xfliu@yzu.edu.cn.
References
- Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, Lewis NS, Davis CT, Vincent AL. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb Perspect Med. 2021;11(3):a038737. doi: 10.1101/cshperspect.a038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antigua KJC, Choi WS, Baek YH, Song MS. The emergence and decennary distribution of clade 2.3.4.4 HPAI H5Nx. Microorganisms. 2019;7(6):156. doi: 10.3390/microorganisms7060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asdaq SMB, Rabbani SI, Alkahtani M, Aldohyan MM, Alabdulsalam AM, Alshammari MS, Alajlan SA, Binrokan A, Mohzari Y, Alrashed A, Alshammari MK, Imran M, Nayeem N. A patent review on the therapeutic application of monoclonal antibodies in COVID-19. Int J Mol Sci. 2021;22(21):11953. doi: 10.3390/ijms222111953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat S, Bhatia S, Sood R, Bhatnagar H, Pateriya A, Venkatesh G. Production and characterization of monoclonal antibodies against nucleoprotein of avian influenza virus. Monoclon Antib Immunodiagn Immunother. 2013;32(6):413–418. doi: 10.1089/mab.2013.0053. [DOI] [PubMed] [Google Scholar]
- Bodewes R, Geelhoed-Mieras MM, Wrammert J, Ahmed R, Wilson PC, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. In vitro assessment of the immunological significance of a human monoclonal antibody directed to the influenza a virus nucleoprotein. Clin Vaccine Immunol. 2013;20(8):1333–1337. doi: 10.1128/CVI.00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss NA, Henderson SJ, McFarlane M, Shenton JM, de Haan L. Monoclonal antibody therapeutics: history and future. Curr Opin Pharmacol. 2012;12(5):615–622. doi: 10.1016/j.coph.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Carnaccini S, Perez DR. H9 influenza viruses: an emerging challenge. Cold Spring Harb Perspect Med. 2020;10(6):a038588. doi: 10.1101/cshperspect.a038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Zheng W, Li M, Bai X, Yang W, Li J, Fan W, Gao GF, Sun L, Liu W. Phosphorylation status of tyrosine 78 residue regulates the nuclear export and ubiquitination of influenza A virus nucleoprotein. Front Microbiol. 2019;10:1816. doi: 10.3389/fmicb.2019.01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YM, Wong FYK, Spirason N, Kaye M, Beazley R, Grau MLL, Shan S, Stevens V, Subbarao K, Sullivan S, Barr IG. Dhanasekaran V (2020) Locally acquired human infection with swine-origin influenza A(H3N2) variant virus, Australia. Emerg Infect Dis. 2018;26(1):143–147. doi: 10.3201/eid2601.191144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet JD, Forget MA, Grange C, Rouxel RN, Arbour N, von Messling V, Lapointe R. Endogenously expressed matrix protein M1 and nucleoprotein of influenza A are efficiently presented by class I and class II major histocompatibility complexes. J Gen Virol. 2011;92(Pt 5):1162–1171. doi: 10.1099/vir.0.029777-0. [DOI] [PubMed] [Google Scholar]
- Ecker DM, Jones SD, Levine HL. The therapeutic monoclonal antibody market. MAbs. 2015;7(1):9–14. doi: 10.4161/19420862.2015.989042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y, Tomioka Y, Takakuwa H, Uechi GI, Yabuta T, Ozaki K, Suyama H, Yamamoto S, Morimatsu M, Mai LQ, Yamashiro T, Ito T, Otsuki K, Ono E. Cross-protective potential of anti-nucleoprotein human monoclonal antibodies against lethal influenza A virus infection. J Gen Virol. 2016;97(9):2104–2116. doi: 10.1099/jgv.0.000518. [DOI] [PubMed] [Google Scholar]
- Gao Y, Huang X, Zhu Y, Lv Z. A brief review of monoclonal antibody technology and its representative applications in immunoassays. J Immunoassay Immunochem. 2018;39(4):351–364. doi: 10.1080/15321819.2018.1515775. [DOI] [PubMed] [Google Scholar]
- Ge Z, Gu M, Cai T, Liu K, Gao R, Liu D, Sun W, Li X, Shi L, Liu J, Wang X, Hu J, Liu X, Hu S, Chen S, Peng D, Jiao X, Liu X. Phylogenetic tracing and biological characterization of a novel clade 2.3.2.1 reassortant of H5N6 subtype avian influenza virus in China. Transbound Emerg Dis. 2021;68(2):730–741. doi: 10.1111/tbed.13736. [DOI] [PubMed] [Google Scholar]
- Ge Z, Xu L, Hu X, Zhu S, Zhao Y, Li Y, Liu K, Gao R, Wang X, Hu J, Liu X, Hu S, Peng D, Gu M, Liu X. Phylogenetic and phenotypic characterization of two novel clade 2.3.2.1 H5N2 subtype avian influenza viruses from chickens in China. Infect Genet Evol. 2022;98:105205. doi: 10.1016/j.meegid.2022.105205. [DOI] [PubMed] [Google Scholar]
- Gu M, Xu L, Wang X, Liu X. Current situation of H9N2 subtype avian influenza in China. Vet Res. 2017;48(1):49. doi: 10.1186/s13567-017-0453-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X, Ge P, Wang X, Yang K, Yu H, Zhao Q, Chen Y, Xia N. Identification of a highly conserved and surface exposed B-cell epitope on the nucleoprotein of influenza A virus. J Med Virol. 2014;86(6):995–1002. doi: 10.1002/jmv.23812. [DOI] [PubMed] [Google Scholar]
- Huang X, Huang J, Yin G, Cai Y, Chen M, Hu J, Feng X. Identification of NP protein-specific B-cell epitopes for H9N2 subtype of avian influenza virus. Viruses. 2022;14(6):1172. doi: 10.3390/v14061172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EC. Influenza Virus. Trends Microbiol. 2018;26(9):809–810. doi: 10.1016/j.tim.2018.05.013. [DOI] [PubMed] [Google Scholar]
- Jespersen MC, Peters B, Nielsen M, Marcatili P (2017) BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 45(W1):W24-W29 [DOI] [PMC free article] [PubMed]
- Joseph U, Su YC, Vijaykrishna D, Smith GJ. The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respir Viruses. 2017;11(1):74–84. doi: 10.1111/irv.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Bertran K, Kwon JH, Swayne DE. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J Vet Sci. 2017;18(S1):269–280. doi: 10.4142/jvs.2017.18.S1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Criado MF, Swayne DE. Pathobiological origins and evolutionary history of highly pathogenic avian influenza viruses. Cold Spring Harb Perspect Med. 2021;11(2):a038679. doi: 10.1101/cshperspect.a038679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yu M, Zheng W, Liu W. Nucleocytoplasmic shuttling of influenza A virus proteins. Viruses. 2015;7(5):2668–2682. doi: 10.3390/v7052668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun L, Zheng W, Madina M, Li J, Bi Y, Wang H, Liu W, Luo TR. Phosphorylation and dephosphorylation of threonine 188 in nucleoprotein is crucial for the replication of influenza A virus. Virology. 2018;520:30–38. doi: 10.1016/j.virol.2018.05.002. [DOI] [PubMed] [Google Scholar]
- Li YT, Linster M, Mendenhall IH, Su YCF, Smith GJD. Avian influenza viruses in humans: lessons from past outbreaks. Br Med Bull. 2019;132(1):81–95. doi: 10.1093/bmb/ldz036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gu M, Liu K, Gao R, Sun W, Liu D, Jiang K, Zhong L, Wang X, Hu J, Hu S, Liu X, Shi W, Ren H, Peng D, Jiao X, Liu X. Amino acid substitutions in antigenic region B of hemagglutinin play a critical role in the antigenic drift of subclade 2.3.4.4 highly pathogenic H5NX influenza viruses. Transbound Emerg Dis. 2020;67(1):263–275. doi: 10.1111/tbed.13347. [DOI] [PubMed] [Google Scholar]
- Liu F, Jin M, Zhang A, Dan B, Chen H. Preparation and partial charaterization of monoclonal antibodies specific for nuclear protein of avian influenza virus. Chin J Cell Mol Immunol. 2006;22(5):648–649. [PubMed] [Google Scholar]
- Liu WJ, Xiao H, Dai L, Liu D, Chen J, Qi X, Bi Y, Shi Y, Gao GF, Liu Y. Avian influenza A (H7N9) virus: from low pathogenic to highly pathogenic. Front Med. 2021;15(4):507–527. doi: 10.1007/s11684-020-0814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CY, Tang YS, Shaw PC. Structure and function of influenza virus ribonucleoprotein. Subcell Biochem. 2018;88:95–128. doi: 10.1007/978-981-10-8456-0_5. [DOI] [PubMed] [Google Scholar]
- Long JS, Mistry B, Haslam SM, Barclay WS. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol. 2019;17(2):67–81. doi: 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- Luo S, Deng X, Xie Z, Huang J, Zhang M, Li M, Xie L, Li D, Fan Q, Wang S, Zeng T, Zhang Y, Xie Z. Production and identification of monoclonal antibodies and development of a sandwich ELISA for detection of the H3-subtype avian influenza virus antigen. AMB Express. 2020;10(1):49. doi: 10.1186/s13568-020-00988-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan B, Govindaraj RG, Shin TH, Kim MO, Lee G. iBCE-EL: a new ensemble learning framework for improved linear B-Cell epitope prediction. Front Immunol. 2018;9:1695. doi: 10.3389/fimmu.2018.01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachbagauer R, Palese P. Is a universal influenza virus vaccine possible? Annu Rev Med. 2020;71:315–327. doi: 10.1146/annurev-med-120617-041310. [DOI] [PubMed] [Google Scholar]
- Ng AK, Zhang H, Tan K, Li Z, Liu JH, Chan PK, Li SM, Chan WY, Au SW, Joachimiak A, Walz T, Wang JH, Shaw PC. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008;22(10):3638–3647. doi: 10.1096/fj.08-112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AK, Wang JH, Shaw PC. Structure and sequence analysis of influenza A virus nucleoprotein. Sci China C Life Sci. 2009;52(5):439–449. doi: 10.1007/s11427-009-0064-x. [DOI] [PubMed] [Google Scholar]
- Nutter S, Cheung M, Adler-Shohet FC, Krusel K, Vogel K, Meyers H. Evaluation of indirect fluorescent antibody assays compared to rapid influenza diagnostic tests for the detection of pandemic influenza A (H1N1) pdm09. PLoS One. 2012;7(3):e33097. doi: 10.1371/journal.pone.0033097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parys A, Vandoorn E, King J, Graaf A, Pohlmann A, Beer M, Harder T. Van Reeth K (2021) Human infection with Eurasian avian-like swine influenza A(H1N1) virus, the Netherlands. Emerg Infect Dis. 2019;27(3):939–943. doi: 10.3201/eid2703.201863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova VN, Russell CA. The evolution of seasonal influenza viruses. Nat Rev Microbiol. 2018;16(1):60. doi: 10.1038/nrmicro.2017.146. [DOI] [PubMed] [Google Scholar]
- Phuong NH, Kwak C, Heo CK, Cho EW, Yang J, Poo H. Development and characterization of monoclonal antibodies against nucleoprotein for diagnosis of influenza A virus. J Microbiol Biotechnol. 2018;28(5):809–815. doi: 10.4014/jmb.1801.01002. [DOI] [PubMed] [Google Scholar]
- Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C, Wei Y, Wang D, Zhu B, Lemmon G, Jiao Y, Duan S, Wang Q, Du Q, Sun M, Bao J, Sun Y, Zhao J, Zhang H, Wu G, Liu J, Webster RG. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A. 2015;112(2):548–553. doi: 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Qiu H, Hao S, Zhu F, Huang Y, Xu K, Yu H, Wang D, Zhou L, Dai Q, Zhou Y, Wang S, Huang H, Yu S, Huo X, Chen K, Liu J, Hu J, Wu M, Bao C. Human infection with an avian-origin influenza A (H10N3) Virus. N Engl J Med. 2022;386(11):1087–1088. doi: 10.1056/NEJMc2112416. [DOI] [PubMed] [Google Scholar]
- Saha S, Raghava GP. Prediction methods for B-cell epitopes. Methods Mol Biol. 2007;409:387–394. doi: 10.1007/978-1-60327-118-9_29. [DOI] [PubMed] [Google Scholar]
- Sangesland M, Lingwood D. Antibody focusing to conserved sites of vulnerability: the immunological pathways for 'universal' influenza vaccines. Vaccines (Basel) 2021;9(2):125. doi: 10.3390/vaccines9020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santak M, Matic Z. The role of nucleoprotein in immunity to human negative-stranded RNA viruses-not just another brick in the viral nucleocapsid. Viruses. 2022;14(3):521. doi: 10.3390/v14030521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Pantin-Jackwood MJ, Swayne DE, Suarez DL. An evaluation of avian influenza diagnostic methods with domestic duck specimens. Avian Dis. 2009;53(2):276–280. doi: 10.1637/8520-111708-Reg.1. [DOI] [PubMed] [Google Scholar]
- Sun Z, Shi B, Meng F, Ma R, Hu Q, Qin T, Chen S, Peng D, Liu X. Development of a colloidal gold-based immunochromatographic strip for rapid detection of H7N9 influenza viruses. Front Microbiol. 2018;9:2069. doi: 10.3389/fmicb.2018.02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Xiao Y, Liu J, Wang D, Li F, Wang C, Li C, Zhu J, Song J, Sun H, Jiang Z, Liu L, Zhang X, Wei K, Hou D, Pu J, Sun Y, Tong Q, Bi Y, Chang KC, Liu S, Gao GF, Liu J. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc Natl Acad Sci U S A. 2020;117(29):17204–17210. doi: 10.1073/pnas.1921186117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MP, Tan WS, Mohamed Alitheen NB, Yap WB. M2e-based influenza vaccines with nucleoprotein: a review. Vaccines (Basel) 2021;9(7):739. doi: 10.3390/vaccines9070739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrell L, Hutchinson EC, Vreede FT, Fodor E. Regulation of influenza A virus nucleoprotein oligomerization by phosphorylation. J Virol. 2015;89(2):1452–1455. doi: 10.1128/JVI.02332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderven HA, Esterbauer R, Jegaskanda S, Tan HX, Wheatley AK, Kent SJ. Poor protective potential of influenza nucleoprotein antibodies despite wide prevalence. Immunol Cell Biol. 2022;100(1):49–60. doi: 10.1111/imcb.12508. [DOI] [PubMed] [Google Scholar]
- Wu Y, Hu J, Jin X, Li X, Wang J, Zhang M, Chen J, Xie S, Qi W, Liao M, Jia W. Accelerated evolution of H7N9 subtype influenza virus under vaccination pressure. Virol Sin. 2021;36(5):1124–1132. doi: 10.1007/s12250-021-00383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, Yao L, Bi Z, Lei J, Yan Y, Yan L. Preparation and identification of monoclonal antibodies against NP protein of avian influenza virus. Animal Husbandry & Veterinary Medicine. 2018;50(12):53–58. [Google Scholar]
- Yang H, Chen Y, Qiao C, He X, Zhou H, Sun Y, Yin H, Meng S, Liu L, Zhang Q, Kong H, Gu C, Li C, Bu Z, Kawaoka Y, Chen H. Prevalence, genetics, and transmissibility in ferrets of Eurasian avian-like H1N1 swine influenza viruses. Proc Natl Acad Sci U S A. 2016;113(2):392–397. doi: 10.1073/pnas.1522643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Deng G, Zeng X, Cui P, Hou Y, Liu Y, Fang J, Pan S, Wang D, Chen X, Zhang Y, Wang X, Tian G, Li Y, Chen Y, Liu L, Suzuki Y, Guan Y, Li C, Shi J, Chen H. Genetic and biological properties of H7N9 avian influenza viruses detected after application of the H7N9 poultry vaccine in China. PLoS Pathog. 2021;17(4):e1009561. doi: 10.1371/journal.ppat.1009561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CM, Chen IC, Tung CP, Peng HP, Jian JW, Chiu YK, Tsou YL, Chen HS, Huang YJ, Hsiao WW, Wang YA, Yang AS. A panel of anti-influenza virus nucleoprotein antibodies selected from phage-displayed synthetic antibody libraries with rapid diagnostic capability to distinguish diverse influenza virus subtypes. Sci Rep. 2020;10(1):13318. doi: 10.1038/s41598-020-70135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu D, Sun W, Liu J, He L, Hu J, Gu M, Wang X, Liu X, Hu S, Chen S, Peng D, Liu X. Multiplex one-step Real-time PCR by Taqman-MGB method for rapid detection of pan and H5 subtype avian influenza viruses. PLoS One. 2017;12(6):e0178634. doi: 10.1371/journal.pone.0178634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed in the present study are available from the corresponding author on reasonable request.